Abstract
Small looped mispairs are efficiently corrected by mismatch repair. The situation with larger loops is less clear. Repair activity on large loops has been reported as anywhere from very low to quite efficient. There is also uncertainty about how many loop repair activities exist and whether any are conserved. To help address these issues, we studied large loop repair in Saccharomyces cerevisiae using in vivo and in vitro assays. Transformation of heteroduplexes containing 1, 16 or 38 nt loops led to >90% repair for all three substrates. Repair of the 38 base loop occurred independently of mutations in key genes for mismatch repair (MR) and nucleotide excision repair (NER), unlike other reported loop repair functions in yeast. Correction of the 16 base loop was mostly independent of MR, indicating that large loop repair predominates for this size heterology. Similarities between mammalian and yeast large loop repair were suggested by the inhibitory effects of loop secondary structure and by the role of defined nicks on the relative proportions of loop removal and loop retention products. These observations indicate a robust large loop repair pathway in yeast, distinct from MR and NER, and conserved in mammals.
INTRODUCTION
DNA mismatch repair (MR) is a powerful mutation avoidance pathway for base pair substitution and small insertion/deletion mutations (reviewed in 1–4). Highly conserved from bacteria to humans, MR acts on base–base mispairs and small loops, ranging up to 8–13 nt in eukaryotes (5–7). MR action eliminates these pre-mutagenic lesions before they can be fixed as mutations. Cells lacking MR exhibit mutation rates that are typically 100–1000 times higher than normal, indicating that MR is a major activity for eliminating mispairs and small loops.
The correction of large looped mispairs presents a more complicated picture. Large loops are defined here as containing 15 nt or more and thus they are probably outside the realm of MR. In prokaryotes, there is little (8,9) or no detectable (10–12) processing of large loops. In yeast, some experiments also suggest poor processing of large loops (6,13–15). Other reports indicate three different loop repair activities in yeast that are distinguishable by their genetic requirements. One activity corrects loops of 26–30 nt during meiotic recombination and requires the MR gene MSH2 and the nucleotide excision repair (NER) gene RAD1 (16). A second pathway (17) functions on very large loops (>2 kb) in mitotic recombination, and activity is reduced by mutations in the MR genes MSH2 or PMS1. A third activity, requiring MSH3 and RAD1, prevents 92 bp deletions that result from replicational slippage errors (18,19). It has been suggested (15,20) that yeast loop processing activities might be of low capacity and thus easily saturable under conditions where excess loops are created, such as in certain replication mutant strains (15,20–24). Alternatively, some of these mutants could be deficient in loop repair activity.
There is also evidence of multiple loop repair pathways in mammalian cells. Transfection studies demonstrated efficient correction of heteroduplexes with loops up to 283 nt (25,26). Repair in this system can either remove the loop or retain it, in proportions of about 2:1 (25). Similar conclusions about the effectiveness of large loop repair (LLR) in mammalian cells have been drawn from recombination experiments (27–29), although the loops tested were smaller. Biochemical experiments using extracts of human cells also showed active repair of loops of 12–216 nt (5,30). Repair in vitro is strongly nick-dependent, with most repair occurring on the nicked strand, regardless of which strand contained the loop (30). This activity is independent of MR and NER genes. It is possible that the loop repair activities seen by transfection and in vitro are due to the same pathway. A third loop repair function was observed during mitotic recombination in rodent cells (31). Correction by this activity is unusual in that both palindromic and non-palindromic sequences are efficiently repaired, suggesting that secondary structure associated with palindromic sequences can be overcome. In most other situations, secondary structure within the loop inhibits repair (27,32–35).
In contrast to other loop repair activities in yeast, we showed previously (36) that yeast nuclear extracts support an efficient LLR activity. This repair pathway functions on loops of 16–216 nt and there is full activity in extracts from msh2 msh3 and mlh1 pms1 mutant strains, although the role of the NER gene RAD1 was not clearly determined. Our yeast LLR system appears to mirror activities seen in mammalian cell culture (25,26) and in extracts of human cells (30), judging by the high efficiency of repair, the loop sizes that are corrected and the independence of MR.
The biochemical results with yeast LLR encouraged us to examine this activity in vivo. We show in this study that LLR is very active in yeast, approaching 100% efficiency in transformation experiments. Tests with mutant strains demonstrated that LLR in vivo is independent of MR and NER genes, proving that LLR is distinct from the other yeast loop repair systems (16–19). We also found that secondary structure in the loop inhibits LLR both in vivo and in vitro, suggesting a difference from a palindrome processing activity found in rodent cells (31). In addition to the in vivo experiments, we report new biochemical experiments showing preferential nick-directed repair when the nick is located 5′ to the loop, compared to a 3′ nicked substrate. Together, these experiments provide new genetic and biochemical evidence for a robust, conserved large loop processing pathway among eukaryotes.
MATERIALS AND METHODS
Reagents and enzymes
Standard reagents, including molecular biology grade CsCl and low melting point agarose, were obtained from Sigma. Hydroxyapatite resin was a product of Bio-Rad. Vistra Green dye for DNA quantitation was from Amersham Life Sciences. All restriction enzymes were obtained from New England Biolabs or Stratagene. Exonuclease V was from US Biochemical Corp. Enzymatic reactions were performed as recommended by the manufacturers.
Heteroduplex construction
The large loop substrates used in the in vivo assays are derivatives of one parent plasmid, pHD5. pHD5 was created in our laboratory by cloning the HindIII–BamHI fragment of the ADE8 gene into the pRS313 vector (37). The resulting plasmid contains: (i) an origin of replication and the bla gene for propagation in Escherichia coli; (ii) an f1 origin which allows for the production of ssDNA by superinfection with a helper phage; (iii) the HIS3 gene for selection of transformed yeast cells; and (iv) a centromere region and an ARS element for chromosome-like segregation in yeast cells. Expression of the ADE8 gene is under the endogenous promoter. Derivatives of pHD5 were created in our laboratory by insertion of duplex oligomers at the HpaI site, 135 bp from the ATG start codon, in the ADE8 gene. In all cases, this is an out-of-frame disruption which produces an ade8 mutation. The resulting plasmids contain the following inserted sequences and underlined diagnostic restriction sites: pHD16, 5′-AATTGCTAGCAAGCTT-3′, NheI; pHD38, 5′-CGACCCCTGTTGCTGCCTCGAGGGCCGCGTCTTTGTCG-3′, XhoI; pCTG38, 5′-CGACCTGCTGCTGCTGGAATTCCTGCTGCTGCTGGTCG-3′, EcoRI; pHPN38, 5′-CGACCTCGTCCTGCTCGCATGCGTGCTGGTCGTGGTCG-3′, SphI. These sequences are on the viral strand of the phage, as confirmed by sequencing. DNA from these phage was used to create heteroduplex molecules as described below. A one base deletion at the middle nucleotide of codon 46, used to create a one base loop heteroduplex (14), was from W. Kramer (University of Göttingen).
