Abstract
Respiratory syncytial virus (RSV) has been identified as a main cause of hospitalisation in infants and children. To date, the current therapeutic arsenal is limited to ribavirin and palivizumab with variable efficacy. In this work, starting from a number of in-house series of previously described anti-RSV agents based on the benzimidazole scaffold, with the aim at gaining a better understanding of the related chemical features involved in potency and safety profiles, we applied a computational study including two focussed comparative molecular fields analysis (CoMFA) and comparative molecular similarity indices analysis (CoMSIA). The results allowed us to derive useful suggestions for the design of derivatives and also to set up statistical models predicting the potency and selectivity index (SI = CC50/EC50) of any new analogue prior to synthesis. Accordingly, here, we discuss preliminary results obtained through the applied exhaustive QSAR analyses, leading to design and synthesise more effective anti-RSV agents.
Keywords: Benzimidazoles, CoMFA, CoMSIA, respiratory syncytial virus, 3D-QSAR
Introduction
Respiratory syncytial virus (RSV) is the aetiological agent of serious and widespread respiratory tract infections that represent the major cause of hospitalisation of infants and children. The virus is highly contagious and frequent re-infections may cause morbidity in elderly people affected by chronic illnesses and in immuno-compromised individuals. Epidemiologic studies proved RSV as a ubiquitous contributor to pulmonary exacerbation, particularly in cystic fibrosis (CF) patients, causing severe clinical symptoms and promoting comorbidities with bacterial pathogens1.
Despite the huge economic impact and the medical needs associated with severe RSV infection, therapy is restricted to ribavirin, whose effectiveness, however, is highly questionable2. Humanised monoclonal antibody palivizumab (Synagis®) is used for the prevention in high-risk infants, with partial efficacy in reducing RSV hospitalisation rates3. Insufficient clinical observation exist to concretely support whether this drug is cost-effective and safe4,5. Thus, there is a clear need for new drugs to prevent and treat RSV infection.
Member of Paramyxoviridae family, RSV has a negative-stranded RNA genome that encodes 11 viral proteins6. The main antigenic determinants, F (fusion) and G (attachment) glycoproteins, are expressed on the virion surface and play a key role in viral entry into host cells; therefore, they represent interesting targets of small molecule inhibitors.
Case in point as a future RSV therapeutic is the orally administered GS-58067 that was found as a potent inhibitor of a broad range of RSV clinical isolates by blocking the virus–cell fusion process8. GS-5806 needs to be improved for its low efficacy against the emergent mutant strains, an issue that elicits concern for efficacy of the current antiviral therapies, in general9.
Benzimidazole and its derivatives are important bioactive molecules class in drugs and pharmaceuticals fields10: they exhibit significant activity against several viruses and are also endowed with antimicrobial, anti-inflammatory, antitumour, antiparasitic, antiprotozoal properties.
The activity against RSV is peculiar for several benzimidazole-based chemotypes11, which were demonstrated to target viral replication machinery by blocking fusion and entry processes12,13. Continue efforts are devoted to design new RSV inhibitors, by decorating the benzimidazole core structure, from which promising leads reached clinical trials14. The benzimidazole derivative, TMC353121, has been shown to inhibit the viral fusion process in an African green monkeys model15: administered under continuous intravenous infusion, it achieved a complete inhibition of RSV entry into host cells in a dose-dependent manner, in contrast to previously described fusion inhibitors that exhibited only a reduction in viral load.
Recently, we identified three principle classes of anti-RSV benzimidazole derivatives, such as 2-benzylbenzimidazoles16, 2-phenylbenzimidazoles17 and [(benzotriazol-1/2-yl)methyl]benzimidazoles18; among them, the most active compounds showed even submicromolar potency associated with a high safety profile (SI = CC50/EC50). In fact, the 75% of the compound library displayed a SI value from >14.3 up to >3333, comparing favourably with the drug ribavirin (SI >14.3), used as positive control.
In this work, starting from these previously studied series of benzimidazole-based anti-RSV agents16–18 including 156 compounds (Figure 1), aiming to gain a better understanding of the related chemical features involved in potency and safety profiles, we applied a computational study including exhaustive quantitative structure–activity relationship (3D-QSAR) analyses. Indeed, the ligand-based approach represents a useful tool guiding for rational drug design, as shown in the literature19–21.
Figure 1.
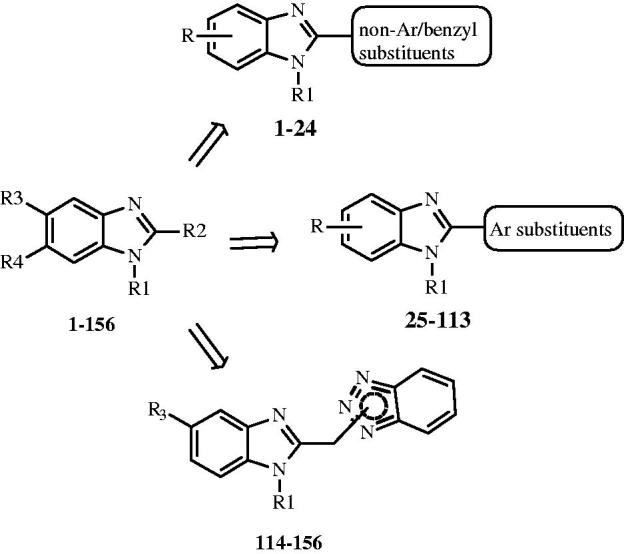
General structures of benzimidazole-based anti-RSV agents
In detail, we developed two comparative molecular fields analysis (CoMFA) and comparative molecular similarity indices analysis (CoMSIA). The first one was performed around the ligands potency values (pEC50), the second one using the pCC50 parameter (cytotoxicity) against human MT-4 cell line. The results allowed us to derive useful suggestions for the design of new derivatives and also to set up statistical models predicting the potency and selectivity index (SI = CC50/EC50) of any new analogue prior to synthesis.
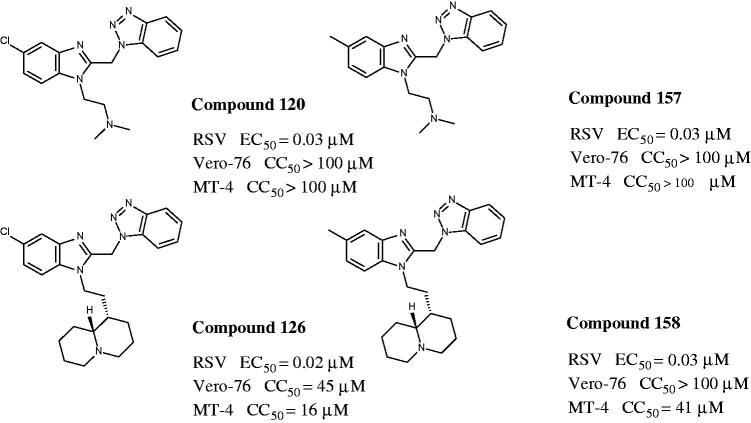
Basing on CoMFA and CoMSIA analyses, we deemed interesting to design and synthesise two new compounds (157, 158), that bear in position 1 of the (benzotriazol-1-yl)methyl]benzimidazole scaffold the 2-(N,N-dimethylamino)ethyl or homo-lupinyl chains, which were previously found as the best suited substitutions, responsible for a high potency profile. These chemical features have been now associated with the new investigated apolar CH3 group in position 5 of the benzimidazole ring, with the aim at improving the activity and even more at reducing the cytotoxicity. Compound were evaluated against RSV in Vero-76 cell cultures in order to validate the computational analyses results.
Methods
Data set
All the compounds here evaluated were built in silico and energy minimised within MOE using MMFF94 force field22. Therefore, all the compounds were also parameterised by means of the Gasteiger–Hückel method. All calculations were carried out using a PC running the Windows XP operating system.
3D-QSAR analyses
The benzimidazoles 1–156 have been aligned and submitted to 3D-QSAR studies through CoMFA and CoMSIA analyses, by means of Sybyl-X1.023.
Model A and model B CoMFA and CoMSIA analyses were performed to analyse the impact played by steric, electrostatic, hydrophobic, H-bond acceptor and H-bond donor features around the potency as anti-RSV agents and the cytotoxicity trend of these series of benzimidazoles.
Training set and test set
Starting from all compounds, a number of benzimidazoles were grouped into a training set, for model generation, and a test set, for model validation, containing: (i) 50 and 11 compounds for model A; (ii) 123 and 32 derivatives for model B, respectively. In any model, the molecules selected for the training and the test set pools were chosen manually, based on representative criteria of the overall biological activity trend and structural variations.
For model A and model B analyses, the RSV EC50 values and the human MT-4 cell line CC50 ones have been transformed into pEC50 and pCC50 values, respectively, and then used as response variables.
CoMFA and CoMSIA models and statistical evaluation
CoMFA24 and CoMSIA25 methods are widely used 3D-QSAR techniques being useful to relate any variation of an experimentally determined parameter (dependent variables), related to a set of molecules, with respect to specific descriptors which are considered as independent variables. In particular, the steric and electrostatic fields and especially the hydrophobic, H-bond donor, H-bond acceptor ones were calculated by CoMFA and CoMSIA analysis, respectively. Starting from a proper molecule alignment within a 3D cubic lattice (with a 2 Å grid spacing), any descriptor was calculated, using the standard Tripos force field method. Successively, the reliability of the derived models can be evaluated using specific statistical tools, such as partial least square (PLS) analysis and cross-validation methods.
Finally, the predictive ability about those compounds included in the test set (r2pred) was also calculated, by means of the following equation:
being SD and PRESS the sum of the squared deviations between the biological activities of the test set molecules and the mean activity of the training set compounds, and the squared deviation between the observed and the predicted activities of the test set compounds, respectively.
Any further detail concerning the (standard) CoMFA and CoMSIA procedures and the statistical and predictive evaluation we applied, were previously elucidated in a consistent number of our works26–29.
Chemistry
General synthetic methods
Melting points were taken in open glass capillaries on a Büchi apparatus and were uncorrected. Elemental analyses were performed on a Carlo Erba EA-1110 CHNS-O instrument in the Microanalysis Laboratory of the Department of Pharmaceutical Sciences of Genoa University. The analytical results are within ±0.4% of calculated values. 1H NMR and 13C NMR spectra (reported as supplemental material S1–S2) were recorded in CDCl3 or DMSO-d6 on Varian Gemini-200 spectrometer; d in ppm rel. to Me4Si as internal standard. J in Hz. Q = quinolizidine ring. Results of elemental analyses, thin-layer chromatography (TLC) and nuclear magnetic resonance (NMR) spectra indicated that the purity of all compounds was ≥95%.
Chemicals, solvents and commercially available intermediates were purchased from Aldrich (Milan). The non-commercially available intermediates were prepared according to the literature, or as follows, when not previously known.
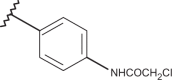
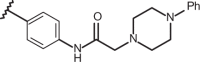
2-[(1H-1,2,3-benzotriazol-1-yl)methyl]-1-[2-(N,N-dimethylamino)ethyl]-5-methylbenzimidazole (157) and 2-[(1H-1,2,3-benzotriazol-1-yl)methyl]-1-[(1S,9aR)-(octahydro-2H-quinolizin-1-yl)ethyl]-5- methylbenzimidazole (158)
General synthetic method. A mixture of the proper N-substituted 1,2-phenylenediamine (3.6 mmol) and (1H-1,2,3-benzotriazol-1-yl)acetic acid (1.28 g, 7.2 mmol) was heated at 180 °C under N2 for 90 min with manual stirring. After cooling, 1N HCl (20 mL) was added, filtering and washing with H2O an amount of unreacted acid. The aqueous solutions were basified with a solution of 6N NaOH and extracted with Et2O. After drying (Na2SO4) the solvent was evaporated, leaving a spongy residue that was cromatographed or crystallised with dry Et2O.
157. Yield: 66%. CC(Al2O3/Et2O). M.p. 125–126 °C. 1H-NMR (200 MHz, CDCl3): 2.29 (s, 6H, N(CH3)2); 2.38 (t, J = 7.0, 2H, CH2CH2N(CH3)2); 2.51 (s, 3H, CH3Ar); 4.31 (t, J = 7.2, 2H, CH2CH2N(CH3)2); 6.27 (s, 2H, CH2-benzotriazole); 7.12–7.25 (m, 2 arom. H); 7.33–7.48 (m, 2 arom. H); 7.60–7.67 (m, 1 arom. H); 7.76–7.84 (m, 1 arom. H); 8.03–8.10 (m, 1 arom. H). 13C NMR (50 MHz, CDCl3): 145.72, 145.20, 141.51, 132.33, 131.98, 131.43, 126.96, 124.22, 123.36, 118.99, 118.72, 109.62, 108.40, 56.81, 45.02, 44.55, 41.22, 20.45. Anal. calc. for C19H22N6: C 68.24, H 6.63, N 25.13; Found: C 68.05, H 6.55, N 25.17.
158. Yield: 40%. M.p. 171–172 °C (Et2O). 1H-NMR (200 MHz, CDCl3): 1.00–2.05 (m, 16H of CH2-Q); 2.51 (s, CH3); 2.68–2.88 (m, 2H, Hα near N of Q); 4.07–4.31 (m, 2H of CH2-CH2-Q); 6.09–6.35 (AB syst., 2H, CH2-benzotriazole); 7.08–7.46 (m, 4 arom. H); 7.60–7.82 (m, 2 arom. H); 8.04 (d, J = 9.0, 1 arom. H). 13C NMR (50 MHz, CDCl3): 145.22, 144.74, 141.46, 132.35, 131.90, 131.36, 127.05, 124.24, 123.38, 118.91, 118.61, 109.68, 108.59, 45.24, 42.25, 34.95, 23.03, 20.47, 19.81. Anal. calc. for C26H32N6: C 72.87, H 7.53, N 19.61; Found: C 72.92, H 7.64, N 19.29.
