Abstract
In HIV infected macrophages, a large population of viral genomes persists as the unintegrated form (uDNA) that is transcriptionally active. However, how this transcriptional activity is controlled remains unclear. In this report, we investigated whether Tat, the viral transactivator of transcription, is involved in uDNA transcription. We demonstrate that de novo Tat activity is generated from uDNA, and this uDNA-derived Tat (uTat) transactivates the uDNA LTR. In addition, uTat is required for the transcriptional persistence of uDNA that is assembled into repressive episomal minichromatin. In the absence of uTat, uDNA minichromatin is gradually silenced, but remains highly inducible by HDAC inhibitors (HDACi). Therefore, functionally, uTat antagonizes uDNA minichromatin repression to maintain persistent viral transcription in macrophages. uTat-mediated viral persistence may establish a viral reservoir in macrophages where uDNA were found to persist.
Keywords: HIV, Unintegrated HIV DNA, Macrophage, Transcription, Tat, Persistence, Reservoir, HDACi
Introduction
The natural process of HIV infection leads to the accumulation of a large amount of unintegrated viral DNA (uDNA) in CD4 T cells, macrophages, lymphoid tissues, and the brain (Chun et al., 1997; Pang et al., 1990; Pauza et al., 1990; Shaw et al., 1984; Teo et al., 1997). In cell culture conditions, uDNA has been found to possess transcriptional activity, which can generate all three classes of viral transcripts: the multiply spliced, the singly spliced, and the non-spliced (Kelly et al., 2008; Wu and Marsh, 2001, 2003). Nevertheless, only viral early proteins such as Nef are selectively translated in primary T cells and macrophages in the absence of cellular stimulation (Kelly et al., 2008; Wu and Marsh, 2001, 2003). This selective transcription has been suggested to be a normal early process of HIV infection of human T cells and macrophages (Kelly et al., 2008; Wu, 2004; Wu and Marsh, 2001, 2003). In particular, in cultured macrophages, uDNA can persist for at least 30 days and remain transcriptionally active at a low level (Gillim-Ross et al., 2005a; Kelly et al., 2008).
Functionally, the production of Nef from uDNA has been suggested to facilitate HIV-1 infection by lowering the threshold of T cell activation (Fenard et al., 2005; Schrager and Marsh, 1999; Wu and Marsh, 2001). This early Nef from uDNA can also effectively downmodulate surface receptors such as CD4, CXCR4/CCR5, and MHC class I (Gillim-Ross et al., 2005b; Sloan et al., 2011a; Sloan et al., 2011b). In addition, uDNA can stimulate the production of numerous inflammatory chemokines such as CXCL8 (IL-8), CXCL9, and CXCL10 (IP-10) in macrophages (Kelly et al., 2008). The persistence of uDNA in immune cells, such as blood resting CD4 T cells and macrophages, has also been suggested to serve as alternative form of viral reservoirs, which can be readily reactivated by T cell activation, cytokines, gut-associated short chain fatty acids, or other latency reversing agents for gene expression, and even low level viral replication (Chan et al., 2016; Gelderblom et al., 2008; Kantor et al., 2009; Petitjean et al., 2007; Trinite et al., 2013; Wu, 2008). In addition, genetic complementation between unintegrated and integrated viral genomes can occur, and this process has been suggested to be essential to prevent possible losses of viral genetic diversity (Gelderblom et al., 2008; Wu, 2008). Thus, the two transcriptional processes, occurring before and after HIV integration, are intimately linked to facilitate viral infection, latency, and persistence (Wodarz et al., 2014; Wu, 2008).
Recent studies have also suggested that preintegration transcription occurs at a large scale, and the number of transcribing uDNA approximates to those of proviral genomes (Iyer et al., 2009). However, each uDNA template transcribes at a homogenously low level (Iyer et al., 2009). This raises the question of whether this low level transcription is regulated by Tat, the viral transactivator of transcription. Tat stimulates viral gene expression by binding to the transactivation response (TAR) element on RNA and recruits factors such as cyclin T1 and CDK9 (P-TEFb) for transcriptional initiation and polymerase processivity (Herrmann and Rice, 1993, 1995; Mancebo et al., 1997; Wei et al., 1998; Zhu et al., 1997). Tat can also directly bind to components of the ATP-dependent chromatin remodeling complex SWI-SNF (Agbottah et al., 2006; Mahmoudi et al., 2006; Treand et al., 2006) and recruits the complex to the HIV promoter (Agbottah et al., 2006; Mahmoudi et al., 2006; Treand et al., 2006). In addition, Tat can bind to HAT (histone acetyltransferase) such as p300/CBP, pCAF, and hGCN5 (Benkirane et al., 1998; Col et al., 2001; Hottiger and Nabel, 1998; Kamine et al., 1996; Marzio et al., 1998), which may promote acetylation of nucleosomes and Tat itself to enhance Tat-mediated transactivation (Gatignol, 2007; Kiernan et al., 1999). Nevertheless, given the low transcriptional activity of uDNA, the role of Tat in activating uDNA transcription remained uncertain. In this article, we examined the role of Tat in regulating uDNA transcription and viral persistence in primary macrophages.
Materials and Methods
Approvals from IRB and IACUC
Peripheral blood was drawn from HIV-negative donors. All protocols involving human subjects were reviewed and approved by the George Mason University (GMU) IRB. Informed written consents from the human subjects were obtained in this study.
Plasmids and DNA cloning
Plasmids HIV-1NL4-3 and its integrase mutant HIV-1NL4-3/D116N, HIV-1AD8 and its integrase mutant HIV-1AD8/D116N were kindly provided by Dr. Malcolm A. Martin. The HIV-1 envelope mutant plasmid pNL4-3(KFS) was kindly provided by Dr. Eric Freed. The integrase mutant plasmid pNL4-3(KFS)(D116N) was constructed by introducing a point mutation (Asp 116 to Asn) into the integrase catalytic domain of pNL4-3(KFS) as previously described (Iyer et al., 2009). The LTR-driven expression plasmids pLTR-GFP (pEV658) and pLTR-Tat-IRES-GFP (pEV731) were kindly provided by Dr. Eric Verdin. pCMVΔR8.2, pCMVΔR8.2(D116N), and pHCMV-G expressing vesicular stomatitis virus (VSV) glycoprotein have been described previously (Wang et al., 2010). pTRE-Tat86 and pTet-On were kindly provided by Dr. Ashok Chauhan. pLTR-Luc was constructed from pNL-Luc-RRE-SA and pNL-RRE-SA (Wu et al., 2007b) by inserting the luciferase gene (firefly) within the XhoI cloning site of pNL-RRE-SA. pLTR-Tat-IRES-Luc was generated from pLTR-TAT-IRES-GFP (pEV731) and pLTR-Luc. A PCR product was generated from using pLTR-TAT-IRES-GFP (pEV731) as the template. The PCR product contains the tat gene, IRES, and two external EcoRI sites. The PCR product and pLTR-Luc were both digested with EcoRI and then ligated to generate plasmid pLTR-Tat-IRES-Luc.
Viruses, viral vectors, and viral infection
HIV-1NL4-3, HIV-1NL4-3/D116N, HIV-1AD8, and HIV-1AD8/D116N were generated by transfection of plasmid DNAs into HEK293T cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) as previously described (Wu and Marsh, 2001). Supernatant was harvested at 48 hours and filtered through a 0.45 µm nitrocellulose membrane. Virus titer (TCID50) was measured by infection of Rev-dependent GFP and GFP/Luc (green fluorescent protein/Luciferase) duo reporter cell lines, Rev-CEM-GFP and Rev-CEM-GFP-Luc (Wu et al., 2007a, b) (Virongy, Manassas, VA). NL4-3(KFS) and NL(KFS)(D116N) were produced by cotransfection of HEK293T cells with pHCMV-G and either pNL4-3(KFS) or pNL4-3(KFS)(D116N) in a 1:1 ratio. vLTR-Tat-IRES-GFP and vLTR-Tat-IRES-GFP(D116N) were produced by cotransfection of HEK293T cells with pLTR-Tat-IRES-GFP, pHCMV-G, and either pCMVΔR8.2 or pCMVΔR8.2(D116N) in a ratio of 4:1:3 respectively. vLTR-GFP and vLTR-GFP(D116N) were produced by cotransfection of HEK293T cells with pLTR-GFP, pHCMV-G, and either pCMVΔR8.2 or pCMVΔR8.2(D116N) in a ratio of 4:1:3, respectively. vNL-Luc and virus vNL-Luc(D116N) were constructed by co-transfection of HEK293T cells with pLTR-Luc, pHCMV-G, and either pCMVΔR8.2 or pCMVΔR8.2(D116N) in a 4:1:3 ratio, respectively. vLTR-Tat-IRES-Luc and vLTR-Tat-IRES-Luc(D116N) were generated from co-transfection of HEK293T cells with pLTR-Tat-IRES-Luc, pHCMV-G, and either pCMVΔR8.2 or pCMVΔR8.2(D116N) in a 4:1:3 ratio, respectively. Viral supernatant was harvested at 48 h postcotransfection, filtered through a 0.45-µm filter, and stored at −80°C. Levels of p24 in viral supernatant were measured using a Perkin Elmer Alliance p24 antigen enzyme-linked immunosorbent assay kit (Perkin Elmer, Waltham, MA) or an in-house p24 antigen ELISA kit. Viral infection of T cells and macrophages were carried out as previously described (Kelly et al., 2008; Yoder et al., 2008). HIV infection was also carried through spinoculation (Guo et al., 2011), in which viral particles were added to the cells and subjected to centrifugation for 2 hours at 600 × g (Guo et al., 2011). After centrifugation, the medium was replaced with 1 ml fresh RPMI 1640 medium supplemented with 10% heat-inactivated FBS and incubated at 37° C for another 48 hours.
