Key Points
TED is defective in patients with MDS.
TED is an independent prognostic factor for survival in MDS.
Abstract
Anemia is the defining feature in most patients with myelodysplastic syndromes (MDS), yet defects in erythropoiesis have not been well characterized. We examined freshly obtained bone marrow (BM) samples for stage-specific abnormalities during terminal erythroid differentiation (TED) from 221 samples (MDS, n = 205 from 113 unique patients; normal, n = 16) by measuring the surface expression of glycophorin A, band 3, and integrin α-4. Clinical and biologic associations were sought with presence or absence of TED and the specific stage of erythroid arrest. In 27% of MDS samples (56/205), there was no quantifiable TED documented by surface expression of integrin α-4 and band 3 by terminally differentiating erythroblasts. Absence of quantifiable TED was associated with a significantly worse overall survival (56 vs 103 months, P = .0001) and SRSF2 mutations (7/23, P < .05). In a multivariable Cox proportional hazards regression analysis, absence of TED remained independently significant across International Prognostic Scoring System-Revised (IPSS-R) categories, myeloid/erythroid ratio, and mutations in several genes. In 149/205 MDS samples, the proportion of cells undergoing TED did not follow the expected 1:2:4:8:16 doubling pattern in successive stages. Absence of TED emerged as a powerful independent prognostic marker of poor overall survival across all IPSS-R categories in MDS, and SRSF2 mutations were more frequently associated with absence of TED.
Visual Abstract
Introduction
Myelodysplastic syndromes (MDS) are essentially incurable primary hematopoietic stem cell disorders except for patients who receive allogeneic transplants.1 The natural history of MDS is highly variable, with survival ranging from months to decades. Identification of accurate prognostic variables is important, and the International Prognostic Scoring System-Revised (IPSS-R) is the most universally used prognostic classification.2 Despite its general success, the IPSS-R has its limitations. For example, within the group of patients identified as having lower risk MDS, there exist at least 3 subcategories with distinct survival patterns ranging from 2.6 to 9.4 years and risk of transformation to acute myeloid leukemia ranging from 5% to 25%.3 Further refinement of the existing classifications through the addition of mutational and gene expression profiling data are being attempted.4,5 Because anemia is the predominant cytopenia, we focused on a study of erythropoiesis to determine its clinical and biologic role in defining the natural history of MDS.
Erythroid differentiation is a complex cellular process that includes both early and late stages of erythroid differentiation. The early stage refers to a process by which pluripotent hematopoietic stem cells proliferate and differentiate into erythroid progenitors, erythroid burst-forming units, and then erythroid colony-forming units that generate proerythroblasts. The proerythroblast undergoes 4 to 5 mitoses to produce reticulocytes by a process termed terminal erythroid differentiation (TED) consisting of 5 distinct phases: proerythroblasts (pros), early basophilic erythroblasts (EBs), late basophilic erythroblasts (LBs), polychromatic erythroblasts (polys), to orthochromatic erythroblasts (orthos), which, upon enucleation, generate reticulocytes.6 Each daughter cell during TED is characterized by changes in expression of membrane proteins in that although the expression of major red cell membrane proteins increases, that of adhesion molecules decreases.7-13 By examining the dynamic changes in the expression of 3 salient marker proteins: glycophorin A (GPA), band 3, and integrin α-4, we quantified erythroblasts at distinct stages of their terminal differentiation in freshly obtained BM samples and successfully defined the stage-specific defects in morphologically and genetically well-defined subgroups of MDS patients. The resulting insights into the biology of MDS and the relationship of TED to mutational profiles and survival are reported here.
Materials and methods
Informed consent was obtained from subjects who participated in the study, which was approved by the institutional review board of Columbia University and was in accordance with the Declaration of Helsinki. Samples were obtained from patients seen at New York Presbyterian Hospital/Columbia University Medical Center and from 16 normal individuals (New York Presbyterian Cornell Hospital). World Health Organization (WHO) 2008 and IPSS-R were used in the classification of MDS patients. Ring sideroblasts (RSs) were detected by Prussian Blue staining and counted by 2 authors (D.H. and R.F.P.). Cytogenetic analysis was performed using standard G-banding. Clinical data were obtained via Columbia University Crown electronic medical and clinic records by researchers blinded to study results. The details of age, sex, WHO, IPSS-R, myeloblast, RS, hemoglobin, platelets, absolute neutrophil count, myeloid/erythroid (M:E) ratio, and percentage of cells quantified in various stages of TED in patients are described in supplemental Table 1. Percentage of cells quantified in various stages of TED in normal individuals is provided in supplemental Table 2.