Intermediates. N-substituted 1,2-phenylendiamines
General synthetic method. To a stirred solution of the above nitroderivative (1.3 mmol) in EtOH (7 mL), a solution of SnCl2.2H2O (3.9 mmol, 0.88 g) in conc. HCl (10 mL) was slowly added. The mixture was refluxed for 6 h and then concentrated in vacuo. The residue was taken up in H2O, alkalinised with a solution of 6N NaOH and then extracted with Et2O. The organic layer was dried (Na2SO4), filtered and then evaporated, affording a yellow oil. A small amount was converted into dihydrochloride salt with a 1N ethanolic solution of HCl for performing elemental analysis.
2-Amino-N-[2-(N’,N’-dimethylamino)ethyl]-4-methylbenzeneamine
Yield: 88%. 1H-NMR (200 MHz, CDCl3): 2.26 (s, 6H, N(CH3)2); 2.28 (s, ArCH3); 2.60 (t, J = 7.2, 2H, CH2CH2N(CH3)2); 3.16 (t, J = 7.2, 2H, CH2CH2N(CH3)2); 3.30–3.60 (br s, 3H, NH2 and NH, collapse with D2O); 6.55–6.70 (m, 3 arom. H). Dihydrochloride: M.p. 186–189 °C (EtOH). Anal. calc. for C11H19N3 + 2HCl: C 49.63, H 7.95, N 15.78; Found: C 49.66, H 7.85, N 15.60.
2-Amino-N-[2-(1S,9aR)-(octahydro-2H-quinolizin-1-yl)ethyl]-4-methylbenzeneamine
Yield: 82%. 1H-NMR (200 MHz, CDCl3): 1.20–2.03 (m, 16 H of Q); 2.21 (s, 3 H, CH3Ar); 2.76–2.85 (m, 2 H, Hα near N of Q); 2.95–3.04 (m, 1 H of CH2NH); 3.07–3.18 (m, 1 H of CH2NH); 3.31 (br. s, 3 H, NH and NH2, collapse with D2O); 6.53–6.62 (m, 3 arom. H). Dihydrochloride: M.p. 106–109 °C (with swelling). Anal. calc. for C18H29N3 +2HCl: C 59.99, H 8.68, N 11.66; Found: C 59.87, H 8.76, N 11.62.
N-[2-(1S,9aR)-(octahydro-2H-quinolizin-1-yl)ethyl]-4-methyl-2-nitrobenzeneamine
A solution of homo-lupinylamine (5 mmol) and 4-chloro-3-nitrotoluene (5 mmol) in DMF (3 mL) was heated in a pressure tube at 120 °C with stirring for 9 h. At room temperature, H2O was added and the mixture alkalinised with a solution of 6N NaOH and extracted with Et2O. The organic phase was extracted with diluted HCl; the acid solution was alkalised and extracted with Et2O. After evaporation, the oily residue was purified by CC(SiO2/Et2O + 2%MeOH). A small amount was converted into monohydrochloride salt (M.p. 176–179 °C) with 1N ethanolic solution of HCl for performing elemental analysis.
Yield: 40%. 1H-NMR (200 MHz, CDCl3): 1.18–2.06 (m, 16 of Q); 2.25 (s, 3H, CH3Ar); 2.77–2.84 (m, 2H, Hα near N of Q); 3.17–3.25 (m, 1H, CH2NH); 3.28–3.40 (m, 1H, CH2NH); 6.78 (d, J = 9.0, 1 arom. H); 7.24 (dd, J = 8.6, 1.2, 1 arom. H); 7.84 (br. s, 1H, NH, collapses with D2O); 8.00 (d, J = 9.2, 1 arom. H). Anal. calc. for C18H27N3O2 + HCl: C 61.09, H 7.97, N 11.87; Found: C 61.03, H 8.23, N 11.84.
Biological assays
Cells and viruses
Cell lines and RSV were purchased from American Type Culture Collection (ATCC). The absence of mycoplasma contamination was checked periodically by the Hoechst staining method. Cell line supporting the multiplication of RSV was the Monkey kidney (Vero 76) [ATCC CRL 1587 Cercopithecus Aethiops], while CD4+ human T cells containing an integrated HTLV-1 genome (MT-4) was used as human cellular model. Human respiratory syncytial virus (RSV) [strain A2 (ATCC VR-1302)].
Cytotoxicity assays
Exponentially growing MT-4 cells were seeded at an initial density of 1 × 105 cells/mL in 96-well plates in RPMI-1640 medium supplemented with 10% foetal bovine serum (FBS), 100 units/mL penicillin G and 100 µg/mL streptomycin. Cell cultures were then incubated at 37 °C in a humidified, 5% CO2 atmosphere in the absence or presence of serial dilutions of test compounds. Cell viability was determined after 96 h at 37 °C by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT) method30.
Vero-76 cells were seeded at an initial density of 4 × 105 cells/mL in 24-well plates, in culture medium (Dulbecco’s modified Eagle medium (D-MEM) with L-glutamine, supplemented with foetal bovine serum (FBS), 0.025 g/L kanamycin). Cell cultures were then incubated at 37 °C in a humidified, 5% CO2 atmosphere in the absence or presence of serial dilutions of test compounds. Cell viability was determined after 48–96 h at 37 °C by the crystal violet staining method.
The results are expressed as CC50, which is the concentration of compound necessary to inhibit cell growth by 50%. Each CC50 value is the mean and standard deviation of at least three separate experiments performed in duplicate.
Antiviral assays
Antiviral activity against RSV was determined by plaque reduction assays in infected cell monolayers. To this end, Vero 76-cells were seeded in 24-well plates at a density of 2 × 105 cells/well and were allowed to form confluent monolayers by incubating overnight in growth medium (Dulbecco’s modified Eagle medium (D-MEM) with L-glutamine and 4500 mg/L D-glucose and 0.025 g/L kanamycin, supplemented with 10% FBS) at 37 °C in a humidified CO2 (5%) atmosphere. Then, monolayers were infected for 2 h with 250 µL of proper viral dilutions to give 50 to 100 PFU/well. Following removal of unadsorbed virus, 500 µL of maintenance medium [D-MEM with L-glutamine and 4500 mg/L D-glucose, supplemented with 1% inactivated FBS] containing 0.75% methylcellulose, without or with serial dilutions of test compounds, were added. Cultures were incubated at 37 °C for 5 days and then fixed with PBS containing 50% ethanol and 0.8% crystal violet, washed and air-dried. Plaques were then counted.
Linear regression analysis
The extent of cell growth/viability and viral multiplication, at each drug concentration tested, were expressed as percentage of untreated controls. Concentrations resulting in 50% inhibition (CC50 or EC50) were determined by linear regression analysis.
Results
Chemistry
Preparations of compounds 157 and 158 were accomplished by the general synthetic steps in Scheme 1.
Scheme 1.
Reagents and conditions: (a) 120 °C, 9 h; (b) SnCl2 . 2H2O, conc. HCl, EtOH, 6 h at reflux; (c) 180°, N2, 90 min.
While the N-[2-(N′,N′-dimethylamino)ethyl]-4-methyl-2-nitrobenzeneamine was obtained according to the literature31, the N-(homo-lupinyl)-4-methyl-2-nitrobenzeneamine was synthesised by reacting the 1-chloro-4-methyl-2-nitrobenzene with homo-lupinylamine (2-((1S,9aR)-octahydro-2H-quinolizin-1-yl)ethanamine)32 at 120 °C for 9 h. Then, the two nitrobenzene amines were reduced with SnCl2*2H2O in concentrated HCl to give the corresponding 1,2-phenylendiamines that were heated to fusion at 180 °C for 90 min with 1H-1,2,3-benzotriazol-1-ylacetic acid33, providing the title compounds.
Biological activity
The two newly synthesised benzimidazole derivatives were assayed for in vitro antiviral activity against the respiratory syncytial virus (RSV). Cytotoxicity was evaluated in parallel with the antiviral activity against the primate Vero76 and human MT-4 cell lines. As reference inhibitors were used ribavirin (pEC50 = 5.15 MT-4 pCC50 = 4.51), NM299 (6-azauridine; pEC50 = 5.92 MT-4 pCC50 = 5.70) and M5255 (mycophenolic acid; pEC50 = 6.22 MT-4 pCC50 = 6.70).
3D-QSAR analyses
Starting from the in-house compounds 1–156 (Table 1), CoMFA and CoMSIA analyses here reported were used to explore, through quantitative methods, the main features responsible for the anti-RSV activity (model A) of benzimidazole-based derivatives and also for the related cytotoxicity profile (model B).
Table 1.
Chemical structure of benzimidazoles 1–156* and the related anti-RSV potency and cytotoxicity profiles (evaluated against MT-4 and VERO-76 cell lines)
| Comp. | R1 | R2 | R3 | R4 | RSV pEC50 | MT-4 pCC50 | VERO-76 pCC50 |
|---|---|---|---|---|---|---|---|
| 1 |  |
–CH3 | –CF3 | –H | <4.00 | 4.00 | 4.00 |
| 2 |  |
–CF3 | –CF3 | –H | 4.66 | 4.27 | 4.00 |
| 3 | -H |  |
–H | –H | <4.00 | 4.00 | 4.00 |
| 4 |  |
 |
–CF3 | –H | <4.00 | 4.96 | 4.00 |
| 5 | –H |  |
–H | –H | <4.00 | 4.00 | 4.00 |
| 6 | –H |  |
–CF3 | –H | < 4.00 | 4.00 | 4.15 |
| 7 | –H |  |
–CF3 | –H | <4.00 | 4.16 | 4.04 |
| 8 | –H |  |
–CF3 | –H | <4.00 | 4.00 | 4.00 |
| 9 | –C4H9(n) |  |
–CF3 | –H | 5.05 | 4.00 | 4.00 |
| 10 | –CH2CH2OCH3 |  |
–CF3 | –H | <4.00 | 4.00 | 4.00 |
| 11 | –H |  |
–Cl | –Cl | 5.30 | 4.54 | 4.00 |
| 12 | –H | 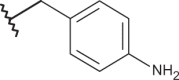 |
–Cl | –Cl | 4.70 | 4.15 | 4.10 |
| 13 | –(CH2)2N(C2H5)2 | 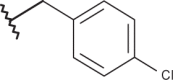 |
–H | –H | <4.00 | 4.38 | 4.00 |
| 14 | –(CH2)2N(C2H5)2 | 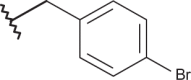 |
–H | –H | 4.12 | 4.31 | 4.00 |
| 15 | –(CH2)2N(C2H5)2 | 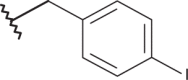 |
–H | –H | 4.40 | 4.60 | 4.00 |
| 16 | –(CH2)2N(CH3)2 |  |
–CF3 | –H | <4.00 | 4.00 | 4.00 |
| 17 | –(CH2)3N(C2H5)2 |  |
–CF3 | –H | <4.00 | 4.40 | 4.00 |
| 18 |  |
 |
–CF3 | –H | <4.00 | 4.00 | 4.03 |
| 19 | 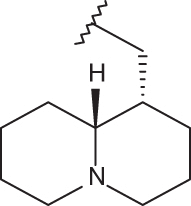 |
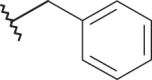 |
–CF3 | –H | <4.00 | 4.00 | 4.00 |
| 20 | 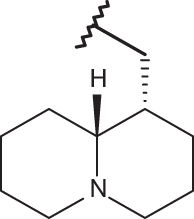 |
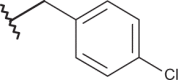 |
–CF3 | –H | 4.82 | 5.22 | 4.00 |
| 21 | 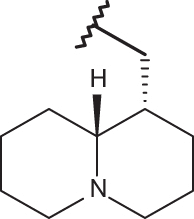 |
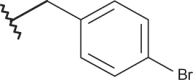 |
–CF3 | –H | 4.60 | 4.89 | 4.00 |
| 22 | 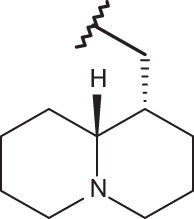 |
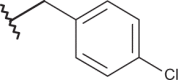 |
–Cl | –Cl | <4.00 | 4.77 | 4.00 |
| 23 | 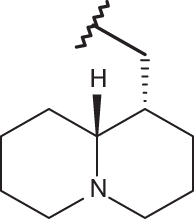 |
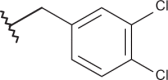 |
–Cl | –Cl | 4.92 | 5.05 | 4.00 |
| 24 | 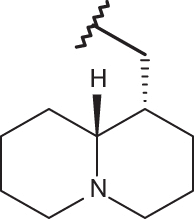 |
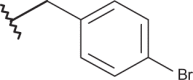 |
–Cl | –Cl | 5.05 | 4.92 | 4.00 |
| 25 | –H |  |
–H | –H | 4.40 | 4.22 | 4.00 |
| 26 | –H |  |
–H | –H | <4.00 | 5.15 | 4.00 |
| 27 | –H |  |
–CF3 | –H | 4.70 | 4.77 | 4.00 |
| 28 | –H |  |
–CF3 | –H | <4.82 | 5.70 | 4.82 |
| 29 | –H | 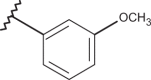 |
–CF3 | –H | 5.15 | 4.72 | 4.00 |
