Abstract
Objective
We analyzed histone deacetylase 10 (HDAC10) for function in the context of the DNA damage response in BRCA1-null ovarian cancer cells as well as evaluated the potential of general HDAC inhibitors in primary ovarian carcinoma cells. HDAC10 had previously been shown to be highly stimulatory to the process of homology directed repair in HeLa cells, and in this study we investigated whether HDAC10 could impact in vitro the response to anticancer therapies. We hypothesized that the loss of HDAC10 would sensitize cells to platinum therapy.
Methods
We combined informatics analysis of large DNA sequencing datasets from ovarian cancer tumors with tissue culture based assays of primary and established cell lines to test for sensitivity to platinum therapy if HDAC10 activity was inhibited or depleted.
Results
Using The Cancer Genome Atlas (TCGA) dataset, we found that deep deletions in HDAC10 occurred in 5–10% of ovarian cancer tumors. From the TCGA data we found that low HDAC10 mRNA levels correlated with platinum sensitivity of the tumors. Cell proliferation and DNA damage assays in a BRCA1-null ovarian carcinoma cell line demonstrated reduced DNA repair capacity and sensitization of platinum therapy. Similarly, primary ovarian carcinoma cells demonstrated a sensitization to platinum therapies when treated with HDAC inhibitors.
Conclusions
From the results of this study, we suggest that the inhibition of HDAC10 may potentiate the effects of platinum therapies in ovarian tumors.
Introduction
Histone modifications have been central in the understanding of post-translational modifications and their effects on the regulation of gene expression [1]. Histone acetylation is a reversible process, governed by two classes of enzymes: histone acetyl transferases (HATs) and histone deacetylases (HDACs). The acetylation of lysine on histone proteins is generally stimulatory to mRNA synthesis [2]. One suggested mechanism is the neutralization of the positively charged lysine residue by acetylation loosens the interaction of the DNA with the underlying nucleosome, providing access to transcriptional machinery [3]. Alternatively, the transcriptional control could be modulated through direct protein interactions, or a combination of the above [4]. HDACs catalyze the reverse reactions, contributing to transcriptional repression by rendering the chromatin to a state that is functionally silenced [5]. In this study we focus specifically on one of these histone deacetylases, HDAC10.
Although originally classified as ‘histone’ deacetylases, these enzymes are not limited to histone substrates. Acetylated lysine on non-histone proteins have been shown to regulate multiple cellular functions, which can be reversed by HDACs [6]. Most findings about HDACs focus on these factors as repressors of transcription, but our understanding of the roles of HDACs continues to expand [7, 8]. As an example, HDAC1, HDAC2, and HDAC3 have all shown tumor-suppressive genomic stability function [9, 10]. HDAC9 and HDAC10 have been shown to stimulate homologous recombination in HeLa cells [11]. The DNA repair function of HDAC10 prompted us to explore a possible association with cancer.
Ovarian cancer is in many cases associated with defects in the repair of DNA double strand breaks (DSB). Mutation in either BRCA1 or BRCA2 is associated with ovarian cancer, and the proteins encoded by these genes are essential regulators of DSB repair [12, 13]. Tumor cells that are BRCA1 or BRCA2 deficient are sensitive to inhibitors of Poly-ADP-ribose Polymerase (PARP) [14], and PARP inhibitors are increasingly found to be effective in killing ovarian cancer cell lines and as a potential therapy for ovarian cancer [15]. Other proteins that stimulate the DSB repair pathway may also contribute to tumorigenesis when mutated and may provide targets for therapy.
In this study we find that HDAC10 is either expressed at low level or deleted in a subset of ovarian cancers. Additionally, we find a significant correlation with sensitivity to platinum-based therapy and low levels of HDAC10 mRNA within the same tumor samples. Based on our results from the in vitro studies, we suggest that inhibition of HDAC10 may potentiate the response to platinum-based therapy in ovarian cancer.
Materials and Methods
Cell Culture and Reagents
HeLa DR-13-9 cells utilized for homology directed repair have been previously described [16] and cultured using standard HeLa culturing protocols. UWB1.289 ovarian carcinoma cells were purchased from ATCC (Manassas, VA) and cultured according to manufacturer specifications. HDAC inhibitors trichostatin A (TSA) and suberanilohydroxamic acid (SAHA) were purchased from Sigma-Aldrich (St. Louis, MO). HDAC10 and control siRNAs were synthesized and purchased from Integrated DNA Technologies (Coralville, IA). Sequences for the siRNAs are listed in Table 1. MTT reagent, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide and comet assay lysis buffer were purchased from Trevigen (Gaithersburg, MD). SYBR Green used in the comet assay was purchased from Bio-Rad (Hercules, CA).
Table 1.
siRNA sequences for HDAC10 study
| Name | Sequence |
|---|---|
| GL2 sense | 5’- CGU ACG CGG AAU ACU UCG ATT - 3’ |
| GL2 antisense | 5’- UCG AAG UAU UCC GCG UAC GTT - 3’ |
| HDAC10-1 sense | 5’ - CGG AGU CAG UGU GCA UGA CAG UAC A - 3’ |
| HDAC10-1 antisense | 5’- UGU ACU GUC AUG CAC ACU GAC UCC G - 3’ |
| HDAC10-2 sense | 5’- UCA CUG CAC UUG GGA AGC UCC UGU A - 3’ |
| HDAC10-2 antisense | 5’ - UAC AGG AGC UUC CCA AGU GCA GUG A - 3’ |
Sequences of siRNAs in study. dTdT are present on the 3’ end of every oligo.
Primary ovarian carcinoma cell population isolation
Primary ovarian carcinoma cell populations used in this study were isolated from patient ascites at the time of primary surgery or collected at the time of tumor recurrence from patients with advanced stage ovarian carcinoma. All patients signed consent forms, and the use of patient samples was approved under the Ohio State University Human Investigations Committee (IRB # 2004C0124). Cells were grown using MCDB/M199 media as previously described [17, 18].
Homology Directed Repair Assay (HDR)
The HDR assay we utilized has been previous described and characterized [16, 19, 20]. 300,000–400,000 HeLa cells were plated in each well of a 6-well tissue culture dish (~9.5 cm2) on day 1. On Day 2, media in wells was replaced with media containing 0, 75, 150, or 300 nM of trichostatin A (TSA) or 0, 0.5, 1.0, or 2.0 µM of suberanilohydroxamic acid (SAHA). Cisplatin was added to each of the TSA and SAHA treatments at 0, 100, and 300 nM concentrations. On Day 3, in fresh media and in the absence of the drug, 3 µg of pCBASce1 plasmid containing I-Sce1 endonuclease was transfected using 3–5 µl of Lipofectamine 2000, first diluted in 125 µl Opti-MEM each. Four to six hours later, the culture media was replaced with fresh media containing 0, 75, 150, or 300 nM of trichostatin A (TSA) or 0, 0.5, 1.0, or 2.0 µM of SAHA, as appropriate. Cisplatin was added to each of the TSA and SAHA treatments at 0, 100, and 300 nM concentrations. Cells were then incubated for 72 hours. The cells in each treatment sample were then treated with trypsin and resuspended in 0.5–1 mL PBS. 10,000 cells were counted using a BD Biosciences FACSCalibur instrument available at the Ohio State University Comprehensive Cancer Center Analytical Flow Cytometry core laboratory. The 10,000 cells were gated using an area of the forward scatter-side scatter plot to optimize live cell counting. The number of cell expressing green fluorescent protein (GFP) was recorded for each sample and the GFP percentage was normalized against untreated samples. The experiment was repeated in quadruplicate.