Double-stranded DNA from the pHD5 vector and derivatives was purified from the E.coli strain JM101 [F′ traD36 lacIq Δ(lacZ)M15 proA+B+/supE thi Δ(lac-proAB)] using published procedures (38). Single-stranded DNA was produced in the E.coli X-90 strain [ara Δlac-pro nalA argEamb rifR thi1–/F′ lac+ pro+ lacI(Q1)] by superinfection with the helper phage R408 (from K. Knight, University of Massachusetts Medical Center) at a multiplicity of infection of 10 and isolated using a CsCl purification method (38). Heteroduplexes for the transformation assay were prepared by the method of Lu et al. (39) with the following slight modifications. Double-stranded DNA harboring the desired sequence on the complementary (C) strand was linearized with BssHII. The linear product was mixed with a 10-fold excess (w/w) of single-stranded circular DNA containing the viral (V) strand sequence. NaOH denaturation and subsequent neutralization resulted in heteroduplex formation. Isolation of the heteroduplex form required two consecutive purifications by electrophoresis on a 1% TPE (36 mM Tris base, 30 mM NaH2PO4, 1 mM EDTA) agarose gel. A final purification was performed by electrophoresis on a 1% TAE (40 mM Tris–acetate, 1 mM EDTA) low melting point agarose gel (Sigma). The DNA was subsequently released from the gel slice by treatment with β-agarase (New England Biolabs) according to the manufacturer’s recommendations. Final heteroduplex preparations were ≥98% pure, as determined by agarose gel electrophoresis. Heteroduplexes were created by combining the respective C and V strands from the following pHD vector derivatives: V1, pHD8 + pHD5; V16, pHD5 + pHD16; V38, pHD5 + pHD38; V38 CTG, pHD5 + pCTG38; V38 hairpin, pHD5 + pHPN38.
For in vitro assays, f1 phage MR9 and MR11 were kindly provided by P. Modrich (Duke University). Additional phage variants were created in our laboratory by insertion of duplex oligonucleotides. The resulting phage contain the following inserted sequences and underlined diagnostic restriction sites: MR9+30, 5′-CCCTGTTGCTGCCTCGAGGGCCGCGTCTTT-3′, XhoI; MR9+CTG, 5′-CTGCTGCTGCTGGAATTCCTGCTGCTGCTG-3′, EcoRI; MR9+HPN, 5′-CTCGTCCTGCTCGCATGCGTGCTGGTCGTG-3′, SphI. These sequences are on the viral strand of the phage, as confirmed by sequencing. Both single-stranded and double-stranded DNA from the f1 phage were purified from JM101 cells using published procedures (38). Heteroduplexes were prepared by the method of Lu et al. (39) with slight modifications (38). To create the 3′ nicked substrate, MR9 double-stranded DNA was linearized with Sau96I and combined with a 5-fold excess (w/w) of MR11 circular single-stranded DNA. The C27 heteroduplex was formed using the method described above and treated with DNA ligase. The covalently closed molecules were purified by electrophoresis on a 1% TAE low melting point agarose gel in the presence of 1 µg/ml ethidium bromide. The DNA was subsequently released from the gel slice by treatment with β-agarase as described above. A site-specific nick 3′ to the loop mispair was introduced on the viral strand using gpII protein (40), kindly provided by J. Genschel and P. Modrich (Duke University). The resulting nicked substrate was again purified by electrophoresis and treatment with β-agarase. Final preparations were ≥98% pure, as judged by agarose gel electrophoresis. Heteroduplexes were created by combining the respective C and V strands from the following phage variants: C27, MR9 + MR11; V27, MR11 + MR9; V30, MR9 + (MR9+30); V30 CTG, MR9 + (MR9+CTG); V30 hairpin, MR9 + (MR9+HPN).
Yeast strains
Yeast strains used for the MR-deficient and the loop secondary structure transformation assays were either MW3317-21A (MATα trp1 ade8ΔKpn ura3-52 hom3-10 met13 met4 ade2 his3-Kpn) or isogenic derivatives. MW3317-21A and MW3317-21A-pms1Δ were from S. Fogel (University of California at Berkeley). Gene disruptions to yield msh2::Tn10LUK, msh3::TRP1, mlh1::URA3 and rad1::URA3 derivatives were performed by a single round of disruption. Disruption plasmids were kindly provided by R. Kolodner (University of California at San Diego) for MSH2, M. Liskay (Oregon Health Sciences University) for MLH1 and L. Prakash (University of Texas Health Sciences) for RAD1. The disruption plasmid for MSH3 was created in our laboratory. All derivatives were confirmed by Southern blotting and appropriate genetic tests.
To study LLR in exonuclease-deficient backgrounds, W303-1a (MATa leu2-3 112 trp1-1 ura3-1 can1-100 ade2-1 his3-11,13) and the isogenic derivatives rad2::TRP1 (LSY381-3D) and rad27::TRP1 (YBL3) were kindly provided by L. Symington (Columbia University). The above strains were modified to complement our assay system. An ade8 derivative was created by a 326 bp deletion (ClaI–Eco47III) 54 nt from the start codon in the ADE8 gene, resulting in the following strains: BL494 (wild-type), BL217 (rad2) and BL218 (rad27). For biochemical experiments, extracts were prepared from the yeast strain DY6 (MATa ura3-52 leu2 trp1 prb1-1122 pep4-3 prc1-407; from B. Jones, Carnegie Mellon University, via T. Hsieh, Duke University).
In vivo loop repair assay
Approximately 100 ng purified heteroduplex DNA was used to transform yeast cells by a modified lithium acetate electroporation procedure (41). This procedure yielded an average of 0.1–1 × 105 transformants/µg heteroduplex DNA, depending on the strain. A 25 ml culture of the strain to be transformed was grown overnight in YPD medium at 30°C to an OD600 of 3.0. The cells were washed once in sterile water and resuspended in 0.5 ml each of 10× LTE (1 M lithium acetate, 100 mM Tris base, 100 mM EDTA) and 1 M lithium acetate. The cells were incubated at 30°C with gentle shaking (50 r.p.m.) for 45 min, then 125 µl of 1 M dithiothreitol (DTT) was added and the cells incubated for an additional 15 min. Two consecutive washes with sterile water and one wash with 1 M sorbitol were necessary after the lithium acetate/DTT treatment. The cells were resuspended in 250 µl of 1 M sorbitol and were transformed immediately. The heteroduplex was incubated with 40 µl of competent cells for 5 min on ice, electroporated at 1.5 kV and resuspended in 1 ml of cold 1 M sorbitol. Approximately 5–10 µl of the transformation mixture was plated onto selective plates (SC his– limited adenine + 1 M sorbitol) and grown at 30°C for 5–6 days. The proportion of red, white and red/white sectored colonies was determined. After the original colony count was performed, the transformant colonies were replica plated to YPG plates to test for petites. Correction for petites did not change the overall percentage of red, white and sectored colonies.
Nuclear extract preparation
Nuclear extracts were prepared essentially as described by Wang et al. (42), with modifications as listed (36). Final extract preparations typically contained 5–6 mg/ml of protein, as measured by the method of Lowry et al. (43) after precipitation with trichloroacetic acid.
