Abstract
Monoclonal antibodies and antibody fragments have recently been developed for use in diverse diagnostic and therapeutic applications. Insect cells can efficiently secrete recombinant proteins such as antibody molecules through post-translational processing and modifications that are similar to those performed in mammalian cells. In eukaryotic cells, the signal sequence in a nascent polypeptide is recognized by the signal recognition particle, and the polypeptide is then folded and modified in the endoplasmic reticulum. The signal sequence consists of three regions, a positively charged N-terminus, a hydrophobic core, and a polar C-terminus. In the present study, we examined the substitutions of the characteristic amino acids of a Drosophila immunoglobulin heavy chain binding protein signal sequence, and investigated the effect on the secretory production of an antibody Fab fragment from lepidopteran insect cells in transient expression. A modification of the signal sequence for the heavy chain resulted in a twofold increase in the secreted Fab fragment, while the modification for the light chain led to a more than 3.6-fold increase.
Electronic supplementary material
The online version of this article (doi:10.1007/s10616-017-0109-0) contains supplementary material, which is available to authorized users.
Keywords: Signal sequence, Secretory production, Antibody, Fab fragment, Insect cell, Recombinant protein production
Introduction
The production of recombinant antibody molecules has improved because monoclonal antibodies and antibody fragments have recently been developed for use in diverse diagnostic and therapeutic applications. The most common host cells to express antibodies are mammalian cells such as CHO cells (Kim et al. 2012; Mohan et al. 2008; Omasa et al. 2010) and 293T cells (Menzel et al. 2008; Puttikhunt et al. 2008). Insect cells can also be used for the production of recombinant antibodies (Furuta et al. 2010; Gilmartin et al. 2012; Sonoda et al. 2012; Yamaji 2011; Yamaji et al. 2008). The advantages of insect cells include the following: ease of handling compared with mammalian cells; CO2 supplementation in the culture atmosphere is not required; insect cells can be grown to a high density in suspension with a serum-free medium; and, the yield of recombinant protein with insect cells is often higher than that with mammalian cells (Drugmand et al. 2012; Yamaji 2011). In addition, the post-translational processing and modifications performed in insect cells are similar to those in mammalian cells (Luckow 1995).
When synthesis of a secretory protein begins on the ribosomes in the cytosol of eukaryotic cells, an endoplasmic reticulum (ER) signal sequence located at the N-terminus of the nascent protein directs the ribosome to the ER membrane. The ER signal sequence of the nascent protein is recognized by the signal recognition particle (SRP), and the growing polypeptide is translocated across the ER membrane. The signal sequence consists of three regions: a hydrophilic N-terminal region that usually contains positively charged amino acid residues, a hydrophobic core region, and a C-terminal region with a cleavage site for a signal peptidase that commonly contains polar amino acid residues. The modification of signal sequences has been extensively examined in microbial cells such as Escherichia coli and yeast (Gennity et al. 1990; Jonet et al. 2012; Klatt and Konthur 2012; Rakestraw et al. 2009). Reportedly, the substitution of amino acid residues in signal sequences for translocation to the periplasm remarkably affects the yield of secreted proteins from E. coli (Gennity et al. 1990). In contrast, only a few studies have reported on signal sequence modification for recombinant protein secretion in mammalian and insect cells (Futatsumori-Sugai and Tsumoto 2010; Haryadi et al. 2015; Tsuchiya et al. 2004, 2005).
Previously, we reported that a Drosophila immunoglobulin heavy chain binding protein (BiP) signal sequence was available for efficient secretory production of recombinant antibody molecules from lepidopteran insect cells (Sonoda et al. 2012; Yamaji et al. 2008). In the present study, we examined the changes of the characteristic amino acids in each region of the BiP signal sequence and the effects that these changes can exert on the secretory production of an antibody Fab fragment from insect cells in transient expression.
Materials and methods
Materials
All reagents were of the highest grade available and were purchased from Nacalai Tesque (Kyoto, Japan) unless otherwise indicated.
Plasmid construction
The transient expression of the Fab fragment of 3A21 mouse anti-bovine RNaseA (Katakura et al. 1996) was examined using the expression vector pIHAneo (Yamaji et al. 2008). The pIHAneo contained the Bombyx mori actin promoter downstream of the B. mori nucleopolyhedrovirus (BmNPV) IE-1 transactivator and the BmNPV HR3 enhancer for high-level expression. A Drosophila BiP signal sequence (Yamaji et al. 2008) was employed upstream of the heavy chain (Hc) and light chain (Lc) genes of the 3A21 Fab. The primers (Eurofins Genomics, Tokyo, Japan; or Life technologies, Tokyo, Japan) used for the plasmid construction are shown in Table S1. Plasmid inserts were confirmed by DNA sequencing.
The numbers contained in the primer names shown in Table S1 are correlated with the names of the modified signal sequences in Table 1. For example, the primer ss1for was used for amplifications via PCR of the Hc gene with the modified signal sequence referred to as Hss1 and the Lc gene with the modified signal sequence referred to as Lss1; and, the primer ssH25/26/27for was used for amplification of the Hc genes with the modified signal sequences Hss25, Hss26, and Hss27.