Comet Assay
The comet assay measures eukaryotic cell DNA damage using gel electrophoresis [21]. Damaged DNA is electrophoresed away from cells suspended in agarose, forming a comet tail shape that can be visualized and quantified. UWB1.289 cells were plated in 6-well tissue plates (~9.5 cm2 per well). Once the cells reached a confluence of approximately 60%, they were transfected with control siRNA targeting luciferase (GL2), or siRNA targeting two different HDAC10 sequences (HDAC10-1 and HDAC10-2). 100 pmol of each siRNA was diluted in 250 µl of Opti-MEM, and 5 µl of Oligofectamine was similarly diluted in 250 µl of Opti-MEM and the transfection was carried out according to manufacturer specifications. 48 hours later, a second transfection was repeated with the same volumes and incubation times. Five days after the initial transfection, the cells were exposed to 4 Gy of x-ray ionizing radiation. The cells were then incubated for four hours, then detached from the plate with trypsin, mixed with culture media, pelleted at low speed, and resuspended in 1 mL of PBS. 5 µl of the suspension was diluted with 45 µl melted low melting point agarose and applied to a microscope slide. Slides were refrigerated to solidify the agarose, and then incubated in comet assay lysis buffer at 4 °C for 45 minutes. The lysis buffer was then aspirated, replaced with electrophoresis buffer, and equilibrated in this buffer for 25 minutes. The cells are then placed in an electric field of 21 volts for 45 minutes. DNA is then precipitated using a precipitation buffer and incubated at room temperature for 25 minutes followed by aspiration and drying overnight. SYBR Green I solution (0.1 ml) was added to each slide for 30 minutes, and the solution was then gently removed and dried for 10–20 minutes. Slides were imaged with an Axiocamera instrument and analyzed with CometScore (TriTek, Wilmington, DE) software. Each treatment and slide was prepared in triplicate.
MTT Assay
UWB1.289 MTT assays follow similar cell culture and transfection procedures as the comet assay until day five. On day five, or 48 hours after the second transfection, 2000 cells of each treatment were counted and plated in a 96-well plate (~0.143 cm2). Cells were cultured for 72 hours in a 96 well plate.
Primary ovarian cancer cells were counted and 1000 cells were plated per well of a 96-well tissue culture plate. 0, 5, 10, and 20 µM cisplatin and 0 and 75 nM TSA were applied to both platinum sensitive and platinum resistant primary ovarian cells.
Three days after the start of the MTT assay, 10 µl of 12 mM MTT stock solution (~10% of the well volume) was added to each well. The cells were incubated at 37 °C for four hours, after which cell membranes were disrupted with a 10% sodium dodecyl sulfate (SDS), 0.01 M hydrochloric acid (HCl) solution. After a final four-hour incubation at 37 °C, the plate is shaken to ensure mixing and the absorbance is measured at 600 nm. Each treatment was performed in triplicate.
Statistics
Error bars represent the standard error of mean. P-values represent two-sided student’s t-tests unless otherwise indicated. Two-way ANOVA analysis was used to test for interaction between TSA and cisplatin treatments in chemotherapy sensitive and resistant primary ovarian tumor cells. ANOVA P-values are interaction P-values.
Results
HDAC10 is deleted in ovarian cancer and associated with sensitivity to cisplatin
We had found that HDAC9 and HDAC10 stimulate the repair of DNA double strand breaks (DSBs) by homologous recombination [11]. We utilized the Database of Genomic Variants (DGV) [22], to detect common deletions in the loci. The HDAC10 gene is in the middle of a large multi-gene deletion that has been observed as heterozygous in 3 out of 443 normal individuals investigated [23] and in 34 cases in 6533 samples [24]. The HDAC10 locus on chromosome 22 is indicated with the deletions (Figure 1A). When looking at the incidence of mutations in the genes encoding these proteins in tumor samples, using The Cancer Genome Atlas (TCGA) (http://cancergenome.nih.gov/) and the web tool cBioPortal for visualization and analysis [25, 26], we found that HDAC10 was deleted in a set of serous ovarian cancers (Figure 1B). We initially screened genetic changes to HDAC10 across multiple tumor types, including a large dataset for serous ovarian cancer. This ovarian dataset had two different gene copy analyses and indicated a high rate of HDAC10 deletion. From a TCGA provisional dataset with 311 samples, 10% of the tumors had a deep deletion of the HDAC10 gene. Deep deletion indicates that more than one allele is deleted, and if there are only two copies of the chromosome, then the locus would be homozygous deleted. A similar dataset analyzed in 2011 with 316 samples indicated about 5% of ovarian cancers with a deep deletion of HDAC10. Both gene copy analyses indicate ovarian tumors had the highest HDAC10 deletion rates out of all the available cancer datasets. Certainly, the frequency of deletion of HDAC10 was higher among ovarian cancers than observed in the general population using DGV. The dataset was also analyzed for loss of BRCA1. Although deep deletion of BRCA1 was relatively rare, approximately 10% of the tumors had a nonsense BRCA1 mutation. Two tumor samples had both an HDAC10 deletion and BRCA1 nonsense mutation.
Figure 1. HDAC10 is deleted in many ovarian tumors, and loss of HDAC10 correlated with sensitivity to cisplatin.
