Abstract
Intestinal colonization by the foodborne pathogen Escherichia coli O157:H7 leads to serious disease symptoms, including hemolytic uremic syndrome (HUS) and hemorrhagic colitis (HC). Synthesis of one or more Shiga toxins (Stx) is essential for HUS and HC development. The genes encoding Stx, including Stx2a, are found within a lambdoid prophage integrated in the E. coli O157:H7 chromosome. Enhanced Stx2a expression was reported when specific non-pathogenic E. coli strains were co-cultured with E. coli O157:H7, and it was hypothesized that this phenotype required the non-pathogenic E. coli to be sensitive to stx-converting phage infection. We tested this hypothesis by generating phage resistant non-pathogenic E. coli strains where bamA (an essential gene and Stx phage receptor) was replaced with an ortholog from other species. Such heterologous gene replacement abolished the ability of the laboratory strain E. coli C600 to enhance toxin production when co-cultured with E. coli O157:H7 strain PA2, which belongs to the hypervirulent clade 8. The extracellular loops of BamA (loop 4, 6, 7) were further shown to be important for infection by stx2a-converting phages. However, similar gene replacement in another commensal E. coli, designated 1.1954, revealed a bamA-independent mechanism for toxin amplification. Toxin enhancement by 1.1954 was not the result of phage infection through an alternative receptor (LamB or FadL), lysogen formation by stx2a-converting phages, or the production of a secreted molecule. Collectively, these data suggest that non-pathogenic E. coli can enhance toxin production through at least two mechanisms.
Keywords: E. coli O157:H7, commensal E. coli, Shiga toxin, Stx2a, BamA
Introduction
Shiga toxin-producing Escherichia coli (STEC) are estimated to cause more than 265,000 illnesses annually in United States, with 3,600 hospitalizations and 30 deaths (Centers for Disease Control and Prevention (CDC), 2012). The foodborne pathogen E. coli O157:H7 is a notorious serotype of STEC which continues to cause various multistate outbreaks. Ingestion of a low infectious dose of <100 cells (Tilden et al., 1996) leads to outcomes ranging from asymptomatic carriage, bloody diarrhea, to life-threatening renal complications of hemolytic uremic syndrome (HUS) (Rangel et al., 2005; Tarr et al., 2005). Cattle are the natural reservoir of E. coli O157:H7 and asymptomatic carriers (Borczyk et al., 1987). Accordingly, beef is the primary food linked to outbreaks, however, E. coli O157:H7's persistence in water, soil, and manure enhances its transmission to food such as fresh produce (Hilborn et al., 1999).
Shiga toxin (Stx) is required for progression of disease to severe outcomes including HUS. It is an AB5 toxin, whose B pentamer binds to globotriaosylceramide (Gb3) on host cell membranes (Waddell et al., 1988). The enzymatic A subunit is delivered into the cytoplasm of eukaryotic cells, and inhibits protein synthesis, resulting in apoptosis and cell death (Saxena et al., 1989; Sandvig and Van Deurs, 1992). Stx has two immunologically distinct isoforms, Stx1 and Stx2. They share 56% identity at the amino acid sequence level (Jackson et al., 1987), however Stx2 is 400-fold more potent than Stx1 (Tesh et al., 1993) and more likely to be associated with severe disease outcomes (Kawano et al., 2008). Seven allelic variants of Stx2, from Stx2a to Stx2g have been described (Scheutz et al., 2012). Epidemiological investigation showed that Stx2a is more frequently found in strains causing HUS (Friedrich et al., 2002; Persson et al., 2007).
The genes encoding Stx are present in temperate prophages (Hayashi et al., 2001; Perna et al., 2001). During the lytic cycle, the prophage excises from the host chromosome, utilizes the host machinery to replicate, assembles new virons, and eventually lyses the host. Conversely, in the lysogenic state, the prophage replicates along with the host without causing substantial cell lysis. The switch between cycles is controlled by cI. During the lysogenic state, the repressor CI dimerizes and inhibits transcription from the promoters PL and PR. However, when the SOS response is triggered by DNA damage, activated RecA cleaves CI repressor and de-represses PL and PR, leading to prophage induction, Stx expression and cell lysis (Waldor and Friedman, 2005).
The progeny stx-converting phages may infect other E. coli strains after adsorbing to the outer membrane proteins including BamA (Watarai et al., 1998; Smith et al., 2007). BamA is essential for outer membrane protein biogenesis (Wu et al., 2005) and exists in all members of the Enterobacteriaceae family. While the amino acid sequence of BamA is nearly invariant between strains of E. coli, the extracellular loops 4, 6, and 7 exhibit heterogeneity between different species (Smith et al., 2007). Genetic experiments supporting BamA as a stx2-converting phage receptor have been difficult to perform since it is an essential gene. However, Ruhe et al. (2013) developed an approach for deleting the chromosomal copy of bamA by complementing in trans with that from E. coli or other Enterobacteriaceae. This system was used to identify extracellular loops 6 and 7 of BamA as critical for cell-to-cell contact of the CdiA contact-dependent growth inhibition (CDI) system.
Enhanced Stx2 production by O157:H7 can be triggered by the addition of antibiotics. Ciprofloxacin, for instance, can increase Stx production more than 40-fold (Zhang et al., 2000). It has also been proposed that Stx production can be modulated by other members of the gut microflora (de Sablet et al., 2009; Thévenot et al., 2015). Other E. coli such as laboratory strain C600 were shown to produce Stx2 upon addition of stx2a-converting phages, leading to a model that C600 enhancement of Stx2a production requires phage infection and replication (Gamage et al., 2003). Our own previous study supported this by showing C600 increased Stx2a production of O157:H7 when the two bacteria were co-cultured, and during growth the viable cell counts for C600 decreased; this phenotype has been validated in vitro and in vivo (Goswami et al., 2015). In that study, O157:H7 strain PA2 was used as a model as it belongs to the hypervirulent clade 8 (Manning et al., 2008; Hartzell et al., 2011), and was the highest Stx2a producer of strains tested when co-cultured with C600 (Goswami et al., 2015).
Genetic evidence demonstrating that toxin amplification acts through phage infection of C600 has been lacking. Additionally, it is unknown whether this model represents the main mechanism by which commensal E. coli enhance Stx2a production. To further study this toxin amplification phenotype, we hypothesized that other strains of E. coli would amplify Stx2a production in a manner that is distinct from the one described for C600. Our objectives were: (1) to take a genetic approach to confirm that toxin amplification requires C600 to be sensitive to stx2a-converting phages; (2) to characterize the mechanism of Stx2a amplification by a commensal E. coli strain designated 1.1954, which functions through a mechanism distinct from that described for C600.
Materials and methods
Strains and culture conditions
All the strains and plasmids used in the study are listed in Table 1. The O157:H7 strains with “PA” designations were from the Pennsylvania Department of Health collection and were characterized previously (Hartzell et al., 2011). The commensal E. coli strains were obtained from the E. coli Reference Center (ECRC) at The Pennsylvania State University. The bacteria were routinely grown in Lysogeny-Broth (LB) broth at 37°C, and their culture stocks were kept in 10% glycerol at −80°C. The modified LB broth and modified LB agar used for co-culture experiments were additionally supplemented with 10 mM CaCl2. Working concentrations for antibiotics used in LB broth or agar were 100 μg/mL for ampicillin (Amp), 50 μg/mL for kanamycin (Kan), 30 μg/mL for nalixidic acid (Nal), 10 μg/mL for chloramphenicol (Cam), and 10 μg/ml for tetracycline (Tet). Spontaneous Nal resistant (NalR) mutants of C600 and 1.1954 were generated by spreading centrifuged cells harvested from 10 mL of overnight cultures onto LB agar plates with Nal at 37°C for 16 h. The NalR colonies were purified by re-streaking twice on similar media.
Table 1.
Strains, plasmids, and primers used in this study.
