Abstract
Recognition of a thymine-adenine base pair in DNA by triplex-forming oligonucleotides can be achieved by a guanine through the formation of a G.TA triad within the parallel triple helix motif. In the present work, we provide the first characterization of the stability of individual base pairs and base triads in a DNA triple helix containing a G.TA triad. The DNA investigated is the intramolecular triple helix formed by the 32mer d(AGATAGAACCCCTTCTATCTTATATCTGTCTT). The exchange rates of imino protons in this triple helix have been measured by nuclear magnetic resonance spectroscopy using magnetization transfer from water and real-time exchange. The exchange rates are compared with those in a homologous DNA triple helix in which the G.TA triad is replaced by a canonical C+.GC triad. The results indicate that, in the G.TA triad, the stability of the Watson–Crick TA base pair is comparable with that of AT base pairs in canonical T.AT triads. However, the presence of the G.TA triad destabilizes neighboring triads by 0.6–1.8 kcal/mol at 1°C. These effects extend to triads that are two positions removed from the site of the G.TA triad. Therefore, the lower stability of DNA triple helices containing G.TA triads originates, in large part, from the energetic effects of the G.TA triad upon the stability of canonical triads located in its vicinity.
INTRODUCTION
The applications of DNA triple-helical structures in biotechnology and molecular medicine rely upon the ability of triplex-forming oligonucleotides to target specific base sequences in large DNA molecules (1). Formation of the triple-helical structure at the targeted sites inhibits the sequence-specific binding of proteins to DNA (2–5), and can mediate sequence-specific DNA cleavage (6). Generally, the targeted sites consist of homopurine-homopyrimidine sequences, which are recognized by bases in the third strand through specific Hoogsteen hydrogen bonds. In pyrimidine–purine–pyrimidine (YRY) triplexes the third strand consists of protonated cytosines and thymines, which recognize, respectively, GC and AT base pairs in the DNA duplex by forming C+.GC and T.AT triads. Alternatively, in purine–purine–pyrimidine (RRY) triplexes, GC and AT base pairs are recognized by a purine-rich third strand through formation of A.AT, T.AT and G.GC triads. One feature common to both YRY and RRY families of triple helices is that the triplex-forming oligonucleotide recognizes only the purine bases in DNA duplex. This fact greatly restricts the range of DNA sequences that can be targeted by triplex formation (1). In the last few decades, sustained efforts have been devoted to extending the triplex recognition code to all four Watson–Crick base pairs in duplex DNA (1,7). A significant advance towards this goal has been made by the demonstration that a TA base pair can be specifically recognized by a guanine in the third strand (8,9). This new G.TA triad has thus been of great interest for the triplex recognition code. The structures of several DNA triple helices containing the G.TA triad have been characterized by nuclear magnetic resonance (NMR) spectroscopy (10). The structures have revealed the pattern of Hoogsteen hydrogen bonding between the guanine in the third strand and the targeted TA base pair as well as the distortions induced in the triplex structure by the presence of a guanine in an otherwise homopyrimidine strand (11,12). Extensive investigations have also been devoted to the characterization of the thermodynamic stability of G.TA-containing triple helices. They have shown that the overall stability of these triple helices is, in general, lower than that of canonical triplexes, and depends on an array of molecular factors such as base sequence, length of the triple helix and number of G.TA triads (8,9,13–16). Moreover, environmental conditions such as pH, ionic strength and temperature also affect the specificity and the energetics of the G.TA recognition (17,18).
In order to understand the molecular origin of the stability of these triple helices, in the present work, we have undertaken a study of a G.TA-containing triple helix using NMR spectroscopy and proton exchange. The DNA triplex of interest is shown in Figure 1B and is henceforth abbreviated GTA triplex. The structure of this triple helix has been previously characterized in solution state by Feigon and co-workers using NMR spectroscopy (11). Our present investigation addresses the question of the effects of the G.TA triad upon the stability of the other, canonical triads in this DNA triplex. The approach taken consists of measuring the rates of exchange of imino protons in individual triads of the triplex structure. The effects of the G.TA triad are distinguished by comparing the exchange rates in the GTA triplex to those in a homologous DNA triplex in which the G.TA triad is replaced by a canonical C+.GC triad (henceforth abbreviated C+GC triplex; Fig. 1C). The imino proton exchange and the base-pair opening in the latter DNA triplex have been recently characterized by this laboratory (19).