In vitro loop repair assays
All loop repair assays were performed as described in Corrette-Bennett et al. (36) unless otherwise noted. All restriction digests for C27 and V27 contained 7 U Bsp106I (Stratagene) to linearize the DNA. Repair that led to removal of the loop was evaluated by addition of 5 U EcoRI for substrates 3′ C27 and 5′ V27. Repair in favor of the loop was evaluated with 2.5 U NheI. In the V30, V30 CTG and V30 hairpin substrates, loop removal was assessed by restriction with 5 U PvuII, whereas loop retention was evaluated with 6 U XhoI, 5 U EcoRI or 5 U SphI, respectively. Following incubation at 37°C for 60 min, restriction analysis was performed on a 1% agarose gel. Unless otherwise noted, the gels were stained in a 1:10 000 dilution of Vistra Green dye (Amersham Life Sciences) and densitometric analysis of repair efficiencies was performed using a Molecular Dynamics Storm 860 PhosphorImager using the Blue Fluorescence/Chemiluminescence mode. Repair activity was measured as DNA present in the 3.1 + 3.3 kb bands divided by the total heteroduplex recovered and then converted to fmol loop repaired/mg yeast nuclear protein/h.
RESULTS
Experimental rationale
The major purpose of this study was to examine LLR in yeast in a way that paralleled our biochemical experiments and also studies done in several mammalian systems. An in vivo assay was chosen as the major focus because it is a sensitive and biologically relevant system. The genetic assay also facilitates the use of MR and NER mutant strains to help distinguish between different reported loop repair systems. We monitored the correction of defined, preformed heteroduplexes. These assays focus directly on repair of the loop and therefore differ from other genetic assays that rely on formation of the loop via recombination or replication slippage, prior to repair. The use of pre-existing looped substrates also allows direct comparison with our previous in vitro work and with several mammalian studies.
In vivo assay for loop repair
Loop repair was examined (14) by creating loop-containing DNA molecules with a defined size and location of the heterology, followed by transformation into cells (Fig. 1A). The two strands of the substrate contain either a wild-type ADE8 sequence or a frameshifted ade8 allele. Placement of the loop in the ADE8 gene allows for phenotypic monitoring of repair, as follows. The heteroduplex substrate is transformed into yeast ade2 ade8 mutants and transformants are selected on synthetic complete medium lacking histidine, but containing limited adenine for color development. Correction of the heteroduplex prior to replication results in all red or all white colonies. In this genetic background, the appearance of a red colony indicates repair that removes the loop (ADE8), whereas repair that retains the loop results in a white colony (ade8). If there is no repair, the colony shows a sectored red and white phenotype. The chromosomal ade8 allele in this strain has a 1.3 kb KpnI deletion that encompasses the site of the mispair for at least 250 bp on either side (14). Therefore, repair in these assays is unlikely to result from recombination between the heteroduplex substrate and the chromosome. It is possible that a sectored colony can arise by transformation of more than one heteroduplex molecule into a single cell. Repair to opposite strands in this case would result in a sectored phenotype. Co-transformation frequencies were estimated as the percentage of sectored colonies that appear when equal amounts of the plasmids pHD5 (ADE8) and pHD8 (ade8) were transformed into this strain. Co-transformation levels are estimated to be <5% based on these trials, similar to a previous report (14). Thus, co-transformation is negligible under these assay conditions.
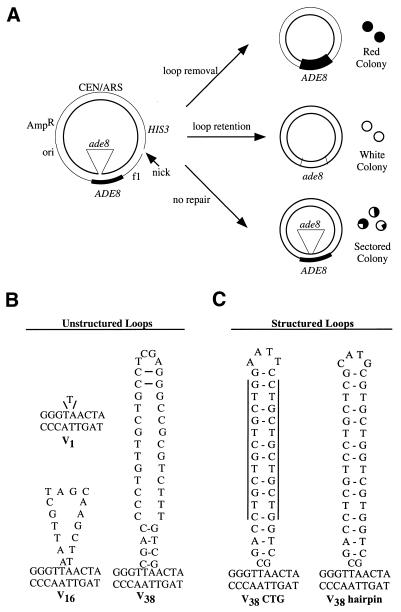
Figure 1.
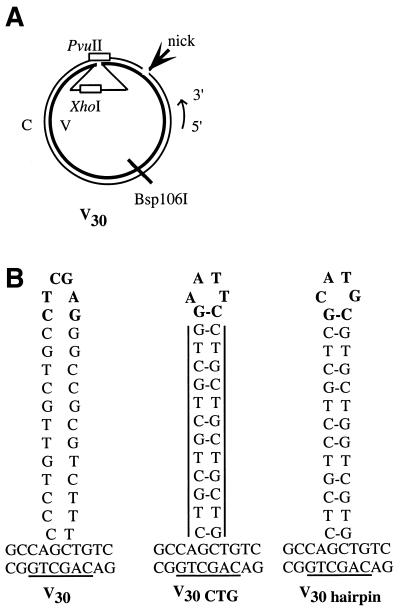
In vivo assay for LLR. The heteroduplex substrate, similar to that used by Kramer et al. (14), is diagrammed schematically in (A). The ori and AmpR genes are for propagation of the parental plasmids in E.coli. An f1 origin allows for the production of viral strand ssDNA by superinfection with a helper phage. The HIS3 gene is used for selection of transformants. The CEN/ARS region provides an origin of replication and centromere function. In this example, a heteroduplex molecule was constructed in which the inserted sequence creates an ade8 mutation in the inner, viral (V) strand, while the outer, complementary (C) strand of the substrate retains the wild-type ADE8 sequence. A nick corresponding to a BssHI site is placed 2.7 kb 3′ from the loop mispair on the complementary strand. The possible repair outcomes are described in the text. The looped heterologies used in this study are shown in (B). In the unstructured loops, the V indicates that the loop is located on the viral strand of the heteroduplex while the numeral indicates the number of nucleotides in the loop. The structured loops both have the potential to form secondary structures with 13 base pairings within the loop. The vertical lines for V38 CTG highlight the location of the CTG repeats.
Several different loops were used to examine LLR (Fig. 1B). We use a nomenclature in which V indicates that the loop is on the inner (viral) strand of the substrate. The numeral denotes the size of the loop in nucleotides. Thus, the V16 substrate contains a 16 nt loop on the inner strand (Fig. 1). The construction of the V16 and V38 substrates places the ade8 gene on the looped strand, whereas a 1 nt deletion in the complementary strand of V1 results in the viral strand harboring both the loop and the wild-type ADE8 sequence (Fig. 1B). Previous in vitro work showed that efficient repair stimulated by a nick 114 bases 5′ of the loop mispair was reduced 3-fold when the nick was moved to 787 bases 5′ (36). This distance effect suggests that a nick engineered 2.7 kb 3′ of the heterology (Fig. 1A) is unlikely to initiate nick-stimulated repair in our transformation assays. Therefore, we used preformed heteroduplex substrates to directly address the role of the loop in signaling repair. V38 does have the potential to form a 4 bp hairpin at the base of the loop, but as shown by Petes and colleagues (33) and confirmed below, such limited structure-forming capacity does not impede correction.