| Characteristic(s) | Reference or source | |
|---|---|---|
| BACTERIA STRAINS | ||
| E. coli O157:H7 | ||
| PA2 | stx2a; clade 8 | Hartzell et al., 2011 |
| PA8 | stx2a; clade 8 | Hartzell et al., 2011 |
| PA28 | stx2a, stx2c; clade 8 | Hartzell et al., 2011 |
| Sakai | stx1a, stx2a; clade 1 | Hayashi et al., 2001 |
| EDL933 | stx1a, stx2a; clade 3 | Perna et al., 2001 |
| Non-pathogenic E. coli | ||
| C600 | K12 derivative | Appleyard, 1954 |
| JM109 | recA−, indicator strain for plaque assay | Yanisch-Perron et al., 1985 |
| ZK1526 | microcinB17 producer | Genilloud et al., 1989 |
| 1.0328 | A phylogroup; O147 | ECRC |
| 1.0342 | D phylogroup; O11 | ECRC |
| 1.0322 | B2 phylogroup; O6 | ECRC |
| 1.1954 | B2 phylogroup; O6 | ECRC |
| 1.1968 | B2 phylogroup; O21 | ECRC |
| 1.0326 | D phylogroup; O77 | ECRC |
| bamA Derivatives | ||
| C600EE | C600ΔbamA::cam +pZS21::bamAE.coli, CamR KanR | This study |
| C600EC | C600ΔbamA::cam +pZS21::bamAEnterobacter cloacae, CamR AmpR | This study |
| C600ST | C600ΔbamA::cam +pZS21::bamASalmonellaTyphimurium,CamR AmpR | This study |
| C600DD | C600ΔbamA::cam +pZS21::bamAD.dadantii, CamR AmpR | This study |
| D4 | C600ΔbamA::cam +pZS21::bamAΔ4E.coli, CamR AmpR | This study |
| D6 | C600ΔbamA::cam +pZS21::bamAΔ6 E.coli, CamR AmpR | This study |
| I4 | C600ΔbamA::cam + pZS21::bamAHA4 E.coli, CamR AmpR | This study |
| I6 | C600ΔbamA::cam + pZS21::bamAHA6 E.coli, CamR AmpR | This study |
| I7 | C600ΔbamA::cam + pZS21::bamAHA7 E.coli, CamR AmpR | This study |
| C4 | C600ΔbamA::cam+ pZS21-bamAEc4E.cloacae, CamR AmpR | This study |
| C7 | C600ΔbamA::cam+ pZS21-bamAEc6 E.cloacae, CamR AmpR | This study |
| C8 | C600ΔbamA::cam+ pZS21-bamAEc7 E.cloacae, CamR AmpR | This study |
| C47 | C600ΔbamA::cam+ pZS21-bamAEc4/7 E.cloacae, CamR AmpR | This study |
| C67 | C600ΔbamA::cam+ pZS21-bamAEc6/7 E.cloacae, CamR AmpR | This study |
| 4EE | 1.1954ΔbamA::cam + pZS21::bamAE.coli, CamR KanR | This study |
| 4EC | 1.1954ΔbamA::cam+pZS21::bamAS.Typhimurium, CamR AmpR | This study |
| fadL Derivatives | ||
| 4F | 1.1954ΔfadL, NalR | This study |
| 4FEE | 1.1954ΔfadLΔbamA::cam + pZS21::bamAE.coli, NalR CamR KanR | This study |
| 4FST | 1.1954ΔfadLΔbamA::cam+pZS21::bamAS.Typhimurium, NalRCamRAmpR | This study |
| lamB Derivatives | ||
| 4L | 1.1954ΔlamB, NalR | This study |
| 4LEE | 1.1954ΔlamBΔbamA::cam + pZS21::bamAE.coli, NalR CamR KanR | This study |
| 4LST | 1.1954ΔlamBΔbamA::cam+pZS21::bamAS.Typhimurium, NalRCamRAmpR | This study |
| stx2 Derivatives | ||
| PA2T | PA2Δstx2::tet, TetR | This study |
| PA8T | PA8Δstx2::tet, TetR | This study |
| PA28T | PA28Δstx2::tet, TetR | This study |
| EDL933T | EDL933Δstx2::tet, TetR | This study |
| SakaiT | SakaiΔstx2::tet, TetR | This study |
| PLASMIDS | ||
| pZS21::bamAE.coli | pZS21 derivative that expresses E. coli bamA, KanR | Ruhe et al., 2013 |
| pZS21::bamAE.cloacae | Expresses bamA from Enterobacter cloacae ATCC 13047 (bamA E.cloacae), AmpR | Ruhe et al., 2013 |
| pZS21bamAS.Typhimurium | Expresses bamA from Salmonella enterica serovar Typhimurium strain LT2 (bamALT2), AmpR | Ruhe et al., 2013 |
| pZS21::bamAD.dadantii | Expresses bamA from Dickeya dadantii 3937 (bamADd3937), AmpR | Ruhe et al., 2013 |
| pZS21::bamAΔ4 E.coli | pZS21amp-bamA+ derivative that deletes residues Pro556 – Asn563 within loop 4 of BamAE.coli, AmpR | Ruhe et al., 2013 |
| pZS21::bamAΔ6 E.coli | pZS21amp-bamA+ derivative that deletes residues Phe675 – Lys701 within loop 6 of BamAE.coli, AmpR | Ruhe et al., 2013 |
| pZS21-bamAHA4 E.coli | pZS21-bamA+ derivative that introduces an HA epitope into extracellular loop 4 of BamAE.coli, AmpR | Ruhe et al., 2013 |
| pZS21-bamAHA6 E.coli | pZS21amp-bamA+ derivative that introduces an HA epitope into extracellular loop 6 of BamAE.coli, AmpR | Ruhe et al., 2013 |
| pZS21-bamAHA7E.coli | pZS21-bamA+ derivative that introduces an HA epitope into extracellular loop 7 of BamAE.coli, KanR | Ruhe et al., 2013 |
| pZS21-bamAEc4 E.coli | Expresses chimeric bamAE.cloacae in which the coding sequence for Asp550-Ala567is replaced with Tyr550-Thr567 from bamAE.coli AmpR | Ruhe et al., 2013 |
| pZS21-bamAEc6 E.coli | Expresses chimeric bamAE.cloacae in which the coding sequence for Tyr675 – Ser693 is replaced with Phe675 – Lys701 from bamAE.coli, AmpR | Ruhe et al., 2013 |
| pZS21-bamAEc7 E.coli | Expresses chimeric bamAE.cloacae in which the coding sequence for Ala739 – Val752 is replaced with Thr747 – Tyr757 from bamAE.coli, AmpR | Ruhe et al., 2013 |
| pZS21-bamAEc4/7 E.coli | Expresses chimeric bamA E.cloacae in which the coding sequence for Asp550 – Ala567 and Ala739 – Val752 is replaced with Tyr550 – Thr567 and Thr747 – Tyr757 from bamAEcoli, AmpR | Ruhe et al., 2013 |
| pZS21-bamAEc6/7E.coli | Expresses chimeric bamA E.cloacae in which the coding sequence for Tyr675 – Ser693 and Ala739 – Val752 is replaced with Phe675 – Lys701 and Thr747 – Tyr757 from bamAEcoli, AmpR | Ruhe et al., 2013 |
| PRIMERS | ||
| BamA-cam-For | aatgatttctctcggttatgagagttagttaggaagaacgcataataacgatggcg GTGTAGGCTGGAGCTGCTTC |
This study |
| BamA-cam-Rev | attgatcgcctaaagtcatcgctacactaccactacattcctttgtggagaacactta ATGGGAATTAGCCATGGTCC |
This study |
| Stx2-tet-For | atctgcgccgggtctggtgctgattacttcagccaaaaggaacacctgtat CATGTTTGACAGCTTATCATCG |
This study |
| Stx2-tet-Rev | ttgtgacacagattacacttgttacccacataccacgaatcaggttatgcc TTTGCGCATTCACAGTTCTC |
This study |
| Stx2-VF | cattagctcatcgggacaga | This study |
| Stx2-VR | gccttggtatatgcctaatctct | This study |
| FadL-UF | TTTTTTtctagaCCAGTTGTTCAATCACTTCAGC | This study |
| FadL-UR | GTAGTTAAAGTTAGTAAACAGGGTTTTCTGGCTCAT | This study |
| FadL-DF | CAGAAAACCCTGTTTACTAACTTTAACTACGCGTTCTGA | This study |
| FadL-DR | TTTTTTtctagaGCGTTTGCCTTTTTCTGTTT | This study |
| FadL-VF | TGCAGTCGGAGTTGTCCATA | This study |
| FadL-VR | CGCTTGGTCATTATGGTGTG | This study |
| LamB-UF | AAAAAAtctagaGGGCTTGAGACGATCACC | This study |
| LamB-UR | CCAGATTTCCATCTGTTTGCGCAGAGTAATCATCAT | This study |
| LamB-DF | ATTACTCTGCGCAAACAGATGGAAATCTGGTGGTAA | This study |
| LamB-DR | AAAAAAtctagaCGTGTTGCCTACCGTAACC | This study |
| LamB-VF | GCAATCGATCAAGTGCAGGT | This study |
| LamB-VR | ACATCGGCAAGACTGATTCC | This study |
ECRC, Penn State E. coli Reference Center; AmpR, ampicillin resistant; CamR, chloramphenicol resistant; KanR, kanamycin resistant; TetR, tetracycline resistant; Stx2a: Shiga toxin 2a; Stx2c: Shiga toxin 2c; The species source for bamA in mutants is represented as superscript and the modification within bamA loop is represented as subscript. Mutants with bamA from E. cloacae, S. Typhimurium or D. dadantii were designated with “EC”, “ST,” or “DD”, respectively. The loop variants which had in-frame deletions are named with “D”; ones having insertions are named with “I”; chimeric bamAE.cloacae with individual loop replaced by the corresponding one from bamAE.coli is named with “C”. The lower case letter in primers designated with “For” or “Rev” represents homologous region while upper case letter for primer used to generate antibiotic resistant cassette; the lower case letter in primers named with “UF” or “DR” stands for overlapping XbaI site.
Co-culture experiment
The co-culture assay was adapted from Gamage et al. (2003). Overnight cultures of PA2 or non-pathogenic E. coli strains were separately diluted in LB broth to an OD600 of 0.05. One hundred and seventy microliters of each strain (OD600 = 0.05) was mixed and added to modified LB broth to a final volume of 1,020 μL. The mixture was placed in a six-well plate (BD Biosciences Inc., Franklin Lakes, NJ). PA2 or non-pathogenic E. coli strains alone were used as controls. The six-well plates had 2 mL modified LB agar serving as the bottom base. Co-culture of C600 and O157:H7 was selected as the positive control (Gamage et al., 2003; Goswami et al., 2015). Stx2a level and cell density were determined after 16 h incubation at 37°C. Polymyxin B (PMB) was added to bacteria samples to final concentration of 6 mg/mL, and incubated at 37°C for 10 min for intracellular Stx2a release. PMB was used to ensure quantification of total Stx2a synthesized by bacteria (Shimizu et al., 2009; Laing et al., 2012; Ogura et al., 2015). After centrifuging at 8,000 × g for 2 min, the supernatants were collected for immediate usage or stored at −80°C for later Stx2a measurement. The Stx2a production was evaluated by a receptor based enzyme-linked immunosorbent assay (R-ELISA) as described below. Viable cell counts were calculated by spreading serial dilutions in phosphate buffer saline (PBS) onto Sorbitol MacConkey agar (SMAC). On this medium, non-O157:H7 and O157:H7 formed red and white colonies, respectively. Cell counts and toxin levels reported are the average from three biological replicates. The relative abundance was reported as percentage of commensal E. coli in the total population after co-culture, calculated by the equation:
Stx2a quantification using R-ELISA
For each R-ELISA run, supernatants from O157:H7 strain PA24 which produces only Stx1 was used as the negative control, while the lysate from high Stx2a-produing strain O157:H7 PA11 served as the positive control (Hartzell et al., 2011). The standard curves were generated using 2-fold serially diluted PA11 lysate or pure Stx2 (BEI resources, Manassas, VA). Any A450 above 0.2 was considered positive. Total protein in each unknown sample was measured by the Bradford assay (VWR Life Science, Philadelphia, PA), following the manufacturer's recommended protocol. Stx2a quantities were reported as μg Stx2a/mg total protein.
