Abstract
Hydration water on the surface of a protein is thought to mediate the thermodynamics of protein-ligand interaction. For hydration water to play a role beyond modulating global protein solubility or stability, the thermodynamic properties of hydration water must reflect on the properties of the heterogeneous protein surface, and thus spatially vary over the protein surface. A potent read-out of local variations in thermodynamic properties of hydration water is its equilibrium dynamics spanning picosecond to nanosecond timescales. In this study, we rely on Overhauser dynamic nuclear polarization (ODNP) to probe the equilibrium hydration water dynamics on the globular protein Chemotaxis Y (CheY), in dilute solution, at select sites located on the protein surface. ODNP reports on site-specific hydration dynamics within 5–10 Å of a label tethered to the biomolecular surface on two separate timescales of motion, corresponding to diffusive water (DW) and protein-water coupled motions referred to as bound water (BW). We find DW dynamics to be highly heterogeneous across the surface of CheY, while also finding significant populations of BW. We identify a significant correlation between DW dynamics and the local hydropathy of the CheY protein surface, as empirically determined by molecular dynamics (MD) simulations, and find the more hydrophobic sites to be hydrated with slower diffusing water. We furthermore compare the DW dynamics and BW population on the surface of CheY to that of another globular protein Annexin XII (Anx), two intrinsically disordered proteins (IDPs) ΔTau-187 and α-synuclein, CheY-inspired 5 residue peptides, polyproline-based peptides with systematic charge variation, and DPPC/DOPC liposomes. The DW dynamics on Anx is similarly heterogeneous as on CheY, and there is significant BW population on both Anx and CheY. In contrast, DW dynamics is relatively homogeneous on IDP and liposome surfaces, while BW is entirely absent. The heterogeneity in hydration water properties suggests that a structured protein surface has the capacity to encode information into its hydration water to mediate the free energy of interactions involving the protein surface.
Introduction
Water is thought to be directly involved in tuning the structure, stability, dynamics and function of biological macromolecules. One proposed avenue by which water participates in molecular recognition of proteins is by thermodynamically mediating the interactions between protein-protein or protein-ligand binding partners1. This suggests that the protein extends its range of interaction by encoding structural or dynamic information in the layer of water molecules that directly hydrate the protein surface1–4. It follows that there must be spatial diversity in the water environments on the surface of a protein to enhance or impose the properties of a protein surface. However, experimental data on surface-, region- or site-specific water properties on protein surfaces in native solution state is scarce5–10. Consequently, the nature or even existence of the relationship between the molecular makeup of the protein surface and variation in hydration water environments by which the protein may encode functional properties continues to elude scientific understanding. To gain an understanding of the role that chemical or geometrical topology of the protein surface plays in tuning the hydration water properties, it is necessary to probe hydration water properties with both spatial and temporal resolution.
The thermodynamics of solvation dictate the hydration water dynamics on a protein surface, thus measuring the dynamics of hydration water around the protein surface under equilibrium conditions serves as a proxy for probing the thermodynamics of solvation. Overhauser dynamic nuclear polarization (ODNP) offers an experimental approach to characterize hydration water dynamics with site-specificity under ambient solution state conditions over ns to ps timescales. The timescale of motions for water on the protein surface varies over decades from bound water (BW) – with lifetimes of few to 10’s of ns - to highly diffusive water (DW) – with lifetimes of 1–100’s of ps. The BW population on the surface has the potential to constitute an entropic reservoir, as it provides a mechanism to increase the total entropy through its release and expulsion from the binding surface of the protein, compensating for the negative entropy change resulting from protein-ligand binding. The retardation of DW may reflect on the enthalpic cost to disrupt, and ultimately dehydrate, the hydration layer at the interface, while the slower population of DW may also constitute an entropic reservoir, whose liberation to bulk water increases the net entropy. To capture the relevant properties of hydration water, it is necessary to probe the water dynamics over this range of timescales. There is already convincing evidence in the literature that the distribution of BW is spatially heterogeneous across a protein surface9,10. However, the spatial heterogeneity in the translational diffusion of hydration water on the protein surface remains in question. Here we report on site-specific ODNP measurements on the Chemotaxis Y (CheY) protein at room temperature in dilute solution state to address this question.
ODNP selectively amplifies the 1H NMR signal of the local hydration water in close proximity (5–10 Å) to the spin label by transferring the larger magnetic polarization from the unpaired electron spin of the label to the 1H nuclear spin of the adjacent water molecules. The dipolar coupling between the electron spin of the nitroxide radical label attached to the biomolecular surface and the 1H of the surrounding water molecules is modulated by the relative motion of the electron spin and the 1H nuclear spin. Transfer of polarization from the electron spins to 1H spins occurs through electron-1H dipolar coupling via cross relaxation and leads to the amplification of the 1H signal through the Overhauser effect11,12. ODNP measurements quantify the dipolar coupling factor (ξ) between the unpaired electron spin and the 1H nuclear spin that is dependent on the dipolar correlation time (τ) of electron-1H inter-spin motions. Thus, ξ and τ serve as a read-out of the water’s 1H motion, given that the relative motion of the spin label is negligible compared to that of water. As discussed previously13, ξ and τ are sensitive to motions on both the picosecond (ps) and nanosecond (ns) timescales which on the protein surface correspond to DW and BW respectively. Thus, it is necessary to expand on the standard ODNP approach to separately analyze the ps and ns timescales of motion to access the DW and BW on the protein surface. Following the method proposed by Franck et al. we separate ξ into the cross-relaxation rate (kσ) that is selectively proportional to the spectral density function for a ~9.8 GHz transition frequency (corresponding to the Larmor frequency of the electron spin) and the electron spin-induced self-relaxation rate (kLow) of the 1H spin that is selectively proportional to the spectral density function for a ~14MHz transition frequency (corresponding to the Larmor frequency of the 1H nuclear spin). This allows us to separately analyze the timescales of motions, as kσ is only sensitive to the 10–100’s of ps timescale and kLow only to the 10–100’s of ns timescale motions. The access to spatial and temporal resolution is critical from an experimental perspective, because it allows for a multiplexed study of water dynamics around many different sites, of the same protein, in the same solution system, and under otherwise identical experimental conditions.
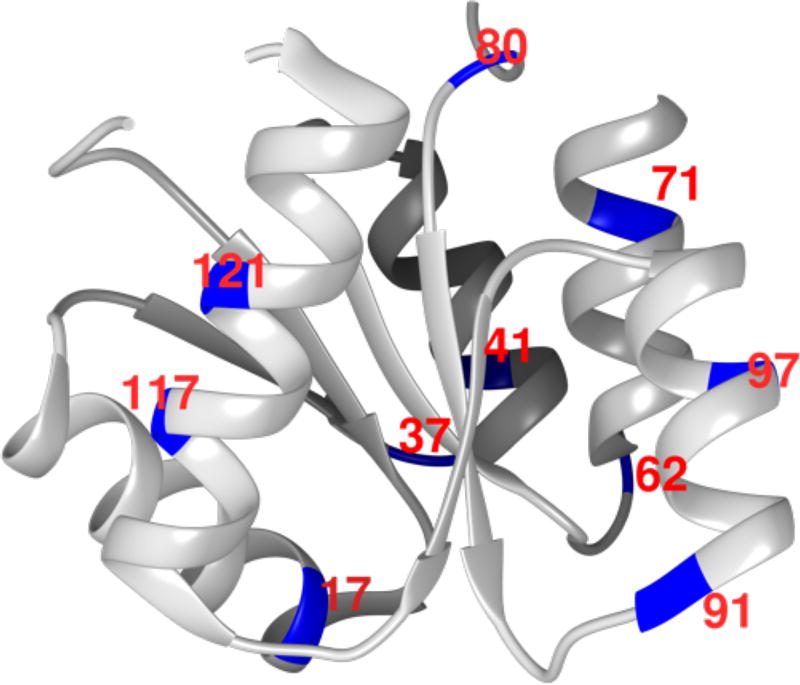
CheY is a globular protein with 12kDa molecular weight (PDB ID 1JBE), with its structure shown in Figure 1. The protein is natively cysteine free to which we introduced a single cysteine residue by site-directed mutagenesis14 at the sites shown in Figure 1, and covalently tether a stable nitroxide radical-based spin label S-(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate (MTSL). We compared both the DW and BW dynamics at 10 different locations on the surface of the CheY protein to that of 10 different CheY-derived 5-residue peptides composed of the nearest and next-nearest neighbors of each site studied on the CheY protein surface. The 5-residue CheY-inspired peptides have a molecular weight of 700 ± 50 Da, and are too short to form any stable secondary structure. Therefore, the difference in the geometrical topology between the different peptides should be negligible in comparison to differences in the surface hydropathy. These peptide systems serve as a control surface to exclusively evaluate the effect of the average hydropathy on the variation in DW and BW dynamics. To further substantiate our experimental data, we determined the local protein surface hydropathy and excluded volumes for CheY by all-atom molecular dynamics (MD) simulation, and compared the results to the trends in both the DW and BW dynamics. Finally, we compared the surface water properties of the CheY protein and the derived peptides to that of a series of other biomolecular surfaces, focusing on identifying defining characteristics of DW and BW contributions on the surface of structured proteins in comparison to that of intrinsically disordered proteins (IDPs) and liposomes.