Table 1.
Amino acid sequences of a modified Drosophhila BiP signal peptide
| Sequence | |
|---|---|
| HssWT, LssWT (native sequence) |
MKLCILLAVVAFVGLSLG |
| Hss1, Lss1 | MKKLCILLAVVAFVGLSLG |
| Hss2, Lss2 | MRRLCILLAVVAFVGLSLG |
| Hss3, Lss3 | MKKKKLCILLAVVAFVGLSLG |
| Hss4, Lss4 | MRRRRLCILLAVVAFVGLSLG |
| Hss5, Lss5 | MKRKRLCILLAVVAFVGLSLG |
| Hss6, Lss6 | MRLCILLAVVAFVGLSLG |
| Hss7, Lss7 | MKRLCILLAVVAFVGLSLG |
| Hss8, Lss8 | MRKLCILLAVVAFVGLSLG |
| Lss9 | MRKRLCILLAVVAFVGLSLG |
| Hss10, Lss10 | MKRKLCILLAVVAFVGLSLG |
| Hss11, Lss11 | MRKRRLCILLAVVAFVGLSLG |
| Hss12, Lss12 | MELCILLAVVAFVGLSLG |
| Lss12/27 | MELCILLAVVAFVGLHSLG |
| Hss13, Lss13 | MDLCILLAVVAFVGLSLG |
| Lss14 | MKLCILLVGLSLG |
| Hss15, Lss15 | MKLCILLAVVAFLVLVGLSLG |
| Hss16, Lss16 | MKLCILVAFVGLSLG |
| Hss17, Lss17 | MKLCILLAVLAVVAFVGLSLG |
| Hss18, Lss18 | MKLCILLAVVVGLSLG |
| Lss19 | MKLCILLAVVVVLAFVGLSLG |
| Hss20, Lss20 | MKLCILLVAFVGLSLG |
| Hss21, Lss21 | MKLCILLAAFVGLSLG |
| Hss22, Lss22 | MKLCILLAVVAFVGLTLG |
| Lss23 | MKLCILLAVVAFVGLNLG |
| Lss24 | MKLCILLAVVAFVGLQLG |
| Hss25, Lss25 | MKLCILLAVVAFVGLQCLG |
| Hss26, Lss26 | MKLCILLAVVAFVGLHCLG |
| Hss27, Lss27 | MKLCILLAVVAFVGLHSLG |
| Lss28 | MKLCILLAVVAFVGLQSLG |
| Hss29 | MKLCILLAVVAFVGLKLG |
| Hss30 | MKLCILLAVVAFVGLDLG |
| Hss31 | MKLCILLAVVAFVGLPLG |
| Hss32 | MKLCILLAVVAFVGLALG |
Altered amino acid residues are underlined. Mw 1600–2400
The DNA fragments encoding each of the modified signal sequences ranging from Hss1 to Hss13 and the 3A21 Hc gene were amplified via PCR using pIHAneo/Hc/myc (Ohmuro-Matsuyama et al. 2016) as a template. Each of the primers ranging from ss1for to ss12/13for was used as the forward primer, while pIHAneorev served as the reverse primer. Each amplified fragment was digested with SacII (New England Biolabs, Ipswich, MA, USA) and XbaI (New England Biolabs) and inserted into pIHAneo/Hc/myc between the SacII and XbaI sites.
To amplify the Hc gene downstream of each of the modified signal sequences of Hss14–Hss18, Hss20–22, Hss25–Hss27, and Hss29–Hss32, PCR was first performed using pIHAneo/Hc/myc as a template with pIHAneofor as the forward primer and each of the primers ss14rev, ss15rev, ss16/17rev, ss18/19rev, ss20/21rev, and ss22/23/24/25/26/27/28/29/30/31/32rev as the reverse primer; or, using pIHAneo/Hc/myc as a template with each of the forward primers of ss14/15for, ss16for, ss17for, and ssH22/29/30/31/32for and pIHAneorev as the reverse primer. The overlap extension PCR was then performed using two PCR-amplified fragments as the templates with a pair of primers: pIHAneofor and pIHAneorev. The following procedure was the same as that for the Hc gene with Hss1–Hss13.
The DNA fragments encoding each of the modified signal sequences ranging from Lss1 to Lss13 and the Lc gene were each amplified via PCR using pIHAneo/Lc/His (Ohmuro-Matsuyama et al. 2016) as a template, along with each of the forward primers ranging from ss1for to ss13for and pIHAneorev as the reverse primer. Each amplified fragment was digested with SacII and XbaI and inserted into pIHAneo/Lc/His between the SacII and XbaI sites.
To amplify the Lc gene downstream of each of the modified signal sequences of Lss14–Lss28, PCR was first performed using pIHAneo/Lc/His as a template with pIHAneofor as the forward primer, and each of ss14rev, ss15rev, ss16/17rev, ss18/19rev, ss20/21rev, and ss22/23/24/25/26/27/28/29/30/31/32rev as the reverse primer; or, using pIHAneo/Lc/His as a template with each of ss14/15for, ss16for–ss21for, ssLfor22–ss24for, ssL25/26for, and ssL27/28for as the forward primer and pIHAneorev as the reverse primer. The overlap extension PCR was then performed using two PCR-amplified fragments as the templates, pIHAneofor as the forward primer, and pIHAneorev as the reverse primer. The following procedure was the same as that for the Lc gene with Lss1–Lss13.