Looped heteroduplex substrates are efficiently corrected in vivo
The V38, V16 and V1 unstructured loops were transformed into yeast cells and the resulting colonies were classified as red, white or sectored. Repair efficiency is determined by summing the number of red and white colonies and dividing by total colonies. In wild-type cells, repair of all three substrates is >90%, indicating that the loop is efficiently corrected (Fig. 2). Thus, preformed heteroduplex loops up to 38 bases are excellent repair substrates in yeast.
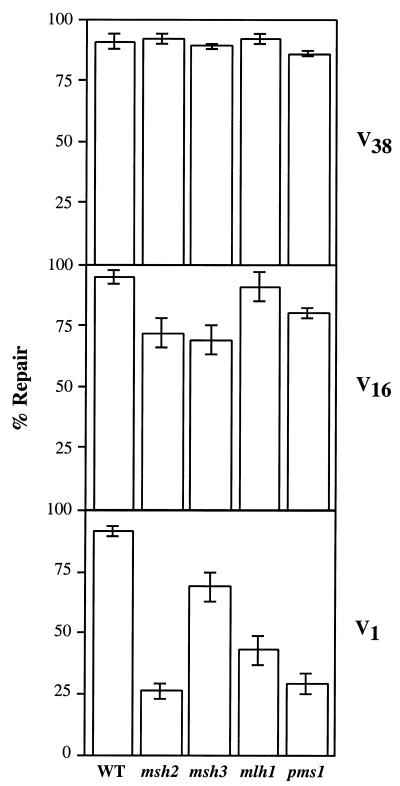
Figure 2.
Loop repair activity in wild-type and MR-deficient strains. Repair activity was assayed on the V1, V16 and V38 substrates as described in Materials and Methods. Repair of each heteroduplex was examined in the strain MW3317-21A (wild-type) and its isogenic derivatives containing null alleles of msh2, msh3, mlh1 or pms1. Percent repair was determined as the sum of unsectored red and white colonies divided by total colonies. The error bars represent one standard deviation from at least three independent measurements.
An advantage of this assay system is that we can determine repair efficiency to either strand, based on the red:white ratio. Both the V16 and V38 large loop substrates exhibit a correction bias, with a preference for loop removal over loop retention at a ratio of 2:1. In contrast, the substrate containing the 1 nt loop showed no strand preference, resulting in nearly equal numbers of red and white colonies, similar to the results of Kramer et al. (14). This difference in strand bias between the different size loops suggested that there is a unique mechanism directing repair of large loops.
Genetic independence of LLR from MR
In addition to assays in a wild-type background, the unstructured loop substrates were tested in isogenic mutant strains containing null alleles of the MR genes msh2, msh3, mlh1 or pms1. V38 was designed as a substrate expected to be corrected by the LLR pathway, as determined previously by in vitro assays (30,36). The 16 nt loop size is near the border where MR and LLR may overlap and was used to determine the contributions of each of these pathways. The 1 nt loop was used as a control, as its repair efficiency is expected to decrease significantly when MR is inactivated.
The correction efficiency of the 38 nt loop in the MR-deficient strains remains essentially unchanged (85–95%) compared to wild-type, indicating that the loop is corrected by a pathway distinct from mismatch repair (Fig. 2). Since repair of the 38 nt substrate does not require MSH2, MSH3 or PMS1, it differentiates LLR from other reported loop repair activities in yeast (16–19). The V16 substrate showed a small but significant decrease in repair in the msh2, msh3 and pms1 strain backgrounds. These data indicate that ∼70–75% of total repair is by the LLR pathway, with the remaining 25–30% of repair presumably due to MR. Loss of mlh1 appears to have no effect on the correction of V16. The 1 nt loop, as predicted, is affected by mutations in MR genes. The loss of msh2 and pms1 show the greatest deficiencies, with repair reduced to 26 and 29%, respectively. These results are consistent with those of Kramer et al. (14), where a pms1 mutation reduced correction of a 1 nt loop to 19%, compared to a wild-type repair efficiency of 94%. In our experiment (Fig. 2), the mlh1 strain exhibits a less severe repair defect, with correction reduced to 43%. In the msh3 background, repair of the V1 substrate is still 75% of wild-type. This limited dependence on MSH3 was expected; the yeast Msh2p–Msh3p heterodimer primarily corrects loops 2–13 nt in size, playing only a small role in repair of 1 nt loops (44,45). This substrate specificity is reflected in our in vivo assay.
Genetic independence of loop repair from NER
In yeast meiotic recombination, LLR requires both MSH2 and the nucleotide excision repair nuclease RAD1 (16). Replicational slippage assays indicate that certain deletion events are inhibited by MSH3 and RAD1 (18,19). In our assays, MSH2 and MSH3 are clearly dispensable (Fig. 2; 36). To determine if components of the NER pathway are required for mitotic loop repair, we tested the V16 substrate in backgrounds deficient for the nucleases rad1 and rad2 (Fig. 3). Correction of the 16 nt loop is indistinguishable in two wild-type strains, suggesting that slight differences between the strain backgrounds do not affect LLR. Mutations in rad1 or rad2 do not show a significant effect on repair efficiency. These data suggest that repair of large loops is not dependent on these NER genes in mitotic cells.
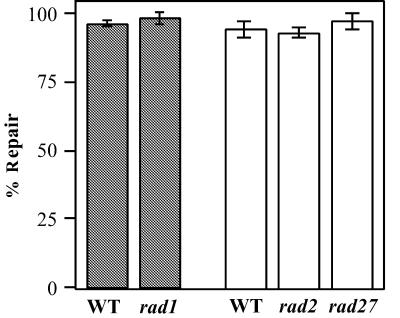
Figure 3.
Loop repair activity in exonuclease-deficient strains. Repair activity in vivo was assayed on the V16 substrate as described in Materials and Methods. Analysis of repair for rad1 was performed in the MW3317-21A strain background (shaded bars); rad2 and rad27 assays were in the BL494 background (white bars). Percent repair was determined as the sum of unsectored red and white colonies divided by total colonies. The error bars represent the standard deviation from at least three independent measurements.
A yeast strain harboring a rad27Δ mutation exhibits genomic instability which manifests as an increased number of large insertion and deletion mutations (21). This phenotype could be indicative of the loss of an LLR component. In addition, biochemical assays have indicated that LLR shows a preference for repair from a 5′ nick (30). The role of Rad27p as a 5′ nuclease, in combination with the mutator phenotye, led us to examine the effect of a rad27Δ mutation on LLR activity (Fig. 3). This mutant strain showed no loss of repair efficiency, suggesting that LLR is independent of RAD27. We cannot rule out the possibility that there are nucleases with redundant function that mask the effect of this single mutant, as seen for bacterial MR (46,47).