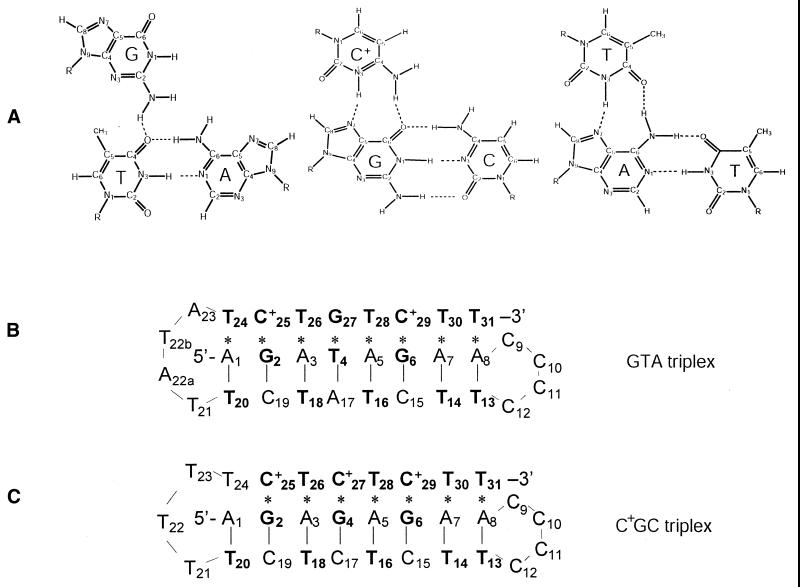
Figure 1.
(A) Structures of G.TA, C+.GC and T.AT triads (11,27). (B) Base sequence and folded conformation of the GTA triplex investigated (11). (C) Base sequence and folded conformation of the C+GC triplex investigated (24). (B and C) Watson–Crick hydrogen bonds are indicated by vertical bars, Hoogsteen hydrogen bonds are indicated by asterisks, and the bases containing hydrogen-bonded imino protons are shown in bold.
MATERIALS AND METHODS
DNA samples
The DNA oligonucleotides were synthesized on an automated DNA synthesizer (Applied Biosystems 381A) using the solid-support phosphoramidite method. They were purified by reverse-phase HPLC on a PRP-1 column (Hamilton) in 50 mM triethylamine acetate buffer at pH 7 (with a gradient of 10–20% acetonitrile in 32 min). The counterions were replaced with Na+ ions by repeated centrifugation through Centricon YM-3 tubes (Amicon Inc.). The final samples were in 100 mM NaCl and 5 mM MgCl2 at pH 5.3 (measured at 20°C). The samples contained ∼200 OD260 units of DNA.
NMR experiments
The NMR experiments were performed at 1°C on a Varian INOVA 500 spectrometer operating at 11.75 T. One-dimensional NMR spectra were obtained using the Jump-and-Return pulse sequence (20). Proton exchange rates were measured in experiments of transfer of magnetization from water and in real-time exchange experiments.
In transfer of magnetization experiments, the exchange was initiated by inverting selectively the water proton resonance using a Gaussian 180° pulse (6.4 ms). During the exchange delay following water inversion a weak gradient (0.21 gauss/cm) was applied to prevent the effects of radiation damping upon the recovery of water magnetization to equilibrium. At the end of the exchange delay, before observation with the Jump-and-Return sequence, a second Gaussian pulse (1.8 ms) was applied to bring the water magnetization back to the oz axis. Twenty-seven values of the exchange delay in the range from 2 to 600 ms were used in each experiment. The intensity of an exchangeable proton resonance depends on the exchange delay τ as (21):
where I0 is the intensity at equilibrium, R1 is the longitudinal relaxation rate and kex is the exchange rate for the proton of interest; Rw is the longitudinal relaxation rate of water, and q = Iw(0)/I0w expresses the efficiency of water inversion (e.g. q = –1 for perfect inversion). Rw and q were measured in separated experiments. The intensity of each imino proton resonance was fitted as a function of the exchange delay τ to equation 1, using a non-linear least-squares program. Equation 1 shows that the time-dependence of the magnetization of an imino proton reflects both the transfer of magnetization process and the longitudinal relaxation of water and DNA imino protons. Due to this fact, the lowest exchange rate that can be measured accurately using the transfer of magnetization method is ∼0.5 s–1 at 1°C.
In real-time exchange experiments, the exchange was initiated by adding D2O to a concentrated DNA solution in H2O. The final volume fraction of D2O was ∼70%. The time elapsed between the initiation of exchange and the first NMR spectrum was 5–15 min. The acquisition time for each spectrum was 4 min. The intensity of each resonance was fitted as a function of the real-time exchange delay t to the equation:
I(t) = [I(0) – I(∞)]exp(–kex × t) + I(∞) 2
where I(∞) is the intensity in a fully exchanged sample. The range of exchange rates that can be measured in these experiments is determined by the time elapsed between the initiation of exchange and the acquisition of the first NMR spectrum. For example, if the first NMR spectrum is acquired at 5 min after starting the exchange, the fastest exchange rate that can be measured reliably is ∼10–3 s–1.