Cells and cell culture
Rev-dependent cell line Rev-CEM-GFP (Wu et al., 2007a, b), CEM-SS cells (Virongy, Manassas, VA), and U937 cells (from NIH AIDS Reference and Reagent Program) were cultured in RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA), penicillin (50 U/ml), and streptomycin (50 mg/ml). The integrase inhibitor, 118-D-24, was from NIH AIDS Reference and Reagent Program (Svarovskaia et al., 2004). For infection, 2 × 105 cells were incubated with 100 to 500 ng (p24) of virus for 2 hours at 37°C. Cells were washed, and then resuspended in fresh culture medium. 1G5 cells (from NIH AIDS Reference and Reagent Program) were cultured in RPMI 1640 medium supplemented with 10% heat-inactivated. For infection, 5 × 105 cells were either pre-treated with etravirine (NIH AIDS Reference and Reagent Program) or not treated for 1 h at 37°C. The pre-treated cells were cultured in medium with the continuous presence of the drug. Cells were infected with 300–500 ng (p24) of virus for 4 hours at 37°C, washed, resuspended in fresh culture medium, and then cultured for another 48 hours. HeLa Tet-On Advanced cells (Clontech Laboratories, Inc., Mountain View, CA) were cultured in DMEM medium supplemented with 10% Tet System approved FBS (Clontech Laboratories, Inc., Mountain View, CA) with G418 (100 µg/ml). Macrophages were differentiated from human monocytes in the peripheral blood mononuclear cells (PBMC). Briefly, 2.5 × 106 PBMCs were plated into each well of a 6-well plate in serum-free RPMI medium for 1 hour. Adherent cells were cultured in RPMI plus 10% heat-inactivated FBS and 10 ng/ml macrophage colony stimulating factor (M-CSF) (R&D System, Minneapolis, MN) for 2 weeks with medium change every 2 days. For infection, equal p24 input virus was added to the cells and incubated at 37°C for 4 hours. Cells were washed twice and cultured in RPMI 1640 supplemented with 10% heat-inactivated FBS, penicillin (50 U/ml), and streptomycin (50 µg/ml). Primary human microglia cells were provided by Dr. Nazira El-Hage. Cells were cultured in DMEM medium supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA), penicillin (50 U/ml), and streptomycin (50 mg/ml).
Flow cytometry and cell sorting
For flow cytometry, 2 × 105 CEM-SS cells were infected with equal p24 input virus (5 ng) and incubated for 4 hours at 37°C. The cells were washed once and resuspended in 1 ml fresh RPMI 1640 supplemented with 10% heat-inactivated FBS. GFP expression was measured at 48 hours post infection. Data analysis was performed using CellQuest (BD Biosciences, San Jose, CA) and FlowJo (Tree Star, San Carlos, CA).
HDAC inhibitor stimulation
Apicidin (Sigma-Aldrich, St. Louis, MD), Sodium butyrate (Merck, Darmstadt, Germany), and Valproic acid (Merck, Darmstadt, Germany) were used to induce gene expression from the LTR of luciferase reporter viruses. HDACis (histone deacetylase inhibitors) were dissolved at different concentrations in RPMI 1640 medium. Cells were infected with equal amount of viral particles, cultured for various periods, stimulated with HDACis for 24 hours, harvested, and then analyzed for the luciferase activity, using the amount of viral DNA in the lysates for normalization. Viral DNA was quantified by quantitative real-time PCR as described previously (Yoder et al., 2008). For luciferase assay, cells were rinsed twice with cold 1 × PBS. After the second rinse, cells were lysed in 500 µl of 1 × reporter lysis buffer (Promega, Madison, Wisconsin) or 1 × cell culture lysis reagent (Promega, Madison, Wisconsin). Firefly luciferase activity was measured using the Promega Glomax Multi Detection system.
Doxycycline induction
HeLa Tet-On cells were transfected with 2 µg of either pTRE-Tat86 or pcDNA3.1+. After 5 to 6 hours, the medium was replaced with fresh DMEM supplemented with 10% Tet-On approved FBS, penicillin (50 U/ml), and streptomycin (50 mg/ml). After 24 hours, cells were infected with 45 ng (p24) of reporter virus vLTR-Luc or vLTR-Luc(D116N) for 2 hours at 37°C. Cells were washed with DMEM supplemented with 10% Tet-On approved FBS and with or without the 1 µg/ml of doxycycline. Cells were harvest after 24 hours for analyzing luciferase activity.
Viral DNA and RNA quantification
Total cellular DNA and RNA were purified using SV total RNA isolation kit as recommended by the manufacture (Promega, Madision, WI). Purified RNA was further treated with DNase I (DNA-free kit, Invitrogen, Carlsbad, CA) as recommended by the manufacture. HIV DNA was quantified by real-time PCR as described previously (Yoder et al., 2008). For RNA reverse transcription, M-MLV reverse transcriptase (Invitrogen, Carlsbad, CA) was used. For each reaction, 2 µl of random decamers (50 µM) and 10 µl of RNA (100 – 200 ng) template were mixed, and incubated at 80°C for 3 minutes, and then cooled down to 4°C for 5 minutes. Reaction was carried out in 20 µl containing 2 µl of 10 × complete PCR buffer, 4 µl of dNTPs (2.5 mM), 10 U of RNase inhibitor, 100 U M-MLV reverse transcriptase. For the real-time PCR assay, Taqman Gene Expression Master Mix (Invitrogen, Carlsbad, CA) was used. The reaction mixture consisted of 25 µL of 2 × Gene Expression Master Mix, 5 µL of 10 × (2 µM) probe, 5 µL of 10 × (3 µM) forward primers, 5 µL 10 × reverse primers, and 10 µL of DNA. For quantification of the nef transcripts, the follow probe and primers were used: FAM-Nef/Rev, 5’(56-FAM) CGGAGACAGCGACGAAGAGCTCATC (3 BHQ-1) 3’; 5’ Nef, 5’GGCGGCGACTGGAAGAA3’; 3’Rev, 5’AGGTGGGTTGCTTTGATAGAGAAG3’. Quantitative real-time PCR analysis was carried out using Bio-Rad iQ5 real-time PCR detection system.
ChIP assay
CHIP was carried as described previously (Tyagi and Karn, 2007). Briefly, 30 to 100 millions of human monocytic cell line, U937, were infected with vLTR-Luc or vLTR-Luc(D116N), and cells were harvested at 4, 12, and 24 hours. Antibodies were purchased from Santa Cruz, including anti-RNAP II (N-20), HDAC-1 (H-51), and p65 (C-20). Anti-acetylated histone-3 and -4 antibodies were obtained from Upstate Biotech (Fisher Scientific). Each sample (5%) was analyzed by quantitative real-time PCR to access the amount of sample immunoprecipitated by individual antibody. A non-antibody control value was subtracted from each sample value to remove the background counts.
Results
Preintegration transcription occurs at a homogenously low level
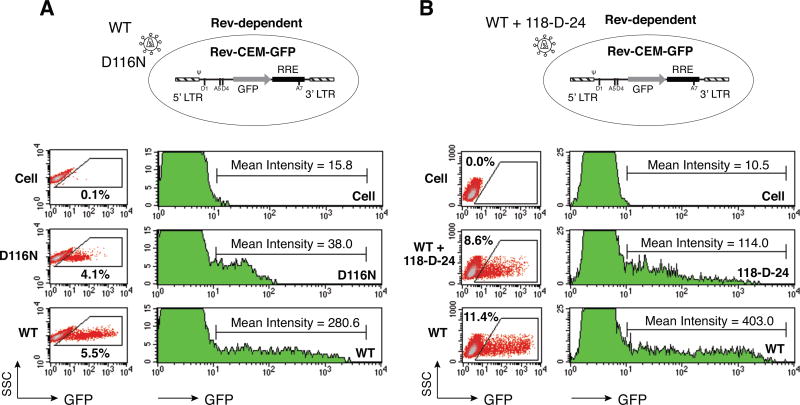
Preintegration transcription from uDNA has been shown to produce a viral early protein Nef (Wu and Marsh, 2001, 2003). The two other viral early proteins, Tat and Rev, are presumably also produced. Both proteins are difficult to detect even in wild-type (WT) HIV-infected cells by Western blot. To measure the activity of Tat and Rev from uDNA, we used an HIV Tat and Rev-dependent GFP (green fluorescent protein) indicator CD4 T cell line, Rev-CEM-GFP, which expressed GFP upon HIV infection and Tat/Rev expression (Wu et al., 2007a, b). Rev-CEM-GFP was infected with WT HIV-1NL4-3 or its integrase mutant, HIV-1NL4-3(D116N). As shown in Fig. 1A, both WT and D116N infection of Rev-CEM-GFP resulted in GFP positive cells, suggestive of the expression of Tat and Rev. Further quantification revealed that the GFP cell percentage in D116N infection (4.1%) was comparable to that of WT infection (5.5%) (using an equal p24 level for infection), indicating a similar number of transcribing DNA templates with or without integration (Iyer et al., 2009). However, when the GFP intensity was quantified, in D116N-infected cells, GFP expression was universally at a low level (mean intensity = 38.0) (Fig. 1A, right panel). This was in great contrast to the WT infection, which generated GFP-positive cells of varied intensities across two orders of magnitude (mean intensity = 280.6). Similar results were also observed when an integrase inhibitor, 118-D-24, was used to block WT HIV integration (Fig. 1B). Given this homogenously low GFP activity from uDNA in D116N infection, it is uncertain whether Tat is indeed generated early and functionally involved in stimulating uDNA transcription.
