Abstract
The end of the RNA polymerase II (Pol II) transcription cycle is strictly regulated to prevent interference between neighbouring genes and safeguard transcriptome integrity1. Pol II accumulation downstream of the cleavage and polyadenylation signal (CPS) can facilitate recruitment of factors involved in mRNA 3’-end formation and termination2, but how this sequence is initiated remains unclear. In a chemical-genetic screen, we identified human protein phosphatase 1 (PP1) isoforms as substrates of positive transcription elongation factor b (P-TEFb), the cyclin-dependent kinase 9 (Cdk9)-cyclin T1 complex3. Here we show that Cdk9 and PP1 govern phosphorylation of the conserved elongation factor Spt5 in the fission yeast Schizosaccharomyces pombe. Cdk9 phosphorylates both Spt5 and a negative regulatory site on the PP1 isoform Dis24. Sites targeted by Cdk9 in the Spt5 carboxy-terminal domain (CTD) can be dephosphorylated by Dis2 in vitro, and dis2 mutations retard Spt5 dephosphorylation after Cdk9 inhibition in vivo. Chromatin immunoprecipitation and sequencing (ChIP-seq) analysis indicates that Spt5 is dephosphorylated as transcription complexes traverse the CPS, concomitant with accumulation of Pol II phosphorylated at CTD repeat residue Ser25. A conditionally lethal Dis2-inactivating mutation attenuates the drop in Spt5 phosphorylation (pSpt5) on chromatin, promotes transcription beyond the normal termination zone detected by precision run-on transcription and sequencing (PRO-seq)6, and is genetically suppressed by ablation of Cdk9 target sites in Spt5. These results suggest that the transition from elongation to termination by Pol II coincides with Dis2-dependent reversal of Cdk9 signaling—a switch analogous to a Cdk1-PP1 circuit that controls mitotic progression4.
In metazoans and fission yeast, the inhibitory phosphorylation of PP1 by Cdk1 and its abrupt reversal promote orderly progression through mitosis4. In human extracts, analogue-sensitive (AS) Cdk9 modified PP1β and PP1γ on conserved, carboxy-terminal sites analogous to the PP1α residue labeled by Cdk13,7. Of the two fission yeast PP1 isoforms, Dis2 and Sds21, only Dis2 has the potential for inhibition by CDKs, through phosphorylation of Thr316; the sole budding yeast PP1 catalytic subunit, Glc7, lacks this regulatory site (Fig. 1a)8,9. Purified S. pombe Cdk9 phosphorylated Dis2 but not Sds21 in vitro (Fig. 1b); labeling was diminished by a T316A substitution, but not by a Dis2-inactivating point mutation10. Dis2 is regulated specifically by Cdk9 in vivo; treatment of cdk9as cells—but not wild-type cells or ones with AS versions of the transcriptional CDKs Mcs6 (orthologue of metazoan Cdk7) or Lsk1 (orthologue of Cdk12)—with the bulky adenine analogue 3-MB-PP1, which inhibits all three AS CDKs11, decreased Thr316 phosphorylation of chromatin-associated Dis2 (Fig. 1c, Extended Data Fig. 1a).
Figure 1. A Cdk9-Dis2-Spt5 circuit.
a, Alignment of C-termini of human and fungal PP1 isoforms. Thr320 of PP1α was identified as a target of Cdk1, and the analogous residues in PP1β and PP1γ as targets of Cdk9. This site (indicated by dot) is conserved in fission yeast Dis2 but not in Sds21, or in the budding yeast PP1 catalytic subunit Glc7. b, Phosphorylation of Dis2-Thr316 by Cdk9 in vitro. Purified, insect-cell derived Cdk9/Pch1 complexes were incubated at indicated molar ratios with purified, bacterially expressed GST-PP1 or GST-Spt5801-990 (containing the CTD), after activation by CDK-activating kinase Csk1 (incubated alone in indicated lanes). In addition to wild-type Dis2, we tested Dis2T316A and the Dis2R245Q variant encoded by dis2-11. Autophosphorylation occurs on both Cdk9 and Pch1. (Top: autoradiogram; bottom: Coomassie-stained gel to confirm equal loading.) c, Cdk9 phosphorylates chromatin-bound Dis2 on Thr316 in vivo. Cells of indicated strains (mcs6as5, cdk9as or lsk1as), with GFP-tagged Dis2 expressed from chromosomal dis2+ locus, were treated for 10 min with 10 µM 3-MB-PP1 or mock-treated with DMSO at 30°C, as indicated. Chromatin extracts were immunoprecipitated with anti-GFP antibodies and probed with antibodies specific for Dis2 phosphorylated at Thr316 (Dis2-T316P) or GFP. In b and c, experiments were performed twice with similar results. d, Phosphorylation by Cdk9 decreases activity of Dis2. Anti-Dis2 immunoprecipitates from wild-type (dis2+) or mutant (dis2Δ, dis2-T316A or dis2-T316D) extracts were treated with Cdk9 or mock treated prior to phosphatase assay with Spt5-pT1 phosphopeptide. Immunoblotting was performed to verify recovery (IB: Dis2) and phosphorylation (IB: Dis2-pT316) of immunoprecipitated Dis2 (n = 3 biological replicates; one-sided error bars, s.d. of mean). e, A Cdk9-Dis2-Spt5 circuit diagram.
The previously known target of S. pombe Cdk9 is Thr1 in the nonapeptide CTD repeat T1P2A3W4N5S6G7S8K9 of Spt512,13. A phosphopeptide with this sequence was dephosphorylated in vitro by PP1s purified from bacteria (Extended Data Fig. 1b) or immunoprecipitated from yeast (Extended Data Fig. 1c-e). We recovered Dis2 in similar amounts from dis2+ and dis2-11 cold-sensitive mutant cells, but detected activity only with the former, consistent with a previous observation that the enzyme encoded by dis2-11 was severely impaired even at permissive temperature10. Dis2 recovered from sds21Δ cells also had reduced activity, perhaps suggesting a contribution by Sds21 to Dis2 activation. Cdk9 treatment of wild-type Dis2 increased its reactivity with anti-Dis2-pT316 antibodies and diminished its phosphatase activity, whereas the same treatment had little effect on the constitutively higher activity of Dis2T316A or the lower activity of Dis2T316D (Fig. 1d, Extended Data Fig. 1d). Together, the results indicate that Dis2 is a target of negative regulation by Cdk9 and a potential Spt5 phosphatase, although it could also promote dephosphorylation of transcriptional CDK substrates by activating protein phosphatase 2A (PP2A), as it does in mitosis4. Either arrangement predicts switch-like responses of pSpt5 and Pol II dynamics to changes in Cdk9 activity (Fig. 1e).
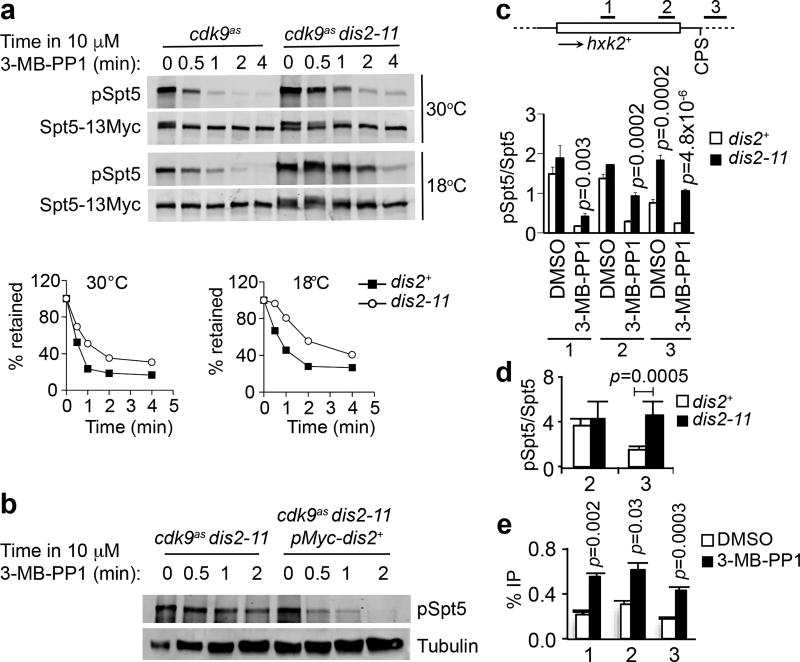
Consistent with this prediction, pSpt5, measured with a phosphospecific antibody against the S. pombe Spt5-derived phosphopeptide described above13, was rapidly lost after 3-MB-PP1 addition to cdk9as cells (T1/2 ≈ 20 sec) (Fig. 2a). The transcription machinery also responds rapidly to Cdk9 shutoff; PRO-seq analysis revealed Pol II slowing within 30 sec of inhibitor addition to cdk9as cells14. The rate of pSpt5 decay after Cdk9 inhibition was ~2-fold slower in dis2-11 relative to dis2+ cells at a permissive temperature of 30°C, and reduced by ~4-fold in dis2-11 cells shifted to 18°C prior to drug addition (Fig. 2a, Extended Data Fig. 2a). Rapid dephosphorylation was restored by expression of active Dis2 in dis2-11 cells (Fig. 2b, Extended Data Fig. 1e). Therefore, the maximal rate of pSpt5 turnover depends on Dis2 activity in vivo.