Theory of imino proton exchange
In nucleic acids, the exchange of imino protons with solvent protons occurs by transient opening of the base pairs. The opening reaction consists of a local conformational change that breaks the hydrogen bond holding the imino proton and moves the proton into a state where its transfer to an acceptor can occur. The acceptors are groups within the same nucleic acid molecule, water, OH– ions or added exchange catalysts (22). The exchange rate observed experimentally depends on the rate of proton transfer from the open state, ktr, as well as on the rates of the opening and closing reactions, kop and kcl, respectively (23):
When the rate of proton transfer is higher than the closing rate (i.e. ktr >> kcl; EX1 regime), kex = kop and the exchange is limited by the rate of base-pair opening. When the rate of closing exceeds the rate of proton transfer (i.e. ktr << kcl; EX2 regime), kex = Kop × ktr where Kop = kop / kcl is the equilibrium constant for the opening reaction. The equilibrium constant Kop defines the free energy change in the opening reaction (23):
ΔGop = –RT lnKop 4
RESULTS
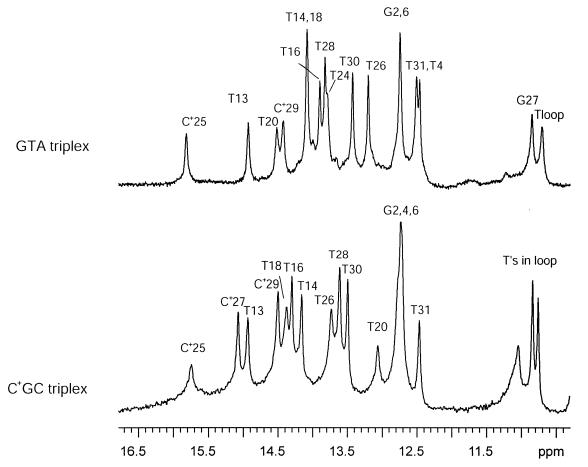
The imino proton resonances of the DNA triplexes investigated are shown in Figure 2. For the GTA triplex, the resonances have been assigned to specific bases by Feigon and co-workers (11). These authors have also shown that eight triads are formed in this structure. Two are canonical C+.GC triads formed by protonated cytosines C+25 and C+29 (Fig. 1). Five are canonical T.AT triads formed by thymines T24, T26, T28, T30 and T31. The remaining is the non-canonical triad G27.T4A17. The imino protons associated with this triad belong to T4 and G27; their resonances are observed, respectively, at 12.46 and, close to the imino proton resonances of thymines in the loop, at 10.85 p.p.m. For the homologous C+GC triplex, the imino proton resonances indicate formation of only seven triads (24). The imino proton resonance of T24 is not observed, indicating that the triad T24.A1T20 is unstable or does not form. This may be due to the fact that the loop connecting the pyrimidine strands in this structure is only three bases long (24).
Figure 2.
NMR resonances of imino protons in the GTA triplex (upper spectrum) and the C+GC triplex (lower spectrum) in 100 mM NaCl and 5 mM MgCl2 at pH 5.3 and at 1°C.
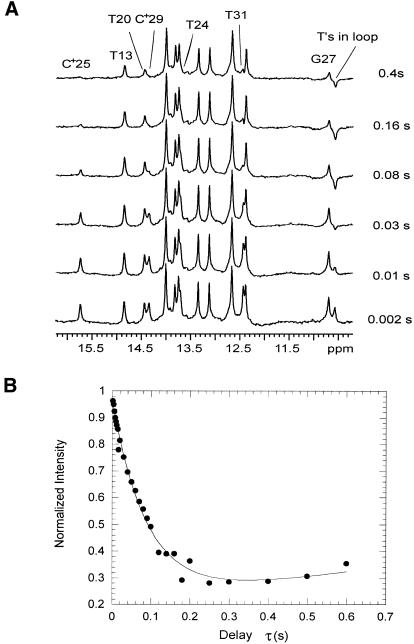
We have measured the rates of exchange of imino protons with solvent in the two DNA triple helices using transfer of magnetization and real-time exchange experiments. A representative series of spectra from experiments of transfer of magnetization from water on the GTA triplex are shown in Figure 3A. Variations in resonance intensities as a result of the magnetization transfer from water are observed for the imino protons of protonated cytosines C+25 and C+29, of terminal thymines T20, T24, T13 and T31, and of the guanine in the G.TA triad (Fig. 3B). For the other imino protons, no transfer of magnetization from water is observed, indicating that their exchange rates are slower than the lowest rate that can be measured in these experiments (namely, kex ∼ 0.5 s–1).