Figure 1.
The X-ray crystal structure of CheY (PDB ID code 1JBE). The residues studied in this work are highlighted in blue and labeled in red. The protein structure was prepared in Chimera16.
We found significant heterogeneity in DW dynamics on the surface of globular proteins, compared to relitively uniform DW dynamics on other peptide, IDP, and liposome surfaces. Furthermore, we found significant correlation between the measured local DW retardation and the local hydropathy, as calculated from the partitioning of hydrophobic probe solutes on the adjacent CheY protein surface site according to MD simulations. In comparison, any hydropathy scale from the literature based on counting or rationalizing individual contributions of amino acid residues produced no correlation to the experimental DW dynamics. Lastly, we found that BW is present on only globular protein surfaces, while no BW was detected on IDP, peptide, or liposome surfaces. These experimental findings suggest that proteins with a defined structure have the capacity to encode functional information into the hydration water surrounding the protein surface.
Methods
Sample Preparation
We investigate the influence of surface sites of E. Coli chemotaxis response regulator protein (CheY), a 14 kDa globular protein (PDB ID 1JBE) shown in Figure 1, on the local surface water diffusivity derived from ODNP measurements. We covalently attach a stable nitroxide radical-based spin label – via a di-sulfide bond - to a single cysteine residue introduced to the CheY protein by site-directed mutagenesis14. Cysteine mutants of the CheY protein were prepared via a similar manner used in a previous study15. Specifically, the gene encoding wild-type Escherichia coli (E. coli) CheY (residues 1–129) was cloned into pET28a (Novagen) at the NcoI and XhoI sites in frame with the carboxy-terminal hexahistidine tag. Cysteine mutants were generated with the QuikChange site-directed mutagenesis and transformed into E. coli BL21(DE3) (Novagen). The expression was induced at OD600 = 0.4 by the addition of 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) in Lysogeny broth (LB) medium. The proteins were then purified by immobilized nickel affinity chromatography column (GE Healthcare). The collected fractions were dialyzed into a buffer consisting of 50 mM sodium phosphate and 150 mM sodium chloride at pH 7.9 and concentrated to about 0.5 mM. A nitroxide radical-based spin label was attached to the CheY mutants as described in the following. CheY cysteine mutants were reduced with 5 mM dithiothreitol (DTT) (Sigma-Aldrich) for 2 hr. Then, Sephadex G-25 spin column (GE-Healthcare) was used to remove DTT prior to the addition of S-(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate (MTSL) at a 20:1 reagent-to-protein molar ratio. The reaction mix was incubated at room temperature for 12 hr, and subsequently a Sephadex G-25 spin column used to remove the unreacted MTSL.
In this report we present and compare ODNP measurements on the surface of ΔTau-18717, α-synuclein18, Annexin XII19, and DOPC/DPPC LUV liposomes20. The hydration dynamics on the surface of ΔTau-187 monomers were measured on sites 313, 316, 322, 400, and 40417, of α-synuclein measured at sites 77, 81, 85, 86, 90, 93, 95, 98, 100, 101, 124, and 13618, and of Annexin XII measured at sites 12, 16, 104, 112, 121, 124, 137, 141, 162, 180, and 260 XII19. The liposomes systems were prepared in the LUV state and composed of purely DOPC, DPPC, and a 50:50 mixture of DOPC/DPPC. The spin label was added to the lipid system by mixing in a phospholipid TEMPO-choline moiety20 in a mol ratio of 2%.
The systematically charge varied peptide sequences are composed of CK, CD, CPPP, CPPPK, CPPPD, CPPPPPK, and CPPPPPD. The cysteine residue was labeled by addition of S-(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate (MTSL) at a 1:3 reagent-to-protein molar ratio. Purification by size exclusion or cation exchange chromatography was unsuccessful. A 1:3 reagent-to-protein molar ratio limited free MTSL to <5% of the total continuous wave EPR spectrum. The samples for ODNP were prepared in 20 mM sodium acetate buffer.
ODNP Measurements
Samples of approximately 3.5 µL volume were placed in a 0.6 mm i.d. and 0.84 o.d. quartz capillary and analyzed by ODNP, as described previously21. A “pass through” NMR probe design built to fit inside a 3mm i.d. 6 mm o.d. quartz tube was used. The quartz tube was inserted into a high sensitivity microwave cavity (ER 4119HS-LC, Bruker Biospin) along with the NMR probe and sample. ODNP experiments were performed using a Bruker EMX CW EPR spectrometer and a Bruker Avance III NMR console. The samples were sealed in a capillary with a protective layer of Critoseal on the top and hot beeswax on the bottom. All ODNP measurements were performed at room temperature. The sample was irradiated with up to 6 W of microwaves at the EPR resonant frequency of the spin labels at ~10 GHz using a home-built microwave amplifier22. The magnetic field was set on resonance at the central electron hyperfine transition, here at 9.8 GHz. The spin label concentration of each sample was determined from the double integral of its cw EPR spectrum measured at 1 mW irradiation power, 0.4 G modulation width. The concentration-dependent ODNP relaxation rates, kσ, kρ, and kLow, are normalized to the sample concentration derived from spin counting per integration of the cw EPR spectrum.
ODNP Theory
The following section details how the Overhauser effect is leveraged to quantify the electron-nuclear inter-spin motion and extract information about the localized water fluctuation. ODNP selectively amplifies the 1H NMR signal of the local hydration water in close proximity (5–10 Å) to the spin label by transferring the larger polarization from the unpaired electron spin of the label to the 1H nuclear spin of the adjacent water molecules. The dipolar coupling between the radical electron and the protons mediates the polarization transfer, and hence the selective amplification of the 1H signal, and is dependent on the inter-spin motion through the Overhauser effect11,23. The ODNP technique quantifies the dipolar coupling factor (ξ) between the unpaired electron spin located on the biomolecular surface and the protons of the surrounding water molecules. The dipolar-coupling factor (ξ) is dependent on the dipolar correlation time modulated by inter-spin motion, given by the spectral density representation in Eqns. 1 and 211.
| (1) |
| (2) |
Here, J is the spectral density function, ωe and ωH are the electron and proton Larmor frequencies respectively, and τc is the electron-1H dipolar correlation time. The correlation time is calculated by interpolating Eq. 1 for a measured dipolar coupling factor, assuming the spectral density function to be given by the force free hard sphere (FFHS) model24 as shown in Eq. 2 for JFFHS. At X-band frequencies ωe = 9.8 GHz and ωH = 14 MHz, therefore the terms with ωe ± ωH are approximately equal to ωe. The coupling factor, and therefore the extracted correlation time, represents two timescales of motion 1/ωe and 1/ωH corresponding to 10–100’s of ps and 10–100’s of ns timescales of inter-spin motion. In the context of evaluating protein surface water hydration dynamics, the convolution of ns and ps timescales of motion can be problematic due to the presence of bound water (fluctuating on the ns timescale). The presence of bound water can lead to an exaggerated apparent retardation factor for surface water diffusivity13. Therefore, it is beneficial and often necessary to probe the ps and ns timescales separately. We separate these timescales of motion following the procedure outlined by Franck et al21. We separate ξ into two relaxation rates kσ and kρ, as given by Eqns. 3 and 4.
| (3) |
| (4) |
Here, k is the dipolar coupling constant for isotropic motion given by , Ne is the number of electron spins adjacent to the proton. Since ωe >>> ωH any term with ωe ± ωH can be approximated as ωe. Thus kσ can be approximated as indicated in the second equality of Eq. 3, showing that kσ only depends on terms of ωe and is only sensitive to motions on the ps timescale. The value of kρ carries both the ωe and ωH terms, and thus the dependence on both the ps and ns timescales. Following the approximation that ωe >>> ωH, we subtract the terms dependent on ωe from kρ to form kLow, a relaxation rate only dependent on ωH as shown in Eq. 5.