Secondary structure within loops reduces repair
In yeast recombination assays, loops predicted to form secondary structure result in an increased frequency of sectored colonies (32–35). This suggests that secondary structure within a loop inhibits repair. In these genetic assays, the formation of the loop is inferred, based on the final repair products. We utilized preformed heteroduplexes to examine directly the effect of loop structure on LLR efficiency. The sequences of our structured loop mispairs (Fig. 1B) were designed to mirror as closely as possible those used by Petes and colleagues (35), so that we could compare our mitotic results to those seen in meiosis. All three 38 nt loop substrates contain similar nucleotide compositions, but vary in sequence and base pairing capacity. The unstructured V38 substrate is used as a positive control and is repaired at ≥90% efficiency. The V38 CTG substrate contains a loop with the trinucleotide sequence CTG repeated eight times, resulting in 13 potential base pairings within the loop. In vivo repair of the V38 CTG substrate is reduced by 50% when compared to the correction efficiency of the unstructured loop (Fig. 4A). Another way to view this result is that the percentage of sectored colonies, corresponding to unrepaired loops, increases 7-fold from 8% for the unstructured loop to 56% for the CTG repeat-containing heteroduplex. This decrease in correction efficiency could be due to secondary structure or to an intrinsic property of the repetitive primary sequence. To determine the potential contributions of structure and sequence, we designed another substrate, V38 hairpin, which contains the same number of G-C base pairings as V38 CTG, but in a non-repeating sequence (Fig. 1C). The V38 hairpin substrate reduced repair by ∼25%. The ratio of loop removal to loop retention was reduced from the 2:1 value seen for the unstructured 38 nt loop to 1:1 for both the V38 CTG and V38 hairpin substrates. It appears that loop removal is more strongly inhibited by secondary structure than loop retention. From this information, we conclude that secondary structure within the loop does reduce LLR activity, however, the primary sequence of the loop may also contribute to the repair deficiency in our system.
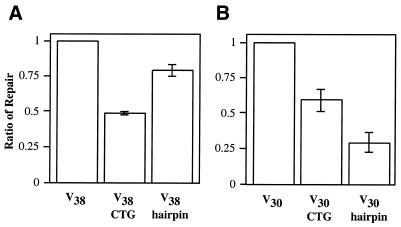
Figure 4.
Secondary structure within the loop affects repair. Repair of large loops was tested in both an in vivo transformation assay (A) and in a biochemical assay using nuclear extracts (B). The transformation assay is performed as described, using the V38, V38 CTG and V38 hairpin heteroduplexes diagrammed schematically in Figure 1B. The ratio of repair is determined by normalizing total repair of the structured loops to that of the unstructured V38 loop. Total repair for V38 was 92%. A schematic representation of the biochemical assay is found in Figure 5A. The in vitro heteroduplexes used in this structural study are diagrammed schematically in Figure 5B. The ratio of repair, as described above, is normalized to the V30 substrate (an absolute value of 185 fmol/mg/h) in this biochemical assay. The error bars for both the in vivo and in vitro assays represent one standard deviation from three independent measurements.
Analysis of loop structure effects in vitro
We also tested the repair of structured loops in vitro, using an assay described previously (Fig. 5A) (36). Briefly, heteroduplex molecules were incubated with yeast nuclear extract, followed by restriction analysis to identify repair products. Three 30 nt substrates were designed to mimic those used in the in vivo assay (Fig. 5B). Whereas the V30 loop has little or no apparent secondary structure, the structured loops (V30 CTG and V30 hairpin) have the potential to form 9 bp within the loop. All three loop mispairs maintain similar nucleotide content. When normalized to the repair of the unstructured V30 loop, correction of the V30 CTG heteroduplex exhibits a 40% decrease in repair efficiency, similar to the results shown in the in vivo assay (Fig. 4B). In contrast, the V30 hairpin shows a 70% decrease in repair. To a first approximation, the inhibition of LLR by structure is similar in vitro to that seen in vivo. However, the relative inhibition by the two structured loops was different in vivo, where the V38 CTG loop was the more strongly inhibiting (Fig. 4A). One possible reason for the difference between the transformation and the biochemical assay involves preparation of the nuclear extracts. Some of the protein components for correction of the hairpin substrate in vivo may not be efficiently released by our extraction protocol. A second possibility is that interactions of the loops with chromatin in vivo may not be recapitulated in the extract. Alternatively, there might be slight differences between the structures of the V30 hairpin and the V38 hairpin loop that affect inhibition of repair. A qualitative analysis suggests that both loop removal and loop retention were reduced in vitro, but the inhibited levels of loop retention with the structured substrates made it difficult to quantitate this effect. Taken together, the evidence from the in vivo and in vitro assays points to an inhibition of LLR by secondary structure within the loop.
Figure 5.
In vitro assay for LLR. The heteroduplex substrate with a loop on the viral (V) strand is diagrammed schematically in (A). Heteroduplex DNA molecules were constructed with a site-specific nick on the complementary (C) strand 114 bp 5′ to the loop. The V strand was covalently closed. The loop renders the heteroduplex resistant to both XhoI and PvuII. If loop repair occurs, the DNA becomes sensitive to PvuII if repair removes the loop, while the DNA is rendered sensitive to XhoI if the loop is retained. (B) The looped heterologies used in this study. A complete description of these molecules is included in the text.
Orientation of the nick affects nick-stimulated but not nick-independent repair
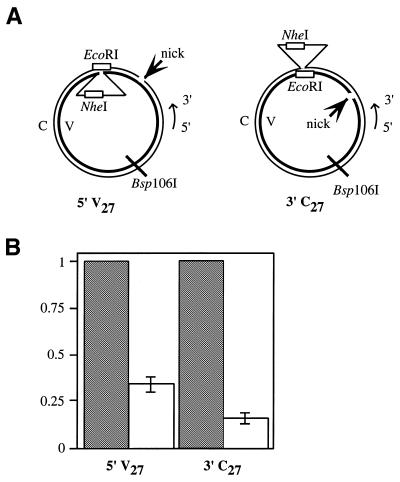
LLR in human cell extracts demonstrated that one of the distinct features of this repair pathway is a strong preference for a 5′ nick, compared to a 3′ break (30). In contrast, MR can utilize a 5′ or 3′ nick with similar efficiencies (5,48). We evaluated the utilization of 5′ and 3′ nicks in yeast extracts. A schematic of the heteroduplex substrates is found in Figure 6A. The two substrates are designed to be as similar as possible, with the orientation of the nick as the primary variable. Each heteroduplex molecule contains a 27 nt loop on one strand while the nick is placed on the opposing strand. Placing these two biologically important cis-elements on separate strands of the construct allows for monitoring of both possible outcomes of repair (36), because nick-stimulated repair directs excision to the outer, non-looped strand while nick-independent correction results in removal of the loop on the inner strand. The V27 molecule has a single nick 114 bases 5′ of the looped mispair, while a nick was introduced 170 bases 3′ of the heterology in the C27 molecule. Mapping of excision tracts in LLR indicates that the proximity of these nicks to the loop mispair is close enough to activate nick-stimulated repair in our assay system (36). One minor difference between these two substrates is the primary sequence of the loop; V27 carries the viral strand sequence while C27 has the complementary strand sequence. Previous work indicates that, despite variations in the primary sequence, these loops have similar correction efficiencies (30,36).
Figure 6.