The modified signal sequence referred to as Lss12/27 and the Lc gene was amplified using Lss27 and the Lc gene as a template with ss12 as the forward primer and pIHAneorev as the reverse primer. The amplified fragment was digested with SacII and XbaI and inserted into pIHAneo/Lc/His between the SacII and XbaI sites.
Cell culture and transfection
Trichoplusia ni BTI-TN-5B1-4 (High Five) cells (Life Technologies) were maintained at 27 °C in a serum-free medium (Express Five SFM; Life technologies), as described previously (Furuta et al. 2010). Cells were inoculated at 8 × 104 cells/well in 24-well cell culture plates. For cotransfection with the Hc and Lc genes, the plasmids of the Hc (450 ng/well) and Lc (450 ng/well) genes were added to polyethyleneimine “Max” (Mw 40,000; Polysciences, Warrington, PA, USA) (1.8 μg/well) in 150 mM of NaCl and incubated for 5 min. The mixture (20 μl/well) was added to the cells 45 min after the cell inoculation. The supernatants were collected three days after the transfection. Each supernatant from triplicate wells was analyzed by enzyme-linked immunosorbent assay (ELISA). For western blotting, equal amounts of each supernatant were mixed and applied. Each transfection experiment was repeated over twice, and a reproducible trend was observed on repeated runs.
ELISA
Culture supernatants were analyzed by ELISA to identify any Fab fragments with antigen-binding activity, as previously described (Furuta et al. 2010). ELISA plates were coated with bovine RNaseA (Sigma-Aldrich, St. Louis, MO, USA) as the antigen, and peroxidase-conjugated goat anti-mouse IgG (Promega, Madison, WI, USA) was used. The detections were carried out using the ELISA POD substrate TMB kit (Nacalai Tesque) according to the manufacturer’s protocol. The culture supernatant obtained by cotransfection with HssWT and LssWT was diluted 100–800 times and used for ELISA. The supernatants obtained by cotransfection with the native signal sequence and each modified signal sequence were diluted 400 times and used for ELISA. From the fitted curve between the dilution ratio and the absorbance, the binding activity in the supernatant obtained by cotransfection with the native signal sequence and each modified signal sequence was calculated.
Western blot analysis
Equal amounts of the culture supernatants from triplicate wells were mixed and were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) using a 12.5% gel under non-reducing conditions. A dry transfer was conducted using the iBlot 2 dry blotting system (Life Technologies) according to the manufacturer’s protocol. The following reaction was performed with anti-mouse IgG (H&L) AP conjugate (Promega) using the SNAP i.d. 2.0 protein detection system (Merck Millipore, Darmstadt, Germany). The detection was carried out with the BCIP/NBT color development substrate (Promega) according to the manufacturer’s protocol.
Results and discussion
Design of modified signal sequences
A Drosophhila BiP signal sequence was modified, and modified signal sequences were used upstream of the Hc and Lc genes of an Fab fragment of 3A21 mouse anti-bovine RNaseA. Twenty-six types of modified BiP signal sequences were designed for the Hc gene, while 29 types of modified BiP signal sequences were designed for the Lc gene (Table 1).
In ss1–ss13, the positively charged amino acid in the N-terminal region of the BiP signal sequence, lysine, was altered. In ss1–ss11, the native lysine was substituted with arginine and/or lysine. In ss12 and ss13, lysine was replaced with a negatively charged amino acid, aspartic acid and glutamic acid, respectively.
In ss14–ss21, the length of the hydrophobic core in the BiP signal sequence was changed. The leucine count was maintained at more than 3, because reportedly a leucine-rich sequence is necessary for the association between a signal sequence and an SRP (Keenan et al. 1998; Ng et al. 1996; Rothe and Lehle 1998; Zheng and Nicchitta 1999).
In ss22–ss31, the polar amino acid in the C-terminal region, serine, was replaced. Since serine is a neutral amino acid, in ss22–ss24 it was substituted with another neutral polar amino acid. For ss25–ss28, a polar amino acid sequence was employed that was reported to be the sequence contained in the C-terminal region of a signal sequence of human antibody (Haryadi et al. 2015). In ss29–ss31, the native serine was substituted with a nonpolar amino acid.