| (5) |
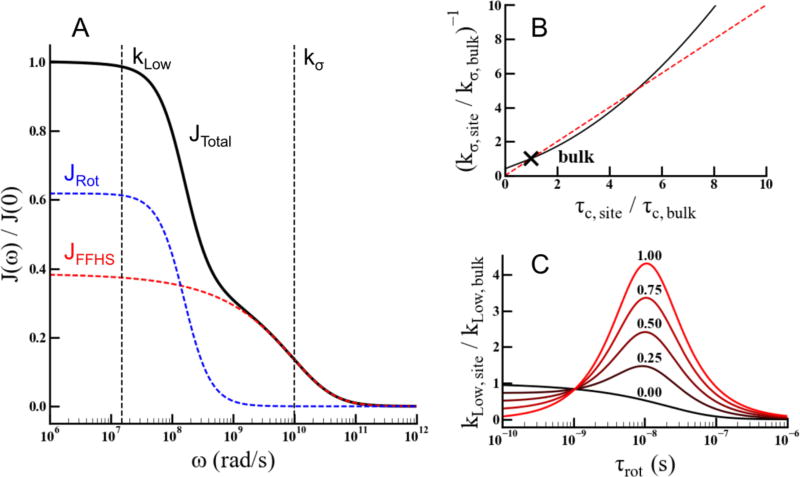
The relaxation rate kσ is sensitive to motions on the 1/ωe (1–100’s of ps) timescale. In the hydration layer of proteins, this timescale corresponds to the translational exchange and diffusive motion of water molecules. As before, we take the FFHS model24, shown as a function of frequency in Eq. 2 as a suitable approximation of the spectral density function for the translational diffusion of water molecules that is probed by kσ in the hydration layer13. This approximation in fact allows us to relate kσ to the correlation time of motion, τc. In Figure 2-B, we show the retardation in the kσ relaxation rate, , as a function of the retardation in correlation time, . The relationship is approximately linear in the retardation factor range up to 6 and still monotonically increases beyond this range, suggesting a two-fold retardation in kσ corresponds to two-fold slower translational diffusion, and so forth. It is worth noting that the FFHS model assumes spherical isotropic motion, while this assumption breaks down for measurements on the protein surface. This breakdown affects the absolute scaling between τc and kσ, however this does not affect the monotonic relationship between τc and kσ. In this manuscript we rely on the relative variation in kσ retardation from protein site to site to serve as a probe for retardation in diffusive water (DW) dynamics near the spin label.
Figure 2.
Shows the relevant relaxation rates extracted from ODNP. A, shows the FFHS spectral density function (red), the lorentzian based rotational spectral density function (blue) and the total spectral density function calculated from Eq 5 assuming an equal weighting between rotational and FFHS spectral densities (black). The frequency in which kσ and kLow probe the total spectral density is shown in A. B, shows the approximate relationship between the retardation in kσ and the retardation in correlation time τc in black, the red dashed line indicates the diagonal, and the bulk water value is indicated with the black (x). B, shows the enhancement in kLow as a function of thef rotational correlation time and the relative weighting (n) between JRot and JFFHS indicated above the respective line.
The kLow relaxation rate is sensitive to motion on the 1/ωH (1–100’s of ns) timescale. In the hydration layer of proteins, this timescale represents water molecules that are bound to and rotating with the protein13. Thus, it is necessary to account for the rotational motion of the protein in the total spectral density function representing kLow as
| (6) |
Here, JRot is the rotational spectral density function that takes the rotation of the protein into account and n is a linear weighting factor that represents the fractional population between bound waters, rotating with the protein, and freely translating waters moving independently of the protein. It is important to note that the translational motion, corresponding to diffusive water, contributes a constant value to kLow that is invariant to changing correlation time because of the frequency that kLow probes. The spectral density function describing the rotational motion of the protein is given by , and shown in Figure 2-A by the blue dashed line alongside the FFHS spectral density function in the red dashed line. The kLow relaxation rate is enhanced on surfaces that tumble on the 1/ωH timescale and feature BW. There are two factors that lead to enhancements in kLow; the fractional amount of bound waters, n, and the timescale of the protein rotational motion, τrot. The enhancement in kLow, , is shown as a function of the protein rotational correlation time, τrot, for n = 0 → 1 in Figure 2-C. When comparing hydration water around surface sites on the same biomolecule, it is reasonable to assume that the rotational correlation time of the biomolecule, τrot, will be similar for each mutant. Thus, differences in kLow enhancement can be attributed to differences in the relative amount of BW, here n. However, similar interpretations of kLow enhancements between biomolecules with significantly different τrot would not be valid, and must be limited as a coarse read out of whether BW is present or not at the given site. To be clear, BW must be present to detect any amount of kLow enhancement.
Recall that kLow is calculated, according to Eq. 5, from the terms kσ and kρ that are directly experimentally accessible from the ODNP and nuclear spin lattice relaxation times, as given by Eqns. 7 and 825.
| (7) |
| (8) |
Here, C is the concentration of the unpaired electron of the nitroxide radical, E(p) is the proton NMR enhancement measured as a function of microwave power p, T1(p) the proton NMR T1 relaxation time measured as a function of microwave power p, S the electron spin saturation factor, ωH and ωe the proton and electron Larmor frequencies, and T1,0 the 1H NMR T1 relaxation time measured in the absence of the nitroxide radicals, and T1 in the presence. The coupling factor is the ratio of these kσ and kρ values according to Eq. 9. The correlation time (τc) is calculated from the experimentally determined ξ by interpolating Eq. 1 for a given τc and assuming the FFHS model shown in Eq. 2 as the spectral density function26.
| (9) |
MD simulations
Molecular dynamics simulations were performed starting from the crystal structure of the CheY protein (3CHY)27. Initial structures of the 5 residue peptides, corresponding to the sequence environment of the experimentally studied sites of CheY, were obtained from the same crystal structure by selecting the atoms of the respective amino acids and completing the N- and C-termini. All simulations were carried using the Gromacs 4.6.1 software package28. The proteins and peptides were modelled with the AMBER29 force field and the SPC/E model30 for water. All simulations were carried out in cubic simulation boxes with periodic boundary conditions. The particle-mesh Ewald31 algorithm was employed to compute long-ranged electrostatic interactions on a real-space periodic grid with 1.2 Å resolution. Short-ranged pair-wise interactions were treated with a real-space cut-off at 9 Å. A constant energy shift of the pairwise potentials was used to ensure zero interaction energies at the cut-off distance. Bond lengths in proteins and peptides were constrained with the LINCS32 algorithm, while the SETTLE33 algorithm was used to constrain the geometry of water molecules.
The protein was solvated with approximately 10500 water molecules, including 147 water molecules resolved in the crystal structure. The protein charge with standard protonation corresponding to a pH of 7 was neutralized with 4 sodium ions. The 5 residue peptides were solvated with approximately 2150 water molecules, and their total charge neutralized with either sodium or chloride ions.
All simulated systems were subjected to an initial energy minimization of 100 steps. This was followed by a 1 nanosecond equilibration simulation with position restraints on protein/peptide non-hydrogen atoms under isothermal/isobaric conditions at 1 bar and 300K and a subsequent 1 nanosecond equilibration without restraints. Both equilibrations were carried out with 1 femtosecond time steps combined with the Berendsen weak coupling algorithm34 for the thermostat and barostat using a 1 picosecond time constant. The equilibrations were then followed by production simulations of 100 nanosecond length in the NPT ensemble. Production simulations used a timestep of 2 femtoseconds. The Nose-Hoover thermostat35 with a reference temperature of 300K and a period time of 1.0 picoseconds was used to describe temperature coupling to an external bath separately for the protein and the surrounding solvent. The Parrinello-Rahman barostat36 with a 1 picosecond period time and a reference pressure of 1 bar was used for pressure coupling.
Local hydration water diffusion dynamics was analyzed in the vicinity of the experimentally studied sites via the mean squared displacements of water oxygens in the production simulations of the solvated CheY protein and the selected 5-residue peptides. For each experimentally studied amino acid in the protein or peptide, water molecules were selected at a given time in the trajectory within 5 Å of any sidechain atom of that residue. The translational motion of these water oxygens was then followed in time via the mean squared displacement (MSD).
| (10) |
MSD time traces were then averaged over the initial time points used to select the water oxygens for the MSD analysis. The average local hydration water MSD observed for delay times of 10 picoseconds, MSD10ps = MSD(t = 10ps), was then used to obtain a local measure of the translational diffusivity of hydration water molecules. The MSD obtained from explicit simulations inherently includes intrinsic effects on the translational mobility of hydration water molecules due to the inaccessible volume occupied by the protein, in addition to other interactions with the protein surface. The local retardation factor for water in the vicinity of a given protein/peptide sidechain was then determined by the local hydration water’s MSD over the MSD of bulk water, , at t = 10ps. The bulk water reference value was obtained from a separate bulk water simulation under equivalent conditions. This system contained 11417 water molecules, which corresponds to a system size equivalent to the simulation box used for the solvated protein. We note that the simulations of proteins and peptides were carried out in absence of the spin label or cysteine mutation to obtain information on the unaltered environment of the specific sites. This is motivated by the observation that local hydration water dynamics is only partially determined by the experimentally labeled site itself, and to a larger degree by its more extended chemical environment. This is suggested, for example, by a previous comparison between simulations of the unlabeled annexin protein with ODNP experiments that are comparable to the ones reported here19. To test for the potential impact of local modifications, we carried out an additional set of simulations for the 5-residue peptides that included the cysteine mutation.