Sample collection and preparation for TED
BM aspirates were assayed for TED using a method we published previously.6,9 Briefly, BM cells were separated on a Ficoll density gradient and mononuclear cells (MNCs) were incubated with CD45 microbeads for negative selection. CD45− cells were stained, analyzed, and quantified as previously described.6,9 Each component of TED at distinct stages of development was evaluated using GPA, band 3, and integrin α-4. The selection of these 3 markers are based on a comprehensive analysis of membrane proteins that established the utility of these markers as necessary and sufficient for identifying various stages of TED in normal samples.6,8-11 The plot of band 3 vs integrin α-4 of GPA+ cells revealed 2 distinct populations in normal controls: an integrin α-4+ population that contained nucleated erythroid cells and an integrin α-4− population that contained enucleated erythroid cells, as previously reported.6,9 Five populations of GPA+ cells were gated on the basis of the expression levels of integrin α-4 and band 3 (Figure 1). The gated cell populations were sorted using fluorescence-activated cell sorter. Populations 1, 2, 3, 4, and 5 represented pro, EB, LB, poly, and ortho, respectively.5 Data from 16 normal donors were subsequently compared with patient samples.
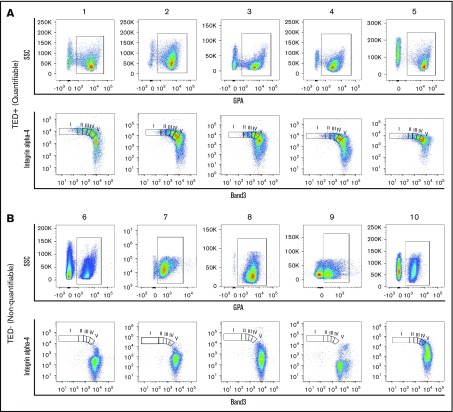
Figure 1.
Flow cytometry profiles of patient samples. Representative dot plots of TED profiles, as observed on flow cytometry, obtained from primary human BM of MDS samples from (A) 5 quantifiable samples and (B) 5 nonquantifiable samples. GPA+ cells (A-B, top) were found in both quantifiable and nonquantifiable samples, but the nonquantifiable samples were characterized by (B, top) low GPA expression and (B, bottom) absence of cells undergoing TED. I, II, III, IV, V, represent pro, EB, LB, poly, ortho stages of TED; SSC, side scatter.
Genetic profiling
Genomic DNA was extracted from BM MNC using Qiagen’s DNAeasy Blood and Tissue kit. In total, 54 genes (supplemental Table 3) that are part of the myeloid/lymphoid/acute leukemia panel at Cancer Genetics Inc. were screened for mutations. All sequencing data were analyzed using Cartagenia pipeline (Agilent Technologies). Mutations were confirmed using Sanger sequencing for a subset of genes and patients.
Statistics
All statistical tests were performed using either GraphPad Prism, version 7, or MedCalc, version 17.8, statistical software. For continuous variables, nonparametric 2-tailed tests were used as described in the legends this article’s figures. For categorical data, patient characteristics were compared using Fisher’s exact test where appropriate. Overall survival (OS) analysis was done using the Kaplan-Meier method. OS was calculated from the date of diagnosis to date of death and censoring data at the time patients were last known to be alive. Survival curves were compared using log-rank test. Cox proportional hazards regression analysis was used for univariate and multivariable analyses and is described in detail in the “Results” section. In survival analysis, for grouping patients with a quantifiable TED profile or those without, the result of TED analysis on first sample analyzed was used irrespective of TED status in subsequent samples. Where appropriate, more details on statistical tests are described in the legends of this article’s figures.
Results
A total of 221 samples were analyzed for TED; 16 normal controls and 205 BM samples (196 MDS including 9 MDS/myeloproliferative neoplasm [MPN] overlap) were from 113 unique patients with myeloid malignancies; breakdown by disease type and WHO classification is provided in Table 1. Successively obtained samples during a given period were studied and do not represent patients at any particular point in their disease.
Table 1.
Number of samples analyzed and distribution of samples in each WHO 2008 MDS subtype
| Samples | ||
|---|---|---|
| Samples analyzed for TED | 205 | |
| Control samples | 16 | |
| Samples with myeloid malignancies (113 unique patients) | 221 | |
| Samples with TED | 149 | |
| Adequate samples but NoTED | 56 | |
| TED (%) | NoTED (%) | |
| MDS | ||
| RCUD | 1 (100) | 0 (0) |
| RA | 11 (73) | 4 (27) |
| RCMD | 73 (70) | 32 (30) |
| RARS | 21 (91) | 2 (9) |
| RAEB-1 | 16 (76) | 5 (24) |
| RAEB-2 | 21 (68) | 10 (32) |
| MDS/MPN | ||
| RARS-T | 6 (66) | 3 (34) |
| Total | 149 | 56 |
RAEB, refractory anemia with excess blasts; RCMD, refractory cytopenia with multilineage dysplasia.
