Abstract
NMR approaches using nucleotide-specific deuterium labeling schemes have enabled structural studies of biologically relevant RNAs of increasing size and complexity. Although local structure is well-determined using these methods, definition of global structural features, including relative orientations of independent helices, remains a challenge. Residual dipolar couplings, a potential source of orientation information, have not been obtainable for large RNAs due to poor sensitivity resulting from rapid heteronuclear signal decay. Here we report a novel multiple quantum NMR method for RDC determination that employs flip angle variation rather than a coupling evolution period. The accuracy of the method and its utility for establishing inter-helical orientations are demonstrated for a 36-nucleotide RNA, for which comparative data could be obtained. Applied to a 78 kDa Rev response element from the HIV-1 virus, which has an effective rotational correlation time of ca. 160 ns, the method yields sensitivity gains of an order of magnitude or greater over existing approaches. Solution-state access to structural organization in RNAs of at least 230 nucleotides is now possible.
Keywords: NMR spectroscopy, RNA, Residual Dipolar Couplings, VF-HMQC, Structure elucidation
Graphical abstract
Authors are required to submit a graphic entry for the Table of Contents (TOC) that, in conjunction with the manuscript title, should give the reader a representative idea of one of the following: A key structure, reaction, equation, concept, or theorem, etc., that is discussed in the manuscript. Consult the journal’s Instructions for Authors for TOC graphic specifications.
INTRODUCTION
RNAs participate in a diverse and growing number of known biological functions.3 Like proteins, RNA function is dependent on structure, both of which can be modulated by effectors such as metabolites, proteins and ions. However, compared to proteins, knowledge about RNA structure and the determinants of folding remain limited. Approximately 4,500 RNA structures have been deposited in the Nucleic Acid Database (NDB), compared to more than 135,000 protein depositions in the Protein Databank (PDB). In part, this relative paucity can be explained by difficulties in applying common biophysical techniques to RNA. Conformational heterogeneity and flexibility can cause crystallization challenges and complicate analysis by electron microscopy. NMR approaches for medium-to-large RNAs are challenging due to low chemical shift dispersion, low proton density, lack of NOEs between secondary structure elements, and large 13C-1H dipolar coupling that severely limits the sensitivity of heteronuclear correlation experiments.5–7
Recent advances in deuterium labeling have provided routes to overcome several of the NMR challenges.9–12 Nucleotide-specific 2Hediting can alleviate spectral crowding and decrease linewidths, allowing chemical shift assignment and determination of inter-proton distances using nuclear Overhauser effect (NOE) spectroscopy. Although NOEs provide high resolution local structural information, they cannot define relative orientations of distinct secondary structure elements, such as distinct helices, except in the rare cases where long-range NOEs can be observed (e.g., at sites of long-range A-minor contacts15). An approach that works well for proteins and has been applied to relatively small RNAs involves NMR detection of residual dipolar couplings (RDCs).9, 15–24 In solution, dipole-dipole interactions between nuclei average to zero due to rapid molecular reorientation. However, upon introduction of a medium that causes a small degree of solute alignment, the dipole-dipole interaction is no longer fully averaged. The sign and magnitude of the resulting RDC are dependent on the time-averaged angle between the internuclear vector and the external field. Given sufficient RDCs, the alignment tensor and the relative orientation of each inter-nuclear vector can be calculated,26–28 thereby providing long-range orientation restraints. Larger RNAs present unique challenges for RDC measurement due to severe 1H NMR line broadening that occurs upon incorporation of 13C nuclei. As such, a recently developed method for measuring RDCs in RNAs from TROSY intensities becomes impractical for RNAs with rotational correlation times longer than 30 ns.29
We have developed an approach that exploits the relatively large chemical shift dispersion and slow relaxation rates of the adenosine H2 protons. Even in large RNAs, the H2 signals remain sharp,9–12 provided that the RNA is dissolved in D2O such that the uridine N3 position is deuterated. Under these conditions the H2 is relatively well isolated from sources of relaxation, with the closest proton ~5 Å away for Watson-Crick base pairs in regular A-helical geometry. Since isotopic enrichment with 13C leads to severe line broadening due to a strong dipolar interaction, our experiments are applied to RNAs that contain uniformly 15N-enriched adenosines. Adenosine-N1 and -N3 nuclei have a negligible effect on relaxation of the H2 proton due to their relatively low gyromagnetic ratio and inter-nuclear separation of ~2 Å. The two-bond H2-N1 and -N3 scalar couplings (~14.5 Hz30) are sufficient to allow recording of high quality 15N-1H SOFAST-HMQC spectra,13–14 which suggested to us that we may be able to utilize the H2-N1 and H2-N3 couplings in adenosines to measure RDCs in large RNAs.