Improved secretion of Fab fragments using modified signal sequences
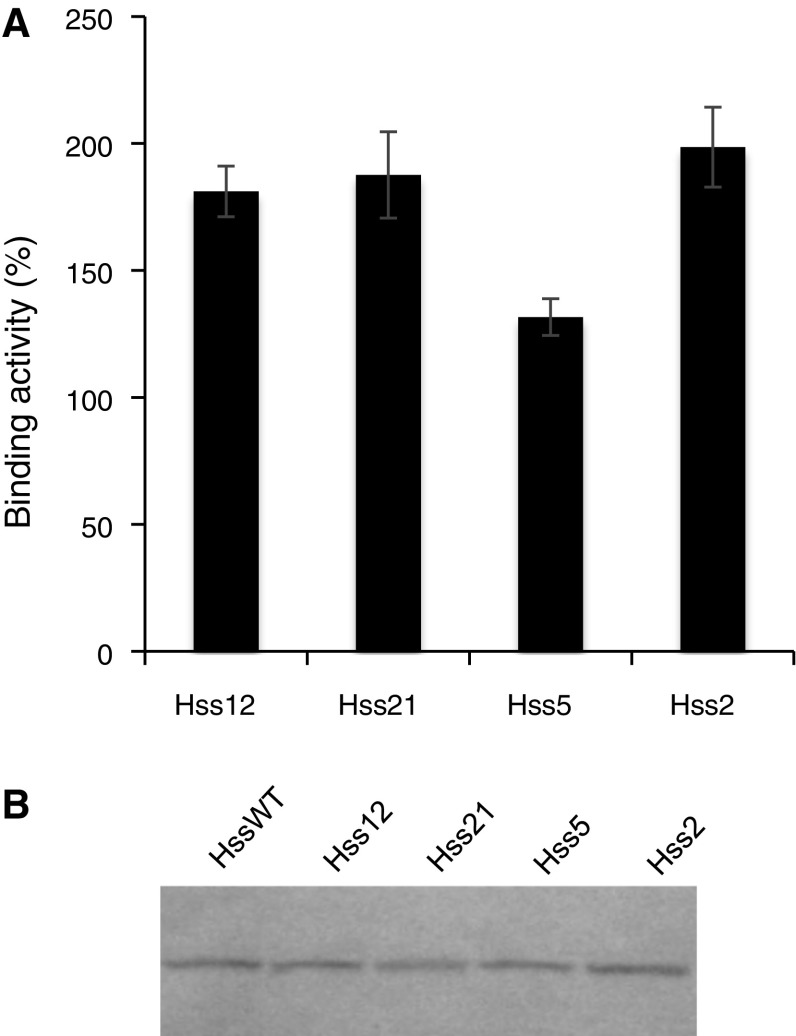
High Five cells were cotransfected with the Hc gene downstream of each of the modified signal sequences and the Lc gene downstream of the native BiP signal sequence. The antigen-binding activity of the Fab fragment secreted in the culture supernatant was measured via ELISA. The use of modified signal sequences with the Hc gene led to varied binding activities. When the modified signal sequence Hss2, Hss5, Hss12, and Hss21 was used, the binding activity significantly increased, along with the maximum binding activity that exhibited a twofold increase (Fig. 1a).
Fig. 1.
Improved secretion of an Fab fragment by using a modified BiP signal sequence upstream of the heavy chain (Hc) gene. a Antigen-binding activity of an Fab fragment, as detected by enzyme-linked immunosorbent assay (ELISA). The binding activity obtained with the native BiP signal sequence was indicated as 100%. Error bar = 1 SD (n = 3). b Fab fragment (Mw 50,300), as detected by western blotting
Western blotting under non-reducing conditions showed that each modified signal sequence was removed by the signal peptidase in the ER, and the Hc without the signal peptide was secreted (Fig. 1b). When Hss21 was used, a lower quantity of the Fab fragment was observed, as shown in Fig. 1b, although the binding activity was increased, as shown in Fig. 1a. This could have been because either the modification decreased the expression level of the nascent polypeptide, or a polypeptide with a different signal sequence was carried into a degradation pathway because the signal peptidase plays the role of ER-associated degradation. This result could also indicate that the use of Hss21 yielded correctly folded Fab fragments, and therefore the modification of the signal sequence might improve both the secreted amount and the quality of the antibody. In addition, a DNA sequence in the 5′-terminal region affects the mRNA structure (Goodman et al. 2013; Kudla et al. 2009) and translation elongation (Ban et al. 2000; Cannarozzi et al. 2010; Tuller et al. 2010; Zhang et al. 2009). Hence, modifications of the signal sequence might have influenced folding of the nascent polypeptide.
