Abstract
Fibrolamellar hepatocellular carcinoma (FLC) is a rare form of primary liver cancer that affects adolescents and young adults without underlying liver disease. Surgery remains the mainstay of therapy, however most patients are either not surgical candidates, or suffer from recurrence. There is no approved systemic therapy and the overall survival remains poor. Historically classified as a subtype of Hepatocellular carcinoma (HCC), FLC has a unique clinical, histological and molecular presentation. At the genomic level FLC contains a single 400kB deletion in chromosome 19, leading to a functional DNAJB1-PRKACA fusion protein. In this review we detail the recent advances in our understanding of the molecular underpinnings of FLC and outline the current knowledge gaps.
Keywords: Liver cancer, Pediatric cancer, Adolescent young adult cancer, Fusion protein, Protein kinase A
Introduction
Fibrolamellar Hepatocellular Carcinoma (FLC) is a primary liver cancer which predominantly affects adolescents and young adults (AYA). FLC is a relatively rare tumor, with an estimated age-adjusted incidence rate of 0.02 per 100,000 in the United States (1, 2). Clinically, FLC most commonly presents as a single large liver mass, in patients without underlying liver disease. Several factors including a paucity of symptoms, an absence of known risk factors, and a lack of serum tumor markers; all contribute to an insidious and delayed presentation and diagnosis. It has a peak age of detection of 21 years, with the vast majority of patients diagnosed between the ages of 5 and 40. However, as a consequence of the delayed diagnosis, the actual age of onset is not known. Based on the existing databases it is slightly more common in women, but otherwise has no ethnic, geographic predilection and is not associated with any known risk factors (3, 4).
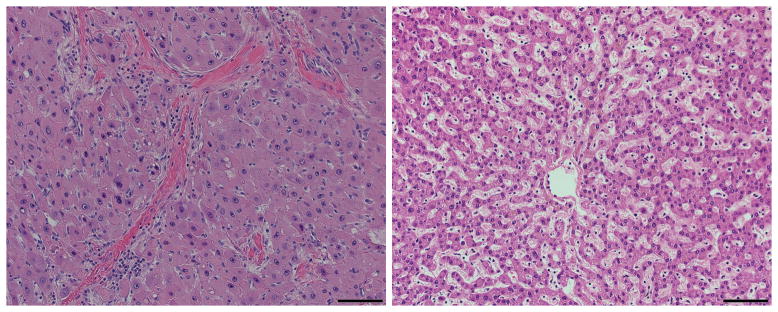
FLC was first described in 1956 based on its unique histology (5), which includes large eosinophilic, hepatocyte-like polygonal cells, with prominent nucleoli and pale inclusion bodies, surrounded by abundant fibrotic bands or “lamellae”, from which the tumor derives its name (6) (Figure 1). Unfortunately, even with a biopsy, the detection of lamellar bands is often unresolved (7), a confounding problem in diagnosis. In radiological scans FLC is seen as a large hypodense mass that enhances with contrast administration in the setting of a normal appearing liver. Like Focal nodular hyperplasia the mass in FLC frequently contains a central scar. It is due to this similarity that focal nodular hyperplasia, was suspected by some to be the premalignant lesion for FLC. Historically, the diagnosis of FLC was based on a combination of the typical presentation (i.e. a single large tumor in a young patient without underlying liver disease) and compatible histological features. However, since some patients lack the typical histological features or exhibit a “mixed” histology, which includes features of both Hepatocellular carcinoma (HCC) and FLC, several immune-stains, including CK7 and CD68, have been recommended to assist with diagnosis (8, 9). Despite these additions, in lieu of a gold standard for diagnosis, it has been an ongoing concern that many reports included misdiagnosed cases, and, conversely, some cases of FLC have been mistakenly diagnosed as other tumors (3, 7, 10).
Figure 1. FLC has a typical histological appearance.
The histology of FLC has several typical features that can be seen on H&E staining. These include: large hepatocyte-like polygonal cells, with an abundant eosinophilic cytoplasm, prominent nucleoli and pale cytoplasmic inclusion bodies. These cells are surrounded by abundant fibrotic bands or “lamellae”.
Based on the size of the tumor at diagnosis and vague or absent abdominal symptoms, FLC is thought to be a slow growing tumor. However, perhaps in part due to delayed diagnosis, over half the patients have metastases to the lymph nodes at diagnosis (3, 11). As of this writing, no systemic chemo- or immunotherapies have found effective and thus there is no standardized treatment (12–15). Current management of FLC is based on surgical resection of the primary tumor, regional lymph nodes, as well as distant metastases in selected cases (16, 17). Surgical resectability has borne out as the main predictor of survival in several case series, and patients often undergo repeated surgeries during the course of their disease (15, 18). However, recurrence after surgical resection, most commonly extra-hepatic, is common.(4, 11, 18) Unfortunately, as a consequence of extensive liver involvement, up to 30% of cases are not considered for resection at the time of diagnosis. Patients not eligible for resection often do not survive a year (16, 19). Even with resection, a majority of patients relapse within the year. Improved survival only correlates with aggressive resection of recurrences (11). (20) As a consequence, the 5-year survival for FLC patients is under 50% (3). Patients with tumors that are confined to the liver but exceed the limit for resection have been considered for orthotopic liver transplantation (OLT). A recent review found that the overall survival of FLC patients receiving OLT was comparable to that of HCC patients (1, 3, and 5 year survival was 96%, 80% and 48% for FLC as compared with 89%, 77%, and 68% for HCC)(21).
Initially FLC was considered to be a subtype of HCC and was codified as such in the ICD O (oncology) and SEER database. As result, it is also referred to as fibrolamellar hepatocellular carcinoma or the fibrolamellar variant of hepatocellular carcinoma. It has been argued to what extent FLC and HCC are separate diseases. While there are often morphological differences, there are times when they cannot be unambiguously differentiated by pathological criteria; and times when the samples appear to have mixtures of the two. Despite the fact that HCC usually occurs in older individuals, and FLC in AYA, non-cirrhotic HCC is still more common in the AYA population than FLC. FLC is found predominantly in the background of an otherwise healthy liver, while HCC usually occurs in patients with chronic viral hepatitis or liver cirrhosis of diverse causes. The question of which is a more indolent and which is more aggressive, is not yet resolved in the literature and may be confounded by the relatively advanced stage at which FLC is diagnosed on the one hand, and the lack of underlying liver disease on the other (4, 22).
The Genomics of Fibrolamellar Carcinoma
Historically, several studies evaluated the genomics of FLC. These studies generally suffered from small sample size and the limitations of the then available technology (restriction fragment length polymorphism, fluorescence in situ hybridization and comparative genomic hybridization). Despite these limitations these studies pointed to a relatively stable genome compared with HCC, with a lower frequency of mutations, and copy number variations. While some of the chromosomal abnormalities were similar (such as alterations in chromosome 1 and 8), by and large the overall pattern differed from that found in HCC. These are summarized nicely in a review by Ward et al. (23). Chromosomal abnormalities were correlated to a more aggressive disease in some of the studies (24–26).