RESULTS AND DISCUSSION
The residual dipolar coupling, DIS, between two spins, I and S, is given in Hz by:31
| (1) |
where the angular brackets denote time or ensemble averaging, μ0 is the vacuum permeability, ℏ is the reduced Planck constant, γ1 is the magnetogyric ratio of nucleus I, rIS is the inter-nuclear distance, and θ is the angle between the inter-nuclear vector and the external magnetic field. The relatively large-inter-nuclear distance (2.06 Å32) means that the dipolar interaction will be small compared to more commonly measured one-bond couplings. Using a commonly targeted 0.1% alignment, the expected range for adenosine 2DHN RDCs is ~5 Hz, although due to the relatively isolated H2, higher degrees of alignment may be possible without the usual complications due to 1H-1H RDCs.33 This will be limited by H2-H2 RDCs in stacked adenosines, which will have similar magnitude to the H2-N1 and H2-N3 couplings. Even though the small amount of 1H-1H dephasing cannot be refocused using band-selective pulses, its effect is the same for both reference and attenuated spectra and therefore does not impact the size of the extracted coupling. The relative angle of H2-N1 and H2-N3 inter-nuclear vectors is fixed by adenine’s geometry at ~72°,32 making them highly complementary.
We initially focused on a 232 nucleotide HIV-1 Rev response element RNA construct engineered to adopt one of two equilibrium conformations (RRE232A).34 Well-dispersed signals indicative of regular secondary structure were readily detected using the SOFAST-HMQC. However, attempts to apply an existing spin-state selective (S3E)25 experiment for measuring H2-N1/N3 couplings were unsuccessful (Figure 1C) due to rapid relaxation of transverse 15N magnetization during the time period necessary for separating antiphase components. The relatively small 2JHN coupling (~14.5 Hz) requires 15N magnetization to be transverse for approximately 17 ms during the S3E element.35 Experimentally we find N3 T1ρ values below 5 ms for RRE232A at 600 MHz and 308 K (SI Figure S1), corresponding to an effective rotational correlation time of ca. 160 ns, under the assumption that chemical shift anisotropy (CSA) is the dominant relaxation mechanism and using a CSA value of 330 ppm.36 We would expect to lose more than 95% of this signal during the S3E element, essentially rendering it undetectable for practical purposes.
Figure 1.
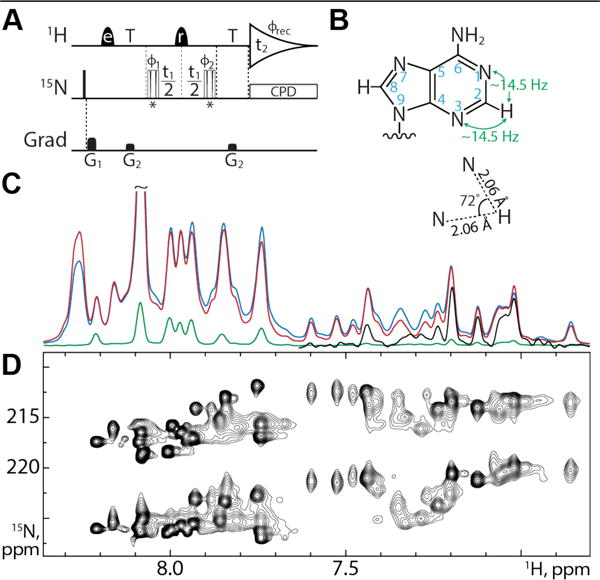
(A) VF-HMQC pulse sequence for measurement of 2DNH in adenine bases. Narrow bars represent 90° pulses, while half-ellipsoids denote shaped 1H pulses, e and r representing EBURP2 and ReBURP pulses respectively1. 15N pulses marked * are BIR-4 pulses2 designed for either 90° or 45° flip angles, applied in an interleaved manner. 1H shaped pulses have a duration of 2.8 ms (EBURP) and 3 ms (ReBURP) at 600 MHz and are centered at 7.4 ppm. 15N pulses are applied at 220 ppm. All pulses have phase x unless otherwise indicated. The delay T is set to 1/3JNH (22.73 ms). Phase cycling: ϕ1 = x,−x, ϕ2 = x,x,−x,−x, ϕrec = x,−x,−x,x. Gradient pulses G1,2 = 5.8, 3.75 G/cm with durations of 1 ms. All gradient pulses are smoothened rectangular shapes. Quadrature detection is achieved using the STATES-TPPI method with ϕ1 incremented by 90° for each FID4. Composite pulse decoupling is achieved using the GARP sequence8. (B) Adenine geometry. Magnetization is transferred via the two-bond coupling between H2 and N1/N3 as indicated. The relative angle between the H2-N1 and H2-N3 inter-nuclear vectors is 72°. (C) VF-HMQC with 90° flip (red), SOFAST-HMQC13–14 (blue) and S3E25 (green) experiments recorded on the 232 nucleotide RRE232A without 15N chemical shift evolution shows the increase in sensitivity of our approach. The upfield region of the S3E experiment is scaled up by 10 times in black. (D) VF-HMQC spectrum of RRE232A showing H2-N1 and H2-N3 correlations.
To address this issue we first considered a quantitative J style experiment.37 However, this is complicated by concurrent evolution of both H2-N1 and H2-N3 couplings during the dephasing / rephasing delays. Selective 15N pulses to determine the individual contribution of N1 and N3 are rendered impractical by their poor frequency separation (Figure 1D) and rapid 15N relaxation during the requisite long shaped pulses. It is possible to reduce the contribution of the passive coupling using a small heteronuclear flip angle.38–40 However, significant reduction requires flip angles as low as 20° (SI Figure S2A), leading to signal loss of nearly 90%.