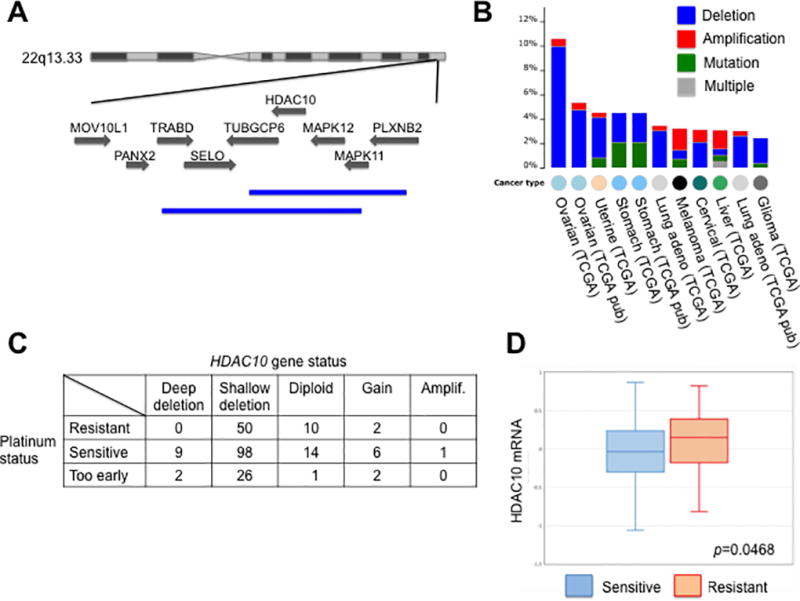
A. The chromosome 22 locus containing the HDAC10 gene is shown, and deletions found as a common variant were shown in blue at the bottom. B. Frequency of HDAC10 alteration in tumor types is indicated. Data were taken from the TCGA database using software from CBioPortal. C. Some of the tumors in the TCGA ovarian cancer dataset were linked with information about cisplatin sensitivity of the tumor. The status of the HDAC10 gene was indicated in columns. D. HDAC10 mRNA abundance in tumor samples from cisplatin-sensitive tumors (blue) was compared to mRNA abundance in cisplatin-resistant tumors (red). The statistical test used was an unpaired student’s t-test.
The uncontrolled cell division of cancers makes DNA a prime target for disrupting the multiple processes needed to sustain the proliferation. Cisplatin is an interstrand DNA crosslinker, interfering with mitosis as well as initiating the apoptosis response of the DNA damage response pathway [27]. Since HDAC10 has been shown to be involved in DNA repair [11], the first characteristic we evaluated was platinum sensitivity. We hypothesized that patients who were deficient in HDAC10 would be more sensitive to platinum therapy. Sensitivity to platinum was known for a subset of ovarian cancers in the TCGA dataset. As shown in Figure 1C, all cancers that had deep deletions of HDAC10 were sensitive to platinum therapy. 66.2% of shallow deletions and 63.6% of diploid or amplified HDAC10 tumors were sensitive to platinum therapy. These results indicated the possibility that the loss of HDAC10 in tumors with deep deletions helps sensitize cells to platinum therapy, and we suggest that when HDAC10 is diploid or amplified other factors influence platinum sensitivity. However the sample size of the deep deletion patients was too small to evaluate statistical significance.
Data regarding HDAC10 DNA copy numbers in cisplatin sensitive tumors were complemented by transcriptional analysis. The HDAC10 mRNA levels correlated with platinum sensitivity in the patients. In the subset of tumors in which platinum status was available, 62 patients were resistant to platinum therapy, while 128 patients were sensitive to platinum therapy. The mean of HDAC10 mRNA levels in the resistant samples was significantly higher than the sensitive samples (Figure 1D). Tumor data suggested that HDAC10 copy loss or low expression were correlated with maintaining sensitivity to cisplatin. This result initiated our investigation of HDAC10 and HDAC inhibitor effects in a number of cell culture assays with an emphasis on ovarian cancer cells and platinum therapy.
HDAC inhibitors enhance the cytotoxicity of primary tumor cells to cisplatin
Since decreased activity of HDAC10 enhanced the sensitivity of cells to cisplatin, we tested whether generally functioning HDAC inhibition enhances cisplatin sensitivity in primary ovarian cancer cells. The primary cells were derived from ascites of patients with ovarian tumors. These cells can only be grown for three passages, making it impossible to use the siRNA transfection; instead, we were able to culture these cells in the presence of HDAC inhibitors and cisplatin. We performed MTT proliferation assays in primary tissues cells, treating cells with various concentrations of cisplatin (0, 5, 10, and 20 µM) and the HDAC inhibitor, Trichostatin A [TSA; 0 nM (red), 75 nM (blue), and 150 nM (green)]. We analyzed three cell lines derived from cisplatin sensitive ovarian tumors, A-195, A2780CA, and OVCAR8 (Figure 2, left), and two cell lines derived from cisplatin resistant ovarian tumors, TR-127 and TR-182 (Figure 2, right); each analysis was done in triplicate. As shown in Figure 2, TSA enhanced the effect of cisplatin on cell proliferation in both sensitive and resistant primary cells. In the case of the TR-182 cells, the effect of the HDAC inhibitor was strongest at low concentration of cisplatin. The results in Figure 2 are consistent with a similar analysis of SAHA and paclitaxel in ovarian cancer cell lines [28].
Figure 2. HDAC inhibitor increases the sensitivity to cisplatin of cells derived from primary ovarian tumors.
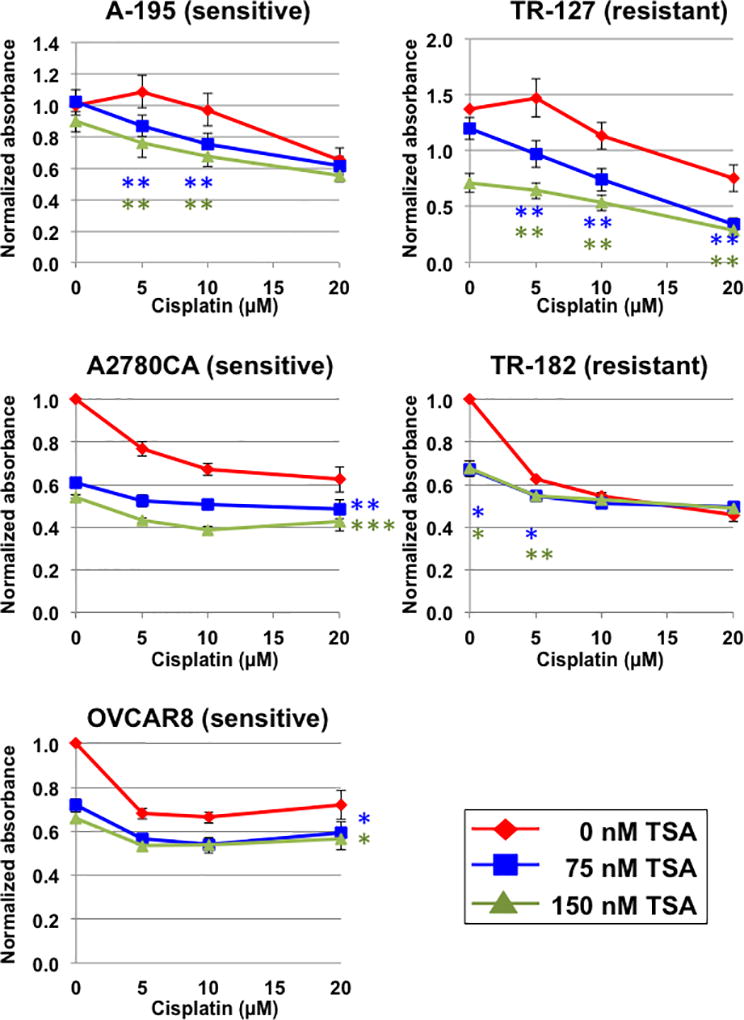
Five primary cell lines derived from the ascites of ovarian tumors were cultured in the presence of cisplatin and the HDAC inhibitor TSA. A-195, A2780CA, and OVCAR8 cells were derived from cisplatin sensitive tumors, and TR-127 and TR-182 were derived from cisplatin resistant tumors. After 3 days, cells were analyzed for proliferation using the MTT assay. For each condition, analyzed in triplicate, cells were normalized to the MTT value of the cells without added cisplatin or TSA. For statistical analysis the unpaired student’s t-test compared the effect of TSA at a given concentration of cisplatin; * p < 0.05, ** p < 0.01, and *** p < 0.001. In the cases of A2780CA and OVCAR8, the statistical analysis was similar for each concentration of cisplatin tested, and the asterisks are indicated to the right of the line.