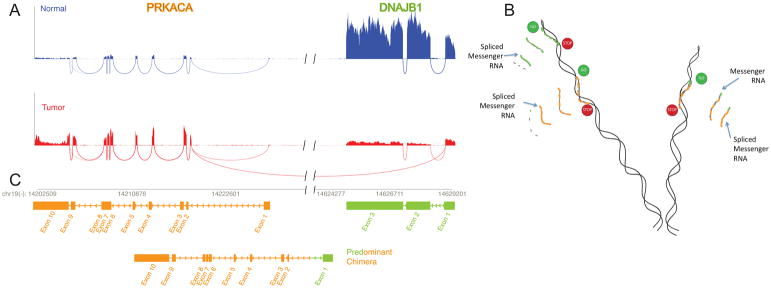
A molecular characterization using next generation sequencing technology, provided unambiguous demonstration that FLC and HCC are very different diseases. As a result of the clear molecular differences, detailed below, many have started referring to FLC as fibrolamellar carcinoma to avoid confusion with hepatocellular carcinoma. In the FLC tumor over 3500 transcripts changed significantly (FDR <0.01 with a |log2|>1) (10). These RNA-Seq results were confirmed for some of the key transcripts using qPCR and they were confirmed at the protein level using Western blot and mass spectrometry (10). The pattern of transcription cleanly segregated from patients with HCC, with a few notable exceptions, to be discussed below (Figure 2) (10). Whole genome sequencing showed a relatively consistent mutational spectrum with low inter-tumor variation; few coding, non-synonymous somatic mutations (median=6, range 0–16) no mutations in any of the known oncogenes and an extremely low level of amplifications, inversions, indels or deletions (27). Each of the kinases that were increased in expression were tested to see if they were using the usual isoform, or an altered isoform, by mapping the RNA reads onto the DNA genome. In all of the kinases, spare one, there were increases in reads for all of the normal exons and for reads that bridged the exons. The one exception was PRKACA, the catalytic subunit of protein kinase A. The number of reads to exons 2–10 and bridging those exons, were all increased. However, there was no increase in the number of reads in exon 1, nor in the reads that were bridging exon 1 and 2. Instead, there were reads that bridged between exon 2 of PRKACA and exon 1 of DNAJB1, a heat shock protein which was ~400 kB upstream (Figure 3A). These results were consistent with a deletion of ~400kb in one copy of chromosome 19 (Figure 3B) (28). This deletion resulted in a chimeric RNA of the first exon of DNAJB1 and the second to final exons of PRKACA (Figure 3C). This was tested by extracting the tumor tissue and demonstrating the chimera RNA by Sanger sequencing. This was further tested by demonstration of a chimera protein which was recognized both by antibodies to the amino terminus of DNAJB1 and the carboxyl terminus of PRKACA. The chimeric DNAJB1-PRKACA protein retained full enzymatic activity (28). This occurred on the background of one normal copy of the chromosome which produced the normal DNAJB1, the normal PRKACA and normal levels of the transcripts for the eight genes in between. An examination of the few HCC patients whose transcriptome clustered with the FLC showed that they had the deletion in chromosome 19 (10). Thus, it was concluded that they were likely to be FLC patients who had been misdiagnosed. While these results were initially demonstrated in 15 patients, all of the observations including the deletion, the formation of the chimera, proper enzymatic activity, the changes in the transcriptome, the segregation between FLC and HCC have since been reproduced in several papers (10, 29–33), although in one study (33) the chimera was found in only 79%. In some studies, RNA-Seq analysis showed that in proposed cases of FLC that did not contain the DNAJB1-PRKACA fusion, the transcriptome clustered with cases of HCC rather than FLC, and these cases where eventually adjudicated as HCCs (10, 32).
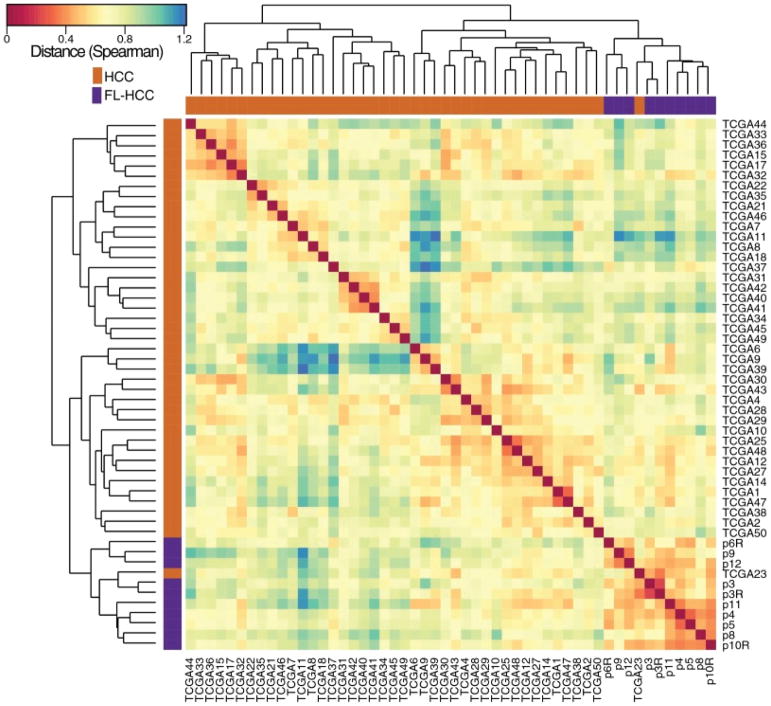
Figure 2. FLC and HCC have distinct molecular characteristics.
The transcriptome of fibrolamellar tumor relative to the adjacent normal and hepatocellular carcinoma tumor, relative to the adjacent normal, were analyzed and characterized by hierarchical clustering (complete method) with a spearman rank correlation matrix for all 3708 protein coding genes that passed the DESeq2 expression filter (|log 2 fold change| > 1, FDR < 0.01, and ranked by moderated t statistic ranking). The transcriptome of the fibrolamellar samples (purple) segregated away from those of hepatocellular carcinoma (orange) [Data shown is for 10 FLC and 42 HCC from over 400 HCC patients analyzed]. An example is shown of one patient out of five found that had been reported to be HCC, but the transcriptome segregated with FLC. This particular example was from a young adolescent who was diagnosed with HCC without a molecular characterization. An analysis showed the presence of a fusion between the genes DNAJB1 and PRKACA consistent with this patient actually having fibrolamellar (see figure 3). A histological sample was not available in this specific case. Figure from (10).