The R-ELISA was performed as described previously (Goswami et al., 2015; Yin et al., 2015). Detachable 96-well polystyrene microtiter strip plates (Thermo Scientific, Waltham, MA) were coated with 2.5 μg per well of Gb3 analog, ceramide trihexoside (CTH), for Stx2a capture. The plate was stored at 4°C overnight with blocking buffer consisting of 4% bovine serum albumin (Sigma-Aldrich, St. Louis, MO) in 0.01 M PBS with 0.05% Tween20 (PBST). Samples were added in triplicate to wells and incubated for 1 h at room temperature (RT). Ten nanograms of monoclonal mouse anti-Stx2 (Santa Cruz Biotech, Santa Cruz CA) which specifically binds to the A subunit of Stx2 was added to each well and incubated at RT for 1 h. Then, 10 ng goat anti-mouse secondary antibody conjugated to horseradish peroxidase (Santa Cruz Biotech, Santa Cruz, CA) was added to each well and incubated at RT for 1 h. Detection was accomplished using the 1-Step Ultra Tetramethylbenzidine (TMB) (Thermo-Fischer, Waltham, MA), which was equilibrated to RT in a foil-wrapped tube for at least 30 min prior to use. Next, 100 μL TMB substrate was added into each well and incubated for 10 min to allow for color development. Finally, 100 μL of stop solution (2 M H2SO4) was added to each well. The reading values of A450 were obtained using a DU®730 spectrophotometer (Beckman Coulter, Atlanta, GA). Between each addition of reagents above, the plate was washed with PBST for five times.
Generations of gene knockouts
To generate bamA mutants in C600 or 1.1954, the approach from a previous study (Ruhe et al., 2013) was followed. The species source for bamA is indicated in the superscript and modifications of bamA loops are indicated in subscripts (Table 1). Target strains were first transformed with pZS21::bamAE.coli(KanR). Next, the chromosomal bamA was deleted through one step recombination (Datsenko and Wanner, 2000) using the primer set of BamA-cam-For/Rev (Table 1). The transformants were selected on LB agar plates supplemented with Cam and Kan. Successful inactivation of chromosomal bamA was verified by PCR using primers BamA-VF/VR. The resulting E. coli ΔbamA::cam carrying plasmid pZS21::bamAE.coli(KanR) was transformed with pZS21 (AmpR) harboring the bamAE.coli variants or bamA from other species (Enterobacter cloacae, Salmonella Typhimurium, Dickeya dadantii). Plasmid exchange was selected on LB agar supplemented with Amp. AmpRKanS colonies were chosen for later experiments.
The in-frame deletion of lamB or fadL in 1.1954 (NalR) was accomplished by marker exchange as previously reported (Chen et al., 2013). PCRs were designed using primer pairs LamB-UF/UR and LamB-DF/DR, which amplified 1,028 bp upstream and downstream of lamB, respectively. The two amplicions overlap by 28 bp including an XbaI site. About 20 ng of each PCR product and primers LamB-UF/DR were used in a second round of PCR. The final PCR product was digested with restriction enzyme XbaI, cloned into the suicide vector pDS132 (Philippe et al., 2004) and transformed into E. coli SM10λpir. CamR colonies were selected. Plasmid (pDS132::ΔlamB) was further transformed into E. coli S17λpir. Conjugation was conducted between E. coli S17λpir (pDS132::ΔlamB) and 1.1954 (NalR) as described before (Dudley et al., 2006). Transconjugants were selected on LB plates lacking NaCl, but supplemented with Cam, Nal and 5% (w/v) sucrose. Colonies were screened for Cam sensitivity and the correct deletion was confirmed by PCR using primers of LamB-VF/VR. The O157:H7 stx2 mutants were generated following the one step recombination method for enterohemorrhagic E. coli strains (Murphy and Campellone, 2003). Primers Stx2-tet-For/Rev were used to replace stx2 with a Tet cassette. The mutants were selected on corresponding LB agar plates and verified by PCR using primers Stx2-VF/VR.
Plaque assay
An overnight culture of PA2 was diluted to an OD600 of 0.05 in LB broth. Ciprofloxacin was added to a final concentration of 45 ng/ml to promote stx2a-converting phage induction. After 8 h, the culture was centrifuged at 4,000 × g for 10 min and the supernatant was filtered through a 0.22 μm cellulose acetate filter (VWR, Radnor, PA). Phage was precipitated by adding one fourth volume of 20% PEG-8000/2.5 M NaCl buffer followed by overnight incubation at 4°C. The lysate was centrifuged at 4,000 × g for 1 h, and serial dilutions of phage suspensions were made in SM buffer [0.1 M NaCl, 50 mM Tris-HCl (pH 7.5), 8 mM MgSO4, and 0.01% gelatin]. Two-hundred liters of the indicator strain C600 was added to 100 μL of phage, and further mixed with 6 mL modified LB soft agar (0.75% agar). This was poured on top of a modified LB agar petri dish, and incubated at 42°C for 16 h followed by plaque quantification.
Lysogenization rate
C600 (NalR) or 1.1954 (NalR) was co-cultured with individual O157:H7 stx2 TetR mutant. After 16 h incubation, a 10-fold diluted culture was spread on LB agar plates containing only Nal to enumerate the total number of non-pathogenic E. coli, or onto plates containing both Tet and Nal to select for the lysogens. The rate of lysogen formation was calculated by using the equation:
Occupancy determination for phage insertion sites
Both C600 and 1.1954 were whole genome sequenced on an Illumina MiSeq (San Diego, CA, USA). The Illumina reads were de novo assembled using SPAdes v3.9 (Bankevich et al., 2012) into contigs to identify potential insertion sites. Previously described primer pairs (Serra-Moreno et al., 2007) were used to locate insertion sites within the assembled genomes, as well as E. coli MG1655 (accession no. CP027060). Visual comparison of these regions in C600 and MG1655, which are known to lack prophage at these sites, with corresponding sequences from 1.1954, was used to assess site occupancy.
Assay for CDI
The CDI assay followed a previously described protocol (Aoki et al., 2005). Polyethylene terephthalate (PET) track-etched membrane inserts (23 mm) of 0.4 μm pore size (Falcon, Corning, NY) were placed in six-well plates to create upper and lower culture wells. Overnight cultures of PA2 and non-pathogenic E. coli strains were diluted to an OD600 of 0.05. Diluted PA2 (3.2 mL) and non-pathogenic E. coli (2.5 mL) were added to the bottom and top chambers, respectively. Plates were incubated at 37°C with shaking at 130 rpm for 6 h. Both top and bottom samples were 10-fold serially diluted in PBS and 100 μL aliquots were plated onto SMAC plates to ensure no cross contamination occurred. After harvesting the cells and treating them with PMB for 5 min at 37°C, samples from the bottom chamber were centrifuged at 10,000 × g for 1 min, and supernatants were stored for immediate use or at −80°C. Stx2a levels were evaluated by R-ELISA.
Data analysis
MS Excel was used to calculate the mean, standard deviation, and standard error; Minitab 18 was used for statistical analysis and GraphPad Prism 8 was used for generating figures.
Results
Commensal E. coli increases Stx2a production of E. coli O157:H7 strain PA2 in co-culture
We began by testing a small collection of non-pathogenic E. coli, including the laboratory strain C600 and five commensal E. coli strains from various O types, for the ability to increase toxin production of O157:H7 strain PA2 when grown in co-culture. Co-culture of PA2+C600 produced the highest amount of Stx2a, reaching to 95.6 ± 8.1 μg Stx2a/mg total protein. Additionally, 1.1954 increased the Stx2a production in co-culture of PA2, producing 40.3 ± 1.3 μg Stx2a/mg total protein, which was significantly higher than the amount of Stx2a that PA2 produced in monoculture of 6.1 ± 0.8 μg Stx2a/mg total protein. The other four commensal E. coli strains, namely, 1.0322, 1.0326, 1.0328, and 1.1968 did not show significantly enhanced toxin production in co-cultures, when compared to PA2 alone (Figure 1A).
Figure 1.