Figure 3.
(A) Selected spectra of the GTA triplex during experiments of transfer of magnetization from water at 1°C. The exchange delay, τ (s), for each spectrum is indicated. The labeled imino proton resonances show changes in intensity during transfer of magnetization from water. (B) Dependence of the intensity of the G27 imino proton resonance on the exchange delay, τ, in experiments of transfer of magnetization from water.
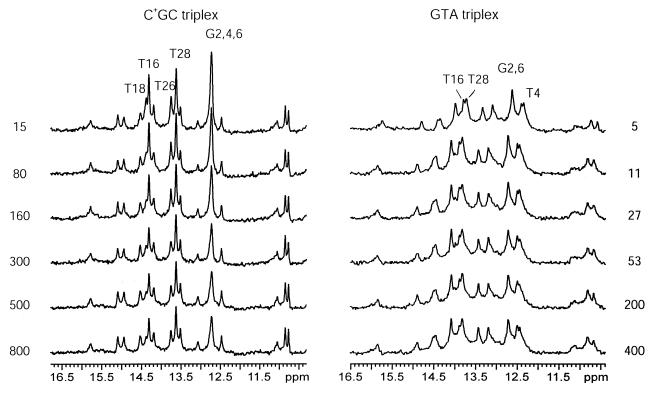
In order to monitor these slowly exchanging protons we have carried out real-time exchange experiments. We have found that, in the GTA triplex, the exchange of imino protons of Watson–Crick guanines and of T16, T28 and T4 can be monitored in real-time (Fig. 4), indicating that their exchange rates are slower than ∼10–3 s–1. For the canonical C+GC triplex, the exchange of two more imino protons, i.e. T18 and T26, is slow enough to be observable in real-time exchange experiments (Fig. 4).
Figure 4.
Selected spectra of the GTA and C+GC triplexes during real-time exchange measurements at 1°C. The exchange time (min) is given for each spectrum. The imino proton resonances, which show changes in intensity during real-time exchange are indicated.
The exchange rates of imino protons in the GTA triplex are compared with those in the C+GC triplex in Table 1. For the imino protons whose exchange rates fall in the range inaccessible to NMR measurements, the values given reflect the fact that the exchange is too fast to be monitored in real-time exchange (kex > ∼10–3 s–1) and too slow to be measurable by transfer of magnetization (kex < ∼0.5 s–1). For these protons we have attempted to define the exact value of the exchange rate by measurements at higher temperatures. The temperature range in which these measurements were possible was from 1 to 15°C. At temperatures >15°C, the resonances of the GTA triplex start decreasing in intensity, indicating that a transition to a different conformation occurs. We have found that, for the imino proton of T30, raising the temperature to 15°C increases the exchange rate in both triplexes into the range measurable by transfer of magnetization, namely, kex = (0.9 ± 0.2) s–1 in the GTA triplex and kex = (0.8 ± 0.2) s–1 in the C+GC triplex.
Table 1. Imino proton exchange rates (s–1) at 1°C: GTA triplex (upper row) and C+GC triplex (lower row).
| T24 | C+25 | T26 | G27/C+27 | T28 | C+29 | T30 | T31 |
| 4.0 ± 0.4 | 7.6 ± 0.6 | 10–3 <kex <0.5 | 4.2 ± 0.2 | (3 ± 1) × 10–4 | 11.6 ± 0.3 | 10–3 <kex <0.5 | 11.2 ± 0.4 |
|
a |
12 ± 1 |
(3.8 ± 0.7) × 10–5 |
0.7 ± 0.4 |
(3.3 + 0.3) × 10–5 |
1.5 ± 0.3 |
10–3 <kex <0.5 |
10.5 ± 0.6 |
| A1 | G2b | A3 | T4/G4b | A5 | G6b | A7 | A8 |
| (9 ± 2) × 10–4 | (2.6 ± 0.8) × 10–4 | (9 ± 2) × 10–4 | |||||
| |
(9.9 ± 0.3) × 10–5 |
|
(9.9 ± 0.3) × 10–5 |
|
(9.9 ± 0.3) × 10–5 |
|
|
| T20 | C13 | T18c | A17/C17 | T16 | C15 | T14c | T13 |
| 1.8 ± 0.2 | 10–3 <kex <0.5 | (4.0 ± 1.0) × 10–4 | 10–3 <kex <0.5 | 1.1 ± 0.2 | |||
| 1.7 ± 0.2 | (3.0 ± 0.3) × 10–4 | (4.7 ± 0.3) × 10–5 | 10–3 <kex <0.5 | 1.1 ± 0.1 |
aThe imino proton resonance of T24 in the C+GC triplex is not observable (see text).