Figure 3. Molecular basis for FLC: A deletion in a chromosome 19 produces a chimeric gene.
Figure A. The reads for the mRNA were mapped onto the genome. An increased number of reads for PRKACA, the catalytic subunit of protein kinase A, in the tumor sample (red) relative to the normal (blue) in exon 2 through 10, and in the reads spanning the exons, demonstrated the increased expression of this gene. In addition in the tumor there were reads that spanned between the start of exon 2 of PRKACA and the end of exon 1 of DNAJB1, a member of the heat shock protein family. Figure B. This pattern of reads demonstrate that in the tumor there is the normal transcript for DNAJB1 (green), and the normal transcript for PRKACA (goldenrod) and a chimeric transcript with the first exon of DNAJB1 and the 2nd to final exons of PRKACA. This chimeric transcript was confirmed by Sanger sequencing (28). Figure C. The formation of the chimeric transcripts, while maintaining the normal transcripts, is consistent with a deletion of ~400 kB in one copy of chromosome 19. This deletion was confirmed by Sanger sequencing (28). Figure adapted from (28).
The genomic data on the formation of the chimeric protein and the highly consistent changes in the transcriptome and proteome of the FLC tumor, relative to the adjacent normal tissue, offered a number of important insights into the disease. One important insight was that FLC was not a genetically inherited disease. This is based on the observation that the alteration was only found in the tumor, not in the adjacent normal tissue. Thus, the driver for FLC was a somatic mutation. This was a relief for families of fibrolamellar patients who were concerned about siblings and children of the patients. These observations also led to a number of insights into therapeutic approaches. The conclusion that FLC is not genetically inherited, indicates that after complete resection there is no genetic disposition that could lead to additional de novo carcinogenesis. Additionally, the results implicate the chimeric protein as a critical therapeutic target. Further therapeutic insights come from the observation that the changes in the tumor of the transcriptome and proteome were highly conserved from patient-to-patient. Thus, on a molecular basis, we can consider FLC as a single disease rather than a collection of syndromes. This suggests that diagnostics, prognostic indicators and therapeutics developed for one patient should work for all patients.
Potential mechanisms of transformation
Two recent studies have supported the role of the chimera in transformation (34, 35). In these studies introducing the chimera into mouse liver via cDNA with the sleeping beauty transposon, or CRISPR induced liver tumors with similar features to human FLC. It is not yet known how expression of the chimera causes transformation. There are a number of clear possibilities all of which are experimentally testable. First, transformation could be the consequence of an increased expression of the chimera over native PRKACA, a result of the promoter for the heat shock protein driving transcription. This may be sufficient. However, if this was the case, additional mutations leading to overexpression of PRKACA should be able to lead to FLC. For example, increased activity of PRKACA could be the result of a mutation-induced increase in the activity of the promoter of PRKACA, a loss of one of the regulatory subunits, or a mutation in the regulatory subunit of PRKACA, which abrogates the regulation of the kinase. There are several human diseases that fit into this category. Carney complex has been reported in families that either have a deletion in the PRKAR1A regulatory subunit (36) or a single point mutation in PRKACA at the 206 position that converts a lysine to an arginine (L206R). This latter point mutation results in a lost interaction with the regulatory subunit and constitutive activation of the PRKACA (37–40). This is a germ line mutation and patients develop multiple neoplasias including: cardiac myxomas, cortisol-producing adrenocortical adenomas, and schwannomas. While there have been a few reports of liver adenomas in these patients, reports of fibrolamellar tumors are extremely rare (3). A second mechanism of transformation could result from loss of myristoylation of the amino terminal glycine. The myristic acid normally sits in an acyl pocket in the large subunit of the kinase. This too may be sufficient. However, if this was the case, one would expect cases of fibrolamellar that are the consequence of a mutation of that glycine. As of yet, these have not been reported. Third, transformation could be driven by loss of the first exon of PRKACA. Once again, there are many different mutations that could result in the loss of the first exon. There are many other genes to which PRKACA could have been fused. However, so far, only fusions to DNAJB1 have been reported in FLC. This suggests that there may be something very specific about the first exon of DNAJB1. Fourth, transformation may be the consequence of an effect of DNAJB1 on localization or reaction partners of the PRKACA. Normally PRKACA is part of a holoenzyme that is localized in the cells through interactions with specific A Kinase Associated Proteins (AKAPs) (41). These AKAPs help form a signaling complex that links protein kinase A with various downstream effectors. The DNAJB1 domain could link PRKACA to new sets of effectors. Fifth, the DNAJB1 domain could be affecting the dynamics or the specificity of the PRKACA. Thus, the possibility remains that FLC could be effected through another alteration, such as dysregulation of PRKACA, and not only as the DNAJB1-PRKACA chimera. Thus, until this is clearly resolved, there are two different strategies going forward. First, we could redefine FLC to be based not on its morphology (which as often proven to be difficult to resolve), but instead define FLC as the tumor driven by the DNAJB1-PRKACA chimera. Second, we continue with the current, albeit loose, definition of FLC but start referring to a DNAJB1-PRKACA-associated variant of FLC, in those tumors where the chimera has been detected.
Unresolved questions in FLC
Despite the rapid progress in the past three years, there are still many critical unresolved questions in FLC: First, the cell type is still unresolved. This is a non-trivial problem, since during certain stress conditions and carcinogenesis hepatocytes have been reported to dedifferentiate into a poorly differentiated state, or trans-differentiate into cholangioccytes (42). In one study, in which ascites from an FLC patient were grown in long-term culture in vitro, after subsequent xenografting into the ice the transcriptome had many similarities to biliary stem cells (43).