| 30 | –H | 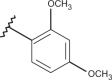 |
–CF3 | –H | 4.00 | 4.00 | 4.00 |
| 31 | –H | 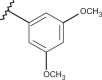 |
–CF3 | –H | 5.00 | 4.72 | 4.00 |
| 32 | –H | 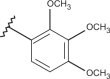 |
–CF3 | –H | <4.15 | 4.03 | 4.15 |
| 33 | –H | 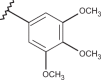 |
–CF3 | –H | <4.54 | 5.40 | 4.54 |
| 34 | –H | 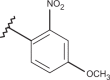 |
–CF3 | –H | <4.07 | 4.70 | 4.07 |
| 35 | –H | 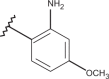 |
–CF3 | –H | <4.30 | 5.30 | 4.30 |
| 36 | –H | 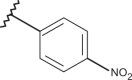 |
–CF3 | –H | <4.35 | 5.22 | 4.35 |
| 37 | –H | 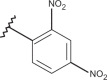 |
–CF3 | –H | <4.60 | 5.05 | 4.60 |
| 38 | –H | 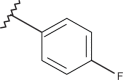 |
–CF3 | –H | <4.22 | 4.66 | 4.22 |
| 39 | –H | 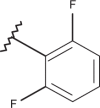 |
–CF3 | –H | <4.60 | 4.00 | 4.60 |
| 40 | –H | 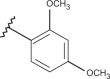 |
–NO2 | –H | <4.00 | 4.00 | 4.00 |
| 41 | –H | 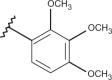 |
–NO2 | –H | <4.00 | 4.00 | 4.00 |
| 42 | –H | 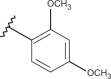 |
–COCH3 | –H | <.00 | 4.00 | 4.00 |
| 43 | –H | 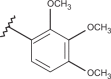 |
–COCH3 | –H | <4.00 | 4.00 | 4.00 |
| 44 | –H | 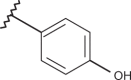 |
–Cl | –Cl | 5.15 | 4.92 | 4.74 |
| 45 | –H | 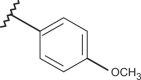 |
–Cl | –Cl | <4.52 | 6.00 | 4.52 |
| 46 | –H | 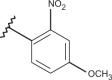 |
–Cl | –Cl | <4.10 | 5.70 | 4.10 |
| 47 | –H | 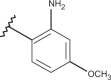 |
–Cl | –Cl | <4.12 | 5.52 | 4.12 |
| 48 | –H | 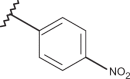 |
–Cl | –Cl | <4.00 | 4.77 | 4.00 |
| 49 | –H | 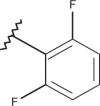 |
–Cl | –Cl | <4.00 | 4.77 | 4.00 |
| 50 | –H | 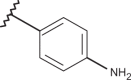 |
–H | –H | n.d | 4.00 | 4.00 |
| 51 | –H | 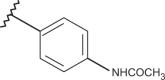 |
–H | –H | n.d | 4.00 | 4.00 |
| 52 | –H | 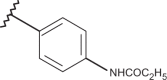 |
–H | –H | n.d | 4.46 | 4.00 |
| 53 | –H | 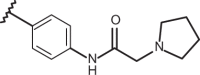 |
–H | –H | n.d | 4.40 | 4.00 |
| 54 | –H | 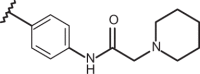 |
–H | –H | n.d | 4.51 | 4.00 |
| 55 | –H | 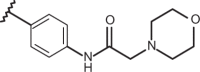 |
–H | –H | n.d | 4.00 | 4.00 |
| 56 | –H | 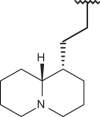 |
–H | –H | n.d | 4.74 | 4.10 |
| 57 | –H |  |
–CF3 | –H | n.d | 4.21 | 4.00 |
| 58 | –H |  |
–CF3 | –H | n.d | 4.72 | 4.30 |
| 59 | –H |  |
–CF3 | –H | n.d | 4.00 | 4.00 |
| 60 | –H |  |
–CF3 | –H | n.d | 5.30 | 4.46 |
| 61 | –H | 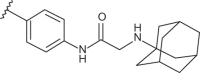 |
–CF3 | –H | n.d | 4.82 | 4.35 |
| 62 | –H | 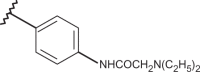 |
–CF3 | –H | n.d | 5.10 | 4.62 |
| 63 | –H | 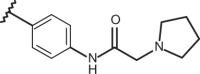 |
–CF3 | –H | n.d | 4.74 | 4.00 |
| 64 | –H | 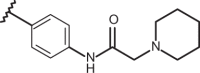 |
–CF3 | –H | n.d | 5.22 | 4.51 |
| 65 | –H | 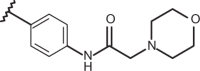 |
–CF3 | –H | n.d | 4.00 | 4.00 |
| 66 | –H | 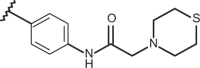 |
–CF3 | –H | n.d | 5.00 | 4.00 |
| 67 | –H | 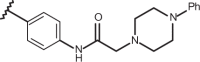 |
–CF3 | –H | n.d | 6.05 | 4.00 |
| 68 | –H | 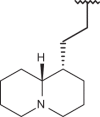 |
–CF3 | –H | n.d | 5.40 | 4.40 |
| 69 | –H | 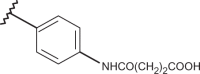 |
–CF3 | –H | n.d | 4.00 | 4.00 |
| 70 | –H | 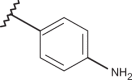 |
–NO2 | –H | n.d | 4.00 | 4.00 |
| 71 | –H | 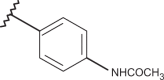 |
–NO2 | –H | n.d | 4.00 | 4.00 |
| 72 | –H | 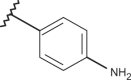 |
–Cl | –Cl | n.d | 4.77 | 4.00 |
| 73 | –H | 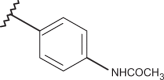 |
–Cl | –Cl | n.d | 4.00 | 4.00 |
| 74 | –H | 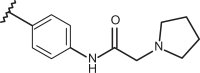 |
–Cl | –Cl | n.d | 5.10 | 4.00 |
| 75 | –H | 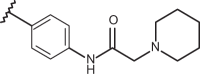 |
–Cl | –Cl | n.d | 4.27 | 4.00 |
| 76 | –H | 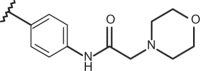 |
–Cl | –Cl | n.d | 4.74 | 4.00 |
| 77 | –H | 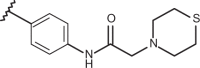 |
–Cl | –Cl | n.d | 4.00 | 4.00 |
| 78 | –H | 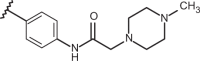 |
–Cl | –Cl | n.d | 4.96 | 4.00 |
| 79 | –CH3 | 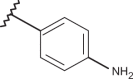 |
–CF3 | –H | n.d | 4.70 | 4.00 |
| 80 | –CH3 | 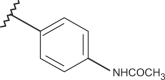 |
–CF3 | –H | n.d | 4.82 | 4.00 |
| 81 | –CH3 | 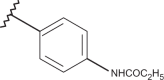 |
–CF3 | –H | n.d | 5.10 | 4.00 |
| 82 | –CH3 | 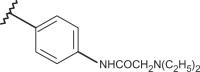 |
–CF3 | –H | n.d | 5.04 | 4.05 |
| 83 | –CH3 | 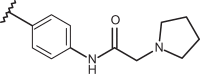 |
–CF3 | –H | n.d | 4.80 | 4.00 |
| 84 | –CH3 | 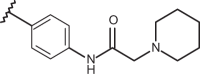 |
–CF3 | –H | n.d | 5.05 | 4.00 |
| 85 | –CH3 | 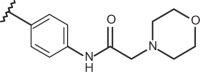 |
–CF3 | –H | n.d | 4.00 | 4.00 |
| 86 | –CH3 | 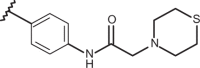 |
–CF3 | –H | n.d | 5.00 | 4.00 |
| 87 | –CH3 | 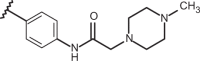 |
–CF3 | –H | n.d | 4.66 | 4.00 |
| 88 | –CH3 | 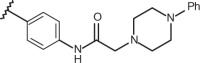 |
–CF3 | –H | n.d | 5.82 | 4.52 |
| 89 |  |
 |
–CF3 | –H | n.d | 4.36 | 4.00 |
| 90 |  |
 |
–CF3 | –H | n.d | 4.62 | 4.52 |
| 91 |  |
 |
–CF3 | –H | n.d | 4.77 | 4.00 |
| 92 |  |
 |
–CF3 | –H | n.d | 4.72 | 4.22 |
| 93 | 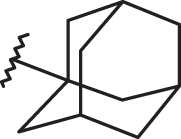 |
 |
–CF3 | –H | n.d | 4.55 | 4.00 |
| 94 | 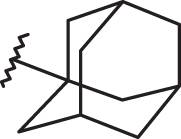 |
 |
–CF3 | –H | n.d | 4.07 | 4.00 |
| 95 | 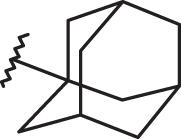 |
 |
–CF3 | –H | n.d | 6.52 | 4.00 |
| 96 | 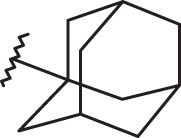 |
 |
–CF3 | –H | n.d | 4.00 | 4.00 |
| 97 | –CH3 |  |
–CF3 | –H | < 4.00 | 4.00 | 4.00 |
| 98 |  |
 |
–CF3 | –H | <4.00 | 4.31 | 4.00 |
| 99 |  |
 |
–H | –H | <4.00 | 4.48 | 4.00 |
| 100 |  |
 |
–H | –H | 5.15 | 4.38 | 4.00 |
| 101 |  |
 |
– H | –H | <4.00 | 4.00 | 4.00 |
| 102 |  |
 |
– H | –CH3 | 4.20 | 4.00 | 4.00 |
| 103 |  |
 |
– H | –Cl | 4.35 | 4.25 | 4.05 |
| 104 |  |
 |
– Cl | –H | <4.00 | 4.00 | 4.00 |
| 105 |  |
 |
–H | –Cl | <4.00 | 4.00 | 4.00 |
| 106 |  |
 |
–CF3 | –H | <4.62 | 4.62 | 4.62 |
| 107 |  |
 |
–CF3 | –H | <4.00 | 4.24 | 4.00 |
| 108 |  |
 |
–CF3 | –H | <4.11 | 4.68 | 4.11 |
| 109 |  |
 |
–Cl | –Cl | <.12 | 4.60 | 4.12 |
| 110 |  |
 |
–Cl | –Cl | <4.00 | 4.38 | 4.00 |
| 111 | –C4H9(n) |  |
–CF3 | –H | <4.10 | 4.00 | 4.10 |
| 112 | –CH2-C6H5 |  |
–H | –H | 4.60 | 4.33 | 4.06 |
| 113 | –H |  |
–H | –H | <4.00 | 4.77 | 4.00 |
| 114 | –(CH2)2N(CH3)2 |  |
–H | –H | 6.15 | 4.00 | 4.00 |
| 115 | –(CH2)2N(C2H5)2 |  |
–H | –H | 5.64 | 4.00 | 4.00 |
| 116 | –(CH2)3N(C2H5)2 |  |
–H | –H | 6.15 | 4.00 | 4.00 |
| 117 |  |
 |
–H | –H | 6.15 | 4.00 | 4.00 |
| 118 |  |
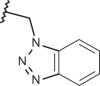 |
–H | –H | 6.52 | 4.04 | 4.00 |
| 119 | 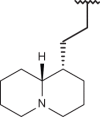 |
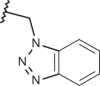 |
–H | –H | 6.82 | 4.19 | 4.00 |
| 120 | –(CH2)2N(CH3)2 | 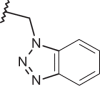 |
–Cl | –H | 7.52 | 4.00 | 4.00 |
| 121 | –(CH2)2N(C2H5)2 |  |
–Cl | –H | 6.15 | 4.00 | 4.00 |
| 122 | –(CH2)3N(CH3)2 |  |
–Cl | –H | 7.22 | 4.00 | 4.00 |
| 123 | –(CH2)3N(C2H5)2 |  |
–Cl | –H | 7.00 | 4.30 | 4.00 |
| 124 |  |
 |
–Cl | –H | 6.05 | 4.43 | 4.22 |
| 125 |  |
 |
–Cl | –H | 7.30 | 4.44 | 4.06 |
| 126 |  |
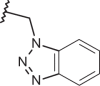 |
–Cl | –H | 7.70 | 4.80 | 4.35 |
| 127 | –(CH2)2N(CH3)2 | 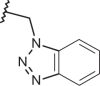 |
–CF3 | –H | 5.00 | 4.00 | 4.00 |
| 128 | –(CH2)2N(C2H5)2 | 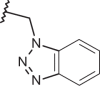 |
–CF3 | –H | 5.15 | 4.00 | 4.00 |
| 129 | –(CH2)3N(C2H5)2 | 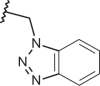 |
–CF3 | –H | 5.72 | 4.30 | 4.00 |
| 130 | 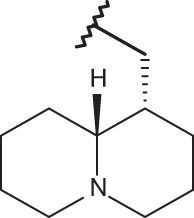 |
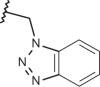 |
–CF3 | –H | <4.44 | 4.04 | 4.44 |
| 131 | –(CH2)2N(CH3)2 | 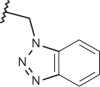 |
–NO2 | –H | 5.05 | 4.00 | 4.00 |
| 132 | –(CH2)2N(C2H5)2 | 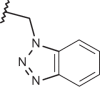 |
–NO2 | –H | 4.96 | 4.00 | 4.00 |
| 133 |  |
 |
–NO2 | –H | 4.64 | 4.41 | 4.08 |
| 134 | –(CH2)2N(CH3)2 |  |
–COCH3 | –H | 5.74 | 4.00 | 4.00 |
| 135 | –(CH2)2N(C2H5)2 |  |
–COCH3 | –H | 5.15 | 4.00 | 4.00 |
| 136 | –(CH2)3N(C2H5)2 |  |
–COCH3 | –H | 5.92 | 4.10 | 4.00 |
| 137 |  |
 |
–COCH3 | –H | 5.60 | 4.11 | 4.10 |
| 138 | –(CH2)3N(C2H5)2 |  |
–H | –H | 5.82 | 4.00 | 4.00 |
| 139 | 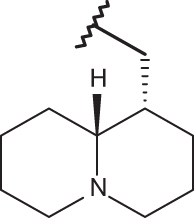 |
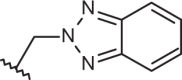 |
–H | –H | 6.15 | 4.00 | 4.00 |
| 140 | 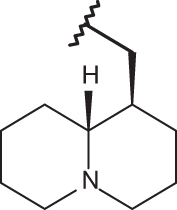 |
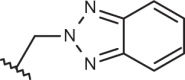 |
–H | –H | 5.92 | 4.03 | 4.00 |
| 141 |  |
 |
–H | –H | 7.00 | 4.15 | 4.00 |
| 142 | –(CH2)2N(CH3)2 |  |
–Cl | –H | 6.52 | 4.00 | 4.00 |
| 143 | –(CH2)2N(C2H5)2 |  |
–Cl | –H | 6.40 | 4.10 | 4.00 |
| 144 | –(CH2)3N(CH3)2 |  |
–Cl | –H | 5.82 | 4.00 | 4.00 |
| 145 | –(CH2)3N(C2H5)2 |  |
–Cl | –H | 6.22 | 4.15 | 4.00 |
| 146 |  |
 |
–Cl | –H | 6.00 | 4.30 | 4.30 |
| 147 | 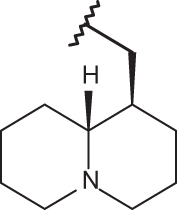 |
 |
–Cl | –H | 6.22 | 4.30 | 4.12 |
| 148 | 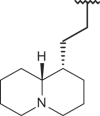 |
 |
–Cl | –H | 7.52 | 4.52 | 4.00 |
| 149 | –(CH2)2N(CH3)2 | 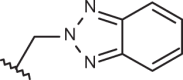 |
–CF3 | –H | 5.60 | 4.00 | 4.00 |
| 150 | –(CH2)2N(C2H5)2 | 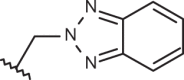 |
–CF3 | –H | 5.70 | 4.05 | 4.00 |
| 151 | 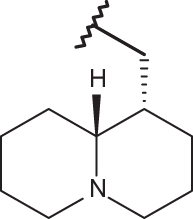 |
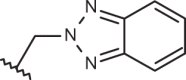 |
–CF3 | –H | 4.96 | 4.40 | 4.10 |
| 152 | –(CH2)2N(CH3)2 |  |
–NO2 | –H | < 4.00 | 4.22 | 4.00 |
| 153 | –(CH2)2N(C2H5)2 |  |
–NO2 | –H | <4.00 | 4.00 | 4.00 |
| 154 |  |
 |
–NO2 | –H | <4.10 | 4.35 | 4.10 |
| 155 | –(CH2)2N(C2H5)2 |  |
–COCH3 | –H | 4.70 | 4.00 | 4.00 |
| 156 |  |
 |
–COCH3 | –H | 5.60 | 4.39 | 4.10 |
All the listed compounds have been previously described16–18.