The intensity of an HMQC signal obtained for a flip anpulses ϕ of the heteronuclear pulses, and de/rephasing delays of duration τ, for the case where the detected spin I is coupled to two heteronuclear spins S and T and assuming JST = 0 is given by:
| (2) |
with and c and s representing cosine and sine functions respectively. Following Fourier transform this yields a signal at (ΩS, ΩI) with intensity given by:
| (3) |
It is apparent that the variable flip angle modulates both the total intensity and the contribution of the passive spin coupling to the final signal. The contribution of the actively coupled spin can therefore be removed by taking the ratio of two experiments where the flip angle is varied (SI Figure S2). For nearly optimal flip angles of 45° and 90° (SI Figure S3), this ratio (RS) is given by:
| (4) |
The couplings can then be found from the intensity ratio using:
| (5) |
The analysis presented thus far assumes that the 15N pulses are perfectly calibrated and homogenous across the sample. In general, the B1 field is not homogenous. We therefore used a pair of BIR-4 adiabatic pulses, designed to be insensitive to B1 miscalibration and inhomogeneity,2, 41 which are exceptionally robust (SI Figure S4). The final pulse sequence used for measurement of the couplings in this work therefore consists of an HMQC in which the 15N pulses are replaced by BIR-4 pulses, with flip angles of 45° and 90° applied in an interleaved fashion. Any 1H-1H RDCs with imino or ribose protons are refocused by the application of a band-selective ReBURP pulse1 at the midpoint of the t1 evolution period. We call this experiment a variable flip HMQC (VF-HMQC) (Figure 1A). The pulse sequence for Bruker spectrometers is included as Supporting Information.
We applied the VF-HMQC sequence to RRE232A and compared the sensitivity to SOFAST-HMQC and S3E experiments, in each case removing the 15N chemical shift evolution period (Figure 1C). As expected on the basis of its similar pulse sequence structure, the sensitivity of our new approach is comparable to the SOFAST-HMQC, whereas the S3E experiment is less sensitive by at least 10-fold, with many signals completely undetectable. A 21 hour VF-HMQC acquisition using 160 μL of ~1.5 mM RRE232A with 15N chemical shift evolution (Figure 1D) gives a S/N for the weakest, fast relaxing signals of ~50:1 in the 90° flip angle experiment.
The uncertainty in RS is given by:
| (6) |
where N is the root-mean-square noise in the spectrum. The uncertainty in J then follows from:
| (7) |
to yield:
| (8) |
as detailed SI Appendix 1. The optimum value for τ to minimize σJ is a function of the total coupling. The most important consideration is to avoid approaching τ = 1/2J where . Graphical analysis indicates that choosing τ = 1/3J yields uncertainties of < 0.2 Hz for the range of expected2(J+D)NH values for a S/N of 50:1, as measured for RRE232A (SI Figure S5). The RDC is calculated from the difference in couplings measured under isotropic and aligned conditions and will therefore have an uncertainty of assuming the same S/N in both experiments, suggesting a total uncertainty of < 0.3 Hz is attainable. This uncertainty is more than an order of magnitude smaller than the expected range of the RDCs, and as such is anticipated to have a negligible effect during fitting compared to the uncertainty in the coordinates of the reference structure.42
From ~80 resolved signals in the RRE232A HMQC we were able to measure 62 2JNH values with uncertainties <0.2 Hz (M = 14.7 Hz, SD = 0.24 Hz; uncertainties for remaining signals were higher due to lower S/N). Following alignment with ~13 mg/mL Pf1 phage we measured 59 RDCs with uncertainties <0.3 Hz, ranging from −0.9 to 2.7 Hz. As there is no comparable method for measuring these couplings in large RNAs, it is not possible to directly validate these measurements. Instead, we designed a 36 nt RNA construct based on stem loop C from the MMLV 5′-Leader (SLCA, Figure 3A). This RNA contains two helices separated by a non-canonical k-turn9 which leads to an inter-helical angle of ~74°.43 We engineered three additional A:U base pairs into the proximal helix so that each helix contained at least 6 H2-N RDCs (Figure 3A). Excellent agreement was observed between 20 15N-1H RDCs calculated using the VF-HMQC (Figure 2E) and S3E (SI Figure S6) approaches (Pearson correlation coefficient = 0.992; RMSD = 0.14 Hz; Figure 2C), which confirmed the accuracy of the VF-HMQC approach.
Figure 3.