A second critical unresolved question is whether there is another factor contributing to FLC. The experimental evidence shows an absolute correlation between FLC and the expression of the DNABJ1-PRKACA chimera. This suggests that the chimera is necessary, but does not mean that it is sufficient. It does not mean that every patient who has the chimera form in the liver will get the tumor. It is possible that all patients have a “second hit”, an additional mutation that is required for the effects of the chimera. This second hit may vary between patients. Alternatively, there may some predisposition in the population. It is possible that there is a polymorphism in the population that is considered innocuous and perhaps insignificant, but in conjunction with the chimera is required to develop the tumor. Amongst the population with whole-genome sequencing, there were several mutations that were found in more than one patient, with the most frequently mutated genes being MUC4 in 4 patients; GOLGA6L2, DSPP, FOXO6, HLA DRB1, and PCSK5 in 3; and FER1L6, CPS1, NEFH, TENM4 and the lincRNA FAM186A, in 2 patients (27). Interestingly, activation of MUC4 can contribute to ERBB2 signaling. Additionally, a duplication in chromosome 22 encompassing the genes USP41 and FAM230A was found in 7 patients, however the transcriptome did not show differential expression of these genes (27). Analysis of a different cohort of patients (n=78) using a combination of array and next generation sequencing technologies, showed low copy number variations compared with HCC and CCA, with focal alterations in LPHN1 (31%), and the tumor suppressor genes STK11 (31%) and CDKN1C (3%). Furthermore, a mutation in BRCA2 was identified in 4.2% of patients (33). Two studies identified additional fusion genes. The first, resulting from a translocation between the CLPTM1L and GLIS3 genes in a single patient (44). The second, identified a fusion between GOLT1A-KISS1 in 14/26 tumors, but also in 2/9 normal liver tissues (45). However, neither of these reported fusions have been replicated in additional studies. An alternative mechanism for a “second hit” may involve noncoding RNAs. LincRNAs AF064858.6, LINC00313, and RP11-157N3.1, were found to be upregulated in FLC relative to HCC and normal liver (Dinh et al., 2017). In a series of 41 patients, 5 of the 71 most commonly deleted miRs target IGF2BP1. The expression of IGF2BP1 was increased nearly 100-fold in primary tumors compared with normal liver. The same study found amplifications (ANKRD11, IFR2BP2, DUSP1, TXNIP, and MCL1) and deletions (FANK1, PRODH, SLC25A1, MIF, EP300, GRAMD4, WNT7B and hsa-mir-33a) in genes affecting the p53 pathway to be common to FLC tumors (45) Another possibility is that the transformation is the result of the loss of genes, similar to the loss of the tumor suppressor Rb. The formation of the chimeric gene is the result of a deletion of ~400kB which eliminates eight coding genes including: ASF1B, ADGRL1, RN7SL231P, ADGRES, DDX39A, PTGER1, GIPC1 and PKN1. Furthermore, 4 putative genes are deleted: ENSG00000160251, ENSG00000224543, ENSG00000266913, ENSG00000267474. There are two reasons to suspect that these do not play a crucial role in tumorigenesis. First, the level of these transcripts is not altered in the FLC tumor relative to the adjacent normal tissue (10). Second, there were no discernable differences in mouse liver tumors that result from using CRISPR/Cas9 to delete the 400 kB region or tumors that result from expressing the chimeric transcript in trans from the sleeping beauty transposon (34).
A third unresolved question is the nature of the age dependence in the disease. Why is it rarely seen in patients younger than ten or older than 40. FLC has been considered a slow growing tumor, is it not seen in younger patients because it takes ten years to develop (46)? Why is it not seen in older patients? If this is the consequence of two breaks and then an appropriate fusion, it would be expected to increase with aging, not decrease. Are there hormones that are lost with aging that are required for tumorigenesis? Alternatively, does it reflect the time course of the loss of the ability of the liver to regenerate, perhaps reflection on the effects of aging on EGF receptor activation(47); or aging on growth hormone induced activation of the pathways involved in hepatocyte proliferation (48). One possible explanation is the formation of the chimera in older livers is either totally innocuous, or so lethal that a tumor is never seen.
A fourth unresolved question is why are tumors driven by the DNAJB1-PRKACA chimera not found in other organs. Thus far the DNAJB1-PRKACA fusion appears to be specific to FLC and has not been demonstrated in any other malignant or non-malignant disease conditions (29) (10, 30). Is there something particular about the way PKA or DNAJB1 is activated in the liver? If the chimera is formed in other organs, do the cells die, or do they grow normally. Alternatively, there could be something about the packing of the DNA in the chromatin in the liver that brings these two domains in cross-proximity, favoring the formation of the chimera in case of breaks. This proximity may be lost with aging, and the loss of the ability to regenerate, contributing to the age dependence of FLC.
A fifth unresolved question is what causes the deletion leading to the chimera. There are no known risk factors associated with FLC, apart from a slight female predominance in some series. One study suggested toxic exposure as a possible cause based on the increased expression of Aryl hydrocarbon receptor in an FLC transplantable cell line (43). It is possible that larger case series and further research may shed light on this point.
A sixth unresolved question is whether the metastases differ from the primary tumor. One possibility is that they are identical, and that the potential for metastasis is encoded in the primary tumor. In this case, an additional genomic change would not be required for metastasis (49). In many primary tumors, given enough time, a small cluster of cells break away and, eventually, successfully colonize another location. An alternative possibility is that there is a discrete difference in the transcriptome, proteome or epigenome between the primary and metastatic tumors. For some cancers differences have been observed in specific non-coding RNA such as micro-RNA or long-non-coding RNA (lncRNA) (50, 51). In melanoma for example, altering the expression of the microRNA that regulates ApoE affected the metastasis (52). Some changes in the non-coding RNA have been reported for FLC (30). However, these are correlations. It remains to be demonstrated that they are causally related to the FLC phenotype. Some epigenomic changes have been characterized in fibrolamellar. These usually take the form of alterations in the methylation of DNA or the histones that affect the expression of genes. Some tumors, such as HCC, demonstrate a global hypomethylation associated with loss of differentiation. There have been some reports that there is no decrease in methylation in FLC (53). However, a slightly lower methylation in FLC has also been reported by others with a third of these tumors hypo-methylated and more aggressive as judged by size, the presence of micro-satellites, vascular invasion, number of tumors and overall survival (32). Other tumors show what is called “methylator phenotype” an increased methylation on CpG rich domains (54). An examination of the methylation status of tumor suppressor gene promoters in FLC compared with HCC and paired non-tumor liver of each did not detect a methylator phenotype (55). However, other studies found an increased methylation in the promoter for RASSF1 (Ras association domain-containing protein 1) in primary and metastatic FLC and not detected in the paired tissue. This increase was also seen in all known primary liver tumors with hepatocyte differentiation, including: hepatoblastoma, hepatic adenoma and HCC (55). The protein is associated with cell cycle control by inhibiting the accumulation of Cyclin-D1 (56). Similar to HCC, increased methylation was also found in Cyclin-D2, RASSF1A, has-mir-9-1 and has-mir-9-2. Finally, two tumor suppressor genes (DLUE7 and ZNF709) and eight putative tumor suppressor genes showed increased methylation (32).
A seventh unresolved question is why doesn’t the immune system attack FLC? On the one hand FLC is at the extreme low end of tumor mutational load. While on the other fusion proteins generate neo-antigens and FLC should therefore have a single clear neo-antigen. There have been a few dozen anecdotal uses of anti-PD1 and anti-PDL1 therapy with FLC with no reported successes. In the case of FLC it remains to be seen what can be done to trigger an effective immune attack.
