Abstract
Precision medicine is an integrative approach to cardiovascular disease prevention and treatment that considers an individual’s genetics, lifestyle, and exposures as determinants of their cardiovascular health and disease phenotypes. This focus overcomes the limitations of reductionism in medicine, which presumes that all patients with the same signs of disease share a common pathophenotype and, therefore, should be treated similarly. Precision medicine incorporates standard clinical and health record data with advanced panomics (i.e., genomics, transcriptomics, epigenomics, proteomics, metabolomics, microbiomics) for deep phenotyping. These phenotypic data can then be analyzed within the framework of molecular interaction (interactome) networks to uncover previously unrecognized disease phenotypes, relationships between diseases, and select pharmacotherapeutics or identify potential protein-drug or drug-drug interactions. In this review, we discuss the current spectrum of cardiovascular health and disease, population averages and the response of extreme phenotypes to interventions, and population-based versus high-risk treatment strategies as a pretext to understanding a precision medicine approach to cardiovascular disease prevention and therapeutic interventions. We also consider the search for resilience and Mendelian disease genes, and argue against the theory of a single causal gene/gene product as a mediator of the cardiovascular disease phenotype as well as an Erlichian ‘magic bullet’ to solve cardiovascular disease. Finally we detail the importance of deep phenotyping and interactome networks, and the use of this information for rational polypharmacy. These topics highlight the urgent need for precise phenotyping to advance precision medicine as a strategy to improve cardiovascular health and prevent disease.
Subject Terms: cardiovascular disease, genetics
Keywords: genomics, interactome networks, polypharmacy, population medicine, precision medicine, systems biology
“It is because we are not exact that we fail.” Paul Ehrlich, 1896 1.
Over the past 50 years, progress towards the eradication of cardiovascular disease has been achieved through the adoption of lifestyle modifications, including dietary, tobacco, and exercise interventions, as well as evidence-based therapies that aim to modify a recognizable and commonly shared cardiovascular or at-risk phenotype. Despite the success of this approach, the related issues of disease prevention and cure have been elusive, presumably because of imprecise deep phenotyping of individuals needed to characterize sub-groups of disease. The magnitude of the problem remains considerable. At present, there are 92.1 million adults (>1 in 3) in the United States who have been diagnosed with cardiovascular disease, with a projection that by 2030, at least 44% of the adult population will have this diagnosis 2. While death rates attributable to cardiovascular disease have declined ~25% from 2004 to 2014 in the United States, cardiovascular disease remains the leading cause of death worldwide accounting for ~32% of all global deaths, with the expectation that this number will rise to > 23.6 million deaths annually by 2030 3, 4. The contribution of attendant comorbidities and traditional risk factors (e.g., hyperlipidemia, hypertension, diabetes mellitus, metabolic syndrome, chronic kidney disease), health behaviors (smoking and tobacco use, physical inactivity, nutrition, overweight and obesity), and other immutable factors (family history) to morbidity and mortality is understood and likely underlies the sex-, age-, race-, ethnic-, regional- and economic-based differences in disease burden (reviewed in 4). A recent meta-analysis stressed the importance of these comorbidities to disease risk. This study, which included data from 9 prospective cohort studies that followed 12,878 individuals, found that working towards optimal cardiovascular health metrics to achieve the greatest possible benefit was associated with a decrease in the risk for major adverse cardiac events, including cardiovascular mortality (RR: 0.25; 95% CI: 0.10–0.63) 5. Taken together, there is broad heterogeneity in the clinical profiles and outcomes of individuals with cardiovascular disease, thereby highlighting the urgent need for precision phenotyping.
The current approach to reducing cardiovascular morbidity and mortality in at-risk individuals or those with established disease is based on a traditional reductionist approach and uses a multi-tiered system with input from patients, the physician and the medical system, and evidence-based medicine at large. The first layer involves patient identification of symptoms that represent a deviation from normal or baseline. This report initiates entry into the medical system where contact with a physician results in a personalized assessment of the physical signs of disease and implementation of evidence-based therapies whose efficacy is supported by results from clinical trials. While this paradigm of care can ameliorate symptoms and affect disease progression, this outcome is not always certain, especially when targeting a chronic complex illness like atherothrombotic cardiovascular disease that does not have a single root cause. To address this issue, it is necessary to understand the totality of cardiovascular disease at a granular and integrative molecular level.
While all patients are fundamentally unique, reductionism in medicine presupposes that patients with common signs and symptoms share the same disease pathophenotype and, therefore, will respond similarly to medical, procedural, and/or behavioral interventions tested in aggregates of like individuals 6–8. It also places the focus squarely on treatment of established cardiovascular disease without addressing health, prevention, timing of disease inception, or cure and eradication 9. Owing to advances in panomics (genomics, transcriptomics, epigenomics metabolomics, proteomics, microbiomics), i.e., technologies and data analysis that provide in-depth clinical, biological, and molecular phenotyping, there is growing awareness that this conventional approach may be an overly simplistic view of the multiple contributors to and complexity of an individual’s cardiovascular disease phenotype. Similarly, it is now understood that other factors, such as exposure to the natural, personal, and social environments, contribute to an individual’s highly personalized disease phenotype (reviewed in 10). Integration of this large body of data points lends itself towards a more exacting (patho)phenotype, one that is amenable to precision medicine.
Why precision medicine? Precision medicine represents a new strategy in the approach to care by targeting prevention and treatment while considering individual differences in genetics, exposures, and lifestyle and health factors that are determinants of a person’s disease phenotype (Figure 1). The goal of precision medicine is to identify optimal care for an individual based on a unique personal profile rather than that of the average population. The power of precision medicine lies in the data and requires the synthesis of rapidly changing datasets, ranging from standard clinical, imaging, and laboratory testing to next-generating sequencing, metabolomics, and proteomic studies, to historical health record data. Data derived from these sources may also be analyzed using advanced systems biology and network analytical methods to uncover new and unbiased relationships between health and disease factors. The optimal system will also evolve rapidly, learn, and be facile enough to be useful for prevention, diagnosis, and treatment across a broad range of cardiovascular health factors, risk factors, and diseases. The evolution of precision medicine and its application to cardiovascular disease holds the promise of improving health as well as revolutionizing prevention and treatment options similar to what has occurred in the field of oncology.
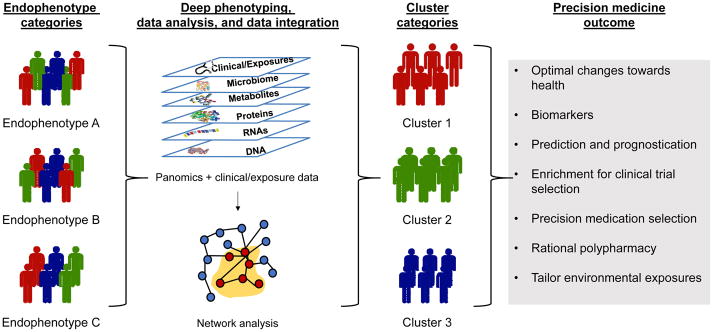
Figure 1. A precision medicine approach to phenotyping.
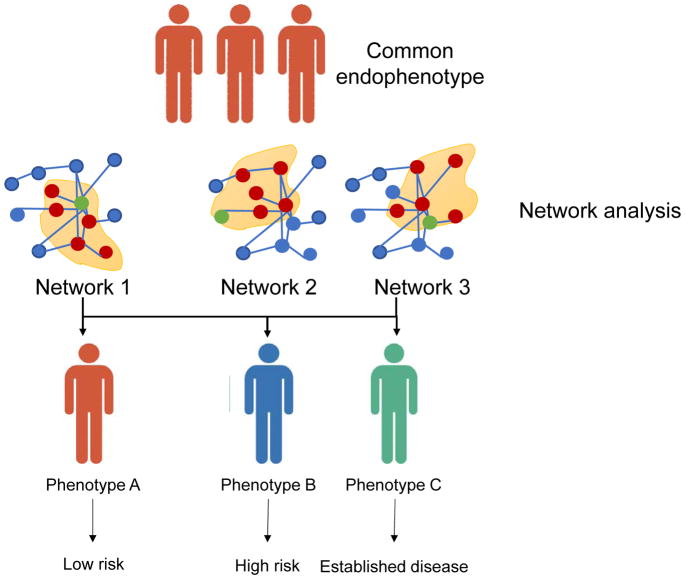
Individuals may have a similar endophenotype but be biologically distinct and have different disease profiles. Using a precision medicine approach, individuals undergo deep phenotyping with data analysis performed using network analysis. This analytical strategy clusters individuals into groups that are different from those based on endophenotype alone. This methodology can be used to optimize medication and behavioral changes to improve health, predict and prognosticate disease, identify biomarkers for disease, enrich clinical trial enrollment, and optimally tailor exposures.
Population averages and probability density function
To move precision medicine into the cardiovascular arena, it is necessary to identify and adopt parameters that reflect what is a considered the normal or ideal range (Figure 2A). This is especially important given the diversity in population age and ethnicity where ‘normal’ standards may change over time. Within the general population, there is variability with some individuals who have ideal health, others who have recognizable disease risk factors, and those who have established disease. Within groups, the transition between these health states tends to be less clear with there generally being a continuum with overlap between categories 9. Examples of this point are blood pressure and blood cholesterol levels. In each case, there are individuals with normal levels who are considered healthy, those with increased levels in the absence of cardiovascular disease, those with high levels and disease, as well those who do not fit into any clear category and are at an overlap of states 9.
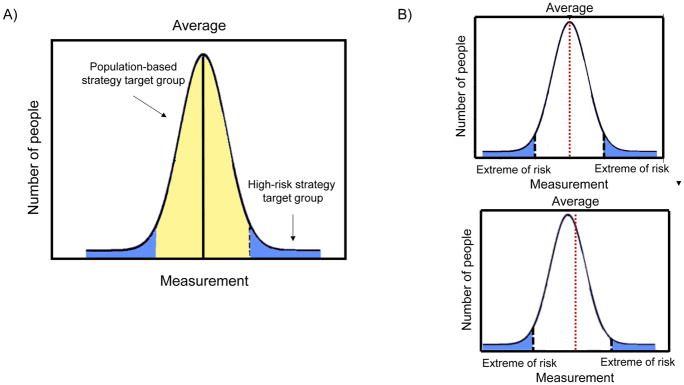
Figure 2. Population distribution and effect of changing the population mean.
