Abstract
Nitrate is required to maintain the growth and metabolism of plant and animals. Nevertheless, in excess amount such as polluted water, its concentration can be harmful to living organisms such as microalgae. Recently, studies on microalgae response towards nutrient fluctuation are usually limited to lipid accumulation for the production of biofuels, disregarding the other potential of microalgae to be used in wastewater treatments and as source of important metabolites. Our study therefore captures the need to investigate overall metabolite changes via NMR spectroscopy approach coupled with multivariate data to understand the complex molecular process under high (4X) and low (1/4X) concentrations of nitrate (). NMR spectra with the aid of chemometric analysis revealed contrasting metabolites makeup under abundance and limited nitrate treatment. By using NMR technique, 43 types of metabolites and 8 types of fatty acid chains were detected. Nevertheless, only 20 key changes were observed and 16 were down regulated in limited nitrate condition. This paper has demonstrated the feasibility of NMR-based metabolomics approach to study the physiological impact of changing environment such as pollution to the implications for growth and productivity of microalgae population.
Introduction
Nitrogen is a primary nutrient regulating growth and metabolism in microalgae, often absorbed in the form of nitrate. Nitrogen is important mainly for tissue growth and protein synthesize1. The ability of microalgae to adapt to varying levels of macronutrients and micronutrients in the environment are proven of an extensive mechanism to holistically reallocate its metabolic components in order to create a response and sustain through the adverse situations2,3.
Although response toward differing levels of nutrients are species-specific, subsequent studies have shown that minor manipulation onto the medium’s nutrient content will induce physiological and biochemical changes in carbohydrate, protein and lipid production. Microalgae adapt to varying levels of nutrients via two strategies; luxury consumption (direct uptake in excess without immediate need) and energy mitigating for nutrient supply (directing use of carbon reserves during starvation periods)4,5. However, exactly how microalgae adapt to one of these two strategies is not fully understood yet. Abundance of nitrate may activate luxury uptake and results in the accumulation of amino acid precursors (NO3− and NH4+) and nitrate reductase3,4. Nitrogen is either assimilated into holding within their cell wall or converted into nitrogen-based reserves such as simple amino acids4,6. Recent molecular studies carried out on Chlamydomonas reinhardtii found a family of three protein transporters NRT1 (NPF), NRT2, and NAR1 are responsible for determining substrate specificity and affinity of nitrate uptake in regards to nitrate concentration7.
Physiologically, nitrate limitation triggers a stress response in microalgae and are often associated with slow growth rates, decreased production in amino acids and declining enzymatic activity leading to reduce photosynthetic capability3,4,8. Respiratory metabolism becomes secondary as more energy is focussed on reallocation of metabolites designed for storage such as nonphosphorylated polyglucans and lipids9.
More notably, nitrogen limitation has been studied for its triacylglycerol (TAG)-enhancing properties and possible formation of economically important secondary metabolites. Many of the microalgae studied reported ~20–40% of increase in total lipid content and a shift towards production of neutral storage fatty acids such as TAG1,6,10–12. Changes in metabolic activities in nitrate and phosphate deplete mediums are also associated with production of reactive oxidative species (ROS) which are believed to trigger the assimilating of nutrients and energy into storage lipids13. This is because storage lipids and TAG synthesis provides an electron sink for excess electrons that occur during photo-oxidative stress thus serving as outlets to detoxify the lipid membrane14,15. Production of secondary carotenoid metabolites (asthaxantin, β-carotene, lutein) are associated with the TAG pathway hence some microalgae like Dunaliella sp. and Hematococcus sp. are able to produce these economically important antioxidants under nutrient stress12,14. Nevertheless, the responses toward nutrient stress is species specific, eg Chlorella sorokiniana and Chlorella minutissima showed only a minor increase in lipid accumulation16, while huge decline of antioxidant activities was detected in Phaeodactylum tricornutum and Chlorella vulgaris12. Therefore, in order to capture the metabolic changes on a wider scale, an analytical method that enables examination of individual metabolite developments rather than total extract changes is needed. Nuclear magnetic resonance (NMR) is a reliable and unbiased spectroscopy technique that can capture what happens metabolically on a wide scale15. Acquisition time for samples is relatively short and hence is time-saving compared to other analytical methods17. NMR coupled with chemomeric analysis is a powerful tool for discriminating between groups of related samples and identifying regions of spectrum that can be dedicated to further analysis. Nutrient limitation and abundance brings out a myriad of reactions that is of great benefit if explored to its full potential. By knowing the cellular changes under different culture conditions, the full potential use of microalgae for different usage, eg pharmaceutical, bioremediation, or as biofuel production can be explored. By using Scenedesmus regularis or synonym of Pectinodesmus regularis18 as microalgae model, this study demonstrated some important biochemical developments and changes in appearance, biomass accumulation, chlorophyll content and metabolites reprogramming when exposed to the presence of different nitrate (N) concentration.