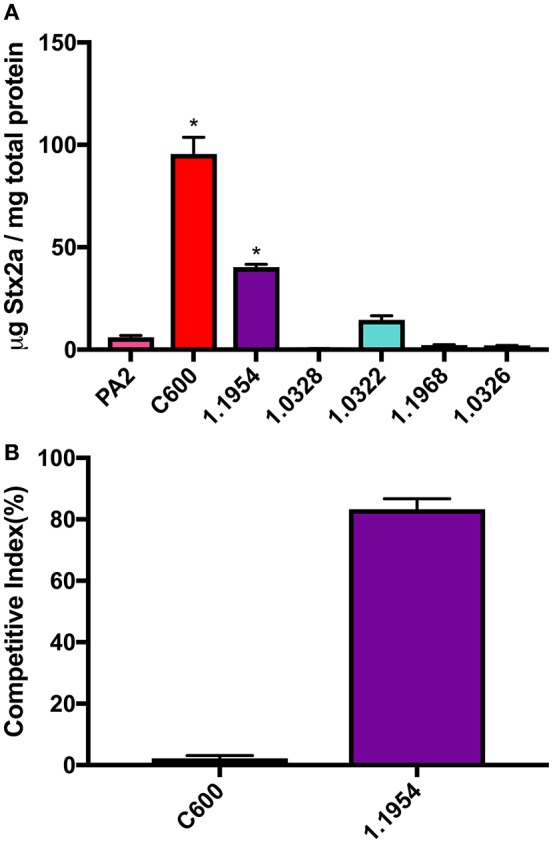
Stx2a concentration (A) and relative abundance of non-pathogenic E. coli strains (B) after co-culture with E. coli O157:H7 strain PA2. Error bars report standard error of the mean from three biological replicates. One-way ANOVA was used and the Stx2a levels in co-cultures marked with an asterisk were significantly higher than that in PA2 monoculture in pink (Dunnett's test, p < 0.05).
In each co-culture, both PA2 and non-pathogenic E. coli were inoculated at the same starting cell density. As reported previously (Goswami et al., 2015), after a 16 h co-culture, C600 abundance was 2.2 ± 0.9% of the total bacterial population, likely due to killing by the stx2a-converting phages produced by PA2. In contrast, an increase in cell counts of 1.1954 to 83.0 ± 2.3% was seen after co-culture with PA2 (Figure 1B). This suggested that the mechanism by which 1.1954 enhances Stx2a production by PA2 differs from that previously described for C600.
E. coli C600 requires BamA for Stx2a enhancement in co-culture with PA2
Several attempts to generate spontaneous phage resistant C600 derivatives were unsuccessful (data not shown), suggesting that disrupting phage adsorption requires changes to BamA beyond what can be achieved by those techniques. Using a previously published method (Ruhe et al., 2013), we generated three derivatives of C600 designated C600EC, C600ST, C600DD, in which a deletion of the chromosomal bamA was constructed through complementing in trans with plasmid-encoded bamA from E. cloacae, S. Typhimurium or D. dadantii, respectively. The deduced amino acid sequences of BamA from these species and E. coli are most divergent within the extracellular loops, which are the portions most likely involved in phage adsorption. All of these derivatives grew similarly to wild type C600 in LB broth (Figure 2A), and did not form any observable plaques when incubated with lysates containing the stx2a-converting phages (Figure 2B). Additionally, the concentrations of Stx2a produced during co-culture of these derivatives with PA2 were indistinguishable from that observed with PA2 alone, and significantly less than that measured in PA2+C600 (p < 0.05) (Figure 2C). Overall, expression of heterologous bamA in phage susceptible C600 rendered it resistant to phage lytic infection, providing further evidence that phage infection of C600 through BamA is required to enhance toxin production by PA2 in co-culture.
Figure 2.
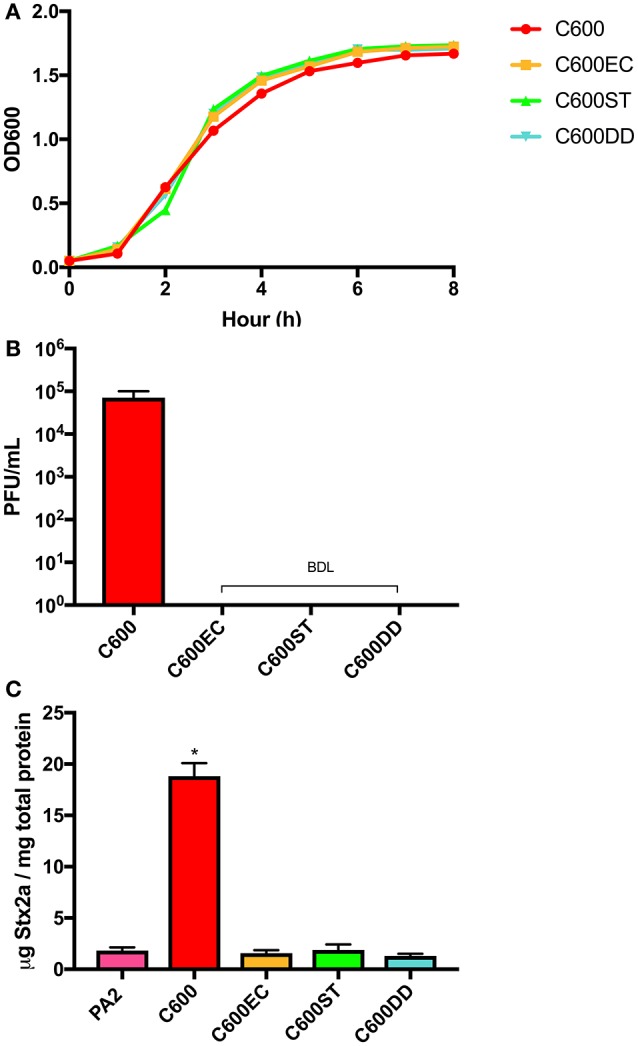
Growth curves (A) and plaque counts for C600 and its three bamA mutants (B) and their Stx2a levels (C) after co-incubating with E. coli O157:H7 strain PA2. C600EC, C600ST, C600DD represents three mutants expressing heterologous bamA from Enterobacter cloacae, Salmonella Typhimurium, and Dickeya dadantii, respectively. Error bars report standard error of the mean from three biological replicates. BDL represented below the detection limit of 10 PFU/mL. One-way ANOVA was used and Stx2a levels in co-culture marked with an asterisk were significantly higher than the PA2 monoculture (Dunnett's test, p < 0.05).
Extracellular loops 4, 6, and 7 of BamA are required for efficient infection of E. coli C600 by stx2a-converting phages
Using the tools developed by Ruhe et al. (2013), we could also address whether BamA loops 4, 6, and 7, which are the longest and least conserved of the extracellular loops, are needed for infection by stx2a-converting phages. Among the 12 variants we generated (Table 2), mutants with in-frame deletions in loop 4 or 6 (D4 and D6) as well as insertions in either loop 4 or 7 (I4 and I7) did not support the formation of detectable plaques by stx2a-converting phages (Figure 3A). Insertion of the HA epitope into loop 6 (I6) decreased plaque numbers by approximately 50% of that seen when C600 or EE was used as the host in the plaque assay. Strains expressing chimeric loops (C6, C7, C47) were also resistant to phage infection. The one exception was mutant C67, in which both loops 6 and 7 from bamAE.cloacae were replaced with the corresponding sequences from bamAE.coli. This restored susceptibility to phage infection, to approximately 25% of the number of plaques seen when either C600 or EE were used as host strains.
Table 2.
Description of loop variants of E. coli C600 used in this studya.
| Strain | BamA | Description |
|---|---|---|
| EE | bamAE.coli | Entire plasmid copy of BamAE.coli |
| EC | bamAE.cloacae | Entire plasmid copy of BamAE.cloacae |
| D4 | bamAΔ4E.coli | Deletion in loop 4 of BamAE.coli |
| D6 | bamAΔ6 E.coli | Deletion in loop 6 of BamAE.coli |
| I4 | bamAHA4E.coli | Insertion in loop 4 of BamAE.coli |
| I6 | bamAHA6E.coli | Insertion in loop 6 of BamAE.coli |
| I7 | bamAHA7E.coli | Insertion in loop 7 of BamAE.coli |
| C4 | bamAEc4E.coli | Chimeric bamAE.cloacaewith loop 4 replaced by that from bamAE.coli |
| C6 | bamAEc6E.coli | Chimeric bamAE.cloacae with loop 6 replaced by that from bamAE.coli |
| C7 | bamAEc7E.coli | Chimeric bamAE.cloacae with loop 7 replaced by that from bamAE.coli |
| C47 | bamAEc4/7E.coli | Chimeric bamAE.cloacae with loop 4 and 7 replaced by those from bamAE.coli |
| C67 | bamAEc6/7E.coli | Chimeric bamAE.cloacae with loop 6 and 7 replaced by those from bamAE.coli |
The plasmids containing above bamA alleles were previously described by Ruhe et al. (2013).
Figure 3.
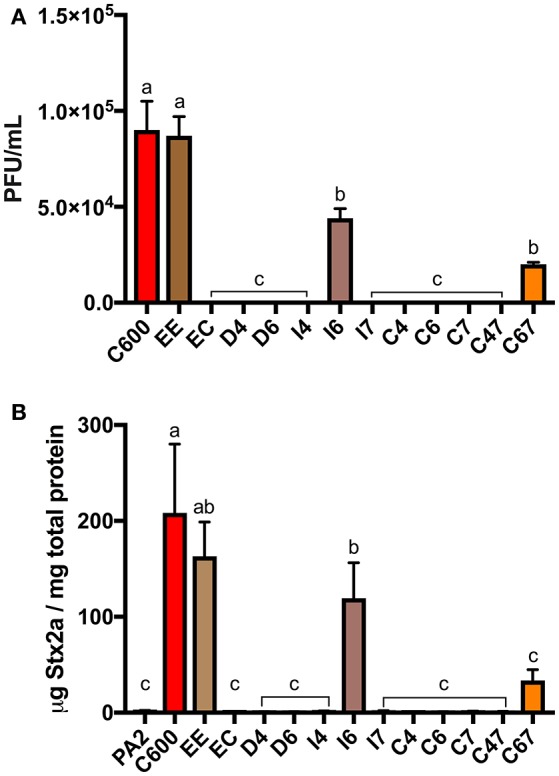
The plaque counts (A) for C600 and its ΔbamA derivatives with modified extracellular loops and Stx2a levels (B) after co-culture with E. coli O157:H7 strain PA2. Error bars were standard error of the mean from three biological replicates. One-way ANOVA was used and groups sharing the same letter (a, b, c) had no significant difference (Tukey's test, p < 0.05).