Fig. 1. Preintegration transcription occurring at a homogenously low level.
(A) An HIV Rev-dependent reporter cell line, Rev-CEM-GFP, was infected with an equal p24 level of HIV-1NL4-3 (Wt) or an integrase mutant, D116N. The percentage of GFP-positive cells was measured at 48 hours post infection with flow cytometry (density plot, left panel). The average mean intensity of GFP in the GFP-positive cells is also shown (histoplot, right panel). (B) The integrase inhibitor, 118-D-24 (50 µM), was used to treat Rev-CEM-GFP cells for 4 hours prior to HIV-1 infection. Cells treated with 118-D-24 or untreated were then infected with an equal p24 level of the Wt virus for 2 hours. Following infection, cells were cultured in the continuous presence of 118-D-24. The percentage (%) and average GFP intensity (M) of GFP-positive cells were measured at 48 hours with flow cytometry. All these experiments have been repeated 3 times.
de novo Tat activity is generated from uDNA
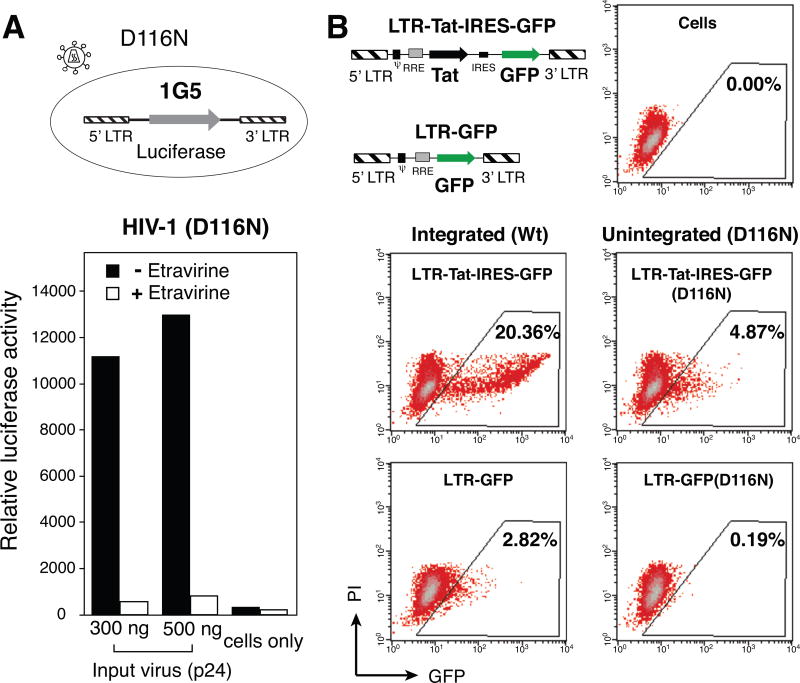
Western blot detection of Tat protein is difficult even with WT infection, but the demonstration of Tat activity is achievable. We used a LTR-driving luciferase reporter cell line, 1G5 (Aguilar-Cordova et al., 1994), to measure Tat stimulation of LTR from the integrase mutant, HIV-1(D116N). We observed a great enhancement of the LTR-driving luciferase expression following infection of 1G5 with D116N (Fig. 2A). However, the intrinsic leakiness of the LTR promoter in indicator cells such as 1G5 made the measurement of low-level HIV transcription problematic, as stimuli such as cytokines, mitogens (Aguilar-Cordova et al., 1994; Akan et al., 1997; Siekevitz et al., 1987; Sweet and Hume, 1995; Swingler et al., 1992; Swingler et al., 1994), or the viral particle binding itself (Merzouki et al., 1995) may also trigger Tat-independent reporter expression. Thus, we pre-treated cells with a reverse transcriptase inhibitor, etravirine, which largely diminished the enhancement of the luciferase activity. This demonstrated that the D116N-stimulated luciferase activity was indeed dependent on newly-synthesized viral DNA, and likely newly-synthesized Tat from uDNA.
Fig. 2. de novo Tat activity from uDNA and its transactivation of preintegration transcription.
(A) de novo Tat activity from uDNA. An HIV Tat-dependent indicator cell line, 1G5 cells were infected with the integrate mutant, HIV-1(D116N). For comparison, cells were also pre-treated with the reverse transcriptase inhibitor etravirine. Equal number of cells were lysed and luciferase activity measured at 48 hours post infection. Uninfected 1G5 cells were used as a control. The experiment has been repeated 4 times. (B) Tat-mediated transactivation of preintegration transcription from uDNA. Lentiviral particles with and without the Tat gene (pLTR-GFP-IRES-Tat and pLTR-GFP), and their integrase mutants, pLTR-GFP-IRES-Tat(D116N) and pLTR-GFP)(D116N), were assembled and used to transduce CEM-SS cells. Equal p24 levels of the Tat+ and Tat− viral particles were used. GFP expressions were measured by flow cytometry at 72 hours post infection. The experiments have been repeated 4 times.
To further confirm the involvement of the uDNA-derived Tat (uTat) in uDNA transcription, we assembled an unintegrating lentiviral vector, pLTR-GFP-IRES-Tat(D116N), that expresses both Tat and GFP from the LTR promoter (Fig. 2B). Infection of human T cells with vLTR-GFP-IRES-Tat(D116N) resulted in the expression of low-level GFP, in comparison with a control integrating viral vector, vLTR-GFP-IRES-Tat. Deletion of Tat from these vectors generated vLTR-GFP(D116N) and vLTR-GFP (Fig. 2B). When cells were similarly infected (using equal p24 of the Tat+ and Tat− viruses), the deletion of Tat greatly diminished the GFP expression in vLTR-GFP(D116N) infection (96% reduction) (Fig. 2B). Similar deletion of Tat from the integrating vector, vLTR-GFP-IRES-Tat, also diminished the GFP cell population, although to a less extent (85%). This result confirmed that Tat is also required to stimulate the low level gene expression from uDNA. Intriguingly and consistent with the result in Fig. 1, even Tat is required, without integration, Tat cannot stimulate LTR to the high levels of GFP expression seen in the integrated vector, suggesting that the sites of LTR integration and local cellular chromatin environment may have a profound impact on the LTR responsiveness to Tat transactivation.
Different responsiveness of uDNA and provirus to Tat stimulation
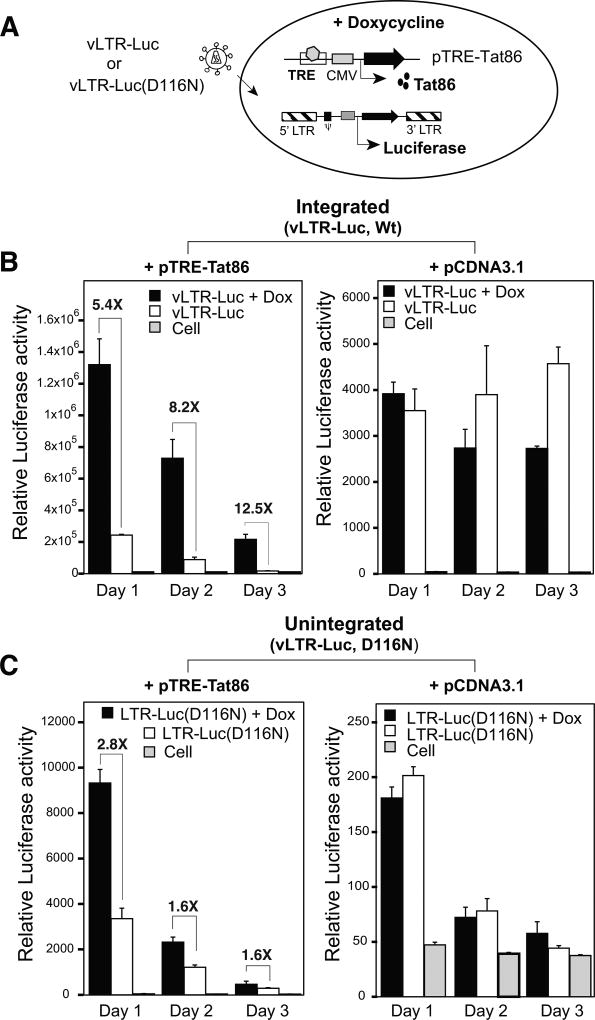
To further quantitatively measure the differences between integrated versus uDNA templates, for their responsiveness to Tat, we established a doxycycline-inducible Tat expression system, in which a doxycycline-responsive transactivator (drTA)-expressing HeLa cell (HeLa Tet-On, Clontech) was transfected with a doxycycline-inducible Tat expression vector, pTRE-Tat86 (Fig. 3A). Following doxycycline induction, activated drTA binds to TRE (Tetracycline Responsive Element) on the promoter region of pTRE-Tat86, activating Tat expression. HeLa Tet-On/pTRE-Tat86 or a control cell, HeLa Tet-On/pCDNA3.1, were infected with a reporter virus, vLTR-Luc, or its unintegrating form, vLTR-Luc(D116N). Both of them express luciferase as the sole protein from the LTR-promoter. Following doxycycline induction of Tat, LTR-driving luciferase activity was measured. As shown in Fig. 3B, for the integrating virus, vLTR-Luc, Tat-induction stimulated a 8.2- to 12.5-fold increase in the luciferase activity (at day 2 and 3), whereas for the non-integrating virus, vLTR-Luc(D116N), similar Tat-induction also led to an increase in the luciferase activity, but it is only 1.9 to 1.6-fold (Fig. 3C) (23% to 13% of integrated). The enhancements were dependent on Tat, as in the absence pTRETat86 doxycycline induction itself does not stimulate the luciferase activity in infected cells (Fig. 3B and 3C). These results confirmed that Tat is capable of stimulating uDNA transcription, but the uDNA LTR is less responsive to Tat than integrated LTR.