Nick orientation affects LLR in vitro. The substrates used in this study are diagrammed schematically in (A). Each heteroduplex molecule contains a 27 nt loop on one strand while the nick is placed on the opposing strand. Nick-independent repair (loop removal) results in a molecule that is EcoRIS. In contrast, nick-stimulated correction (loop retention) produces a NheIS molecule. The V27 molecule has a single nick 114 bp 5′ of the looped mispair, while a nick was introduced 170 bp 3′ of the heterology by gpII protein in the C27 molecule. (B) A comparison of the correction efficiency of nick-stimulated repair (unfilled bars) relative to nick-independent repair (shaded bars). Absolute values for nick-independent correction activities are reported as fmol repaired heteroduplex/mg protein/h, and are as follows: 5′ V27, 75 ± 2; 3′ C27, 87 ± 2. The ratio of repair was determined by normalizing nick-stimulated repair to nick-independent correction. The error bars represent one standard deviation from three independent measurements.
After incubation in nuclear extracts, the DNA was tested for loop removal because this nick-independent reaction should be unaffected by nick orientation. Therefore, it is a convenient internal control. Absolute values for nick-independent loop removal (in fmol repaired heteroduplex/mg protein/h) were 75 ± 2 for 5′ V27 and 87 ± 2 for 3′ C27. The comparable levels of loop removal in the two substrates supports the prediction of a nick-independent reaction. In contrast, the nick-stimulated repair showed a 2-fold preference for a 5′ nick. In Figure 6B, nick-stimulated repair of 5′ V27 was 30% as effective as loop removal, whereas nick-stimulated correction of the 3′ C27 heteroduplex is only 15% as effective as loop removal. This result indicates that while a 3′ nick may be utilized for excision, nick-stimulated LLR is more effective from a 5′ break.
DISCUSSION
One of the major findings of this study is that in vivo repair of large loops in S.cerevisiae can be very efficient. Transformation assays with loops of 1, 16 or 38 nt showed high level, approximately equal correction levels for all three substrates. Under these assay conditions, LLR on the 38 nt loop was about as active as MR was on the 1 nt heterology. This extent of correction indicates that yeast has substantial capacity to correct loop sizes well beyond the range of MR. The second major conclusion from this study is that in vivo LLR in yeast shows close similarity to loop repair activities catalyzed by yeast extracts (36) and those seen in several mammalian systems (5,25,26,30,49). Taken together, these reports indicate a robust repair capacity on loops ranging from 16 to 283 nt. The similarities of LLR in yeast and mammalian cells suggests conservation of function in eukaryotes that is independent of numerous MR or NER components (30,36; this work). The identification of LLR protein components will be an important goal in the field.
LLR can be differentiated genetically from other loop repair activities reported in the yeast literature. For example, our assays show that mutation of either the NER nuclease encoded by RAD1 or the MR MSH2 gene product does not detectably alter repair on a 16 base target. These results indicate that loop repair in mitotic yeast cells occurs by a different pathway than in meiotic recombination (16). Similarly, the fact that LLR activity does not require MSH3 or RAD1 distinguishes it from another loop correction activity that helps overcome certain replication slippage events (18,19). A third alternative pathway for very large loops (≥2 kb) formed during mitotic recombination is at least partially dependent on MSH2 and PMS1 (17). Clearly, the efficient LLR activity observed both in vitro (36) and in vivo (this study) occurs by a unique pathway in yeast. One possible explanation for the presence of multiple loop correction pathways in yeast may involve the manner in which the loops are formed. It has been suggested that accessibility to the loop might be modulated by other proteins, thus controlling which repair pathway can act to correct the loop (17,50). For example, a loop formed during recombination could conceivably be accessed by a different subset of repair proteins than a loop arising from replicational slippage. Our results indicate that, given a preformed looped heteroduplex, the yeast cell is capable of efficiently correcting heterologies up to 216 nt.
What features determine whether the loop is removed or retained? Like our transformation experiments in yeast, transfection experiments with mammalian cells showed that LLR has a preference for removal of the heterology (25,26,49), suggesting that the loop is the predominant cis-factor. The influence of a nick 71 or 125 bases from the loop was also tested in mammalian cells. The effect of the nick appeared to be small, regardless of its orientation (49). However, it could not be ascertained whether the nick persisted after transfection or was ligated. Biochemical experiments clearly indicate that a 5′ nick can be utilized to direct repair to the discontinuous strand. This preference is strong in human cell extracts (30) and also occurs to a lesser extent in yeast extracts (this study). One explanation for the reduced 5′ nick preference in yeast is that endogenous nicking activities in the extract might occasionally cleave the strand that already has the existing nick. If so, some of the apparent nick-stimulated repair could arise from secondary nicking activities, rather than utilization of the existing DNA terminus. This scenario would reduce the apparent 5′ versus 3′ nick preference observed in Figure 6, because some baseline level of nick-stimulated repair on both substrates would be catalyzed by endogenous nicking activity. This line of reasoning also suggests that nick-stimulated and nick-independent repair may not be completely distinct, if one accepts that single-strand cleavage on the continuous strand (leading to loop removal in Fig. 6) might also sometimes occur on the discontinuous strand. To address possible nucleases involved in either creating a nick or utilizing a pre-existing 5′ nick, we tested loop repair efficiency in single mutants of RAD1, RAD2 and RAD27. However, repair of a 16 nt loop was at wild-type levels in these mutant strains, despite the fact that our in vivo assay is suitably sensitive for detection of minor defects in loop repair. Our results suggest that either these genes play no role in LLR or else there is a redundant counterpart.
The results reported here provide two insights into the inhibitory effect of secondary structure within the 38 nt loop. First, reduced loop repair activity on hairpins is distinct from an activity in human cells that can repair palindromic hairpins up to 40 bases in length (31). Secondly, our inhibition studies indicate that LLR is one of the DNA repair pathways that is overcome during expansion of trinucleotide repeats (51). The hairpins we tested, inspired by the work of Moore et al. (35), were designed to mimic replication intermediates thought to be important during the triplet repeat expansion process. The reduced repair of hairpin loops containing only eight CTG repeats suggests that LLR is unable to process mutagenic intermediates in important disease genes. Among trinucleotide repeat expansions in affected human families, an increase of eight repeats is relatively small. Many disease-causing expansions are significantly larger, in some cases increasing the tract by thousands of repeats (52). The fact that an eight CTG repeat loop inhibits LLR by 40–50% suggests that loops with more repeats will be even more inhibitory. Our findings help explain how mutagenic intermediates containing trinucleotide repeat sequences avoid LLR.
Gene conversion studies indicate that most insertions and deletions up to ∼1 kb undergo frequent correction in yeast meiosis (53), suggesting the active repair of large loops. One exception is the ade8-18 allele, a 38 base deletion that shows low gene conversion and high post-meiotic segregation (13). Heteroduplex loops created by annealing wild-type ADE8 to ade8-18 are also refractory to repair upon transformation (14). We suggest that resistance of the ade8-18 allele to loop repair is the exception, rather than the rule, because LLR has been demonstrated for numerous other heterologies (16,25,26,30,35,36,53), including a different 38 nt loop (this study).