TED profiles of MDS samples
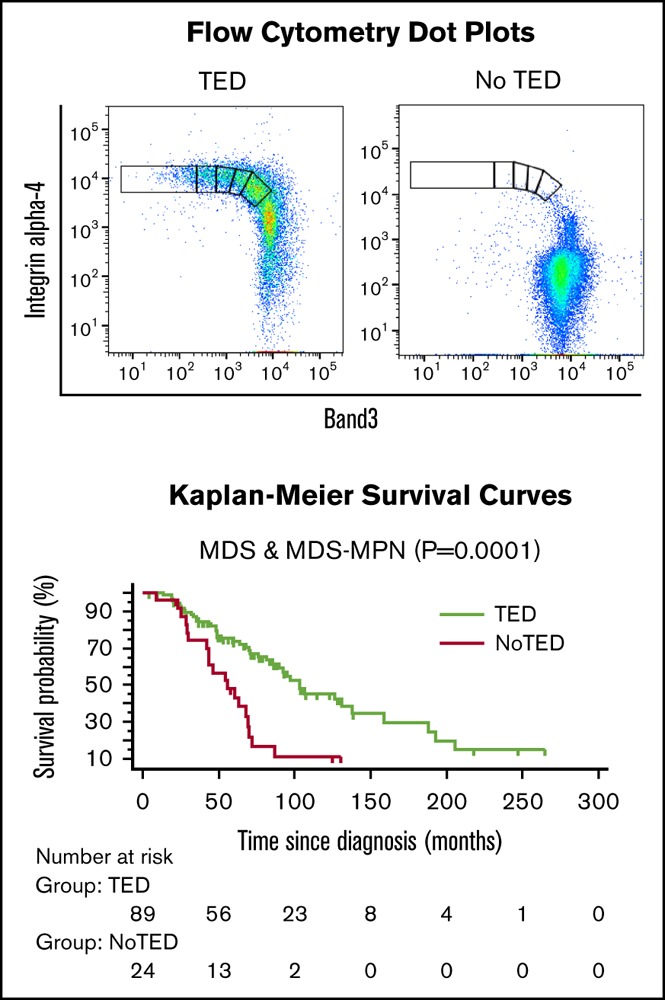
As proerythroblasts (GPA+ cells) undergo TED through 4 successive mitoses, surface expression of band 3 increases and integrin α-4 decreases. Using flow cytometry, we measured cells in pro, EB, LB, poly, and ortho stages of TED. Of 205 BM samples, 149 exhibited a quantifiable TED profile (referred to as TED-positive or simply TED), whereas 56/205 (27%) samples did not yield a reliable estimation of TED because too few cells were positive for both integrin α-4 and band 3 (referred to as TED-negative or NoTED). Both the TED (Figure 1A) and NoTED (Figure 1B) group of samples showed sufficient number of cells marked by expression of GPA (Figure 1A-B, top).
TED followed an expected doubling pattern with little variation between samples from normal individuals (supplemental Figure 1; supplemental Table 2). But in MDS patients, it was clear that TED did not follow the expected doubling pattern from the start, showing significantly fewer cells in pro (P ≤ .01) and EB (P ≤ .05) (Figure 2A; supplemental Table 4). At the LB stage, an equal number of cells appeared in normal and patient samples, but in the poly stage, a significantly higher percentage of cells were detected (P < .001), with a sudden drop off in ortho (Figure 2A). These differences persisted when refractory anemia with ring sideroblasts associated with thrombocytosis (RARS-T) samples were excluded (supplemental Figure 2A) or patients whose samples showed a different TED outcome on repeat sampling at later time in their disease history were excluded and only patients whose samples consistently showed TED on repeat sampling were analyzed (supplemental Figure 2B).This increased number of cells in poly and a decrease in ortho in MDS samples suggest a loss of cells either by apoptosis somewhere between poly and ortho stages or cells normally destined for progression to ortho remained poly either because of cell-cycle and/or maturation arrest.
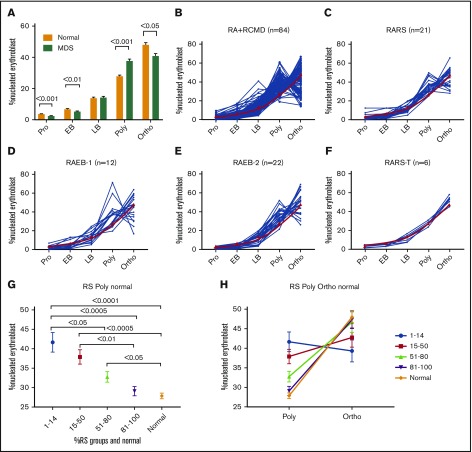
Figure 2.
TED is abnormal in patient samples. (A) Bar diagram showing percentage of nucleated erythroid cells in each stage of TED. When compared with normal (n = 16), MDS/MDS-MPN/secondary acute myeloid leukemia samples (n = 95) were characterized by a significant decrease in percentage of cells in pro and EB stages and a significantly abnormal increase of cells in the poly stage, and a decrease in the ortho stage. Data in bar diagram are presented as mean ± standard error of the mean. Each TED stage was compared between normal and MDS samples using unpaired, 2-tailed, nonparametric t test (Mann-Whitney U). (B-F) Line plot showing percentage of cells quantified in each TED stage in individual samples of WHO subgroups. (B) RA and RCMD, (C) RARS, (D) RAEB-1, (E) RAEB-2, and (F) RARS-T. Red line is the mean percentage of cells observed in normal individuals. (G-H) RS subgroups and TED. (G) Pot showing percentage of nucleated erythroblast cells in the poly stage within various RS subgroups and normal samples. There was significant difference in the poly stage between different RS subgroups; less cell arrest was observed in the poly stage as RS increased. All data are presented as mean ± standard error of the mean. Samples were compared using 1-way analysis of variance (Kruskal-Wallis test); multiple comparisons were done using the uncorrected Dunn test. (H) A graph showing inverse relation between poly and ortho stages within various RS groups and normal. Less arrest in poly is translated into more ortho stages. RA, refractory anemia; RS, ring sideroblast.