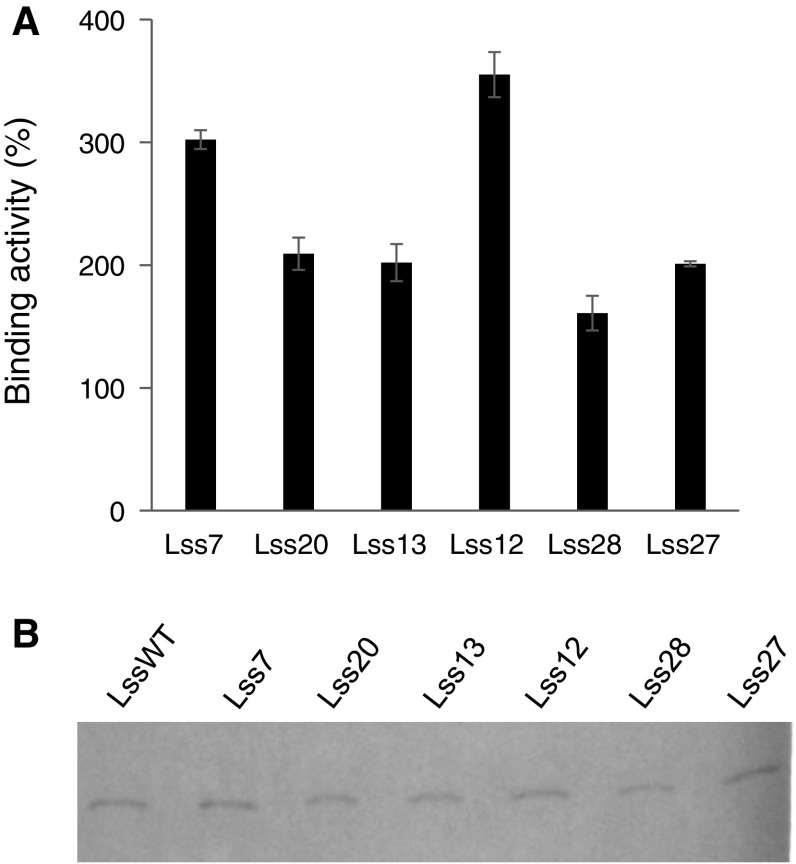
The use of modified signal sequences with the Lc gene also resulted in different binding activities. When the modified signal sequence, Lss7, Lss12, Lss13, Lss20, Lss27, and Lss28, was used with the Lc gene, the binding activity was remarkably improved (Fig. 2a). The maximum binding activity showed a 3.6-fold increase. Western blotting also confirmed that the modified signal sequences were removed (Fig. 2b).
Fig. 2.
Improved secretion of an Fab fragment via a modified BiP signal sequence upstream of the light chain (Lc) gene. a Antigen-binding activity of an Fab fragment, as detected by ELISA. The binding activity obtained with the native BiP signal sequence was indicated as 100%. Error bar = 1 SD (n = 3). b Fab fragment (Mw 50,300), as detected by western blotting
In Hss12, Lss12, and Lss13, each of which increased the secretory production of Fab, the native lysine in the N-terminal region was substituted by either glutamic acid or aspartic acid, although common signal sequences contain a positively charged amino acid rather than a negatively charged amino acid. Interestingly, this is in accord with a recent report showing that some ER signal sequences for human antibodies contained aspartic acid and glutamic acid but not a positively charged amino acid in the N-terminal regions, and that the signal sequences increase the secretory production of other human antibodies (Haryadi et al. 2015). The increased negative charge might prevent protein aggregation and enhance soluble expression as previously reported (Joshi et al. 2012; Kvam et al. 2010).
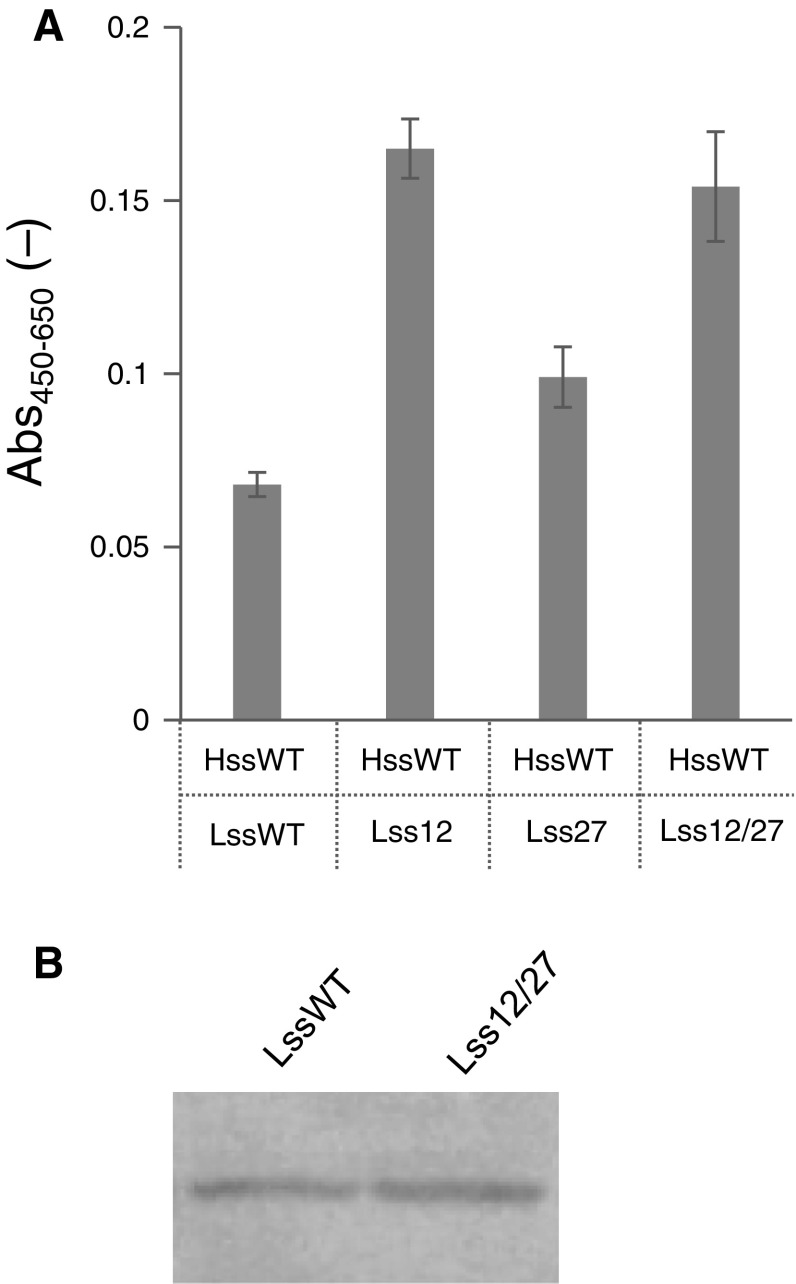
Effect of multiple modifications
The modifications in Lss12 and Lss27 were combined, and a doubly modified signal sequence referred to as Lss12/27 was prepared (Table 1). Lss12/27 showed no synergistic effect on the secretory production of the Fab fragment, although Lss12 and Lss27 both showed increased production (Fig. 3a).