Fig. 3. Different responsiveness of HIV uDNA and provirus to Tat stimulation.
(A) Schematic representation of the doxycycline-inducible Tat expression system. HeLa cells (HeLa Tet-On) were stably transfected with a plasmid vector expressing the doxycycline-responsive transcriptional activator, and then transfected with a doxycycline-inducible Tat expression vector, pTRE-Tat86, or a control empty vector, pCDNA3.1. Cells were subsequently infected with a lentiviral particle, vLTR-Luc or its integrase mutant, vLTR-Luc(D116N) (equal p24 for pTRE-Tat86 and pCDNA3.1 transfected cells). Cells were infected with our without Tat-induction by doxycycline. (B) Cells were infected with vLTR-Luc, and luciferase activity was measured at day 1 to 3 post infection. (C) Cells were infected with vLTR-Luc(D116N), and luciferase activity was measured at day 1 to 3 post infection. The mean luciferase activity was from experimental triplicates.
Tat antagonizes minichromatin repression to promote persistent uDNA transcription in primary macrophages
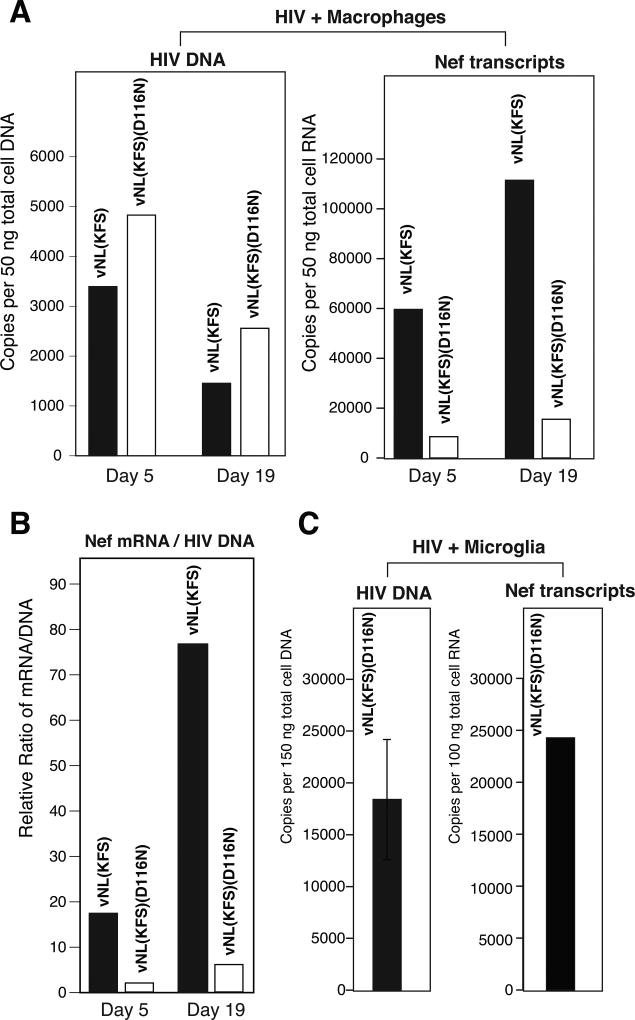
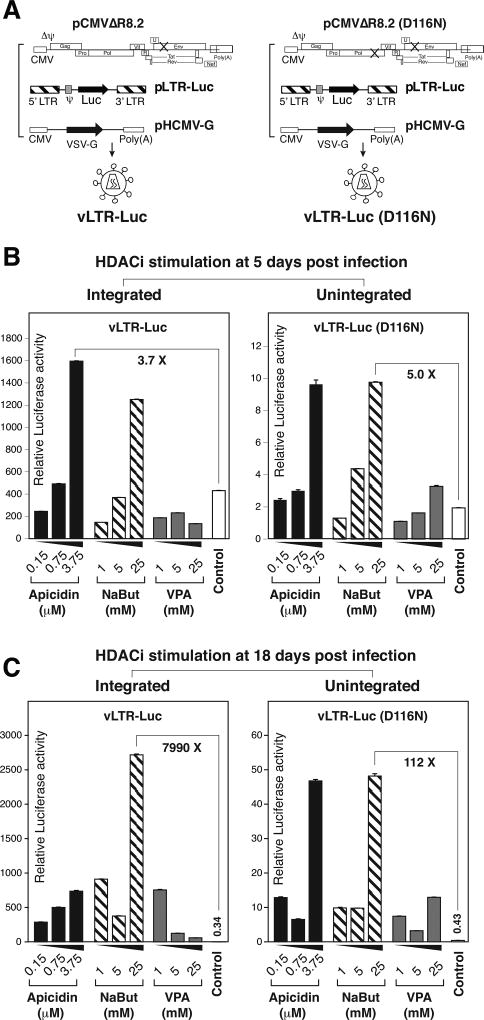
It has been shown that a large amount of uDNA accumulates in brain tissues that are infected through migrating peripheral macrophages (Pang et al., 1990). In cell culture conditions, uDNA and its transcripts can persist in macrophages for as long as 20–30 days (Kelly et al., 2008). We confirmed these previous results by infecting blood monocytes-derived macrophages (MDM) and primary human microglia with an integrating or unintegrated virus, HIV-1(KFS) or HIV-1(KFS(D116N), and observed the persistence of uDNA and its nef transcripts (Fig. 4), consistent with previous results (Kelly et al., 2008). To further determine whether uTat is required for the transcriptional persistence of uDNA in macrophages, we generated a Tat-negative LTR-driving luciferease reporter vector, pLTR-Luc. Integrated and unintegrated lentiviral particles, vLTR-Luc and vLTR-Luc(D116N) respectively, were assembled and used to infect MDM (Fig. 5A). As shown in Fig. 5B, we observed LTR-driving luciferase activity at day 5 post infection with both vNL-Luc and vNL-Luc(D116N) (Fig. 5B, Control). However, without Tat, the activity was largely diminished at day18 post infection (Fig. 5C, Control). This diminished transcriptional activity is in great contrast to the persistent transcription (20–30 days) seen in the Tat+ HIV(D116N) infection of macrophages (Kelly et al., 2008). The loss of transcriptional activity did not result from possible losses of viral DNA templates, as they were readily detectable in cells infected with vLTR-Luc or vLTR-Luc(D116N) (data not shown). It is possible that repressive histones may be assembled onto the LTR region that is gradually silenced in the absence of Tat. Thus, we stimulated infected cells with 3 different histone deacetylase (HDAC) inhibitors, apicidin, sodium butyrate (NaBut), and valproic acid (VPA). As shown in Fig. 5B, at day 5 post infection, apicidin and NaBut are capable of stimulating gene expression from the both integrated and unintegrated LTR. However, VPA had minimal effect, and was slight inhibitory for integrated proviral DNA. Although VPA inhibits histone deacetylase, it can also induce cell differentiation and selectively inhibits the transcription of certain genes such as AKT1/AKT2, which are important for HIV infection (Chen et al., 2006). For the unintegrated LTR, apicidin stimulated a 3.4-fold increase in luciferase expression, while NaBut induced a 4.0-fold increase. For the integrated LTR, we observed up to 2.7- and 2.0-fold enhancement by these two drugs, respectively. At day 18, when similar HDACi stimulation was performed, we observed a dramatic stimulation of the LTR activity. For the uDNA LTR, we observed an approximately 100-fold increase by apicidin and NaBut, whereas for the integrated LTR, we observed an approximately 8,000-fold enhancement by NaBut. This dramatic enhancement was observed in the low transcription background in the absence of Tat, suggesting that repressive chromatin may have been assembled.
Fig. 4. Persistent transcription from HIV uDNA in primary macrophages and microglia.