bIn the GTA triplex the imino proton resonance of G2 and G6 overlap (Fig. 2); similarly, in the C+GC triplex, the imino proton resonance of G2, G4 and G6 overlap (Fig. 2). The values of kex given were obtained from the overlapped resonances.
cIn the GTA triplex the imino proton resonance of T18 and T14 overlap (Fig. 2).
DISCUSSION
In the present work we have used the exchange of imino protons to probe the stability at distinct sites in the GTA triplex, and compare it with the stability in a canonical triplex in which a C+.GC triad replaces the G.TA triad. Our approach is based on the local opening model for proton exchange (23). The main idea of this model is that the exchange of an imino proton with solvent occurs by an opening reaction that transiently perturbs the native structure to allow the transfer away of the proton. Accordingly, the model postulates that the stability of the site containing the imino proton of interest can be described by the free energy change associated with the opening reaction, ΔGop (equation 4). A corollary of the model is that a structural perturbation that affects the stability of the site should also be reflected in a change in the exchange rate of the imino proton contained in that site. Accordingly, the change in stabilization free energy as a result of the structural perturbation can be expressed as:
where the primed symbols refer to the perturbed site. Equation 5 is strictly valid only in the EX2 regime of exchange where kex is directly proportional to the equilibrium constant for opening. When the exchange is rate-limited by the rate of base-pair opening (i.e. the EX1 regime of exchange), equation 5 is valid only if the structural perturbation does not affect the rates of base-pair closing. Previous work from our laboratory has shown that for most of the imino protons in the C+GC triplex and under the experimental conditions investigated (i.e. 5 mM MgCl2 and 100 mM NaCl at pH 5.3), the exchange is in the EX2 regime (19). However, for the imino protons of protonated cytosines, the exchange is in, or close to, the EX1 regime (19). As the data in Table 1 show, the exchange rates of these protons are among the fastest rates measured in both triplexes. The exchange is fast because the cytosine imino proton in the open state of the Hoogsteen C+.G base pair is transferred to the N7 group of the guanine. The efficiency of this internal catalysis is high [ktr = 6 × 108 s–1 (19)] and thus, the exchange is rate-limited by the opening of the base pair. The imino protons investigated provide several markers for the GTA triplex. In the duplex part of the structure these markers include the imino protons of the thymines. As shown in Table 1, the exchange rates of two thymine protons are affected by the presence of the G.TA triad. The first is the imino proton in T16. The exchange rate of this proton increases ∼10-fold in the GTA triplex as compared with the C+GC triplex. The second is the imino proton of T18 for which the exchange rate is at least three times faster in the GTA triplex than in the C+GC triplex. Hence, according to equation 5, the presence of the G.TA triad destabilizes the Watson–Crick AT base pairs adjacent to it by 0.6–1.3 kcal/mol at 1°C. In contrast, for the terminal thymines, T13 and T20, the exchange rates in the GTA triplex are, within experimental errors, the same as those in the control triplex. This suggests that the effects of the G.TA triad are confined to triads located in its close proximity. Another insight into the extent of the perturbation induced by the G.TA triad is provided by the exchange rates for the guanines in the duplex part of the structure. As shown in Figure 2, the imino proton resonances of the guanines in both triplexes overlap. Nevertheless, the exchange of the overlapped resonance in each triplex can be monitored in real-time exchange experiments (Fig. 4), and the exchange is described by a single exponential (data not shown). In the C+GC triplex, the apparent rate of exchange of the guanine resonance is (9.9 ± 0.3) × 10–5 s–1. In the GTA triplex, the rate of exchange increases by one order of magnitude (Table 1), indicating that the destabilizing effects of the G.TA triad may extend to Watson–Crick base pairs which are two triads removed from the G.TA site. In the Hoogsteen strand of the triplex, the pattern of exchange rates parallels that in the duplex part of the structure. A large destabilization is observed for T26 which is located next to the G.TA site: the exchange rate of the imino proton in this base is at least 25 times larger in the GTA than in the C+GC triplex. This corresponds to a destabilization of the Hoogsteen base pair in the T26.A3T18 triad of the GTA triplex by at least 1.8 kcal/mol at 1°C (equation 5). Smaller destabilizations are observed for the Hoogsteen base pairs located on the 3′-side of the G.TA triad (this directionality is defined relative to that of the purine strand of the