Figure 2. Cdk9 and Dis2 regulate Spt5 phosphorylation in vivo.
a, Dis2 inactivation stabilises pSpt5 after Cdk9 inhibition. Fission yeast strains—cdk9as spt5-13Myc or cdk9as spt5-13Myc dis2-11—were grown to mid-log phase at 30°C and shifted to 18°C (bottom) or not shifted (top) for 10 min before addition of 10 µM 3-MB-PP1, after which cultures were sampled at indicated times and subjected to immunoblot analysis with anti-pSpt5 or anti-Myc antibodies. Signals were quantified by fluorescence and mean values plotted as percent retention of pSpt5 relative to Spt5-Myc with time after addition of 3-MB-PP1 in dis2+ and dis2-11 cells (n = 2 biological replicates). b, Ectopic expression of wild-type Dis2 restores rapid Spt5 dephosphorylation kinetics in a dis2 mutant. cdk9as dis2-11 cells were shifted to 18°C and treated with 10 µM 3-MB-PP1 for indicated times, without or with expression of Myc-Dis2 from a plasmid, prior to immunoblot detection of pSpt5 and tubulin (loading control). Experiment was repeated twice independently with similar results. c, Dis2 inactivation stabilises chromatin-associated pSpt5. Either cdk9as spt5-13myc or cdk9as spt5-13myc dis2-11 cells were shifted to 18°C and treated with 10 µM 3-MB-PP1 or mock-treated with DMSO for 2 min and subjected to ChIP-qPCR analysis at hxk2+ for pSpt5 and total Spt5 (anti-Myc). The pSpt5:Spt5 ratio was plotted for dis2+ and dis2-11 cells for each treatment. d, Comparison of pSpt5:Spt5 ratio by ChIP-qPCR analysis upstream and downstream of CPS on hxk2+ gene in dis2+ and dis2-11 cells at 18°C. e, Suppression of Dis2 recruitment to transcribed chromatin by Cdk9. ChIP-qPCR analysis of GFP-Dis2 crosslinking at hxk2+ in cdk9as GFP-dis2 cells treated for 10 min with 10 µM 3-MB-PP1. In c-e, n = 3 biological replicates; one-sided error bars show s.d. of mean; p-values (Student’s t-test) indicated between wild-type (dis2+) and mutant (dis2-11) cells (c, d), or between 3-MB-PP1 and DMSO treatment (e).
Inhibition of Lsk1 had no effect on pSpt5 (Extended Data Fig. 2b) but diminished phosphorylation of the Pol II CTD repeat Y1S2P3T4S5P6S7 on Ser2 (pS2, Extended Data Fig. 2c). This mark was refractory to Lsk1 inhibition at 37°C in cells with a temperature-sensitive mutation in fcp115, which encodes a pS2-specific phosphatase16,17. The rate of pS2 decay was unaffected by dis2-11 (Extended Data Fig. 2d) and, conversely, Fcp1 inactivation had no effect on pSpt5 stability in cdk9as strains (Extended Data Fig. 2e). Similarly, pSpt5 turnover was impervious to genetic inactivation of Ssu72, a Pol II CTD Ser5 phosphatase18 (Extended Data Fig. 2f). We surmise that orthogonal CDK-phosphatase pairs govern Pol II and Spt5 phosphorylations implicated in elongation.
Dis2 also influences pSpt5 turnover on chromatin; by ChIP-quantitative PCR (ChIP-qPCR) analysis, pSpt5 occupancy was nearly abolished after Cdk9 inhibition for 2 min in dis2+ cells (Extended Data Fig. 3a), but was more resistant to 3-MB-PP1 treatment in cdk9as dis2-11 cells at 18°C (Fig. 2c, Extended Data Fig. 3b-d). The dis2-11 mutant also had higher pSpt5:Spt5 ChIP signal ratios when Cdk9 remained active (Fig. 2d). Cdk9 inhibition stimulated Dis2 recruitment to chromatin, whereas Mcs6 or Lsk1 inhibition had no consistent effect on Dis2 occupancy (Fig. 2e, Extended Data Fig. 4a-c). Dis2 ChIP signals were also enhanced by cdk9ΔC, which removes a carboxy-terminal region of Cdk9 that promotes its recruitment to chromatin19, and by cdk9T212A, which prevents Cdk9-activating phosphorylation20 (Extended Data Fig. 4d). Thus Cdk9 can limit both activity and chromatin recruitment of Dis2—mutually reinforcing effects that might contribute to the switch-like behavior of pSpt5.
Dis2 and Ssu72 are components of the cleavage and polyadenylation factor CPF21, as is Glc7, impairment of which led to termination defects in budding yeast22,23. Cdk9 inhibition did not cause ectopic recruitment of the entire CPF to gene bodies; crosslinking of three other CPF subunits was not affected (Extended Data Fig. 5a). Glc7 removes Pol II CTD Tyr1 phosphorylation (pY1) to facilitate pS2-dependent termination factor recruitment23, but pY1 ChIP signals were not increased (and pS2 was unaffected) by a dis2-11 mutation (Extended Data Fig. 5b). Recruitment of the CPF subunit Pfs2 was likewise unaffected by dis2-11, and thermal inactivation of Pfs2 in pfs2-11 cells did not alter pSpt5:Spt5 ratios, although it increased Pol II occupancy downstream of the CPS (Extended Data Fig. 5c, d). These results support specific, direct interactions between Cdk9 and Dis2 in governing pSpt5, rather than an indirect effect of impaired 3’-end processing or loss of CPF complex integrity (while suggesting that Glc7 in budding yeast has different regulators and targets).
In cdk9+ cells with different dis2 alleles, ChIP-qPCR analysis revealed significant increases in pSpt5 in the loss-of-function mutants dis2-11, dis2Δ and dis2T316D, relative to dis2+ and dis2T316A cells (Extended Data Fig. 6a, b). The relative increases due to dis2-11 and dis2∆ correlated with the degree of bulk pSpt5 stabilisation after Cdk9 inhibition (Fig. 2a, Extended Data Fig. 6c). We suspect dis2-11 is more severely affected than dis2Δ because loss of Dis2 protein might allow more effective compensation by other phosphatases. An sds21Δ mutation did not retard pSpt5 decay, however, and Cdk9 inhibition did not increase Sds21 recruitment to chromatin (Extended Data Fig. 6d, e), indicating that Dis2 is the major PP1 isoform that regulates pSpt5 in wild-type cells, in opposition to Cdk9 and possibly in collaboration with downstream phosphatases.