(A) Monocyte-derived macrophages were infected with an equal p24 level of HIV-NL(KFS) or its non-integrating mutant HIV-vNL(KFS)(D116N) for 5 and 19 days. Cells were lysed and total cellular DNA and RNA were extracted and used for the quantification of viral DNA and Nef mRNA by real-time PCR and real-time reverse transcriptase PCR. (B) The relative ratio of Nef mRNA and HIV DNA was plotted. (C) Primary human microglia were infected with the non-integrating HIV-NL(KFS)(D116N) for 3 days. Cells were lysed and total cellular DNA and RNA were extracted and used for the quantification of viral DNA and Nef mRNA by real-time PCR and real-time reverse transcriptase PCR.
Fig. 5. Responses of HIV uDNA to HDAC inhibitor stimulation.
(A) Schematics of lentiviral particle assembly. Integrating lentiviral particle vLTR-Luc and its integrase mutant vLTR-Luc(D116N) were assembled by cotransfection with pCMVΔR8.2 or pCMVΔR8.2(D116N), and a VSV-G expression vector, pHCMV-G. Particles were harvested and used to infect monocyte-derived macrophages (MDM) as shown in (B) and (C). (B) At 5 days post infection with vLTR-Luc or vLTR-Luc(D116N), cells were stimulated with apicidin, sodium butyrate (NaBut), and valproic acid (VPA), and harvested for luciferase assay. Control cells were infected but not treated with HDAC inhibitors. HIV viral genomic DNA in cell lysates was quantified for normalization. (C) At 18 days post infection with vLTR-Luc or vLTR-Luc(D116N), cells were identically stimulated and analyzed. The mean luciferase activity was from experimental triplicates.
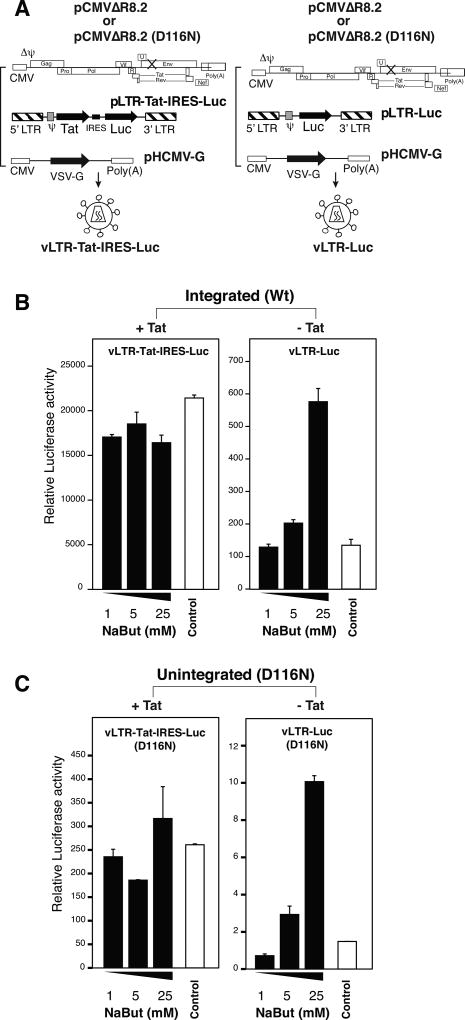
Next, we tested whether Tat is indeed capable of antagonizing repressive chromatin in MDM. Tat has been shown to directly bind to components of the ATP-dependent chromatin remodeling complex SWI-SNF (Agbottah et al., 2006; Mahmoudi et al., 2006; Treand et al., 2006) and recruits the complex to the HIV promoter (Agbottah et al., 2006; Mahmoudi et al., 2006; Treand et al., 2006). In addition, Tat can bind to HAT (histone acetyltransferase) such as p300/CBP, pCAF, and hGCN5 (Benkirane et al., 1998; Col et al., 2001; Hottiger and Nabel, 1998; Kamine et al., 1996; Marzio et al., 1998), which may mediate acetylation of nucleosomes to enhance Tat-mediated transactivation (Gatignol, 2007; Kiernan et al., 1999). To determine impacts of Tat on LTR responsiveness to histone modulation, we constructed a lentiviral vector, pLTR-Tat-IRES-Luc, in which both Tat and luciferase are co-expressed from the LTR. For comparison, we also used the Tat-negative vector, pLTR-Luc, as described above. Intergrated and unintegrated viral particles were assembled, and then used to infect MDM (Fig. 6A). At 5 days post infection, cells were treated with NaBut to stimulate the LTR transcription. As shown in Fig. 6B and 6C, when Tat was expressed from integrated or uDNA templates, it stimulated high luciferase activity. However, NaBut did not stimulate the LTR further when Tat is present. In contrast, when Tat was deleted, upon the low transcripational background, NaBut stimulated an approximately 5-fold enhancement from the integrated and unintegrated LTR (Fig. 6B and 6C, right panel). These results suggest that Tat itself may function to antagonize the chromatin repression to maintain persistent transcription in primary macrophages.
Fig. 6. Effects of Tat on HIV uDNA responses to HDACi stimulation.
(A) Schematics of lentiviral particle assembly. Integrating lentiviral particles, vLTR-Luc or vLTR-Tat-IRES-Luc, and their integrase mutants, vLTR-Luc(D116N) or Tat-IRES-Luc(D116N), were assembled by cotransfection with pCMVΔR8.2 or pCMVΔR8.2(D116N), and a VSV-G expression vector, pHCMV-G. Particles were harvested and used to infect monocyte-derived macrophages (MDM) as shown in (B) and (C). (B and C) MDMs were infected with an equal p24 level of vLTR-Tat-IRES-Luc or vLTR-Luc, either integrating (B) or non-integrating (C). At 5 days post infection, cells were stimulated with sodium butyrate (NaBut), harvested, and analyzed for luciferase activity. Control cells were infected but not treated with HDAC inhibitors. HIV viral genomic DNA in cell lysates was quantified for normalization. The mean luciferase activity was from experimental triplicates.
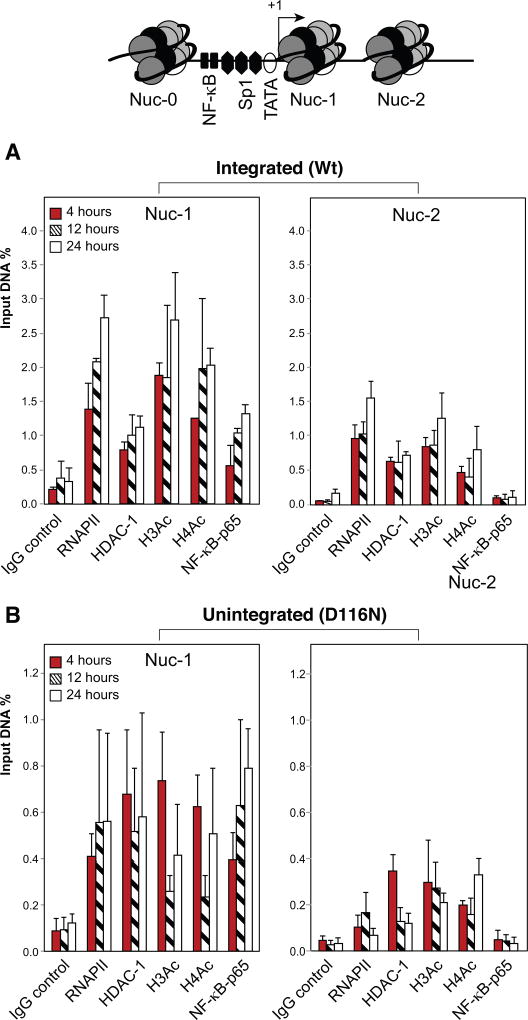
To further confirm that minichromatin is indeed assembled onto uDNA, we infected a monocyte cell, U937, with integrating vLTR-Luc or unintegrating vLTR-LTR (D116N) and performed the chromatin immunoprecipitation (ChIP) assay at 4, 12, and 24 hours post infection. HIV LTR contains two well-ordered nucleosomes, Nucleosome 0 (Nuc-0) and nucleosome 1 (Nuc-1). Nuc-0 is positioned immediately upstream of the enhancer (−415 to −255), while Nuc-1 is very close to the viral RNA start site (+10 to +155). We examined chromatin modifications and the recruitment of responsible epigenetic enzymes at the HIV LTR. As shown in Fig. 7, we observed immediate enrichment of acetylated histones and enhanced recruitment of RNAP II even at 4 hours post infection, showing ongoing gene expression from the LTR promoter. We also noted the higher binding of NF-kB (p65), specifically at its cognate binding sites, which validates specific NF-kB recruitment. In addition, we observed acetylated histone H3 and repressive HDAC-1 on Nuc-1, suggesting that histone deacetylation may also occur in the absence of Tat. Overall, for both integrated and unintegrated HIV genomes, we found chromatin assembly, demonstrating that immediately after entry into cells, HIV DNA interacts with histones and assemble itself into chromatin structures. The levels of recruitment of different factors were relatively less pronounced at unintegrated DNA, suggesting that genomic integration promotes the chromatin organization of proviral DNA, and HIV uDNA may have a different chromatin structure.
Fig. 7. Assembly of minichromatin onto HIV uDNA in macrophages.