TED relationship with RS and WHO subtypes
Except for RARS-T, the significant differences observed in pro, EB, and poly stages in all MDS samples were retained in other MDS subtypes (Figure 2B-E). Given a near-normal TED in the RARS-T subgroup (Figure 2F), we next analyzed all MDS samples with any RS (Figure 2G-H). There were 104 samples in our cohort with accurate quantification of RS ranging from 1% to 100%, irrespective of WHO classification (supplemental Table 1). We divided these samples in 4 groups: 1% to 14%, 15% to 50%, 51% to 80%, and 81% to 100%. No significant differences were observed between the 4 RS groups in pro, EB, LB, and ortho stages but a significant difference was observed in the poly stage (P = .0003) with a strong negative correlation between poly and RS (r = −0.505, P < .0001) (Figure 2G-H). Patients with RS between 1% and 14% showed higher poly than patients with 51% to 80% and 81% to 100% RS (P = .0414 and P = .0007, respectively). Patients with 15% to 50% RS had a higher number of polys than 81% to 100% RS (P = .0096). In general, there was a striking reduction in polys as RS increased, especially remarkable in cases with >50% RS (Figure 2G-H). Interestingly, although the percent of poly stage erythroblasts was lower, the percent of ortho stage cells increased with increasing RS. This implies that RS exhibit lower cell arrest at the poly stage with resulting near normal TED progression.
No significant differences were observed in all 5 TED stages when compared within the 5 IPSS-R categories or the 4 categories of blasts <5%, 5% to 9%, 10% to 19%, and ≥20% blasts (data not shown).
MDS samples with NoTED
Despite adequate cells for flow analysis, 56 samples had too few erythroid cells undergoing TED as defined by expression patterns of integrin α-4 and band 3 to be accurately quantified (Table 1). Because all the patients we assayed for TED using flow cytometry were also analyzed by pathologists at Columbia University Medical Center as part of routine care, we analyzed the manual differential cell count data from BM specimens as reported by pathologists. We reasoned that if the “too few erythroid cells” observed on flow cytometry in these 56 samples (NoTED group) is an artifact, then the manual differential count data on erythroid cell lineage from pathology reports should not be significantly different between the TED and NoTED groups. The manual differential count data on BM specimen reports percentage of at least 16 different cells types, a count made from 500 cells, identifying pronormoblast, baso normoblast, polychromatic, and orthochromatic cells of erythroid lineage, among others, based on their morphology. Interestingly, we saw a significantly low number of all 4 cell types of erythroid lineage in the NoTED group (supplemental Figure 3A-D). Taken together, these data suggest that the flow cytometry method is accurately quantifying the various TED stages and that there is not a complete absence of TED, but that there are too few cells undergoing TED.
We also analyzed the M:E ratio from the pathology reports and found that samples with NoTED had a higher M:E ratio (mean, 5.7:1) compared with samples with TED (mean, 2:1); this trend was barely statistically significant (P = .0506). Most important, the proportion of patients with a >5:1 ME ratio were statistically more significant (P = .012) in NoTED (30%) compared with TED (8%). Also, a more pronounced anemia was observed in NoTED patients, with lower hemoglobin in NoTED (median, 8.9 g/dL) compared with TED (median, 9.75 g/dL), a trend that narrowly fell short of significance (P = .0643). No statistically significantly differences were observed in absolute neutrophil count, blast, and serum EPO levels between TED and NoTED patients (data not shown).
When separated based on their disease subtype, 84% RARS-T showed TED and 16% did not, whereas 70% RCMD showed TED and 30% did not. In RAEB-1/2 cases, 70% showed TED and 30% did not (Table 1). As the severity of IPSS-R risk increased, the proportion of TED-negative cases increased (supplemental Table 5).
Some patients were studied more than once; in 20 patients, repeat sampling gave different results at least at 1 time point. Appearance or disappearance of quantifiable TED in multiple studied cases was not related to any apparent clinical/pathologic change, and exclusion of these 22 cases did not change the overall statistics related to the clinical significance of TED (raw data provided in supplemental Tables 1-11).
TED vs treatment status
Treatments, both approved and experimental, can affect gene expression profiles of cells, which in turn may alter protein expression and or localization, presumably including the surface markers analyzed for TED in this study. We analyzed the proportions of TED and NoTED patients within each treatment group (supplemental Table 6). The majority of patients were not on any treatment at the time of sample collection (70/113), yet 23% (16/70) patients were NoTED. Between TED (60%) and NoTED (66%), the proportions of untreated patients were roughly equal. Of 113 patients, 27 were treated (either ongoing or in the past) with hypomethylating agents (HMAs) and 21 had TED, whereas 6 did not. Among the 7 patients who were on HMA at the time of sample collection, 5 were TED and 2 were NoTED. Given that more HMA-treated patients were TED-positive, the TED-negative outcome is not related to HMA therapy, thus alleviating the concern that the treatment did not alter the markers used in the study. For other treatments, such as an erythropoiesis-stimulating agent and rigosertib, we saw more patients in the TED group than in the NoTED group (supplemental Table 7). Although it is tempting to suggest that treatment may have a role in improving TED, the study was not sufficiently powered to analyze this effect. For example, as noted previously, there were only 7 patients on HMA treatment, but the time of sample collection from the start of treatment was different for each. Similarly, change in repeat sampling from NoTED to TED or vice versa in individual cases could be due to therapeutic interventions, but once again the numbers are low for meaningful analysis.