For both models, calculations were developed using CoMFA steric and electrostatic fields, and CoMSIA steric, electrostatic, hydrophobic, H-bond acceptor and H-bond donor parameters, as independent variables. On the other hand, the RSV pEC50 and the MT-4 pCC50 were employed as dependent variables for model A and B, respectively (experimental section).
Concerning model A, CoMFA and CoMSIA analyses were performed choosing among compounds 1–156 a training set pool molecules (2, 9, 11, 12, 14, 15, 21, 23, 25, 27, 29–31, 44, 100, 102, 114–120, 122–129, 131–134, 136, 138–141, 143, 145, 146, 149–153, 155, 156) for model generation and a test set one (20, 24, 103, 112, 121, 135, 137, 142, 144, 147, 148), for model validation. Model B CoMFA and CoMSIA analyses were performed including compounds 2–4, 6–20, 22–29, 31–33, 35, 37–39, 41–46, 49–51, 53–57, 59–65, 67–72, 74, 75, 77–81, 84, 86, 88–91, 93–95, 97, 98, 100–104, 106–108, 110, 111, 113–115, 118–121, 123, 125–129, 131–137, 139–149, 151–154, 156 into the training set, while compounds 1, 5, 19, 21, 30, 34, 36, 40, 47, 48, 52, 58, 66, 73, 76, 82, 83, 85, 87, 92, 96, 99, 105, 109, 112, 116, 117, 124, 130, 138, 150, 155 were included in the test set.
All statistical parameters supporting the two series of 3D-QSAR analyses are reported in Table 2, while final models A and B experimental and predicted pEC50 (pCC50) values are listed in Tables 3–6. Any detail is described as follows.
Table 2.
Summary of CoMFA and CoMSIA analyses calculated as model A and B.
| Model A |
Model B |
|||
|---|---|---|---|---|
| CoMFA | CoMSIA | CoMFA | CoMSIA | |
| No. compounds | 50 | 50 | 123 | 123 |
| Optimal number of components (ONC) | 5 | 5 | 8 | 8 |
| Leave one out r2 (r2loo) | 0.653 | 0.725 | 0.612 | 0.641 |
| Cross validated r2 (r2cv) | 0.703 | 0.710 | 0.726 | 0.712 |
| Std. error of estimate (SEE) | 0.279 | 0.305 | 0.161 | 0.196 |
| Non-cross validated r2 (r2ncv) | 0.92 | 0.89 | 0.91 | 0.87 |
| F value | 90.457 | 57.341 | 128.631 | 82.855 |
| Steric contribution | 0.574 | 0.131 | 0.597 | 0.151 |
| Electrostatic contribution | 0.426 | 0.203 | 0.403 | 0.196 |
| H-bond acceptor contribution | – | 0.208 | – | 0.156 |
| H-bond donor contribution | – | 0.235 | – | 0.238 |
| Hydrophobicity contribution | – | 0.2223 | – | 0.260 |
| Bootstrap r2 (r2boot) | 0.980 | 0.943 | 0.981 | 0.975 |
| Standard error of estimate r2boot (SEE r2boot) | 0.210 | 0.232 | 0.267 | 0.269 |
| Test set r2 (r2pred) | 0.88 | 0.88 | 0.63 | 0.56 |
Table 3.
Model A CoMFA and CoMSIA analyses experimental and predicted pEC50 values of the training set compounds.
| CoMFA model |
CoMSIA model |
||||
|---|---|---|---|---|---|
| Compound | Exp. pEC50 | Pred. pEC50 | Residual | Pred. pEC50 | Residual |
| 2 | 4.66 | 4.94 | −0.28 | 4.247 | 0.41 |
| 9 | 5.05 | 5.28 | −0.23 | 5.211 | −0.16 |
| 11 | 5.30 | 5.05 | 0.25 | 5.375 | −0.08 |
| 12 | 4.70 | 4.68 | 0.02 | 4.544 | 0.16 |
| 14 | 4.12 | 4.17 | −0.04 | 4.058 | 0.06 |
| 15 | 4.40 | 4.18 | 0.22 | 4.049 | 0.35 |
| 21 | 4.60 | 4.78 | −0.18 | 4.745 | −0.15 |
| 23 | 4.92 | 5.05 | −0.13 | 5.143 | −0.22 |
| 25 | 4.40 | 4.71 | −0.31 | 4.422 | −0.02 |
| 27 | 4.70 | 4.64 | 0.06 | 4.968 | −0.27 |
| 29 | 5.15 | 5.09 | 0.06 | 4.703 | 0.45 |
| 30 | 4.00 | 4.07 | −0.07 | 4.12 | −0.12 |
| 31 | 5.00 | 5.12 | −0.12 | 4.838 | 0.16 |
| 44 | 5.15 | 4.85 | 0.30 | 5.289 | −0.14 |
| 100 | 5.15 | 4.97 | 0.18 | 5.154 | 0.00 |
| 102 | 4.20 | 3.96 | 0.24 | 4.135 | 0.07 |
| 114 | 6.15 | 6.56 | −0.41 | 6.441 | −0.29 |
| 115 | 5.64 | 5.66 | −0.02 | 6.116 | −0.48 |
| 116 | 6.15 | 6.46 | −0.31 | 6.535 | −0.39 |
| 117 | 6.15 | 5.87 | 0.28 | 6.084 | 0.07 |
| 118 | 6.52 | 6.78 | −0.26 | 6.532 | −0.01 |
| 119 | 6.82 | 7.08 | −0.26 | 7.064 | −0.24 |
| 120 | 7.52 | 6.98 | 0.54 | 6.798 | 0.72 |
| 122 | 7.22 | 6.98 | 0.25 | 6.798 | 0.42 |
| 123 | 7.00 | 6.90 | 0.10 | 6.772 | 0.23 |
| 124 | 6.05 | 6.26 | −0.21 | 6.421 | −0.37 |
| 125 | 7.30 | 7.22 | 0.08 | 6.923 | 0.38 |
| 126 | 7.70 | 7.54 | 0.16 | 7.45 | 0.25 |
| 127 | 5.00 | 5.32 | −0.32 | 5.906 | −0.91 |
| 128 | 5.15 | 4.69 | 0.46 | 5.562 | −0.41 |
| 129 | 5.72 | 5.54 | 0.18 | 5.307 | 0.41 |
| 131 | 5.05 | 5.07 | −0.02 | 5.098 | −0.05 |
| 132 | 4.96 | 4.80 | 0.16 | 4.798 | 0.16 |
| 133 | 4.64 | 4.87 | −0.23 | 4.641 | 0.00 |
| 134 | 5.74 | 5.67 | 0.07 | 5.314 | 0.43 |
| 136 | 5.92 | 5.99 | −0.07 | 5.756 | 0.16 |
| 138 | 5.82 | 5.77 | 0.05 | 5.624 | 0.20 |
| 139 | 6.15 | 5.68 | 0.47 | 5.852 | 0.30 |
| 140 | 5.92 | 5.71 | 0.21 | 5.909 | 0.01 |
| 141 | 7.00 | 7.12 | −0.12 | 7.34 | −0.34 |
| 143 | 6.40 | 6.56 | −0.16 | 6.023 | 0.38 |
| 145 | 6.22 | 6.42 | −0.20 | 6.017 | 0.20 |
| 146 | 6.00 | 6.08 | −0.08 | 6.144 | −0.14 |
| 149 | 5.60 | 5.02 | 0.59 | 5.453 | 0.15 |
| 150 | 5.70 | 5.28 | 0.42 | 5.356 | 0.34 |
| 151 | 4.96 | 5.14 | −0.18 | 5.573 | −0.61 |
| 152 | 4.00 | 4.73 | −0.73 | 4.64 | −0.64 |
| 153 | 4.00 | 4.39 | −0.39 | 4.502 | −0.50 |
| 155 | 4.70 | 5.00 | −0.30 | 4.907 | −0.21 |
| 156 | 5.60 | 5.52 | 0.08 | 5.113 | 0.49 |
Table 4.
Model A CoMFA and CoMSIA analyses experimental and predicted pEC50 values of the test set compounds.
| CoMFA model |
CoMSIA model |
||||
|---|---|---|---|---|---|
| Compound | Exp. pEC50 | Pred. pEC50 | Residual | Pred. pEC50 | Residual |
| 20 | 4.82 | 4.99 | −0.17 | 4.75 | 0.07 |
| 24 | 5.05 | 4.91 | 0.14 | 5.08 | −0.03 |
| 103 | 4.35 | 3.90 | 0.45 | 4.04 | 0.31 |
| 112 | 4.60 | 4.77 | −0.17 | 4.39 | 0.21 |
| 121 | 6.15 | 5.87 | 0.28 | 6.48 | −0.32 |
| 135 | 5.15 | 4.95 | 0.20 | 4.98 | 0.17 |
| 137 | 5.60 | 5.93 | −0.33 | 5.18 | 0.42 |
| 142 | 6.52 | 5.97 | 0.55 | 5.95 | 0.57 |
| 144 | 5.82 | 6.32 | −0.50 | 6.29 | −0.47 |
| 147 | 6.22 | 6.12 | 0.10 | 6.19 | 0.03 |
| 148 | 7.52 | 7.68 | −0.16 | 7.76 | −0.24 |
Table 5.