Our work suggests the presence of an active LLR activity in yeast. Yet experiments that examined the mutational spectrum created by certain mutant alleles of POL3 (polymerase δ), POL30 (PCNA) and RAD27 (FEN-1) (15,20–24) show numerous insertion/deletion events of a size that should be prevented by this pathway. One possibility is that the number of strand slippage errors is so great in these mutants as to saturate LLR capacity. Another explanation, outlined earlier, is that accessibility of repair components is somehow modulated by other proteins involved in formation of the loop. A third possibility is that polymerase δ and PCNA may be involved in LLR. Thus it is possible that these mutations may also inhibit efficient correction of the heterologies.
Together, MR and LLR provide complementary correction of loops that range from one to hundreds of nucleotides. There appears to be overlap between the pathways for intermediate size loops. We found that repair of a 16 nt loop showed a partial (25–30%) dependence on MSH2, MSH3 and PMS1. These results suggest that MR has, under certain circumstances, some activity on loops as large as 16 bases. In this respect, we were surprised to see that loss of MLH1 did not affect repair of the 16 nt loop, although previous studies indicate that repair of a different 16 nt heterology occurs readily in extracts from human MLH1–/– cells (5). It seems unlikely that the disrupted mlh1 allele we tested had residual activity, since repair of a 1 nt loop was significantly compromised. A more detailed analysis of the repair of intermediate size loops will be required to resolve these issues. What is clear from these studies is that overlapping substrate specificity of MR and LLR ensures coverage of a wide spectrum of possible loop sizes.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by American Cancer Society Research Grant NP-943 (to R.S.L.), by post-doctoral fellowships GM18922 from the National Institutes of Health (to S.E.C.-B.) and ACS PF-4033 (to B.O.P.), by National Cancer Institute Cancer Center Support Grant P30 CA36727 (to the Eppley Institute) and by funds from the University of Nebraska Medical Center.
References
- 1.Modrich P. and Lahue,R.S. (1996) Mismatch repair in replication fidelity, genetic recombination and cancer biology. Annu. Rev. Biochem., 65, 101–133. [DOI] [PubMed] [Google Scholar]
- 2.Kolodner R. (1996) Biochemistry and genetics of eukaryotic mismatch repair. Genes Dev., 10, 1433–1442. [DOI] [PubMed] [Google Scholar]
- 3.Buermeyer A.B., Deschenes,S.M., Baker,S.M. and Liskay,R.M. (1999) Mammalian DNA mismatch repair. Annu. Rev. Genet., 33, 533–564. [DOI] [PubMed] [Google Scholar]
- 4.Harfe B.D. and Jinks-Robertson,S. (2000) DNA mismatch repair and genetic instability. Annu. Rev. Genet., 34, 359–399. [DOI] [PubMed] [Google Scholar]
- 5.Umar A., Boyer,J.C. and Kunkel,T.A. (1994) DNA loop repair by human cell extracts. Science, 266, 814–816. [DOI] [PubMed] [Google Scholar]
- 6.Sia E.A., Kokoska,R.J., Dominska,M., Greenwell,P. and Petes,T.D. (1997) Microsatellite instability in yeast: dependence on repeat unit size and DNA mismatch repair genes. Mol. Cell. Biol., 17, 2851–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Genschel J., Littman,S.J., Drummond,J.T. and Modrich,P. (1998) Isolation of MutSβ from human cells and comparison of the mismatch repair specificities of MutSβ and MutSα. J. Biol. Chem., 273, 19895–19901. [DOI] [PubMed] [Google Scholar]
- 8.Dohet C., Dzidic,S., Wagner,R. and Radman,M. (1987) Large non-homology in heteroduplex DNA is processed differently than single base pair mismatches. Mol. Gen. Genet., 206, 181–184. [DOI] [PubMed] [Google Scholar]
- 9.Fishel R. and Kolodner,R. (1989) Gene conversion in Escherichia coli: the recF pathway for resolution of heteroduplex DNA. J. Bacteriol ., 171, 3046–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gasc A.M., Garcia,P., Baty,D. and Sicard,A.M. (1987) Mismatch repair during pneumococcal transformation of small deletions produced by site-directed mutagenesis. Mol. Gen. Genet., 210, 369–372. [DOI] [PubMed] [Google Scholar]
- 11.Parker B.O. and Marinus,M.G. (1992) Repair of DNA heteroduplexes containing small heterologous sequences in Escherichia coli. Proc. Natl Acad. Sci. USA, 89, 1730–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carraway M. and Marinus,M.G. (1993) Repair of heteroduplex DNA molecules with multibase loops in Escherichia coli. J. Bacteriol ., 175, 3972–3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.White J.H., DiMartino,J.F., Anderson,R.W., Lusnak,K., Hilbert,D. and Fogel,S. (1988) A DNA sequence conferring high postmeiotic segregation frequency to heterozygous deletions in Saccharomyces cerevisiae is related to sequences associated with eucaryotic recombination hotspots. Mol. Cell. Biol., 8, 1253–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kramer B., Kramer,W., Williamson,M.S. and Fogel,S. (1989) Heteroduplex DNA correction in Saccharomyces cerevisiae is mismatch specific and requires functional PMS genes. Mol. Cell. Biol., 9, 4432–4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tran H.T., Gordenin,D.A. and Resnick,M.A. (1996) The prevention of repeat-associated deletions in Saccharomyces cerevisiae by mismatch repair depends on size and origin of deletions. Genetics, 143, 1579–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirkpatrick D.T. and Petes,T.D. (1997) Repair of DNA loops involves DNA-mismatch and nucleotide-excision repair proteins. Nature, 387, 929–931. [DOI] [PubMed] [Google Scholar]
- 17.Clikeman J.A., Wheeler,S.L. and Nickoloff,J.A. (2001) Efficient incorporation of large (>2 kb) heterologies into heteroduplex DNA: Pms1/Msh2-dependent and -independent large loop mismatch repair in Saccharomyces cerevisiae. Genetics, 157, 1481–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harfe B.D. and Jinks-Robertson,S. (1999) Removal of frameshift intermediates by mismatch repair proteins in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 4766–4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harfe B.D., Minesinger,B.K. and Jinks-Robertson,S. (2000) Discrete in vivo roles for the MutL homologs Mlh2p and Mlh3p in the removal of frameshift intermediates in budding yeast. Curr. Biol., 10, 145–148. [DOI] [PubMed] [Google Scholar]
- 20.Chen C., Merrill,B.J., Lau,P.J., Holm,C. and Kolodner,R.D. (1999) Saccharomyces cerevisiae pol30 (proliferating cell nuclear antigen) mutations impair replication fidelity and mismatch repair. Mol. Cell. Biol., 19, 7801–7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tishkoff D.X., Filosi,N., Gaida,G.M. and Kolodner,R.D. (1997) A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell, 88, 253–263. [DOI] [PubMed] [Google Scholar]