Results and Discussion
Biomass and related growth
Algal biomass gained from each treatment was evaluated in order to understand how growth was affected. Approximately one fold decrease in mass gain was observed in nitrate depleting culture when compared against 4X and control NO3 treatment. The reduction of biomass was correlated with the reduction of growth rate observed in 14 days of treatment (Fig. 1). In nitrate depleting environment, S. regularis was found enter stationary phase mush earlier than in other treatment starting from day 6 (Fig. 1). Nitrate is the most important nutrient depended upon at the initial stage of growth in microalgae thus a lack of NO3 stunted microalgal growth7. Besides primary growth, nitrate is utilised as building blocks of genetic materials (DNA and RNA), proteins for cell expansion and pigment building blocks1,3. Therefore, it is not surprise that under limited nitrate condition, when the nitrate was used up after one week of culture, most of the cell division stop and push S. regularis moved into stationary growth phase. Contrarily, when additional of NO3 was available, carbon fixation occurs and prolong the exponential growth phase, which was in agreement of batch photobioreactors of Scenedesmus and Chlorella cultures19. Biomass accumulation of the control and 4X treatments in both nutrients did not differ significantly might due to the level of NO3 abundance were probably not enough to trigger a sudden “bloom” in its numbers as it has been reported that levels of NO3 have to be over 4.0 mg/L20. Rapidly utilisation of nitrate but not causing bloom phenomenon in high nitrate culture condition may suggest an application in wastewater treatment21. Wastewater from industrial, agricultural or urban can serve as growth substrate to support microalgae biomass production22.
Figure 1.
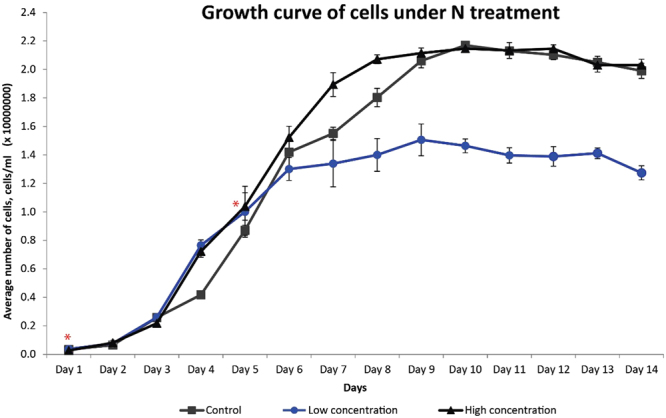
Growth curve of S. regularis under different concentrations of NO3 treatment. Points show average concentration of cells in 1.6 L of liquid culture from each batch of culture. The difference in exponential growth phase of 1/4X NO3 with the control and 4X NO3 treatment is noted with asterisks (*).
Chlorophyll content and total carotenoids
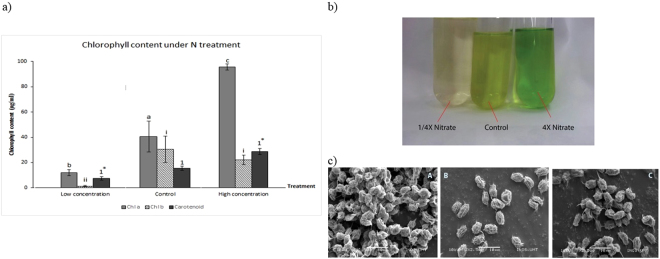
Chlorophyll pigments are important light harvesting proteins that ultimately vary in concentration under the microalgae’s environment conditions. There are evidences shown that nitrate is an important building block for chlorophyll12,13. Chlorophyll pigments in cultures of limited NO3 were drastically reduced in comparison to control and high NO3 cultures (Fig. 2a). Due to very low chlorophyll content, the microalgae in nitrate-limited treatment appear pale green with hints of yellow (Fig. 2b). This condition known as chlorosis or “bleaching” is characteristic of NO3 limitation12,13,23. Chloroplasts serve as large reserves of nitrogen sources and has been shown to be degraded in a contractual autophagy mechanism in times of nitrogen starvation so that the remnants of degraded chlorophylls may be recycled to support metabolic needs that require NO33,4,14,24,25. The highest chlorophyll content was achieved under 4X NO3 treatment with total chlorophyll a and b measured at 107.2 µg/ml. Chl a accumulation was often observed coinciding with high rates of cell division23, similar finding were observed in this study where the highest biomass produced in 4X nitrate treatment. The high Chl content would also coincide with higher photosynthesis activities and production of reactive oxygen species (ROS) as photosynthesis by products. Nevertheless, plants equipped with carotenoids that play photoprotective roles in the photosystem12. As pointed out by Singh, et al.14, nutrient stress activates TAG-related secondary carotenoid formation and can be seen as a way to combat high ROS activity that happens during stressed conditions. Therefore, higher Chl a levels coincided with carotenoid levels that were averagely higher than the control in 4X NO3 treatment. While under limited nitrate condition, photo-oxidative stress occurs with ROS attacking the chlorophyll system but plants fail to trigger more carotenoids to overcome the effect of oxidative stress which would explain no significant increase after being cultured in NO3 limited conditions12,13. Under limited NO3 treatment, cells appeared to be clumped together, shrivelled in appearance and spine formation were observed (Fig. 2c). According to26, the length of the spines represent the nitrate starvation experienced in microalgae. Interestingly, cells cultured in nitrate abundant cultures did not produce any signs of clumping and were observed to be more prone to form unicellular cells instead of paired sets of coenobia.
Figure 2.
(a) Chlorophyll and carotenoid content of S. regularis measured from cultures under NO3 treatment. Alphabets, roman numerals and numbers represent significant different between chl a, chl b and carotenoid respectively. Asterisks (*) mark significant difference between treatments group. (b) Comparison of colour in medium culture of 1/4X NO3, control and 4X NO3. (c) Scanning electron microscope (SEM) images of S. regularis; (A) 1/4X NO3, (B) control and (C) 4X NO3.