A) Within a population, the majority of persons express a phenotype that centers around a mean with a minority of individuals who exhibit an extreme or “high-risk” phenotype. B) A change in the population average affects the number of individuals who are considered to be at the extremes. This may also influence the number of individuals who reach a pre-defined threshold for initiating treatment.
In clinical trials, this continuum of disease is often simplified with the assumption that all participants have the same phenotype, which can be represented using a typical bell-shaped curve with a discernable median measurement. Individuals entered in the clinical trial are considered based on the variable of interest, such as their cholesterol level, blood pressure, or history of myocardial infarction, and not their complex phenotype. This oversimplification allows results to be described in terms of an average finding on a per group basis. While this paradigm has been the norm, the hazard of assuming similarity is that only relatively large differences may be detected as significantly different and the actual range of responses may be undetected 9. This was concept was illustrated when patients with chronic heart failure were “re-phenotyped” using cluster analysis. This analysis revealed significant heterogeneity between patients with individuals segregating into groups that would not have been predicted based on their initial clinical phenotype 11. In precision medicine, similarity in phenotypes is not assumed for the purpose of studying a population behavior or response. Instead, the goal is to deviate from population averages as the metric and focus on the individual’s unique phenotype with an eye towards identifying a response to treatment as one that moves the individual from disease to health and those at health to ideal health 9.
Responses of the “tails” of the distribution and implications
The importance of phenotyping and identifying an ideal level of health within community-based populations has implications for defining what constitutes a high-risk phenotype, or those individuals who fall in the tail(s) of the distribution curve. For example, individuals with blood pressure or blood cholesterol levels at the population extreme are considered high-risk. There have been two strategies considered when deciding how to reduce adverse events: a high-risk strategy and a population-based strategy. The high-risk strategy targets only those individuals who fall within the tail of the distribution curve and have the greatest risk for disease in an attempt to lower levels to what is considered normal or ideal. However, the majority of cases of cardiovascular disease occur in individuals who fall in the average risk group. A population-based strategy targets this latter group, focuses on treating more individuals in the population, and aims for a smaller population-based risk factor reduction. This approach follows the argument put forth by Geoffrey Rose, viz., that shifting the risk distribution curve by only a small amount within a population will have greater societal benefit in reducing or preventing cardiovascular morbidity and mortality than treating only high-risk patients 12. Others have also argued that significant reduction in the population burden of cardiovascular disease can only occur from a population approach shifting the entire population distribution to lower levels 13. Despite the epidemiological cogency of this argument, adopting a population-based strategy that shifts the population mean also shifts the distribution of individuals who fall within the tails and, therefore, cross thresholds to initiate or stop treatment (Figure 2B). The net outcome can be either beneficial or harmful. Consider differences in the threshold for treatment of cholesterol using guidelines established by the ACC/AHA as compared to the US Preventive Services Task Force. Differences in the guidelines suggest that shifting to the US Preventive Services Task Force recommendations from the ACC/AHA guidelines resulted in ~9% of individuals who met ACC/AHA guideline criteria for statin use for primary prevention no longer being recommended for treatment; 55% of this group are younger adults with high mean long-term risk for cardiovascular disease 14.
Using data from the National Health and Nutrition Examination Survey III study, investigators performed a head-to-head comparison of population-based and high risk-based treatment strategies using a low- or moderate-intensity intervention to determine which had the greatest benefit for reducing cardiovascular events and cardiovascular mortality. The study compared a population-based strategy that treated all, a high-risk strategy that selected individuals within the top 25% low-density lipoprotein cholesterol (LDL) level, and a high-risk strategy based on individuals within the top 25% for cardiovascular risk as determined by a risk prediction tool. In this case, the investigators found that the high-risk strategy focused on individuals within the top 25% of cardiovascular risk was the most efficient method for preventing cardiovascular events over the long-term but was comparable to a population-based strategy with a more moderate interventional goal. This finding led to the conclusion that while the high-risk treatment strategy was the most efficacious, the population-based prevention strategy was a reasonable option if the intervention had no or minimal side effects 13. In contrast, in developing low- and middle-income countries, the high-risk strategy should be utilized, especially when there are few resources available 15.
The search for single pathogenic genes/gene products and its flaws
The standard view that all or most cardiovascular diseases have a heritable component has fueled the search for single pathogenic genes for specific disorders. Although cardiovascular disease is significantly broad and encompasses diseases related to blood vessels, the myocardium, heart valves, the conduction system, and developmental abnormalities, there are only a few cardiovascular disorders that can be attributed to a single pathogenic gene. Nonetheless, there are notable examples of monogenic disorders that cause cardiovascular disease, such as a mutation in the low-density lipoprotein receptor (LDLR) gene that causes familial hypercholesterolemia 16, 17; in the beta-myosin heavy chain and other sarcomeric proteins that are causal for hypertrophic cardiomyopathy 18, 19 and a mutation in the fibrillin (FBN1) gene that causes Marfan syndrome among others 20. In contrast, atherosclerosis and myocardial infarction are multifaceted diseases with a complex inheritance, cannot be explained by a single pathogenic gene, and are more likely due to perturbations of large, and possibly phenotypically related, genes and other environmental factors 21.
There are several inherent limitations associated with the search for a single pathogenic gene in complex cardiovascular diseases, and these are exemplified in known monogenic disorders. The first is that the genotype-phenotype relationship can be difficult to ascertain. For example, mutations in the alpha subunit of the type V voltage-gated sodium channel (SCN5A) was initially described as a single causal gene for inherited long QT syndrome 22. Since that time, other phenotypes have been associated with the mutation, including Brugada syndrome and dilated cardiomyopathy 23, 24. The second consideration involves penetrance and expressivity with examples of incomplete penetrance in a pedigree with familial hypercholesterolemia and an LDLR mutation, and differences in disease expression in families with Marfan syndrome and the same heritable mutation in FBN1 25, 26. Incomplete penetrance of these classic Mendelian disorders simply suggests other interacting genetic variants and/or environmental exposures on their pathogenesis.
The limitation of genome-wide association studies (GWAS) as a method designed to implicate a single pathogenic gene, or a limited number of pathogenic genes, is that performing genetic mapping by association only identifies genomic regions that may contribute to the disease process. While coverage of the genome is improving, the singular information provided by these data are that a pathogenic gene resides nearby with distance determined by the sequencing platform. Results from GWAS support the concept that complex disorders possess genetic heterogeneity of variants (common, low-frequency, or rare), and this is likely the most frequent expression pattern in these types of diseases. This point was exemplified in a GWAS meta-analysis of individuals with myocardial infarction and control subjects. In this study of 60,801 individuals with a myocardial infarction and 123,504 controls, there were common and rare variants identified accounting for genetic heterogeneity 27.
Another argument against a single pathogenic gene for atherothrombotic vascular disease and myocardial infarction is illustrated by the relationship between chromosome 9p21 and coronary artery disease or myocardial infarction. Single nucleotide polymorphisms (SNP) in 9p21 were identified by several independent GWAS and each risk allele was associated with a 29% increased risk of cardiovascular disease 28–30. The findings that the SNPs were in non-coding regions, the nearest genes were > 100 Kb away, and causality between the nearest genes (CDKN2B, CDKN2A) and susceptibility to atherosclerosis has not yet been ascertained all argue against the single pathogenic gene explanation 28–30. Considering the history of GWAS and the observation that these studies have not identified a single pathogenic gene or SNP as the mechanism underlying coronary artery disease or myocardial infarction indicates further that searching for a single element to explain a complex phenotype is not sufficient. Instead, it is more likely that results from GWAS and next-generation sequencing will contribute to a mechanistic exploration of molecular networks (see below) and to the need for concomitant deep phenotyping multifaceted cardiovascular diseases, such as coronary artery disease and myocardial infarction.
Resilience and Mendelian disease genes
Advances in next generation sequencing have expanded our understanding of the genetic basis of cardiovascular diseases in particular and of all human disease in general. There are >150,000 disease-related genetic variants mapped to >6,000 Mendelian disorders (Online Mendelian Inheritance in Man[OMIM]) that have been catalogued in the Human Gene Mutation Database (HGMD)31, 32. Although this information is readily available, there are relatively few therapies developed that effectively treat or cure these Mendelian diseases 33. For common complex diseases, such as atherothrombotic cardiovascular disease, few gene variants have translated directly into predictors of disease risk or severity, thereby limiting their potential to be developed into a diagnostic or therapeutic.
More recently, attention has turned to examining the genetic and environmental factors that predispose to resilience, the ability to adapt successfully to stress. In the context of Mendelian disease, resilience implies the ability to withstand changes to fitness in the presence of disease-associated mutations 34. The role of second site mutations or environmental modifications that promote resilience to disease has also been established in preclinical models 35, 36. In addition, clinical studies that have found highly penetrant disease-causing mutations in individuals who do not manifest the disease phenotype, suggesting that there are either genetic or environmental resilience factors that protect individuals from manifesting disease 37, 38. A healthy lifestyle, defined as the absence of obesity, no current tobacco use, a healthy diet, and regular physical activity, has been shown to serve as an environmental resilience factor and modify genetic risk of cardiovascular disease. In a study of 55,685 individuals who were stratified according to a polygenic risk score comprised of 50 SNPs associated with coronary artery disease, individuals with a high genetic risk with a healthy lifestyle had a 46% reduction in the relative risk of coronary events 39.
While many genetic studies have attempted to identify mutations linked to disease, an alternative is to search for genetic factors that prevent disease, or wellness factors, in the presence of Mendelian disease-causing mutations. While attempts to find resilience factors have been limited, some secondary cardiovascular disease modulators have been identified, including a loss-of-function mutation in PCSK9 and mutations in zinc transporter 8 that protect obese individuals from diabetes mellitus 35, 40–42.
A more recent study used a different tactic to find resilience factors and studied the genomes of 589,306 individuals who were apparently healthy. Using this approach, they identified 13 adults with what had been viewed as completely penetrant Mendelian disease mutations for 8 different severe childhood diseases, including cystic fibrosis, atelosteogenesis, and Smith-Lemli-Opitz syndrome, but who were without any clinical manifestations of disease. This finding suggested that these individuals have some form of “genetic protection” or resilience against even these classical Mendelian disorders; however, it was not possible to isolate the resilience factor(s) due to the data available and study design 43.