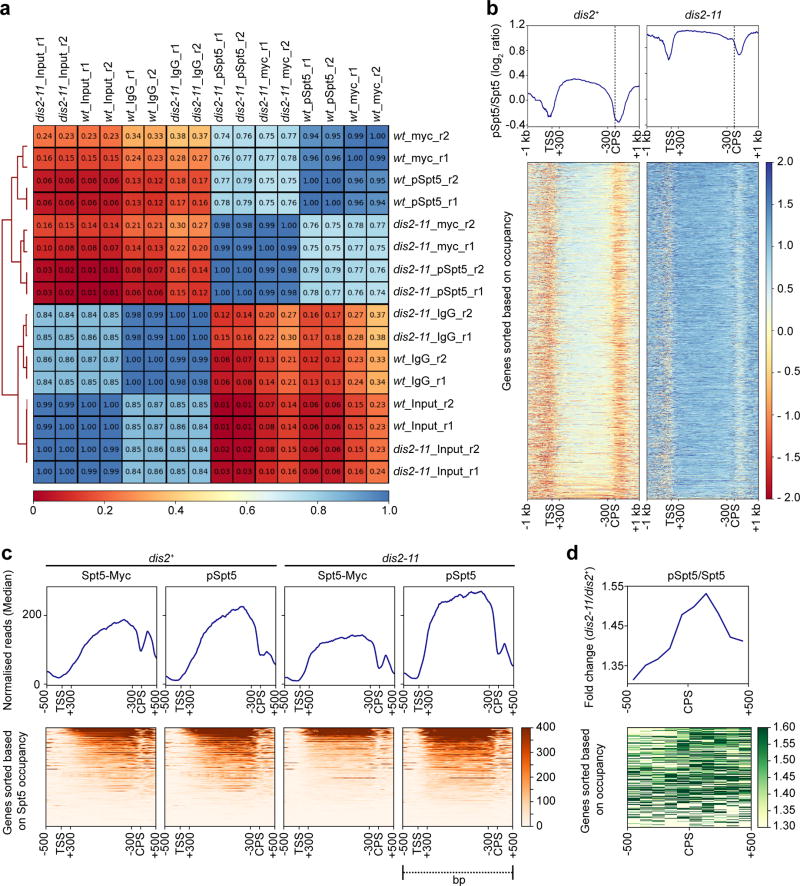
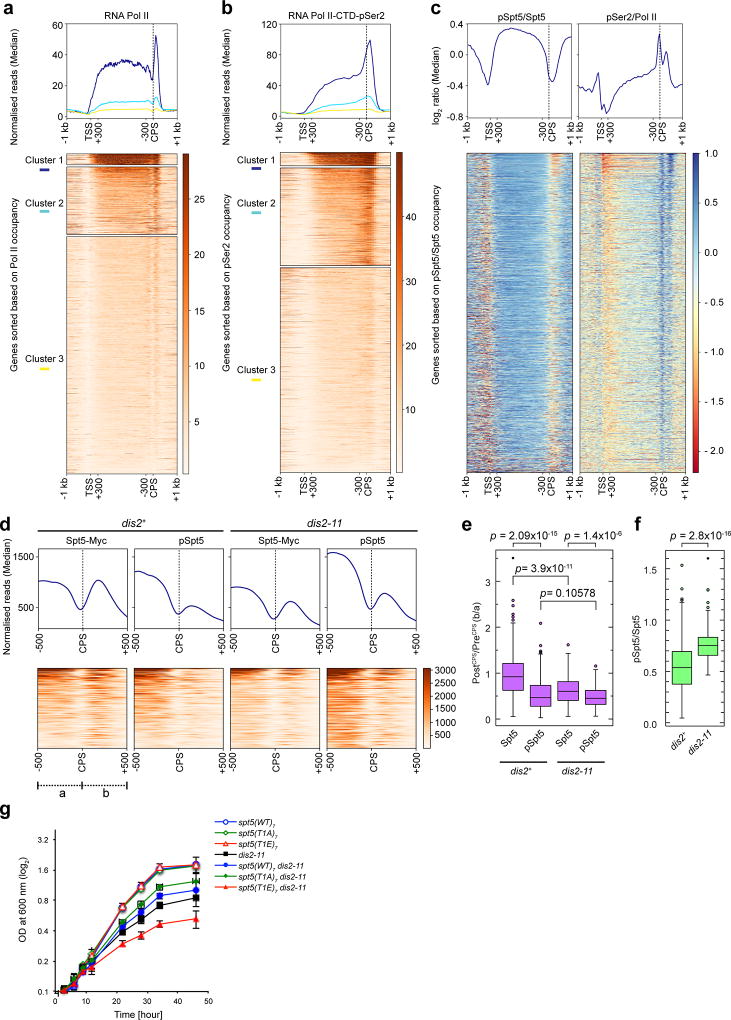
Cdk9 plays a rate-limiting role in Pol II elongation14, likely to depend in part on Spt5, depletion of which caused Pol II accumulation in upstream gene regions in fission yeast5 and disrupted coupling between 3’-processing and termination in budding yeast24. ChIP-seq analysis of dis2+ cells revealed similar distributions of total Spt5 and pSpt5 in gene bodies (Fig. 3a, b, Extended Data Fig. 7a). The patterns diverged beyond the CPS, where a prominent peak of total Spt5 corresponds to a much smaller peak of pSpt5. In dis2-11 cells, pSpt5 signals were elevated throughout gene bodies, but more so downstream of the CPS (Fig. 3a, b, Extended Data Figs. 7b, c). This increased pSpt5 retention was most obvious on genes selected for high Spt5 occupancy (cluster 1 in Fig. 3a), or filtered to minimize signals from neighbouring genes (Extended Data Fig. 7c, d). A metagene analysis comparing pSpt5:Spt5 ratios between dis2+ and dis2-11 strains reveals a sharp, CPS-centred increase in the mutant (Fig. 3c, Extended Data Fig. 7d), suggesting a Dis2-dependent, switch-like transition as elongation complexes traverse this landmark. The trough of pSpt5 near the CPS in dis2+ cells coincides with a peak of Ser2-phosphorylated Pol II in a recent ChIP-seq analysis5 (Fig. 3d, Extended Data Fig. 8a-c)—a reciprocal relationship consistent with independent pSpt5 and pS2 regulation.
Figure 3. Dis2-dependent loss of pSpt5 downstream of the CPS.
a, Genome-wide distribution of Spt5 and pSpt5 on transcribed genes. Metagene analyses (top) and heatmaps (bottom) of ChIP-seq data reveal patterns of Spt5-Myc and pSpt5 occupancy (n = 3054 Pol II-transcribed genes) in dis2+ and dis2-11 cells. Each condition was analyzed in duplicate. Genes were sorted based on Spt5 occupancy and partitioned into 3 different clusters with nearest mean occupancy by k-means clustering (Note: The regions between +300 bp relative to TSS and -300 bp relative to CPS were scaled to allow comparisons among genes of different lengths.) b, Spt5 dephosphorylation downstream of the CPS. Individual gene tracks show accumulation of Spt5-myc downstream of CPS, with lower levels of pSpt5 in dis2+ cells; this drop is attenuated in Dis2-deficient (dis2-11) cells. c, Stabilisation of Spt5 phosphorylation upon inactivation of Dis2 is centred around CPS. Metagene (top) and heatmap (bottom) analyses show fold-change of pSpt5:Spt5 ratio across Pol II-transcribed genes (n = 3054) in dis2-11 versus dis2+ cells. d, Distribution of pSpt5 and Pol II-pSer2 near CPS is inversely correlated. Metagene plots of log2 pSpt5:Spt5 and pSer2:Pol II ratios reveal reciprocal distribution of pSpt5 and Pol II-pSer2 on transcribed genes. e, Suppression of dis2-11 by an Spt5 CTD mutant. Serial dilutions of indicated strains grown at 30°C, 20°C and 18°C. The mutant alleles tested were: spt5-(WT)7, spt5-(T1A)7 and spt5-(T1E)7. Experiment was repeated three times independently with similar results.
There is a transient drop in total Spt5 occupancy near the CPS (Fig. 3a, b), also seen in metagene plots of Pol II5 (Extended Data Fig. 8a) and of Spt5 in budding yeast, where it corresponds to a peak of Spt5 cross-linking to the nascent transcript24. Although an exchange of phosphorylated for unphosphorylated Spt5 is formally possible, active dephosphorylation near the CPS seems more likely a priori, given the similarities in Pol II and Spt5 distribution and Spt5’s tight association with the Pol II clamp25. Consistent with a phosphatase-dependent mechanism, the dis2-11 mutation caused significantly greater pSpt5 stabilisation in the 500-bp region downstream of the CPS than in the 500 bp upstream (Extended Data Fig. 8d-f). Moreover, the dis2-11 phenotype is due in part to Spt5; replacement of Thr1 with alanine in a truncated, seven-repeat Spt5 CTD—spt5-(T1A)7, which by itself imparts cold-sensitivity26—partially suppressed the cold-sensitive lethality of dis2-11, whereas a phosphomimetic spt5-(T1E)7 mutation (which did not affect growth on its own26) exacerbated it (Fig. 3e, Extended Data Fig. 8g).
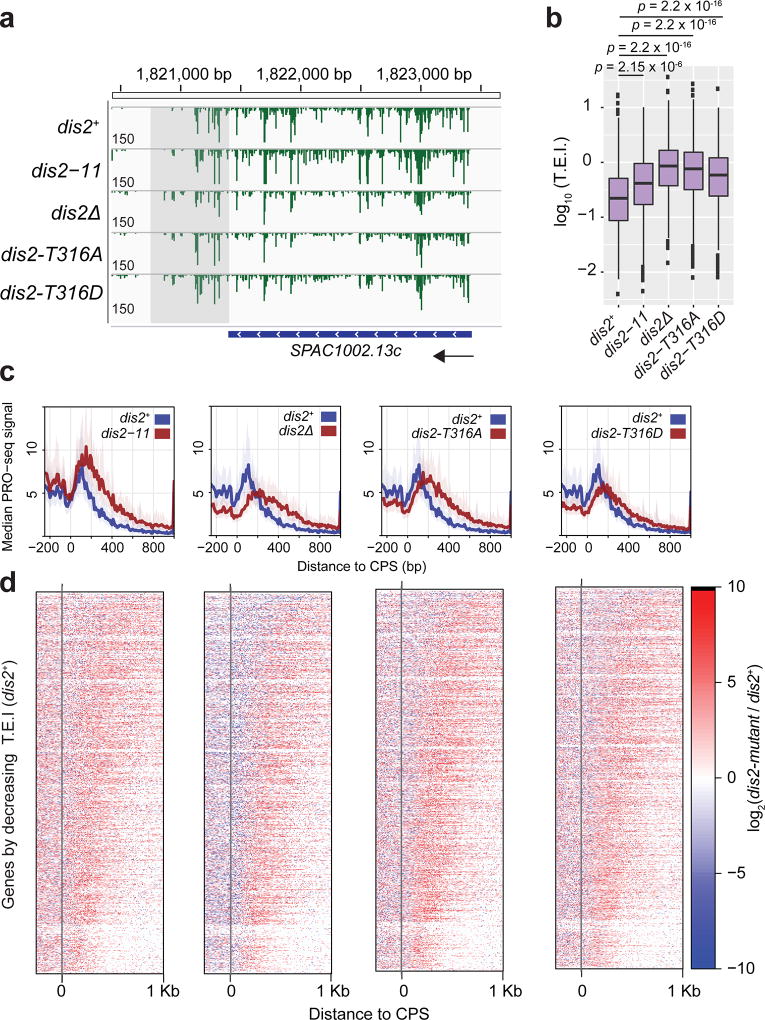
We performed PRO-seq analysis6 to uncover effects of Dis2 impairment on the distribution of transcribing Pol II (Fig. 4, Extended Data Fig. 9a). On individual genes, PRO-seq reads decreased within a narrowly defined zone following the CPS in dis2+ cells, but this zone extended ~500 bp further downstream in dis2-11 cells. Alignment of PRO-seq and ChIP-seq read distributions suggested correlation between zones of Dis2-dependent termination and Spt5 dephosphorylation (Fig. 4a). Metagene analysis of PRO-seq data revealed increased transcription beyond the CPS, both in absolute terms and relative to transcription of upstream regions, in the dis2-11 mutant (Fig. 4b, c, Extended Data Fig. 9b, c). This defect was apparent at both 18°C and 30°C (and in both cdk9+ and mock-treated cdk9as cells), indicating that effects of Dis2 impairment on Pol II distribution are constitutive. Loss of viability occurs only at low temperatures, however, perhaps suggesting heightened dependence on elongation control under conditions of cold stress (although Dis2 functions in cell division might also become essential only at low temperature8).