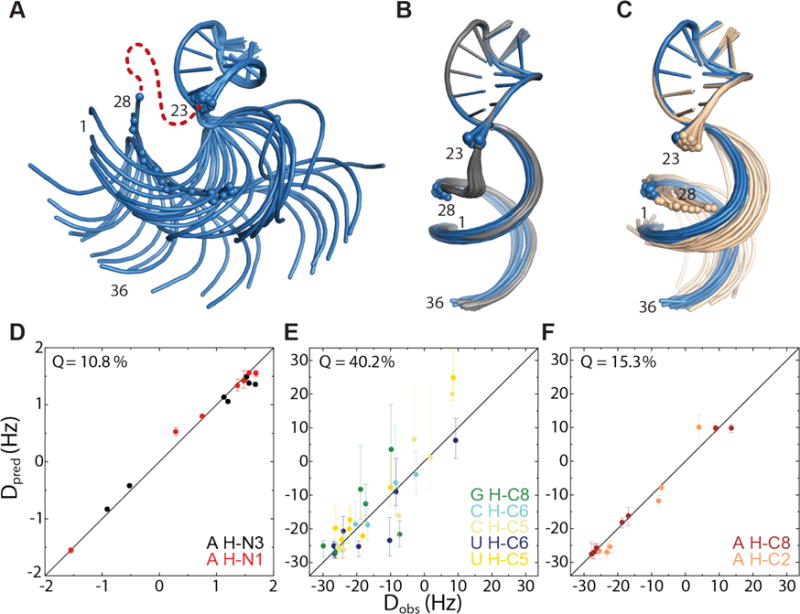
Ensemble of 20 lowest energy SLCA structures calculated using NOEs but without residues from the inter-helical bulge. The relative orientation of the helices is not well-defined, with inter-helical angles ranging from −83° to 110°. (B) In blue, as (A) but with incorporation of 47 13C-1H RDC restraints. The inter-helical angle is between 76° and 84° (M = 81.5°, SD =2.6°). This agrees well with the ensemble calculated for the full RNA, shown in gray. (C) In cream, as (A) but with addition of 14 15N-1H RDCs measured with the VF-HMQC approach. These RDCs are sufficient to restrain the inter-helical angle between 77° and 94° (M = 84°, SD = 4.1°), in agreement with (B), overlaid in blue. (D) Observed 15N-1H RDCs plotted against those back-calculated for the ensemble calculated with 13C-1H RDCs, shown in blue in (B). Error bars for Dpred represent the maximum and minimum RDCs calculated for the ensemble. (E) Observed 13C-1H RDCs plotted for guanosine, cytidine and uridine residues and (F) Observed 13C-1H RDCs for adenosine residues, plotted against those back-calculated for the ensemble calculated with 15N-1H RDCs, shown in cream in (C).
Figure 2.
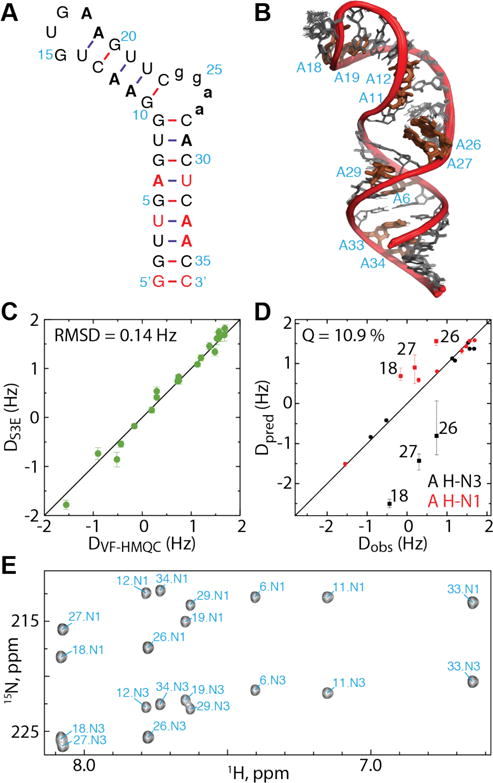
(A) SLCA RNA construct used to validate the VF-HMQC method. Adenosines are shown in bold. Non-native residues are shaded in red. Residues from the inter-helical bulge omitted in some calculations are shown in lower case. (B) Ensemble of 20 lowest energy structures of SLCA, calculated using NOE and 13C-1H RDC restraints. (C) Comparison of 2DNH for SLCA adenosines measured using the VF-HMQC and S3E approaches show good correlation (RMSD = 0.14 Hz). Error bars for each experiment indicate uncertainty due to noise and linewidth. (D) Observed 2DNH for SLCA obtained using VF-HMQ are well correlated with back-calculated RDCs from the ensemble shown in B (Q = 10.9%). Loop and bulge adenosines are labeled and omitted from the Q value calculation. Error bars for Dpred represent the maximum and minimum RDCs calculated for the ensemble. (E) Assigned VF-HMQC spectrum for SLCA.
We next wished to determine if the H2-N1/3 RDCs were sufficient to independently establish the inter-helical angle in SLCA. A total of 47 H2-C2, H8-C8, H5-C5 and H6-C6 RDCs were measured using the ARTSY approach29, 44 (SI Figure S7), which were used together with 262 distance restraints to calculate a structural ensemble for this construct (Figure 2B). Back-calculated RDCs from this structure ensemble correlate exceptionally well with 15N-1H RDCs measured using the VF-HMQC approach for the helical regions (Q = 10.9 %, Figure 2D), despite not having been used as input parameters.