To characterize the local hydropathy of the protein surface of CheY, we carried out an additional simulation of the solvated CheY protein in which 75 hydrophobic Lennard-Jones (LJ) particles were added. The parameters of these particles were chosen according to a previous study of Acharya et al.37 (σ = 3.855 Å and ε = 0.694 kJ/mol) that correspond roughly to a united atom representation of a methane molecule. The relative local concentration of LJ particles within 4 Å of any given protein atom, in comparison to the bulk solution, was used to determine the local excess chemical potential, , which we interpret as a measure of the local hydropathy of the protein surface37. To compare the local and bulk solution concentrations of the LJ particles, we simply divide the average number of LJ particles by the average number of water molecules in the respective reference volume, i.e. within 4 Å of a given protein atom or within the total system.
| (11) |
In this definition, negative values for correspond to an increased concentration of LJ particles compared to the bulk solution and thus to a more hydrophobic character of the protein surface site. Positive values correspond to preferred hydration and a resulting expulsion of LJ particles compared to the bulk solution and therefore a more hydrophilic character of the protein surface site. With this definition, we follow previous work by Garde and co-workers37 that utilizes the local excess chemical potential of LJ probe molecules as a reference for an approach to quantify the hydropathy of a protein surface. For a given amino acid, the average local hydropathies were computed as an average over atomic hydrophilicities obtained for the atoms of the amino acid itself, and atoms of nearby protein residues within varying distance cut-offs of R ranging from 0 to 10 Å. This procedure then resulted in the cut-off distance dependent averaged local hydropathy .
Local protein surface accessibilities were analyzed by counting the average number of water molecules within 4 Å of each protein atom, , and comparing it to the expectation value for a sphere of the same volume in bulk water, . The volume fraction excluded by the protein in the vicinity of that atom provides us with a measure of the site’s accessibility, which is dictated by the protein geometrical topology:
| (12) |
Expected values for fexcl are ≈ 0.5 in case of primarily flat surfaces, ≈ 1 for completely buried atoms and <0.5 for highly solvent-exposed atoms and/or at positive surface curvatures. For the experimentally studied sites of the CheY protein, we computed the average values for fexcl for the respective atoms of the respective amino acid.
Results and Discussion
Retardation of surface water dynamics on CheY proteins and peptides
We investigated the hydration water dynamics near the surface of a small globular protein, CheY, with site specificity. The hydration water dynamics were sampled around 10 residues located on the CheY protein surface, specifically residues 17, 37, 41, 62, 71, 80, 91, 97, 117, and 121, see Figure 1. To probe the local environment, the respective amino acid was mutated to a cysteine for spin labeling, making this a measurement of the local environment around the site, not of the respective residue. To evaluate the effect of the protein surface on hydration water dynamic properties around these 10 surface sites of CheY, we compared these to measurements of 5 residue peptides with the same primary sequence as the protein segment around the CheY site in question. In other words, the 5 residue peptides are composed of the nearest and next-nearest neighbor of each of the select 10 cysteine mutated sites of CheY, as listed in Table SI-1. The experiment was designed to separately evaluate the effects of the complex surface environment of a globular folded protein vs the average hydrophilicity given by the amino acid composition on the local hydration water dynamics around the protein surface site.
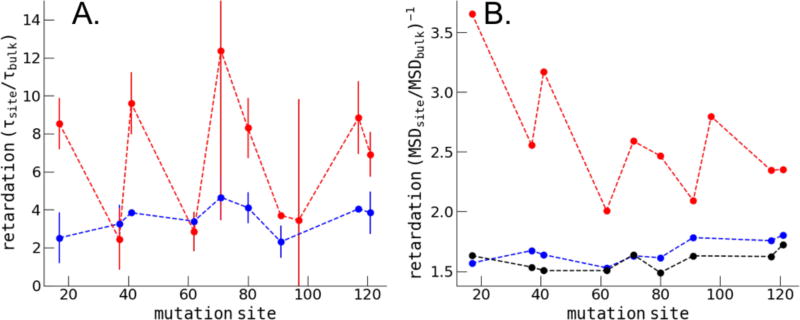
The ODNP-derived electron-1H dipolar correlation time for hydration water, τc, modulated by the diffusive dynamics of water within 5–10 Å of the respective spin label is shown in Figure 3 for the CheY protein (red) and the 5-residue peptides (blue). We observed that the surface water dynamics (in-depth discussion to follow) as reflected in the τc value measured on the CheY surface relative to bulk water is significantly varied from site to site. The τc value measured on some sites increased up to 10–12 fold relative to that of bulk water (henceforth referred to as retardation factor in correlation time of τc) while other sites reveal a modest retardation factor in τc of 2–3 fold. In contrast, the retardation factor of τc for all peptide surface sites was modest, and ranged between 2–4 fold. This trend is generally reproduced by the computationally derived retardation factor for local hydration water diffusion, as obtained from the slope of water oxygen MSD from our MD simulations (see Methods). The corresponding results are shown in Figure 3-B, for the CheY protein in red, the 5-residue peptide in blue and for the 5-residue peptide including the CYS mutation in black. The simulated retardation factor of hydration water diffusion for the CheY protein varies between 2 – 3.5, while it ranges narrowly around 1.5 for all peptides. Qualitatively the ODNP and MD simulation results agree, in that the hydration water of the protein surface displayed greater overall retardation, as well as a greater dispersion of retardation values from surface site to site of the protein compared to that of the peptides. However, the magnitude of the hydration water retardation factor reported by the ODNP-derived τc was significantly larger than found by MD simulations—an observation that will be further dissected.
Figure 3.
The hydration water retardation of the CheY protein (red) and 5-mer peptide segments (blue) as a function of the site of cysteine mutation. (A) shows the ODNP experimental results for the protein and peptide comparison, (B) show the MD simulation results for the protein and peptide comparison. MD derived retardation factors for 5-mer peptides including the cysteine mutation are shown in black.
The τc value measured by ODNP is modulated by two timescales of inter-spin motion between the hydration water and the spin label on the protein surface. We separately analyzed these timescales of inter-spin motion, and so access the dynamic properties of DW and BW. From the ODNP measurements we computed the cross-relaxation rate (kσ) and the self-relaxation rate (kLow), as described in detail in Methods (see Eq 4, 6, and 7). The cross-relaxation rate (kσ) and self-relaxation rate (kLow) are sensitive to 1–100’s of ps and 1–100’s of ns timescale motions, respectively, that occur within the 5–10 Å sphere about the nitroxide spin label. The kσ value increases monotonically with decreasing correlation time of water fluctuations, thus an increase in kσ represents an increase in the rate for translational diffusion of hydration waters. Consequently, the retardation of DW is presented as the retardation of the kσ relaxation rate, (see Methods). This correlation is valid as the kσ retardation over the range of dynamics present on the surface of the protein is approximately linear with τc retardation, as shown in Figure 2-B. While the kσ relaxation rate is strongly modulated by the timescale of motion in the 1–100’s of ps range, the kLow relaxation rate is both modulated by the timescale of biomolecule rotational motion on the 1–100’s of ns timescale and by the number of nearby BW molecules. As discussed in Methods, it is reasonable to assume that the rotational correlation time of each mutant protein is similar, so that any difference in the kLow relaxation rate can be attributed to differences in the relative population of BW. Thus, on surface sites that harbor BW, we observe an enhancement in the kLow relaxation rate relative to bulk solvent, as presented as .