Metabolites detected from aqueous solvent
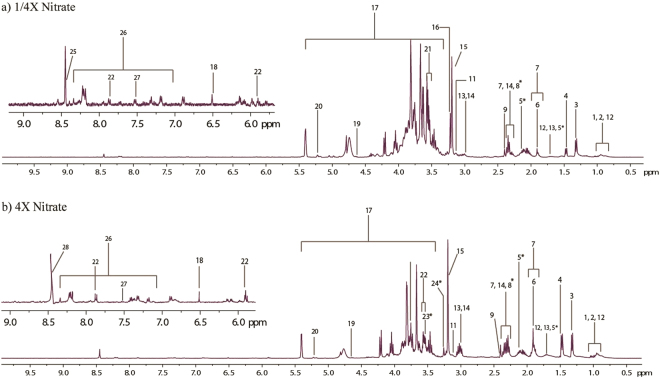
1H-NMR spectroscopy carried out on aqueous extracts returned rich spectra. Figure 3 shows a representative 400 Mhz NMR spectra of a microalgae sample from aqueous extracts of treated sample. The spectra was utilised for the identification of several endogenous metabolites (Table 1). Comparing spectra data from aqueous extracts of treated samples (Fig. 3a and b), some new peaks (trimethylamine, betaine and glycine) were observed in comparison to the control sample. This indicates an elicited response for certain metabolites when exposed to nutrient fluctuations. Choline-2 produces a prominent singlet at 3.19 ppm and is related to resonance from free choline while Choline-1 singlet at 3.20 ppm is a culmination of glycerophosphocholine, phosphorylcholine and minute influences from free choline27. Choline production is an aliphatic monoamine commonly partitioned metabolically as phospholipid- related compounds with the most common being phosphotidylcholine (PC) in microalgae. Ethanolamine produced a visible peak only in NO3 abundant cultures. Ethanolamine is often related as precursors of betaine-type lipids besides being incorporated as phosphotidylethanolamine (PE) into cell membranes28. Betaine lipids are naturally occurring ether-linked glycerolipids containing a betaine moiety with no phosphate group. Betaine was detected in control and NO3 abundance treatment but was absent in NO3 limited cultures (Fig. 3a). The structural capacity of cell membranes in microalgae typically contain diacylglyceryl-3-O-4′-(N,N,N-trimethyl)- homoserine (DGTS), a betaine lipid and PC in interchangeable amounts. It has however, been observed by Boroda, et al.29 that the cell membrane constitution does not distinguish between PC or DGTS as both lipids probably have equivalent functions. Trimethylamine is a constituent for the production of betaine lipids and can be correlated to an increase in ethanolamine production. Glycerol moiety was detected at δ 3.55. Glycerol is an important constituent in regards to both neutral and phospholipid-type lipids and is procured from the direct glycerol pathway or from the fermentation of starch14,30.
Figure 3.
1H NMR spectra of S. regularis extracted using aqueous solvent from (a) 1/4X Nitrate and (b) 4X Nitrate treatment. Metabolites marked with an asterisk appear only in the treatment sample and not in control. Key for spectra refer to Table 1.
Table 1.
Proton chemical shifts observed in Scenedesmus regularis treated with different nitrate concentration (a) metabolite and (b) fatty acid fraction.
| Metabolites | H Group | δH+ (Multiplicity) | Solvent |
|---|---|---|---|
| Lipoproteins | CH3 | 0.91 (m) | Aqueous |
| Leucine | Terminal-CH3, γ-CH3 | 0.95 (t), 1.72 (m) | Aqueous |
| Valine | Terminal CH3, Terminal CH3, β-CH3, α-CH3 | 0.98 (d), 1.03 (d), 2.2 (m), 3.63 (d) | Aqueous |
| Lactate | CH3, CH | 1.33 (d), 4.1 (q) | Aqueous |
| Alanine | CH3, CH | 1.48 (d), 3.76 (qt) | Aqueous |
| Lysine | γ-CH2, δ-CH2, β-CH2, ɛ-CH2 | 1.5 (m), 1.72 (m), 1.9 (m), 3.03 (t) | Aqueous |
| Spermine | H8, H3, (H2, H9, H6, H11) | 1.71 (m), 2.12 (m), 3.14 (m) | Aqueous |
| Acetic acid | CH3 | 1.91 (s) | Aqueous |
| Gamma-amino-N-butyrate | β-CH2, α-CH2, | 1.91 (q), 2.31 (t) | Aqueous |
| Butyric acid-4-amino | 1.9 (m), 2.3 (t), 3.03 (t) | Aqueous | |
| Glutamate | β-CH2, γ-CH2, α-CH | 2.1 (m), 2.33 (dt), 3.77 (t) | Aqueous |
| Succinate | 2xCH2 | 2.40 (s) | Aqueous |
| Ethanolamine | CH2NH2, CH2OH | 3.14 (s), 3.85 (s) | Aqueous |
| Choline-1 | CH3, | 3.19 (s) | Aqueous |
| Choline-2 | CH3, CH2NH | 3.20 (s), 3.55 (m) | Aqueous |
| Betaine | CH3, CH2 | 3.26 (s), 3.91 (s) | Aqueous |
| Sucrose | H10; H9; OH; H17,19; H5, H3, H7 | 3.46 (t), 3.54 (d), 3.66 (s), 3.81 (m), 4.05 (t), 4.21 (d), 5.4 (d) | Aqueous |
| Glycine | CH | 3.55 (s) | Aqueous |
| Glycerol | CHOH, CHOH, COH | 3.55 (d), 3.65 (d), 3.77 (m) | Aqueous |
| α-glucose | CH-1, CH-6 | 3.81 (m), 5.22 (d) | Aqueous |
| β-glucose | CH-1 | 4.63 (d) | Aqueous |
| Uracil | H5, H6 | 5.8 (d), 7.52 (d) | Aqueous |
| Uridine | H(a), H-6 | 5.89 (t), 7.86 (d) | Aqueous |