Figure 4. Loss of Dis2 function impairs termination.
a, Transcription beyond normal termination zone in dis2-11 cells under multiple conditions. Representative gene browser tracks show termination zones (shaded) in dis2+ or dis2-11 cells at 18°C or 30°C. Alignment with ChIP-seq tracks reveals correlation between pSpt5 loss and Dis2-dependent termination. All analyses were performed in the absence of 3-MB-PP1; under this condition, PRO-seq read distributions were not significantly different between cdk9+ and cdk9as cells14. Track values reflect the maximum displayed signal (some peaks exceed these values). b, Genome-wide effects on Pol II distribution due to dis2-11 mutation. Metagene analysis of PRO-seq read distributions reveals differences in dis2-11, relative to dis2+ cells. (Note: To compare genes of different lengths, regions between +300 bp relative to TSS and -300 bp relative to CPS were scaled.) c, CPS-centred metagene analysis comparing PRO-seq read distributions in wild-type and dis2-11 cells reveals relative increase in transcription downstream of CPS in the mutant at 30°C. Peak heights (y axis) were scaled as a fraction of maximum signal in each condition; position along the gene (x axis) was unscaled. d, Two metrics of termination efficiency, shown schematically: Termination Index (T.I.), the ratio of signals in the regions 500 bp downstream and upstream of CPS (b/a); and Termination Elongation Index (T.E.I.), the ratio of signals in the region 250-750 bp downstream of CPS and the region 500 bp upstream of CPS (c/a). In b, c, solid lines represent averaged data plot and shaded regions represent s.d. of median. e, The dis2-11 mutation causes a significant increase in T.E.I. in both cdk9as (left) and cdk9+ (WT, right) backgrounds, and in T.I. in cdk9+ cells (p-values calculated by Welch’s two sample t-test). f, Heatmaps showing change in PRO-seq read distribution due to dis2-11 mutation. Genes were ranked by decreasing T.E.I. in cdk9as dis2-11 at 18°C, a measure of termination-window size. All genes in b–f were required to be active and at least 1 kb from nearest genes on same strand to eliminate effects of nearby initiation and run-through transcription (n = 939). In a–f, n = 2 biological replicates. g, A transcription exit network comprising Cdk9, Dis2 and Spt5. At or near CPS, Dis2 becomes active due to a drop in Cdk9 activity and triggers Spt5 dephosphorylation, to facilitate 3’-pausing and termination.
To quantify termination defects, we defined two metrics based on PRO-seq read distribution: Termination Index (T.I.), the signal ratio in the regions 500 bp downstream or upstream of the CPS; and Termination Elongation Index (T.E.I.), the ratio in regions 250-750 bp downstream and 500 bp upstream of the CPS (Fig. 4d). The dis2-11 mutation caused statistically significant increases in T.I. in cdk9+, and in T.E.I. in both cdk9+ and cdk9as cells (Fig. 4e). A heatmap analysis of genes ranked by T.E.I. revealed termination-zone expansion consistent with imprecise termination upon loss of Dis2 function (Fig. 4f, Extended Data Fig. 9d). We also tested effects of other dis2 alleles on Pol II distribution. Both the dis2∆ and dis2T316D loss-of-function mutations produced similar termination defects (Extended Data Fig. 10a-d). Unexpectedly, so did dis2T316A, which encodes an active enzyme refractory to negative regulation by Cdk9 (Fig. 1d), suggesting that an orderly transition from elongation to termination requires both phosphorylated and unphosphorylated forms of Dis2. Cdk9 inhibition had the opposite effect—decreasing both T.I. and T.E.I.14—consistent with antagonism between the kinase and phosphatase.
Recent studies suggest an ordered series of events at 3’ ends of mammalian and fission yeast protein-coding genes: 1) Pol II pausing, leading to 2) increased pS2, which promotes 3) recruitment of cleavage factors to the CTD, 4) transcript cleavage and 5) termination by the 5’→3’ “torpedo” exoribonuclease XRN2/Rat12,3,24,27,28. This sequence can be initiated by blocks to transcription imposed in cis2, and pS2 is elevated in 5’ gene regions by mutations that decrease the intrinsic rate of Pol II29, but a physiologic trigger remains unknown. A priori both Spt5 and PP1 are likely participants in this transition—the former as a regulator of Pol II processivity and rate5,24,30, the latter as a CPF component21,23. Here we place Spt5 downstream of PP1 signaling in S. pombe, where both Spt5 and a PP1 isoform are substrates of Cdk9, itself a positive regulator of elongation14. We define enzyme-substrate relationships among Cdk9, Dis2 and Spt5 that recapitulate a cell-cycle regulatory module4, and suggest a model of “transcriptional exit” (Fig. 4g): Dis2-dependent dephosphorylation of Spt5 and possibly other Cdk9 targets to reverse elongation-rate enhancement and facilitate Pol II capture by the torpedo.
METHODS
Yeast strains and standard methods
Fission yeast strains used in this study are listed in Supplementary Table 131,32. New strains were generated by standard techniques33. Cells were grown in YES medium at 30°C unless otherwise specified.
Immunological methods
Antibodies used in this study recognised pSpt5 or total Spt513, Dis2-pT3164 or total Dis234, Myc epitope (EMD Millipore, 05-724), total Pol II (BioLegend, MMS-126R), Pol II pSer2 (Abcam, ab5095), Pol II pTyr1 (Active Motif, 61383, clone 3D12), α-tubulin (Sigma, T-5168) or green fluorescent protein (GFP) (Invitrogen, rabbit polyclonal (A11122) or Santa Cruz, mouse monoclonal (sc-9996)). Proteins were visualised in immunoblot analysis either by enhanced chemiluminescence (ECL, HyGLO HRP detection kit, Denville Scientific, E250) or with Odyssey Imaging System (LI-COR Biosciences).
Kinase and phosphatase assays
Kinase assays were performed with purified proteins (Cdk9/Pch1 complex and GST-Spt5801-990) and [γ-32P] ATP (PerkinElmer, BLU002A250UC), as described previously20. GST-PP1s were expressed in E. coli at 16°C for 16 h and purified with Glutathione Sepharose 4 Fast Flow beads35. To measure protein phosphatase activity, PP1 isoforms (GST-Dis2 or GST-Sds21) purified from E. coli (2 µg) or immunoprecipitated from fission yeast extracts (4 mg total protein) were incubated with 50 µM peptide (Spt5-NP, Spt5-pT1 or H3pS10) at 37°C for 1 h. Colorimetric assays were performed in triplicate using BioMOL Green (Enzo Life Sciences, BML-AK111) in 25 mM HEPES, pH 7.5, 100 mM NaCl, 1 mM MnCl2 and 1 mM DTT, in 96-well plates as described in manufacturer’s protocol. To test Dis2 activity after phosphorylation by Cdk9, Dis2 immunoprecipitated from fission yeast extract (8 mg total protein) was incubated with Cdk9 (activated by Csk1) or mock-treated (no kinase added) in kinase assay buffer (25 mM HEPES, pH 7.5, 10 mM MgCl2, 1 mM ATP, 1 mM DTT) for 1 h at 25°C, washed three times with phosphatase assay buffer and tested for activity as described above.