Model B CoMFA and CoMSIA analyses experimental and predicted pCC50 values of the training set compounds.
| CoMFA model |
CoMSIA model |
||||
|---|---|---|---|---|---|
| Compound | Exp. pCC50 | Pred. pCC50 | Residual | Pred. pCC50 | Residual |
| 2 | 4.27 | 4.41 | −0.14 | 4.56 | −0.29 |
| 3 | 4.00 | 3.85 | 0.15 | 3.92 | 0.08 |
| 4 | 4.96 | 5.13 | −0.17 | 4.85 | 0.12 |
| 6 | 4.00 | 4.06 | −0.06 | 4.12 | −0.12 |
| 7 | 4.16 | 4.09 | 0.07 | 4.16 | 0.00 |
| 8 | 4.00 | 3.90 | 0.11 | 3.86 | 0.14 |
| 9 | 4.00 | 3.98 | 0.02 | 3.96 | 0.04 |
| 10 | 4.00 | 3.87 | 0.13 | 3.92 | 0.08 |
| 11 | 4.54 | 4.37 | 0.17 | 4.48 | 0.06 |
| 12 | 4.15 | 4.38 | −0.23 | 4.17 | −0.02 |
| 13 | 4.38 | 4.34 | 0.04 | 4.15 | 0.23 |
| 14 | 4.31 | 4.38 | −0.07 | 4.23 | 0.08 |
| 15 | 4.60 | 4.36 | 0.24 | 4.37 | 0.23 |
| 16 | 4.00 | 3.97 | 0.03 | 4.30 | −0.30 |
| 17 | 4.40 | 4.29 | 0.11 | 4.39 | 0.01 |
| 18 | 4.00 | 4.21 | −0.21 | 4.07 | −0.07 |
| 20 | 5.22 | 4.96 | 0.26 | 4.72 | 0.50 |
| 22 | 4.77 | 5.03 | −0.26 | 5.00 | −0.23 |
| 23 | 5.05 | 4.84 | 0.21 | 4.92 | 0.13 |
| 24 | 4.92 | 5.08 | −0.16 | 5.09 | −0.17 |
| 25 | 4.22 | 4.29 | −0.07 | 3.96 | 0.27 |
| 26 | 5.15 | 5.32 | −0.17 | 5.02 | 0.13 |
| 27 | 4.77 | 4.65 | 0.12 | 4.74 | 0.03 |
| 28 | 5.70 | 5.59 | 0.11 | 5.31 | 0.39 |
| 29 | 4.72 | 4.73 | −0.01 | 4.53 | 0.19 |
| 31 | 4.72 | 4.82 | −0.10 | 4.73 | −0.01 |
| 32 | 4.03 | 3.86 | 0.17 | 4.17 | −0.14 |
| 33 | 5.40 | 5.57 | −0.17 | 5.12 | 0.28 |
| 35 | 5.30 | 5.32 | −0.02 | 5.42 | −0.12 |
| 37 | 5.05 | 5.10 | −0.05 | 4.80 | 0.25 |
| 38 | 4.66 | 4.55 | 0.11 | 4.58 | 0.09 |
| 39 | 4.00 | 4.41 | −0.41 | 4.47 | −0.47 |
| 41 | 4.00 | 4.06 | −0.06 | 4.26 | −0.26 |
| 42 | 4.00 | 3.98 | 0.02 | 3.97 | 0.04 |
| 43 | 4.00 | 4.02 | −0.01 | 4.10 | −0.10 |
| 44 | 4.92 | 4.79 | 0.14 | 5.05 | −0.13 |
| 45 | 6.00 | 5.73 | 0.27 | 5.61 | 0.39 |
| 46 | 5.70 | 5.79 | −0.09 | 5.93 | −0.23 |
| 49 | 4.77 | 4.61 | 0.16 | 4.78 | −0.01 |
| 50 | 4.09 | 4.01 | 0.09 | 3.99 | 0.10 |
| 51 | 4.00 | 3.99 | 0.01 | 4.17 | −0.17 |
| 53 | 4.40 | 4.49 | −0.09 | 4.30 | 0.10 |
| 54 | 4.51 | 4.42 | 0.09 | 4.31 | 0.20 |
| 55 | 4.00 | 4.16 | −0.16 | 4.04 | −0.04 |
| 56 | 4.74 | 5.04 | −0.30 | 4.83 | −0.09 |
| 57 | 4.21 | 4.48 | −0.27 | 4.37 | −0.16 |
| 59 | 4.00 | 4.02 | −0.02 | 4.52 | −0.52 |
| 60 | 5.30 | 4.93 | 0.38 | 4.95 | 0.35 |
| 61 | 4.82 | 4.90 | −0.08 | 4.78 | 0.04 |
| 62 | 5.10 | 5.07 | 0.03 | 5.10 | 0.00 |
| 63 | 4.74 | 4.85 | −0.11 | 4.83 | −0.09 |
| 64 | 5.22 | 4.79 | 0.43 | 5.21 | 0.01 |
| 65 | 4.00 | 4.51 | −0.51 | 4.00 | 0.00 |
| 67 | 5.22 | 5.20 | 0.02 | 5.44 | −0.22 |
| 68 | 5.40 | 5.40 | 0.00 | 5.20 | 0.20 |
| 69 | 4.00 | 4.15 | −0.15 | 4.22 | −0.22 |
| 70 | 4.00 | 4.17 | −0.17 | 4.12 | −0.12 |
| 71 | 4.00 | 4.16 | −0.16 | 4.29 | −0.29 |
| 72 | 4.77 | 4.63 | 0.14 | 4.69 | 0.08 |
| 74 | 5.10 | 5.11 | −0.01 | 5.01 | 0.09 |
| 75 | 4.27 | 4.13 | 0.14 | 4.32 | −0.05 |
| 77 | 4.00 | 4.01 | −0.01 | 4.19 | −0.19 |
| 78 | 4.96 | 4.88 | 0.08 | 4.63 | 0.33 |
| 79 | 4.70 | 4.38 | 0.32 | 4.37 | 0.33 |
| 80 | 4.82 | 5.09 | −0.27 | 4.82 | 0.00 |
| 81 | 5.10 | 4.89 | 0.22 | 4.80 | 0.30 |
| 84 | 5.05 | 5.04 | 0.01 | 5.28 | −0.23 |
| 86 | 5.00 | 4.98 | 0.02 | 5.13 | −0.13 |
| 88 | 5.82 | 5.66 | 0.16 | 5.63 | 0.19 |
| 89 | 4.36 | 4.36 | 0.00 | 4.37 | −0.01 |
| 90 | 4.62 | 4.81 | −0.19 | 4.90 | −0.28 |
| 91 | 4.77 | 4.53 | 0.24 | 4.86 | −0.09 |
| 93 | 4.55 | 4.35 | 0.20 | 4.42 | 0.13 |
| 94 | 4.07 | 4.11 | −0.04 | 4.51 | −0.44 |
| 95 | 6.52 | 6.50 | 0.02 | 6.59 | −0.07 |
| 97 | 4.00 | 4.17 | −0.17 | 4.07 | −0.07 |
| 98 | 4.31 | 4.31 | 0.00 | 4.11 | 0.20 |
| 100 | 4.38 | 4.65 | −0.27 | 4.46 | −0.08 |
| 101 | 4.00 | 3.86 | 0.14 | 3.81 | 0.19 |
| 102 | 4.00 | 4.18 | −0.18 | 3.91 | 0.10 |
| 103 | 4.25 | 4.24 | 0.01 | 4.25 | 0.00 |
| 104 | 4.00 | 4.11 | −0.11 | 4.27 | −0.27 |
| 106 | 4.62 | 4.67 | −0.05 | 4.54 | 0.09 |
| 107 | 4.24 | 4.76 | −0.52 | 4.47 | −0.23 |
| 108 | 4.68 | 4.46 | 0.23 | 4.82 | −0.14 |
| 110 | 4.38 | 4.26 | 0.12 | 4.41 | −0.03 |
| 111 | 4.00 | 4.02 | −0.02 | 4.06 | −0.05 |
| 113 | 4.77 | 4.80 | −0.03 | 4.60 | 0.17 |
| 114 | 4.00 | 3.84 | 0.17 | 3.80 | 0.20 |
| 115 | 4.00 | 3.78 | 0.22 | 3.91 | 0.09 |
| 118 | 4.04 | 4.13 | −0.09 | 4.00 | 0.04 |
| 119 | 4.19 | 4.32 | −0.13 | 4.42 | −0.23 |
| 120 | 4.00 | 4.01 | −0.01 | 4.09 | −0.09 |
| 121 | 4.00 | 4.05 | −0.05 | 4.20 | −0.20 |
| 123 | 4.30 | 4.22 | 0.08 | 4.24 | 0.06 |
| 125 | 4.44 | 4.37 | 0.07 | 4.32 | 0.12 |
| 126 | 4.80 | 4.73 | 0.07 | 4.75 | 0.05 |
| 127 | 4.00 | 4.12 | −0.12 | 4.16 | −0.16 |
| 128 | 4.00 | 4.13 | −0.13 | 4.26 | −0.26 |
| 129 | 4.30 | 4.21 | 0.09 | 4.30 | 0.00 |
| 131 | 4.04 | 4.20 | −0.16 | 3.89 | 0.16 |
| 132 | 4.00 | 4.02 | −0.02 | 4.03 | −0.03 |
| 133 | 4.41 | 4.47 | −0.06 | 4.52 | −0.11 |
| 134 | 4.00 | 3.98 | 0.02 | 3.74 | 0.26 |
| 135 | 4.00 | 3.96 | 0.04 | 3.84 | 0.16 |
| 136 | 4.00 | 4.15 | −0.15 | 3.90 | 0.11 |
| 137 | 4.11 | 4.35 | −0.24 | 4.34 | −0.23 |
| 139 | 4.00 | 3.97 | 0.03 | 4.02 | −0.02 |
| 140 | 4.03 | 4.00 | 0.04 | 4.02 | 0.01 |
| 141 | 4.15 | 4.12 | 0.03 | 4.28 | −0.13 |
| 142 | 4.00 | 3.98 | 0.02 | 4.10 | −0.10 |
| 143 | 4.10 | 4.09 | 0.01 | 4.22 | −0.12 |
| 144 | 4.00 | 4.14 | −0.14 | 4.07 | −0.07 |
| 145 | 4.15 | 4.17 | −0.02 | 4.12 | 0.04 |
| 146 | 4.30 | 4.24 | 0.06 | 4.30 | 0.00 |
| 147 | 4.30 | 4.20 | 0.10 | 4.29 | 0.01 |
| 148 | 4.52 | 4.52 | 0.00 | 4.62 | −0.10 |
| 149 | 4.00 | 4.07 | −0.07 | 4.21 | −0.21 |
| 151 | 4.40 | 4.34 | 0.06 | 4.39 | 0.01 |
| 152 | 4.22 | 4.01 | 0.21 | 4.01 | 0.21 |
| 153 | 4.00 | 4.18 | −0.18 | 4.05 | −0.05 |
| 154 | 4.35 | 4.19 | 0.17 | 4.17 | 0.18 |
| 156 | 4.39 | 4.30 | 0.09 | 4.31 | 0.08 |
Table 6.
Model B CoMFA and CoMSIA analyses experimental and predicted pCC50 values of the test set compounds.
| CoMFA model |
CoMSIA model |
||||
|---|---|---|---|---|---|
| Compound | Exp. pCC50 | Pred. pCC50 | Residual | Pred. pCC50 | Residual |
| 1 | 4.00 | 4.10 | −0.10 | 4.10 | −0.10 |
| 5 | 4.00 | 3.92 | 0.08 | 3.98 | 0.02 |
| 19 | 4.00 | 4.24 | −0.24 | 4.23 | −0.23 |
| 21 | 4.89 | 4.97 | −0.08 | 4.82 | 0.07 |
| 30 | 4.00 | 4.40 | −0.40 | 4.33 | −0.33 |
| 34 | 4.70 | 5.00 | −0.30 | 4.76 | −0.06 |
| 36 | 5.22 | 4.80 | 0.42 | 5.18 | 0.04 |
| 40 | 4.00 | 4.02 | −0.01 | 4.12 | −0.12 |
| 47 | 5.52 | 5.52 | 0.00 | 5.61 | −0.09 |
| 48 | 4.77 | 4.51 | 0.26 | 4.77 | 0.00 |
| 52 | 4.46 | 4.55 | −0.09 | 4.34 | 0.13 |
| 58 | 4.72 | 4.46 | 0.26 | 4.65 | 0.07 |
| 66 | 5.00 | 4.59 | 0.41 | 4.76 | 0.24 |
| 73 | 4.00 | 4.30 | −0.30 | 4.27 | −0.27 |
| 76 | 4.74 | 4.40 | 0.34 | 4.22 | 0.52 |
| 82 | 5.04 | 5.05 | 0.00 | 5.29 | −0.25 |
| 83 | 4.80 | 4.87 | −0.07 | 4.97 | −0.17 |
| 85 | 4.00 | 4.30 | −0.30 | 4.43 | −0.43 |
| 87 | 4.66 | 5.20 | −0.54 | 5.13 | −0.47 |
| 92 | 4.72 | 5.05 | −0.33 | 5.23 | −0.51 |
| 96 | 4.00 | 4.53 | −0.53 | 4.42 | −0.42 |
| 99 | 4.48 | 4.55 | −0.07 | 4.21 | 0.27 |
| 105 | 4.00 | 4.49 | −0.49 | 4.34 | −0.34 |
| 109 | 4.60 | 4.63 | −0.03 | 4.86 | −0.26 |
| 112 | 4.33 | 4.19 | 0.14 | 3.52 | 0.81 |
| 116 | 4.00 | 3.89 | 0.11 | 3.95 | 0.05 |
| 117 | 4.00 | 4.37 | −0.37 | 4.23 | −0.23 |
| 124 | 4.43 | 4.57 | −0.14 | 4.70 | −0.27 |
| 130 | 4.04 | 4.23 | −0.19 | 4.32 | −0.28 |
| 138 | 4.00 | 4.00 | 0.00 | 3.94 | 0.06 |
| 150 | 4.05 | 4.08 | −0.03 | 4.08 | −0.03 |
| 155 | 4.00 | 4.18 | −0.18 | 4.14 | −0.14 |
The final model A CoMFA was generated by employing non-cross-validated PLS analysis with the optimum number of components (ONC = 5) to give a non-cross validated r2 (r2ncv) = 0.92, a test set r2 (r2pred) = 0.88, standard error of estimate (SEE) = 0.279, steric contribution = 0.574 and electrostatic contribution = 0.426.
The related CoMSIA analysis was derived using a statistical PLS analysis leading to the following results: ONC = 5, a non-cross validated r2 (r2ncv) = 0.89, a test set r2 (r2pred) = 0.88, standard error of estimate (SEE) = 0.305, steric contribution = 0.131, electrostatic contribution = 0.203, hydrophobic contribution = 0.223, H-bond acceptor = 0.208 and H-bond donor = 0.235.
An overall overview of the predictive ability of model A study can be obtained from graphical distributions of the predicted pEC50 values of the training set and test compounds, as shown in Supplemental materials S3–S4.