triplex). Increases in the exchange rates are observed for the Hoogsteen bases in T28.A5T16 and C+29.G6C15 triads (Table 1). However, for the terminal thymines in the Hoogsteen strand, T30 and T31, the imino proton exchange rates in the GTA triplex are the same as those in the C+GC triplex (Table 1 and Results). This finding, together with the exchange rates for T13, indicates that the two triads at the 3′-end of the structure have similar stabilities in the two triplexes. Interestingly, for the 5′-end of the structures, the stability appears to be higher in the GTA than the C+GC triplex. This is indicated by the exchange rate of the imino proton in C+25, which is ∼1.6-fold lower in the GTA triplex. This enhanced stability may not reflect, however, an effect of the G.TA triad. It is more likely that C+25 is stabilized in the GTA triplex due to the fact that, in this triplex, the loop connecting the two pyrimidine strands is longer than in the C+GC triplex. This interpretation is supported by the fact that, in the GTA triplex, the imino proton resonance of T24 is observable in the NMR spectrum (Fig. 2), and the T24.A1 Hoogsteen base pair is formed (11). In contrast, in the C+GC triplex, the absence of the T24 imino proton resonance from the spectrum suggests that the T24.A1 Hoogsteen base pair does not form, possibly due to the steric constraints imposed by the short, three-base loop at the 5′-end of the structure (24). The structure of the GTA triplex in solution state has been previously characterized by Feigon and co-workers (11). These authors have observed significant changes in the intensities of NOESY cross-peaks between protons in the G.TA triad and neighboring protons; for example, the NOEs between G27 imino proton and T26 methyl protons, between imino protons of T4 and T16, and between base/sugar protons of A5 and A17. These variations indicated that the helix parameters (e.g. twist and/or slide) in the G.TA triad and its two neighboring triads are different from those in the rest of the structure. These structural observations are consistent with the higher exchange rates that we observe for imino protons in T26.A3T18 and T28.A5T16 triads (Table 1). However, the NMR structural studies did not reveal any large conformational changes beyond the triads flanking the G.TA site. In contrast, our results indicate that the exchange rates of the imino protons in the C+29.G6C15 triad and the G2C13 base pair are increased by the presence of the G.TA triad (Table 1). These observations, therefore, suggest that subtle changes in the stability of local interactions within the DNA triple helix can exist in the absence of large alterations in structure. Our present investigation also allows a direct characterization of the exchange of imino protons in the non-canonical G.TA triad itself. The imino proton of T4 (resonance at 12.46 p.p.m. in Fig. 2) is involved in a Watson–Crick hydrogen bond with A17 in the G.TA triad. The exchange of this proton can be monitored in real-time exchange experiments (Fig. 4). Its exchange rate [(2.6 ± 0.8) × 10–4 s–1, Table 1] is the same as, or slightly faster than, that of Watson–Crick imino protons in canonical T.AT triads (for example, T16 and T18 imino protons in the C+GC triplex, Table 1). This result suggests that the stability of the Watson–Crick TA base pair in the G.TA triad is comparable with that of an AT base pair in a canonical T.AT triad. This may seem unexpected as formation of a canonical T.AT triad involves two Hoogsteen hydrogen bonds whereas in a G.TA triad, only one Hoogsteen hydrogen bond exists, from the guanine H2(2) proton to the O4 atom of thymine (Fig. 1A). One should note, however, that the opening reactions responsible for the exchange of the thymine imino proton in the Watson–Crick base pairs of these two triads could be very different. In a T.AT triad, there is no steric hindrance for opening of the Watson–Crick thymine, in either the major or the minor groove (Fig. 1). However, as suggested recently by our laboratory (19), this opening reaction most likely involves larger perturbations in the structure that also affect the hydrogen-bonded adenine. In the G.TA triad, one expects the opening of the Watson–Crick thymine to involve concerted motions of both the guanine and the adenine. The energetic cost of these motions should be dependent on factors other than the inter-base hydrogen bonding; for example, perturbations in the stacking interactions and in the phosphate backbone conformation at the G.TA site. Thus, the net free energies of opening and the resulting exchange rates for the Watson–Crick thymines in G.TA and T.AT triads could have comparable values. For the Hoogsteen base of the G.TA triad (G27), the exchange rate is only 20-fold lower than that in free guanine under comparable experimental conditions [namely, kex = 80 s–1 at 1°C and at pH 5.5 (25)]. NMR structural studies (11,12) have demonstrated that, in the G.TA triad, the