Chromatin immunoprecipitation analysis
Immunoprecipitation and chromatin immunoprecipitation (ChIP) were carried out by published methods19,36,37. Quantitative PCR was performed with USB VeriQuest SYBR Green qPCR Master Mix (2x) (Affymetrix, 75600) in 386-well plates. ChIP was performed with 3 or 4 biological replicates and qPCR was performed in triplicate for each sample. Oligonucleotides used as primers are listed in Supplementary Table 2. In ChIP-qPCR calculations, input Ct values were first corrected by dilution factor (Raw CtInput – log2 of 100; 100 is dilution factor), ChIP signals were normalised against input (ΔCt=CtIP –CtInput), and ChIP yield was expressed as percent input (%IP) and calculated by the equation: %IP=2-∆Ct x100%. Fold enrichment of specific (S) ChIP-signal over nonspecific (NS) control (rabbit IgG or ChIP signals from untagged strains) was calculated by the equation: 2-ΔΔCt (ΔΔCt=∆CtS-∆CtNS). To measure dependency of Dis2-Thr316 phosphorylation on CDKs in vivo, cells expressing GFP-Dis2 from the chromosomal dis2 locus in wild-type, cdk9as, mcs6as5 or lsk1as backgrounds were grown at 30°C to a density of ~ 1.5×107 ml−1, treated with either DMSO or 10 µM 3-MB-PP1 for 10 min, and crosslinked with 1% (v/v) formaldehyde for 15 min at 25°C. Chromatin extracts were prepared according to a standard protocol for ChIP sample preparation19,36,37 and 5 mg total protein was subjected to immunoprecipitation with anti-GFP antibody (sc-9996) and immunoblot analysis with either anti-GFP or anti-Dis2-pT316 antibody4.
Chemical genetics
To measure rates of Spt5 and Rpb1 dephosphorylation after CDK inhibition, cells were grown at 30°C in YES medium to a density of ~1.2 x107 cells/ml, harvested by centrifugation at 25°C, resuspended in fresh YES (preincubated at desired temperature) and incubated for 10 min before addition of DMSO or 3-MB-PP1 (10 or 20 µM). At intervals thereafter, cells (~0.6×108) were transferred directly to tubes containing 500 µl 100% (w/v) trichloroacetic acid (TCA) and harvested by centrifugation. Protein extracts were prepared in the presence of 20% (w/v) TCA38 and processed for immunoblot analysis with appropriate antibodies.
ChIP-seq
The dis2+ or dis2-11 cells were grown at 30°C prior to crosslinking in 1% (v/v) formaldehyde. ChIP was performed with 250 µg of total protein and 1 µg of antibody. Multiplexed ChIP-seq libraries were prepared using the NEBNext® Ultra™ II DNA Library Preparation kit (E7103S) with 25 ng of input or IP DNA and barcode adaptors (NEBNext Multiplex Oligos for Illumina [Set 1, E7335] & [Set 2, E7500]). Paired-end sequencing (50-nt reads) was performed on an Illumina NextSeq 500. Quality control and adapter trimming of FASTQ (raw sequencing) files were done in Galaxy using FastQC Read Quality reports (Galaxy Version 0.69) and trimmomatic flexible read trimming tool for Illumina NGS data (Galaxy Version 0.36.3), respectively. Processed FASTQ files were aligned to the S. pombe genome using Bowtie239 (Galaxy Version 2.2.6.2). Aligned sequences of each biological replicate were fed into MACS240 (Galaxy Version 2.1.1.20160309.0) to call peaks from alignment results. Generated “bedgraph treatment” files were converted to bigwig using “Wig/BedGraph-to-bigWig converter” (Galaxy Version 1.1.0), concatenated (Galaxy Version 1.0.1) to combine replicates of each sample, converted to bigwig using “Wig/BedGraph-to-bigWig converter” (Galaxy Version 1.1.0) and processed using computeMatrix (Galaxy Version 2.3.6.0) in DeepTools41 to prepare data for plotting heatmaps and/ or profiles of given regions. Heatmap and Metagene plots were generated using “plotHeatmap” (Galaxy Version 2.5.0.0) and “plotProfile” (Galaxy Version 2.5.0.0) tools, respectively. The pSpt5:Spt5 signal ratios and fold change of the signal ratios in dis2-11 versus dis2+ were calculated using bigwigCompare (Galaxy Version 2.5.0.0). First, all ChIP-seq signals were normalised by subtracting non-specific IgG signals generated in each strain. Next, pSpt5 signals were normalised over Spt5-Myc signals in corresponding strains. Finally, the fold-change of pSpt5/Spt5 in dis2-11 versus dis2+ cells was calculated using bigwigCompare (Galaxy Version 2.5.0.0). For metagene analysis, all Pol II active and filtered genes were used (n=3122)6.
PRO-seq
Two batches of PRO-seq42 experiments were performed, each with slight differences in run-on procedure and library preparations. For each strain biological replicates were derived from separately picked colonies. Cultures were grown in YES medium at 30 °C overnight and diluted to an OD600= 0.2. After reaching approximate OD600 = 0.5, an equal number of cells (based on OD) was set aside for all treatments (~10 mL culture). At this point, a fixed amount of thawed S. cerevisiae (50 µL, OD = 0.68) was spiked into each sample. Cultures were then immediately spun down at 4 °C and subjected to permeabilisation and library preparation. In the batch of experiments with temperature shifts, cells were spun down and resuspended in fresh YES medium, pre-conditioned at the desired temperature (30 °C or 18 °C) and allowed to incubate for 10 min before permeabilisation and library preparation.
Samples from the temperature-varied experiments were prepared according to the standard procedure, which uses each of the four biotinylated NTPs during the run-on reaction as well as previously described adaptors43. For the batch of experiments lacking temperature shifts (Extended Data Fig. 10), precision run-on reactions were performed with only two biotinylated NTPs (biotin-11-C and biotin-11-U) in addition to an equal concentration of rATP and rGTP (the two-biotin run-on approach saves cost but modestly reduces the resolution of the assay). Additionally, for these libraries a novel 3’ RNA adaptor was used. Here, an adaptor with a distinct hexanucleotide sequence preceded by a guanine at the 5’ end is used for each library prepared (5’ -/5Phos/ GNNNNNNGAUCGUCGGACUGUAGAACUCUGAAC-/Inverted dT/). This results in a distinct barcode for each library and permits the pooling of all samples after the 3’ end ligation step. After pooling, the remainder of the library preparation was carried out as described previously43, but in a single tube. After sequencing, the inline barcode was used to parse reads based on their sample of origin. Reads were processed and aligned for analysis as described14.
Experimental batches
With the exception of wild-type data used in Fig. 4 (taken from ref. 14), two batches of experiments were performed to generate the precision run-on data in this work. In order to minimise noise introduced from across-batch comparisons, all analyses were restricted to PRO-seq libraries prepared within the same batch. However, several features of the second batch of samples (used in Extended Data Fig. 10) limited our further use of these data: 1) There were subtle profile differences that complicated direct comparisons between batches of experiments, possibly due to the different run-on procedure described above. 2) Raw read length was much shorter in the second batch, producing fewer uniquely aligning reads, possibly reflecting lesser quality of the preparations. 3) Spike-in based normalisations did not reproducibly capture changes in transcription profiles, thus limiting direct quantitative comparisons between strains. Nonetheless, results using these samples clearly and independently recapitulated the Dis2-dependent influence on termination described in Fig. 4.
Code availability
Custom scripts and alignment pipelines have been made publicly available through the following GitHub repository: https://github.com/gregtbooth/Pombe_PROseq.
Data availability
Source data for results plotted in Figs. 1d, 2a, c-e, and in Extended Data Figs. 1b-e, 3a-d, 4a-d, 5a-d, 6a, b, e, and 8g are available online. The raw and processed sequencing files have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) (Accession number GSE102590).
Extended Data
Extended Data Figure 1. A Cdk9-Dis2-Spt5 regulatory circuit.
a, Cdk9-dependence of Dis2-T316 phosphorylation in vivo. Cells of wild-type (wt) or cdk9as strains, with or without GFP-tagged Dis2 expressed from the chromosomal dis2+ locus, were treated for 10 min with 20 µM 3-MB-PP1 or mock-treated, as indicated. Chromatin extracts were immunoprecipitated with anti-GFP antibodies and probed with antibodies specific for Dis2 phosphorylated at Thr316 (Dis2-T316P) or GFP (n = 2 independent repeats). b, Spt5 dephosphorylation by purified PP1 in vitro. Purified GST-Dis2 and GST-Sds21 were incubated with a control phosphopeptide derived from histone H3 (“H3pS10”), an Spt5 CTD consensus phosphopeptide (“Spt5-pT1”), or a non-phosphorylated peptide of the same sequence (“Spt5-NP”). c, Spt5 dephosphorylation by PP1 isolated from fission yeast. A polyclonal anti-Dis2 antibody immunoprecipitates pSpt5 phosphatase activity from extracts of dis2+ but not dis2Δ or dis2-11 mutant cells. (Note: The antibody cross-reacts with Sds21 in immunoblots but does not efficiently immunoprecipitate Sds21.) d, Loss of Dis2 activity upon Cdk9-dependent phosphorylation. As in Fig. 1d, results show activity of Dis2, isolated from yeast and phosphorylated by Cdk9 or mock-treated, towards H3pS10. e, Top: Anti-Myc immunoprecipitates from Myc-Dis2-expressing cells were tested for phosphatase activity towards Spt5-pT1 and H3pS10 peptides. Bottom: immunoblot to verify expression and immunoprecipitation of Myc-Dis2. In b–e, n = 3 biological replicates; one-sided error bars show s.d. of mean.