The search for the Ehrlichian ‘magic bullet’ and its flaws
The complexities of the cardiovascular disease phenotype also highlight the fact that there is no single therapy, or Ehrlichian ‘magic bullet,’ that will cure the disease. In 1900, the German Nobel laureate Paul Erlich, a pioneer of chemotherapy, put forth the concept of the magic bullet. He hypothesized that there would be a way to target specifically disease-causing microbes in the same way a bullet fired from a gun hits its target. He referred to this hypothetical agent as Zauberkugel or magic bullet. His continued work to cure syphilis using a magic bullet led to the discovery of Salvarsan (an arsenical compound) on August 31, 1909, the 606th compound he tested that was efficacious and free of important side effects 44.
The magic bullet concept was soon popularized and its meaning expanded to include a perfect drug to cure a specific disease that acted without side effects. The goal would be achieved by identifying a compound that targets a specific disease-related molecule and does so in a highly specific manner with no off-target effects. This idea has become an enduring part of cardiovascular (and all of ) medicine even though there are numerous examples of therapies or interventions for cardiovascular disease that have been hailed as a ‘magic bullet,’ yet have failed to provide a cure, including stem cells and statins. In fact, this is not surprising as cardiovascular diseases are complex pathophenotypes with significant biologic diversity owing to numerous genetic, metabolic, and environmental mediators.
Thus, the idea of a single target in the original, strictest definition of a magic bullet is unlikely to have broad applicability in cardiovascular diseases. In other words, the use of a mono-specific drug(s) to target a multifactorial disease will not be successful. This has led to the idea of targeting multiple targets at once (e.g., the polypill). While this strategy may improve the pharmacological profile, there is also the increased risk of adverse effects. Biologicals such as monoclonal antibodies are less likely to have off-target effects, but may show variations in efficacy and unknown off-target effects or on-target toxicity (e.g., abciximab and severe bleeding or immune checkpoint inhibitors and autoimmune diseases). This approach has evolved further to fall within the rubric of precision medicine via network-targeted therapy that uses a combination therapy strategy targeting multiple steps in a signaling pathway or network identified by deep phenotyping. This more complex approach could be considered a modern interpretation of Ehrlich’s magic bullet theory in the contemporary era 44, 45.
Importance of molecular interaction (“interactome”) networks
Using data from genomic and other panomic profiling, data can be analyzed using molecular interaction or interactome networks to discover disease associations and possible therapeutic targets (figure 3). Interactome networks play an increasingly important role in the discovery of novel relationships between genes or proteins that may have broad applicability across a spectrum of human diseases. The interactome defines a comprehensive, unbiased set of biologically relevant molecular (i.e., gene, protein, metabolite) interactions in a cell, organ, or person. The advantage of examining the interactome to understand disease pathobiology is that the interactome allows for the simultaneous consideration of (ideally) all, or a large number of relevant genes, metabolites, and/or proteins as well as their interactions as depicted through network representation. These networks and sub-networks allow for discovery of new interactions, ascertainment of pathways governing phenotype, identification of critical regulatory checkpoints in a biological system, and prediction of the consequences of disease in network associations or pathway functions on phenotype. The interactome eliminates the bias introduced by analyzing a dataset using a reductionist approach, which examines a limited number of interactions in the dataset to find a differentially expressed gene or protein associated with a phenotype without consideration of the network context within which it operates 46, 47. The current human protein-protein interactome contains 13,460 proteins that have 141,296 physical interactions (protein-protein, metabolic pathways, kinase-substrate)48. The interactome network is scale-free, indicating that interactions are not random, and the proteins within it exhibit emergent behavior such that detailed information about one protein does not predict its functional response within the context of the interactome or of the interactome itself 49. Although the human interactome is “incomplete” owing to the fact that the majority (80%) of pairwise protein interactions have not been analyzed using currently available high-throughput methodology, it remains a valuable tool for discovery of disease-related phenomena 48.
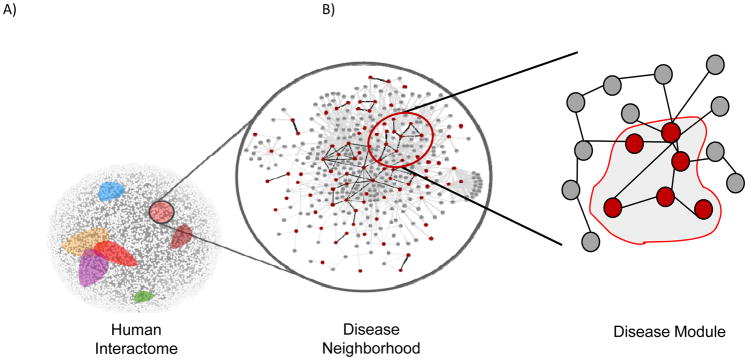
Figure 3. Molecular interaction networks (“interactome”).
The protein-protein interactome describes interactions between proteins in a scale-free (i.e., interactions are not random but clustered) manner. Within the interactome, groups of proteins reside in disease neighborhoods (colored areas) that may overlap indicating common mediators (left, center). Within the neighborhood, there are a number of disease modules, or groups of protein interactions that are related. Analysis of the interactome may uncover new or previously unrecognized relationships. Gray circles, nodes (i.e., proteins in a protein-protein interaction network); black lines, edges (i.e., interactions).
The human interactome has been shown to contain sub-networks for specific diseases, so-called disease modules. Certain disease modules overlap with one another, and these diseases typically have similar symptoms, patterns of co-expression, and comorbidities, while diseases that reside in separate neighborhoods in the network do not share these characteristics and are clinically distinct. Analysis of disease relationships has shown that 7% of disease pairs have overlapping disease neighborhoods in the interactome with unexpected disease relationships, such as that between asthma and celiac disease, which may share a molecular phenotype despite differences in their pathobiology. Interrogation of the shared factors between asthma and celiac disease revealed pathways for immunoglobulin A production as a common intermediary 33. Interactome-based network analysis has been used to inform GWAS data. For example, the great majority of genes associated with type 2 diabetes mellitus have very small effect size and limited statistical significance when analyzed in isolation; however, mapping these alleles to the interactome leads to the identification of novel interactions in pathway clusters that provide unique, highly statistically significant information on mechanism not appreciated by conventional GWAS (Figure 4). Another capability of the interactome is to improve comprehension of the molecular actions of drugs with a goal of identifying drug targets, the potential for adverse reactions, and the possibility of repurposing drugs based on newly identified relevant molecular targets. This concept was investigated for myocardial infarction using a network that incorporated myocardial infarction disease genes, drugs, drug targets, and drug target interactors or binding partners in the interactome to reveal that many myocardial infarction-related drug targets and disease proteins have close relationships within the disease module in the interactome, and some of the drug target intermediaries are themselves targets for other drugs suggesting potential drug repurposing strategies 50. Studies of the interactome have also been utilized to identify tissue specificity of modules associated with human disease. This has shown that an entire functional sub-network or pathway incorporating disease gene products, or a disease module, must be expressed within a particular tissue for a disease to be manifest in that tissue, not simply expression of the disease gene product itself. This type of analysis also leads to the creation of a disease-tissue network, and allows for the discovery of unexpected disease-tissue associations 51.
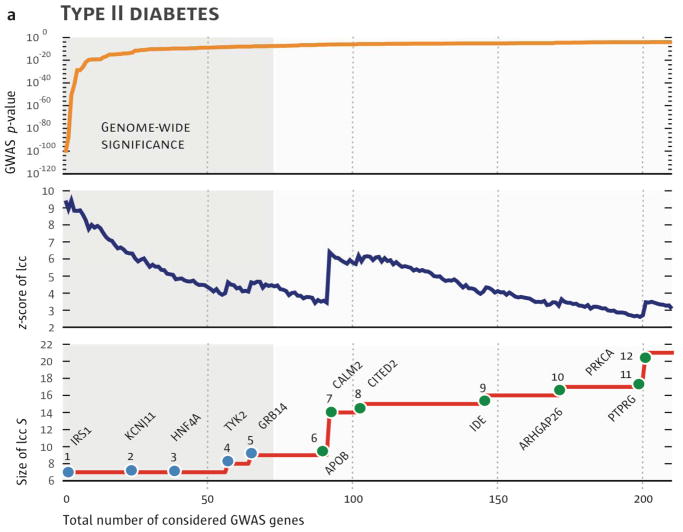
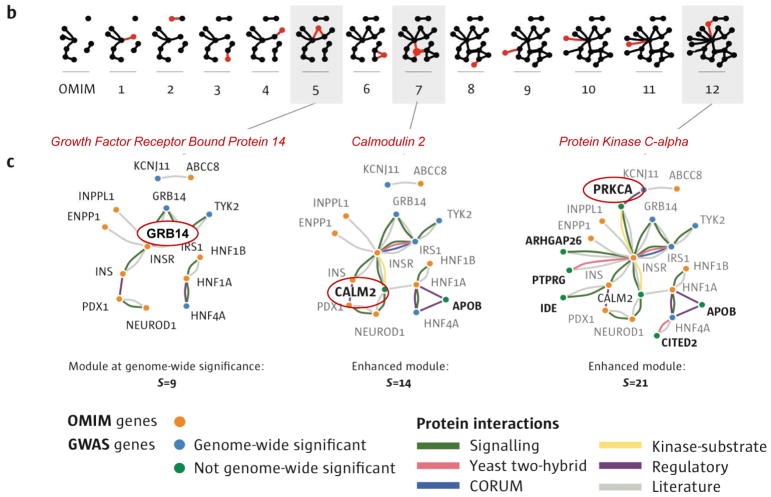
Figure 4. Identifying GWAS genes that have biological relevance.
Analysis of GWAS data reveals that there are a large number of genes that, while having genome-wide significance have very modest effect sizes. A) Using data from patients with type 2 diabetes mellitus as an example and starting with a module that includes six genes of greatest statistical significance, GWAS genes are added to the module in decreasing order of their p-value (top). The statistical significance of the growing network as compared to a random network is shown (middle) as well as the size of the resulting module (bottom). B,C) Of 77 significant GWAS genes, only 5 connect to the interactome cluster initially; however, the addition of the interactor genes (product), CALM2, joins disconnected parts of the network and increases both its size and significance. Another disconnected cluster is then joined to the module through consideration of 200 GWAS genes, including the KCNJ11 (GWAS significant gene) and ABCC8. Reproduced with permission from 48.