The selected CoMFA model B was generated by employing non-cross-validated PLS analysis with the optimum number of components (ONC = 8) to give a non-cross validated r2 (r2ncv) = 0.91, a test set r2 (r2pred) = 0.63, standard error of estimate (SEE) = 0.161, steric contribution = 0.597 and electrostatic contribution = 0.403. The CoMSIA model B was obtained with the following statistical results: ONC = 8, a non-cross validated r2 (r2ncv) = 0.87, a test set r2 (r2pred) = 0.56, standard error of estimate (SEE) = 0.196, steric contribution = 0.151, electrostatic contribution = 0.196, hydrophobic contribution = 0.260, H-bond acceptor = 0.156 and H-bond donor = 0.238.
The derived distributions of the predicted pCC50 values of the training set and test compounds are as Supplemental material S5–S6.
The CoMFA and CoMSIA model A and B reliability thus generated was supported also by bootstrapping results (Table 2).
Discussion
CoMFA and CoMSIA contour maps
In order to deeply discuss the contribution of any feature displayed within the series of benzimidazoles here investigated with respect to their potency as anti-RSV agents, and also to their cytotoxicity profile, the description of the related 3D-QSAR models (A and B, respectively) was organised in two sections separately.
According to the CoMFA steric map descriptors, green polyhedra represent those areas that should be decorated with bulky groups, while yellow maps highlight those regions related to an unfavourable or slightly allowed presence of substituents. On the other hand, the CoMFA electrostatic descriptors are shown as blue areas around those regions predicted to be beneficial for electropositive or electron donor moieties, while red polyhedra occupy any area recommended for much more electronegative or electron withdrawing.
Concerning CoMSIA analysis, the hydrophobic map reveals through yellow and white polyhedra those ligand features predicted to be favoured for lipophilic and polar groups, respectively.
The introduction of H-bond acceptor and H-bond donor moieties results to be encouraged or discouraged by the presence of magenta and cyan and of red and purple areas, respectively.
Model a CoMFA and CoMSIA analyses (benzimidazoles anti-RSV profile)
On the basis of model A, for all the data set, the steric contour map predicts favourable substitutions at the benzimidazole position 5 (green polyhedral), while the presence of any bulky group at the position 6 results to be detrimental for the anti-RSV activity (yellow polyhedral). The reliability of these information is supported by the inactivity of the 5,6-dichloro-benzimidazoles 44–49 and by the poor anti-RSV potency values of the 6-substituted benzimidazoles 102 (pEC50 = 4.20) and 103 (pEC50 = 4.35).
Notably, the presence of a (even small) substituent linked to the benzimidazole position 2 is favoured. Accordingly, compound 2 (R2 = trifluoromethyl; pEC50 = 4.66) display a better potency profile if compared with the analogue 21 (R2 = 4-bromobenzyl; pEC50 = 4.60), suggesting that a proper decoration on the benzimidazole position 5 and at the R1 group could be successfully accompanied by a small moiety in R2.
On the other hand, for those 2-substituted benzimidazoles bearing a rigid phenyl group in R2 (such as 27, 29–31 and 102, 103), any further substitution at the ortho positions proves to be encouraged, falling in a green area, while any other at the meta and para ones are disfavoured, being surrounded by yellow polyhedra (Figure 2(a)).
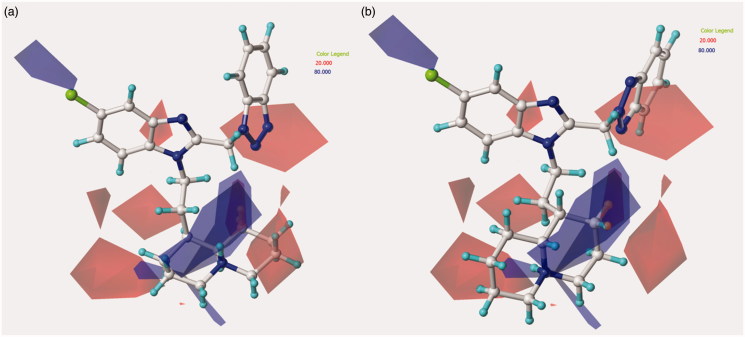
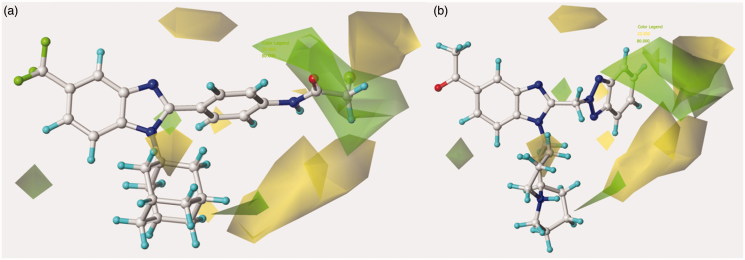
Figure 2.
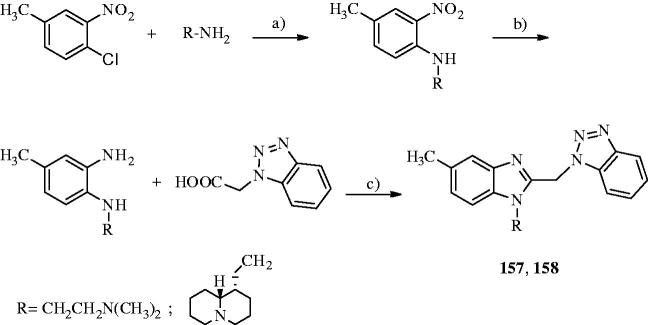
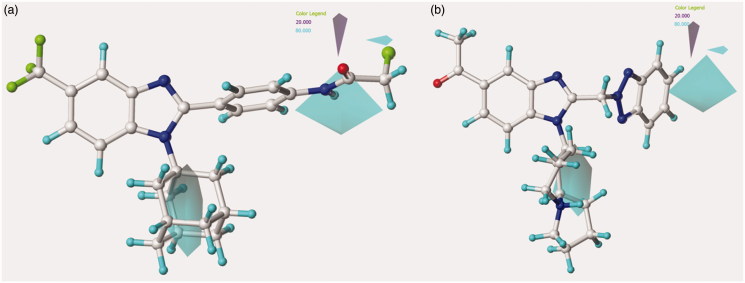
Contour map of model A CoMFA steric regions are shown around the anti-RSV agent 44 (a) and 11 (b). The compounds are displayed in ball and stick mode.
These results are confirmed by the inactivity of compounds 40–43 (pEC50 < 4.00), which are poly-substituted at the phenyl ring in R2. Moreover, for this series of compounds, any group eventually placed in R1 results to be disfavoured. Indeed, also compounds 97 (R1 = methyl; pEC50 < 4.00) and 98 (R1 = cyclohexyl; pEC50 < 4.00) are not interesting as anti-RSV agents.
On the contrary, those 2-substituted benzimidazoles bearing a flexible group in R2 such as the benzyl one (compounds 5–24), seems to properly affect the R1 substituent, moving it towards a sterically favoured green area (Figure 2(b)). These results are supported by the better anti-RSV activity of compound 24 (R1 = lupinyl-; pEC50 = 5.05) with respect to compound 14 (R1 = N,N-diethylaminoethyl; pEC50 = 4.12).
Notably, this information is also in accordance with the promising potency profile displayed within the series of 2-benzotriazolyl-methyl-benzimidazoles, such as the analogues 120, 122 and 125 (pEC50 = 7.22–7.52).
In addition, for the 2-benzyl-benzimidazoles here discussed, the introduction of any decoration onto the benzyl ortho and para positions is encouraged, as supported by the higher pEC50 value of 15 (R2 = 4-iodophenyl-; pEC50 = 4.40) than that of 14 (R2 = 4-bromophenyl-; pEC50 = 4.12) and by the pEC50 value of 11 (R2 = 4-methoxyphenyl-; pEC50 = 5.30).
Based on an overall analysis of the biological assays about the whole dataset, those analogues bearing a (benzotriazol-1/2-yl)methyl in R2 result to be the most promising. Accordingly, the most of them display a successful behaviour with respect to the 3D-QSAR maps. In particular, in the case of steric contribution, the N(1)-substituted benzotriazoles much more properly fit the two green favourable areas placed around the bicyclic ring (Figure 3(a)). Accordingly, compound 2c (R2 = N(1)-benzotriazolyl; pEC50 = 5.64) shows a higher potency value than that of 28b (R2 = 4-iodophenyl-; pEC50 = 4.40) and 27b (R2 = 4-bromophenyl-; pEC50 = 4.12). In addition, the most effective compounds of all these series (7c, 9c, 10c, 12c and 13c; pEC50 = 7.00–7.70) are decorated with a the (benzotriazol-1-yl)methyl moiety in R2.
Figure 3.
Contour map of model A CoMFA steric regions are shown around the anti-RSV agent 126 (a) and 148 (b). The compounds are displayed in ball and stick mode.
Concerning the N(2)-substituted analogues, they display a quite switched orientation, falling in any case within the allowed steric contour map Figure 3(b)). Indeed, 148 (R2 = N(2)-benzotriazolyl; pEC50 = 7.52) shows a higher potency values than 20 (R2 = 4-chlorobenzyl-; pEC50 = 4.82).
Finally, it should be noticed that the presence of the N(1)-substituted benzotriazole ring rather than the N(2)-substituted benzotriazole promote a favourable positioning of the R1 group, such as that shown by the homolupinyl chain of 126 and 148, depicted in Figure 3. Indeed, the R1 of 126 much more effectively fall in a beneficial green steric map.
Consequently, those compounds bearing a (benzotriazol-1-yl)methyl nucleus in R2, such as 135 (pEC50 = 5.15), 136 (pEC50 = 5.92), 118 (pEC50 = 6.52) and 126 (pEC50 = 7.70), are characterised by an increased anti-RSV activity values than the related N(2)-benzotriazolyl-substituted analogues 150 (pEC50 = 4.70), 138 (pEC50 = 5.82), 140 (pEC50 = 5.92) and 148 (pEC50 = 7.52), respectively.
On all these basis, the 3D-QSAR study confirms that the introduction of an N(1)-substituted benzotriazole ring in R2 represents the better choice at the benzimidazole position 2, in comparison with the other chemical moieties explored within the dataset.
For model A, the electrostatic CoMFA map revealed for all the molecules here investigated a blue area (beneficial for electropositive moieties) in proximity of the R3 substituent, while two other polyhedral favourable for electropositive functions would involve a (bulky) substituent eventually present in R1. Notably, these results suggest the introduction of an electron-donor group onto the benzimidazole position 5 (R3) rather than an electron-withdrawing one, and also strongly promote the presence of a basic chain or substituent properly connected with the benzimidazole N1 nitrogen atom.
In addition, a consistent blue region is placed near one of the ortho positions of the R2 phenyl- and benzyl-substituted benzimidazoles, as shown for compounds 44 and 11 in Figure 4. The reliability of this information is confirmed by the poor activity as anti-RSV agents of compounds 25 (R2 = 2-nitrophenyl-; pEC50 = 4.40) and 49 (R2 = 2,6-difluorophenyl-; pEC50 <4.00), bearing highly electron-withdrawing functions at the R2 phenyl ortho positions.
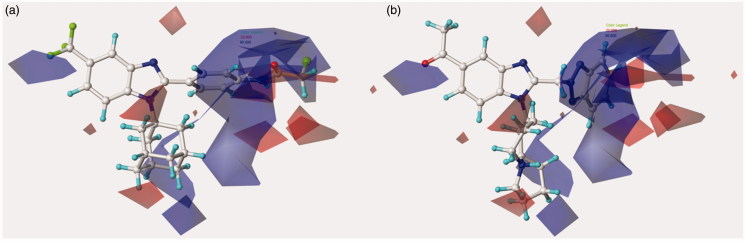
Figure 4.
Contour maps of model A CoMFA electrostatic regions are shown around the anti-RSV agents 44 (a) and 11 (b). The compounds are displayed in ball and stick mode.
On the other hand, the introduction of much more electronegative groups is recommended in proximity of the benzimidazole core and of the R2 phenyl and benzyl ring in R2 of 44 and 11. Consequently, the presence of electron-donor groups properly decorating the aromatic R2 substituents results to be the most beneficial. Indeed, among those compounds being unsubstituted in R1, compound 11 (R2 = 4-methoxybenzyl-; pEC50 = 5.30) results to be more effective than 48 (R2 = 4-nitrophenyl-; pEC50 < 4.00) and 49 (R2 = 2,6-difluorophenyl-; pEC50< 4.00).
In addition, contour maps predicted to be favoured for electronegative moieties are also shown around the electropositive core eventually involving R1, and in the area placed between the benzimidazole positions 6 and 7, even opening the possibility to design new derivatives based on a tricyclic heteroaromatic ring in place of the benzimidazole scaffold.
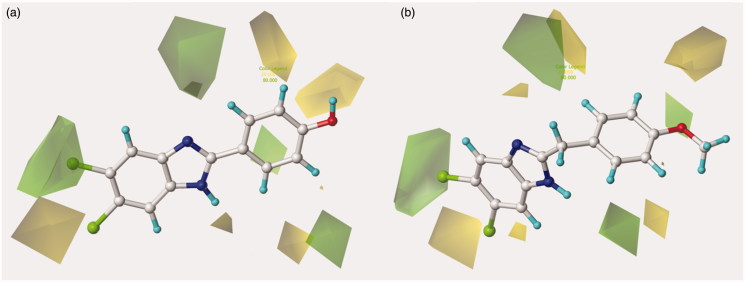
The most effective benzotriazole-based benzimidazoles follow the aforementioned preferred electrostatic profile, being in particular the N(1)-substituted benzotriazoles, rather than the N(2)-substituted ones, more able to fulfil the electronegative area occupied by the triazole ring, as depicted in Figure 5 around 126 and 148.
Figure 5.
Contour maps of model A CoMFA electrostatic regions are shown around the anti-RSV agents 126 (a) and 148 (b). The compounds are displayed in ball and stick mode.