U937 cells, a monocytic cell line, were infected with the integration and non-integrating reporter particle vNL-Luc (A) or vNL-Luc(D116N) (B). Cells were cross-linked at 4, 12, and 24 hours post infection, and ChIP assay was performed. On every run, 5% of each sample was analyzed by quantitative real-time PCR to determine the amount of sample immunoprecipitated by individual antibodies. Specific primer sets were used to amplify different regions of the LTR. The reading obtained with preimmune sera was subtracted as background counts.
Discussion
In this article, we provided evidence that de novo Tat activity is generated from HIV uDNA, and this uTat can stimulate HIV uDNA LTR. Functionally, this uTat is important in antagonizing repressive minichromatin that is assembled onto uDNA, permitting persistent transcription to occur in immune cells such as macrophages (Kelly et al., 2008). Given the accumulation of a large amount of uDNA in lymphoid tissues and in the brain, this uTat-mediated low level transcription may contribute to the persistence of viral reservoirs in tissues.
Levy and co-authors has suggested that HIV uDNA transcription is important for the maintenance of viral genetic diversity (Gelderblom et al., 2008; Wodarz et al., 2014; Wu, 2008), as only a small percentage of viral genomes can integrate. Most viral genomes exist as uDNA that can transcribe genomic RNAs (Kelly et al., 2008; Wu and Marsh, 2001, 2003). These RNA genomes are also rescuable for co-packaging, and capable of supporting continuous viral replication (Gelderblom et al., 2008). In the absence of immune stimulation, uDNA transcription has also been shown to produce a viral early protein Nef (Kelly et al., 2008; Wu and Marsh, 2001, 2003), which can modulate resting T cell activity to favor viral replication (Wu and Marsh, 2001). Remarkably, recent results have also demonstrated that in resting CD4 T cells and primary macrophages, this low-level transcriptional activity can be greatly up-regulated by cytokines, T cell activation, or latency reversing agents, leading to low-level production of replication-competent viruses (Chan et al., 2016; Kantor et al., 2009; Trinite et al., 2013). These studies support the possibility that viral uDNA genomes can serve as alternative reservoirs different from integrated proviral reservoirs in blood T cells and tissues (Petitjean et al., 2007), where a large amount of uDNA accumulates to form the predominant species of viral DNA (Chun et al., 1997; Pang et al., 1990; Pauza et al., 1990; Shaw et al., 1984; Teo et al., 1997).
In a previous animal monotherapy trial of the integrase inhibitor, L870812, the drug effectively inhibited acute SIV replication in rhesus macaques following early treatment (day 10 after infection) (Hazuda et al., 2004). However, the inhibitor was not effective in controlling viremia when used at a later time (day 87 post infection); the drug failed to persistently suppress viral load in 5 of the 6 animals (Hazuda et al., 2004). This is in great contrast to reverse transcriptase inhibitors in viremia control. Importantly, the recovered viruses were found to remain wild type in the integrase sequence (Hazuda et al., 2004). Given this intrinsic viral resistance to L-870812, it could suggest that the accumulation of HIV uDNA following L870812 treatment may lead to low level viral replication from the uDNA, especially in later disease stages when cytokines and other immune factors are dysregulated (Chan et al., 2016; Kantor et al., 2009; Trinite et al., 2013). In cell culture conditions, uDNA accumulation does not lead to viral replication in resting CD4 T cells and macrophages (Kelly et al., 2008; Wu and Marsh, 2001). However, if resting T cells or macrophage are cultured in cytokines or histone deacetylase inhibitors, and then stimulated, low level viral replication can occur from uDNA (Chan et al., 2016; Kantor et al., 2009; Trinite et al., 2013).
Consistent with the animal trial (Hazuda et al., 2004), in recent human HAART intensification studies, although the integrase inhibitor raltegravir (RAL) increased the amounts of unintegrated 2-LTR circles, it did not reduce the residual viremia, nor the total amount of viral DNA (Gandhi et al., 2012; Gandhi et al., 2010; Shi et al., 2014). Compared with RAL, the second generation integrase inhibitor dolutegravir (DTG) has been found to bring faster plasma viral load decay when used in combination with reverse transcriptase (RT) inhibitors (Clotet et al., 2014; Raffi et al., 2013; Walmsley et al., 2013). It is possible that the integrase inhibitor may block integration of residue uDNA. Nevertheless, multiple clinical studies have found that the integrase inhibitor resistant viruses from at least half of the patients do not have mutations in the integrase region (Fourati et al., 2015; Malet et al., 2017; White et al., 2015). Consistently, a recent study has isolated a replication competent virus that is highly resistant to all clinical integrase inhibitors (Malet et al., 2017); intriguingly, sequencing of the viral genome showed no mutations in the integrase gene, and the integrase enzymatic activity remains sensitive to the integrase inhibitors. These studies support the hypothesis that uDNA could contribute to viral persistence and residual viremia that may not be reduced by blocking integration. A recent mathematical modeling has suggested that uDNA could contribute to 20% of total viremia in HIV infection (Lau et al., 2015).
Our results suggest that HIV Tat plays an important role in the persistence of viral transcription from uDNA. Tat stimulates uDNA transcription likely through various mechanisms; Tat promotes the processivity of polymerase II through P-TEFb, and antagonizes repressive chromatin through binding to chromatin remodeling complexes and HAT (Benkirane et al., 1998; Col et al., 2001; Hottiger and Nabel, 1998; Kamine et al., 1996; Marzio et al., 1998). Intriguingly, there is a clear distinction in response to Tat stimulation between integrated provirus and uDNA; although Tat is required for uDNA transcription, uDNA is less responsive to Tat than provirus (Fig. 3 and Fig. 5). The mechanism regulating such differences is not currently understood. It is possible that the chromatin structure assembled onto uDNA may be different from that assembled onto integrated provirus. HIV can integrate into both heterochromatin and euchromatin, leading to different levels of basal transcription. The LTR responses to Tat would be greater in the heterochromatin than in the euchromatin. Such structural diversity may not be present in uDNA.
In conclusion, our study demonstrated a critical role of Tat in mediating persistent transcription from uDNA, which could serve as a residual viral reservoir. Targeting Tat is critical, and may offer a way to silence uDNA and reduce its persistence in immune cells.
Research Highlights.
large amounts of unintegrated HIV DNA (uDNA) accumulate and persist in infected macrophages.
uDNA can persistently transcribe viral early genes in macrophages.
Tat is produced from unintegrated HIV genome early in HIV infection (uTat, uDNA-derived Tat).
In the absence uTat, uDNA that is assembled into repressive episomal minichromatin.
uTat antagonizes uDNA minichromatin repression to maintain persistent viral transcription in macrophages.
uTat-mediated viral persistence may establish a viral reservoir in macrophages
Acknowledgments
The authors wish to thank the George Mason University Student Health Center for blood donations; the NIH AIDS Research and Reference Reagent Program for reagents. BM performed experiments on DNA cloning, viral infection, gene expression quantification, Tat induction, and HDAC inhibitor stimulation studies. JG and DD performed data analyses. MT, FK, and DD designed and performed the ChIP assay. YW conceived, directed, and supervised the study, analyzed the data, and wrote the manuscript.
The Content is solely the responsibility of the authors and does not necessarily represent the official view of the National Institute of Health.