| Fumarate | CH | 6.51 (s) | Aqueous |
| Nucleoside/Nucleoside | 7.1–7.9 | Aqueous | |
| Formate | 8.45 (s) | Aqueous | |
| Phosphatidylcholine | N(CH3)3, CH2OP | 3.22 (m), 4.27 (m) | Chloroform |
| Glyceroglycolipids | CH3-sn3 | 4.05 (m) | Chloroform |
| Phosphatidylglycerol | Glycerol moiety | 3.61 (m) | Chloroform |
| Phospholipid | 3.1–3.9 | Chloroform | |
| Phosphatidylserine | CH2OP | 4.28 (m) | Chloroform |
| Diacylglycerophospholipid | CH3-sn3 | 4.05 (m) | Chloroform |
| Triglyceride | CH sn1, 3(a), CH sn1, 3(b), CH sn2 | 4.31 (m), 4.14 (m), 5.26 (m) | Chloroform |
| Diglyceride | CH sn1, CH sn2 | 4.31 (m), 5.13 (m) | Chloroform |
| Squalene | 4xCH3, 2x terminal CH3,=CHCH2 | 1.61(d), 1.69 (d), 2.04(m) | Chloroform |
| Sterol | CH3–18 | 0.53 (m) | Chloroform |
| Carotenoids | C=CH, CH=CH, CH-C(CH3) | 6.14 (m), 6.64 (m), 6.35 (m) | Chloroform |
| Phaeophytin | N-H (pophyrin ring) | −1.41 (s), −1.61 (s) | Chloroform |
| Olefinic protons (alkene) | CH=CH/−C=CH | 5.8–6.8 | Chloroform |
| Aldehyde | CH=O | 8.0 (m), 8.55 (m), 9.40 (m), 9.5 (m), 9.53 (m), 10.4 (m) | Chloroform |
| Ester | CH2C=O | 2.32 (triplet of doublets) | Chloroform |
| Free fatty acid | CH2C=O | 2.35 (doublet of singlet) | Chloroform |
| PUFA | Bis-allylic chains | i) C 18:2–2.77 (m) ii) C 18:3–2.81(m) iii) C 22:6–2.84(m) | Chloroform |
| b) Fatty Acid Chains | |||
| Methyl end | n-CH3 | 0.88 (m) | Chloroform |
| PUFA methyl end | n-CH3 | 0.98 (m) | Chloroform |
| Alkyl chain | (-CH2-)n | 1.25 (m) | Chloroform |
| Carboxylic end | CH2COO_ | 1.61 (m) | Chloroform |
| Alkyl chain (allylic) | CH2CH=CH…CH=CHCH2 | 2.06 (m) | Chloroform |
| Carboxylic end* | CH2COO_ | 2.34 (m) | Chloroform |
| Alkyl chain (bisallylic) | -CH=CH(CH2CH=CH)n | 2.81 (m) | Chloroform |
| Double bonds | -CH=CH- | 5.4 (m) | Chloroform |
| PUFA methyl end | n-CH3 | 0.98 (m) | Chloroform |
| Alkyl chain | (-CH2-)n | 1.25 (m) | Chloroform |
| Double bonds | -CH=CH- | 5.4 (m) | Chloroform |
Amine based metabolites are nitrogenous compounds and it is not surprising to observe the reduction of amine and amino acids compounds in limited nitrate condition. Chlorophytes usually contain mono, di- and polyamines in varying amounts. These often contribute either to odour, toxins or play ecologically important roles within the microalgae31. Amines also exhibit certain biological activity and hold a potential for developing value-added products; spermine as an example involved in nucleic acid synthesis and cell division31. Choline, ethanolamine and trimethylamine are common amines often included into cell wall constitution to maintain fluidity and selectivity of the cell membrane.
Carbon source such as alpha- and beta-glucose were also observed in spectra (Fig. 3). Microalgae have adapted to storing carbon in the form of a disaccharide because it is more stable chemically thus allowing the cell to also use sucrose as an osmolyte32. Spectra obtained from 1/4X NO3 cultures were not as sharp as spectra obtained from other treatment cultures. Broad spectra are indicative of heavy molecular weight components inside the extract sample15,33,34. A possible higher relative concentration of sugar or starch granules (in comparison to other metabolites) may have accumulated within the microalgae cell as microalgae cells have been acknowledged to synthesis starch under nitrate starved conditions8,35,36.
Simple osmolytes (sucrose δ 3.66, betaine δ 3.25 and glycine δ 3.55) identified from the spectra show different intensity peaks among treatments. Two intermediates of the tricarboxylic-cycle (TCA) were also detected from all spectra. Fumarate produced a single singlet at δ 6.51 while succinate produced a singlet at δ 2.40 which can interconvert and produce succinyl-CoA, another TCA intermediate. Glutamate, TCA intermediate, was also detected across all spectra. The TCA cycle is therefore indicative of an aerobic route used by S. regularis for procuring its energy currency and subsequent metabolites. Furthermore, metabolites indicative of fermentative pathways were also identified (lactate at δ 1.33; formate at δ 8.45; acetic acid at 1.91).