Finally, there is the question of whether there is any consequence to a second DNAJB1-PRKACA chimera that is formed. Some of the patients have a break between the first intron of DNAJB1 and first intron of PRKACA and these tumors only produce a chimeric transcript of the 1st exon of DNAJB1 and the 2nd to 10th exons of PRKACA. A subgroup of patients have one break in the middle of the second exon of DNAJB1 and their second break in the first intron of PRKACA. These patients predominantly make a chimeric transcript that is like all the other FLC tumors with the 1st exon of DNAJB1 and the 2nd to 10th exons of PRKACA. However, they also make a smaller amount of a second chimera with the 1st exon of DNAJB1, part of the second exon of DNAJB1 and the 2nd to 10th exons of PRKACA. This minority chimeric transcript also produces a chimeric protein. So far no features have been found that distinguish these two tumors. Thus, it is not known if the minority chimera is of significance.
Therapeutic directions for Fibrolamellar Carcinoma
Of greatest consequence to the patients are new therapeutics on the horizon. The recently published work suggests at least two new therapeutic directions: Inhibition of the DNAJB1-PRKACA chimera and targeting the pathways that are consistently altered in FLC.
Targeting the chimera
There are a number of advantages to this approach, one of which is that the DNAJB1-PRKACA kinase is specific to FLC and has not been found in any normal tissue; this would potentially minimize side-effects. There are a few different strategies for targeting the chimera. One approach is to target the kinase pocket. An industry has built up around designing inhibitors for kinases, however the kinase pocket is very similar between the DNAJB1-PRKACA and the native PRKACA. The fusion junction is on the side of the protein opposite the kinase pocket so it may not be possible to design a selective blocker. Even a small inhibition of the native PRKACA could be lethal as a consequence of effects on lymphocytes (57), platelet signaling (58), the heart (59) or the brain (60). It is possible that an inhibitor that blocks both the native and the chimeric protein kinase A may have a sufficient therapeutic window if the level of activity of PKA is high enough in the tumor cells and if it is dependent on the chimera. However, it would still be challenging to design an inhibitor that is specific for PKA over other kinases. A second approach is to find an allosteric inhibitor. In the chimeric kinase Bcr-Abl, responsible for chronic myeloma leukemia, an inhibitor into an acyl pocket, normally occupied by a myristoyl group, can allosterically block activity (61). In FLC, the amino terminus is not myristoylated, leaving the acyl pocket accessible to inhibitors.
Targeting the activated pathways
Many years of characterization of change in the FLC proteome (3, 10) and transcriptome (10, 62), have helped identify a number of oncogenic proteins or pathways that are activated in FLC. A critical issue is to determine which of these are merely correlated with FLC and which are driving the tumor. Aromatase, which is considered oncogenic in breast cancer (63), is increased at the transcript level 62-fold in FLC (10). However, a recent clinical trial failed to show a therapeutic effect on FLC (64). Aurora Kinase A, which is oncogenic in breast cancers, bladder cancer, colorectal cancer, neuroblastoma, and hepatocellular carcinoma (65–68) is increased in the transcriptome 4-fold in FLC and it is the subject of an on-going clinical trial (NCT02234986). There are a number of other oncogenic pathways that are upregulated in FLC including the EGF receptor pathway for which both the ligands (EGF, EREG, AREG, NRG2), and the receptors EGFR and ErbB2 are significantly increased (10). Many inhibitors have been developed and approved for members of this pathway. Other suspects of interest include the Wnt, PAK21 and the mTOR pathway. There are numerous members of the Wnt pathway significantly increased in FLC including DKK4 (log2=6.52, FZD10 (log2=6.13) wnt16 (log2=4.4) (10). Alterations in β-catenin (CTNNB1), the key effector in the Wnt pathway, have been found in a subtype of hepatic adenomas, HCC and hepatoblastoma. However, several series did not find similar alterations in FLC (69). In HCC there have been conflicting reports regarding the effect of CTNNB1 mutations on tumor phenotype. In one study HCCs with missense mutations in exon-3 of CTNNB1 displayed a more aggressive histological phenotype, including increased tumor size and vascular invasion, compared to those without CTNNB1 mutations. FLC tumors (n=5) in this series did not have CTNNB1 mutations, but showed elevated levels of total beta-catenin, similar to HCC, compared to controls. (70); and higher levels of Y654 phosphorylation of beta-catenin (Tyrosine kinase) compared with HCC. This was associated with elevated levels of Cyclin–D1 (71). Interestingly EGF and HGF lead to Y654 phosphorylation of beta-catenin, and EGFR in turn is regulated by PKA. Expression of the CTNNB1 mutation in mouse liver in conjugation with expression of the DNAJB1-PRKACA chimera increases the penetrance of the tumor, the size of the tumor and accelerates demise of the animal (34).
The mTOR pathway integrates nutritional signals, and is a central regulator of cellular homeostasis. It has been implicated in HCC, where rapamycin inhibitors have shown anti-tumor activity. In FLC in phosphorylation of S6, the downstream target of mTORC1, coupled with low levels of p-Akt, suggested primary activation of mTORC1 (72). However, a clinical trial using the mTORC1 inhibitor Everolimus failed to show therapeutic effect in FLC (NCT01642186).
As altered PKA activity is expected to alter the transcriptome and proteome of cells, it becomes critical to resolve which of these pathways are important for tumorigenesis and fitness of FLC and which are epiphenomenon of the overactivation of protein kinase A. An effective model system for distinguishing between these would help accelerate bringing therapeutics to the patients.
Summary
The application of next generation sequencing has led to a dramatic advancement in our understanding of FLC. FLC has immerged as a distinct molecular entity driven by a novel DNAJB1-PRKACA fusion protein, with a unique and relatively homogeneous transcriptome and proteome. The fusion protein now serves as the main focus of drug discovery efforts, while the differentially regulated oncogenic pathways may serve as a framework for research aimed at drug repurposing. As inhibitors of many of these targets/pathways already exist and either in development or already approved this may facilitate a more rapid path to an effective therapeutic.
Contributor Information
Gadi Lalazar, The Laboratory for Cellular Biophysics, The Rockefeller University.
Sanford Simon, The Laboratory for Cellular Biophysics, The Rockefeller University.