imino proton of the guanine is not hydrogen-bonded (Fig. 1). Moreover, in free guanine, in the pH range of interest, the exchange of the imino proton is acid-catalyzed by protonation of the N7 group (25). Therefore, it is likely that the exchange of the imino proton of G27 in the GTA triplex is slowed down by the restricted accessibility of the guanine N7 and/or N1 groups to H+ ions and to proton acceptors. This interpretation is supported by the temperature dependence of the exchange rate of the G27 imino proton in the GTA triplex that yields an activation enthalpy of 12 ± 1 kcal/mol (data not shown). This value is the same as that in free guanine (25), indicating that the exchange of the imino proton in the triplex involves the same activated state as in free guanine. The stability of DNA triplexes containing G.TA triads has been characterized previously using optical spectroscopy, calorimetry, gel mobility shift and quantitative affinity cleavage (8,9,13–18). By and large, all these studies have shown that DNA triplexes containing the G.TA triad are less stable than canonical YRY triplexes. For example, for the triplex investigated here, the melting temperature is lowered to 55°C from a value of 68°C in an identical triplex in which the G.TA triad is replaced by a C+.GC triad (11). The destabilization of DNA triplexes containing the G.TA triad has also been demonstrated by quantitative affinity cleavage titration (9). These studies have shown that the free energy of formation of the triple-helical structure decreases by 1.5–1.7 kcal/mol (at 22°C) when either one of the canonical triads is replaced by a G.TA triad. The formation free energy is a global measure of the stability of the triple helix and includes many more interactions than those probed here by proton exchange. Therefore, the decrease in formation free energy cannot be directly compared with the changes in stabilization free energy detected in this work at individual sites. Nevertheless, our results suggest that the global destabilization of G.TA-containing triplexes originates, in large part, from the effects that the G.TA triad has on neighboring triads, one and two positions removed from the GTA site. It is also interesting to note that thermodynamic studies have shown that the stability of G.TA-containing triplexes is strongly influenced by near neighbor effects (15). Specifically, G.TA-containing triplexes are most stable when the G.TA triad is flanked by two T.AT triads, like in the triplex investigated here. The stability is lowered when the G.TA triad is flanked by one or two C+.GC triads (13–15,26). In light of these findings, characterization of imino proton exchange in DNA triplexes with other sequences flanking the G.TA site is clearly of interest and this work is currently in progress in our laboratory.
Acknowledgments
ACKNOWLEDGEMENT
This work was supported by a grant from the National Science Foundation.
References
- 1.Soyfer V.N. and Potaman,V.N. (1995) Triple-Helical Nucleic Acids. Springer, New York, NY.
- 2.Cooney M., Czernuszewicz,G., Postel,E.H., Flint,S.J. and Hogan,M.E. (1988) Site-specific oligonucleotide binding represses transcription of the human c-myc gene in vitro. Science, 241, 456–459. [DOI] [PubMed] [Google Scholar]
- 3.Postel E.H., Flint,S.J., Kessler,D.J. and Hogan,M.E. (1991) Evidence that a triplex-forming oligodeoxyribonucleotide binds to the c-myc promoter in HeLa cells, thereby reducing c-myc mRNA levels. Proc. Natl Acad. Sci. USA, 88, 8227–8231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Orson F.M., Thomas,D.W., McShan,W.M., Kessler,D.J. and Hogan,M.E. (1991) Oligonucleotide inhibition of IL2Rα mRNA transcription by promoter region collinear triplex formation in lymphocytes. Nucleic Acids Res., 19, 3435–3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vasquez K.M. and Wilson,J.H. (1998) Triplex-directed modification of genes and gene activities. Trends Biochem. Sci., 23, 4–9. [DOI] [PubMed] [Google Scholar]
- 6.Strobel S.A., Doucette-Stamm,L.A., Riba,L., Housman,D.E. and Dervan,P.B. (1991) Site-specific cleavage of human chromosome 4 mediated by triple-helix formation. Science, 254, 1639–1642. [DOI] [PubMed] [Google Scholar]
- 7.Gowers D.M. and Fox,K.R. (1999) Towards mixed sequence recognition by triple helix formation. Nucleic Acids Res., 27, 1568–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Griffin L.C. and Dervan,P.B. (1989) Recognition of thymine adenine base pairs by guanine in a pyrimidine triple helix motif. Science, 245, 967–971. [DOI] [PubMed] [Google Scholar]
- 9.Best G.C. and Dervan,P.B. (1995) Energetics of formation of sixteen triple helical complexes which vary at a single position within a pyrimidine motif. J. Am. Chem. Soc., 117, 1187–1193. [Google Scholar]
- 10.Wang E. and Feigon,J. (1999) Structures of nucleic acid triplexes. In Neidle,S. (ed.), Oxford Handbook of Nucleic Acid Structure. Oxford University Press, New York, NY, pp. 355–388.