The presence of multiple long-range NOEs from residues in the inter-helical bulge to helix 2 leads to a relatively well defined inter-helical angle, even in the absence of RDC restraints. To better assess the ability of the RDCs to determine inter-helical angles, we calculated an ensemble of SLCA structures in which the bulge residues were substituted by a long chain of pseudoatoms, thus removing inter-helical distance and geometric constraints. In the absence of RDCs, the relative orientation of the two helices is poorly defined (Figure 3A, SI Figure S8). Upon refinement with 13C-1H RDC restraints (Figure 3B), the inter-helical angle is well-defined (between 76° and 84°) and shows a good correlation with the measured 15N-1H RDCs (Q=10.8%, Figure 3D). Importantly, refinement with the 14 15N-1H RDCs measured using the VF-HMQC approach (and without the 13C-1H RDCs) also affords structures with a well-defined inter-helical angle (between 77° and 94°) that is in good agreement with both the models derived using 13C-1H RDCs and the original SLCA structure (Figure 3C). Agreement with the measured 13C-1H RDCs remains good (overall Q=34.8%), although the correlation is weaker for non-adenosine residues (Q=40.2%, Figure 3D) than for the adenosines (Q=15.3%, Figure 3E) as may be expected, as our 15N-1H RDCs do not provide restraints for non-adenosine residues.
We have presented a new experiment that significantly extends the size limit for RNA RDC measurement. Studies with a 36-nucleotide RNA show that this method gives results consistent with existing methods that utilize an S3E element and affords data sufficient to define the inter-helical angle for RNAs containing as few as 3 adenosines (6 RDCs) per helix. In most cases, large RNAs should contain sufficient adenosine signals in each secondary structure element (e.g. RRE232A, SI Figure S1D). For unfavorable sequences, additional A:U base pairs may be introduced by mutagenesis, perhaps in combination with lrAID sequences,11 to ensure maximal signal dispersion and resolution. We expect the VF-HMQC approach be applicable to RNAs on the order of hundreds of nucleotides, thereby providing access to higher quality structures of larger RNAs than previously possible.
MATERIALS AND METHODS
In vitro transcription
RNA molecules were produced by in vitro transcription using T7 RNA polymerase45 in 7.5 mL reactions, containing 50 μg of PCR-amplified DNA template (RRE232A) or 2.5 nmol annealed DNA template (SLCA), 2 mM spermidine, 80 mM Tris·HCl (pH 8.5), 2 mM DTT, 20% (vol/vol) DMSO, 0.5 mg T7 RNA polymerase, 10–20 mM MgCl2, and 3–6 mM NTPs. DNA templates are 2′-O-Methyl-modified at the last two nucleotides of the 5′ end to improve 3′ end homogeneity of transcribed RNA.46-47 15N-labelled samples were prepared with [U-98-99% 15N]-ATP, [U-97%+ 2H]-CTP, [U-97%+ 2H]-GTP, and [U-97%+ 2H]-UTP (CIL). 13C-labelled samples were prepared with [U 13C]-ATP, [U 13C]-CTP, [U 13C]-GTP, and [U 13C]-UTP (CIL). Reactions were incubated at 37 °C for four hours before quenching by addition of EDTA. RNA was purified by electrophoresis on urea-containing polyacrylamide denaturing gels (SequaGel, National Diagnostics) using FisherBiotech DNA sequencing system at 20 W overnight, before electroelution using the Elutrap system (Whatman) at 120 V overnight. The eluted RNAs were washed with 2 M NaCl and then desalted using a 30 kDa (RRE232A) or 3-kDa (SLCA) MWCO Amicon Ultra-4 Centrifugal Filter Device (Millipore). The concentration of each sample was determined by measuring the optical absorbance at 260 nm. Samples were exchanged into D2O (99.96%; CIL) by two rounds of lyophilization before dissolution in NMR buffer.
NMR sample preparation
RRE232A NMR samples [160 μL of ~1.5 mM RNA in D2O] were prepared in a 3 mm NMR tube with 20 mM Tris–d11 buffer (pH = 7.4), 140 mM KCl, and 1 mM MgCl2. SLCA NMR samples [500 μL of ~500 μM RNA in D2O] were prepared in a 5 mm NMR tube with 20 mM Tris–d11 buffer (pH = 7.4). For aligned samples, Pf1 phage (ASLA Biotech) was exchanged into D2O by repeated washing using a 30 kDa MWCO Amicon Ultra-4 Centrifugal Filter Device (Millipore). Phage concentration was estimated by the quadrupolar splitting of the D2O signal.48 Lyophilized RNA was resuspended in 10-15 mg/mL of Pf1 in D2O. To simplify analysis, 15N and 13C-labeled SLCA were combined [500 μL total volume with ~250 μM of each RNA] for the aligned sample.
NMR experiments
All experiments were performed at 308 K on a Bruker AVANCE III HD spectrometer at 600.13 MHz. SOFAST-HMQC,13–14 S3E25 and ARTSY29 experiments were performed as previously described. Spectra were processed using NMRFx49 and NMRPipe.50 Chemical shifts and NOEs were assigned using NMRViewJ51 with a combination of 1H-1H NOESY, 1H-1H TOCSY and 13C-1H HMQC spectra, utilizing predicted chemical shifts52–53 and in-house scripts. Relaxation data was processed and analyzed using Bruker TopSpin v3.5.