Extended Data Figure 2. Distinct kinase-phosphatase circuits regulate phosphorylation of Spt5-Thr1 and Rpb1-Ser2 in vivo.
a, Rapid dephosphorylation of Spt5 after Cdk9 inhibition and stabilisation of pSpt5 by Dis2 inactivation occur in spt5+ strain and do not depend on the C-terminal Myc-epitope tag. b, Spt5 dephosphorylation kinetics in single, pairwise and triple cdkas mutants treated with 3-MB-PP1 indicate that Cdk9 is the sole kinase needed to phosphorylate this site in vivo. c, Fcp1 inactivation stabilises Rpb1 Ser2 phosphorylation after Lsk1 inhibition. Fission yeast strains—lsk1as or lsk1as fcp1-452—were grown at 30°C and shifted to 37°C (or not shifted), treated for the indicated time with 20 µM 3-MB-PP1, and analyzed by immunoblotting for Pol II Ser2 phosphorylation. Note: CTD dephosphorylation leads to increased reactivity with 8WG16 antibody used to detect total Pol II. d, Dis2 activity is dispensable for Pol II CTD Ser2 dephosphorylation. As in Fig. 2a except that experiment was performed in lsk1as cells, 20 µM 3-MB-PP1 was added, and extracts were probed for Pol II Ser2 phosphorylation (or tubulin as a loading control). e, Fcp1 activity is dispensable for Spt5 Thr1 dephosphorylation. As in (c) except that strains carried a cdk9as allele and were tested for both pSpt5 (unaffected by Fcp1 inactivation) and pSer2 (unaffected by Cdk9 inhibition). f, CPF-associated Pol II pSer5 phosphatase Ssu72 is dispensable for Spt5 Thr1 dephosphorylation. Fission yeast strains (cdk9as, cdk9as ssu72-C13S and ssu72-C13S) were grown at 30°C, treated for the indicated time with 10 µM 3-MB-PP1, and analysed by immunoblotting for pSpt5 and total Spt5. In a-f, n = 2 independent repeats.
Extended Data Figure 3. Chromatin-associated Spt5 is dephosphorylated rapidly upon Cdk9 inhibition and stabilised in dis2-11 cells.
a, Rapid pSpt5 turnover on chromatin. ChIP-qPCR analysis of pSpt5 versus total Spt5 crosslinking at the eng1+ gene after 3-MB-PP1 treatment for various times. Left: absolute ChIP signals for anti-Myc; middle: absolute signals for anti-pSpt5; right: ratio of phospho- to total Spt5, expressed as a percentage of ratio in the absence of the inhibitor. b, Loss of Dis2 function stabilises pSpt5 on chromatin. Either cdk9as spt5-13Myc dis2+ or cdk9as spt5-13Myc dis2-11 cells were shifted to 18°C and treated with 10 µM 3-MB-PP1 or mock-treated with DMSO for 2 min and subjected to ChIP-qPCR analysis at the eng1+ locus for Spt5-Myc (left), pSpt5 (middle) and the pSpt5:Spt5 signal ratios in 3-MB-PP1- versus DMSO-treated samples (right). The pSpt5:Spt5 signal ratios between treatments (DMSO and 3-MB-PP1) were plotted for each condition (dis2+ or dis2-11). (Note: Higher residual pSpt5 in cdk9as dis2+ cells, compared to those analyzed in (a), may reflect less efficient dephosphorylation at 18°C, relative to 30°C.) c, Same as in (b) at aro1+ gene. d, Same as in (b) at hxk2+ gene (raw data for phospho- and total Spt5 from which ratios in Fig. 2c were calculated). In a-d, n = 3 biological replicates; one-sided error bars show s.d. of mean. In b-d, p-values (Student’s t-test) are indicated between wild-type (dis2+) and mutant (dis2-11) cells.
Extended Data Figure 4. A specific link between Cdk9 activity and Dis2 recruitment to chromatin.
a, Increased recruitment of Dis2 to chromatin is a specific consequence of Cdk9 inhibition. ChIP-qPCR analysis of Myc-Dis2 (left), Pol II (middle) and Myc-Dis2:Pol II signal ratios (right) at the eng1+ locus in cdk9as cells, expressing Myc-Dis2 from a plasmid, treated with 20 µM 3-MB-PP1 or DMSO for 10 min at 30°C. b, c, Mcs6 and Lsk1 activities do not influence Dis2 recruitment to chromatin. Same as (a), except cells containing mcs6as5 (b) or lsk1as (c) allleles were treated with 20 µM 3-MB-PP1 for 10 min at 30°C. In a-c, n = 3 biological replicates; one-sided error bars show s.d. of mean. d, Constitutive cdk9 loss-of-function mutations increase GFP-Dis2 recruitment to chromatin. Dis2 occupancy at the eng1+ locus analysed in cdk9+ (“wt”) cells, a cdk9ΔC mutant and a cdk9T212A mutant (n = 1; one-sided error bars show s.d. of mean in technical duplicates).
Extended Data Figure 5. Regulation of pSpt5 by Cdk9 and Dis2 occurs independently of CPF recruitment and upstream of CPF function.
a, Core CPF recruitment to chromatin is unaffected by Cdk9 inhibition. Cells with different CPF subunits GFP-tagged and expressed from their respective chromosomal loci (cdk9as pfs2-GFP-HA, cdk9as pla1-GFP-HA or cdk9as cft1-GFP) were grown at 30°C and treated with 10 µM 3-MB-PP1 or DMSO for 10 min. ChIP-qPCR analysis of GFP:Pol II signal ratios was performed at hxk2+ gene (n = 3 biological replicates; one-sided error bars, s.d. of mean). b, Wild-type (dis2+) and mutant (dis2-11) cells were grown at 30°C and shifted to 18°C (or not shifted) for 10 min. ChIP-qPCR analysis of Pol II-pTyr1:Pol II (top), pSpt5:Spt5 (middle) and Pol II-pSer2:Pol II (bottom) signal ratios was performed at psu1+ gene. c, Loss of Dis2 activity does not affect chromatin recruitment of a core CPF subunit, Pfs2. Cells of dis2+ and dis2-11 strains with Pfs2-GFP-HA expressed from the chromosomal pfs2+ locus were grown at 30°C and shifted to 18°C (or not shifted) for 10 min prior to formaldehyde crosslinking and chromatin isolation. ChIP-qPCR analysis of GFP:Pol II signal ratios was performed at psu1+ gene at 30°C (top) and 18°C (bottom). In b, c, n = 2 biological replicates; one-sided error bars show s.d. of mean. d, Spt5 phosphorylation is not affected by thermal inactivation of an essential CPF subunit. Cells of pfs2+ and pfs2-11 (temperature-sensitive) strains were grown at 30°C, shifted to 37°C (or not shifted) and incubated for various times as indicated. ChIP-qPCR analysis of pSpt5, total Spt5, pSpt5:Spt5 signal ratios, Pol II and Pol II signal ratios (pfs2-11 over pfs2+), as indicated, was performed at hxk2+ locus (n = 2 biological replicates; one-sided error bars, s.d. of mean).
Extended Data Figure 6. PP1 allele- and isoform-specific stabilisation of Spt5 phosphorylation on chromatin.
a, ChIP-qPCR analysis at rps17a+ gene. Comparison of pSpt5:Spt5 ratio in the indicated strains upstream and downstream of CPS at 30°C (left) and comparison of the ratio between dis2+ and dis2-11 cells at 18°C (right). b, ChIP-qPCR analysis at hxk2+ gene. Comparison of pSpt5:Spt5 ratio in the indicated strains upstream and downstream of CPS at 30°C. In a, b, n = 3 biological replicates; one-sided error bars show s.d. of mean; p-values (Student’s t-test) are between wild-type (dis2+) and mutant (dis2Δ, dis2-11, dis2T316A or dis2T316D). c, Dephosphorylation of Spt5 after Cdk9 inhibition is retarded in a dis2Δ strain, relative to a dis2+ strain. d, Spt5 dephosphorylation kinetics after Cdk9 inhibition are unaffected by sds21 deletion in a dis2+ strain. In c, d, n = 2 independent repeats. e, Cdk9 does not restrict chromatin recruitment of Sds21. Anti-GFP ChIP-qPCR analysis at eng1+ in a cdk9as GFP-sds21 strain treated for 10 min with 10 µM 3-MB-PP1 reveals unchanged (or slightly decreased) Sds21 occupancy when Cdk9 is inhibited (n = 1; one-sided error bars, s.d. of mean in technical duplicates).
Extended Data Figure 7. Spt5 and pSpt5 ChIP-seq analysis.
a, Correlation between ChIP-seq samples. Paired-end sequencing reads were mapped to the fission yeast genome using Bowtie2 (Galaxy Version 2.2.6.2). Mapped reads of each biological replicate were used to calculate correlation between pairs of replicates. Values in boxes represent Pearson correlation coefficient between corresponding samples (n = 2 biological replicates). b, Metagene (top) and heatmap (bottom) analyses show genome-wide (n = 3054 genes) comparison of pSpt5:Spt5 ratios (log2) between dis2+ and dis2-11 cells (raw data from which fold-change in Fig. 3c was calculated). c, Metagene plots (top) and heatmaps (bottom) show Spt5-Myc and pSpt5 distribution in dis2+ and dis2-11 cells, as indicated, across Pol II-transcribed genes (n = 175), filtered to include only genes separated from nearest neighbours by >500 bp at both ends, on both strands. d, Metagene plot (top) and heatmap (bottom) represent fold-change of pSpt5:Spt5 ratio in dis2-11 over dis2+ around CPS of the genes analysed in (c). In b, c, regions between +300 bp relative to the TSS and −300 bp relative to the CPS were scaled to allow comparisons among genes of different lengths.