HDAC inhibitors in combination with cisplatin inhibit DNA repair by homologous recombination
Cisplatin has multiple mechanisms of action through which it modulates its cytotoxic effect, including DNA crosslinking, and we tested whether cisplatin inhibited DNA repair by homology directed repair (HDR) and whether HDAC inhibition would enhance the effect cisplatin had on HDR. HDR activity was measured in HeLa cells, and we infer that effects of HDAC inhibitors in this assay would relate to ubiquitous function and not be specific to ovarian cells. We focused on inhibitors that are effective with HDAC10. Trichostatin A (TSA) and suberanilohydroxamic acid (SAHA) both inhibit HDAC10, though SAHA inhibits HDAC10 to a lesser degree than the other HDACs. There is currently no HDAC10 specific inhibitor [29]. Cisplatin, alone, inhibited homologous recombination in our HDR assay at concentrations as low as 100 nM cisplatin. The effect of cisplatin inhibition of HDR had been observed before [30]. Interestingly, the inhibition of HDR was at concentrations lower than those at which cisplatin is cytotoxic. Importantly, HDAC inhibition with TSA concentrations of 150 nM or 300 nM further reduced the HDR activity of these cells, even when already inhibited by cisplatin (Figure 3A). Similarly, when SAHA was included in the medium at 1.0 µM or 2.0 µM, the HDR activity was also reduced when in the presence of cisplatin (Figure 3B).
Figure 3. HDAC inhibitors, in combination with cisplatin, potently inhibit the homology directed repair of DNA double strand breaks.
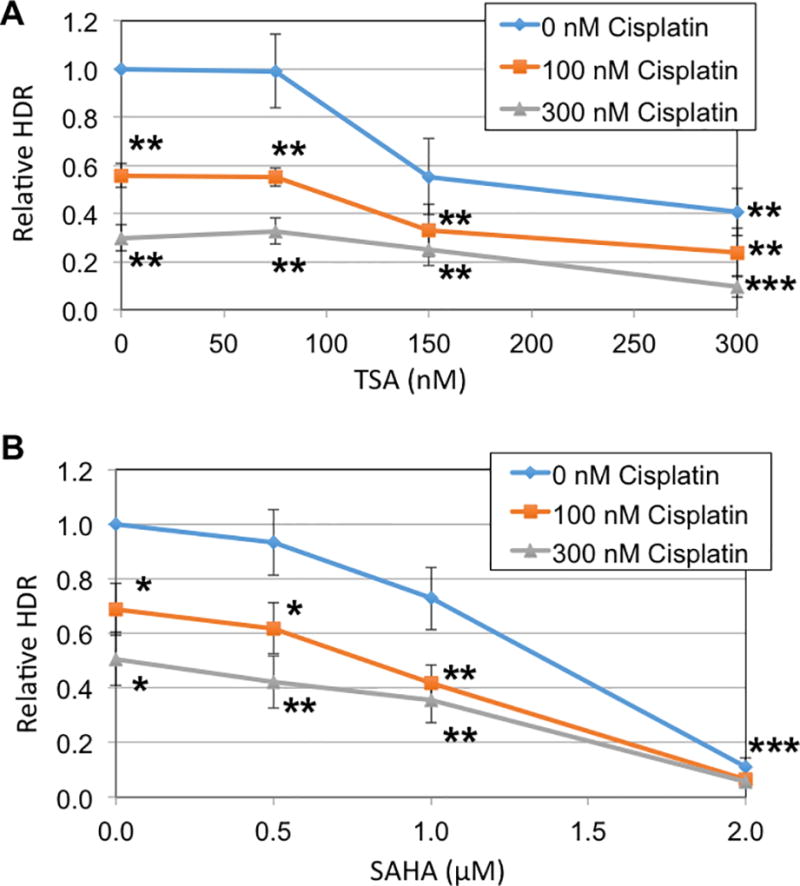
A. HeLa-DR cells, which have stably integrated a DNA for measuring HDR activity, were treated with cisplatin and TSA at the indicated concentrations. HDR activity was measured as the percentage of cells that convert to GFP positive, and results were normalized relative to the sample without drug added. Statistical analysis compared each data point with the indicated drug to the results in the absence of added drug; the student’s two-tailed t-test was used, and asterisks represent *, p < 0.05; **, p < 0.01; ***, p < 0.001. B. HeLa-DR cells were tested as in panel A using the HDAC inhibitor SAHA. Statistical tests were done as in Panel A; for the results in the presence of 2.0 µM SAHA, all three curves had p < 0.001.
We have previously established that HDAC10 stimulates HDR in HeLa cells, consistent with the results of the effects of cisplatin and HDAC inhibition in the HDR assay. In the following experiment we tested whether HDAC10 was involved in DNA repair with ovarian cancer cells. To study this, we utilized a comet assay in an ovarian carcinoma cell line, UWB1.289. We exposed cells treated with HDAC10 siRNA to 4 Gy ionizing radiation, allowed four hours for recovery, and embedded treated cells in agarose with neutral pH buffer. When exposed to an electric field, DNA with double strand breaks (DSBs) electrophorese out and form a comet tail, which can be quantified for the level of unrepaired DNA. The tail moments detected in irradiated cells that had been transfected with either of two different non-overlapping siRNAs targeting HDAC10 were significantly larger than the tail moments from cells transfected with the control siRNA. The larger tail moment indicated more unrepaired DNA damage four hours post irradiation than in control transfected cells (Figure 4). The UWB1.289 cells are null for the BRCA1 gene, and both BRCA1 and HDAC10 function in the repair of DSBs by homology directed repair [11]. Since both factors participate in the same pathway, it might be expected that depletion of HDAC10 would be epistatic with the BRCA1 deletion, but instead the DSB repair defect was worsened when HDAC10 was depleted. It is possible that this ovarian cancer cell line has partially compensated for the deficiency in BRCA1 by activating a bypass pathway, and HDAC10 inhibition caused the compensation to fail. UWB1.289 cells did not show significant DNA damage after ionizing radiation under control conditions despite being BRCA1 negative, while other cell lines that have BRCA1 deficiency exhibit significant DNA damage in a comet assay [31].