In accordance with these results, the co-cultures of PA2+C600 and PA2+EE produced similar levels of Stx2a, however expression of bamAE.cloacae in place of bamAE.coli (PA2+EC) decreased Stx2a expression to the baseline level (Figure 3B). An increase in Stx2a level was observed for PA2+C67 which had a chimeric bamAE.cloacae with loop 6 and 7 replaced by those from bamAE.coli, but this was not significantly different from the PA2 monoculture. The PA2+I6 combination produced half of Stx2a level in PA2+C600. Together, these results suggest that the three extracellular loops (4, 6, and 7) of bamAE.coli are essential for optimal infection of C600 by stx2a-converting phages.
Commensal E. coli 1.1954 uses a bamA-independent mechanism for toxin enhancement in co-culture with PA2
Since commensal 1.1954 was a Stx2a amplifier as well (Figure 1), we utilized the above approach to test whether bamA was necessary for 1.1954 mediated Stx2a amplification of PA2 in co-culture. Two bamA mutants were generated for 1.1954, in which the chromosomal bamA was deleted and complemented in trans by plasmid-encoded bamA from either E. coli (4EE) or S. Typhimurium (4ST). As expected, strains carrying bamAE.coli (C600 or C600EE) produced significantly more Stx2a in co-cultures with PA2 than co-culture with the phage resistant strain C600ST (p < 0.05) (Figure 4). To the contrary, Stx2a concentrations in co-cultures of PA2+4ST and PA2+4EE were indistinguishable from that in PA2+1.1954, producing an average of 40 μg Stx2a/mg total protein (Figure 4). This suggests that commensal 1.1954 uses a bamA-independent mechanism for toxin enhancement in co-culture with PA2.
Figure 4.
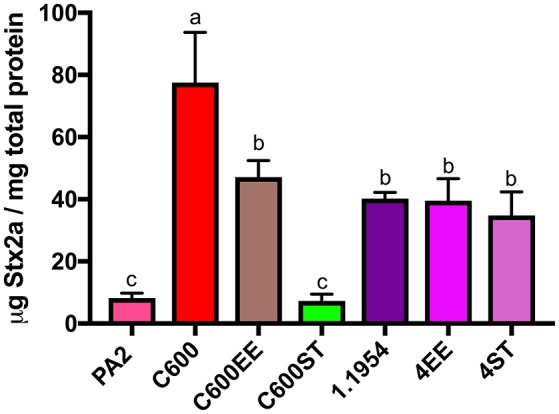
Stx2a levels after co-culture of C600 and commensal 1.1954 expressing heterologous BamA with E. coli O157:H7 strain PA2. ST stands for ΔbamA mutant with heterologous BamA from either S. Typhimurium and EE for ΔbamA mutant with homologous BamA from E. coli. Error bars report standard error of the mean from three biological replicates. One-way ANOVA was used and groups sharing the same letter (a, b, c) had no significant difference (Tukey's test, p < 0.05).
Commensal E. coli 1.1954 is likely not infected by stx2a-converting phages
Attempts to test whether 1.1954 bamAS.Typhimurium is phage-susceptible by standard plaque assays were unsuccessful, as 1.1954 does not form a bacterial lawn when grown on the antibiotic-containing medium (data not shown). Others have suggested LamB and FadL could serve as alternative receptors for stx-converting phages (Watarai et al., 1998). Therefore, we generated mutants of 1.1954 with in-frame deletion in fadL or lamB (4F, 4L), or in combination with either homologous (4FEE, 4LEE) or heterologous bamA (4FST, 4LST). In the absence of FadL or LamB or BamA, the single knockouts (4F, 4L, 4ST) produced statistically indistinguishable levels of Stx2a when compared to PA2+1.1954 (Figure 5). Similarly, the double knockouts (4FST, 4LST) lacking BamA plus either LamB or FadL, still exhibited the toxin amplification phenotype as wild type 1.1954. These results indicate that 1.1954 does not require BamA, FadL or LamB to enhance toxin production of O157:H7.
Figure 5.
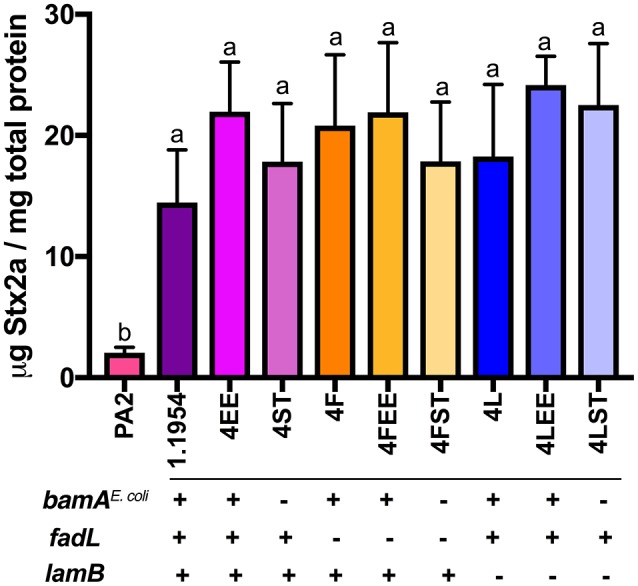
Stx2a levels in the mutants of 1.1954 after co-culture with E. coli O157:H7 strain PA2. In strain name, F represents ΔfadL, L for ΔlamB, EE for ΔbamA::cam+pZS21::bamAE.coli, ST for ΔbamA::cam+pZS21::bamAS.Typhimurium. The genotype is listed below the mutant names which is displayed across X axis. Error bars report standard error of the mean from three biological replicates. One-way ANOVA was used and groups sharing the same letter (a, b) had no significant difference (Tukey's test, p < 0.05).
As an indirect measure of whether stx2a-converting phage infect the strain 1.1954, the PA2 mutant (PA2T) whose stx2 was replaced with a tetracycline resistance marker was used to monitor lysogenized rates when co-culturing with either C600 or 1.1954. The average lysogen forming rate in C600 was 0.016% (Figure 6), however, no lysogen formation was observed in 1.1954. We also monitored the rate for 1.1954 at different time points during the 16 h co-culture, and no lysogens were observed at any time point (data not shown). In order to test if phage type affected lysogen formation during co-culture, several other E. coli O157:H7 strains carrying genetically diverse stx2a-converting phages (Yin et al., 2015) were also tested. In the C600 background, SakaiT had the lowest average lysogen forming rate at <0.008%, while EDL933T gave the highest of 0.021%. No difference was observed for the lysogen forming rates among PA2T, PA8T, PA28T, and EDL933T (p < 0.05). However, no lysogens formed in the 1.1954 background by any tested stx2a-converting phage (data not shown). This suggested that 1.1954 does not undergo lysogenic conversion during co-culture with O157:H7.
Figure 6.
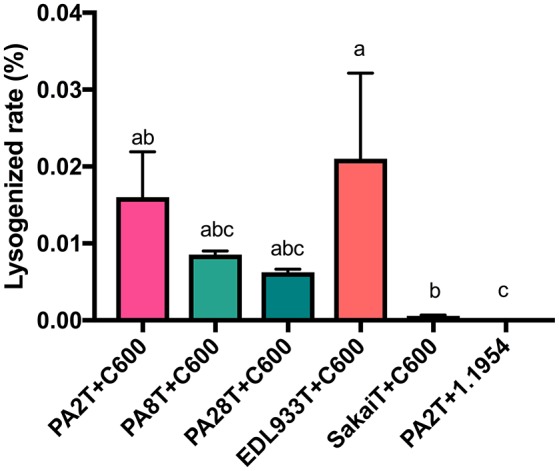
Lysogen forming rates of C600 and 1.1954 after co-culture with stx2::tet mutants of different E. coli O157:H7 strains. The E. coli O157:H7 strain was designated with the letter T indicate where a tetracycline resistant marker replaces the stx2 gene. No lysogens were detected when any of the E. coli O157:H7 strains were co-cultured with E. coli 1.1954. Error bars report standard error of the mean from three biological replicates. One-way ANOVA was used and groups sharing the same letter (a, b, c) were not significantly different (Tukey's test, p < 0.05).
It was reported earlier that if the primary phage insertion site in the host strain is occupied, the stx2-converting phages will integrate at alternative sites (Serra-Moreno et al., 2007). Five stx2a-converting phage insertion sites (sbcB, yehV, argW, yecE, and z2577) were checked for occupancy in both C600 and 1.1954, the first three of which are preferred by stx-converting phages. Four out of five were available in 1.1954, with only z2577 occupied. For C600, all five phage insertion sites were unoccupied. Collectively, these data suggest that stx2a-converting phages do not infect 1.1954.
Commensal E. coli 1.1954 does not secrete DNA damaging agents that increase Stx2a of PA2
To test whether secreted factors produced by 1.1954 could trigger toxin amplification of PA2, we used a modified CDI assay (Aoki et al., 2005), where non-pathogenic E. coli strains and PA2 were grown together while separated by a membrane. The E. coli strain ZK1526, which produces DNA gyrase inhibitor—microcin B17 (Genilloud et al., 1989), was selected as the positive control. As shown in Figure 7, ZK1526 promoted significantly more Stx2a production of PA2 than the negative control, PA2 alone. The toxin levels for PA2 in setups of PA2+C600 or PA2+1.1954 were as similar as the baseline level in PA2+LB. This indicated that neither C600 or 1.1954 secreted a soluble enhancer for Stx2a production of PA2.