Moreover, the N(1)-substituted benzotriazoles better arrange the basic core included in R1 towards the electropositive blue area, being in accordance with the behaviour previously discussed about the steric map. Interestingly, within the R1 basic moieties here explored, the bulkier homolupinyl and the more flexible dialkylaminoalkyl chains prove to be the most effective, optimising any contacts with the CoMFA contour maps. Accordingly, compound 126 is the most active of the whole series (R1 = homolupinyl-; R2 = N(1)-benzotriazolyl; pEC50 = 7.70).
The information derived by the CoMSIA hydrophobic map highlights the relevance of lipophilic group in R3 (yellow regions), while a small yellow polyhedral is also located in proximity of the substituent eventually placed in R1. Conversely, more polar function (white regions) are predicted to be favoured at R4 and at the R2 group, as shown around the phenyl and benzyl functions of 44 and 11 (Supplemental material S7).
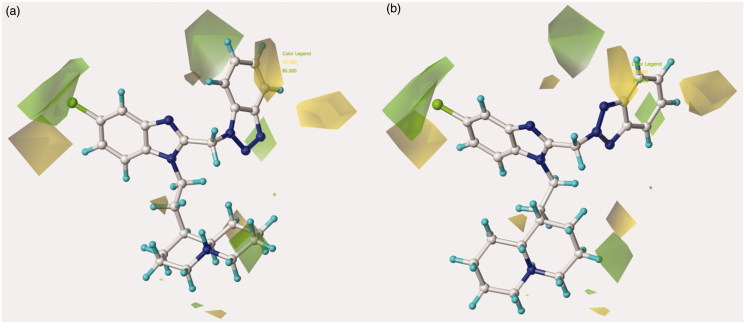
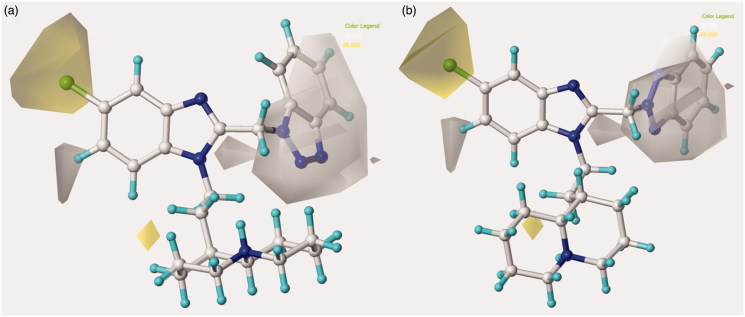
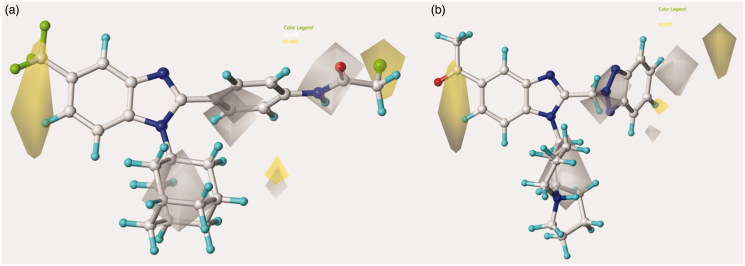
The related hydrophobic map depicted around the benzotriazole-based benzimidazoles 126 and 148 points out the beneficial role played by a high lipophilic substituent falling in R1, such as the homolupinyl and N,N-diethylamino-ethyl ones (Figure 6).
Figure 6.
Model A CoMSIA hydrophobic favoured and disfavoured regions are shown around the anti-RSV agents 126 (a) and 148 (b). The compounds are displayed in ball and stick mode.
The anti-RSV activity of benzimidazoles is enhanced by H-bond acceptor functions (magenta polyhedral) near the ortho positions of those analogues decorated with a phenyl group in R1, and at the para position of those compounds bearing a benzyl moiety (Supplemental material S8). On the contrary, it results to be disfavoured by any H-bond acceptor function eventually placed in R3 or R4 (green areas).
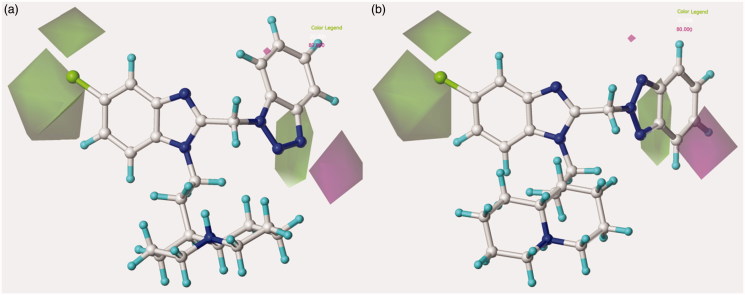
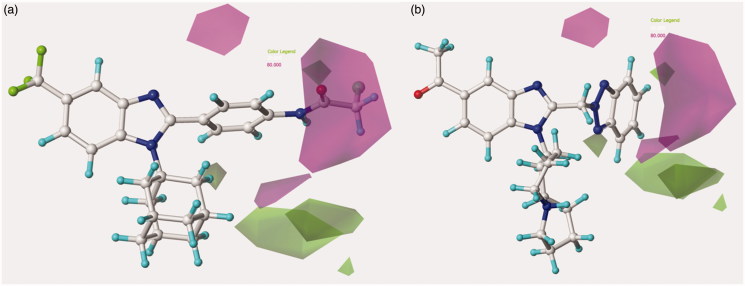
Concerning the most promising benzotriazole-based benzimidazoles, they also follow the aforementioned H-bond acceptor preferences, being in particular the N(1)-substituted benzotriazoles, rather than the N(2)-substituted ones, more able to arrange near the favoured magenta area involving R2, as depicted in Figure 7.
Figure 7.
Model A CoMSIA H-bond acceptor favoured and disfavoured contour maps are displayed around benzimidazoles 126 (a) and 148 (b). The compounds are displayed in ball and stick mode.
Finally, the H-donor map discloses the relevance of favoured H-bond donor groups (cyan polyhedral) only at the central core of the area occupied by R1, being disfavoured at the surrounding region (see Figure 8 depicted around 126 and 148 and Supplemental material S9 concerning the phenyl and benzyl derivatives 44 and 11). Notably, this information is probably related to the presence of the key basic chain, as we previously highlight through the CoMFA electrostatic map. In particular, the presence in R2 of the quinolizidine ring (lupinyl, epilupinyl and honolupinyl) let the topology of the benzotriazole nucleus be arranged in allowed conformations.
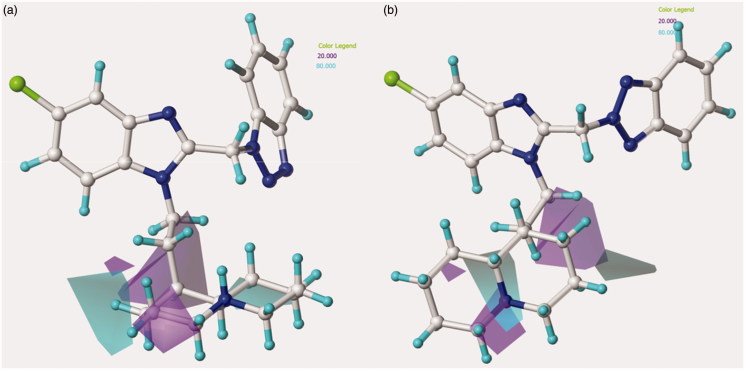
Figure 8.
Model A CoMSIA H-bond donor favoured and disfavoured contour maps are shown around the anti-RSV agents 126 (a) and 148 (b). The compounds are displayed in ball and stick mode.
Model B CoMFA and CoMSIA analyses (benzimidazoles cytotoxicity profile)
Model B CoMFA and CoMSIA analyses allowed us to highlight any key feature eventually increasing the cytotoxicity against the human MT-4 cell line.
In detail, the CoMFA steric map reveals, for all the compounds, a probable higher toxicity profile related to the presence of substituents near R3 or in the area placed between the positions 6 and 7 of the benzimidazole ring (green area; Figure 9), and also in the case of even small groups linked in R2 (compare compounds 2 (R3 = trifluoromethyl-; pCC50 = 4.66) and 4 (R2 = cyclopentylmethyl; pCC50 = 4.96) with 19 (R2 = benzyl; pCC50 = 4.00)).
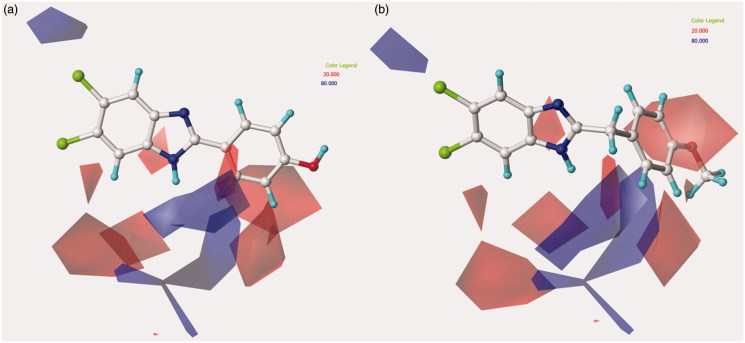
Figure 9.
Contour map of model B CoMFA steric regions are shown around the anti-RSV agent 95 (a) and 156 (b). The compounds are displayed in ball and stick mode.
In the case of bulkier decorations in R2, the most flexible ones, such as the benzyl-substituted benzimidazoles and the N(1)-substituted benzotriazoles-based derivatives, are projected towards yellow disfavoured regions, being therefore characterised by an adequate safety profile. Conversely, the more rigid R2 phenyl- or N(2)-substituted benzotriazoles-based analogues much more overlap a large green area, thus confirming their related possible cytotoxic role. Interestingly, the reliability of these results is supported by the lower MT-4 pCC50 value of 11 (R2 = 4-methoxybenzyl-; pCC50 = 4.54) if compared with the analogue 45 (R2 = 4-methoxyphenyl-; pCC50 = 6.00). On the other hand, based on the experimental data about the pattern of cytotoxicity investigated within the benzotriazole analogues, they proved to display a quite comparable profile.
Finally, for all the compounds, the introduction of a small substituent in R1 seems to be disfavoured in terms of pCC50 values. Consequently, 6 (R1 = H; pCC50 = 4.00) is found less toxic than 20 (R1 = lupinyl-; pCC50 = 5.22), and 9–10 (pCC50 = 4.00), characterised by a longer (basic) chain in R1, are more safe than 17 (pCC50 = 4.40).
The CoMFA electrostatic contour map depicted in Figure 10 describes an overall incisive effect played by electropositive functions included in a bulky R1 group, at the central core of the R2 substituent and in the area near R3. On the other hand, electronegative functions or electron-withdrawing moieties are favoured as small R1 group, at the latter portion of R2 and in proximity of the N2 nitrogen atom of the benzimidazole ring. In particular, the R2 phenyl or benzyl analogues, further decorated with electro-withdrawing or electron-rich functions especially at the para position, represent those compounds which better fit the electrostatic map, while the N(1)-substituted benzotriazoles, followed by the N(2)-substituted benzotriazoles ones (lacking any further group on the heterocyclic system) are projected elsewhere. Notably, these information allow to retain the series including benzotriazole-based decoration as the most safety ones with respect to the others shown within the data set.
Figure 10.
Contour maps of model B CoMFA electrostatic regions are shown around the anti-RSV agents 95 (a) and 156 (b), represented in stick mode.
In agreement with these results, those compounds bearing nitro-substituted benzyl moieties show high pCC50 values, such as 36, 37 (pCC50 = 5.05–5.22).
Concerning the effect played by hydrophobic substitutions around the benzimidazole scaffold with respect to the related cytotoxicity profile, a critical role proves to be determined by lipophilic groups at the latter part of R1 and in R3 and R4 (Figure 11). As a consequence, most of the compounds bearing a trifluoromethyl in R3 or a dichloro substitution at R3 and R4 led to quite toxic compounds [see 44–49 (pCC50 = 4.77–6.00)].
Figure 11.
Model B CoMSIA hydrophobic favoured and disfavoured regions are shown around the anti-RSV agents 95 (a) and 156 (b), represented in stick mode.
As we previously discussed for the electrostatic map, even in this case the R2 phenyl- or benzyl-group analogues, rather than the N(1)-substituted benzotriazoles- and N(2)-substituted benzotriazoles-based ones, are those that fully occupy the hydrophobic map. In details, the R1 aromatic ring falls in a disfavoured hydrophobic region (white polyhedral), while any substituent eventually placed at the phenyl- or benzyl-para position is also in contact with a favourable yellow area. Interestingly, these data underline that the benzimidazole cytotoxicity profile could be enhanced by halogens in R3 and/or R4 and also by the presence of para-substituted phenyl- or benzyl-carboxamide groups in R2, falling this moiety in a white area.
These results could be verified comparing 50 (pCC50 = 4.00) with 51–55 (pCC50 = 4.00–4.51). Moreover, the presence of less hydrophobic moieties in R1 also promotes the MT-4 pCC50 values (compare 90 (R1 = cyclohexyl-; pCC50 = 4.62), 91 (R1 = cyclohexyl-; pCC50 = 4.77) with 93 (R1 = 1-adamantyl-; pCC50 = 4.55), 94 (R1 = 1-adamantyl-; pCC50 = 4.07), respectively).
As shown in Figure 12, H-bond acceptor functions result detrimental for the benzimidazole selectivity index by increasing cytotoxicity especially when they are placed around the (benzotriazol-2-yl) scaffold (magenta areas) and at the R2 phenyl- or benzyl-para positions and, confirming the critical role of the carboxamide function at this position.
Figure 12.
Model B CoMSIA H-bond acceptor favoured and disfavoured contour maps are displayed around benzimidazoles 95 (a) and 156 (b), depicted in stick mode.
Accordingly, also the H-donor contour map points out the presence of favoured (cyan areas) and disfavoured (purple areas) donor functions in R2, representing the aforementioned carboxamide moiety (Figure 13).
Figure 13.