Extended Data Figure 8. Relative distributions of Pol II and Spt5.
a, Pol II distribution on chromatin. Metagene plot (top) and heatmap (bottom) of Pol II ChIP-seq distributions (data from ref. 5) across Pol II-transcribed genes (n = 3054) in wild-type cells. Genes were sorted based on Pol II occupancy and k-means clustering done to partition genes into 3 clusters with nearest median occupancy. b, Pol II pSer2 distribution on chromatin. Metagene plot (top) and heatmap (bottom) of Pol II pSer2 (data from ref. 5) across Pol II-transcribed genes (n = 3054) in wild-type cells. c, Distribution of pSpt5 and pSer2 on chromatin. Metagene analyses (top) and heatmaps (bottom) show genome-wide comparison between log2 ratios of pSpt5:Spt5 and pSer2:Pol II for Pol II-transcribed genes (n = 3054) in wild-type cells (separate plots of data superimposed in Fig. 3d). In a-c, regions between +300 bp relative to TSS and −300 bp relative to CPS were scaled to allow comparisons among genes of different lengths. d, Spt5 and pSpt5 distribution around CPS of Pol II-transcribed genes. Metagene plots (top) and heatmaps (bottom) represent distribution of Spt5-Myc and pSpt5 around CPS (-500 to +500 bp) of highly active genes (n = 137; cluster 1 of Fig. 3a) in dis2+ and dis2-11 cells. e, Statistical analysis of Spt5-Myc and pSpt5 occupancy around CPS. Box plots represent occupancy of Spt5 and pSpt5 in region 500 bp downstream of CPS (b; “PostCPS”) versus region 500 bp upstream of CPS (a; “PreCPS”), for genes in cluster 1 of Fig 3a (n=137), in dis2+ and dis2-11 cells (Spt5, dis2+: High = 3.506, Low = 5.5312×10-2, Median = 0.9199, 95% confidence interval; pSpt5, dis2+: High = 2.085, Low = 2.8570×10-2, Median = 0.4655, 95% confidence interval; Spt5, dis2-11: High = 1.617, Low = 5.7597×10-2, Median = 0.6042, 95% confidence interval; pSpt5, dis2-11: High = 1.155, Low = 6.2344×10-2, Median = 0.4530, 95% confidence interval). f, Box plots represent statistical significance of increases in pSpt5:Spt5 ratios in PostCPS regions in dis2-11 versus dis2+ cells (n = 137) (dis2+: High = 1.533, Low = 4.5765×10-2, Median = 0.5356, 95% confidence interval; dis2-11: High = 1.601, Low = 0.4651, Median = 0.7482, 95% confidence interval). In e, f, p-values were calculated by two-sided Student’s t-test. g, A non-phosphorylatable Spt5 suppresses conditional lethality of dis2-11. Growth kinetics in liquid culture of indicated strains after shift to 18°C (n = 2 biological replicates; two-sided error bars, s.d. of mean).
Extended Data Figure 9. The dis2-11 mutation affects global transcription properties independent of temperature.
a, PRO-seq experiments are reproducible. Scatter plots comparing PRO-seq libraries from two biological replicates for each experiment. Values represent log10 (normalised reads) within gene body (TSS + 200 bp to CPS) of all filtered genes (n = 3383). Colours indicate numbers of genes represented by each point. Spike-in based normalisation should centre scatter about the diagonal line x = y (magenta, dotted). Correlation values represent Spearman’s rank correlation. b, Comparison of composite PRO-seq profiles of dis2-11 mutant alone (top panels) or cdk9as dis2-11 (bottom panels) at 18°C and 30°C. Profiles are centred either on TSS (left) or CPS (right). Shaded areas on composite profiles represent the 12.5 and 87.5% quantiles at each position. c, Composite PRO-seq profiles comparing dis2+ strain (cdk9as) with dis2-11 strain, both at 18°C. Genes were scaled to a common length by fixing the middle gene body region (TSS + 300 bp to CPS – 300 bp) to 60 windows. In b, c, solid lines represent averaged data plot and shaded regions represent s.d. of median. d, Heat maps of spike-in normalised PRO-seq signal (log10) within 10-bp windows relative to the CPS (-250 to +1000) for cdk9as (left) and cdk9as dis2-11 (right) strains at 18°C. Genes were ranked by decreasing T.E.I. in cdk9as dis2-11 at 18°C, a measure of termination-window size. Each panel represents data from filtered genes that are at least 1 kb from neighbouring genes on the same strand (n = 939). In a-d, n = 2 biological replicates.
Extended Data Figure 10. Multiple dis2 mutations cause termination defects.
a, Browser image displaying normalised PRO-seq signal at the SPAC1002.13c gene locus. Track values reflect the maximum displayed signal (some peaks exceed these values). b, Boxplots displaying the distribution of termination elongation index (T.E.I.) values in each strain for all filtered genes separated from same-strand neighbours by at least 1 Kb (n = 939). Significant differences (p-value; Welch’s two sample t-test) in mean T.E.I. for each strain compared with dis2+ are indicated (dis2+: High = 1.2304, Low = −2.4005, Median = −0.6532; dis2-11: High = 1.0066, Low = −2.3483, Median = −0.3802; dis2Δ: High = 1.5563, Low = −1.8325, Median = −0.06695; dis2-T316A: High = 1.4314, Low = −2.1004, Median = −0.1176; dis2-T316D: High = 1.3424, Low = −2.0934, Median = −0.2310; each box shows 25th-75th percentiles). c, Composite PRO-seq profiles of each dis2 mutant strain (red) compared with dis2+ (blue). Profiles reflect region from −250 bp to +1000 bp around the CPS. Shaded areas on composite profiles represent the 12.5 and 87.5% quantiles at each position. Each panel represents data from filtered genes that are at least 1 kb from neighbouring genes on the same strand (n = 939). Solid lines represent averaged data plot of median. d, Heatmaps displaying log2(Mutant / WT PRO-seq signal) within 10-bp windows from −250 bp to +1000 bp around the CPS for all genes used in c sorted by decreasing T.E.I. values in dis2+ (top to bottom). In a-d, n = 2 biological replicates.
Supplementary Material
Acknowledgments
This work is dedicated to the memory of Günter Blobel (1936-2018), a visionary scientist and inspiring mentor. We thank I.M. Hagan, B. Schwer, S. Shuman, V. Vanoosthuyse, M. Yanagida, M.J. O’Connell and the National BioResource Project/Yeast Genetic Resource Center for providing yeast strains and/or antibodies; K. M. Shokat for providing 3-MB-PP1; C. Zhang for guidance in AS-allele optimisation; and N. Steinbach and R. Parsons for assistance in phosphatase activity measurements. J.C.T. was supported by Canadian Institutes of Health Research grant MOP-130362 and by a fellowship from Fond de recherche Quebec Santé (3315). This work was supported by National Institutes of Health grants GM25232 to G.T.B. and J.T.L., and GM104291 to R.P.F. Next-generation sequencing was supported in part by grant P30 CA196521 to the Tisch Cancer Institute.
Footnotes
Author Contributions P.K.P. and M.S. identified PP1 isoforms as Cdk9 substrates. P.K.P conducted enzymologic studies, measured Spt5 dephosphorylation rates in wild-type and PP1 mutant backgrounds, performed ChIP-qPCR analysis and characterised genetic interactions between dis2 and spt5 mutant alleles. P.K.P., M.S., B.B. and J.C.T. performed ChIP-seq analysis to map distribution of pSpt5. G.T.B. performed PRO-seq analysis and developed metrics to quantify termination defects in dis2 mutants . P.K.P., G.T.B., J.T.L. and R.P.F. prepared the manuscript.
Author Information The authors declare no competing financial interests.