Fig. 3.
Effect of double modifications in the BiP signal sequence upstream of the Lc gene. a Antigen-binding activity of an Fab fragment, as detected by ELISA. Error bar = 1 SD (n = 3). b Fab fragment (Mw 50,300), as detected by western blotting
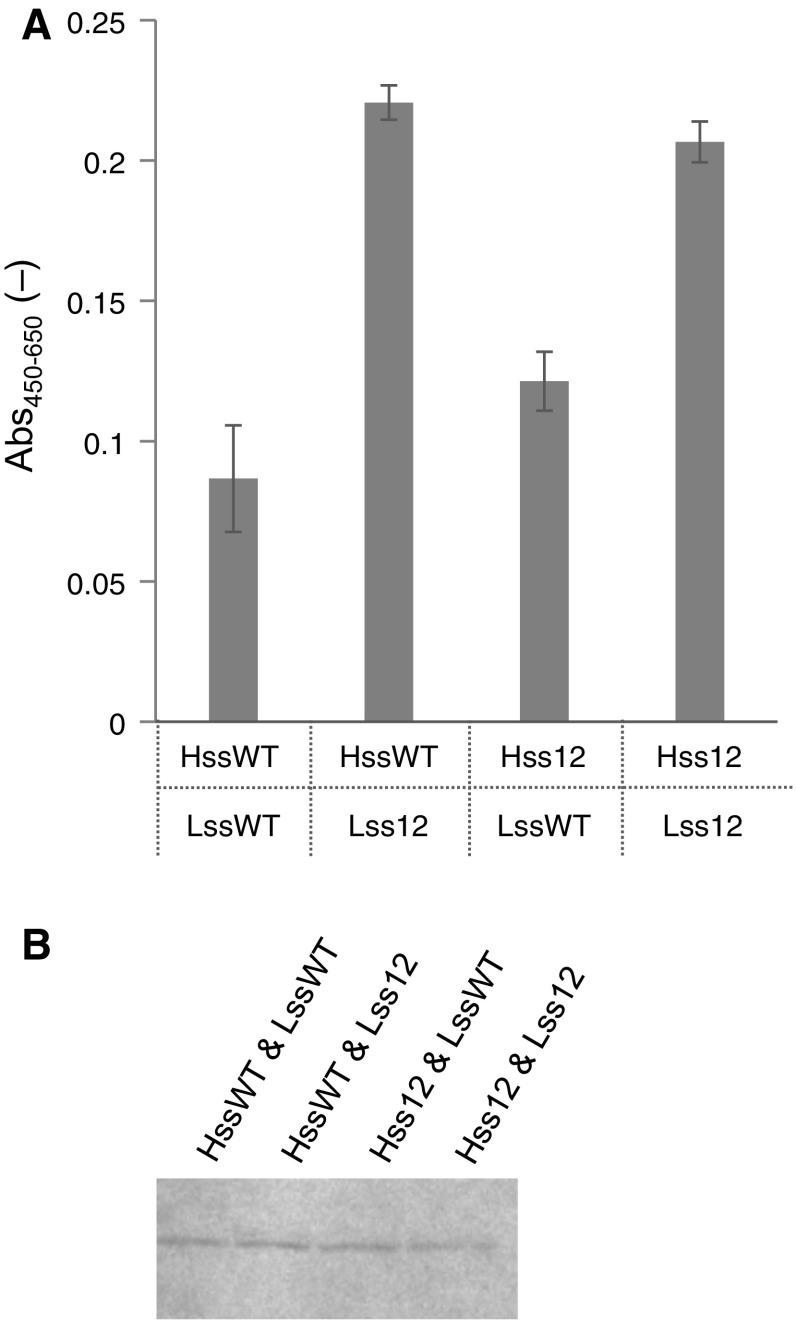
As described above, Hss12 and Lss12, respectively, increased the secretory production of Fab. When Hss12 and Lss12 were simultaneously used with the Hc and Lc genes, respectively, they did not improve the secretory production (Fig. 4a). It remains unclear why additive effects were not obtained in these examinations, and this should be elucidated. In both cases, western blotting showed that each modified signal sequence was removed (Figs. 3b, 4b).