References
- 1.El-Serag HB, Davila JA. Is fibrolamellar carcinoma different from hepatocellular carcinoma? A US population-based study. Hepatology. 2004;39(3):798–803. doi: 10.1002/hep.20096. [DOI] [PubMed] [Google Scholar]
- 2.Eggert T, McGlynn KA, Duffy A, Manns MP, Greten TF, Altekruse SF. Epidemiology of fibrolamellar hepatocellular carcinoma in the USA, 2000–10. Gut. 2013;62(11):1667–8. doi: 10.1136/gutjnl-2013-305164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Torbenson M. Fibrolamellar carcinoma: 2012 update. Scientifica. 2012;2012:743790. doi: 10.6064/2012/743790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mavros MN, Mayo SC, Hyder O, Pawlik TM. A systematic review: treatment and prognosis of patients with fibrolamellar hepatocellular carcinoma. Journal of the American College of Surgeons. 2012;215(6):820–30. doi: 10.1016/j.jamcollsurg.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 5.Edmondson HA. Differential diagnosis of tumors and tumor-like lesions of liver in infancy and childhood. AMA J Dis Child. 1956;91(2):168–86. doi: 10.1001/archpedi.1956.02060020170015. [DOI] [PubMed] [Google Scholar]
- 6.Craig JR, Peters RL, Edmondson HA, Omata M. Fibrolamellar carcinoma of the liver: a tumor of adolescents and young adults with distinctive clinico-pathologic features. Cancer. 1980;46(2):372–9. doi: 10.1002/1097-0142(19800715)46:2<372::aid-cncr2820460227>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 7.Malouf G, Falissard B, Azoulay D, Callea F, Ferrell LD, Goodman ZD, et al. Is histological diagnosis of primary liver carcinomas with fibrous stroma reproducible among experts? Journal of clinical pathology. 2009;62(6):519–24. doi: 10.1136/jcp.2008.062620. [DOI] [PubMed] [Google Scholar]
- 8.Abdul-Al HM, Wang G, Makhlouf HR, Goodman ZD. Fibrolamellar hepatocellular carcinoma: an immunohistochemical comparison with conventional hepatocellular carcinoma. Int J Surg Pathol. 2010;18(5):313–8. doi: 10.1177/1066896910364229. [DOI] [PubMed] [Google Scholar]
- 9.Ross HM, Daniel HDJ, Vivekanandan P, Kannangai R, Yeh MM, Wu T-T, et al. Fibrolamellar carcinomas are positive for CD68. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2011;24(3):390–5. doi: 10.1038/modpathol.2010.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simon EP, Freije CA, Farber BA, Lalazar G, Darcy DG, Honeyman JN, et al. Transcriptomic characterization of fibrolamellar hepatocellular carcinoma. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(44):E5916–25. doi: 10.1073/pnas.1424894112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamashita S, Vauthey JN, Kaseb AO, Aloia TA, Conrad C, Hassan MM, et al. Prognosis of Fibrolamellar Carcinoma Compared to Non-cirrhotic Conventional Hepatocellular Carcinoma. J Gastrointest Surg. 2016;20(10):1725–31. doi: 10.1007/s11605-016-3216-x. [DOI] [PubMed] [Google Scholar]
- 12.Fonseca GM, Varella AD, Coelho FF, Abe ES, Dumarco RB, Herman P. Downstaging and resection after neoadjuvant therapy for fibrolamellar hepatocellular carcinoma. World J Gastrointest Surg. 2014;6(6):107–11. doi: 10.4240/wjgs.v6.i6.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fakih M. A case of fibrolamellar cancer with a palliative response and minor radiographic regression with erlotinib and bevacizumab combination therapy. Am J Ther. 2014;21(6):e207–10. doi: 10.1097/MJT.0b013e3182840fa6. [DOI] [PubMed] [Google Scholar]
- 14.Gras P, Truant S, Boige V, Ladrat L, Rougier P, Pruvot F-R, et al. Prolonged Complete Response after GEMOX Chemotherapy in a Patient with Advanced Fibrolamellar Hepatocellular Carcinoma. Case Rep Oncol. 2012;5(1):169–72. doi: 10.1159/000338242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ang CS, Kelley RK, Choti MA, Cosgrove DP, Chou JF, Klimstra D, et al. Clinicopathologic Characteristics and Survival Outcomes of Patients With Fibrolamellar Carcinoma: Data From the Fibrolamellar Carcinoma Consortium. Gastrointestinal Cancer Research: GCR. 2013;6(1):3–9. [PMC free article] [PubMed] [Google Scholar]
- 16.Moreno-Luna LE, Arrieta O, Garcia-Leiva J, Martinez B, Torre A, Uribe M, et al. Clinical and pathologic factors associated with survival in young adult patients with fibrolamellar hepatocarcinoma. BMC Cancer. 2005;5:142. doi: 10.1186/1471-2407-5-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eggert T, McGlynn K, Greten TF, Altekruse S. Response to fibrolamellar hepatocellular carcinoma versus conventional hepatocellular carcinoma: better 5-year survival or artefactual result of research methodology? Gut. 2013 doi: 10.1136/gutjnl-2013-306407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mayo SC, Mavros MN, Nathan H, Cosgrove D, Herman JM, Kamel I, et al. Treatment and prognosis of patients with fibrolamellar hepatocellular carcinoma: a national perspective. Journal of the American College of Surgeons. 2014;218(2):196–205. doi: 10.1016/j.jamcollsurg.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stipa F, Yoon SS, Liau KH, Fong Y, Jarnagin WR, D’Angelica M, et al. Outcome of patients with fibrolamellar hepatocellular carcinoma. Cancer. 2006;106(6):1331–8. doi: 10.1002/cncr.21703. [DOI] [PubMed] [Google Scholar]
- 20.Hemming AW, Langer B, Sheiner P, Greig PD, Taylor BR. Aggressive surgical management of fibrolamellar hepatocellular carcinoma. J Gastrointest Surg. 1997;1(4):342–6. doi: 10.1016/s1091-255x(97)80055-8. [DOI] [PubMed] [Google Scholar]
- 21.Atienza LG, Berger J, Mei X, Shah MB, Daily MF, Grigorian A, et al. Liver transplantation for fibrolamellar hepatocellular carcinoma: A national perspective. J Surg Oncol. 2017;115(3):319–23. doi: 10.1002/jso.24515. [DOI] [PubMed] [Google Scholar]
- 22.Njei B, Konjeti VR, Ditah I. Prognosis of Patients With Fibrolamellar Hepatocellular Carcinoma Versus Conventional Hepatocellular Carcinoma: A Systematic Review and Meta-analysis. Gastrointestinal Cancer Research: GCR. 2014;7(2):49–54. [PMC free article] [PubMed] [Google Scholar]
- 23.Ward S, Waxman S. Fibrolamellar Carcinoma: A Review with Focus on Genetics and Comparison to Other Malignant Primary Liver Tumors. Semin Liver Dis. 2011;31(01):061–70. doi: 10.1055/s-0031-1272835. [DOI] [PubMed] [Google Scholar]
- 24.Lowichik A, Schneider NR, Tonk V, Ansari MQ, Timmons CF. Report of a complex karyotype in recurrent metastatic fibrolamellar hepatocellular carcinoma and a review of hepatocellular carcinoma cytogenetics. Cancer genetics and cytogenetics. 1996 Jun 01;1996(2):170–4. doi: 10.1016/0165-4608(95)00314-2. [DOI] [PubMed] [Google Scholar]