Figure 4. Depletion of HDAC10 from the UWB1.289 ovarian cancer cell line resulted in delay in repair of DSBs.
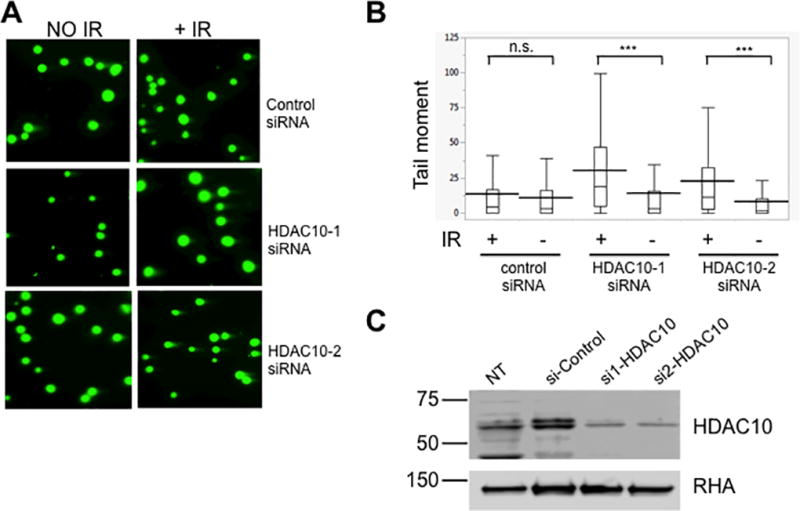
A. UWB1.289 cells were transfected with two rounds of the indicated siRNA, and 48 h post transfection irradiate with 4 Gy of x-ray. After 4 h of recovery, cells were subjected to a comet assay in the presence of neutral agar. B. The amount of unrepaired DSBs in each sample from panel A was quantified by measuring the tail moment for each cell. Statistical analysis used the unpaired student’s t-test; *** indicates p < 0.0001. C. Immunoblot analysis of the depletions of HDAC10 in samples used in panel A (top) and analysis of RNA Helicase A (RHA; bottom) to evaluate equal loading of the samples. Samples were whole cell extracts from non-transfected (NT), control siRNA (si-Control), HDAC10 specific siRNA-1 (si1-HDAC10), and HDAC10 specific siRNA-2 (si2-HDAC10) cells.
We hypothesized that the inhibition of HDAC10 would sensitize the UWB1.289 cells to the DNA damage and cytotoxic effects of platinum therapy. To test this hypothesis, we performed an MTT assay. MTT is a measure of metabolically active cells, which we use to measure proliferative activity. We transfected the cells with two rounds of siRNA specific for HDAC10 or control, and 48 hours after the second transfection, we incubated the cells in the presence of different concentrations of cisplatin. After three days of treatment with cisplatin at a range of concentrations from 0 to 80 µM, we performed the MTT assay. When treating UWB1.289 with cisplatin, cells transfected with HDAC10 siRNA had significantly lower proliferation as compared to cells transfected with control siRNA (Figure 5A) consistent with a decrease in abundance of the HDAC10 protein analyzed in these cells (Figure 5B). This result demonstrated the potential enhancement the inhibition of HDAC10 can have on cisplatin in BRCA1 deficient ovarian tumors.
Figure 5. Depletion of HDAC10 potentiates cytotoxicity of cisplatin on UWB1.289 cells.
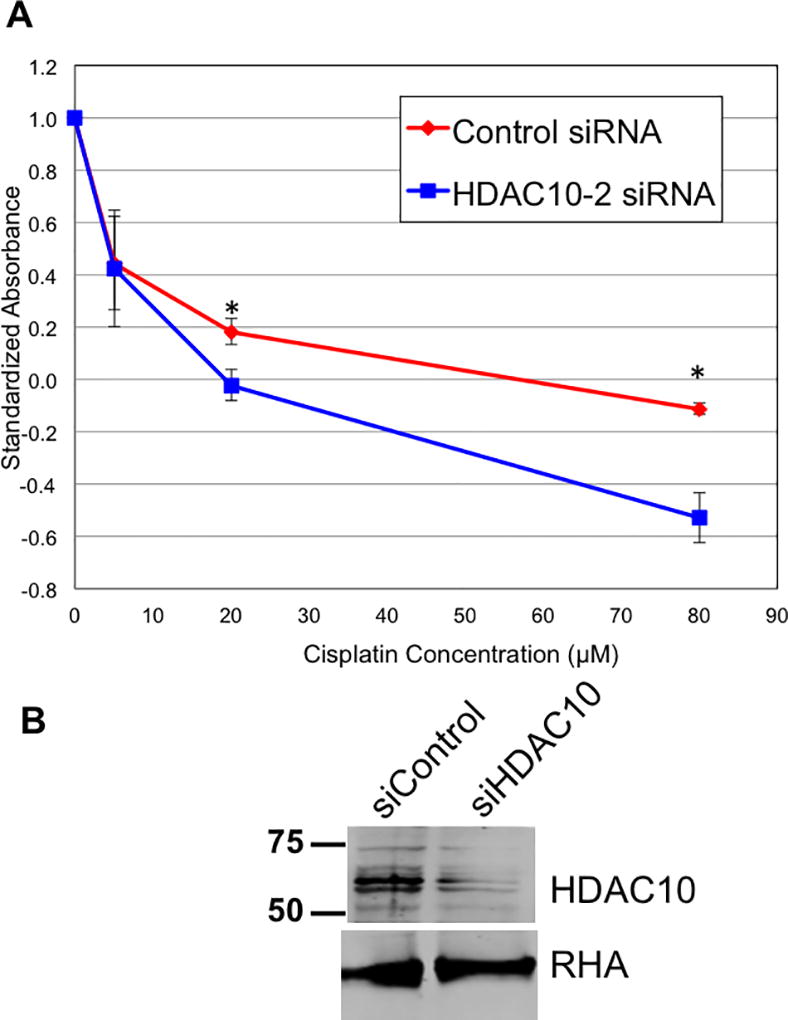
A. UWB1.289 cells were subjected to two rounds of siRNA transfection with control siRNA (red) or HDAC10 specific siRNA (blue) and analyzed for the effect of cisplatin on proliferation using the MTT assay. Statistically different datapoints were determined by student’s t-test and indicated by * (p < 0.05). B. Immunoblot analysis of the depletions of HDAC10 in samples used in panel A (top) and analysis of RNA Helicase A (RHA; bottom) to evaluate equal loading of the samples.