Fig. 4.
Effect of the simultaneous use of Hss12 and Lss12. a Antigen-binding activity of an Fab fragment, as detected by ELISA. Error bar = 1 SD (n = 3). b Fab fragment (Mw 50,300), as detected by western blotting
In conclusion, the modification of a signal sequence is highly effective in promoting efficient secretory production of recombinant proteins including antibodies using cultured cells. After selection of the best signal sequence for the production of a target protein in each cell system (Kober et al. 2013), modification of the signal sequence could further improve both the yield and the quality of the secreted protein.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This research was partially supported by the programs for developing key technologies for discovering and manufacturing pharmaceuticals used for next-generation treatments and diagnoses both from the Ministry of Economy, Trade and Industry, Japan (METI) and from the Japan Agency for Medical Research and Development (AMED).
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s10616-017-0109-0) contains supplementary material, which is available to authorized users.
References
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- Cannarozzi G, Schraudolph NN, Faty M, von Rohr P, Friberg MT, Roth AC, Gonnet P, Gonnet G, Barral Y. A role for codon order in translation dynamics. Cell. 2010;141:355–367. doi: 10.1016/j.cell.2010.02.036. [DOI] [PubMed] [Google Scholar]
- Drugmand J-C, Schneider Y-J, Agathos SN. Insect cells as factories for biomanufacturing. Biotechnol Adv. 2012;30:1140–1157. doi: 10.1016/j.biotechadv.2011.09.014. [DOI] [PubMed] [Google Scholar]
- Furuta T, Ogawa T, Katsuda T, Fuji I, Yamaji H. Efficient production of an antibody fragment using the baculovirus–insect cell system. J Biosci Bioeng. 2010;110:577–581. doi: 10.1016/j.jbiosc.2010.06.001. [DOI] [PubMed] [Google Scholar]
- Futatsumori-Sugai M, Tsumoto K. Signal peptide design for improving recombinant protein secretion in the baculovirus expression vector system. Biochem Biophys Res Commun. 2010;391:931–935. doi: 10.1016/j.bbrc.2009.11.167. [DOI] [PubMed] [Google Scholar]
- Gennity J, Goldstein J, Inouye M. Signal peptide mutants of Escherichia coli. J Bioenerg Biomembr. 1990;22:233–269. doi: 10.1007/BF00763167. [DOI] [PubMed] [Google Scholar]
- Gilmartin AA, Lamp B, Rumenapf T, Persson MA, Rey FA, Krey T. High-level secretion of recombinant monomeric murine and human single-chain Fv antibodies from Drosophila S2 cells. Protein Eng Des Sel. 2012;25:59–66. doi: 10.1093/protein/gzr058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman DB, Church GM, Kosuri S. Causes and effects of N-terminal codon bias in bacterial genes. Science. 2013;342:475–479. doi: 10.1126/science.1241934. [DOI] [PubMed] [Google Scholar]
- Haryadi R, Ho S, Kok YJ, Pu HX, Zheng L, Pereira NA, Li B, Bi X, Goh LT, Yang Y, Song Z. Optimization of heavy chain and light chain signal peptides for high level expression of therapeutic antibodies in CHO cells. PLoS ONE. 2015;10:e0116878. doi: 10.1371/journal.pone.0116878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonet MA, Mahadi NM, Murad AMA, Rabu A, Bakar FDA, Rahim RA, Low KO, Illias RM. Optimization of a heterologous signal peptide by site-directed mutagenesis for improved secretion of recombinant proteins in Escherichia coli. J Mol Microbiol Biotechnol. 2012;22:48–58. doi: 10.1159/000336524. [DOI] [PubMed] [Google Scholar]
- Joshi SN, Butler DC, Messer A. Fusion to a highly charged proteasomal retargeting sequence increases soluble cytoplasmic expression and efficacy of diverse anti-synuclein intrabodies. MAbs. 2012;4:686–693. doi: 10.4161/mabs.21696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katakura Y, Kobayashi E, Kurokawa Y, Omasa T, Fujiyama K, Suga KI. Cloning of cDNA and characterization of anti-RNase A monoclonal antibody 3A21. J Ferment Bioeng. 1996;82:312–314. doi: 10.1016/0922-338X(96)88826-X. [DOI] [Google Scholar]
- Keenan RJ, Freymann DM, Walter P, Stroud RM. Crystal structure of the signal sequence binding subunit of the signal recognition particle. Cell. 1998;94:181–191. doi: 10.1016/S0092-8674(00)81418-X. [DOI] [PubMed] [Google Scholar]
- Kim JY, Kim YG, Lee GM. CHO cells in biotechnology for production of recombinant proteins: current state and further potential. Appl Microbiol Biotechnol. 2012;93:917–930. doi: 10.1007/s00253-011-3758-5. [DOI] [PubMed] [Google Scholar]