Analysis of highly diffusive surface water
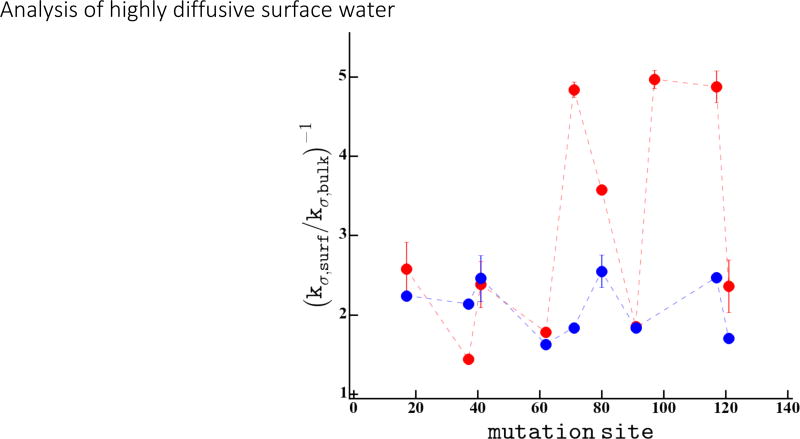
We now focus on the influence of the protein surface on the DW dynamics. We present kσ retardation, , found on the surface of the CheY protein and the 5-residue peptides in Figure 4. We found on average a 3 fold retardation with a distribution ranging between 1.5 – 5 on the CheY protein surface, while the peptide surfaces displayed a much narrower distribution of 2.3 ± 0.5 fold. The relatively modest retardation of ~3 fold for DW dynamics reported here for the CheY protein is en par with experimental values in the literature of diffusing water on protein surfaces2,6,9,10 as well as the calculated values by MD simulation in this study (Figure 3-B) and the literature.38,39
Figure 4.
kσ is shown normalized to the bulk water relaxation rate for each site measured on the CheY protein in red and for the 5-mer peptides in blue.
The observation that DW retardation on the peptide surfaces converges to a narrow range of values around ~2.3 suggests that a common property of all peptides is dictating the DW retardation. The excluded volume of each peptide is similar given their comparable molecular weights (see Table SI-1), and as such is the likely common factor that determines the average DW retardation value. Looking to the small, yet distinct, variation in DW retardation, an intuitive cause for this is the variation in hydropathy between the peptides. The average hydropathy of each peptide, calculated by the GRand Average of hydropathY (GRAVY) score (see Table SI-2), varies from −2 to 2. However, we find no correlation between the experimental DW retardation from kσ relaxation and the GRAVY score, as shown in Figure SI-1. This indicates that the hydration water properties of even simple peptide systems are not well described by an average over individual amino acid residue properties. In order to describe the local hydration water properties even qualitatively, it is necessary to take the local chemical and geometrical topologies into account40.
In contrast, the dispersion in DW retardation on the CheY protein surface is much greater than on the peptide surfaces, while the average value of 3 is not much greater than the 2.3 found for the peptides. In other words, DW dynamics is slower around some, but faster around other protein surface sites compared to that of the peptide counterparts. This variation may come from differences in local excluded volume between topologically different protein surface sites or variations in local surface hydrophilicity and chemical topology. To examine the molecular basis of the DW retardation, we compute the local surface hydrophilicity and excluded volume of the CheY protein surface by MD simulation.
MD Simulation: Local Hydrophilicity and Excluded Volume
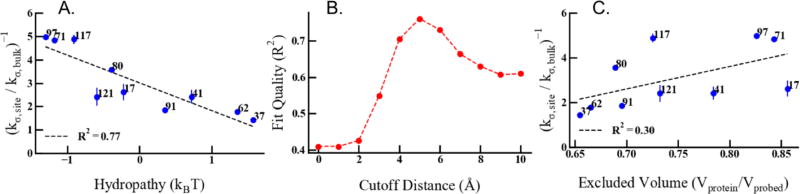
The surface hydrophilicity was determined by analyzing the local concentration of small methane-sized LJ particles (added to the simulated system at a low concentration) in the vicinity of protein surface atoms (see Methods and Eq. 11 for details). Variations in the local concentration of the LJ particles describe changes in the excess chemical potential, , which we use to quantify the local hydrophobicity. Averages are computed over atoms belonging to a respective amino acid, , and also including additional atoms in the vicinity within a cutoff-radius R, , which includes influences of the extended environment. Negative values for or correspond to hydrophobic sites with increased local concentrations of the LJ probe particles, while positive values are found for hydrophilic sites that are preferentially hydrated by water. The simulation results show that the local hydropathy of the protein surface varies roughly from −2 kBT (hydrophobic) to +2 kBT (hydrophilic), spanning the x-axis of Figure 5-A. We varied the cutoff distance for averaging over atomic hydropathies around a labelled site, and observe the strongest correlation with kσ retardation of DW when we account for contributions within 5 Å of a given site. In this case, the coefficient of determination amounts to R2 = 0.76 (see Figure 5-A). The R2 coefficient of determination is severely reduced to 0.4 when considering only local contributions of the atoms of the individual amino acids (Figure 5-B). When accounting for increasingly non-local contributions of adjacent fractions of the protein surface beyond 5 Å distances, the correlation of the surface hydrophathy with the DW retardation plateaus at R2 = 0.6 at > 8 Å. This finding indicates that properties of the protein surface, which determine chemical properties such as the effective hydropathy and the dynamic properties of hydration water, exert their main influence on a length scale of around 5 Å, a distance that corresponds roughly to the diameter of two water molecules.
Figure 5.
(a) shows the correlation between surface hydrophilicity shown on the x-axis with the DW represented by kσ retardation on the y-axis. (b) shows the quality of the fit between DW retardation and surface hydrophilicity shown in (a) as R2 as a function of the cutoff distance for averaging atomic hydropathies at the surface site. (c) shows the correlation between excluded volume on the protein surface and DW retardation.
The correlation reveals that DW is least retarded in the vicinity of hydrophilic surfaces and most retarded on hydrophobic surfaces. This might seem counter-intuitive as hydrophilic surfaces are expected to form hydrogen bonds with water molecules that are associated with the slowdown in water dynamics39. On the other hand, water molecules can form clathrate-like structures around hydrophobic residues with a high surface curvature41, which may also result in a slowdown. However, on low surface curvatures, i.e. flat hydrophobic surfaces, increasing numbers of defects in the water hydrogen bond network with increasing hydrophobicity should result in an increase in water dynamics42,43. Neither of these mechanisms directly apply to a protein surface that displays heterogeneous topological and chemical features, making our finding a non-obvious result. We note that the average local hydropathy determined from our simulations is derived from the preferential partitioning of hydrophobic probe particles, and therefore does not simply rely on the presence of polar or non-polar groups. Instead, combined effects due to the geometrical and chemical topology of the protein surface are captured empirically, which is apparent from the absence of any correlation between the computed hydrophilicity values and the chemical nature of the central amino acid sidechain for a selected site.
The experimentally derived DW retardation and the computed local excluded volume around the CheY protein surface sites are plotted in Figure 5-C, where we find a weak to negligible correlation between kσ and the surface excluded volume with R2 = 0.30. In other words, variation in DW retardation shows no clear dependence on the local excluded volume determined by the protein surface geometrical topology, i.e. surface curvature and accessibility. This result does not imply that the excluded volume of the protein surface has no effect on DW retardation. Instead, the variation in the local protein’s geometric surface topology is not sufficient to determine the variation in DW dynamics, and additional chemical information needs to be taken into account. These effects are collectively and inherently included in our empirical hydropathy measure that hence provides a significantly better predictor of local DW dynamics, with faster DW dynamics found in the vicinity of hydrophilic, i.e. preferentially hydrated, regions of the protein surface and slower DW dynamics in the vicinity of hydrophobic surface sites.
Importantly, kσ does not probe the translational diffusion of one water molecule, but rather the average translational diffusion of several waters within a 5–10 Å sphere around the nitroxide oxygen. This sphere includes a number of water molecules that are within the first, second and even third hydration layer on the surface of the protein. This implies that the DW dynamics probed by kσ that is sensitive to the strength of the extended hydrogen bond network in the hydration layer, not merely a single water molecule most adjacent to and directly interacting with the protein surface site, is systematically slowed with increasing hydrophobicity. Overall, our result is consistent with a clathrate-like behavior in which water molecules adjacent to hydrophobic surface sites form stronger hydrogen bonds to the neighboring water molecules of the hydration layer than the hydration waters adjacent to hydrophilic surface sites. This effect has been studied by Raman spectroscopy and MD simulation on small hydrophobic solutes that find increased water ordering around hydrophobic solutes compared to hydrophilic solutes41,44. However, it was unknown to date what to expect from hydrophobic sites on net hydrophilic protein surfaces in equilibrium solution state.
Survey of highly diffusive surface water on other biological systems
Next, we examined other biomolecular surfaces with the goal of exploring the possible role that DW plays in specific binding. We analyzed the DW on a range of surfaces that can be separated into two categories: ones that take part in highly specific binding, such as folded globular proteins, and ones that tend to take part in less specific binding, such as intrinsically disordered proteins (IDPs), small peptides, and liposomes. Here, we analyzed the DW retardation on the folded globular protein surfaces of Annexin XII (Anx)19 and CheY, the intrinsically disordered proteins of α-Synuclein (α-Syn)18 and ΔTau18717, CheY-inspired 5-residue peptides, poly proline peptides with systematically varied charges (discussed in SI), and model LUV liposome surfaces made of DPPC and DOPC20. The description for each system is found in Methods. While ΔTau-187 and α-synuclein also participate in specific binding, the binding region extends over a significant portion of the IDP and/or these systems adopt a defined structure either just before binding or after binding to its target has occurred. Thus ΔTau-187 and α-synuclein in the disordered solution state does not represent the structure that participates in site-specific binding.