Structure Calculation
Structures were calculated with CYANA54 (v2.1 and 3.97) using NOE, H-bond, and database derived inter-phosphate distance restraints, as described previously.16 Structures with low target functions were then subjected to rounds of conjugate gradient minimization using RDC restraints with increasing weight coefficients (to a maximum of 0.1).15N-1H RDCs were weighted 14.45 times greater than 13C-1H restraints.
Supplementary Material
Acknowledgments
The authors would like to acknowledge Daniel Morris, Stanley Wang, Colin O’Hern and Sophia Abbot for preparation of RNA used in this work, together with HHMI staff at UMBC for their general assistance.
Funding Sources
Funding from the NIH (U54 GM103297 to JM; GM42561 to MFS; Intramural Research Program of the NIDDK and the Intramural Antiviral Target Program of the Office of the Director, NIH, to AB) and HHMI is gratefully acknowledged.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
Supplementary Figures 1-8 and Appendix 1 (PDF)
VF-HMQC pulse sequence (TXT)
SLCA chemical shifts (TXT)
SLCA atomic coordinates (PDB)
Author Contributions
The manuscript was written through contributions of all authors.
Notes
The authors declare no competing financial interest.
References
- 1.Geen H, Freeman R. Band-selective radiofrequency pulses. J Magn Reson. 1991;93:93–141. [Google Scholar]
- 2.Garwood M, Ke Y. Symmetric pulses to induce arbitrary flip angles with compensation for RF inhomogeneity and resonance offsets. J Magn Reson. 1991;94:511–525. [Google Scholar]
- 3.Caprara MG, Nilsen TW. RNA: versatility in form and function. Nat Struct Biol. 2000;7(10):831–3. doi: 10.1038/82816. [DOI] [PubMed] [Google Scholar]
- 4.Marion D, Ikura M, Tschudin R, Bax A. Rapid recording of 2D NMR spectra without phase cycling. Application to the study of hydrogen exchange in proteins. J Magn Reson. 1989;85:393–399. [Google Scholar]
- 5.Alvarado LJ, LeBlanc RM, Longhini AP, Keane SC, Jain N, Yildiz ZF, Tolbert BS, D’Souza VM, Summers MF, Kreutz C, Dayie TK. Regio-selective chemical-enzymatic synthesis of pyrimidine nucleotides facilitates RNA structure and dynamics studies. Chembiochem. 2014;15(11):1573–7. doi: 10.1002/cbic.201402130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allain FH-T, Varani G. How accurately and precisely can RNA structure be determined by NMR. J Mol Biol. 1997;267:338–351. doi: 10.1006/jmbi.1996.0855. [DOI] [PubMed] [Google Scholar]
- 7.Lukavsky PJ, Puglisi JD. Structure determination of large biological RNAs. Methods in Enzymology. 2005;394:399–415. doi: 10.1016/S0076-6879(05)94016-0. [DOI] [PubMed] [Google Scholar]
- 8.Shaka AJ, Barker PB, Freeman R. Computer-optimized decoupling scheme for wideband applications and low-level operation. J Magn Reson. 1985;64:1985. [Google Scholar]
- 9.D’Souza V, Dey A, Habib D, Summers MF. NMR structure of the 101-nucleotide core encapsidation signal of the Moloney murine leukemia virus. J Mol Biol. 2004;337(2):427–42. doi: 10.1016/j.jmb.2004.01.037. [DOI] [PubMed] [Google Scholar]
- 10.Miyazaki Y, Irobalieva RN, Tolbert BS, Smalls-Manty A, Iyalla K, Loeliger K, D’Souza V, Khant H, Schmid MF, Garcia E, Telesnitsky A, Chiu W, Summers MF. Structure of a conserved retroviral RNA packaging element by NMR spectroscopy and cryo-electron tomography. J Mol Biol. 2010;404:751–772. doi: 10.1016/j.jmb.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu K, Heng X, Garyu L, Monti S, Garcia E, Kharytonchyk S, Dorjsuren B, Kulandaivel G, Jones S, Hiremath A, Sachin Divakaruni S, LaCotti C, Barton S, Tummillo D, Hosic A, Edme K, Albrecht S, Telesnitsky A, Summers MF. NMR detection of structures in the HIV-1 5′-leader RNA that regulate genome packaging. Science. 2011;344:242–245. doi: 10.1126/science.1210460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keane SC, Heng X, Lu K, Kharytonchyk S, Ramakrishnan V, Carter G, Barton S, Hosic A, Florwick A, Santos J, Bolden NC, McCowin S, Case DA, Johnson BA, Salemi M, Telesnitsky A, Summers MF. Structure of the HIV-1 RNA packaging signal. Science. 2015;348(6237):917–21. doi: 10.1126/science.aaa9266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schanda P, Brutscher B. Very fast two-dimensional NMR spectroscopy for real-time investigation of dynamic events in proteins on the time scale of seconds. J Am Chem Soc. 2005;127(22):8014–5. doi: 10.1021/ja051306e. [DOI] [PubMed] [Google Scholar]