TED vs mutational profiles in MDS
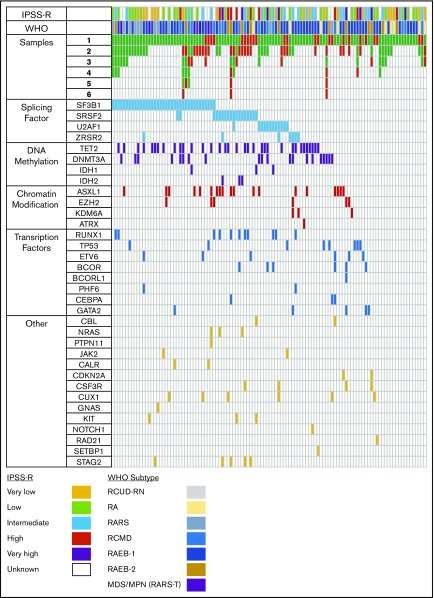
Figure 3 is a graphic representation of each sample examined for TED; the mutational profile (n = 112). Supplemental Table 7 lists all the mutations. No mutations were detected in 16% of patients, and TED-negative cases had slightly more, but not statistically significant, mutations than TED-positive cases (mean, 2.3 vs 2.0). The most frequently mutated individual gene in our cohort was SF3B1 followed by TET2, DNMT3A, SRSF2, and ASXL1 (Figure 3). The most common mutations were in splicing factor (SF) genes (61%) followed by TET2 (30%). Although SF mutations as a group were equally distributed between TED and NoTED patients (45/89 or 50% with TED and 12/24 50% with NoTED), the distribution of specific SF genes was highly skewed. The proportion of SRFS2 mutations were significantly more in the NoTED (30%) compared with TED (12%) groups (P = .0497). On the other hand, SF3B1 mutation was more common in the TED (35%) compared with NoTED (13%) groups, but the difference was not statistically significant (P = .0701). SF3B1 mutation was seen in 75% samples with >15% RS and was associated with quantifiable TED.
Figure 3.
Mutational analysis. Graphic representation of number of samples analyzed for each patient (N = 112) and mutations observed in the first sample analyzed. Green box, sample on whom TED profile was obtained; red box, the sample was adequate but no cells were found undergoing TED. Each column represents 1 unique patient. Although for some patients, every sample analyzed at different time points showed TED, there were others who failed to show cells in TED at every point analyzed; there were some samples that failed to show cells in TED at 1 or more point. Colored boxes colored according to their functional group in mutation panels show presence of mutation in that gene and patient. Splicing factor mutations were more common followed by DNA methylation. Bottom, color codes for the IPSSR and WHO categories.
TED vs OS
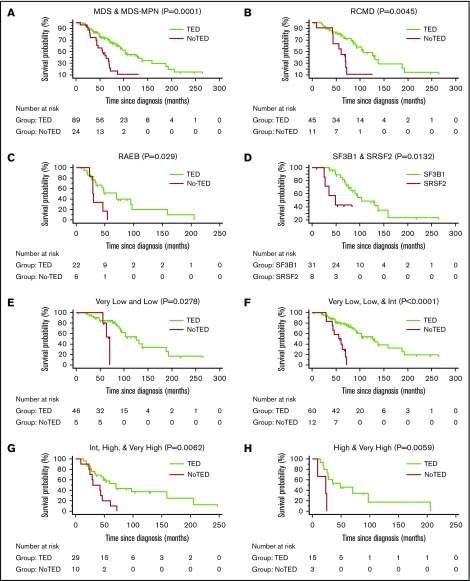
Supplemental Table 8 shows data used for survival analysis; supplemental Table 9 shows patient characteristics and their association with median survival. For all survival analyses, only the results of the first sample of each patient was used. There was a highly significant difference in OS between TED-positive (median, 103 months) vs TED-negative (median, 60 months) patients (P = .0001; Figure 4A). The difference in median survival remained unchanged when 6 RARS-T patients were excluded (supplemental Figure 4A).
Figure 4.
Survival curves. Kaplan-Meier survival curves were generated to calculate OS. OS among patients in whom TED profile was obtained (TED) compared with patients in whom sample was adequate but no cells were found undergoing TED (NoTED). There was a significant difference in OS between TED and NoTED among (A) all patients: MDS (n = 108) and RARS-T (n = 6) (P = .0001), (B) RCMD patients (P = .0045), and (C) RAEB (RAEB-1 and RAEB-2) patients (P = .029). (D) We also found a difference in OS between patients with TED and SF3B1 or SRSF2 mutations (P = .0132). (E) IPSS-R very low and low categories (P = .0278); (F) very low, low, and intermediate categories (P < .0001); (G) intermediate, high, and very high categories (P = .0062); and (H) high and very high categories (P = .0059). Log-rank tests were used to compare the curves. The tables below the curves indicate the number of patients at risk in each group.