Metabolites detected from chloroform solvent
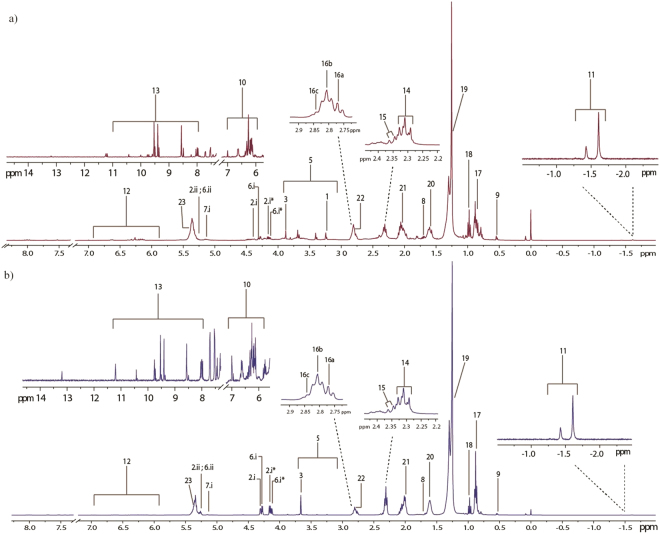
Lipids accumulated by microalgae can be divided into polar and non-polar lipids (neutral) lipids (Fig. 4, Table 1). Both types of lipids follow very clear and distinct accumulation strategies and may transform from one to another depending on the needs of cells37,38.
Figure 4.
1H NMR spectra of S. regularis extracted using chloroform solvent from (a) 1/4X Nitrate and (b) 4X Nitrate treatment. Metabolites marked with an asterisk appear only in the treatment sample and not in control. Key for spectra refer to Table 1.
Microalgae primarily store lipid since formation of lipid requires powerful reducing cofactors and ATP39. These lipids can then be used to either stabilize cell due to its hydrophobic nature or broken down to release energy when required. The signals produced by the protons linked on double bonds from polyunsaturated fatty acids (PUFA) culminate in a multiplet signal at δ 5.4. The allylic chain protons give rise to signals at δ 2.06 and δ 2.81 while protons located near the carboxylic end of the chain produce signals at δ 1.61 and δ 2.34. Protons situated along the long saturated carbon chain gives a very obvious multiplet at δ 1.25. It is interesting to note that fatty acid chains ending with a methyl- end attached with a double bond at an omega position gives rise to signals at δ 0.98 while non-omega methyl end protons culminate in a signal at δ 0.88. Recent studies have designated signals at the multiplet δ 2.81 to correspond to double bonds of three types of PUFA; docosahexaenoic acid, linoleic acid, α-linolenic acid40. The corresponding signals, marked 16a, b and c, were also observed in this study at: (i) δ 2.77 for linoleic acid (C 18:2); (ii) δ 2.81 for α-linolenic acid (C 18:3); (iii) δ 2.84 for Docosahexaenoic acid (C 22:6/DHA). Signals related to carotenoid were found in the region of δ 6.0–δ 6.7. According to Sobolev, et al.41, salient signals appearing in the region between δ 6.0–δ 6.7 can be attributed to a fragment of conjugated trans double bonds, represented chemically as –(CH3)-C=CH-CH-CH-C–(CH3)-, common to all carotenoids. The protons (in bold) give rise to signals of multiplets at δ 6.14, δ 6.64 and δ 6.35 in the literature. A structure related to chlorophyll, i.e. phaeophytin, produces two prominent singlets at δ −1.43 and δ −1.61 in a solvent of CDCL341. Two prominent singlets were also observed at δ −1.41 and δ −1.61 in this study thus proving presence of chlorophyll-related molecules in the non-aqueous mixture. Carotenoids are important pigments of the photosynthetic apparatus and also act as powerful antioxidant to relief the cell from excess photo-oxidation42.
Methyl group of C-18 sterol structures of different sterols often resonate at different ppm. β-sitosterol for example, resonates at δ 0.68 while stigmasterol produces a signal at δ 0.740,41. As a general rule, C-18 moiety of methyl groups belonging to sterol compounds resonates at δ 0.6–δ 0.841. Our study furnished a signal at δ 0.53, pointing to presence of ergosterol-like sterols in the mixture. Microalgae may synthesis either glycerides or fatty alcohols as their main neutral lipid storage43. Triacylglycerides (TAG) are the major storage lipids in a majority of microalgae44. Spectral ID returns glycerol moieties detected at δ 4.31, δ 4.14 and δ 5.26.
Chemical shifts coinciding with signals related to phospholipids are usually detected in the region δ 3.0–δ 4.0540,45. Higher intensity signals (characteristic of phospholipid head group signals) were detected between the region of δ 3.1–δ 4.05 in nutrient abundant and control spectra in comparison to nutrient limited spectra suggesting reduced phospholipid production under nutrient limitation. Signals corresponding to phosphate head groups (CH2OP) connected to phosphatidylcholine and phosphatidylserine were also detected at δ 4.27 and δ 4.28 respectively. The phosphate head group signal at δ 4.02 corresponding to phosphatidylethnolamine41,46 however was not detected. Diacylglycerophospholipids (glyceroglycolipids) presented very weak signals at δ 4.05 and is almost not visible in nutrient limited spectra. This shows the flexibility of the microalgae’s metabolome to shift from membrane-lipid production to storage lipid and vice versa under different environmental cues.