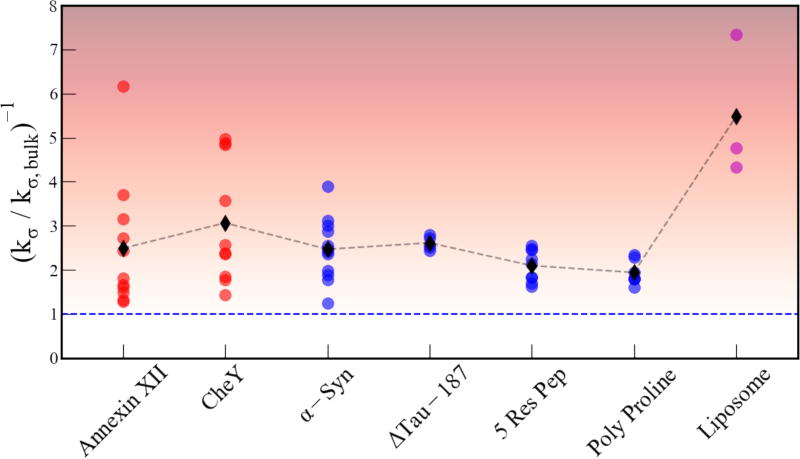
The surface DW dynamics for each system was evaluated by the DW retardation as expressed in and shown in Figure 6. Each data point along a vertical line from the axis represents a different location on the respective surface. The globular proteins CheY and Anx displayed the largest distribution in DW retardation factors ranging from 1–6. The IDP α-Synuclein also had a relatively large distribution ranging from 1 – 4. The other IDP ΔTau187 displayed a narrow range of DW retardation around 2.7 ± 0.3, which is similar to that of the CheY-derived 5-residue peptides that displayed values 2.1 ± 0.5 around and the poly proline-based peptides of around 2.1 ± 0.4. Even so, the average DW dynamics was found to be rather similar for any of these protein and peptide-based surfaces with the average retardation factor, , falling between 2–3.
Figure 6.
The kσ retardation for each system studied. Each data point corresponding to a given system represents the value at a given site on the surface for the given system. The color shading represents fast or bulk like water as white and the maximally retarded water as black. The blue dashed line indicates the bulk value.
To evaluate whether there is a dominant effect of charges on the DW dynamics on surfaces, we test poly proline-based peptides with systematically added charged residues. The sequences studied were CK, CD, CPPP, CPPPK, CPPPD, CPPPPPK, and CPPPPPD, where the lysine (K - positively charged) or aspartate (D – negatively charged) residue was systematically moved further from the spin label attached to the cysteine residue by the addition of proline residues. The DW retardation for each peptide was compared to the GRAVY score for each peptide and the composition of each peptide, as shown in Figure SI-1. We do not observe any consistent trend between DW retardation and the GRAVY score or the net charge of each peptide. This implies that the variation in local charges on a disordered peptide surface does not exert any persistent modulation in the DW dynamics. The effects of charges on structured protein surfaces may be highly different.
Compared to IDP and peptide surfaces the LUV liposome surfaces showed significantly greater average DW retardation factors with ~4.5 on DOPC, ~ 5 on DPPC, and ~7.5 on mixed DOPC/DPPC liposomes. While the three values are co-plotted under the common categorization as “liposomes”, the DW retardation for a given LUV composition is uniform as there are no site-specific variations on the synthetic LUV surface studied here. What is clear is that the DW retardation on the liposome surfaces is significantly larger than that of the peptide and protein surfaces, including that of the globular proteins.
As introduced earlier, the retardation of DW dynamics can be viewed as reflecting on the enthalpic cost for disrupting and displacing the hydration water that is collectively strengthened by the attractive interaction between the protein surface and the hydrogen bonds of the local water network. When using DW retardation as a proxy in this way, the broad distribution of DW retardation observed on Anx and CheY surfaces suggests that some sites have a high enthalpic cost (most retarded) and other sites have a low enthalpic cost (least retarded) for displacing the local hydration water. In contrast, ΔTau187, the CheY peptides and the liposomes display narrow distributions in DW retardation, implying that displacing hydration waters has an approximately equal enthalpic cost for any position on these biomolecular surfaces. It is interesting to note that α-Syn—characterized under conditions known to populate an IDP state—displays a rather broad distribution in DW retardation, which may indicate that this IDP tends to populate a more persistent residual or transient solution state secondary structure and is prone to participate in specific binding, as has been postulated45.
Recall that we found DW retardation in kσ to inversely correlate with the local hydrophilicity of the protein surface sites (Figure 5-A). This would suggest that the enthalpic cost for disrupting and displacing the collective hydration water layer is higher near the more hydrophobic protein sites. The other finding is that the enthalpic cost for disrupting and displacing the collective hydration water layer of, and thus the enthalpic cost for solutes to approach, the liposome surface is by far greater compared to the cost for solutes to approach and bind to protein or peptide surfaces. In other words, liposome surfaces display greater protection from solute interactions than protein or peptide surfaces.
Analysis of bound surface water
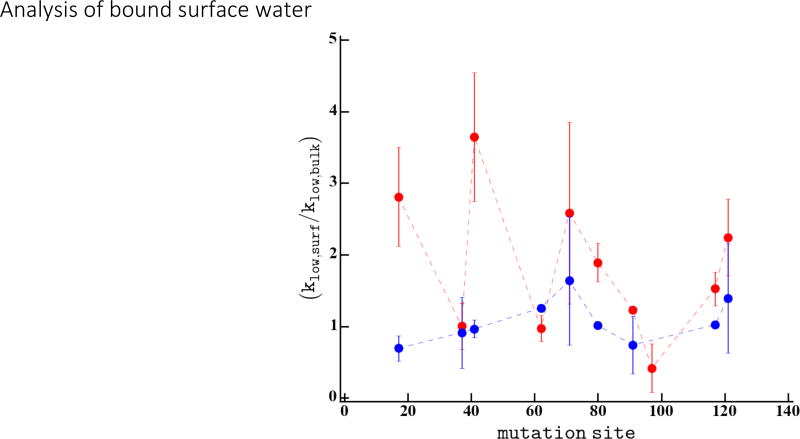
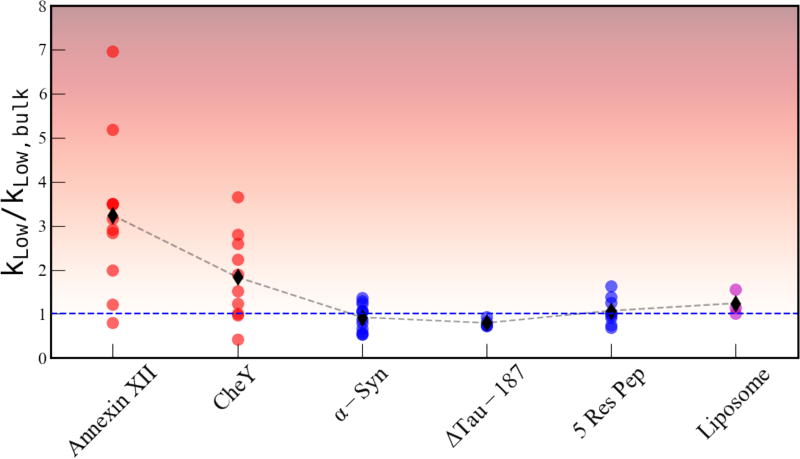
Next, we focus on the influence of the protein surface on the BW population and dynamics, as measured by the kLow relaxation rate. As introduced in Methods, the kLow relaxation rate is enhanced by inter electron (spin label)-nuclear (water proton) spin fluctuations on the 1–100’s of ns timescale. The total enhancement in the kLow relaxation rate is a function of the number of water molecules fluctuating on the 1–100’s of ns timescale and the correlation time of each fluctuation (see Methods, Eq. 5). As discussed in the Methods section, we limit our interpretation of the kLow enhancement to a measure of the increase in bound water population, as expressed as an enhancement of kLow, . The variation in kLow relaxation enhancement measured on the CheY protein surface (red) and the 5 residue CheY peptides (blue) is shown in Figure 7. The kLow enhancement on the surface of CheY varies from 1 – 4, where many sites have a kLow enhancement above 2, indicating the presence of bound water at these sites. In contrast, the 5 residue peptides all have a negligible enhancement (1±0.5) in the kLow relaxation rate. The negligible kLow enhancement may be due to the absence of BW population or the rapid tumbling time of the peptide system which prevents the detection of slow BW water.