- 22.Kokoska R.J., Stefanovic,L., Tran,H.T., Resnick,M.A., Gordenin,D.A. and Petes,T.D. (1998) Destabilization of yeast micro- and minisatellite DNA sequences by mutations affecting a nuclease involved in Okazaki fragment processing (rad27) and DNA polymerase δ (pol3-t). Mol. Cell. Biol., 18, 2779–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kokoska R.J., Stefanovic,L., Buermeyer,A.B., Liskay,R.M. and Petes,T.D. (1999) A mutation of the yeast gene encoding PCNA destabilizes both microsatellite and minisatellite DNA sequences. Genetics, 151, 511–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kokoska R.J., Stefanovic,L., DeMai,J. and Petes,T.D. (2000) Increased rates of genomic deletions generated by mutations in the yeast gene encoding DNA polymerase δ or by decreases in the cellular levels of DNA polymerase δ. Mol. Cell. Biol., 20, 7490–7504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weiss U. and Wilson,J.H. (1987) Repair of single-stranded loops in heteroduplex DNA transfected in mammalian cells. Proc. Natl Acad. Sci. USA, 84, 1619–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ayares D., Ganea,D., Chekuri,L., Campbell,C.R. and Kucherlapati,R. (1987) Repair of single-stranded DNA nicks, gaps and loops in mammalian cells. Mol. Cell. Biol., 7, 1656–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bollag R.J., Elwood,D.R., Tobin,E.D., Godwin,A.R. and Liskay,R.M. (1992) Formation of heteroduplex DNA during mammalian intrachromosomal gene conversion. Mol. Cell. Biol., 12, 1546–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deng W.P. and Nickoloff,J.A. (1994) Mismatch repair of heteroduplex DNA intermediates of extrachromosomal recombination in mammalian cells. Mol. Cell. Biol., 14, 400–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taghian D.G., Hough,H. and Nickoloff,J.A. (1998) Biased short tract repair of palindromic loop mismatches in mammalian cells. Genetics, 148, 1257–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Littman S.J., Fang,W.-h. and Modrich,P. (1999) Repair of large insertion/deletion heterologies in human nuclear extracts is directed by a 5′-single-strand break and is independent of the mismatch repair system. J. Biol. Chem., 274, 7474–7481. [DOI] [PubMed] [Google Scholar]
- 31.Bill C.A., Taghian,D.G., Duran,W.A. and Nickoloff,J.A. (2001) Repair bias of large loop mismatches during recombination in mammalian cells depends on loop length and structure. Mutat. Res., 485, 255–265. [DOI] [PubMed] [Google Scholar]
- 32.Nag D.K., White,M.A. and Petes,T.D. (1989) Palindromic sequences in heteroduplex DNA inhibit mismatch repair in yeast. Nature, 340, 318–320. [DOI] [PubMed] [Google Scholar]
- 33.Nag D.K. and Petes,T.D. (1991) Seven-base-pair inverted repeats in DNA form stable hairpins in vivo in Saccharomyces cerevisiae. Genetics, 129, 669–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weng Y.-s. and Nickoloff,J.A. (1998) Evidence for independent mismatch repair processing on opposite sides of a double-strand break in Saccharomyces cerevisiae. Genetics, 148, 59–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore H., Greenwell,P.W., Liu,C.-P., Arnheim,N. and Petes,T.D. (1999) Triplet repeats form secondary structures that escape DNA repair in yeast. Proc. Natl Acad. Sci. USA, 96, 1504–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Corrette-Bennett S.E., Parker,B.O., Mohlman,N.L. and Lahue,R.S. (1999) Correction of large mispaired DNA loops by extracts of Saccharomyces cerevisiae. J. Biol. Chem., 274, 17605–17611. [DOI] [PubMed] [Google Scholar]
- 37.Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Corrette-Bennett S. and Lahue,R.S. (1999) Mismatch repair assay. Methods Mol. Biol., 113, 121–132. [DOI] [PubMed] [Google Scholar]
- 39.Lu A.-L., Clark,S. and Modrich,P. (1983) Methyl-directed repair of DNA base pair mismatches in vitro. Proc. Natl Acad. Sci. USA, 80, 4639–4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fang W.-h., Li,G.-m., Longley,M., Holmes,J., Thilly,W. and Modrich,P. (1993) Mismatch repair and genetic stability in human cells. Cold Spring Harb. Symp. Quant. Biol., 58, 597–603. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 42.Wang Z., Wu,X. and Friedberg,E.C. (1993) Nucleotide-excision repair of DNA in cell-free extracts of the yeast Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 90, 4907–4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lowry O.H., Rosebrough,N.J., Farr,A.L. and Randall,R.J. (1951) Protein measurement with the Folin phenol reagent. J. Biol. Chem., 193, 265–275. [PubMed] [Google Scholar]
- 44.Johnson R.E., Kovvali,G.K., Prakash,L. and Prakash,S. (1996) Requirement of the yeast MSH3 and MSH6 genes for MSH2-dependent genomic stability. J. Biol. Chem., 271, 7285–7288. [DOI] [PubMed] [Google Scholar]
- 45.Marsischky G.T., Filosi,N., Kane,M.F. and Kolodner,R. (1996) Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev., 10, 407–420. [DOI] [PubMed] [Google Scholar]
- 46.Burdett V., Baitinger,C., Viswanathan,M., Lovett,S.T. and Modrich,P. (2001) In vivo requirement for RecJ, ExoVII, ExoI and ExoX in methyl-directed mismatch repair. Proc. Natl Acad. Sci. USA, 98, 6765–6770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Viswanathan M., Burdett,V., Baitinger,C., Modrich,P. and Lovett,S.T. (2001) Redundant exonuclease involvement in Escherichia coli methyl-directed mismatch repair. J. Biol. Chem., 276, 31053–31058. [DOI] [PubMed] [Google Scholar]
- 48.Fang W.-h. and Modrich,P. (1993) Human strand-specific mismatch repair occurs by a bidirectional mechanism similar to that of the bacterial reaction. J. Biol. Chem., 268, 11838–11844. [PubMed] [Google Scholar]
- 49.Weiss U. and Wilson,J.H. (1989) Effects of nicks on repair of single-stranded loops in heteroduplex DNA in mammalian cells. Somat. Cell Mol. Genet., 15, 13–18. [DOI] [PubMed] [Google Scholar]
- 50.Nicholson A., Hendrix,M., Jinks-Robertson,S. and Crouse,G.F. (2000) Regulation of mitotic homeologous recombination in yeast: functions of mismatch repair and nucleotide excision repair genes. Genetics, 154, 133–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McMurray C.T. (1999) DNA secondary structure: a common and causative factor for expansion in human disease. Proc. Natl Acad. Sci. USA, 96, 1823–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cummings C.J. and Zoghbi,H.Y. (2000) Fourteen and counting: unraveling trinucleotide repeat diseases. Hum. Mol. Genet., 9, 909–916. [DOI] [PubMed] [Google Scholar]
- 53.Petes T.D., Malone,R.E. and Symington,L.S. (1991) In Broach,J.R., Pringle,J.R. and Jones,E.W. (eds), The Molecular and Cellular Biology of the Yeast Saccharomyces. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, Vol. 1, pp. 407–521.