Literature data show alkenes/fatty alcohols (olefinic protons) and aldehydes producing characteristic signals at δ 5.8–δ 6.8 and δ 8.0–δ 11.0 respectively40. These signals are a mixture of signals from fatty hydrocarbons and conjugated/non-conjugated protons of oxylipins of PUFAs such as DHA40. Fatty esters and free fatty acids were also detected in the spectra. In regards to their physiological roles, not much is known47. Some secondary fatty aldehydes and alkenes from microalgae have been studied for their antimicrobial properties and thus may have been synthesised by microalgae as an ecological defence system48,49.
Multivariate analysis
The supervised PLS-DA plot the variation that occurs by separating between groups observed and segregation of the studied group. Both aqueous and chloroform sample showed similar pattern, in which clear cluster were observed between three treatment (Fig. 5). Clustering among groups of treatments was more apparent in the chloroform extracts of nitrate (Fig. 5b; R2X = 0.896). Lower R2X values in aqueous extracts of nitrate treatment (Fig. 5a; R2X = 0.76), signifies a partial diversion in the degree of response towards different nutrient conditions. High predictability values from the PLS-DA score plots (>0.7) shows high reproducibility of data from treatments.
Figure 5.
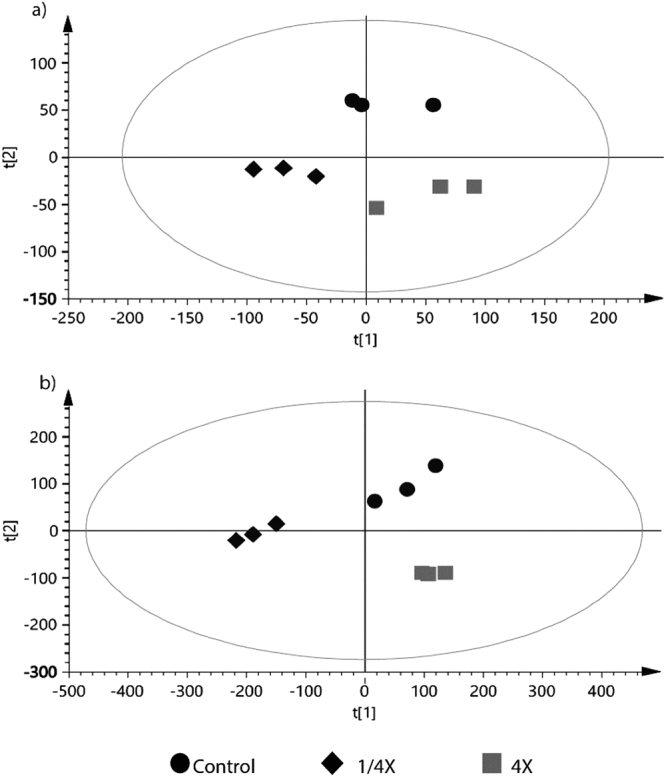
PLS-DA grouped according to different nitrate concentrations. (a) Metabolome extracted from aqueous (b) metabolome extracted from chloroform. Clear separation is observed among the different levels of nutrient treatment, eclipse on the plot represents the 95% confidence interval.
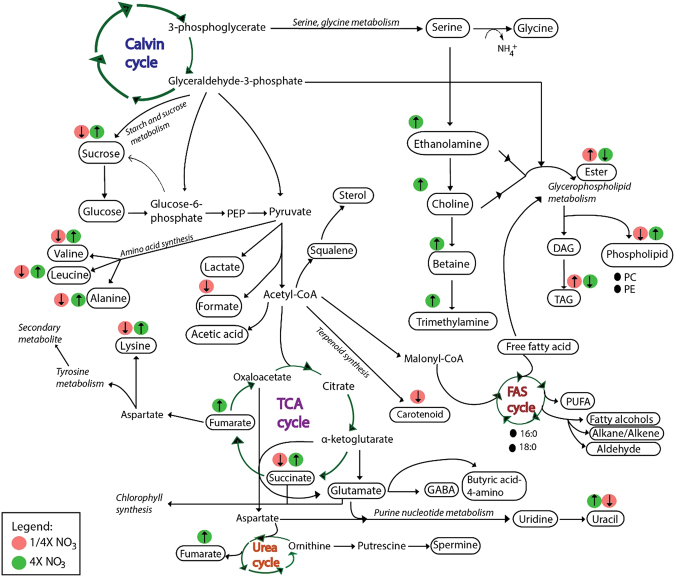
Following PLSDA analysis, the data represent each metabolites were all log2 transformed and tested with student T-test between treatments. Metabolite that show significant different p < 0.05 were then plotted as relative to control into bar charts (Fig. 6). Significant difference between treatments and control were observed in membrane-lipid related metabolites (Choline-2, ethanolamine, betaine), amine compounds and amino acids (trimethylamine, alanine, valine, leucine, aminobutyric acid), TCA cycle intermediates (succinate, fumarate, uracil), energy-related/osmolytes metabolites (sucrose, formate, glycerol) and lipid fragments (ester, fatty acids, TAG, phosphatidycholine, carotenoid and phospholipid) (Fig. 6). These metabolites are involved in Calvin cycle, TCA cycle, Urea cycle and Fatty acid synthesis cycle (Fig. 7). Fluctuations in aforementioned metabolites prove that different nutrient regimes are able to affect the up-regulation and down-regulation of several important pathways.