Figure 7.
The kLow enhancement, the site specific kLow relaxation rate normalize to the bulk water relaxation rate, is shown for each site measured on the CheY protein in red and for the 5-mer peptide segments in blue.
Comparison to MD simulation
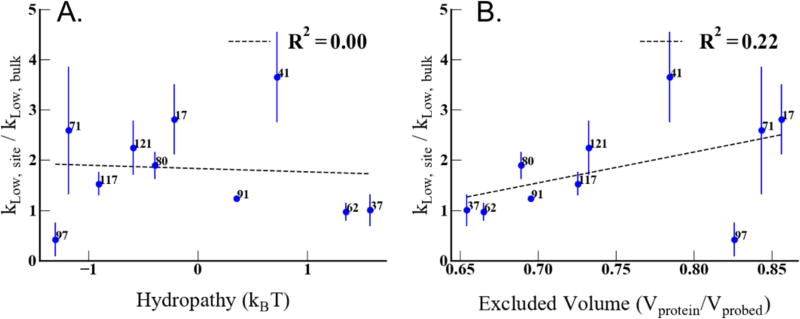
Finally, we compare the presence of BW on the CheY protein surface to the estimated surface hydrophilicity and excluded volume, as calculated from MD simulation, in Figure 8. The correlation between kLow and the surface hydropathy is non-existent, with an R2 = 0.00 (Figure 8-A). There is a weak to negligible correlation between kLow and the excluded volume of the protein surface, with an R2 = 0.22 (Figure 8-B). The lack of any significant correlation between kLow and the surface hydropathy or excluded volume does not necessarily imply that the BW population is not modulated by the properties of the protein surface. It is more likely that the BW population is determined by a combination of factors modulated by surface chemistry and geometry.
Figure 8.
(a) shows the correlation between surface hydrophilicity shown on the x-axis with the BW represented by kLow enhancement on the y-axis. (b) shows the correlation between the excluded volume of the surface sites and the BW represented as kLow enhancement.
Survey of bound surface water on other biological systems
We next examine the presence and variation in kLow enhancements on the other biomolecular surfaces that were characterized earlier in terms of kσ retardation. The relaxation enhancement in kLow for each system was normalized to the respective bulk water values, as shown in Figure 9. The two globular proteins, Anx and CheY showed significant kLow enhancements, ranging from 7 to 1 with an average of ~3 for Anx and ranging from 4 to 0.5 with an average of ~2 for CheY. The IDPs, ΔTau187 and α–Syn, and the 5-residue peptides showed negligible enhancements in kLow, with all three systems averaging around 1 within error (± 0.5) of the bulk water value for kLow. It is important to note that both ΔTau187 and α-Syn are large enough and tumble with a slow enough correlation time (τrot > 1 ns)46,47 to produce an enhancement in kLow. Thus, the lack of enhancement in kLow on either of these IDP surfaces shows that the BW population must indeed be absent. The liposome surfaces also showed kLow values around 1 as for bulk water. Taken together, our data shows that only the two globular protein surfaces of Anx and CheY display significant kLow enhancements, and thus bear significant BW populations. It is noteworthy that both Anx and CheY in their native state take part in specific binding interactions48,49. Our data shows that the most significant and clear-cut differences between folded vs intrinsically disordered proteins is the presence vs lack of BW harbored on the biomolecular surface. It is also noteworthy that liposome surfaces lack contributions from BW entirely, which in fact is consistent with the conclusions of the seminal study by Bryant and coworkers by NMR relaxation dispersion50. Given that the kLow enhancement is an experimental variable, not typically used to characterize BW population, finding consistency between different measurement methods and across different literature report is highly valuable. Uniquely, the measurement of kLow enhancements by means of ODNP provides a site-specific view into the presence of BW.
Figure 9.
The kLow relaxation rate for each system studied shown relative to the value of kLow in the respective bulk solvent. The color shading represents fast or bulk like water as white and the maximally enhanced ns water motion as black. The blue dashed line indicates the bulk value.
Discussion
This study demonstrates that the freely exchanging surface water translational diffusivity is strongly modulated by the surface of proteins in dilute solution state and under ambient solution condition. This is shown by the significant spatial variation in DW retardation factors found on the surface of the globular proteins, CheY and Anx, in contrast to the relatively homogeneous distribution in DW retardation found on CheY peptides, polyproline-based peptides and ΔTau187. The comparison of DW dynamics measured on the CheY surface to the surface hydrophilicity and excluded volume calculated by MD simulation suggest that the DW dynamics are modulated much more significantly by variation in the average local surface hydrophilicity, and not by the variation in the local excluded volume as determined by surface geometrical topology. We discover that DW is systematically retarded with increasing hydrophobicity and decreasing hydrophilicity around local sites on the CheY protein surface. This is in contrast to what one would expect if the surface water retardation was simply due to the average chemistry of the individual nearby residues, in the absence of geometrical or chemical topology or second order solution effects. Our result agrees with what one would expect from considerations of surface clathrates where the water hydrogen bond network is stronger around hydrophobic solutes, leading to more structure and greater retardation in the water network near hydrophobic solutes compared to hydrophilic solutes41,44.
Efforts to rationalize the factors of the protein surface that modulate the relatively fast (ps and below) hydration water dynamics have been made both experimentally6 and by MD simulations38,39, but simple rules did not emerge. Laage and co-workers offer an in-depth analysis showing that the majority of hydration water molecules is retarded due to the excluded volume of the surface, suggesting that the average retardation observed on the peptide and protein surface is primarily due to the excluded volume. Laage and co-workers also find that the rate of breaking and reforming of the hydrogen bond between a water molecule and the protein surface is modulated by the strength of that specific hydrogen bond. This leads to the conclusion that the stronger the hydrogen bond between a residue on the protein surface and a hydration water, the slower is the rate of the hydrogen bond breaking and reforming process. At first glance, this contradicts the findings in this study. However, it is likely that this apparent discrepancy with our finding of more retarded DW dynamics around hydrophobic surfaces is due to the difference in the types of water motion and population probed. Laage and co-workers track the femtosecond timescale breaking and reforming of individual hydrogen bonds, while our study tracks the translational exchange of a hydration water ensemble within a 5 – 10 Å radius sphere on a picosecond timescale. DW dynamics measured by ODNP will not depend on the strength of any single hydrogen bond between a water molecule and a surface residue, but reflects on the strength of the hydrogen bonds in the extended hydration water network around the local site of interest. The range of ODNP measurements thus includes both water-water and water-protein hydrogen bonds, encompassing up to three layers of water molecules.
Finally, we find that BW is present on folded protein surfaces of CheY and Anx and absent on IDPs, including peptides, as well as on liposome surfaces. We find that there is no measurable correlation between the BW present on the protein surface to the hydrophilicity and excluded volume of the protein surface. Likely, the factors that govern the presence of BW must lie in the three-dimensional fold of the protein surface, while the molecular and structural determinants of the population of BW remain to be uncovered. It is important to note that we do not observe any correlation between DW retardation and BW populations, shown in Figure SI-2. This implies that not only are the two timescales of motion decoupled, but so is the origin for the variation in DW and BW contributions.
Conclusion
The contributions from and variation in DW and BW are thought to play a key role in protein ligand interactions through the modulation of the local thermodynamics of the interaction. Specific binding requires that some locations on the protein surface become thermodynamically more favorable for ligand binding than other locations on the surface. While the connection between the diffusive and bound water dynamics and the relevant enthalpic cost and entropic gain associated for expelling surface water is not clearly delineated and understood to date, our study finds that both DW and BW waters display significant spatial heterogeneity. This demonstrates that both bound and diffusive water dynamics are potentially effective modulators of specific and localized interactions involving the protein. This is further corroborated by our discovery that the most significant difference between structured proteins that engage in highly specific binding events and intrinsically disordered proteins that do not is the presence or absence of spatial variation in diffusive surface water dynamics and the presence or absence of bound surface water populations.
Supplementary Material
Acknowledgments
This work is supported by the 2011 NIH Innovator award, the NSF award #CHE 1505038 awarded to S. Han, and by the DFG, Cluster of Excellence RESOLV (EXC 1069). This project made use of the UCSB MRL Shared Experimental Facilities, which are supported by the MRSEC Program of the National Science Foundation under award NSF DMR 1121053; a member of the NSF-funded Materials Research Facilities Network (www.mrfn.org). Molecular graphics and analyses were performed with the UCSF Chimera package. Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311).