Model B CoMSIA H-bond donor favoured and disfavoured contour maps are shown around the anti-RSV agents 95 (a) and 156 (b), represented in stick mode.
Based on an overall analysis of all the information coming from model A and model B, all the three much more consistent series of benzimidazoles, being substituted in R2 with a phenyl, benzyl or benzotriazole ring, could be optimised through the introduction of small group in R3 endowed with electropositive profile and a less hydrophobic one than that displayed by the trifluoromethyl group or chlorine atom. Concerning R1 it should be noticed that model A underlines a key role played by H-bond acceptor moieties included in bulky and hydrophobic appendages. On the contrary, the corresponding map developed around the MT-4 pCC50 values does not reveal any effect of H-bond acceptor functions at this level. Consequently, all these series of derivatives should be decorated with proper basic chain, such as the effective homolupinyl and dialkylaminoalkyl ones (see the model A CoMFA electrostatic results). In particular, new 2-phenyl-substituted benzimidazoles could particularly take advantage from such a decoration in R1. Finally, the most promising substituent in R2 explored within the dataset proved to be the (benzotriazolyl)methyl one, opening the possibility to be optimised through a series of isostere and/or by a proper selection of other R3 substituents.
Concerning this issue, it should be noticed that the anti-RSV clinical candidate BMS-433771 was designed starting from a benzimidazolone-based compound, being bioisostere of the here discussed benzotriazol-1-yl moiety (Figure 14)34.
Figure 14.
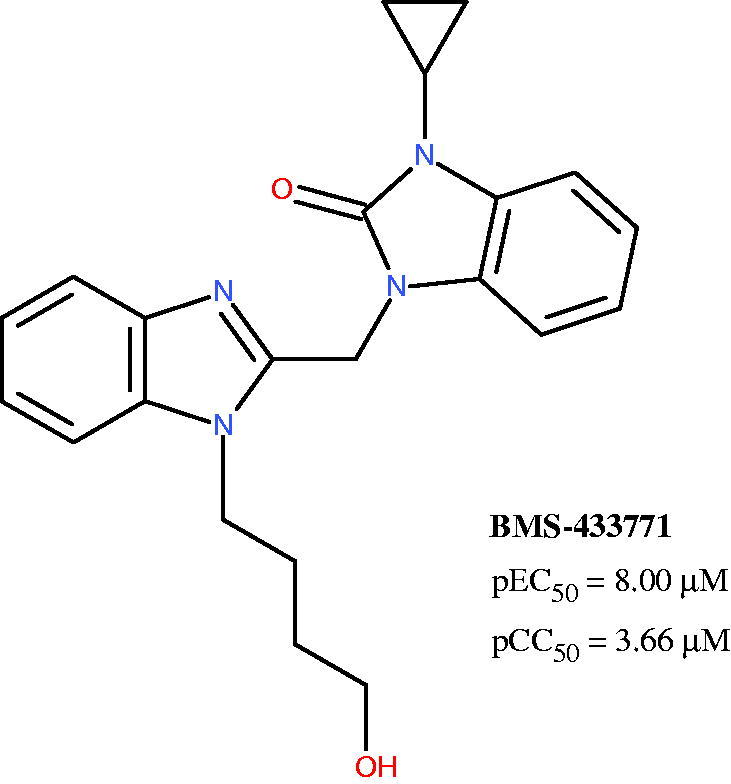
Chemical structure of the anti-RSV agent BMS-433771.
Notably, BMS-433771 qualitatively fulfils any key features recommended by the 3D-QSAR maps here proposed, also about the substituent linked at the benzimidazole position 1, giving a further validation of the applied computational protocol.
In this context, in order to gain a preliminary evaluation of the reliability of the applied 3D-QSAR study, we synthesised two new compounds exploring the effectiveness of the apolar and electrodonor CH3 group in position 5 of the 2-[(benzotriazol-1-yl)methyl]benzimidazole framework.
Biological data
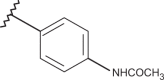
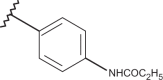
As shown in Figure 15, the biological result of compounds 157 (pEC50= 7.52 MT-4 pCC50 <4.00) and 158 (pEC50= 7.52 MT-4 pCC50 = 4.39) confirmed the same anti-RSV activities with respect to the previously synthesised prototype 120 (pEC50= 7.52 MT-4 pCC50 <4.00), characterised by a N,N-dimethylaminoethyl chain linked at the benzimidazole position 1.
Figure 15.
Chemical structures and biological data about the prototypes 120, 126 and the 5-methyl newly synthesised analogues 157, 158.
The presence of (quinolizidinyl)alkyl residue associated with the 5-Cl group of benzimidazole derivative 126 ring (pEC50= 7.70, MT-4 pCC50= 4.35) contributed to the best activity against RSV, also leading to an increased cytotoxicity. Notably, based on these data and on the information coming from 3D-QSAR studies, the proposed 5-methyl analogue 158 proved to be endowed with the same potency, but lower toxicity against both the cell lines; underlying that 5-methyl substitution is fruitful for modulating the safety profile (CC50/EC50).
Conclusions
The computational studies here presented highlight and discuss the role played by the steric and electrostatic features and also by hydrophobic, H-bond acceptor and donor moieties, in terms of anti-RSV activity and cytotoxicity around the benzimidazole scaffold.
Through focussed CoMFA and CoMSIA analyses, any pattern of requirements able to specifically discriminate much more effective antiviral agents endowed with an increased selectivity index was addressed and deeply discussed. Based on information coming from CoMFA and CoMSIA studies, we designed and synthesised two new benzimidazole derivatives (157, 158) in order to verify the reliability of computational studies. These compounds, being actually endowed with the same high potency but improved safety profiles, allowed us to pave the way for a more focussed rational design process. A much more consistent number of more drug-like anti-RSV agents will be disclosed in due course.
Supplementary Material
Acknowledgements
Authors would like to thank Mr. O. Gagliardo for performing elemental analysis and Mr. V. Ruocco for the informatic support to calculations.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Funding
This work was financially supported by the University of Genoa.
References
- 1.Kong M, Maeng P, Hong J, et al. Respiratory syncytial virus infection disrupts monolayer integrity and function in cystic fibrosis airway cells. Viruses 2013;9:2260–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Clercq E.Chemotherapy of respiratory syncytial virus infections: the final breakthrough. Int J Antimicrob Agents 2015;45:234–7. [DOI] [PubMed] [Google Scholar]
- 3.Committee on Infectious Diseases From the American Academy of Pediatrics: policy statements-modified recommendations for use of palivizumab for prevention of respiratory syncytial virus infections. Pediatrics 2009;124:1694–701. [DOI] [PubMed] [Google Scholar]
- 4.Joffe S, Ray GT, Escobar GJ, et al. Cost-effectiveness of respiratory syncytial virus prophylaxis among preterm infants. Pediatrics 1999;104:419–27. [DOI] [PubMed] [Google Scholar]
- 5.Thomas G.A cost-benefit analysis of the immunisation of children against respiratory syncytial virus (RSV) using the English Hospital Episode Statistics (HES) data set. Eur J Health Econ 2015;1–11. doi: 10.1007/s10198-014-0662-9. [DOI] [PubMed] [Google Scholar]
- 6.Collins PL, Huang YT, Wertz GW.. Identification of a tenth mRNA of respiratory syncytial virus and assignment of polypeptides to the 10 viral genes. J Virol 1984;49:572–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeVincenzo JP, Whitley RJ, Mackman RL, et al. Oral GS-5806 activity in a respiratory syncytial virus challenge study. N Engl J Med 2014;8:711–22. [DOI] [PubMed] [Google Scholar]
- 8.Perron M, Stray K, Kinkade A, et al. GS-5806 inhibits a broad range of respiratory syncytial virus clinical isolates by blocking the virus-cell fusion process. Antimicrob Agents Chemother 2015;60:1264–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perron M, Stray K, Kinkade A, et al. GS-5806 inhibits a broad range of respiratory syncytial virus clinical isolates by blocking the virus-cell fusion process. Antimicrob Agents Chemother 2015;pii:AAC.01497-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keri RS, Hiremathad A, Budagumpi S, Nagaraja BM.. Comprehensive review in current developments of benzimidazole-based medicinal chemistry. Chem Biol Drug Des 2015;86:19–65. [DOI] [PubMed] [Google Scholar]
- 11.Boido V, Paglietti G, Tonelli M, Vitale G, Non-nucleoside benzimidazoles as antiviral drugs against HCV and RSV infections. In Carta, A., ed. RNA-viruses. Enzymatic and receptor inhibitors; Research Signpost: Kerala, 2009:40–91. ISBN: 978-81-308-0329-6. [Google Scholar]
- 12.Cianci C, Langley DR, Dischino DD, et al. Targeting a binding pocket within the trimer-of-hairpins: small-molecule inhibition of viral fusion . Proc Natl Acad Sci USA 2004;101:15046–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Battles MB, Langedijk JP, Furmanova-Hollenstein P, et al. Molecular mechanism of respiratory syncytial virus fusion inhibitors. Nat Chem Biol 2016;12:87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun Z, Pan Y, Jiang S, Lu L.. Respiratory syncytial virus entry inhibitors targeting the F protein. Viruses 2013;5:211–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ispas G, Koul A, Verbeeck J, et al. Antiviral activity of TMC353121, a respiratory syncytial virus (RSV) fusion inhibitor, in a non-human primate model. PLoS One 2015;10:e0126959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tonelli M, Novelli F, Tasso B, et al. Antiviral activity of benzimidazole derivatives. III. Novel anti-CVB-5, anti-RSV and anti-Sb-1 agents. Bioorg Med Chem 2014;22:4893–909. [DOI] [PubMed] [Google Scholar]
- 17.Tonelli M, Simone M, Tasso B, et al. Antiviral activity of benzimidazole derivatives. II. antiviral activity of 2-phenylbenzimidazole derivatives. Bioorg Med Chem 2010;18:2937–53. [DOI] [PubMed] [Google Scholar]
- 18.Tonelli M, Paglietti P, Boido V, et al. Antiviral activity of benzimidazole derivatives. I. Antiviral activity of 1-substituted-2-[(Benzotriazol-1/2-yl)methyl]benzimidazoles. Chem Biodivers 2008;5:2386–401. [DOI] [PubMed] [Google Scholar]
- 19.Putz MV, Duda-Seiman C, Duda-Seiman D, et al. Chemical structure-biological activity models for pharmacophores' 3D-interactions. Int J Mol Sci 2016;17:E1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duda-Seiman C, Duda-Seiman D, Dragos D, et al. Design of Anti-HIV ligands by means of minimal topological difference (MTD) method. Int J Mol Sci 2006;7:537–55. [Google Scholar]
- 21.Duda-Seiman D, Speranţa A, Mancaş S, et al. MTD-comsia modelling of HMG-CoA reductase inhibitors. J Serbian Chem Soc 2011;76:85–99. [Google Scholar]
- 22. MOE: Chemical Computing Group Inc. Montreal. H3A 2R7 Canada. Available from: www.chemcomp.com.
- 23.Sybyl-X 1.0. Tripos Inc 1699 South Hanley Road. St Louis, Missouri 63144, USA [Google Scholar]
- 24.Cramer RD III, Patterson DE, Bunce JD.. Comparative molecular field analysis (CoMFA). Effect of shape on binding of steroids to carrier proteins. J Am Chem Soc 1988;110:5959–67. [DOI] [PubMed] [Google Scholar]
- 25.Klebe G, Abraham U, Mietzner T.. Molecular similarity indices in a comparative analysis (CoMSIA) of drug molecules to correlate and predict their biological activity. J Med Chem 1994;37:4130–46. [DOI] [PubMed] [Google Scholar]
- 26.Cichero E, Buffa L, Fossa P.. 3,4,5-Trisubstituted-1,2,4-4H-triazoles as WT and Y188L mutant HIV-1 non-nucleoside reverse transcriptase inhibitors: docking-based CoMFA and CoMSIA analyses. J Mol Model 2011;7:1537–50. [DOI] [PubMed] [Google Scholar]
- 27.Cichero E, Cesarini S, Mosti L, Fossa P.. CoMFA and CoMSIA analyses on 1,2,3,4-tetrahydropyrrolo[3,4-b]indole and benzimidazole derivatives as selective CB2 receptor agonists. J Mol Model 2010;16:1481–98. [DOI] [PubMed] [Google Scholar]
- 28.Cichero E, Cesarini S, Fossa P, et al. Acylthiocarbamates as non-nucleoside HIV-1 reverse transcriptase inhibitors: docking studies and ligand-based CoMFA and CoMSIA analyses. J Mol Model 2009;15:871–84. [DOI] [PubMed] [Google Scholar]
- 29.Cichero E, Fossa P.. Docking-based 3D-QSAR analyses of pyrazole derivatives as HIV-1 non-nucleoside reverse transcriptase inhibitors. J Mol Model 2012;18:1573–82. [DOI] [PubMed] [Google Scholar]
- 30.Pauwels R, Balzarini J, Baba M, et al. Rapid and automated tetrazolium-based colorimetric assay for the detection of anti-HIV compounds. J Virol Methods 1988;20:309–21. [DOI] [PubMed] [Google Scholar]
- 31.Hara H, Maruyama T, Saito M, et al. Yamanouchi Pharmaceutical Co., Ltd. Patent: EP0454330A1, 1991. [Google Scholar]
- 32.Boido V, Boido A, Canu C, Sparatore F. [Quinolozidinylalkylamines with antihypertensive activity]. Farmaco Sci 1979;34:673–87. [PubMed] [Google Scholar]
- 33.Sparatore F, Pagani F, Some benzotryazolylalkanoic acids and their amide derivatives Farmaco Sci. 1964;19:55–75. [PubMed] [Google Scholar]
- 34.Cianci C, Yu KL, Combrink K, et al. Orally active fusion inhibitor of respiratory syncytial virus. Antimicrob Agents Chemother 2004;48:413–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.