Discussion
The standard front-line adjuvant therapy for ovarian cancer includes platinum-based chemotherapy [32]. Initial response is often promising, where 80% of patients have a response to platinum. Unfortunately, most patients relapse and go on to die of chemotherapy resistant disease [33]. As long as ovarian tumors are sensitive to cisplatin, progression of the cancer is arrested; thus any mechanism for maintaining cisplatin sensitivity of tumors is desirable. In this study we discovered: 1) serous ovarian cancers more commonly have HDAC10 deletions than the general population; 2) in ovarian cancers homozygous deletion of HDAC10 correlated with tumor sensitivity to cisplatin; 3) HDAC inhibitors potentiated the cytotoxicity of cisplatin in primary ovarian cancer cell lines derived from tumor ascites; 4) HDAC inhibitors potentiated the inhibition by cisplatin of DNA repair by homologous recombination; and 5) HDAC10 was required for DNA repair and survival in cells damaged by ionizing radiation or cisplatin treatment.
Our results indicated that HDAC inhibition of HDR could enhance the first line platinum therapy and thus could improve survival in patients with ovarian cancer. Several clinical trials are already underway of HDAC inhibitors (including SAHA) in combination with platinum-based chemotherapy in ovarian cancer [34]. Preclinical studies corroborate our finding of platinum sensitization in ovarian cancer cells using other ovarian cells lines when treated with SAHA [28]. We propose, based on our findings, that HDAC10 inhibition enhances the platinum sensitization of ovarian carcinoma cells. Strikingly, even in the ovarian cell line with BRCA1 deleted, HDAC10 stimulated DSB repair.
A major issue with platinum therapy and ovarian cancer is relapse as a result of development of platinum resistance. One strategy to potentiate the effectiveness of platinum therapy is to use a combination therapy including HDAC inhibitors along with the platinum; however, such combination therapy is associated with significant drug toxicities [35, 36]. Since most HDAC inhibitors do not inhibit, or have only low levels of inhibition of HDAC10 [29], results from this study suggest that an HDAC10 isoform-specific inhibitor could improve the platinum sensitivity in ovarian carcinoma tumors and possibly with lower toxicity.
HDAC inhibitors have become an important class of drugs in the search for new therapies against cancer among other diseases. Several current clinical trials analyze HDAC inhibitors as anti-tumor agents [29]. Most of these inhibitors are broad spectrum inhibitors, affecting both class I and class II HDACs. Although many HDAC inhibitor studies are in progress, most of these are based on empirical data, and the exact mechanisms through which these agents are producing their cytotoxic effects are currently not well understood.
Valproic acid (VPA) has been used to treat neurological disease such as epilepsy for more than 30 years. However, VPA is specific to class I HDACs, and has no effect on the class II HDAC10 [29]. VPA had no effect in the HDR assay, supporting the concept that HDAC10 is the HDAC protein associated with the DNA repair activity [11]. VPA is currently in phase I and phase II clinical trials for leukemia and cervical cancer [37]. However, VPA does have neurological toxicity side effects, which limits its therapeutic window.
Another currently FDA approved HDAC inhibitor is SAHA, also known as vorinostat. Approved for the treatment of CTCL, SAHA is currently being studied in multiple phase I and phase II studies [29, 34]. SAHA is a broad spectrum HDAC inhibitor, affecting both class I and II HDACs. Unlike the other FDA approved HDAC inhibitors, it does inhibit HDAC10, albeit to a reduced effect compared to other HDACs [29]. Out of the current clinical trials, SAHA has potential to sensitize cancer cells to platinum therapy mediated by inhibition of HDAC10 [38, 39].
Although our analysis was limited to ovarian cancer cells, the DNA repair mechanisms through which HDAC10 potentially modulates cisplatin sensitivity could be present in other cell lines. A recent clinical trial demonstrated SAHA enhanced the efficacy of platinum therapy in patients with non-small-cell lung cancer [38, 39]. Similarly, HDAC inhibitor potentiation of platinum sensitivity is not mediated solely by HDAC10. This is evidenced by the multiple trials where HDAC inhibition is in combination with platinum therapy as well as preclinical studies demonstrating other HDACs mediating platinum sensitivity [37, 40].
HDAC inhibition has tremendous potential in the search for pharmacological solutions against cancer. HDAC inhibitors affect a large number of pathways and have already been demonstrated to improve patient outcomes in certain cancers. Although there is an ever-growing library of work that is analyzing the mechanisms through which HDAC and their inhibition modulate their effects, many of their functions are still unknown [37]. HDAC10 in particular is poorly understood, only recently being discovered to be involved in DNA repair [11]. Similarly, few of the current pharmacological solutions affect HDAC10 as strongly as other HDACs [29]. HDAC inhibitors are associated with significant side effects, but if an inhibitor were developed that could target specifically HDAC10 and not HDAC1 or any of the ten other HDACs, then perhaps these significant side effects could be avoided. Our results provide evidence to explore HDAC10 specific inhibition as an adjuvant therapy to platinum therapies, especially for BRCA1-deficient ovarian carcinoma patients.
Acknowledgments
This work was supported by an intramural research grant (J.D.P. and D.C.) and by a Pelotonia training program fellowship (to M.M.I. and C.Z.P.).