- 14.Sathyamoorthy B, Lee J, Kimsey I, Ganser LR, Al-Hashimi H. Development and application of aromatic [(13)C, (1)H] SOFAST-HMQC NMR experiment for nucleic acids. J Biomol NMR. 2014;60(2-3):77–83. doi: 10.1007/s10858-014-9856-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis JH, Tonelli M, Scott LG, Jaeger L, Williamson JR, Butcher SE. RNA helical packing in solution: NMR structure of a 30 kDa GAAA tetraloop-receptor complex. J Mol Biol. 2005;351:371–382. doi: 10.1016/j.jmb.2005.05.069. [DOI] [PubMed] [Google Scholar]
- 16.Tolbert BS, Miyazaki Y, Barton S, Kinde B, Starck P, Singh R, Bax A, Case DA, Summers MF. Major groove width variations in RNA structures determined by NMR and impact of 13C residual chemical shift anisotropy and 1H-13C residual dipolar coupling on refinement. J Biomol NMR. 2010;47:205–219. doi: 10.1007/s10858-010-9424-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mollova ET, Hansen MR, Pardi A. Global structure of RNA determined with residual dipolar couplings. J Am Chem Soc. 2000;122:11561–11562. [Google Scholar]
- 18.Ying J, Grishaev A, Latham MP, Pardi A, Bax A. Magnetic field induced residual dipolar couplings of imino groups in nucleci acids from measurements at a single magnetic field. J Biomol NMR. 2007;39:91–96. doi: 10.1007/s10858-007-9181-7. [DOI] [PubMed] [Google Scholar]
- 19.Lukavsky PJ, Kim I, Otto GA, Puglisi JD. Structure of HCV IRES domain II determined by NMR. Nature Struct Biol. 2003;10:1033–1038. doi: 10.1038/nsb1004. [DOI] [PubMed] [Google Scholar]
- 20.Staple DW, Butcher SE. Solution structure and thermodynamic investigation of the HIV-1 frameshift inducing element. J Mol Biol. 2005;349:1011–1023. doi: 10.1016/j.jmb.2005.03.038. [DOI] [PubMed] [Google Scholar]
- 21.Kim NK, Zhang Q, Zhou J, Theimer CA, Peterson RD, Feigon J. Solution structure and dynamics of the wild-type pseudoknot of human telomerase RNA. J Mol Biol. 2008;384(5):1249–61. doi: 10.1016/j.jmb.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skrisovska L, Bourgeois CF, Stefl R, Grellscheid SN, Kister L, Wenter P, Elliott DJ, Stevenin J, Allain FH. The testis-specific human protein RBMY recognizes RNA through a novel mode of interaction. EMBO Rep. 2007;8(4):372–9. doi: 10.1038/sj.embor.7400910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Q, Sun X, Watt ED, Al-Hashimi HM. Resolving the motional modes that code for RNA adaptation. Science. 2006;311:653–656. doi: 10.1126/science.1119488. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Q, Stelzer AC, Fisher CK, Al-Hashimi HM. Visualizing spatially correlated dynamics that directs RNA conformational transitions. Nature. 2007;450:1263–1267. doi: 10.1038/nature06389. [DOI] [PubMed] [Google Scholar]
- 25.Zidek L, Wu H, Feigon J, Sklenar V. Measurement of small scalar and dipolar couplings in purine and pyrimidine bases. J Biomol NMR. 2001;21(2):153–60. doi: 10.1023/a:1012435106858. [DOI] [PubMed] [Google Scholar]
- 26.Tjandra N, Bax A. Direct measurement of distances and angles in biomolecules by NMR in a dilute liquid crystalline medium. Science. 1997;278(5340):1111–1114. doi: 10.1126/science.278.5340.1111. [DOI] [PubMed] [Google Scholar]
- 27.Prestegard JH, al-Hashimi HM, Tolman JR. NMR structures of biomolecules using field oriented media and residual dipolar couplings. Q Rev Biophys. 2000;33(4):371–424. doi: 10.1017/s0033583500003656. [DOI] [PubMed] [Google Scholar]
- 28.Bax A, Kontaxis G, Tjandra N. Dipolar couplings in macromolecular structure determination. Methods Enzymol. 2001;339:127–74. doi: 10.1016/s0076-6879(01)39313-8. [DOI] [PubMed] [Google Scholar]
- 29.Ying J, Wang J, Grishaev A, Yu P, Wang YX, Bax A. Measurement of (1)H-(15)N and (1)H-(13)C residual dipolar couplings in nucleic acids from TROSY intensities. J Biomol NMR. 2011;51(1-2):89–103. doi: 10.1007/s10858-011-9544-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wijmenga SS, van Buurehn BNM. The use of NMR methods for conformational sutdies of nucleic acids. Prog NMR Spec. 1998;32:287–387. [Google Scholar]
- 31.Prestegard JH. New techniques in structural NMR — anisotropic interactions. Nat Struct Biol. 1998:517–522. doi: 10.1038/756. Supplement July. [DOI] [PubMed] [Google Scholar]
- 32.Fonseca Guerra C, Matthias Bickelhaupt F, Snijders JG, Baerends EJ. Hydrogen bonding in DNA base pairs: Reconcilliation of theory and experiment. J Am Chem Soc. 2000;122:4117–4128. [Google Scholar]
- 33.Ward JM, Skrynnikov NR. Very large residual dipolar couplings from deuterated ubiquitin. J Biomol NMR. 2012;54(1):53–67. doi: 10.1007/s10858-012-9651-4. [DOI] [PubMed] [Google Scholar]