Figure 7.
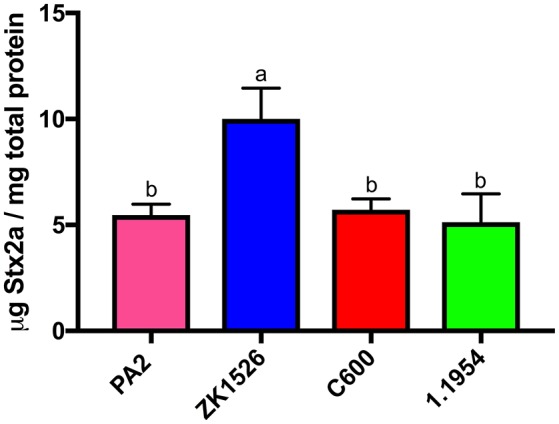
Stx2a levels of PA2 after 6 h incubation in CDI setups where PA2 grew on the bottom chamber and LB/non-pathogenic E. coli (ZK1526, C600, and 1.1954) on the top. Error bars report standard error of the mean from three biological replicates. One-way ANOVA was used and groups shared the same letter (a, b) had no significant difference (Tukey's test, p < 0.05).
With this result, physical contact between 1.1954 and PA2 seemed to be required for toxin amplification. Thus, we also considered a role for cdiA-encoding CDI systems (Aoki et al., 2005). Using BLAST, we found that 1.1954 harbors a typical cdiBAI operon while PA2 does not (data not shown). The deduced amino acid sequences of the carbon terminal (CT) of CdiA in 1.1954 shares 99% identity to that of uropathogenic E. coli (UPEC) 536, and the immunity protein CdiI shared 100% homology. Given the close relationship, we speculated the CdiA of 1.1954 is a tRNA anticodon nuclease as it is in UPEC 536 (Diner et al., 2012). If true, it seems unlikely that a tRNase is involved in increasing toxin expression (Toshima et al., 2007), and additionally this CDI system in UPEC 536 is repressed when grown in LB broth at 37°C (Aoki et al., 2010).
Discussion
The microbiota of the human gastrointestinal (GI) tract is estimated to contain 1014 bacteria belonging to over 2,000 species (Thursby and Juge, 2017). E. coli is one species in this population, which colonizes to about 108 organisms per gram of feces in healthy individuals (Tenaillon et al., 2010). Symptoms of O157:H7 infection can vary in degree of severity, and it is thought that the gut microbiota is responsible in part for modulating expression of virulence factors (de Sablet et al., 2009; Curtis et al., 2014). Commensal E. coli also impact toxin production of O157:H7, and a previous study reported 10% of commensal E. coli increased toxin produced by O157:H7 when grown in co-culture (Gamage et al., 2003). We reported previously that this phenomenon could be recapitulated in vivo, as mice inoculated with both O157:H7 and the non-pathogenic E. coli laboratory strain C600 exhibited greater signs of kidney damage and a higher mortality rate than those fed O157:H7 alone (Goswami et al., 2015). The observation that toxin production is enhanced by only a subset of commensal E. coli, combined with the diversity of E. coli strains found between individuals (Gordon et al., 2015), provides one possible explanation for individual difference in disease outcome. This study is part of a larger effort to describe commensal E. coli and O157:H7 interactions that alter Stx levels.
The role for phage in toxin amplification during co-culture of E. coli O157:H7 with other strains was previously suggested by demonstrating that addition of stx2-converting phages to C600 increased toxin production more than two-orders of magnitude over that seen when using a phage-resistant C600 strain (Gamage et al., 2003). Consistent with the hypothesis that toxin production requires phage to initiate a lytic infection, Goswami et al. (2015) showed that cell counts of C600 decreased upon co-culturing with E. coli O157:H7. Adding anti-BamA antibodies decreases phage adsorption up to 50% in a dose-dependent manner (Smith et al., 2007) and the overexpression of BamA increased the rate of lysogen formation approximately 2- to 4-fold (Islam et al., 2012), arguing that this outer membrane protein is the target for phage adsorption. Despite these data, evidence that BamA is the only receptor for stx2a-converging phages has been elusive, as E. coli bamA mutants are generally not viable (Werner and Misra, 2005). Here, we provide genetic evidence conclusively demonstrating that, at least for C600, stx2a-converting phages infect non-pathogenic E. coli exclusively through BamA. BamA from other Enterobacteriaceae share 73–93% identity to BamAE.coli with the largest variation within the central region of predicted extracellular loops 4, 6, and 7 (Ruhe et al., 2013). Expression of heterologous bamA from E. cloacae, S. Typhimurium, or D. dadantii in place of that from C600 was sufficient to impart phage resistance, suggesting that stx2a-converting phage tail fibers initially bind to one or more of these loops (Figure 2). In our study, all three loops appeared to be important for phage lytic infection (Figure 3), suggesting they may come into contact with the stx2a-converting phage tail. In contrast, CdiA, the component of the CDI system from E. coli EC93 that is responsible for recognition and pore formation, recognizes target cells through BamA, in a manner that involves only extracellular loops 6 and 7 (Ruhe et al., 2013).
Although Smith et al. (2007) suggested BamA was the receptor specifically for short-tailed stx2-converting phage, we lack visual evidence for PA2 phage being short-tailed due to poor resolution of our transmission electron microscopy (TEM) results. Two lines of genetic evidence suggest that it is. First, Yin et al. (2015) reported that the stx2a-converting phage of PA2 belongs to phage type PST2-1, which is similar to phage from the German outbreak strain E. coli O104:H4. TEM classified the stx2a-convering phage of E. coli O104:H4 as short-tailed (Beutin et al., 2012). Secondly, Mondal et al. (2016) identified nine genes responsible for phage morphogenesis of short-tailed phage designated SP5, and these were found by BLAST to be nearly identical (>99%) on amino acid level to the homologs from phage PA2. Notably, one of these genes, ECs1228, is predicted to encode a phage tail fiber and 100% identical to the PA2 phage homolog on amino acid level.
As BamA from C600 and 1.1954 share 100% identity to one another on the amino acid level, we were surprised to find that replacement of BamAE.coli with heterologous BamAS.Typhimurium in 1.1954 did not abolish its toxin amplifying ability in co-culture with O157:H7 (Figure 4). The additional deletion of other stx2a-converting phage receptors (LamB or FadL) in the 1.1954ΔbamA background had no effects on toxin amplification as well (Figure 5). This suggests that the mechanism behind toxin amplification in 1.1954 either does not involve phage infection, or a novel phage receptor exists in 1.1954 that is absent in C600.
We investigated other mechanisms to explain why 1.1954 increases Stx production, focusing on those previously described or hypothesized given our knowledge of toxin regulation in O157:H7. One study (Iversen et al., 2015) found that 39% of commensal E. coli were lysogenized by the stx2a-converting phage φ734, from a highly virulent strain of STEC O103:H25. Of the 13 lysogens studied, 12 produced more phage when grown in the absence of inducing agents (mitomycin C or H2O2) than did the O103:H25 parent strain, suggesting that Stx2a levels would be higher in these strains as well. One lysogen, C600::φ734, was tested for Stx2a production and indeed this was found to be the case. Therefore, Iversen et al. (2015) proposed that lysogenization of commensal E. coli during an O157:H7 infection enhances overall toxin production. To the contrary, none of the five genetically distinct stx2a-converting phages tested in our study formed detectable lysogens in 1.1954. Lysogen formation can be inhibited if insertion sites are occupied by other phage, however analysis of the 1.1954 genome indicated most of the preferred insertion sites are unoccupied. Another study demonstrated that the DNase-colicins E8 and E9 can activate the SOS response, leading to greater toxin production when strains producing either are grown in co-culture with E. coli O157:H7 (Toshima et al., 2007). Our data also shows that microcin B17, which activates the SOS response through inhibition of DNA gyrase (Herrero and Moreno, 1986; Yorgey et al., 1994) does the same (Figure 7). Our data argues against the hypothesis that 1.1954 secretes DNA damaging molecules or other soluble factors known to regulate Stx2a production such as autoinducer 2 (Sperandio et al., 2001), and to the contrary suggests that physical contact between 1.1954 and PA2 is required (Figure 7). Although our bioinformatics analysis revealed that 1.1954 possesses a CDI system which may function as a tRNA anticodon nuclease, it may not function under our current laboratory condition. Future work should also consider whether other CDI systems previously described (type IV, V, and VI) may be involved (Aoki et al., 2005; Hood et al., 2010; MacIntyre et al., 2010; Souza et al., 2015).
Although SOS-mediated induction of Stx and phage production is the best understood pathway, several other mechanisms have been reported which serve as hypotheses for future experiments. For example, earlier reports revealed that lambdoid phage production is regulated through the capsular polysaccharide proteins RscA and DsrA (Rozanov et al., 1998), polynucleotide phosphorylase (Hu and Zhu, 2015), and RNA polyadenylation (Nowicki et al., 2015), and whether regulation of any of these are changed in O157:H7 upon co-culture with 1.1954 should be explored. Additionally, lambdoid phage production is under environmental control including salt concentration (Shkilnyj and Koudelka, 2007), pH and cationic chelators (Imamovic and Muniesa, 2012). The mechanisms behind these observations remain largely unexplored, however none of these were altered in our co-culture experiments and thus are not believed to contribute to differences in Stx production observed here.