References
- 1.Allfrey VG, Faulkner R, Mirsky AE. Acetylation and Methylation of Histones and Their Possible Role in the Regulation of Rna Synthesis. Proc Natl Acad Sci U S A. 1964;51:786–94. doi: 10.1073/pnas.51.5.786. doi. PMC300163. http://www.ncbi.nlm.nih.gov/pubmed/14172992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84(6):843–51. doi: 10.1016/s0092-8674(00)81063-6. doi. http://www.ncbi.nlm.nih.gov/pubmed/8601308. [DOI] [PubMed] [Google Scholar]
- 3.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21(3):381–95. doi: 10.1038/cr.2011.22. PMC3193420. http://www.ncbi.nlm.nih.gov/pubmed/21321607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Torok MS, Grant PA. Histone acetyltransferase proteins contribute to transcriptional processes at multiple levels. Adv Protein Chem. 2004;67:181–99. doi: 10.1016/S0065-3233(04)67007-0. http://www.ncbi.nlm.nih.gov/pubmed/14969728. [DOI] [PubMed] [Google Scholar]
- 5.Taunton J, Hassig CA, Schreiber SL. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272(5260):408–11. doi: 10.1126/science.272.5260.408. doi. http://www.ncbi.nlm.nih.gov/pubmed/8602529. [DOI] [PubMed] [Google Scholar]
- 6.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325(5942):834–40. doi: 10.1126/science.1175371. http://www.ncbi.nlm.nih.gov/pubmed/19608861. [DOI] [PubMed] [Google Scholar]
- 7.Glozak MA, Sengupta N, Zhang X, Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. http://www.ncbi.nlm.nih.gov/pubmed/16289629. [DOI] [PubMed] [Google Scholar]
- 8.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393(6683):386–9. doi: 10.1038/30764. http://www.ncbi.nlm.nih.gov/pubmed/9620804. [DOI] [PubMed] [Google Scholar]
- 9.Bhaskara S, Knutson SK, Jiang G, Chandrasekharan MB, Wilson AJ, Zheng S, Yenamandra A, Locke K, Yuan JL, Bonine-Summers AR, Wells CE, Kaiser JF, Washington MK, Zhao Z, Wagner FF, Sun ZW, Xia F, Holson EB, Khabele D, Hiebert SW. Hdac3 is essential for the maintenance of chromatin structure and genome stability. Cancer Cell. 2010;18(5):436–47. doi: 10.1016/j.ccr.2010.10.022. PMC3004468. http://www.ncbi.nlm.nih.gov/pubmed/21075309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heideman MR, Wilting RH, Yanover E, Velds A, de Jong J, Kerkhoven RM, Jacobs H, Wessels LF, Dannenberg JH. Dosage-dependent tumor suppression by histone deacetylases 1 and 2 through regulation of c-Myc collaborating genes and p53 function. Blood. 2013;121(11):2038–50. doi: 10.1182/blood-2012-08-450916. PMC3596963. http://www.ncbi.nlm.nih.gov/pubmed/23327920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kotian S, Liyanarachchi S, Zelent A, Parvin JD. Histone deacetylases 9 and 10 are required for homologous recombination. J Biol Chem. 2011;286(10):7722–6. doi: 10.1074/jbc.C110.194233. PMC3048658. http://www.ncbi.nlm.nih.gov/pubmed/21247901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Metcalfe KA, Lynch HT, Ghadirian P, Tung N, Olivotto IA, Foulkes WD, Warner E, Olopade O, Eisen A, Weber B, McLennan J, Sun P, Narod SA. The risk of ovarian cancer after breast cancer in BRCA1 and BRCA2 carriers. Gynecologic oncology. 2005;96(1):222–6. doi: 10.1016/j.ygyno.2004.09.039. http://www.ncbi.nlm.nih.gov/pubmed/15589605. [DOI] [PubMed] [Google Scholar]
- 13.Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266(5182):66–71. doi: 10.1126/science.7545954. doi http://www.ncbi.nlm.nih.gov/pubmed/7545954. [DOI] [PubMed] [Google Scholar]
- 14.Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434(7035):917–21. doi: 10.1038/nature03445. http://www.ncbi.nlm.nih.gov/pubmed/15829967. [DOI] [PubMed] [Google Scholar]
- 15.Fleury H, Carmona E, Morin VG, Meunier L, Masson JY, Tonin PN, Provencher D, Mes-Masson AM. Cumulative defects in DNA repair pathways drive the PARP inhibitor response in high-grade serous epithelial ovarian cancer cell lines. Oncotarget. 2016 doi: 10.18632/oncotarget.10308. http://www.ncbi.nlm.nih.gov/pubmed/27374179. [DOI] [PMC free article] [PubMed]
- 16.Parvin J, Chiba N, Ransburgh D. Identifying the effects of BRCA1 mutations on homologous recombination using cells that express endogenous wild-type BRCA1. J Vis Exp. 2011;(48) doi: 10.3791/2468. PMC3197403. http://www.ncbi.nlm.nih.gov/pubmed/21372787. [DOI] [PMC free article] [PubMed]
- 17.Shepherd TG, Theriault BL, Campbell EJ, Nachtigal MW. Primary culture of ovarian surface epithelial cells and ascites-derived ovarian cancer cells from patients. Nat Protoc. 2006;1(6):2643–9. doi: 10.1038/nprot.2006.328. http://www.ncbi.nlm.nih.gov/pubmed/17406520. [DOI] [PubMed] [Google Scholar]
- 18.Saini U, Naidu S, ElNaggar AC, Bid HK, Wallbillich JJ, Bixel K, Bolyard C, Suarez AA, Kaur B, Kuppusamy P, Hays J, Goodfellow PJ, Cohn DE, Selvendiran K. Elevated STAT3 expression in ovarian cancer ascites promotes invasion and metastasis: a potential therapeutic target. Oncogene. 2016 doi: 10.1038/onc.2016.197. http://www.ncbi.nlm.nih.gov/pubmed/27292260. [DOI] [PMC free article] [PubMed]
- 19.Ransburgh DJ, Chiba N, Ishioka C, Toland AE, Parvin JD. Identification of breast tumor mutations in BRCA1 that abolish its function in homologous DNA recombination. Cancer Res. 2010;70(3):988–95. doi: 10.1158/0008-5472.CAN-09-2850. PMC2943742. http://www.ncbi.nlm.nih.gov/pubmed/20103620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Towler WI, Zhang J, Ransburgh DJ, Toland AE, Ishioka C, Chiba N, Parvin JD. Analysis of BRCA1 variants in double-strand break repair by homologous recombination and single-strand annealing. Hum Mutat. 2013;34(3):439–45. doi: 10.1002/humu.22251. PMC3906639. http://www.ncbi.nlm.nih.gov/pubmed/23161852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tice RR, Agurell E, Anderson D, Burlinson B, Hartmann A, Kobayashi H, Miyamae Y, Rojas E, Ryu JC, Sasaki YF. Single cell gel/comet assay: guidelines for in vitro and in vivo genetic toxicology testing. Environ Mol Mutagen. 2000;35(3):206–21. doi: 10.1002/(sici)1098-2280(2000)35:3<206::aid-em8>3.0.co;2-j. doi. http://www.ncbi.nlm.nih.gov/pubmed/10737956. [DOI] [PubMed] [Google Scholar]