- 25.Wilkens L, Bredt M, Flemming P, Kubicka S, Klempnauer J, Kreipe H. Cytogenetic aberrations in primary and recurrent fibrolamellar hepatocellular carcinoma detected by comparative genomic hybridization. Anatomic Pathology. 2000;114(6):867–74. doi: 10.1309/BMTT-JBPD-D13H-1UVD. [DOI] [PubMed] [Google Scholar]
- 26.Kakar S, Chen X, Ho C, Burgart LJ, Sahai V, Dachrut S, et al. Chromosomal changes in fibrolamellar hepatocellular carcinoma detected by array comparative genomic hybridization. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2008;22(1):134–41. doi: 10.1038/modpathol.2008.178. [DOI] [PubMed] [Google Scholar]
- 27.Darcy DG, Chiaroni-Clarke R, Murphy JM, Honeyman JN, Bhanot U, LaQuaglia MP, et al. The genomic landscape of fibrolamellar hepatocellular carcinoma: whole genome sequencing of ten patients. Oncotarget. 2015;6(2):755–70. doi: 10.18632/oncotarget.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Honeyman JN, Simon EP, Robine N, Chiaroni-Clarke R, Darcy DG, Lim II, et al. Detection of a recurrent DNAJB1-PRKACA chimeric transcript in fibrolamellar hepatocellular carcinoma. Science. 2014;343(6174):1010–4. doi: 10.1126/science.1249484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Graham RP, Jin L, Knutson DL, Kloft-Nelson SM, Greipp PT, Waldburger N, et al. DNAJB1-PRKACA is specific for fibrolamellar carcinoma. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2015;28(6):822–9. doi: 10.1038/modpathol.2015.4. [DOI] [PubMed] [Google Scholar]
- 30.Dinh TA, Vitucci EC, Wauthier E, Graham RP, Pitman WA, Oikawa T, et al. Comprehensive analysis of The Cancer Genome Atlas reveals a unique gene and non-coding RNA signature of fibrolamellar carcinoma. Scientific reports. 2017;7:44653. doi: 10.1038/srep44653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu L, Hazard FK, Zmoos A-F, Jahchan N, Chaib H, Garfin PM, et al. Genomic analysis of fibrolamellar hepatocellular carcinoma. Human molecular genetics. 2015 doi: 10.1093/hmg/ddu418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malouf GG, Tahara T, Paradis V, Fabre M, Guettier C, Yamazaki J, et al. Methylome sequencing for fibrolamellar hepatocellular carcinoma depicts distinctive features. Epigenetics. 2015;10(9):872–81. doi: 10.1080/15592294.2015.1076955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cornella H, Alsinet C, Sayols S, Zhang Z, Hao K, Cabellos L, et al. Unique genomic profile of fibrolamellar hepatocellular carcinoma. Gastroenterology. 2015;148(4):806–18 e10. doi: 10.1053/j.gastro.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kastenhuber ER, Lalazar G, Tschaharganeh DF, Houlihan SL, Baslan T, Chen C, et al. DNAJB1-PRKACA fusion kinase drives tumorigenesis and interacts with β-catenin and the liver regenerative response. BioRxiv. 2017 doi: 10.1073/pnas.1716483114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Engelholm LH, Riaz A, Serra D, Dagnaes-Hansen F, Johansen JV, Santoni-Rugiu E, et al. CRISPR/Cas9 Engineering of Adult Mouse Liver Demonstrates That the Dnajb1-Prkaca Gene Fusion is Sufficient to Induce Tumors Resembling Fibrolamellar Hepatocellular Carcinoma. Gastroenterology. 2017 doi: 10.1053/j.gastro.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kirschner LS, Carney JA, Pack SD, Taymans SE, Giatzakis C, Cho YS, et al. Mutations of the gene encoding the protein kinase A type I-alpha regulatory subunit in patients with the Carney complex. Nature genetics. 2000;26(1):89–92. doi: 10.1038/79238. [DOI] [PubMed] [Google Scholar]
- 37.Beuschlein F, Fassnacht M, Assié G, Calebiro D, Stratakis CA, Osswald A, et al. Constitutive activation of PKA catalytic subunit in adrenal Cushing’s syndrome. The New England journal of medicine. 2014;370(11):1019–28. doi: 10.1056/NEJMoa1310359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cao Y, He M, Gao Z, Peng Y, Li Y, Li L, et al. Activating hotspot L205R mutation in PRKACA and adrenal Cushing's syndrome. Science (New York, NY) 2014;344(6186):913–7. doi: 10.1126/science.1249480. [DOI] [PubMed] [Google Scholar]
- 39.Di Dalmazi G, Kisker C, Calebiro D, Mannelli M, Canu L, Arnaldi G, et al. Novel Somatic Mutations in the Catalytic Subunit of the Protein Kinase A as a Cause of Adrenal Cushing’s Syndrome: A European Multicentric Study. The Journal of clinical endocrinology and metabolism. 2014;99(10):E2093–100. doi: 10.1210/jc.2014-2152. [DOI] [PubMed] [Google Scholar]
- 40.Sato Y, Maekawa S, Ishii R, Sanada M, Morikawa T, Shiraishi Y, et al. Recurrent somatic mutations underlie corticotropin-independent Cushing’s syndrome. Science. 2014;344(6186):917–20. doi: 10.1126/science.1252328. [DOI] [PubMed] [Google Scholar]
- 41.Scott JD, Dessauer CW, Tasken K. Creating order from chaos: cellular regulation by kinase anchoring. Annu Rev Pharmacol Toxicol. 2013;53:187–210. doi: 10.1146/annurev-pharmtox-011112-140204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sia D, Villanueva A, Friedman SL, Llovet JM. Liver Cancer Cell of Origin, Molecular Class, and Effects on Patient Prognosis. Gastroenterology. 2017;152(4):745–61. doi: 10.1053/j.gastro.2016.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oikawa T, Wauthier E, Dinh TA, Selitsky SR, Reyna-Neyra A, Carpino G, et al. Model of fibrolamellar hepatocellular carcinomas reveals striking enrichment in cancer stem cells. Nature communications. 2015;6:8070. doi: 10.1038/ncomms9070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu L, Hazard FK, Zmoos AF, Jahchan N, Chaib H, Garfin PM, et al. Genomic analysis of fibrolamellar hepatocellular carcinoma. Human molecular genetics. 2015;24(1):50–63. doi: 10.1093/hmg/ddu418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sorenson EC, Khanin R, Bamboat ZM, Cavnar MJ, Kim TS, Sadot E, et al. Genome and transcriptome profiling of fibrolamellar hepatocellular carcinoma demonstrates p53 and IGF2BP1 dysregulation. PloS one. 2017;12(5):e0176562. doi: 10.1371/journal.pone.0176562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pinna AD, Iwatsuki S, Lee RG, Todo S, Madariaga JR, Marsh JW, et al. Treatment of fibrolamellar hepatoma with subtotal hepatectomy or transplantation. Hepatology. 