MDS patients with SF3B1 mutations show a better prognosis compared with SF3B1 wild-type patients.14-16 In our dataset (supplemental Figure 4B), we saw a similar better survival rate of MDS patients with SF3B1 mutations (median, 94.9 months) compared with SF3B1 wild-type (median, 71.8 months). To alleviate the concern that the better OS of patients with an SF3B1 mutation might explain the better prognosis of TED-positive patients (supplemental Figure 4B), because most of the SF3B1-mutated patients were TED-positive, we did an OS analysis excluding patients with mutations in the SF3B1 gene (supplemental Figure 4C). OS between TED-positive and TED-negative groups was still significantly different after excluding SF3B1 patients (P = .0015; supplemental Figure 4C). The median survival of the TED-positive group (106 months) was not much different after excluding the SF3B1 group, suggesting that the observed differences were not because of SF3B1-mutated patients. The difference persisted within each subgroup examined: RCMD (P = .0041; Figure 4B) and RAEB-1/2 (P = .029; Figure 4C). When TED-positive samples were divided by those with mutations in SF3B1 or SRSF2, OS was worse for those with TED and SRSF2 (P = .0132; Figure 4D). OS was significantly different within lower and higher risk IPSS-R categories (Figure 4E-H; supplemental Table 10). OS analysis of a subset of patients who were not on any therapy at the time of sample collection still showed a significant difference between TED and NoTED (P = .0015) (supplemental Figure 4D). The survival of patients with M:E ratio ≥5 was worse than those with patient with a <5 M:E ratio (P = .0005).
TED vs red blood cell transfusion dependence
Transfusion data were available on all 113 unique patients. Patients were considered transfusion dependent if they have received at least 2 units of red blood cells within the 56 days before their first sample was collected for this study. Fifty-seven percent (64/113) were transfusion dependent. Among the NoTED, 88% (21/24) were transfusion dependent, whereas among the TED, only 46% (43/89) were transfusion dependent (P = .0005). Within the transfusion-dependent patients, transfusion requirements were higher in the NoTED group (supplemental Figure 5). The median survival of transfusion-dependent patients (72 months) was lower than transfusion-independent patients (103 months; supplemental Figure 4E), but this difference was not statistically significant. Within the transfusion-dependent patients, we saw a significant difference in survival based on their TED status (P = .0017; supplemental Figure 4F).
Multivariable survival model using presence or absence of TED
In a univariate analysis, absence of TED, IPSS-R, and M:E ratio (≥5) and presence of mutation in CEBPA, CUX1, IDH2, NRAS, RUNX1, SRSF2, and STAG2 was significantly associated with survival in MDS patients (supplemental Table 11). To determine the contribution of significant factors affecting survival, we generated a multivariable Cox proportional hazards regression model, using a stepwise variable selection procedure, incorporating variables noted previously except CEBPA, IDH2, and NRAS, which were present in <3% of patients. TED and the IPSS-R risk categories high and very high and mutations in CUX1 and STAG2 remained significant (Table 2). We obtained the same final model using a forward selection method.
Table 2.
Hazard ratios for death in a multivariable model
| Covariate | P | HR | 95% CI of HR |
|---|---|---|---|
| TED (quantifiable vs NoTED) | .0001 | 4.9 | 2.1-11.0 |
| IPSS-R (high vs low) | .0035 | 3.7 | 1.5-9.0 |
| IPSS-R (very high vs low) | .031 | 2.7 | 1.0-6.8 |
| CUX1 (absent vs present) | .0157 | 3.5 | 1.2-9.6 |
| STAG2 (absent vs present) | .045 | 3.0 | 1.0-9.1 |
The model includes TED (quantifiable or not), IPSS-R (very low, low, intermediate, high, and very high), M:E ratio (<5 and ≥5), and presence or absence of mutation in CUX1, RUNX1, SRSF2, and STAG2.
CI, confidence interval; HR, hazard ratio.
Discussion
This study represents the first attempt to accurately quantify cells in various stages of TED from freshly obtained BM samples of patients with MDS and MDS/MPN overlap syndromes (RARS-T). One-third of the cases examined did not have quantifiable TED. TED-negative cases had a markedly shorter survival across all IPSS-R categories. The mechanism and biologic significance of fewer cells undergoing TED remain to be defined, but the clinical value of this observation is of immediate practical use. In MDS patients, treatment choices, as well as the timing of intervention, are guided by an accurate assessment of prognosis, yet the risk of death, especially in the lower risk group, can be underestimated. The median survival for TED-negative cases was significantly worse in both the lower (very low, low, and intermediate; median, 56 vs 126 months, P < .0001) and higher risk (high and very high; median, 23 vs 48 months, P = .0059) IPSS-R groups. Given that it remained a powerful independent variable for OS in a multivariable Cox regression model, assessment and quantification of TED by established erythroid cell surface markers can improve the prediction of prognosis within the various IPSS-R categories. Further striking associations emerged when these cases were found to be more frequently associated with mutations in SRSF2 and more profound anemia.