Importance of phenotype
A core principle of precision medicine is establishing the precise phenotype for any given disorder (Figure 5). The ability to understand and recognize the relationship between the elements that comprise health phenotypes and disease (patho)phenotypes is required for precision medicine to improve individual and population outcomes 52, 53. The foray into precision phenotyping has faced some obstacles, including the unknown function of many genes or metabolites, the incomplete descriptors used to categorize diversity within disease phenotypes, and inadequate data collection or extraction from medical records 52, 54. These shortcomings have led to the characterization of most phenotype descriptions as imprecise and highlight the necessity of a unified descriptive ontology that is widely adopted. This widely recognized need has led to the proposal that the Human Phenotype Ontology (HPO) be utilized as a standard to characterize disease phenotypes using a specific vocabulary; however, it remains to be seen if this is system will achieve widespread use 55.
Figure 5. The importance of phenotype and its molecular network underpinning.
Individuals who share a common endophenotype, such as hypertension or hypercholesterolemia, may have a very different phenotype when examined at a molecular level. Using molecular phenotyping, such as genome sequencing, and network analysis, individuals may cluster into distinct phenotypes. These phenotypes have important implications for disease risk, prognosis, or response to medication. These important differences are not discoverable when relying on endophenotype alone. (Blue nodes, normal or major alleles; red nodes, variant or minor alleles in protein-protein interaction network).
The importance of obtaining a more detailed granular phenotype for cardiovascular diseases is underscored by the fact that phenotypic profiling is lagging with respect to panomics technologies, and, that phenotypic information is necessary to deconvolute the results of genomic, transcriptomic, metabolic, and proteomic studies. Only recently has the approach to clinical phenotyping moved beyond historical metrics, such as blood pressure, to improve stratification of cardiovascular diseases. Current phenotypic assessments are beginning to incorporate standard clinical traits, such as blood pressure, as well as other complex characteristics like social, environmental, and personal exposures 10, 56. Phenotypes are also being assessed continually using personal sensors and wearable devices, illustrating the importance of time trajectories in monitoring early identifiers of disease progression 57, 58.
There are several recent examples of the use of advanced phenotyping methodologies to discover differences within individuals with a clinical syndrome that has underlying heterogeneity. Clinical phenomapping of patients with heart failure and preserved ejection fraction using unbiased hierarchical clustering analysis revealed three distinct groups of patients, with one phenogroup having an increased risk of heart failure hospitalization (HR: 4.2, 95% CI: 2.0–9.1, p<0.001) 58. Similarly, cluster analysis of 1,619 chronic heart failure patients identified heterogeneity within the group as a whole and 4 phenotypic clusters of patients with differences in their risk of all-cause mortality or all-cause hospitalization 11.
The concept of an unbiased or undirected phenome-wide assessment of an individual is also entering the realm of possibility, and has been conducted in a single person who was extensively phenotyped over the course of 18 months. In the MyConnectome project, brain function over time was assessed with imaging, functional studies, health surveys, and genomic and metabolomic profiling with data from the latter examined using network analysis. Using this platform, gene and metabolite expression were related to dynamic changes in brain connectivity over a timescale of days to months. This pioneering study illustrates the feasibility of this deep temporal phenotyping approach and provides a strategy for translating these types of studies into the cardiovascular space 59.
Endophenotype diversity and pre-emptive/preventive strategies
The endophenotype, or intermediate phenotype, is a biological trait that is quantitative and reflects the function of a biological system. It is heritable and is more closely related to the root cause of a disease than the broad clinical phenotype 60, 61. GWAS that examine conventional phenotypes can be improved upon by focusing on individual endophenotypes 62. Endophenotypes are themselves believed to be subject to natural selection leading to the concept that there are evolutionarily established molecular mechanisms, which regulate endophenotypes, and that these factors translate into genetic predisposition for the correlate downstream clinical phenotypes 63–65. This view leads to the expectation that complex molecular mechanisms link genes responsible for both endophenotypes and phenotypes 66. Thus, genetic effects on endophenotype and phenotype do not necessarily have to be the same in different populations, and genetic predisposition to an endophenotype does not necessarily translate into predisposition for a downstream phenotype.
A number of GWAS studies have illustrated the diverse relationships between endophenotypes and phenotypes. For example, one study examined the association of the rs693 and rs562338 polymorphisms in the apolipoprotein B locus with total cholesterol and high-density lipoprotein cholesterol (endophenotype) with myocardial infarction and survival (phenotypes) across 4 population-based studies (MESA, ARIC, FHS, Cardiovascular Health Study [CHS]). The endophenotypes were investigated to determine if they could mediate the associations of the polymorphisms with myocardial infarction or mortality. In this analysis, the SNPs rs693 and rs562338 were robustly associated with the endophenotype (total cholesterol) but had a trivial association with the phenotype (myocardial infarction and survival) despite the well-known relationship between lipids and myocardial infarction 67.
Network analysis has been used to examine endophenotypes and their overlapping relationships to different diseases. Inflammation, thrombosis, and fibrosis are endophenotypes that are linked pathologically and common to virtually all disease states, both in terms of response to injury and to repair 68–71. To explore biological and topological crosstalk between these endophenotypes, network analysis of the inflammasome, thrombosome, and fibrosome was undertaken. Mapping these networks to the human interactome identified inflammation, thrombosis, and fibrosis sub-regions that were highly overlapping and contained disease genes associated with complex diseases and cardiovascular risk factors. These sub-regions were found to be integral to the structure of the basic interactome network thereby highlighting the key role of these processes in (health and) disease 72.
With the advent of panomics profiling, endophenotypes and their association with complex disease phenotypes will be refined further. To ensure the integrity of these relationships, however, there is an implicit need for standardization of methodology and platforms, as well as adoption of Clinical Laboratory Improvement Amendments (CLIA)-grade laboratory testing with strict adherence to quality control. Analytical methods to integrate endophenotypic data should favor unbiased approaches, such as network analysis, and account for issues related to multiple testing, which is an inherent limitation of large dataset analysis.
Genetic diversity and rational polypharmacy
Although randomized clinical trials have determined the efficacy of many new therapeutics across large populations of individuals with similar clinical phenotypes, heterogeneity in the response to drugs remains a relevant and challenging issue. Aggregating patients with similar disease characteristics into a cohort deemed to be putative therapeutic responders with equal effect obviously ignores this heterogeneity for clinical trial design and therapeutic simplicity, even though there is no phenotypic basis to support such an oversimplified categorization (Figure 6A) 56. Pharmacogenetic variation due to single and cumulative drug-gene interactions can lead to adverse drug interactions 73, 74; these types of reactions increase with polypharmacy. Approximately 66% of adults age 65 years or older have at least one or more prescription medications that require daily use 75–77. It is estimated that ~35% of geriatric patients have adverse drug events, at least half of which are deemed preventable; and 10–17% of hospitalizations in this population are related to drug reactions leading to 51% of adverse drug-related deaths 77–79. The application of precision medicine to this problem has revealed that cardiovascular pharmacogenetics can be used to identify genetic diversity markers associated with poor response or adverse reactions to drugs. In fact, drug interactions related to predisposing genetic factors lead to ~47% of warnings of potential interactions associated with side effects 79.
Figure 6. Precision medicine approach to rational polypharmacy.
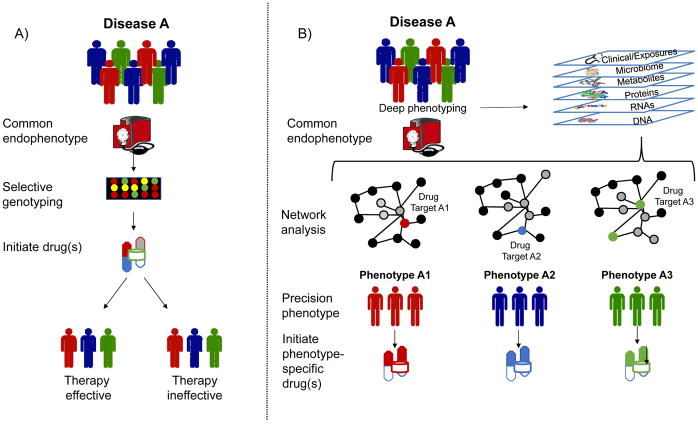
A) The current approach to selecting pharmacologic therapy involves identifying a group with a common endophenotype, using genomic profiling in select cases to identify potential drug (non)responders, and then testing a medication based on results from clinical trials. B) Using a precision medicine approach, individuals with a common endophenotype undergo deep phenotyping, including panomics, exposures, and clinical assessments, and the data are analyzed using network analysis to identify more precise phenotypes and their molecular determinants in the interactome. (Gray nodes, disease-determining module on network; red blue, and green nodes, drug targets by phenotype). This information is then used to select agents that target key pathways or proteins.
There are numerous examples of genetic diversity and DNA variants as determinants of response to a drug. In cardiovascular disease, platelet aggregation promotes atherothrombotic vascular disease and stent thrombosis following percutaneous coronary intervention. The P2Y12 inhibitor clopidogrel, which is part of a typical dual antiplatelet regimen, has a range of inter-individual variability. Some individuals are known to be clopidogrel “non-responders,” a phenotype that has been associated with genetic determinants and an increase in ischemic events 80–83. Loss-of-function alleles in CYP2C19 (CYP2C19*2 and CYP2C19*3) have been associated with poor drug responsiveness, while the gain-of-function allele CYP2C19*17 is associated with increased bleeding risk 84, 85,86. Attempts to utilize this information for tailored therapy in patients with drug-eluting stents and high on-treatment platelet reactivity, however, did not affect cardiovascular death, myocardial infarction, or stent thrombosis 87.