- 22.MacDonald JR, Ziman R, Yuen RK, Feuk L, Scherer SW. The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res. 2014;42(Database issue):D986–92. doi: 10.1093/nar/gkt958. PMC3965079. http://www.ncbi.nlm.nih.gov/pubmed/24174537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jakobsson M, Scholz SW, Scheet P, Gibbs JR, VanLiere JM, Fung HC, Szpiech ZA, Degnan JH, Wang K, Guerreiro R, Bras JM, Schymick JC, Hernandez DG, Traynor BJ, Simon-Sanchez J, Matarin M, Britton A, van de Leemput J, Rafferty I, Bucan M, Cann HM, Hardy JA, Rosenberg NA, Singleton AB. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451(7181):998–1003. doi: 10.1038/nature06742. http://www.ncbi.nlm.nih.gov/pubmed/18288195. [DOI] [PubMed] [Google Scholar]
- 24.Xu H, Poh WT, Sim X, Ong RT, Suo C, Tay WT, Khor CC, Seielstad M, Liu J, Aung T, Tai ES, Wong TY, Chia KS, Teo YY. SgD-CNV, a database for common and rare copy number variants in three Asian populations. Human mutation. 2011;32(12):1341–9. doi: 10.1002/humu.21601. http://www.ncbi.nlm.nih.gov/pubmed/21882294. [DOI] [PubMed] [Google Scholar]
- 25.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–4. doi: 10.1158/2159-8290.CD-12-0095. PMC3956037. http://www.ncbi.nlm.nih.gov/pubmed/22588877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. PMC4160307. http://www.ncbi.nlm.nih.gov/pubmed/23550210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jung Y, Lippard SJ. Direct cellular responses to platinum-induced DNA damage. Chem Rev. 2007;107(5):1387–407. doi: 10.1021/cr068207j. http://www.ncbi.nlm.nih.gov/pubmed/17455916. [DOI] [PubMed] [Google Scholar]
- 28.Dietrich CS, 3rd, Greenberg VL, DeSimone CP, Modesitt SC, van Nagell JR, Craven R, Zimmer SG. Suberoylanilide hydroxamic acid (SAHA) potentiates paclitaxel-induced apoptosis in ovarian cancer cell lines. Gynecol Oncol. 2010;116(1):126–30. doi: 10.1016/j.ygyno.2009.09.039. http://www.ncbi.nlm.nih.gov/pubmed/19875160. [DOI] [PubMed] [Google Scholar]
- 29.West AC, Johnstone RW. New and emerging HDAC inhibitors for cancer treatment. J Clin Invest. 2014;124(1):30–9. doi: 10.1172/JCI69738. PMC3871231. http://www.ncbi.nlm.nih.gov/pubmed/24382387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raaphorst GP, Leblanc M, Li LF. A comparison of response to cisplatin, radiation and combined treatment for cells deficient in recombination repair pathways. Anticancer Res. 2005;25(1A):53–8. doi. http://www.ncbi.nlm.nih.gov/pubmed/15816518. [PubMed] [Google Scholar]
- 31.Chen BY, Huang CH, Lin YH, Huang CC, Deng CX, Hsu LC. The K898E germline variant in the PP1-binding motif of BRCA1 causes defects in DNA Repair. Sci Rep. 2014;4:5812. doi: 10.1038/srep05812. PMC4108927. http://www.ncbi.nlm.nih.gov/pubmed/25056273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agarwal R, Kaye SB. Ovarian cancer: strategies for overcoming resistance to chemotherapy. Nat Rev Cancer. 2003;3(7):502–16. doi: 10.1038/nrc1123. http://www.ncbi.nlm.nih.gov/pubmed/12835670. [DOI] [PubMed] [Google Scholar]
- 33.Greenlee RT, Hill-Harmon MB, Murray T, Thun M. Cancer statistics, 2001. CA Cancer J Clin. 2001;51(1):15–36. doi: 10.3322/canjclin.51.1.15. doi. http://www.ncbi.nlm.nih.gov/pubmed/11577478. [DOI] [PubMed] [Google Scholar]
- 34.Modesitt SC, Sill M, Hoffman JS, Bender DP Gynecologic Oncology G. A phase II study of vorinostat in the treatment of persistent or recurrent epithelial ovarian or primary peritoneal carcinoma: a Gynecologic Oncology Group study. Gynecol Oncol. 2008;109(2):182–6. doi: 10.1016/j.ygyno.2008.01.009. http://www.ncbi.nlm.nih.gov/pubmed/18295319. [DOI] [PubMed] [Google Scholar]
- 35.Matulonis U, Berlin S, Lee H, Whalen C, Obermayer E, Penson R, Liu J, Campos S, Krasner C, Horowitz N. Phase I study of combination of vorinostat, carboplatin, and gemcitabine in women with recurrent, platinum-sensitive epithelial ovarian, fallopian tube, or peritoneal cancer. Cancer Chemother Pharmacol. 2015;76(2):417–23. doi: 10.1007/s00280-015-2813-9. http://www.ncbi.nlm.nih.gov/pubmed/26119093. [DOI] [PubMed] [Google Scholar]
- 36.Mendivil AA, Micha JP, Brown JV, 3rd, Rettenmaier MA, Abaid LN, Lopez KL, Goldstein BH. Increased incidence of severe gastrointestinal events with first-line paclitaxel, carboplatin, and vorinostat chemotherapy for advanced-stage epithelial ovarian, primary peritoneal, and fallopian tube cancer. Int J Gynecol Cancer. 2013;23(3):533–9. doi: 10.1097/IGC.0b013e31828566f1. http://www.ncbi.nlm.nih.gov/pubmed/23385285. [DOI] [PubMed] [Google Scholar]
- 37.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006;6(1):38–51. doi: 10.1038/nrc1779. http://www.ncbi.nlm.nih.gov/pubmed/16397526. [DOI] [PubMed] [Google Scholar]
- 38.Ramalingam SS, Maitland ML, Frankel P, Argiris AE, Koczywas M, Gitlitz B, Thomas S, Espinoza-Delgado I, Vokes EE, Gandara DR, Belani CP. Carboplatin and Paclitaxel in combination with either vorinostat or placebo for first-line therapy of advanced non-small-cell lung cancer. J Clin Oncol. 2010;28(1):56–62. doi: 10.1200/JCO.2009.24.9094. PMC2799233. http://www.ncbi.nlm.nih.gov/pubmed/19933908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramalingam SS, Parise RA, Ramanathan RK, Lagattuta TF, Musguire LA, Stoller RG, Potter DM, Argiris AE, Zwiebel JA, Egorin MJ, Belani CP. Phase I and pharmacokinetic study of vorinostat, a histone deacetylase inhibitor, in combination with carboplatin and paclitaxel for advanced solid malignancies. Clin Cancer Res. 2007;13(12):3605–10. doi: 10.1158/1078-0432.CCR-07-0162. http://www.ncbi.nlm.nih.gov/pubmed/17510206. [DOI] [PubMed] [Google Scholar]
- 40.Kim MG, Pak JH, Choi WH, Park JY, Nam JH, Kim JH. The relationship between cisplatin resistance and histone deacetylase isoform overexpression in epithelial ovarian cancer cell lines. J Gynecol Oncol. 2012;23(3):182–9. doi: 10.3802/jgo.2012.23.3.182. PMC3395014. http://www.ncbi.nlm.nih.gov/pubmed/22808361. [DOI] [PMC free article] [PubMed] [Google Scholar]