References
- 1.Proudfoot NJ. Transcriptional termination in mammals: Stopping the RNA polymerase II juggernaut. Science. 2016;352:aad9926. doi: 10.1126/science.aad9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davidson L, Muniz L, West S. 3' end formation of pre-mRNA and phosphorylation of Ser2 on the RNA polymerase II CTD are reciprocally coupled in human cells. Genes Dev. 2014;28:342–356. doi: 10.1101/gad.231274.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanso M, et al. P-TEFb regulation of transcription termination factor Xrn2 revealed by a chemical genetic screen for Cdk9 substrates. Genes Dev. 2016;30:117–131. doi: 10.1101/gad.269589.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grallert A, et al. A PP1-PP2A phosphatase relay controls mitotic progression. Nature. 2015;517:94–98. doi: 10.1038/nature14019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shetty A, et al. Spt5 Plays Vital Roles in the Control of Sense and Antisense Transcription Elongation. Mol Cell. 2017;66:77–88. e75. doi: 10.1016/j.molcel.2017.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Booth GT, Wang IX, Cheung VG, Lis JT. Divergence of a conserved elongation factor and transcription regulation in budding and fission yeast. Genome Res. 2016;26:799–811. doi: 10.1101/gr.204578.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blethrow JD, Glavy JS, Morgan DO, Shokat KM. Covalent capture of kinase-specific phosphopeptides reveals Cdk1-cyclin B substrates. Proc Natl Acad Sci U S A. 2008;105:1442–1447. doi: 10.1073/pnas.0708966105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohkura H, Kinoshita N, Miyatani S, Toda T, Yanagida M. The fission yeast dis2+ gene required for chromosome disjoining encodes one of two putative type 1 protein phosphatases. Cell. 1989;57:997–1007. doi: 10.1016/0092-8674(89)90338-3. [DOI] [PubMed] [Google Scholar]
- 9.Yamano H, Ishii K, Yanagida M. Phosphorylation of dis2 protein phosphatase at the C-terminal cdc2 consensus and its potential role in cell cycle regulation. EMBO J. 1994;13:5310–5318. doi: 10.1002/j.1460-2075.1994.tb06865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kinoshita N, Ohkura H, Yanagida M. Distinct, essential roles of type 1 and 2A protein phosphatases in the control of the fission yeast cell division cycle. Cell. 1990;63:405–415. doi: 10.1016/0092-8674(90)90173-c. [DOI] [PubMed] [Google Scholar]
- 11.Viladevall L, et al. TFIIH and P-TEFb coordinate transcription with capping enzyme recruitment at specific genes in fission yeast. Mol Cell. 2009;33:738–751. doi: 10.1016/j.molcel.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pei Y, Shuman S. Characterization of the Schizosaccharomyces pombe Cdk9/Pch1 protein kinase: Spt5 phosphorylation, autophosphorylation, and mutational analysis. J Biol Chem. 2003;278:43346–43356. doi: 10.1074/jbc.M307319200. [DOI] [PubMed] [Google Scholar]
- 13.Sansó M, et al. A Positive Feedback Loop Links Opposing Functions of P-TEFb/Cdk9 and Histone H2B Ubiquitylation to Regulate Transcript Elongation in Fission Yeast. PLoS Genet. 2012;8:e1002822. doi: 10.1371/journal.pgen.1002822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Booth GT, Parua PK, Sanso M, Fisher RP, Lis JT. Cdk9 regulates a promoter-proximal checkpoint to modulate RNA polymerase II elongation rate in fission yeast. Nat Commun. 2018;9:543. doi: 10.1038/s41467-018-03006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sajiki K, et al. Genetic control of cellular quiescence in S. pombe. J Cell Sci. 2009;122:1418–1429. doi: 10.1242/jcs.046466. [DOI] [PubMed] [Google Scholar]
- 16.Cho EJ, Kobor MS, Kim M, Greenblatt J, Buratowski S. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev. 2001;15:3319–3329. doi: 10.1101/gad.935901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hausmann S, Shuman S. Characterization of the CTD phosphatase Fcp1 from fission yeast. Preferential dephosphorylation of serine 2 versus serine 5. J Biol Chem. 2002;277:21213–21220. doi: 10.1074/jbc.M202056200. [DOI] [PubMed] [Google Scholar]
- 18.Schwer B, Ghosh A, Sanchez AM, Lima CD, Shuman S. Genetic and structural analysis of the essential fission yeast RNA polymerase II CTD phosphatase Fcp1. Rna. 2015;21:1135–1146. doi: 10.1261/rna.050286.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.St Amour CV, et al. Separate Domains of Fission Yeast Cdk9 (P-TEFb) Are Required for Capping Enzyme Recruitment and Primed (Ser7-Phosphorylated) Rpb1 Carboxyl-Terminal Domain Substrate Recognition. Mol Cell Biol. 2012;32:2372–2383. doi: 10.1128/MCB.06657-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pei Y, et al. Cyclin-dependent kinase 9 (Cdk9) of fission yeast is activated by the CDK-activating kinase Csk1, overlaps functionally with the TFIIH-associated kinase Mcs6, and associates with the mRNA cap methyltransferase Pcm1 in vivo. Mol Cell Biol. 2006;26:777–788. doi: 10.1128/MCB.26.3.777-788.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vanoosthuyse V, et al. CPF-associated phosphatase activity opposes condensin-mediated chromosome condensation. PLoS Genet. 2014;10:e1004415. doi: 10.1371/journal.pgen.1004415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nedea E, et al. The Glc7 phosphatase subunit of the cleavage and polyadenylation factor is essential for transcription termination on snoRNA genes. Mol Cell. 2008;29:577–587. doi: 10.1016/j.molcel.2007.12.031. [DOI] [PubMed] [Google Scholar]
- 23.Schreieck A, et al. RNA polymerase II termination involves C-terminal-domain tyrosine dephosphorylation by CPF subunit Glc7. Nat Struct Mol Biol. 2014;21:175–179. doi: 10.1038/nsmb.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baejen C, et al. Genome-wide Analysis of RNA Polymerase II Termination at Protein-Coding Genes. Mol Cell. 2017;66:38–49. e36. doi: 10.1016/j.molcel.2017.02.009. [DOI] [PubMed] [Google Scholar]
- 25.Bernecky C, Herzog F, Baumeister W, Plitzko JM, Cramer P. Structure of transcribing mammalian RNA polymerase II. Nature. 2016;529:551–554. doi: 10.1038/nature16482. [DOI] [PubMed] [Google Scholar]
- 26.Schneider S, Pei Y, Shuman S, Schwer B. Separable functions of the fission yeast Spt5 carboxyl-terminal domain (CTD) in capping enzyme binding and transcription elongation overlap with those of the RNA polymerase II CTD. Mol Cell Biol. 2010;30:2353–2364. doi: 10.1128/MCB.00116-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fong N, et al. Effects of Transcription Elongation Rate and Xrn2 Exonuclease Activity on RNA Polymerase II Termination Suggest Widespread Kinetic Competition. Mol Cell. 2015;60:256–267. doi: 10.1016/j.molcel.2015.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glover-Cutter K, Kim S, Espinosa J, Bentley DL. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol. 2008;15:71–78. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fong N, Saldi T, Sheridan RM, Cortazar MA, Bentley DL. RNA Pol II Dynamics Modulate Co-transcriptional Chromatin Modification, CTD Phosphorylation, and Transcriptional Direction. Mol Cell. 2017 doi: 10.1016/j.molcel.2017.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamada T, et al. P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol Cell. 2006;21:227–237. doi: 10.1016/j.molcel.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 31.Hayashi A, et al. Localization of gene products using a chromosomally tagged GFP-fusion library in the fission yeast Schizosaccharomyces pombe. Genes Cells. 2009;14:217–225. doi: 10.1111/j.1365-2443.2008.01264.x. [DOI] [PubMed] [Google Scholar]
- 32.Saiz JE, Fisher RP. A CDK-activating kinase network is required in cell cycle control and transcription in fission yeast. Curr. Biol. 2002;12:1100–1105. doi: 10.1016/s0960-9822(02)00903-x. [DOI] [PubMed] [Google Scholar]
- 33.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 34.Stone EM, Yamano H, Kinoshita N, Yanagida M. Mitotic regulation of protein phosphatases by the fission yeast sds22 protein. Curr Biol. 1993;3:13–26. doi: 10.1016/0960-9822(93)90140-j. [DOI] [PubMed] [Google Scholar]
- 35.Parua PK, Mondal A, Parrack P. HflD, an Escherichia coli protein involved in the lambda lysis-lysogeny switch, impairs transcription activation by lambdaCII. Arch Biochem Biophys. 2010;493:175–183. doi: 10.1016/j.abb.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 36.Sanso M, et al. Gcn5 facilitates Pol II progression, rather than recruitment to nucleosome-depleted stress promoters, in Schizosaccharomyces pombe. Nucleic Acids Res. 2011;39:6369–6379. doi: 10.1093/nar/gkr255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanny JC, Erdjument-Bromage H, Tempst P, Allis CD. Ubiquitylation of histone H2B controls RNA polymerase II transcription elongation independently of histone H3 methylation. Genes Dev. 2007;21:835–847. doi: 10.1101/gad.1516207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kao CF, Osley MA. In vivo assays to study histone ubiquitylation. Methods. 2003;31:59–66. doi: 10.1016/s1046-2023(03)00088-4. [DOI] [PubMed] [Google Scholar]
- 39.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feng J, Liu T, Qin B, Zhang Y, Liu XS. Identifying ChIP-seq enrichment using MACS. Nat Protoc. 2012;7:1728–1740. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramirez F, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwak H, Fuda NJ, Core LJ, Lis JT. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science. 2013;339:950–953. doi: 10.1126/science.1229386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mahat DB, et al. Base-pair-resolution genome-wide mapping of active RNA polymerases using precision nuclear run-on (PRO-seq) Nat Protoc. 2016;11:1455–1476. doi: 10.1038/nprot.2016.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data for results plotted in Figs. 1d, 2a, c-e, and in Extended Data Figs. 1b-e, 3a-d, 4a-d, 5a-d, 6a, b, e, and 8g are available online. The raw and processed sequencing files have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) (Accession number GSE102590).