It has been suggested that ~50% of patients taking statins stop taking their medications owing to side effects or adverse events 88. Of the related genes, only the gene for the soluble carrier organic anion transporter 1B1 (SLCO1B1), which regulates statin influx and metabolism in the liver, has been associated consistently with myopathy 88. Patients who are either homozygous or heterozygous for the rs4263657 polymorphism have an increased risk for rhabdomyolysis with statin use, and it has been suggested that individuals with this SNP avoid statin drugs 89.
Warfarin, one of the most commonly prescribed anticoagulants, has a narrow therapeutic window but wide inter-individual variation 90. Warfarin is metabolized primarily in the liver by oxidation by cytochrome P450 2C9 (CYP2C9), and inhibits the protein vitamin K epoxide reductase complex subunit 1 (VKORC1) 91, 92. Studies have attributed the ~10%–50% variability in dose requirements to genotypes, notably SNPs in CYP2C9 (CYP2C9*2, CYP2C9*3) and VKORC1 (rs9923231) 93–95. The Food and Drug Administration has recognized the importance of these genetic variants and updated the drug packaging to include information on dosing based on CYP2C9 and VKORC1 genotypes 96, 97. Two randomized clinical trials, the Clarification of Optimal Anticoagulation through Genetics (COAG) trial and the European Pharmacogenetics and Anticoagulant Therapy-Warfarin (EU-PACT) study, evaluated genotype-guided warfarin dosing 98, 99. These studies incorporated CYP2C9 and VKORC1 genotyping into the clinical algorithm for selecting the warfarin dose. The 1,015 patient COAG trial reported that genotyping had no effect on the percent of time in the therapeutic International Normalized Ratio (INR) range, while the 455 patient EU-PACT study found that genotyping was useful, resulted in fewer episodes of excess anticoagulation (INR levels >=4.0), and led to more rapid achievement of the therapeutic range (21 vs 29 days) than those who were not guided by genotype 98, 99. The differences observed between these trials has been attributed to trial design, prevalence of the SNPs in the study population, and the demographics of the study populations 100.
Genetic variants have also been identified that modify the response to other relevant cardiovascular drugs, including beta-blockers (ADRB1, ADRB2, GRK5, GRK4); angiotensin converting enzyme inhibitors (ACE, AGTR1); diuretics (ADD1, NPPA, NEDD4L); and calcium channel blockers (CACNB2, CACNA1C) (reviewed in 101). To date, however, genetic testing has not been employed routinely to guide selection of these drugs, and many genetic variants require confirmation in larger studies.
Other pilot studies have examined the efficacy of implementing pharmacogenetic profiling coupled with a clinical decision support tool in polypharmacy patients to reduce adverse drug reactions and emergency department visits. In a small study of 110 patients randomized to genetic profiling of CYP 450 genes and prescribed relevant medications, the use of pharmacogenetic profiling was associated with a reduction in the number of emergency department visits (RR 0.58; 95% CI: 0.34–0.99, p=0.045) and hospitalizations at 60 days (RR, 0.48; 95% CI: 0.27–0.82, p-=0.0007). Interestingly, of 124 recommendations on drug therapy based on pharmacogenetic profiling that were passed on to the patient’s clinician, only 96 (77%) were followed. While exploratory, this study highlights the benefits of identifying genetic diversity to achieve rational polypharmacy and clinician attitudes towards this approach 102.
Pharmacogenetic profiling has also been used to determine if individuals treated with polypharmacy tend to express more rare variants than the general population and, therefore, are less likely to respond to individual medications. Using a de-identified electronic medical record and corresponding samples from a biorespository, investigators identified 326 “frequently medicated” individuals (defined as prescribed clopidogrel or warfarin in addition to more than five medications from select classes with at least one drug known to have a pharmacogenetic interaction). Analysis found that most of the marker allele frequencies in polypharmacy patients did not differ from what was reported for individuals of European descent in the 1000 Genomes Pilot with a single exception in CYP2D6 (rs1080985) 103. The relevance of this finding was deemed unclear as it was not certain if it was the result of sequencing errors in the 1000 Genomes Pilot or a true difference 104.
Precision medicine will advance this field by using enhanced phenotyping for disease stratification to identify groups of patients that differ in drug responsiveness, will benefit from existing pharmacotherapies, or will benefit from rational polypharmacy [i.e., the use of multiple drugs chosen based on knowledge of their interactome-based network location and effects on key pathways in the interactome that govern pathophenotype (Figure 6B)] 105. This strategy can also be used to identify biomarkers to guide drug dosing, continue therapy, or limit side effects and adverse drug reactions 101. When and how pharmacogenetic profiling will become a mainstay of clinical practice to guide polypharmacy and limit adverse drug reactions remains to be determined 106. In part, the slow uptake of clinically applied pharmacogeneticcs is a reflection of statistical uncertainty, the abundance of genomic variants (even those previously believed to be causative), and the typical association analysis rather than network analysis of the functional consequences of the variants 107.
Precision versus personalized medicine
Although the terms precision medicine and personalized medicine have been used interchangeably, they are not identical but, rather, related and overlapping disciplines. Precision medicine identifies the unique aspects of an individual related to health and disease to select appropriate and effective therapy. Personalized medicine may refer to implementation of these data into a person-specific treatment plan or, simply, the creation of a treatment plan for an individual based on a known biomarker(s). Personalized medicine is not, however, meant to suggest that results from detailed phenotyping will be used to create new drug or other therapies.
Precision medicine is increasingly considered as the art and science of creating individualized assessments of health and disease that are derived from established clinical and pathological indices integrated with state-of-the-art panomic (i.e., genomic, transcriptomic, metabolomic, and proteomic) profiling 108, 109. Thus, precision medicine aims to drive the field away from the implementation of the same pharmacotherapy, lifestyle intervention, or behavioral modification to a group of individuals presumed to share the same phenotype and towards personalizing treatments that are aimed at prevention, promotion of health, and amelioration of disease. While this concept has been embraced with success in the field of oncology, the issue of patient adherence with a personalized therapy identified using precision medicine remains a concern 109. Estimates indicate that approximately half of all medications are not taken as prescribed and that patients with chronic conditions such as cardiovascular disease only take 50%–60% of the medications prescribed 110. While this is a valid point and developments in precision and personalized medicine must be matched by patient education and acceptance of these strategies, it is also plausible that poor adherence rates are the result of imprecise medication use leading to limited efficacy and/or increased side effect risk in a given individual. This issue will clearly require further study.
Future considerations
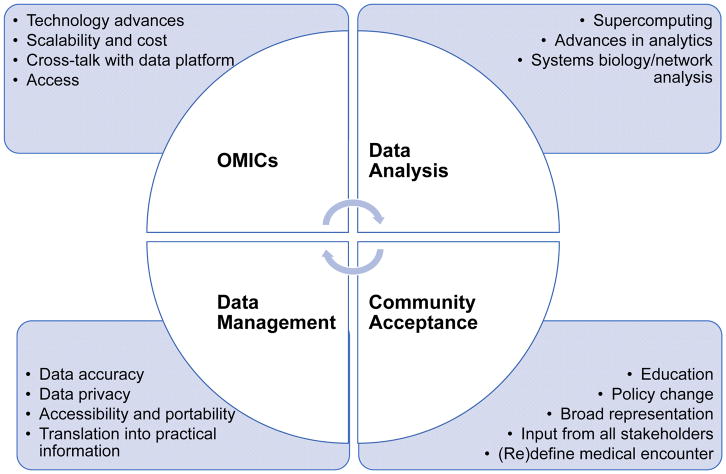
Precision medicine is poised to become the next great revolution in the practice of medicine, as well as in the maintenance of cardiovascular health and the prevention and cure of cardiovascular disease. Precision medicine disrupts standard practice and draws from clinical testing, electronic health records, panomics profiling, big datasets, and novel analytical methods, such as systems biology and network science, to create a person-specific phenotype that can then be used to identify an optimal intervention with minimal risk. The obvious benefits of this approach to patients, clinicians, and researchers are numerous and include: individual phenotype specificity, identification of individuals with a similar molecular phenotype, selection of best drugs or therapies with maximal efficacy and no or limited adverse reactions, efficient selection and enrichment of clinical trial participants, potential to improve adherence and reduce costs, and creating a paradigm shift in how cardiovascular care is delivered. To accomplish this laudable goal, the medical community at large and other stakeholders will need to overcome barriers to implementation that range from technical to sociopolitical (Figure 7) 111. At the very least, precision medicine will need to demonstrate that phenotype-based person-specific interventions are superior to the current standard of care and, ultimately, have a population effect by moving the mean on the disease spectrum towards health 111. Other barriers are related to large dataset collection and focuses on methods to ensure data accuracy, computational power, security and privacy of datasets, renewal of accruing data, and continuous development and refinement of analytical methods. Finally, education, affordability, and public acceptance of the strategy all play key roles in its ultimate implementation 111.
Figure 7. Challenges in precision medicine.
Precision medicine relies on integration and dynamic adaptability of panomic analysis, overall data analysis and data management; and acceptance by the public, medical, research, big pharma, and policy-maker stakeholders.
Despite a clear path forward toward mainstream application of precision medicine, there continues to be debate about whether a precision medicine approach will have a global impact on cardiovascular disease prevention and treatment, or will only serve a small group of patients and be relegated to a highly selected niche role. This skepticism has arisen, in part, based on progress in the field to date and perceived challenges in maintaining the physician-patient relationship 112, 113. While these concerns are valid points, they are not insurmountable and are currently being addressed in visions as how best to implement precision medicine in the practice of cardiovascular medicine. The nascent field of precision medicine fully embraces Paul Ehrlich’s concept that imprecision leads to failure, and harnesses our most precise technologies and methodologies to improve cardiovascular health and treat disease.
Acknowledgments
Sources of Funding
This work was supported in part by the American Heart Association and NIH grants U01 125215 (JAL) and HL61795, HG007640, and GM107618 (JL).