- 11.Wang E., Malek,S. and Feigon,J. (1992) Structure of a G.T.A triplet in an intramolecular DNA triplex. Biochemistry, 31, 4838–4846. [DOI] [PubMed] [Google Scholar]
- 12.Radhakrishnan I. and Patel,D.J. (1994) Solution structure of a pyrimidine.purine.pyrimidine DNA triplex containing T.AT, C+.GC and G.TA triples. Structure, 2, 17–32. [DOI] [PubMed] [Google Scholar]
- 13.Roberts R.W. and Crothers,D.M. (1991) Specificity and stringency in DNA triplex formation. Proc. Natl Acad. Sci. USA, 88, 9397–9401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mergny J.-L., Sun,J.-S., Rougee,M., Montenay-Garestier,T., Barcelo,F., Chomilier,J. and Helene,C. (1991) Sequence specificity in triple-helix formation: experimental and theoretical studies of the effect of mismatches on triplex stability. Biochemistry, 30, 9791–9798. [DOI] [PubMed] [Google Scholar]
- 15.Kiessling L.L., Griffin,L.C. and Dervan,P.B. (1992) Flanking sequence effects within the pyrimidine triple-helix motif characterized by affinity cleaving. Biochemistry, 31, 2829–2834. [DOI] [PubMed] [Google Scholar]
- 16.Yoon K., Hobbs,C.A., Koch,J., Sardaro,M., Kutny,R. and Weis,A.L. (1992) Elucidation of the sequence-specific third-strand recognition of four Watson–Crick base pairs in a pyrimidine triple-helix motif: T.AT,C.GC,T.CG and G.TA. Proc. Natl Acad. Sci. USA, 89, 3840–3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Plum G.E., Pilch,D.S., Singleton,S.F. and Breslauer,K.J. (1995) Nucleic acid hybridization: triplex stability and energetics. Annu. Rev. Biophys. Biomol. Struct., 24, 319–350. [DOI] [PubMed] [Google Scholar]
- 18.Plum G.E. (1997) Thermodynamics of oligonucleotide triple helices. Biopolymers, 44, 241–256. [Google Scholar]
- 19.Powell S., Jiang,L. and Russu,I.M. (2001) Proton exchange and base-pair opening in a DNA triple helix. Biochemistry, 40, 11065–11072. [DOI] [PubMed] [Google Scholar]
- 20.Plateau P. and Gueron,M. (1982) Exchangeable proton NMR without base-line distortion, using new strong-pulse sequences. J. Am. Chem. Soc., 104, 7310–7311. [Google Scholar]
- 21.Ernst R.R., Bodenhausen,G. and Wokaun,A. (1987) Principles of Nuclear Magnetic Resonance in One and Two Dimensions. Clarendon Press, Oxford, UK.
- 22.Gueron M. and Leroy,J.L. (1995) Studies of base pair kinetics by NMR measurement of proton exchange. Methods Enzymol., 261, 383–413. [DOI] [PubMed] [Google Scholar]
- 23.Englander S.W. and Kallenbach,N.R. (1984) Hydrogen exchange and structural dynamics of proteins and nucleic acids. Q. Rev. Biophys., 16, 521–655. [DOI] [PubMed] [Google Scholar]
- 24.Macaya R., Wang,E., Schultze,P., Sklenar,V. and Feigon,J. (1992) Proton nuclear magnetic resonance assignments and structural characterization of an intramolecular DNA triplex. J. Mol. Biol., 225, 755–773. [DOI] [PubMed] [Google Scholar]
- 25.Nonin S., Leroy,J.-L. and Gueron,M. (1996) Acid-induced exchange of the imino proton in GC pairs. Nucleic Acids Res., 24, 586–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Belotserkovskii B.P., Veselkov,A.G., Filippov,S.A., Dobrynin,V.N., Mirkin,S.M. and Frank-Kamenetskii,M.D. (1990) Formation of intramolecular triplex in homopurine-homopyrimidine mirror repeats with point substitutions. Nucleic Acids Res., 18, 6621–6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mirkin S.M. and Frank-Kamenetskii,M.D. (1994) H-DNA and related structures. Annu. Rev. Biophys. Biomol. Struct., 23, 541–576. [DOI] [PubMed] [Google Scholar]