- Klatt S, Konthur Z. Secretory signal peptide modification for optimized antibody-fragment expression-secretion in Leishmania tarentolae. Microb Cell Fact. 2012;11:97. doi: 10.1186/1475-2859-11-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kober L, Zehe C, Bode J. Optimized signal peptides for the development of high expressing CHO cell lines. Biotechnol Bioeng. 2013;110:1164–1173. doi: 10.1002/bit.24776. [DOI] [PubMed] [Google Scholar]
- Kudla G, Murray AW, Tollervey D, Plotkin JB. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324:255–258. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvam E, Sierks MR, Shoemaker CB, Messer A. Physico-chemical determinants of soluble intrabody expression in mammalian cell cytoplasm. Protein Eng Des Sel. 2010;23:489–498. doi: 10.1093/protein/gzq022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckow VA. Protein production and processing from baculovirus expression vectors. In: Shuler ML, Wood HA, Granados RR, Hammer DA, editors. Baculovirus expression systems and biopesticides. New York: Wiley; 1995. pp. 51–90. [Google Scholar]
- Menzel C, Schirrmann T, Konthur Z, Jostock T, Dubel S. Human antibody RNase fusion protein targeting CD30 + lymphomas. Blood. 2008;111:3830–3837. doi: 10.1182/blood-2007-04-082768. [DOI] [PubMed] [Google Scholar]
- Mohan C, Kim YG, Koo J, Lee GM. Assessment of cell engineering strategies for improved therapeutic protein production in CHO cells. Biotechnol J. 2008;3:624–630. doi: 10.1002/biot.200700249. [DOI] [PubMed] [Google Scholar]
- Ng DT, Brown JD, Walter P. Signal sequences specify the targeting route to the endoplasmic reticulum membrane. J Cell Biol. 1996;134:269–278. doi: 10.1083/jcb.134.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmuro-Matsuyama Y, Mori K, Hamada H, Ueda H, Yamaji H. Electrostatic engineering of the interface between heavy and light chains promotes antibody Fab fragment production. Cytotechnology. 2016 doi: 10.1007/s10616-016-9955-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omasa T, Onitsuka M, Kim WD. Cell engineering and cultivation of Chinese hamster ovary (CHO) cells. Curr Pharm Biotechnol. 2010;11:233–240. doi: 10.2174/138920110791111960. [DOI] [PubMed] [Google Scholar]
- Puttikhunt C, Keelapang P, Khemnu N, Sittisombut N, Kasinrerk W, Malasit P. Novel anti-dengue monoclonal antibody recognizing conformational structure of the prM-E heterodimeric complex of dengue virus. J Med Virol. 2008;80:125–133. doi: 10.1002/jmv.21047. [DOI] [PubMed] [Google Scholar]
- Rakestraw JA, Sazinsky SL, Piatesi A, Antipov E, Wittrup KD. Directed evolution of a secretory leader for the improved expression of heterologous proteins and full-length antibodies in Saccharomyces cerevisiae. Biotechnol Bioeng. 2009;103:1192–1201. doi: 10.1002/bit.22338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothe C, Lehle L. Sorting of invertase signal peptide mutants in yeast dependent and independent on the signal-recognition particle. Eur J Biochem. 1998;252:16–24. doi: 10.1046/j.1432-1327.1998.2520016.x. [DOI] [PubMed] [Google Scholar]
- Sonoda H, Kumada Y, Katsuda T, Yamaji H. Production of single-chain Fv–Fc fusion protein in stably transformed insect cells. Biochem Eng J. 2012;67:77–83. doi: 10.1016/j.bej.2012.05.005. [DOI] [Google Scholar]
- Tsuchiya Y, Morioka K, Shirai J, Yoshida K. Structural requirements of signal peptide in insect cell. Nucleic Acids Symp Ser Oxf. 2004;48:181–182. doi: 10.1093/nass/48.1.181. [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y, Morioka K, Taneda I, Shirai J, Yoshida K. Gene design of signal sequence for the effective secretion of recombinant protein using insect cell. Nucleic Acids Symp Ser Oxf. 2005;49:305–306. doi: 10.1093/nass/49.1.305. [DOI] [PubMed] [Google Scholar]
- Tuller T, Carmi A, Vestsigian K, Navon S, Dorfan Y, Zaborske J, Pan T, Dahan O, Furman I, Pilpel Y. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell. 2010;141:344–354. doi: 10.1016/j.cell.2010.03.031. [DOI] [PubMed] [Google Scholar]
- Yamaji H. Production of antibody in insect cells. In: Al-Rubeai M, editor. Antibody expression and production. Cell engineering. Dordrecht: Springer Science + Business Media; 2011. pp. 53–76. [Google Scholar]
- Yamaji H, Manabe T, Watakabe K, Muraoka M, Fujii I, Fukuda H. Production of functional antibody Fab fragment by recombinant insect cells. Biochem Eng J. 2008;41:203–209. doi: 10.1016/j.bej.2008.04.017. [DOI] [Google Scholar]
- Zhang G, Hubalewska M, Ignatova Z. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat Struct Mol Biol. 2009;16:274–280. doi: 10.1038/nsmb.1554. [DOI] [PubMed] [Google Scholar]
- Zheng T, Nicchitta CV. Structural determinants for signal sequence function in the mammalian endoplasmic reticulum. J Biol Chem. 1999;274:36623–36630. doi: 10.1074/jbc.274.51.36623. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.