Non-standard Abbreviations and Acronyms
- ACE
Angiotensin converting enzyme gene
- ADD1
Adducin1 gene
- ADRB1
β1-adrenergic receptor gene
- ADRB2
β2-adrenergic receptor gene
- AGTR1
Angiotensin II receptor, type 1 gene
- ARIC
Atherosclerosis Risk in Communities study
- CACNA1C
Calcium voltage-gated channel, subunit α1 C gene
- CACNB2
Calcium voltage-gated channel subunit β2 gene
- CDKN2A
Cyclin-dependent kinase inhibitor 2A gene
- CDKN2B
Cyclin-dependent kinase inhibitor 2B gene
- CHS
Cardiovascular Health Study
- CLIA
Clinical Laboratory Improvement Amendments
- COAG
Clarification of Optimal Anticoagulation through Genetics study
- CYP2C19
Cytochrome P450 2C19 gene
- CYP2D6
Cytochrome P450 2D6 gene
- CYP450
Cytochrome P450 gene
- EU-PACT
European Pharmacogenetics for Anticoagulant Therapy
- FBN1
Fibrillin1 gene
- FHS
Framingham Heart Study
- GRK4
G protein-coupled receptor kinase 4 gene
- GRK5
G protein-coupled receptor kinase 5 gene
- GWAS
Genome wide association study
- HGMD
Human Gene Mutation Database
- HPO
Human Phenotype Ontology
- INR
International Normalized Ratio
- LDL
Low-density lipoprotein
- LDLR
Low-density lipoprotein receptor gene
- MESA
Multi-Ethnic Study of Atherosclerosis
- NEDD4L
Neural precursor cell expressed, developmentally down-regulated 4-like gene
- NPPA
Natriuretic peptide A gene
- OMIM
Online Mendelian Inheritance in Man database
- P2Y12
Purinergic receptor P2Y12 gene
- PCSK9
Proprotein convertase subtilisin/kexin type 9 gene
- SCN5A
Sodium voltage-gated channel, α subunit 5 gene
- SLC01B1
Solute carrier organic anion transporter family member 1B1 gene
- SNP
Single nucleotide polymorphism
- VKORC1
Vitamin K epoxide reductase complex subunit 1 gene
Footnotes
Disclosures
None
References
- 1.de Kruif P. Microbe hunters. New York, NY: Houghton Mifflin Harcourt; 1954. [Google Scholar]
- 2.Heidenreich PA, Trogdon JG, Khavjou OA, et al. Forecasting the future of cardiovascular disease in the united states: A policy statement from the american heart association. Circulation. 2011;123:933–944. doi: 10.1161/CIR.0b013e31820a55f5. [DOI] [PubMed] [Google Scholar]
- 3.Roth GA, Forouzanfar MH, Moran AE, Barber R, Nguyen G, Feigin VL, Naghavi M, Mensah GA, Murray CJ. Demographic and epidemiologic drivers of global cardiovascular mortality. N Engl J Med. 2015;372:1333–1341. doi: 10.1056/NEJMoa1406656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benjamin EJ, Blaha MJ, Chiuve SE, et al. American Heart Association Statistics C, Stroke Statistics S. Heart disease and stroke statistics-2017 update: A report from the american heart association. Circulation. 2017;135:e146–e603. doi: 10.1161/CIR.0000000000000485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fang N, Jiang M, Fan Y. Ideal cardiovascular health metrics and risk of cardiovascular disease or mortality: A meta-analysis. Int J Cardiol. 2016;214:279–283. doi: 10.1016/j.ijcard.2016.03.210. [DOI] [PubMed] [Google Scholar]
- 6.Snyderman R, Meade C, Drake C. Value of personalized medicine. JAMA. 2016;315:613. doi: 10.1001/jama.2015.17136. [DOI] [PubMed] [Google Scholar]
- 7.Snyderman R, Dreke CD. Personalized health care: Unlocking the potential of genomic and precision medicine. Journal of Precision Medicine. 2015;1:38–41. [Google Scholar]
- 8.Naylor S. What’s in a name? The evolution of p-medicine. Journal of Precision Medicine. 2015;1:15–29. [Google Scholar]
- 9.Antman EM, Loscalzo J. Precision medicine in cardiology. Nat Rev Cardiol. 2016;13:591–602. doi: 10.1038/nrcardio.2016.101. [DOI] [PubMed] [Google Scholar]
- 10.Bhatnagar A. Environmental determinants of cardiovascular disease. Circ Res. 2017;121:162–180. doi: 10.1161/CIRCRESAHA.117.306458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahmad T, Pencina MJ, Schulte PJ, O’Brien E, Whellan DJ, Pina IL, Kitzman DW, Lee KL, O’Connor CM, Felker GM. Clinical implications of chronic heart failure phenotypes defined by cluster analysis. J Am Coll Cardiol. 2014;64:1765–1774. doi: 10.1016/j.jacc.2014.07.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rose G. Sick individuals and sick populations. Int J Epidemiol. 1985;14:32–38. doi: 10.1093/ije/14.1.32. [DOI] [PubMed] [Google Scholar]
- 13.Zulman DM, Vijan S, Omenn GS, Hayward RA. The relative merits of population-based and targeted prevention strategies. Milbank Q. 2008;86:557–580. doi: 10.1111/j.1468-0009.2008.00534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pagidipati NJ, Navar AM, Mulder H, Sniderman AD, Peterson ED, Pencina MJ. Comparison of recommended eligibility for primary prevention statin therapy based on the us preventive services task force recommendations vs the acc/aha guidelines. JAMA. 2017;317:1563–1567. doi: 10.1001/jama.2017.3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Babu RB, Alam M, Helis E, Fodor JG. Population-based versus high-risk strategies for the prevention of cardiovascular diseases in low- and middle-income countries. Indian Heart J. 2012;64:439–443. doi: 10.1016/j.ihj.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lehrman MA, Goldstein JL, Brown MS, Russell DW, Schneider WJ. Internalization-defective ldl receptors produced by genes with nonsense and frameshift mutations that truncate the cytoplasmic domain. Cell. 1985;41:735–743. doi: 10.1016/s0092-8674(85)80054-4. [DOI] [PubMed] [Google Scholar]
- 17.Lehrman MA, Schneider WJ, Sudhof TC, Brown MS, Goldstein JL, Russell DW. Mutation in ldl receptor: Alu-alu recombination deletes exons encoding transmembrane and cytoplasmic domains. Science. 1985;227:140–146. doi: 10.1126/science.3155573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jarcho JA, McKenna W, Pare JA, Solomon SD, Holcombe RF, Dickie S, Levi T, Donis-Keller H, Seidman JG, Seidman CE. Mapping a gene for familial hypertrophic cardiomyopathy to chromosome 14q1. N Engl J Med. 1989;321:1372–1378. doi: 10.1056/NEJM198911163212005. [DOI] [PubMed] [Google Scholar]
- 19.Geisterfer-Lowrance AA, Kass S, Tanigawa G, Vosberg HP, McKenna W, Seidman CE, Seidman JG. A molecular basis for familial hypertrophic cardiomyopathy: A beta cardiac myosin heavy chain gene missense mutation. Cell. 1990;62:999–1006. doi: 10.1016/0092-8674(90)90274-i. [DOI] [PubMed] [Google Scholar]
- 20.Dietz HC, Cutting GR, Pyeritz RE, Maslen CL, Sakai LY, Corson GM, Puffenberger EG, Hamosh A, Nanthakumar EJ, Curristin SM, et al. Marfan syndrome caused by a recurrent de novo missense mutation in the fibrillin gene. Nature. 1991;352:337–339. doi: 10.1038/352337a0. [DOI] [PubMed] [Google Scholar]
- 21.Kathiresan S, Srivastava D. Genetics of human cardiovascular disease. Cell. 2012;148:1242–1257. doi: 10.1016/j.cell.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Q, Shen J, Splawski I, Atkinson D, Li Z, Robinson JL, Moss AJ, Towbin JA, Keating MT. Scn5a mutations associated with an inherited cardiac arrhythmia, long qt syndrome. Cell. 1995;80:805–811. doi: 10.1016/0092-8674(95)90359-3. [DOI] [PubMed] [Google Scholar]
- 23.Chen Q, Kirsch GE, Zhang D, Brugada R, Brugada J, Brugada P, Potenza D, Moya A, Borggrefe M, Breithardt G, Ortiz-Lopez R, Wang Z, Antzelevitch C, O’Brien RE, Schulze-Bahr E, Keating MT, Towbin JA, Wang Q. Genetic basis and molecular mechanism for idiopathic ventricular fibrillation. Nature. 1998;392:293–296. doi: 10.1038/32675. [DOI] [PubMed] [Google Scholar]
- 24.McNair WP, Ku L, Taylor MR, Fain PR, Dao D, Wolfel E, Mestroni L Familial Cardiomyopathy Registry Research G. Scn5a mutation associated with dilated cardiomyopathy, conduction disorder, and arrhythmia. Circulation. 2004;110:2163–2167. doi: 10.1161/01.CIR.0000144458.58660.BB. [DOI] [PubMed] [Google Scholar]
- 25.Hobbs HH, Leitersdorf E, Leffert CC, Cryer DR, Brown MS, Goldstein JL. Evidence for a dominant gene that suppresses hypercholesterolemia in a family with defective low density lipoprotein receptors. J Clin Invest. 1989;84:656–664. doi: 10.1172/JCI114212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Faivre L, Collod-Beroud G, Loeys BL, et al. Effect of mutation type and location on clinical outcome in 1,013 probands with marfan syndrome or related phenotypes and fbn1 mutations: An international study. Am J Hum Genet. 2007;81:454–466. doi: 10.1086/520125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nikpay M, Goel A, Won HH, et al. A comprehensive 1,000 genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47:1121–1130. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Helgadottir A, Thorleifsson G, Manolescu A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 29.McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, Hinds DA, Pennacchio LA, Tybjaerg-Hansen A, Folsom AR, Boerwinkle E, Hobbs HH, Cohen JC. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Samani NJ, Erdmann J, Hall AS, et al. Wtccc, the Cardiogenics C. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKusick VA. Mendelian inheritance in man and its online version, omim. Am J Hum Genet. 2007;80:588–604. doi: 10.1086/514346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stenson PD, Mort M, Ball EV, Shaw K, Phillips A, Cooper DN. The human gene mutation database: Building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet. 2014;133:1–9. doi: 10.1007/s00439-013-1358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dietz HC. New therapeutic approaches to mendelian disorders. N Engl J Med. 2010;363:852–863. doi: 10.1056/NEJMra0907180. [DOI] [PubMed] [Google Scholar]