In summary, our findings further define the bamA-dependent mechanism by which C600 increases Stx production. Although our study has not identified the exact mechanism for 1.1954, we provide evidence to indicate that a new mechanism exists which does not require BamA or act via secretion of a DNA-damaging molecule. This and other studies (Figler and Dudley, 2016; Matamouros et al., 2018) highlight the importance of appreciating strain-level diversity of E. coli when assessing how this organism affects health and disease outcomes.
Author contributions
LX designed and performed all experiments, collected and analyzed data, and wrote the manuscript; HF contributed in manuscript writing; KG contributed in co-culture experiment; CH contributed in providing technical assistances; ED advised experimental design and manuscript writing.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Zachary C. Ruhe at University of California, Santa Barbara for providing bamA related plasmids, Dr. Roberto Kolter at Harvard University for providing strain ZK1526, Andrea Keefer and Rebecca Abelman for manuscript proofreading. HF is supported by USDA National Needs Grant 2914-38420-21822. This work was supported by the USDA National Institute of Food and Agriculture Federal Appropriations under Project PEN04522 and Accession number 0233376.
References
- Aoki S. K., Diner E. J., Roodenbeke C. T., Burgess B. R., Poole S. J., Braaten B. A., et al. (2010). A widespread family of polymorphic contact-dependent toxin delivery systems in bacteria. Nature, 468, 439–442. 10.1038/nature09490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki S. K., Pamma R., Hernday A. D., Bickham J. E., Braaten B. A., Low D. A. (2005). Contact-dependent inhibition of growth in Escherichia coli. Science 309, 1245–1248. 10.1126/science.1115109 [DOI] [PubMed] [Google Scholar]
- Appleyard R. K. (1954). Segregation of new lysogenic types during growth of a doubly lysogenic strain derived from Escherichia coli K12. Genetics 39, 440–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutin L., Hammerl J. A., Strauch E., Reetz J., Dieckmann R., Kelner-Burgos Y., et al. (2012). Spread of a distinct Stx2-encoding phage prototype among Escherichia coli O104:H4 strains from outbreaks in Germany, Norway, and Georgia. J. Virol. 86, 10444–10455. 10.1128/JVI.00986-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borczyk A. A., Karmali M. A., Lior H., Duncan L. M. C. (1987). Bovine reservoir for verotoxin-producing Escherichia coli O157:H7. Lancet 329:98 10.1016/S0140-6736(87)91928-3 [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC) (2012). National Shiga Toxin-Producing Escherichia coli (STEC) Surveillance Overview. Atlanta: US Department of Health and Human Services, CDC. [Google Scholar]
- Chen C., Lewis C. R., Goswami K., Roberts E. L., DebRoy C., Dudley E. G. (2013). Identification and characterization of spontaneous deletions within the Sp11-Sp12 prophage region of Escherichia coli O157:H7 Sakai. Appl. Environ. Microbiol. 79, 1934–1941. 10.1128/AEM.03682-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis M. M., Hu Z., Klimko C., Narayanan S., Deberardinis R., Sperandio V. (2014). The gut commensal Bacteroides thetaiotaomicron exacerbates enteric infection through modification of the metabolic landscape. Cell Host Microbe 16, 759–769. 10.1016/j.chom.2014.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko K. A., Wanner B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sablet T., Chassard C., Bernalier-Donadille A., Vareille M., Gobert A. P., Martin C. (2009). Human microbiota-secreted factors inhibit shiga toxin synthesis by enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 77, 783–790. 10.1128/IAI.01048-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diner E. J., Beck C. M., Webb J. S., Low D. A., Hayes C. S. (2012). Identification of a target cell permissive factor required for contact-dependent growth inhibition (CDI). Genes Dev. 26, 515–525. 10.1101/gad.182345.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley E. G., Thomson N. R., Parkhill J., Morin N. P., Nataro J. P. (2006). Proteomic and microarray characterization of the AggR regulon identifies a pheU pathogenicity island in enteroaggregative Escherichia coli Mol. Microbiol. 61, 1267–1282. 10.1111/j.1365-2958.2006.05281.x [DOI] [PubMed] [Google Scholar]
- Figler H. M., Dudley E. G. (2016). The interplay of Escherichia coli O157:H7 and commensal E. coli: the importance of strain-level identification. Expert Rev. Gastroenterol. Hepatol. 10, 415–417. 10.1586/17474124.2016.1155449 [DOI] [PubMed] [Google Scholar]
- Friedrich A. W., Bielaszewska M., Zhang W. L., Pulz M., Kuczius T., Ammon A., et al. (2002). Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J. Infect. Dis. 185, 74–84. 10.1086/338115 [DOI] [PubMed] [Google Scholar]
- Gamage S. D., Strasser J. E., Chalk C. L., Weiss A. A. (2003). Nonpathogenic Escherichia coli can contribute to the production of Shiga toxin. Infect. Immun. 71, 3107–3115. 10.1128/IAI.71.6.3107-3115.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genilloud O., Moreno F., Kolter R. (1989). DNA sequence, products, and transcriptional pattern of the genes involved in production of the DNA replication inhibitor microcin B17. J. Bacteriol. 171, 1126–1135. 10.1128/jb.171.2.1126-1135.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D. M., O'brien C. L., Pavli P. (2015). Escherichia coli diversity in the lower intestinal tract of humans. Environ. Microbiol. Rep. 7, 642–648. 10.1111/1758-2229.12300 [DOI] [PubMed] [Google Scholar]
- Goswami K., Chen C., Xiaoli L., Eaton K. A., Dudley E. G. (2015). Coculture of Escherichia coli O157:H7 with a nonpathogenic E. coli strain increases toxin production and virulence in a germfree mouse model. Infect. Immun. 83, 4185–4193. 10.1128/IAI.00663-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzell A., Chen C., Lewis C., Liu K., Reynolds S., Dudley E. G. (2011). Escherichia coli O157:H7 of genotype lineage-specific polymorphism assay 211111 and clade 8 are common clinical isolates within Pennsylvania. Foodborne Pathog. Dis. 8, 763–768. 10.1089/fpd.2010.0762 [DOI] [PubMed] [Google Scholar]
- Hayashi T., Makino K., Ohnishi M., Kurokawa K., Ishii K., Yokoyama K., et al. (2001). Complete genome sequence of enterohemorrhagic Eschelichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8, 11–22. 10.1093/dnares/8.1.11 [DOI] [PubMed] [Google Scholar]
- Herrero M., Moreno F. (1986). Microcin B17 blocks DNA replication and induces the SOS system in Escherichia coli. Microbiology 132, 393–402. 10.1099/00221287-132-2-393 [DOI] [PubMed] [Google Scholar]
- Hilborn E. D., Mermin J. H., Mshar P. A., Hadler J. L., Voetsch A., Wojtkunski C., et al. (1999). A multistate outbreak of Escherichia coli O157:H7 infections associated with consumption of mesclun lettuce. Arch. Intern. Med. 159, 1758–1764. 10.1001/archinte.159.15.1758 [DOI] [PubMed] [Google Scholar]
- Hood R. D., Singh P., Hsu F., Güvener T., Carl M. A., Trinidad R. R., et al. (2010). A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7, 25–37. 10.1016/j.chom.2009.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Zhu M. J. (2015). Defects in polynucleotide phosphorylase impairs virulence in Escherichia coli O157:H7. Front. Microbiol. 6:806. 10.3389/fmicb.2015.00806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamovic L., Muniesa M. (2012). Characterizing RecA-independent induction of Shiga toxin2-encoding phages by EDTA treatment. PLoS ONE 7:e32393. 10.1371/journal.pone.0032393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M. R., Ogura Y., Asadulghani M., Ooka T., Murase K., Gotoh Y., et al. (2012). A sensitive and simple plaque formation method for the Stx2 phage of Escherichia coli O157:H7, which does not form plaques in the standard plating procedure. Plasmid 67, 227–235. 10.1016/j.plasmid.2011.12.001 [DOI] [PubMed] [Google Scholar]
- Iversen H., L' Abée-Lund T. M., Aspholm M., Arnesen L. P., Lindbäck T. (2015). Commensal E. coli Stx2 lysogens produce high levels of phages after spontaneous prophage induction. Front. Cell. Infect. Microbiol. 5:5. 10.3389/fcimb.2015.00005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson M. P., Newland J. W., Holmes R. K., O'Brien A. D. (1987). Nucleotide sequence analysis of the structural genes for Shiga-like toxin I encoded by bacteriophage 933J from Escherichia coli. Microb. Pathog. 2, 147–153. 10.1016/0882-4010(87)90106-9 [DOI] [PubMed] [Google Scholar]
- Kawano K., Okada M., Haga T., Maeda K., Goto Y. (2008). Relationship between pathogenicity for humans and stx genotype in Shiga toxin-producing Escherichia coli serotype O157. Eur. J. Clin. Microbiol. Infect. Dis. 27, 227–232. 10.1007/s10096-007-0420-3 [DOI] [PubMed] [Google Scholar]