References
- 1.Ball P. Chem. Rev. 2008;108(1):74. doi: 10.1021/cr068037a. [DOI] [PubMed] [Google Scholar]
- 2.Ebbinghaus S, Kim SJ, Heyden M, Yu X, Heugen U, Gruebele M, Leitner DM, Havenith M. Proc. Natl. Acad. Sci. 2007;104(52):20749. doi: 10.1073/pnas.0709207104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cui D, Ou S, Patel S. Proteins Struct. Funct. Bioinforma. 2014;82(12):3312. doi: 10.1002/prot.24683. [DOI] [PubMed] [Google Scholar]
- 4.Oleinikova A, Smolin N, Brovchenko I, Geiger A, Winter R. J. Phys. Chem. B. 2005;109(5):1988. doi: 10.1021/jp045903j. [DOI] [PubMed] [Google Scholar]
- 5.Armstrong BD, Choi J, López C, Wesener DA, Hubbell W, Cavagnero S, Han S. J. Am. Chem. Soc. 2011;133(15):5987. doi: 10.1021/ja111515s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qiu W, Kao Y-T, Zhang L, Yang Y, Wang L, Stites WE, Zhong D, Zewail AH. Proc. Natl. Acad. Sci. U. S. A. 2006;103(38):13979. doi: 10.1073/pnas.0606235103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang L, Yang Y, Kao Y-T, Wang L, Zhong D. J. Am. Chem. Soc. 2009;131(30):10677. doi: 10.1021/ja902918p. [DOI] [PubMed] [Google Scholar]
- 8.King JT, Arthur EJ, Brooks CL, Kubarych KJ. J. Phys. Chem. B. 2012;116(19):5604. doi: 10.1021/jp300835k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mattea C, Qvist J, Halle B. Biophys. J. 2008;95(6):2951. doi: 10.1529/biophysj.108.135194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nucci NV, Pometun MS, Wand AJ. Nat. Struct. Mol. Biol. 2011;18(2):245. doi: 10.1038/nsmb.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hausser K, Stehlik D. Advances in Magnetic Resonance. 1968;3:79–139. [Google Scholar]
- 12.Kaminker I, Barnes R, Han S. Methods in Enzymology. In: Warncke PZQ, Electron K, editors. Paramagnetic Resonance Investigations of Biological Systems by Using Spin Labels, Spin Probes, and Intrinsic Metal Ions, Part B. Vol. 564. Academic Press; 2015. pp. 457–483. [DOI] [PubMed] [Google Scholar]
- 13.Franck JM, Ding Y, Stone K, Qin PZ, Han S. J. Am. Chem. Soc. 2015;137(37):12013. doi: 10.1021/jacs.5b05813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Columbus L, Hubbell WL. Trends Biochem. Sci. 2002;27(6):288. doi: 10.1016/s0968-0004(02)02095-9. [DOI] [PubMed] [Google Scholar]
- 15.Mo G, Zhou H, Kawamura T, Dahlquist FW. Biochemistry (Mosc.) 2012;51(18):3786. doi: 10.1021/bi300147m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. J. Comput. Chem. 2004;25(13):1605. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 17.Pavlova A, Cheng C-Y, Kinnebrew M, Lew J, Dahlquist FW, Han S. Proc. Natl. Acad. Sci. 2016;113(2):E127. doi: 10.1073/pnas.1504415113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng C-Y, Varkey J, Ambroso MR, Langen R, Han S. Proc. Natl. Acad. Sci. 2013;110(42):16838. doi: 10.1073/pnas.1307678110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fisette O, Päslack C, Barnes R, Isas JM, Langen R, Heyden M, Han S, Schäfer LV. J. Am. Chem. Soc. 2016 doi: 10.1021/jacs.6b07005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng C-Y, Song J, Pas J, Meijer LHH, Han S. Biophys. J. 2015;109(2):330. doi: 10.1016/j.bpj.2015.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Franck JM, Sokolovski M, Kessler N, Matalon E, Gordon-Grossman M, Han S, Goldfarb D, Horovitz A. J. Am. Chem. Soc. 2014;136(26):9396. doi: 10.1021/ja503501x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Armstrong BD, Lingwood MD, McCarney ER, Brown ER, Blümler P, Han S. J. Magn. Reson. 2008;191(2):273. doi: 10.1016/j.jmr.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 23.Abragam A. Phys. Rev. 1955;98(6):1729. [Google Scholar]
- 24.Hwang L, Freed JH. J. Chem. Phys. 1975;63(9):4017. [Google Scholar]
- 25.Franck JM, Pavlova A, Scott JA, Han S. Prog. Nucl. Magn. Reson. Spectrosc. 2013;74:33. doi: 10.1016/j.pnmrs.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Armstrong BD, Soto P, Shea J-E, Han S. J. Magn. Reson. 2009;200(1):137. doi: 10.1016/j.jmr.2009.05.013. [DOI] [PubMed] [Google Scholar]
- 27.Volz K, Matsumura P. J. Biol. Chem. 1991;266(23):15511. doi: 10.2210/pdb3chy/pdb. [DOI] [PubMed] [Google Scholar]
- 28.Hess B, Kutzner C, van der Spoel D, Lindahl E. J. Chem. Theory Comput. 2008;4(3):435. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 29.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. J. Am. Chem. Soc. 1995;117(19):5179. [Google Scholar]
- 30.Berendsen HJC, Grigera JR, Straatsma TP. J. Phys. Chem. 1987;91(24):6269. [Google Scholar]
- 31.Darden T, York D, Pedersen L. J. Chem. Phys. 1993;98(12):10089. [Google Scholar]
- 32.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. J Comput Chem. 1997;18:18. [Google Scholar]
- 33.Miyamoto S, Kollman PA. J. Comput. Chem. 1992;13(8):952. [Google Scholar]
- 34.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. J. Chem. Phys. 1984;81(8):3684. [Google Scholar]
- 35.Nosé S. Mol. Phys. 1984;52(2):255. [Google Scholar]
- 36.Parrinello M, Rahman A. J. Appl. Phys. 1981;52(12):7182. [Google Scholar]
- 37.Acharya H, Vembanur S, Jamadagni SN, Garde S. Faraday Discuss. 2010;146(0):353. doi: 10.1039/b927019a. [DOI] [PubMed] [Google Scholar]
- 38.Pizzitutti F, Marchi M, Sterpone F, Rossky PJ. J. Phys. Chem. B. 2007;111(26):7584. doi: 10.1021/jp0717185. [DOI] [PubMed] [Google Scholar]
- 39.Fogarty AC, Laage D. J. Phys. Chem. B. 2014;118(28):7715. doi: 10.1021/jp409805p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chong S-H, Ham S. Angew. Chem. Int. Ed. 2014;53(15):3961. doi: 10.1002/anie.201309317. [DOI] [PubMed] [Google Scholar]
- 41.Chandler D. Nature. 2005;437(7059):640. doi: 10.1038/nature04162. [DOI] [PubMed] [Google Scholar]
- 42.Weiß RG, Heyden M, Dzubiella J. Phys. Rev. Lett. 2015;114(18):187802. doi: 10.1103/PhysRevLett.114.187802. [DOI] [PubMed] [Google Scholar]
- 43.Niehues G, Heyden M, Schmidt DA, Havenith M. Faraday Discuss. 2011;150(0):193. doi: 10.1039/c0fd00007h. [DOI] [PubMed] [Google Scholar]
- 44.Davis JG, Gierszal KP, Wang P, Ben-Amotz D. Nature. 2012;491(7425):582. doi: 10.1038/nature11570. [DOI] [PubMed] [Google Scholar]
- 45.Wang C, Zhao C, Li D, Tian Z, Lai Y, Diao J, Liu C. Front. Mol. Neurosci. 2016;9 doi: 10.3389/fnmol.2016.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee JC, Langen R, Hummel PA, Gray HB, Winkler JR. Proc. Natl. Acad. Sci. U. S. A. 2004;101(47):16466. doi: 10.1073/pnas.0407307101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Theillet F-X, Binolfi A, Bekei B, Martorana A, Rose HM, Stuiver M, Verzini S, Lorenz D, van Rossum M, Goldfarb D, Selenko P. Nature. 2016;530(7588):45. doi: 10.1038/nature16531. [DOI] [PubMed] [Google Scholar]
- 48.Patel DR, Jao CC, Mailliard WS, Isas JM, Langen R, Haigler HT. Biochemistry (Mosc.) 2001;40(24):7054. doi: 10.1021/bi010345+. [DOI] [PubMed] [Google Scholar]
- 49.Dyer CM, Dahlquist FW. J. Bacteriol. 2006;188(21):7354. doi: 10.1128/JB.00637-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hodges MW, Cafiso DS, Polnaszek CF, Lester CC, Bryant RG. Biophys. J. 1997;73(5):2575. doi: 10.1016/S0006-3495(97)78286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.