- 34.Soule T. Resilient individuals improve evolutionary search. Artif Life. 2006;12:17–34. doi: 10.1162/106454606775186437. [DOI] [PubMed] [Google Scholar]
- 35.Hartman JLt. Buffering of deoxyribonucleotide pool homeostasis by threonine metabolism. Proc Natl Acad Sci U S A. 2007;104:11700–11705. doi: 10.1073/pnas.0705212104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Friend SH, Schadt EE. Translational genomics. Clues from the resilient. Science. 2014;344:970–972. doi: 10.1126/science.1255648. [DOI] [PubMed] [Google Scholar]
- 37.Cooper DN, Krawczak M, Polychronakos C, Tyler-Smith C, Kehrer-Sawatzki H. Where genotype is not predictive of phenotype: Towards an understanding of the molecular basis of reduced penetrance in human inherited disease. Hum Genet. 2013;132:1077–1130. doi: 10.1007/s00439-013-1331-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sosnay PR, Siklosi KR, Van Goor F, et al. Defining the disease liability of variants in the cystic fibrosis transmembrane conductance regulator gene. Nat Genet. 2013;45:1160–1167. doi: 10.1038/ng.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khera AV, Emdin CA, Drake I, et al. Genetic risk, adherence to a healthy lifestyle, and coronary disease. N Engl J Med. 2016;375:2349–2358. doi: 10.1056/NEJMoa1605086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Louie RJ, Guo J, Rodgers JW, et al. A yeast phenomic model for the gene interaction network modulating cftr-deltaf508 protein biogenesis. Genome Med. 2012;4:103. doi: 10.1186/gm404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cohen J, Pertsemlidis A, Kotowski IK, Graham R, Garcia CK, Hobbs HH. Low ldl cholesterol in individuals of african descent resulting from frequent nonsense mutations in pcsk9. Nat Genet. 2005;37:161–165. doi: 10.1038/ng1509. [DOI] [PubMed] [Google Scholar]
- 42.Flannick J, Thorleifsson G, Beer NL, et al. Loss-of-function mutations in slc30a8 protect against type 2 diabetes. Nat Genet. 2014;46:357–363. doi: 10.1038/ng.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen R, Shi L, Hakenberg J, et al. Analysis of 589,306 genomes identifies individuals resilient to severe mendelian childhood diseases. Nat Biotechnol. 2016;34:531–538. doi: 10.1038/nbt.3514. [DOI] [PubMed] [Google Scholar]
- 44.Strebhardt K, Ullrich A. Paul ehrlich’s magic bullet concept: 100 years of progress. Nat Rev Cancer. 2008;8:473–480. doi: 10.1038/nrc2394. [DOI] [PubMed] [Google Scholar]
- 45.Bosch F, Rosich L. The contributions of paul ehrlich to pharmacology: A tribute on the occasion of the centenary of his nobel prize. Pharmacology. 2008;82:171–179. doi: 10.1159/000149583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barabasi AL, Gulbahce N, Loscalzo J. Network medicine: A network-based approach to human disease. Nat Rev Genet. 2011;12:56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vidal M, Cusick ME, Barabasi AL. Interactome networks and human disease. Cell. 2011;144:986–998. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Menche J, Sharma A, Kitsak M, Ghiassian SD, Vidal M, Loscalzo J, Barabasi AL. Disease networks. Uncovering disease-disease relationships through the incomplete interactome. Science. 2015;347:1257601. doi: 10.1126/science.1257601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chan SY, Loscalzo J. The emerging paradigm of network medicine in the study of human disease. Circ Res. 2012;111:359–374. doi: 10.1161/CIRCRESAHA.111.258541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang RS, Loscalzo J. Illuminating drug action by network integration of disease genes: A case study of myocardial infarction. Mol Biosyst. 2016;12:1653–1666. doi: 10.1039/c6mb00052e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kitsak M, Sharma A, Menche J, Guney E, Ghiassian SD, Loscalzo J, Barabasi AL. Tissue specificity of human disease module. Sci Rep. 2016;6:35241. doi: 10.1038/srep35241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Delude CM. Deep phenotyping: The details of disease. Nature. 2015;527:S14–15. doi: 10.1038/527S14a. [DOI] [PubMed] [Google Scholar]
- 53.Robinson PN, Mungall CJ, Haendel M. Capturing phenotypes for precision medicine. Cold Spring Harb Mol Case Stud. 2015;1:a000372. doi: 10.1101/mcs.a000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wei WQ, Denny JC. Extracting research-quality phenotypes from electronic health records to support precision medicine. Genome Med. 2015;7:41. doi: 10.1186/s13073-015-0166-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kohler S, Vasilevsky NA, Engelstad M, et al. The human phenotype ontology in 2017. Nucleic Acids Res. 2017;45:D865–D876. doi: 10.1093/nar/gkw1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.MacRae CA, Vasan RS. The future of genetics and genomics: Closing the phenotype gap in precision medicine. Circulation. 2016;133:2634–2639. doi: 10.1161/CIRCULATIONAHA.116.022547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McConnell MV, Shcherbina A, Pavlovic A, et al. Feasibility of obtaining measures of lifestyle from a smartphone app: The myheart counts cardiovascular health study. JAMA Cardiol. 2017;2:67–76. doi: 10.1001/jamacardio.2016.4395. [DOI] [PubMed] [Google Scholar]
- 58.Shah SJ, Katz DH, Selvaraj S, Burke MA, Yancy CW, Gheorghiade M, Bonow RO, Huang CC, Deo RC. Phenomapping for novel classification of heart failure with preserved ejection fraction. Circulation. 2015;131:269–279. doi: 10.1161/CIRCULATIONAHA.114.010637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Poldrack RA, Laumann TO, Koyejo O, et al. Long-term neural and physiological phenotyping of a single human. Nat Commun. 2015;6:8885. doi: 10.1038/ncomms9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gottesman II, Gould TD. The endophenotype concept in psychiatry: Etymology and strategic intentions. Am J Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- 61.Cannon TD, Keller MC. Endophenotypes in the genetic analyses of mental disorders. Annu Rev Clin Psychol. 2006;2:267–290. doi: 10.1146/annurev.clinpsy.2.022305.095232. [DOI] [PubMed] [Google Scholar]
- 62.Willer CJ, Schmidt EM, Sengupta S, et al. Global Lipids Genetics C. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Willer CJ, Sanna S, Jackson AU, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Teslovich TM, Musunuru K, Smith AV, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nesse RM, Ganten D, Gregory TR, Omenn GS. Evolutionary molecular medicine. J Mol Med (Berl) 2012;90:509–522. doi: 10.1007/s00109-012-0889-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Button KS, Ioannidis JP, Mokrysz C, Nosek BA, Flint J, Robinson ES, Munafo MR. Empirical evidence for low reproducibility indicates low pre-study odds. Nat Rev Neurosci. 2013;14:877. doi: 10.1038/nrn3475-c6. [DOI] [PubMed] [Google Scholar]
- 67.Kulminski AM, Kernogitski Y, Culminskaya I, et al. Uncoupling associations of risk alleles with endophenotypes and phenotypes: Insights from the apob locus and heart-related traits. Aging Cell. 2017;16:61–72. doi: 10.1111/acel.12526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Loscalzo J, Kohane I, Barabasi AL. Human disease classification in the postgenomic era: A complex systems approach to human pathobiology. Mol Syst Biol. 2007;3:124. doi: 10.1038/msb4100163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fox EA, Kahn SR. The relationship between inflammation and venous thrombosis. A systematic review of clinical studies. Thromb Haemost. 2005;94:362–365. doi: 10.1160/TH05-04-0266. [DOI] [PubMed] [Google Scholar]
- 70.Wakefield TW, Strieter RM, Prince MR, Downing LJ, Greenfield LJ. Pathogenesis of venous thrombosis: A new insight. Cardiovasc Surg. 1997;5:6–15. doi: 10.1016/s0967-2109(96)00083-x. [DOI] [PubMed] [Google Scholar]
- 71.Libby P, Simon DI. Inflammation and thrombosis: The clot thickens. Circulation. 2001;103:1718–1720. doi: 10.1161/01.cir.103.13.1718. [DOI] [PubMed] [Google Scholar]
- 72.Ghiassian SD, Menche J, Chasman DI, et al. Endophenotype network models: Common core of complex diseases. Sci Rep. 2016;6:27414. doi: 10.1038/srep27414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wilkinson GR. Drug metabolism and variability among patients in drug response. N Engl J Med. 2005;352:2211–2221. doi: 10.1056/NEJMra032424. [DOI] [PubMed] [Google Scholar]
- 74.Cardelli M, Marchegiani F, Corsonello A, Lattanzio F, Provinciali M. A review of pharmacogenetics of adverse drug reactions in elderly people. Drug Saf. 2012;35(Suppl 1):3–20. doi: 10.1007/BF03319099. [DOI] [PubMed] [Google Scholar]
- 75.Qato DM, Alexander GC, Conti RM, Johnson M, Schumm P, Lindau ST. Use of prescription and over-the-counter medications and dietary supplements among older adults in the united states. JAMA. 2008;300:2867–2878. doi: 10.1001/jama.2008.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hajjar ER, Cafiero AC, Hanlon JT. Polypharmacy in elderly patients. Am J Geriatr Pharmacother. 2007;5:345–351. doi: 10.1016/j.amjopharm.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 77.Safran DG, Neuman P, Schoen C, Kitchman MS, Wilson IB, Cooper B, Li A, Chang H, Rogers WH. Prescription drug coverage and seniors: Findings from a 2003 national survey. Health Aff (Millwood) 2005;(Suppl Web Exclusives) doi: 10.1377/hlthaff.w5.152. W5-152-W155-166. [DOI] [PubMed] [Google Scholar]