- Laing C. R., Zhang Y., Gilmour M. W., Allen V., Johnson R., Thomas J. E., et al. (2012). A comparison of Shiga-toxin 2 bacteriophage from classical enterohemorrhagic Escherichia coli serotypes and the German E. coli O104: H4 outbreak strain. PLoS ONE 7:e37362. 10.1371/journal.pone.0037362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIntyre D. L., Miyata S. T., Kitaoka M., Pukatzki S. (2010). The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc. Natl. Acad. Sci. U.S.A. 107, 19520–19524. 10.1073/pnas.1012931107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning S. D., Motiwala A. S., Springman A. C., Qi W., Lacher D. W., Ouellette L. M., et al. (2008). Variation in virulence among clades of Escherichia coli O157:H7 associated with disease outbreaks. Proc. Natl. Acad. Sci. U.S.A. 105, 4868–4873. 10.1073/pnas.0710834105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matamouros S., Hayden H. S., Hager K. R., Brittnacher M. J., Lachance K., Weiss E. J., et al. (2018). Adaptation of commensal proliferating Escherichia coli to the intestinal tract of young children with cystic fibrosis. Proc. Natl. Acad. Sci. U.S.A. 115, 1605–1610. 10.1073/pnas.1714373115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal S. I., Islam M. R., Sawaguchi A., Asadulghani M., Ooka T., Gotoh Y., et al. (2016). Genes essential for the morphogenesis of the Shiga toxin 2-transducing phage from Escherichia coli O157:H7. Sci. Rep. 6:39036. 10.1038/srep39036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K. C., Campellone K. G. (2003). Lambda Red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli. BMC Mol. Biol. 4:11. 10.1186/1471-2199-4-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowicki D., Bloch S., Nejman-Falenczyk B., Szalewska-Pałasz A., Wegrzyn A., Wegrzyn G. (2015). Defects in RNA polyadenylation impair both lysogenization by and lytic development of Shiga toxin-converting bacteriophages. J. Gen. Virol. 96, 1957–1968. 10.1099/vir.0.000102 [DOI] [PubMed] [Google Scholar]
- Ogura Y., Mondal S. I., Islam M. R., Mako T., Arisawa K., Katsura K., et al. (2015). The Shiga toxin 2 production level in enterohemorrhagic Escherichia coli O157:H7 is correlated with the subtypes of toxin-encoding phage. Sci. Rep. 5:16663. 10.1038/srep16663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perna N. T., Plunkett I. I. I. G., Burland V., Mau B., Glasner J. D., Rose D. J., et al. (2001). Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409, 529–531. 10.1038/35054089 [DOI] [PubMed] [Google Scholar]
- Persson S., Olsen K. E., Ethelberg S., Scheutz F. (2007). Subtyping method for Escherichia coli Shiga toxin (verocytotoxin) 2 variants and correlations to clinical manifestations. J. Clin. Microbiol. 45, 2020–2024. 10.1128/JCM.02591-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe N., Alcaraz J. P., Coursange E., Geiselmann J., Schneider D. (2004). Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid 51, 246–255. 10.1016/j.plasmid.2004.02.003 [DOI] [PubMed] [Google Scholar]
- Rangel J. M., Sparling P. H., Crowe C., Griffin P. M., Swerdlow D. L. (2005). Epidemiology of Escherichia coli O157:H7 outbreaks, United States, 1982–2002. Emerg. Infect. Dis. 11, 603–609. 10.3201/eid1104.040739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozanov D. V., D'Ari R., Sineoky S. P. (1998). RecA-independent pathways of lambdoid prophage induction in Escherichia coli. J. Bacteriol. 180, 6306–6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhe Z. C., Wallace A. B., Low D. A., Hayes C. S. (2013). Receptor polymorphism restricts contact-dependent growth inhibition to members of the same species. MBio 4:e00480-13. 10.1128/mBio.00480-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandvig K., Van Deurs B. (1992). Toxin-induced cell lysis: protection by 3-methyladenine and cycloheximide. Exp. Cell Res. 200, 253–262. 10.1016/0014-4827(92)90171-4 [DOI] [PubMed] [Google Scholar]
- Saxena S. K., O'Brien A. D., Ackerman E. J. (1989). Shiga toxin, Shiga-like toxin II variant, and ricin are all single-site RNA N-glycosidases of 28 S RNA when microinjected into Xenopus oocytes. J. Biol. Chem. 264, 596–601. [PubMed] [Google Scholar]
- Scheutz F., Teel L. D., Beutin L., Piérard D., Buvens G., Karch H., et al. (2012). Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing Stx nomenclature. J. Clin. Microbiol. 50, 2951–2963. 10.1128/JCM.00860-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra-Moreno R., Jofre J., Muniesa M. (2007). Insertion site occupancy by stx2 bacteriophages depends on the locus availability of the host strain chromosome. J. Bacteriol. 189, 6645–6654. 10.1128/JB.00466-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T., Ohta Y., Noda M. (2009). Shiga toxin 2 is specifically released from bacterial cells by two different mechanisms. Infect. Immun. 77, 2813–2823. 10.1128/IAI.00060-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkilnyj P., Koudelka G. B. (2007). Effect of salt shock on stability of λimm434 lysogens. J. Bacteriol. 189, 3115–3123. 10.1128/JB.01857-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. L., James C. E., Sergeant M. J., Yaxian Y., Saunders J. R., McCarthy A. J., et al. (2007). Short-tailed stx phages exploit the conserved YaeT protein to disseminate Shiga toxin genes among enterobacteria. J. Bacteriol. 189, 7223–7233. 10.1128/JB.00824-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza D. P., Oka G. U., Alvarez-Martinez C. E., Bisson-Filho A. W., Dunger G., Hobeika L., et al. (2015). Bacterial killing via a type IV secretion system. Nat. Commun. 6:6453. 10.1038/ncomms7453 [DOI] [PubMed] [Google Scholar]
- Sperandio V., Torres A. G., Girón J. A., Kaper J. B. (2001). Quorum sensing is a global regulatory mechanism in enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 183, 5187–5197. 10.1128/JB.183.17.5187-5197.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarr P. I., Gordon C. A., Chandler W. L. (2005). Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet 365, 1073–1086. 10.1016/S0140-6736(05)71144-2 [DOI] [PubMed] [Google Scholar]
- Tenaillon O., Skurnik D., Picard B., Denamur E. (2010). The population genetics of commensal Escherichia coli. Nat. Rev. Microbiol. 8, 207–217. 10.1038/nrmicro2298 [DOI] [PubMed] [Google Scholar]
- Tesh V. L., Burris J. A., Owens J. W., Gordon V. M., Wadolkowski E. A., O'brien A. D., et al. (1993). Comparison of the relative toxicities of Shiga-like toxins type I and type II for mice. Infect. Immun. 61, 3392–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thévenot J., Cordonnier C., Rougeron A., Le Goff O., Nguyen H. T. T., Denis S., et al. (2015). Enterohemorrhagic Escherichia coli infection has donor-dependent effect on human gut microbiota and may be antagonized by probiotic yeast during interaction with Peyer's patches. Appl. Microbiol. Biotechnol. 99, 9097–9110. 10.1007/s00253-015-6704-0 [DOI] [PubMed] [Google Scholar]
- Thursby E., Juge N. (2017). Introduction to the human gut microbiota. Biochem. J. 474, 1823–1836. 10.1042/BCJ20160510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilden J., Jr., Young W., McNamara A. M., Custer C., Boesel B., Lambert-Fair M. A., et al. (1996). A new route of transmission for Escherichia coli: infection from dry fermented salami. Am. J. Public Health 86, 1142–1145. 10.2105/AJPH.86.8_Pt_1.1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toshima H., Yoshimura A., Arikawa K., Hidaka A., Ogasawara J., Hase A., et al. (2007). Enhancement of Shiga toxin production in enterohemorrhagic Escherichia coli serotype O157:H7. Appl. Environ. Microbiol. 73, 7582–7588. 10.1128/AEM.01326-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell T., Head S., Petric M., Cohen A., Lingwood C. (1988). Globotriosyl ceramide is specifically recognized by the Escherichia coli verocytotoxin 2. Biochem. Biophys. Res. Commun. 152, 674–679. 10.1016/S0006-291X(88)80091-3 [DOI] [PubMed] [Google Scholar]
- Waldor M. K., Friedman D. I. (2005). Phage regulatory circuits and virulence gene expression. Curr. Opin. Microbiol. 8, 459–465. 10.1016/j.mib.2005.06.001 [DOI] [PubMed] [Google Scholar]
- Watarai M., Sato T., Kobayashi M., Shimizu T., Yamasaki S., Tobe T., et al. (1998). Identification and characterization of a newly isolated Shiga toxin 2-converting phage from Shiga toxin-producing Escherichia coli. Infect. Immun. 66, 4100–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner J., Misra R. (2005). YaeT (Omp85) affects the assembly of lipid-dependent and lipid-independent outer membrane proteins of Escherichia coli. Mol. Microbiol. 57, 1450–1459. 10.1111/j.1365-2958.2005.04775.x [DOI] [PubMed] [Google Scholar]
- Wu T., Malinverni J., Ruiz N., Kim S., Silhavy T. J., Kahne D. (2005). Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 121, 235–245. 10.1016/j.cell.2005.02.015 [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. (1985). Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mpl8 and pUC19 vectors. Gene 33, 103–119. 10.1016/0378-1119(85)90120-9 [DOI] [PubMed] [Google Scholar]
- Yin S., Rusconi B., Sanjar F., Goswami K., Xiaoli L., Eppinger M., et al. (2015). Escherichia coli O157:H7 strains harbor at least three distinct sequence types of Shiga toxin 2a-converting phages. BMC Genomics 16:733. 10.1186/s12864-015-1934-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorgey P., Lee J., Kördel J., Vivas E., Warner P., Jebaratnam D., et al. (1994). Posttranslational modifications in microcin B17 define an additional class of DNA gyrase inhibitor. Proc. Natl. Acad. Sci. U.S.A. 91, 4519–4523. 10.1073/pnas.91.10.4519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., McDaniel A. D., Wolf L. E., Keusch G. T., Waldor M. K., Acheson D. W. (2000). Quinolone antibiotics induce Shiga toxin-encoding bacteriophages, toxin production, and death in mice. J. Infect. Dis. 181, 664–670. 10.1086/315239 [DOI] [PubMed] [Google Scholar]