Figure 6.
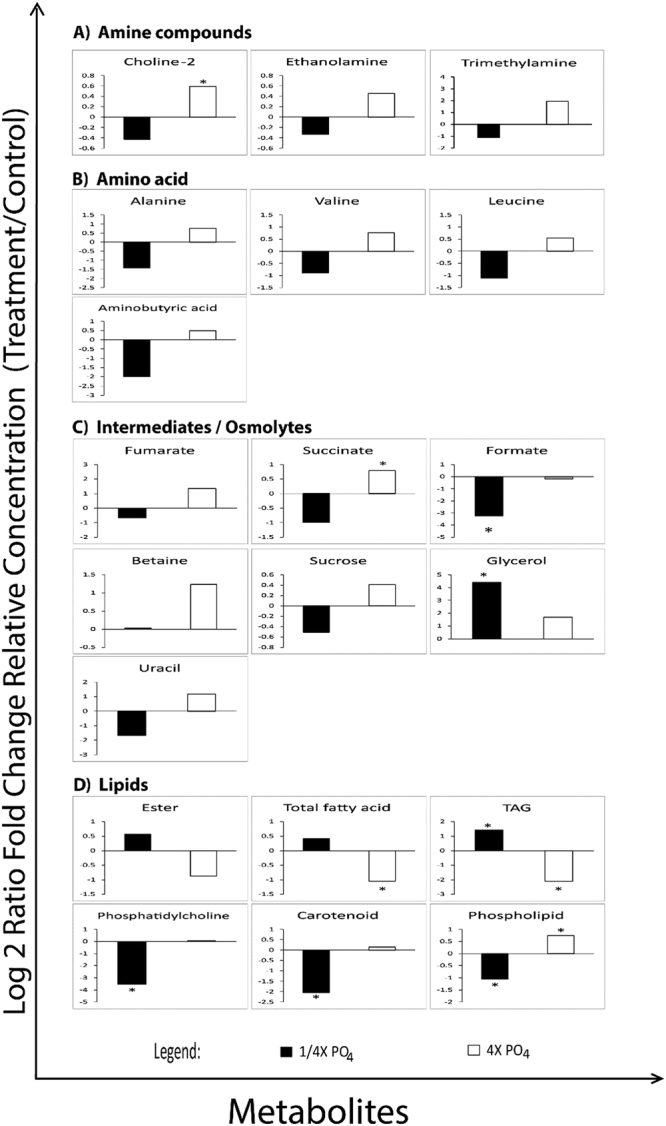
Fold changes of metabolites plotted against relative concentration of the control. Asterisks represents significant different between each nutrient treatment (t-test; P < 0.05).
Figure 7.
Metabolite regulation pathways involved in S. regularis under nitrate treatment. Key for abbreviations: (i) FAS: Fatty acid synthesis; (ii) PC: Phosphatidylcholine; (iii) PE: Phosphatidylethanolamine; (iv) PEP: Phosphoenulpyruvate; (v) TCA: Tricarboxylic cycle; (vi) TAG: Triacylglyceride; (vii) DAG: Diacylglyceride; (viii) GABA: γ-aminobutyric acid.
In 20 metabolites that detected significant versus to control, apart of lipids fragments, all were increased under 4X nitrate treatment, while mostly reduced its content under limit nitrate treatment. Amino acid synthesis pathways were most severely affected in low nitrate treatments. Amino acid contents (alanine, valine, leucine, aminobutyric acid) were significantly reduced in nitrate limited cultures. This in turn would explain that under nitrate limitation, the rate of protein synthesis was greatly reduced thus affecting the synthesis of chlorophyll proteins due to the low nitrate substrate to support synthesis of amino-containing compounds. Example is the reduction of chlorophyll content (Fig. 1) as a direct consequence of reduced chlorophyll protein under nitrate limitation, as the synthesis of chlorophyll requires 4 molecules of nitrogen. However, there seems to be another route of amino acid synthesis and catabolism adopted by microalgae by proteolysis of ribulose-1,5-bisphosphate carboxylase oxygenase (Rubisco) and conversion of other N-containing materials such as nucleotides and their derivatives to maintain amino acids synthesis44. The increment of amino acid and amine compounds were observed in nitrate abundance condition. This is in agreement with treatment of diatom in nitrogen replete treatment, where all amino acids were found increased except histidine and methionine18. Similar explanation for the succinate and fumarate which may be a direct consequence of increased nitrate availability thus providing the cell enough substrate to support photosynthesis, which increases energy output of the cells. This increased energy-production coincides with increased sucrose levels in 4X NO3 cultures. Luxury uptake of nitrate increases photosynthetic rate thus diverting the excess carbon into storage components within the cell.
Nitrate is an important content for the production of RNA material in cells50. Uracil, an amino containing nucleotide, was observed to be reduced considerably under nitrate limitation but significantly increased under nitrate abundance condition. Limitations on DNA/RNA material synthesis may inhibit cell growth or reduce cell division rates in nitrate limited microalgal cultures. Reduction of osmolytes or osmoprotectants may be a common response to nitrate limited stress44. Osmolytes accumulate in the cytoplasm and mainly play an active role in maintaining the ionic balance, help in scavenging ROS, regulating pH and for maintaining the osmotic pressure (osmoregulation) within the microalgae cell43,51. Production of osmolytes are dependent on environmental cues and may be synthesised at any stage of growth51. Betaine levels were significantly elevated in 4X NO3. Betaine may play dual roles as a non-phosphorus containing cell wall component or as an osmolyte in the microalgae cell51.