- 78.Budnitz DS, Lovegrove MC, Shehab N, Richards CL. Emergency hospitalizations for adverse drug events in older americans. N Engl J Med. 2011;365:2002–2012. doi: 10.1056/NEJMsa1103053. [DOI] [PubMed] [Google Scholar]
- 79.Lazarou J, Pomeranz BH, Corey PN. Incidence of adverse drug reactions in hospitalized patients: A meta-analysis of prospective studies. JAMA. 1998;279:1200–1205. doi: 10.1001/jama.279.15.1200. [DOI] [PubMed] [Google Scholar]
- 80.Oqueli E, Hiscock M, Dick R. Clopidogrel resistance. Heart Lung Circ. 2007;16(Suppl 3):S17–28. doi: 10.1016/j.hlc.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 81.Ahmad T, Voora D, Becker RC. The pharmacogenetics of antiplatelet agents: Towards personalized therapy? Nat Rev Cardiol. 2011;8:560–571. doi: 10.1038/nrcardio.2011.111. [DOI] [PubMed] [Google Scholar]
- 82.Collet JP, Hulot JS, Pena A, Villard E, Esteve JB, Silvain J, Payot L, Brugier D, Cayla G, Beygui F, Bensimon G, Funck-Brentano C, Montalescot G. Cytochrome p450 2c19 polymorphism in young patients treated with clopidogrel after myocardial infarction: A cohort study. Lancet. 2009;373:309–317. doi: 10.1016/S0140-6736(08)61845-0. [DOI] [PubMed] [Google Scholar]
- 83.Mega JL, Close SL, Wiviott SD, Shen L, Hockett RD, Brandt JT, Walker JR, Antman EM, Macias W, Braunwald E, Sabatine MS. Cytochrome p-450 polymorphisms and response to clopidogrel. N Engl J Med. 2009;360:354–362. doi: 10.1056/NEJMoa0809171. [DOI] [PubMed] [Google Scholar]
- 84.Mega JL, Hochholzer W, Frelinger AL, 3rd, Kluk MJ, Angiolillo DJ, Kereiakes DJ, Isserman S, Rogers WJ, Ruff CT, Contant C, Pencina MJ, Scirica BM, Longtine JA, Michelson AD, Sabatine MS. Dosing clopidogrel based on cyp2c19 genotype and the effect on platelet reactivity in patients with stable cardiovascular disease. JAMA. 2011;306:2221–2228. doi: 10.1001/jama.2011.1703. [DOI] [PubMed] [Google Scholar]
- 85.Geisler T, Schaeffeler E, Dippon J, Winter S, Buse V, Bischofs C, Zuern C, Moerike K, Gawaz M, Schwab M. Cyp2c19 and nongenetic factors predict poor responsiveness to clopidogrel loading dose after coronary stent implantation. Pharmacogenomics. 2008;9:1251–1259. doi: 10.2217/14622416.9.9.1251. [DOI] [PubMed] [Google Scholar]
- 86.Sibbing D, Koch W, Gebhard D, Schuster T, Braun S, Stegherr J, Morath T, Schomig A, von Beckerath N, Kastrati A. Cytochrome 2c19*17 allelic variant, platelet aggregation, bleeding events, and stent thrombosis in clopidogrel-treated patients with coronary stent placement. Circulation. 2010;121:512–518. doi: 10.1161/CIRCULATIONAHA.109.885194. [DOI] [PubMed] [Google Scholar]
- 87.Price MJ, Berger PB, Teirstein PS, et al. Standard- vs high-dose clopidogrel based on platelet function testing after percutaneous coronary intervention: The gravitas randomized trial. JAMA. 2011;305:1097–1105. doi: 10.1001/jama.2011.290. [DOI] [PubMed] [Google Scholar]
- 88.Wei MY, Ito MK, Cohen JD, Brinton EA, Jacobson TA. Predictors of statin adherence, switching, and discontinuation in the usage survey: Understanding the use of statins in america and gaps in patient education. J Clin Lipidol. 2013;7:472–483. doi: 10.1016/j.jacl.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 89.Group SC, Link E, Parish S, Armitage J, Bowman L, Heath S, Matsuda F, Gut I, Lathrop M, Collins R. Slco1b1 variants and statin-induced myopathy--a genomewide study. N Engl J Med. 2008;359:789–799. doi: 10.1056/NEJMoa0801936. [DOI] [PubMed] [Google Scholar]
- 90.International Warfarin Pharmacogenetics C. Klein TE, Altman RB, Eriksson N, Gage BF, Kimmel SE, Lee MT, Limdi NA, Page D, Roden DM, Wagner MJ, Caldwell MD, Johnson JA. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009;360:753–764. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zabalza M, Subirana I, Sala J, Lluis-Ganella C, Lucas G, Tomas M, Masia R, Marrugat J, Brugada R, Elosua R. Meta-analyses of the association between cytochrome cyp2c19 loss- and gain-of-function polymorphisms and cardiovascular outcomes in patients with coronary artery disease treated with clopidogrel. Heart. 2012;98:100–108. doi: 10.1136/hrt.2011.227652. [DOI] [PubMed] [Google Scholar]
- 92.Johnson JA, Cavallari LH. Warfarin pharmacogenetics. Trends Cardiovasc Med. 2015;25:33–41. doi: 10.1016/j.tcm.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jorgensen AL, FitzGerald RJ, Oyee J, Pirmohamed M, Williamson PR. Influence of cyp2c9 and vkorc1 on patient response to warfarin: A systematic review and meta-analysis. PLoS One. 2012;7:e44064. doi: 10.1371/journal.pone.0044064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cooper GM, Johnson JA, Langaee TY, Feng H, Stanaway IB, Schwarz UI, Ritchie MD, Stein CM, Roden DM, Smith JD, Veenstra DL, Rettie AE, Rieder MJ. A genome-wide scan for common genetic variants with a large influence on warfarin maintenance dose. Blood. 2008;112:1022–1027. doi: 10.1182/blood-2008-01-134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Takeuchi F, McGinnis R, Bourgeois S, Barnes C, Eriksson N, Soranzo N, Whittaker P, Ranganath V, Kumanduri V, McLaren W, Holm L, Lindh J, Rane A, Wadelius M, Deloukas P. A genome-wide association study confirms vkorc1, cyp2c9, and cyp4f2 as principal genetic determinants of warfarin dose. PLoS Genet. 2009;5:e1000433. doi: 10.1371/journal.pgen.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Finkelman BS, Gage BF, Johnson JA, Brensinger CM, Kimmel SE. Genetic warfarin dosing: Tables versus algorithms. J Am Coll Cardiol. 2011;57:612–618. doi: 10.1016/j.jacc.2010.08.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gage BF, Eby C, Johnson JA, et al. Use of pharmacogenetic and clinical factors to predict the therapeutic dose of warfarin. Clin Pharmacol Ther. 2008;84:326–331. doi: 10.1038/clpt.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kimmel SE, French B, Kasner SE, et al. A pharmacogenetic versus a clinical algorithm for warfarin dosing. N Engl J Med. 2013;369:2283–2293. doi: 10.1056/NEJMoa1310669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pirmohamed M, Burnside G, Eriksson N, et al. A randomized trial of genotype-guided dosing of warfarin. N Engl J Med. 2013;369:2294–2303. doi: 10.1056/NEJMoa1311386. [DOI] [PubMed] [Google Scholar]
- 100.Ross S, Nejat S, Pare G. Use of genetic data to guide therapy in arterial disease. J Thromb Haemost. 2015;13(Suppl 1):S281–289. doi: 10.1111/jth.12924. [DOI] [PubMed] [Google Scholar]
- 101.Zaiou M, El Amri H. Cardiovascular pharmacogenetics: A promise for genomically-guided therapy and personalized medicine. Clin Genet. 2017;91:355–370. doi: 10.1111/cge.12881. [DOI] [PubMed] [Google Scholar]
- 102.Elliott LS, Henderson JC, Neradilek MB, Moyer NA, Ashcraft KC, Thirumaran RK. Clinical impact of pharmacogenetic profiling with a clinical decision support tool in polypharmacy home health patients: A prospective pilot randomized controlled trial. PLoS One. 2017;12:e0170905. doi: 10.1371/journal.pone.0170905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Genomes Project C. Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Oetjens MT, Denny JC, Ritchie MD, Gillani NB, Richardson DM, Restrepo NA, Pulley JM, Dilks HH, Basford MA, Bowton E, Masys DR, Wilke RA, Roden DM, Crawford DC. Assessment of a pharmacogenomic marker panel in a polypharmacy population identified from electronic medical records. Pharmacogenomics. 2013;14:735–744. doi: 10.2217/pgs.13.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Garmaroudi FS, Handy DE, Liu YY, Loscalzo J. Systems pharmacology and rational polypharmacy: Nitric oxide-cyclic gmp signaling pathway as an illustrative example and derivation of the general case. PLoS Comput Biol. 2016;12:e1004822. doi: 10.1371/journal.pcbi.1004822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Stingl Kirchheiner JC, Brockmoller J. Why, when, and how should pharmacogenetics be applied in clinical studies?: Current and future approaches to study designs. Clin Pharmacol Ther. 2011;89:198–209. doi: 10.1038/clpt.2010.274. [DOI] [PubMed] [Google Scholar]
- 107.Manrai AK, Funke BH, Rehm HL, Olesen MS, Maron BA, Szolovits P, Margulies DM, Loscalzo J, Kohane IS. Genetic misdiagnoses and the potential for health disparities. N Engl J Med. 2016;375:655–665. doi: 10.1056/NEJMsa1507092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mirnezami R, Nicholson J, Darzi A. Preparing for precision medicine. N Engl J Med. 2012;366:489–491. doi: 10.1056/NEJMp1114866. [DOI] [PubMed] [Google Scholar]
- 109.Fuster V. A first dilemma in cardiovascular medicine: Adherence versus personalized therapy. J Am Coll Cardiol. 2014;64:1059–1060. doi: 10.1016/j.jacc.2014.07.936. [DOI] [PubMed] [Google Scholar]
- 110.Bosworth HB, Granger BB, Mendys P, et al. Medication adherence: A call for action. Am Heart J. 2011;162:412–424. doi: 10.1016/j.ahj.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kohane IS. Health care policy. Ten things we have to do to achieve precision medicine. Science. 2015;349:37–38. doi: 10.1126/science.aab1328. [DOI] [PubMed] [Google Scholar]
- 112.Joyner MJ. Precision medicine, cardiovascular disease and hunting elephants. Prog Cardiovasc Dis. 2016;58:651–660. doi: 10.1016/j.pcad.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 113.Fuster V. A second dilemma in cardiovascular medicine: Personalized medicine versus personal interaction with the patient. J Am Coll Cardiol. 2014;64:1292–1293. doi: 10.1016/j.jacc.2014.08.006. [DOI] [PubMed] [Google Scholar]