- 34.Sherpa C, Rausch JW, Le Grice SF, Hammarskjold ML, Rekosh D. The HIV-1 Rev response element (RRE) adopts alternative conformations that promote different rates of virus replication. Nucleic Acids Res. 2015;43(9):4676–86. doi: 10.1093/nar/gkv313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meissner A, Duus JO, Sorensen OW. Integration of spin-state-selective excitation into 2D NMR correlation experiments with the heteronuclear ZQ/2Q pi rotations for 1JXH-resolved E. COSY-type measurements of heteronuclear coupling constants in proteins. J Biomol NMR. 1997;10(1):89–94. doi: 10.1023/a:1018331001961. [DOI] [PubMed] [Google Scholar]
- 36.Stueber D, Grant DM. 13C and (15)N chemical shift tensors in adenosine, guanosine dihydrate, 2′-deoxythymidine, and cytidine. J Am Chem Soc. 2002;124(35):10539–51. doi: 10.1021/ja012485c. [DOI] [PubMed] [Google Scholar]
- 37.Bax A, Max D, Zax D. Measurement of long-range 13C-13C J coupling in a 20-kDa protein-peptide complex 1992. 1992;114(17):6923–6925. [Google Scholar]
- 38.Griesinger C, Sørensen OW, Ernst RR. Correlation of connected transitions by two-dimensional NMR spectroscopy. Journal of Chemical Physics. 1986;85(12):6837–6852. [Google Scholar]
- 39.Otting G, Messerle BA, Soler LP. 1H-Detected, gradient-enhanced 15N and 13C NMR experiments for the measurement of small heteronuclear coupling constants and isotopic shifts. J Am Chem Soc. 1996;118:5096–5102. [Google Scholar]
- 40.Otting G, Messerle BA, Soler LP. 1H-Detected multinuclear NMR experiments for the measurement of small heteronuclear coupling constants in transition metal complexes. J Am Chem Soc. 1997;119:5425–5434. [Google Scholar]
- 41.Garwood M, DelaBarre L. The return of the frequency sweep: designing adiabatic pulses for contemporary NMR. J Magn Reson. 2001;153(2):155–77. doi: 10.1006/jmre.2001.2340. [DOI] [PubMed] [Google Scholar]
- 42.Zweckstetter M, Bax A. Evaluation of uncertainty in alignment tensors obtained from dipolar couplings. J Biomol NMR. 2002;23(2):127–37. doi: 10.1023/a:1016316415261. [DOI] [PubMed] [Google Scholar]
- 43.Bailor MH, Musselman C, Hansen AL, Gulati K, Patel DJ, Al-Hashimi HM. Characterizing the relative orientation and dynamics of RNA A-form helices using NMR residual dipolar couplings. Nat Protoc. 2007;2(6):1536–46. doi: 10.1038/nprot.2007.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fitzkee NC, Bax A. Facile measurement of (1)H-(1)5N residual dipolar couplings in larger perdeuterated proteins. J Biomol NMR. 2010;48(2):65–70. doi: 10.1007/s10858-010-9441-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Research. 1987;15(21):8783–9798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kao C, Zheng M, Rüdisser S. A simple and efficient method to reduce nontemplated nucleotide addition at the 3 terminus of RNAs transcribed by T7 RNA polymerase. RNA. 1999;5:1268–1272. doi: 10.1017/s1355838299991033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Helmling C, Keyhani S, Sochor F, Furtig B, Hengesbach M, Schwalbe H. Rapid NMR screening of RNA secondary structure and binding. J Biomol NMR. 2015;63(1):67–76. doi: 10.1007/s10858-015-9967-y. [DOI] [PubMed] [Google Scholar]
- 48.Hansen MR, Mueller L, Pardi A. Tunable alignment of macromolecules by filamentous phage yields dipolar coupling interactions. Nat Struct Biol. 1998;5:1065–1074. doi: 10.1038/4176. [DOI] [PubMed] [Google Scholar]
- 49.Norris M, Fetler B, Marchant J, Johnson BA. NMRFx Processor: a cross-platform NMR data processing program. Journal of biomolecular NMR. 2016:1–12. doi: 10.1007/s10858-016-0049-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 51.Johnson BA, Blevins RA. NMRview: a Computer Program for the Visualization and Analysis of NMR Data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 52.Barton S, Heng X, Johnson BA, Summers MF. Database proton NMR chemical shifts for RNA signal assignment and validation. J Biomol NMR. 2013;55(1):33–46. doi: 10.1007/s10858-012-9683-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brown JD, Summers MF, Johnson BA. Prediction of hydrogen and carbon chemical shifts from RNA using database mining and support vector regression. Journal of biomolecular NMR. 2015;63(1):39–52. doi: 10.1007/s10858-015-9961-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Güntert P, Mumenthaler C, Wüthrich K. Torsion angle dynamics for protein structure calculations with a new program, DYANA. J Mol Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