TED studies raise a series of questions in terms of the underlying biological implications and of technological assessment of erythropoiesis. BM aspirate and biopsy slides of TED-positive and TED-negative cases were reviewed by a hematopathology colleague (D.H.) in a blinded manner. No obvious morphologic distinction was found between the 2 cases. A trend for a higher percentage of myeloid cells in TED-negative samples was seen by M:E ratio, but the association did not apply to individual cases. In other words, many samples with large numbers of erythroid cells did not have quantifiable numbers progressing through TED. Were these erythroid cells arrested or undergoing apoptosis in erythroid colony-forming unit or erythroid burst-forming unit stages? Another concern was whether the inability to estimate cells undergoing TED represented a purely technical problem. To satisfy this concern, we compared total cell numbers in TED+/− samples and found that there was no difference between the 2 groups. Although GPA positivity was lower (mean fluorescence intensity) in the TED-negative cases (Figure 1), there were enough cells present in the samples being subjected to flow cytometric analyses. Most striking was our finding of the absence of integrin α-4– and band 3–expressing cells in the TED-negative cases. There is always a possibility that the inability to detect expression resulted from a failure of the antibody to recognize the surface expression because of epitope modification in these cells. If so, what is the pathology driving such epitope alteration?
MDS patients with SRSF2 mutations have distinct clinical and biologic features in that they are older, more often male, and have an inferior OS.17 We have previously reported that SRSF2 mutations cause subtle alterations in RNA-binding affinity and that the magnitude of splicing changes, as a result, is low consistent with the view that the pathogenesis of MDS is a slow process in which small effects of altered splicing gradually give rise to the disease phenotype by causing “death by a thousand cuts.”18 In this study, we identified a predominance of TED absence (7/23 unique patients) in the SRSF2-mutated group.
The case of RARS is curious in that, as with other MDS subtypes, a higher than expected percentage of cells appeared in the poly compartment, but unlike other MDS patients, in some of these cases more cells progressed to the ortho stage. There was also a striking inverse relationship between the percentage of RS and the number of erythroid cells arrested in the poly stage. Even in cases in which RS ranged between 81% and 100% of the BM, cells effectively passed into the penultimate orthochromatic stage, presumably apoptosing before becoming reticulocytes. Conte et al analyzed 5 RARS patients by inducing their CD34+ cells toward erythroid differentiation in vitro and showed that the SF3B1 allelic burden was 1.5 times lower in reticulocytes compared with the corresponding CD34+ cells, suggesting a reduction in the terminal differentiation of mutated erythroblasts.19 These in vitro data also support our in vivo findings using primary BM cells.
Samples were collected over a period of 2.5 years; many patients during this time donated multiple marrow samples for TED studies. Although most patients (66%) showed a consistent TED outcome on repeat sampling (Figure 3), there were some inconsistent results. The “inconsistent” group, although biologically interesting, is too small in size for meaningful analysis (n = 20). Variables influencing erythroid differentiation include treatment and natural evolution of the disease. It is beyond the scope of the current paper, which was designed to characterize TED and its association with OS. Future studies using TED as an outcome will answer such questions. Given that improvement in anemia and transfusion independence, among others, are 2 end points related to erythroid differentiation in any clinical trial on MDS patients, TED can be used to assess biological changes associated with response.
The observable behavior of a cell, or its phenotype, results from a combination of both its genotype and the microenvironment. No studies have previously examined stages of TED in fresh BM cells. One reason could be the lack of technology until the recent past, but another may be the focus on a reductionist strategy in the era of genomics. We have come full circle in understanding cancer in a dense, phenotypic way, a practice more or less abandoned since the introduction of genomic sequencing. In the current study, an attempt is made to develop the phenotypic understanding simultaneously along with examining the genotype, which by itself would be inadequate to give important information regarding overall behavior of cells. Future studies are planned to understand the molecular mechanisms by sequencing the transcriptome of TED and NoTED groups. A recent study by Shiozawa et al5 reported 2 distinct groups of MDS patients, based on unsupervised clustering of transcriptome from BM CD34+ and MNC cells, 1 group with predominant expression of genes related to erythroid/megakaryocyte lineage with better survival and another group with genes related to immature progenitor cells and worst survival. Based on the results of Shiozawa et al, we hypothesize that the TED quantifiable group may be predominantly composed of erythroid/megakaryocyte type and the NoTED group of immature progenitor groups. The real-time clinical applicability of the results presented is apparent. Because MDS is a heterogeneous disease in which even the best of prognostic classifications falter, underestimating the risk of death in a substantial number of cases, quantification of TED can immediately serve as an independent prognostic variable. In addition, the findings open a novel area of research that could lead to identification of new therapeutic targets.
In conclusion, erythroid differentiation is profoundly abnormal across all MDS subtypes and absence of quantifiable cells undergoing TED by well-defined cell surface markers is strongly associated with inferior OS. The addition of this biologic marker to characterize hematopoietic defects in MDS has the potential to further refine the current prognostic classification systems.
Supplementary Material
The full-text version of this article contains a data supplement.