Funding information
This work was funded by the 2010 NYCDC AIDS Ride organized by Marty Rosen, and in part by Public Health Service grants 1R01MH102144 and 1R03AI110174 from NIAID and NIMH to YW, 1R01DA041746-01 and 5R21DA033924-02 from NIDA to MT, and AI078859, AI074410, and AI043894 to FK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agbottah E, Deng L, Dannenberg LO, Pumfery A, Kashanchi F. Effect of SWI/SNF chromatin remodeling complex on HIV-1 Tat activated transcription. Retrovirology. 2006;3:48. doi: 10.1186/1742-4690-3-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar-Cordova E, Chinen J, Donehower L, Lewis DE, Belmont JW. A sensitive reporter cell line for HIV-1 tat activity, HIV-1 inhibitors, and T cell activation effects. AIDS Res Hum Retroviruses. 1994;10:295–301. doi: 10.1089/aid.1994.10.295. [DOI] [PubMed] [Google Scholar]
- Akan E, Chang-Liu CM, Watanabe J, Ishizawa K, Woloschak GE. The effects of vinblastine on the expression of human immunodeficiency virus type 1 long terminal repeat. Leuk Res. 1997;21:459–464. doi: 10.1016/s0145-2126(96)00125-7. [DOI] [PubMed] [Google Scholar]
- Benkirane M, Chun RF, Xiao H, Ogryzko VV, Howard BH, Nakatani Y, Jeang KT. Activation of integrated provirus requires histone acetyltransferase. p300 and P/CAF are coactivators for HIV-1 Tat. J Biol Chem. 1998;273:24898–24905. doi: 10.1074/jbc.273.38.24898. [DOI] [PubMed] [Google Scholar]
- Chan CN, Trinite B, Lee CS, Mahajan S, Anand A, Wodarz D, Sabbaj S, Bansal A, Goepfert PA, Levy DN. HIV-1 latency and virus production from unintegrated genomes following direct infection of resting CD4 T cells. Retrovirology. 2016;13:1. doi: 10.1186/s12977-015-0234-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Ghazawi FM, Bakkar W, Li Q. Valproic acid and butyrate induce apoptosis in human cancer cells through inhibition of gene expression of Akt/protein kinase B. Mol Cancer. 2006;5:71. doi: 10.1186/1476-4598-5-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun TW, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn TC, Kuo YH, Brookmeyer R, Zeiger MA, Barditch-Crovo P, Siliciano RF. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection [see comments] Nature. 1997;387:183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- Clotet B, Feinberg J, van Lunzen J, Khuong-Josses MA, Antinori A, Dumitru I, Pokrovskiy V, Fehr J, Ortiz R, Saag M, Harris J, Brennan C, Fujiwara T, Min S Team, I.N.G.S. Once-daily dolutegravir versus darunavir plus ritonavir in antiretroviral-naive adults with HIV-1 infection (FLAMINGO): 48 week results from the randomised open-label phase 3b study. Lancet. 2014;383:2222–2231. doi: 10.1016/S0140-6736(14)60084-2. [DOI] [PubMed] [Google Scholar]
- Col E, Caron C, Seigneurin-Berny D, Gracia J, Favier A, Khochbin S. The histone acetyltransferase, hGCN5, interacts with and acetylates the HIV transactivator, Tat. Journal of Biological Chemistry. 2001;276:28179–28184. doi: 10.1074/jbc.M101385200. [DOI] [PubMed] [Google Scholar]
- Fenard D, Yonemoto W, de Noronha C, Cavrois M, Williams SA, Greene WC. Nef is physically recruited into the immunological synapse and potentiates T cell activation early after TCR engagement. J Immunol. 2005;175:6050–6057. doi: 10.4049/jimmunol.175.9.6050. [DOI] [PubMed] [Google Scholar]
- Fourati S, Charpentier C, Amiel C, Morand-Joubert L, Reigadas S, Trabaud MA, Delaugerre C, Nicot F, Rodallec A, Maillard A, Mirand A, Jeulin H, Montes B, Barin F, Bettinger D, Le Guillou-Guillemette H, Vallet S, Signori-Schmuck A, Descamps D, Calvez V, Flandre P, Marcelin AG Group, A.A.R.S. Cross-resistance to elvitegravir and dolutegravir in 502 patients failing on raltegravir: a French national study of raltegravir-experienced HIV-1-infected patients. J Antimicrob Chemother. 2015;70:1507–1512. doi: 10.1093/jac/dku535. [DOI] [PubMed] [Google Scholar]
- Gandhi RT, Coombs RW, Chan ES, Bosch RJ, Zheng L, Margolis DM, Read S, Kallungal B, Chang M, Goecker EA, Wiegand A, Kearney M, Jacobson JM, D'Aquila R, Lederman MM, Mellors JW, Eron JJ Team, A.C.T.G.A. No effect of raltegravir intensification on viral replication markers in the blood of HIV-1-infected patients receiving antiretroviral therapy. J Acquir Immune Defic Syndr. 2012;59:229–235. doi: 10.1097/QAI.0b013e31823fd1f2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi RT, Zheng L, Bosch RJ, Chan ES, Margolis DM, Read S, Kallungal B, Palmer S, Medvik K, Lederman MM, Alatrakchi N, Jacobson JM, Wiegand A, Kearney M, Coffin JM, Mellors JW, Eron JJ team, A.C.T.G.A. The effect of raltegravir intensification on low-level residual viremia in HIV-infected patients on antiretroviral therapy: a randomized controlled trial. PLoS Med. 2010;7 doi: 10.1371/journal.pmed.1000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatignol A. Transcription of HIV: Tat and cellular chromatin. Adv Pharmacol. 2007;55:137–159. doi: 10.1016/S1054-3589(07)55004-0. [DOI] [PubMed] [Google Scholar]
- Gelderblom HC, Vatakis DN, Burke SA, Lawrie SD, Bristol GC, Levy DN. Viral complementation allows HIV-1 replication without integration. Retrovirology. 2008;5 doi: 10.1186/1742-4690-5-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillim-Ross L, Cara A, Klotman ME. HIV-1 extrachromosomal 2-LTR circular DNA is long-lived in human macrophages. Viral Immunol. 2005a;18:190–196. doi: 10.1089/vim.2005.18.190. [DOI] [PubMed] [Google Scholar]
- Gillim-Ross L, Cara A, Klotman ME. Nef expressed from human immunodeficiency virus type 1 extrachromosomal DNA downregulates CD4 on primary CD4+ T lymphocytes: implications for integrase inhibitors. J Gen Virol. 2005b;86:765–771. doi: 10.1099/vir.0.80570-0. [DOI] [PubMed] [Google Scholar]
- Guo J, Wang W, Yu D, Wu Y. Spinoculation triggers dynamic actin and cofilin activity facilitating HIV-1 infection of transformed and resting CD4 T cells. J Virol. 2011;85:9824–9833. doi: 10.1128/JVI.05170-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazuda DJ, Young SD, Guare JP, Anthony NJ, Gomez RP, Wai JS, Vacca JP, Handt L, Motzel SL, Klein HJ, Dornadula G, Danovich RM, Witmer MV, Wilson KA, Tussey L, Schleif WA, Gabryelski LS, Jin L, Miller MD, Casimiro DR, Emini EA, Shiver JW. Integrase inhibitors and cellular immunity suppress retroviral replication in rhesus macaques. Science. 2004;305:528–532. doi: 10.1126/science.1098632. [DOI] [PubMed] [Google Scholar]
- Herrmann CH, Rice AP. Specific interaction of the human immunodeficiency virus Tat proteins with a cellular protein kinase. Virology. 1993;197:601–608. doi: 10.1006/viro.1993.1634. [DOI] [PubMed] [Google Scholar]
- Herrmann CH, Rice AP. Lentivirus Tat proteins specifically associate with a cellular protein kinase, TAK, that hyperphosphorylates the carboxyl-terminal domain of the large subunit of RNA polymerase II: candidate for a Tat cofactor. J Virol. 1995;69:1612–1620. doi: 10.1128/jvi.69.3.1612-1620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hottiger MO, Nabel GJ. Interaction of human immunodeficiency virus type 1 Tat with the transcriptional coactivators p300 and CREB binding protein. J Virol. 1998;72:8252–8256. doi: 10.1128/jvi.72.10.8252-8256.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer SR, Yu D, Biancotto A, Margolis LB, Wu Y. Measurement of human immunodeficiency virus type 1 preintegration transcription by using Rev-dependent Rev-CEM cells reveals a sizable transcribing DNA population comparable to that from proviral templates. J Virol. 2009;83:8662–8673. doi: 10.1128/JVI.00874-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamine J, Elangovan B, Subramanian T, Coleman D, Chinnadurai G. Identification of a cellular protein that specifically interacts with the essential cysteine region of the HIV-1 Tat transactivator. Virology. 1996;216:357–366. doi: 10.1006/viro.1996.0071. [DOI] [PubMed] [Google Scholar]
- Kantor B, Ma H, Webster-Cyriaque J, Monahan PE, Kafri T. Epigenetic activation of unintegrated HIV-1 genomes by gut-associated short chain fatty acids and its implications for HIV infection. Proc Natl Acad Sci U S A. 2009;106:18786–18791. doi: 10.1073/pnas.0905859106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly J, Beddall MH, Yu D, Iyer SR, Marsh JW, Wu Y. Human macrophages support persistent transcription from unintegrated HIV-1 DNA. Virology. 2008;372:300–312. doi: 10.1016/j.virol.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiernan RE, Vanhulle C, Schiltz L, Adam E, Xiao H, Maudoux F, Calomme C, Burny A, Nakatani Y, Jeang KT, Benkirane M, Van Lint C. HIV-1 tat transcriptional activity is regulated by acetylation. Embo J. 1999;18:6106–6118. doi: 10.1093/emboj/18.21.6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau JW, Levy DN, Wodarz D. Contribution of HIV-1 genomes that do not integrate to the basic reproductive ratio of the virus. J Theor Biol. 2015;367:222–229. doi: 10.1016/j.jtbi.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoudi T, Parra M, Vries RG, Kauder SE, Verrijzer CP, Ott M, Verdin E. The SWI/SNF chromatin-remodeling complex is a cofactor for Tat transactivation of the HIV promoter. J Biol Chem. 2006;281:19960–19968. doi: 10.1074/jbc.M603336200. [DOI] [PubMed] [Google Scholar]