1997;26(4):877–83. doi: 10.1002/hep.510260412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ohtake Y, Maruko A, Ohishi N, Fukumoto M, Ohkubo Y. Effect of aging on EGF-induced proliferative response in primary cultured periportal and perivenous hepatocytes. J Hepatol. 2008;48(2):246–54. doi: 10.1016/j.jhep.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 48.Schmucker DL, Sanchez H. Liver regeneration and aging: a current perspective. Curr Gerontol Geriatr Res. 2011;2011:526379. doi: 10.1155/2011/526379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nature genetics. 2003;33(1):49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- 50.Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451(7175):147–52. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pencheva N, Tavazoie SF. Control of metastatic progression by microRNA regulatory networks. Nature cell biology. 2013;15(6):546–54. doi: 10.1038/ncb2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pencheva N, Tran H, Buss C, Huh D, Drobnjak M, Busam K, et al. Convergent multi-miRNA targeting of ApoE drives LRP1/LRP8-dependent melanoma metastasis and angiogenesis. Cell. 2012;151(5):1068–82. doi: 10.1016/j.cell.2012.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tränkenschuh W, Puls F, Christgen M, Albat C, Heim A, Poczkaj J, et al. Frequent and distinct aberrations of DNA methylation patterns in fibrolamellar carcinoma of the liver. PloS one. 2010;5(10):e13688. doi: 10.1371/journal.pone.0013688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Issa JP. CpG island methylator phenotype in cancer. Nature reviews Cancer. 2004;4(12):988–93. doi: 10.1038/nrc1507. [DOI] [PubMed] [Google Scholar]
- 55.Vivekanandan P, Torbenson M. Epigenetic instability is rare in fibrolamellar carcinomas but common in viral-associated hepatocellular carcinomas. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2008;21(6):670–5. doi: 10.1038/modpathol.2008.32. [DOI] [PubMed] [Google Scholar]
- 56.Agathanggelou A, Cooper WN, Latif F. Role of the Ras-association domain family 1 tumor suppressor gene in human cancers. Cancer research. 2005;65(9):3497–508. doi: 10.1158/0008-5472.CAN-04-4088. [DOI] [PubMed] [Google Scholar]
- 57.Lang P, Gesbert F, Delespine-Carmagnat M, Stancou R, Pouchelet M, Bertoglio J. Protein kinase A phosphorylation of RhoA mediates the morphological and functional effects of cyclic AMP in cytotoxic lymphocytes. The EMBO journal. 1996;15(3):510–9. [PMC free article] [PubMed] [Google Scholar]
- 58.Graves LM, Bornfeldt KE, Raines EW, Potts BC, Macdonald SG, Ross R, et al. Protein kinase A antagonizes platelet-derived growth factor-induced signaling by mitogen-activated protein kinase in human arterial smooth muscle cells. Proceedings of the National Academy of Sciences of the United States of America. 1993;90(21):10300–4. doi: 10.1073/pnas.90.21.10300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peters KA, Demaille JG, Fischer EH. Adenosine 3′:5′-monophosphate dependent protein kinase from bovine heart. Characterization of the catalytic subunit. Biochemistry. 1977;16(26):5691–7. doi: 10.1021/bi00645a007. [DOI] [PubMed] [Google Scholar]
- 60.Takai Y, Kishimoto A, Inoue M, Nishizuka Y. Studies on a cyclic nucleotide-independent protein kinase and its proenzyme in mammalian tissues. I. Purification and characterization of an active enzyme from bovine cerebellum. The Journal of biological chemistry. 1977;252(21):7603–9. [PubMed] [Google Scholar]
- 61.Adrian FJ, Ding Q, Sim T, Velentza A, Sloan C, Liu Y, et al. Allosteric inhibitors of Bcr-abl-dependent cell proliferation. Nature chemical biology. 2006;2(2):95–102. doi: 10.1038/nchembio760. [DOI] [PubMed] [Google Scholar]
- 62.Malouf GG, Job S, Paradis V, Fabre M, Brugières L, Saintigny P, et al. Transcriptional profiling of pure fibrolamellar hepatocellular carcinoma reveals an endocrine signature. Hepatology. 2014 doi: 10.1002/hep.27018. [DOI] [PubMed] [Google Scholar]
- 63.Johnston SRD, Dowsett M. Aromatase inhibitors for breast cancer: lessons from the laboratory. Nature reviews Cancer. 2003;3(11):821–31. doi: 10.1038/nrc1211. [DOI] [PubMed] [Google Scholar]
- 64.Abou-Alfa G, Mayer RJ, Cosgrove D, Capanu M, Choti MA, Atreya CE, et al. Randomized phase II study of everolimus (E), leuprolide + letrozole (LL), and E + LL (ELL) in patients (pts) with unresectable fibrolamellar carcinoma (FLC) J Clin Oncol. 2015;33:e15149. [Google Scholar]
- 65.Sen S, Zhou H, Zhang R-D, Yoon DS, Vakar-Lopez F, Ito S, et al. Amplification/overexpression of a mitotic kinase gene in human bladder cancer. Journal of the National Cancer Institute. 2002;94(17):1320–9. doi: 10.1093/jnci/94.17.1320. [DOI] [PubMed] [Google Scholar]
- 66.Crane R, Gadea B, Littlepage L, Wu H, Ruderman JV. Aurora A, meiosis and mitosis. Biology of the cell/under the auspices of the European Cell Biology Organization. 2004;96(3):215–29. doi: 10.1016/j.biolcel.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 67.Bischoff JR, Anderson L, Zhu Y, Mossie K, Ng L, Souza B, et al. A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. The EMBO journal. 1998;17(11):3052–65. doi: 10.1093/emboj/17.11.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jeng Y-M, Peng S-Y, Lin C-Y, Hsu H-C. Overexpression and amplification of Aurora-A in hepatocellular carcinoma. Clinical cancer research: an official journal of the American Association for Cancer Research. 2004;10(6):2065–71. doi: 10.1158/1078-0432.ccr-1057-03. [DOI] [PubMed] [Google Scholar]
- 69.Terris B, Pineau P, Bregeaud L, Valla D, Belghiti J, Tiollais P, et al. Close correlation between beta-catenin gene alterations and nuclear accumulation of the protein in human hepatocellular carcinomas. Oncogene. 1999;18(47):6583–8. doi: 10.1038/sj.onc.1203051. [DOI] [PubMed] [Google Scholar]
- 70.Cieply B, Zeng G, Proverbs-Singh T, Geller DA, Monga SPS. Unique phenotype of hepatocellular cancers with exon-3 mutations in beta-catenin gene. Hepatology. 2009;49(3):821–31. doi: 10.1002/hep.22695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303(5663):1483–7. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Riehle KJ, Yeh MM, Yu JJ, Kenerson HL, Harris WP, Park JO, et al. mTORC1 and FGFR1 signaling in fibrolamellar hepatocellular carcinoma. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2015;28(1):103–10. doi: 10.1038/modpathol.2014.78. [DOI] [PubMed] [Google Scholar]