Acknowledgments
This work was supported in part by grants from the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases (DK100810 and DK32094), the Natural Science Foundation of China (81530005), and in part from The Partnership for Cure.
Authorship
Contribution: A.M.A., Y.H., F.X., C.H., and J.H. performed experiments; R.F.P., N.I., D.H., D.C., J.K., B.C., C.H., and N.G. collected data and analyzed the results; A.M.A. collected and analyzed the data and results, made the figures, and wrote paper; J.L. provided normal samples; J.J., N.M., and X.A. designed research and analyzed the data; and A.R. designed the research, analyzed the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Azra Raza, Department of Medicine, Columbia University Medical Center, 177 Fort Washington Ave, 6GN-435, New York, NY 10032; e-mail: azra.raza@columbia.edu.
References
- 1.Raza A, Galili N. The genetic basis of phenotypic heterogeneity in myelodysplastic syndromes. Nat Rev Cancer. 2012;12(12):849-859. [DOI] [PubMed] [Google Scholar]
- 2.Greenberg PL, Tuechler H, Schanz J, et al. . Revised international prognostic scoring system for myelodysplastic syndromes. Blood. 2012;120(12):2454-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pomares H, Sánchez-Ortega I, Alonso E, et al. . Validation of the Low Risk Prognostic Scoring System (LR-PSS) in patients with very low, low and intermediate risk IPSS-R myelodysplastic syndrome. Results from a single center. Blood. 2015;126(23):2902. [Google Scholar]
- 4.Bejar R, Stevenson KE, Caughey BA, et al. . Validation of a prognostic model and the impact of mutations in patients with lower-risk myelodysplastic syndromes. J Clin Oncol. 2012;30(27):3376-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shiozawa Y, Malcovati L, Gallì A, et al. . Gene expression and risk of leukemic transformation in myelodysplasia. Blood. 2017;130(24):2642-2653. [DOI] [PubMed] [Google Scholar]
- 6.Hu J, Liu J, Xue F, et al. . Isolation and functional characterization of human erythroblasts at distinct stages: implications for understanding of normal and disordered erythropoiesis in vivo. Blood. 2013;121(16):3246-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blikstad I, Nelson WJ, Moon RT, Lazarides E. Synthesis and assembly of spectrin during avian erythropoiesis: stoichiometric assembly but unequal synthesis of alpha and beta spectrin. Cell. 1983;32(4):1081-1091. [DOI] [PubMed] [Google Scholar]
- 8.Chang H, Langer PJ, Lodish HF. Asynchronous synthesis of erythrocyte membrane proteins. Proc Natl Acad Sci USA. 1976;73(9):3206-3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen K, Liu J, Heck S, Chasis JA, An X, Mohandas N. Resolving the distinct stages in erythroid differentiation based on dynamic changes in membrane protein expression during erythropoiesis. Proc Natl Acad Sci USA. 2009;106(41):17413-17418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gronowicz G, Swift H, Steck TL. Maturation of the reticulocyte in vitro. J Cell Sci. 1984;71:177-197. [DOI] [PubMed] [Google Scholar]
- 11.Hanspal M, Hanspal JS, Kalraiya R, et al. . Asynchronous synthesis of membrane skeletal proteins during terminal maturation of murine erythroblasts. Blood. 1992;80(2):530-539. [PubMed] [Google Scholar]
- 12.Liu J, Mohandas N, An X. Membrane assembly during erythropoiesis. Curr Opin Hematol. 2011;18(3):133-138. [DOI] [PubMed] [Google Scholar]
- 13.Peters LL, White RA, Birkenmeier CS, Bloom ML, Lux SE, Barker JE. Changing patterns in cytoskeletal mRNA expression and protein synthesis during murine erythropoiesis in vivo. Proc Natl Acad Sci USA. 1992;89(13):5749-5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malcovati L, Karimi M, Papaemmanuil E, et al. . SF3B1 mutation identifies a distinct subset of myelodysplastic syndrome with ring sideroblasts. Blood. 2015;126(2):233-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patnaik MM, Lasho TL, Hodnefield JM, et al. . SF3B1 mutations are prevalent in myelodysplastic syndromes with ring sideroblasts but do not hold independent prognostic value. Blood. 2012;119(2):569-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mangaonkar AA, Lasho TL, Finke CM, et al. . Prognostic interaction between bone marrow morphology and SF3B1 and ASXL1 mutations in myelodysplastic syndromes with ring sideroblasts. Blood Cancer J. 2018;8(2):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu SJ, Kuo YY, Hou HA, et al. . The clinical implication of SRSF2 mutation in patients with myelodysplastic syndrome and its stability during disease evolution. Blood. 2012;120(15):3106-3111. [DOI] [PubMed] [Google Scholar]
- 18.Zhang J, Lieu YK, Ali AM, et al. . Disease-associated mutation in SRSF2 misregulates splicing by altering RNA-binding affinities. Proc Natl Acad Sci USA. 2015;112(34):E4726-E4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Conte S, Katayama S, Vesterlund L, et al. . Aberrant splicing of genes involved in haemoglobin synthesis and impaired terminal erythroid maturation in SF3B1 mutated refractory anaemia with ring sideroblasts. Br J Haematol. 2015;171(4):478-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.