- Malet I, Subra F, Charpentier C, Collin G, Descamps D, Calvez V, Marcelin AG, Delelis O. Mutations Located outside the Integrase Gene Can Confer Resistance to HIV-1 Integrase Strand Transfer Inhibitors. MBio. 2017;8 doi: 10.1128/mBio.00922-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancebo HS, Lee G, Flygare J, Tomassini J, Luu P, Zhu Y, Peng J, Blau C, Hazuda D, Price D, Flores O. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes & Development. 1997;11:2633–2644. doi: 10.1101/gad.11.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzio G, Tyagi M, Gutierrez MI, Giacca M. HIV-1 tat transactivator recruits p300 and CREB-binding protein histone acetyltransferases to the viral promoter. Proc Natl Acad Sci U S A. 1998;95:13519–13524. doi: 10.1073/pnas.95.23.13519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merzouki A, Patel P, Cassol S, Ennaji M, Tailor P, Turcotte FR, O'Shaughnessy M, Arella M. HIV-1 gp120/160 expressing cells upregulate HIV-1 LTR directed gene expression in a cell line transfected with HIV-1 LTR-reporter gene constructs. Cell Mol Biol (Noisy-le-grand) 1995;41:445–452. [PubMed] [Google Scholar]
- Pang S, Koyanagi Y, Miles S, Wiley C, Vinters HV, Chen IS. High levels of unintegrated HIV-1 DNA in brain tissue of AIDS dementia patients. Nature. 1990;343:85–89. doi: 10.1038/343085a0. [DOI] [PubMed] [Google Scholar]
- Pauza CD, Galindo JE, Richman DD. Reinfection results in accumulation of unintegrated viral DNA in cytopathic and persistent human immunodeficiency virus type 1 infection of CEM cells. J Exp Med. 1990;172:1035–1042. doi: 10.1084/jem.172.4.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petitjean G, Al Tabaa Y, Tuaillon E, Mettling C, Baillat V, Reynes J, Segondy M, Vendrell JP. Unintegrated HIV-1 provides an inducible and functional reservoir in untreated and highly active antiretroviral therapy-treated patients. Retrovirology. 2007;4:60. doi: 10.1186/1742-4690-4-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffi F, Jaeger H, Quiros-Roldan E, Albrecht H, Belonosova E, Gatell JM, Baril JG, Domingo P, Brennan C, Almond S, Min S extended, S.-S.G. Once-daily dolutegravir versus twice-daily raltegravir in antiretroviral-naive adults with HIV-1 infection (SPRING-2 study): 96 week results from a randomised, double-blind, non-inferiority trial. Lancet Infect Dis. 2013;13:927–935. doi: 10.1016/S1473-3099(13)70257-3. [DOI] [PubMed] [Google Scholar]
- Schrager JA, Marsh JW. HIV-1 Nef increases T cell activation in a stimulus-dependent manner. Proc Natl Acad Sci U S A. 1999;96:8167–8172. doi: 10.1073/pnas.96.14.8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw GM, Hahn BH, Arya SK, Groopman JE, Gallo RC, Wong-Staal F. Molecular characterization of human T-cell leukemia (lymphotropic) virus type III in the acquired immune deficiency syndrome. Science. 1984;226:1165–1171. doi: 10.1126/science.6095449. [DOI] [PubMed] [Google Scholar]
- Shi M, Liu C, Cook TJ, Bullock KM, Zhao Y, Ginghina C, Li Y, Aro P, Dator R, He C, Hipp MJ, Zabetian CP, Peskind ER, Hu SC, Quinn JF, Galasko DR, Banks WA, Zhang J. Plasma exosomal alpha-synuclein is likely CNS-derived and increased in Parkinson's disease. Acta Neuropathol. 2014;128:639–650. doi: 10.1007/s00401-014-1314-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siekevitz M, Josephs SF, Dukovich M, Peffer N, Wong-Staal F, Greene WC. Activation of the HIV-1 LTR by T cell mitogens and the trans-activator protein of HTLV-I. Science. 1987;238:1575–1578. doi: 10.1126/science.2825351. [DOI] [PubMed] [Google Scholar]
- Sloan RD, Donahue DA, Kuhl BD, Bar-Magen T, Wainberg MA. Expression of Nef from unintegrated HIV-1 DNA downregulates cell surface CXCR4 and CCR5 on T-lymphocytes. Retrovirology. 2011a;7:44. doi: 10.1186/1742-4690-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan RD, Kuhl BD, Donahue DA, Roland A, Bar-Magen T, Wainberg MA. Transcription of preintegrated HIV-1 cDNA modulates cell surface expression of major histocompatibility complex class I via Nef. J Virol. 2011b;85:2828–2836. doi: 10.1128/JVI.01854-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svarovskaia ES, Barr R, Zhang X, Pais GC, Marchand C, Pommier Y, Burke TR, Jr, Pathak VK. Azido-containing diketo acid derivatives inhibit human immunodeficiency virus type 1 integrase in vivo and influence the frequency of deletions at two-long-terminal-repeat-circle junctions. J Virol. 2004;78:3210–3222. doi: 10.1128/JVI.78.7.3210-3222.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet MJ, Hume DA. RAW264 macrophages stably transfected with an HIV-1 LTR reporter gene provide a sensitive bioassay for analysis of signalling pathways in macrophages stimulated with lipopolysaccharide, TNF-alpha or taxol. J Inflamm. 1995;45:126–135. [PubMed] [Google Scholar]
- Swingler S, Easton A, Morris A. Cytokine augmentation of HIV-1 LTR-driven gene expression in neural cells. AIDS Res Hum Retroviruses. 1992;8:487–493. doi: 10.1089/aid.1992.8.487. [DOI] [PubMed] [Google Scholar]
- Swingler S, Morris A, Easton A. Tumour necrosis factor alpha and interleukin-1 beta induce specific subunits of NFKB to bind the HIV-1 enhancer: characterisation of transcription factors controlling human immunodeficiency virus type 1 gene expression in neural cells. Biochem Biophys Res Commun. 1994;203:623–630. doi: 10.1006/bbrc.1994.2228. [DOI] [PubMed] [Google Scholar]
- Teo I, Veryard C, Barnes H, An SF, Jones M, Lantos PL, Luthert P, Shaunak S. Circular forms of unintegrated human immunodeficiency virus type 1 DNA and high levels of viral protein expression: association with dementia and multinucleated giant cells in the brains of patients with AIDS. J Virol. 1997;71:2928–2933. doi: 10.1128/jvi.71.4.2928-2933.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treand C, du Chene I, Bres V, Kiernan R, Benarous R, Benkirane M, Emiliani S. Requirement for SWI/SNF chromatin-remodeling complex in Tat-mediated activation of the HIV-1 promoter. Embo J. 2006;25:1690–1699. doi: 10.1038/sj.emboj.7601074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinite B, Ohlson EC, Voznesensky I, Rana SP, Chan CN, Mahajan S, Alster J, Burke SA, Wodarz D, Levy DN. An HIV-1 replication pathway utilizing reverse transcription products that fail to integrate. J Virol. 2013;87:12701–12720. doi: 10.1128/JVI.01939-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi M, Karn J. CBF-1 promotes transcriptional silencing during the establishment of HIV-1 latency. Embo J. 2007;26:4985–4995. doi: 10.1038/sj.emboj.7601928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walmsley SL, Antela A, Clumeck N, Duiculescu D, Eberhard A, Gutierrez F, Hocqueloux L, Maggiolo F, Sandkovsky U, Granier C, Pappa K, Wynne B, Min S, Nichols G Investigators, S. Dolutegravir plus abacavir-lamivudine for the treatment of HIV-1 infection. N Engl J Med. 2013;369:1807–1818. doi: 10.1056/NEJMoa1215541. [DOI] [PubMed] [Google Scholar]
- Wang Z, Tang Z, Zheng Y, Yu D, Spear M, Iyer SR, Bishop B, Wu Y. Development of a nonintegrating Rev-dependent lentiviral vector carrying diphtheria toxin A chain and human TRAF6 to target HIV reservoirs. Gene Ther. 2010;17:1063–1076. doi: 10.1038/gt.2010.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei P, Garber ME, Fang SM, Fischer WH, Jones KA. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]
- White K, Kulkarni R, Miller MD. Analysis of early resistance development at the first failure timepoint in elvitegravir/cobicistat/emtricitabine/tenofovir disoproxil fumarate-treated patients. J Antimicrob Chemother. 2015;70:2632–2638. doi: 10.1093/jac/dkv149. [DOI] [PubMed] [Google Scholar]
- Wodarz D, Chan CN, Trinite B, Komarova NL, Levy DN. On the laws of virus spread through cell populations. J Virol. 2014;88:13240–13248. doi: 10.1128/JVI.02096-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y. HIV-1 gene expression: lessons from provirus and non-integrated DNA. Retrovirology. 2004;1:13. doi: 10.1186/1742-4690-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y. The second chance story of HIV-1 DNA: Unintegrated? Not a problem! Retrovirology. 2008;5:61. doi: 10.1186/1742-4690-5-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Beddall MH, Marsh JW. Rev-dependent indicator T cell line. Current HIV Research. 2007a;5:395–403. doi: 10.2174/157016207781024018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Beddall MH, Marsh JW. Rev-dependent lentiviral expression vector. Retrovirology. 2007b;4:12. doi: 10.1186/1742-4690-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Marsh JW. Selective transcription and modulation of resting T cell activity by preintegrated HIV DNA. Science. 2001;293:1503–1506. doi: 10.1126/science.1061548. [DOI] [PubMed] [Google Scholar]
- Wu Y, Marsh JW. Early transcription from nonintegrated DNA in human immunodeficiency virus infection. J Virol. 2003;77:10376–10382. doi: 10.1128/JVI.77.19.10376-10382.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder A, Yu D, Dong L, Iyer SR, Xu X, Kelly J, Liu J, Wang W, Vorster PJ, Agulto L, Stephany DA, Cooper JN, Marsh JW, Wu Y. HIV envelope-CXCR4 signaling activates cofilin to overcome cortical actin restriction in resting CD4 T cells. Cell. 2008;134:782–792. doi: 10.1016/j.cell.2008.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Pe'ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews MB, Price DH. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]