Glycerophospholipid pathways leading towards formation of phospholipids and pyrimidine biosynthesis pathways were found up-regulated in nitrate abundance. Contradictory, low nutrient condition stimulates phospholipid hydrolysis and the activation of acyl-hydrolase therefore increasing fatty acid content in the cell52. The cell switches its mode of synthesis from forming polar lipids to storage lipids. Increased in fatty acid and TAG was associated with degradation of phospholipids and phosphatidycholine, suggesting that TAG was recycled from membrane proteins44. Nevertheless, while nitrate limitation has been known to achieve higher lipid to protein ratio within the cell, behaviour of polar metabolites such as carbohydrates, proteins and phosphatases are not popularly known to be affected3,53–56. Alternatively, exogenous bioactive molecules and light intensity can be utilised in growth promotion and lipid content accumulation in microalgae57,58.
Materials and Method
Microalgae cultures and nitrate treatment
Scenedesmus regularis (KS-MC1) was obtained from Institute of Marine Biotechnology (IMB) of Universiti Malaysia Terengganu. The culture and maintenance of Scenedesmus regularis was adopted from34. In nitrate treatment, the concentration for NaNO3 was altered while the other components remained the same as in Guillard’s F2 media. The concentrations for NaNO3 in control cultures is 8.824 × 10−1 mM, in 4X nitrate treatment, concentration of NaNO3 was modified to 3.528 mM while in 1/4X treatment, NaNO3 was modified to 2.205 × 10−1 mM. An inoculum concentration of 1 × 105 cells/ml was transferred to freshly prepared treatment media. The treatment cut off point is on Day 8 of culture. For each treatment, three biological samples were used and the experiment were repeated three time.
Quantification of Chlorophyll Content
Chlorophyll content was extracted by taking 1.5 ml aliquots of culture media containing microalgae from each treatment and centrifuged at 10,000 rpm for 5 minutes to get 200 µl of supernatant. Pellet was rehydrated with 1 ml of methanol (Merk, ACS grade) and left to shake at 300 rpm on heating plate at 60 °C for 5 minutes. Experiments were carried out under dark conditions. Tubes were then removed from the heating plates and left to stand in the dark for 24 hours at 4 °C. The tubes were subsequently centrifuged for 5 minutes at 10 000 rpm and the supernatant were collected. Chlorophyll content was determined using a spectrophotometer and measured the absorbance at wavelength 665 and 652 nm59. Chlorophyll content calculated in µg/ml was determined according to Taylor and Fletcher60 formula as follows:
Biomass yield
The wet biomass were harvested from 1.2 L of liquid culture then frozen overnight at −80 °C before subjected to freeze drying (LABCONCO) at −80 °C until constant weight were obtained. The biomass yield was expressed as difference between final dried weight minus initial fresh weight.
SEM preparation and analysis
Fresh microalgae samples in 50 ml was collected by centrifuged and the pellet was underwent general fixation overnight at room temperature using 2.5% glutaraldehyde in 0.1 M sodium cacodyle. Solution was then discarded and pellet rinsed with 0.1 M sodium cocadylate buffer (pH 7.2). Pellet was then submerged in 1% osmiumtetraoxide prepared in 0.1 M sodium cocadylate buffer and left to stand for 4 hours. The pellet was then rinsed with buffer and underwent a series of dehydration steps using various concentrations of ethanol. Pellet was then mounted on specimen stub and left to dry in a fume hood with the aid of hexamethyldisilazane (HMDS – Merk, ACS grade). Specimen stub was then coated with gold using auto fine coater (INOS, UMT).
Extraction procedure and NMR spectra acquisition
A total of 0.215 g of freeze-dried algal biomass was used for (homogenizer) extraction34. Extraction solvents were prepared by adding Chloroform (99% absolute chloroform, ACS grade) with aqueous solvent that prior mixed from 99% absolute methanol (ACS, ISO, Reag grade) and pure distilled water into 1:1 ratio. Two layers of supernatant formed following each extraction and each layer was collected separately and dried up in freeze drier. The aqueous phase of extract was re-suspended in 600 µl of phosphate buffer (0.1 M, pH 7) containing 10% D2O and the chloroform phase extract was re-suspended in 600 µl of chloroform –d 99.8% containing 0.01% of sodium tetramethysilane (TMS). Both sample were then subjected to 10 min of 10,000 rpm centrifugation before transferred into 5 mm NMR tube34.
NMR analysis and data reduction
Analysis and further furnishing of NMR peaks (baseline correction, phasing, solvent peak removal) were carried out in Mnova (Ver 9.0 – Mestrelab) and Topspin (Ver 3.2 – Bruker). TMS (δ 0.0) was used as the reference point in chloroform spectra while alanine (δ 1.48; doublet) was used as reference peaks in aqueous spectra. The interfering signal of suppressed H2O resonance and CHCl3 in aqueous and chloroform spectra was removed. NMR peaks were then identified based on their integration, chemical shifts and peak multiplicity.
Multi-variant analysis
All variables were log-centred and scaled to unit variance before proceeding with supervised and unsupervised multivariated analysis, namely Principal Component Analysis (PCA) and Partial List Squares Discriminant Analysis (PLS-DA) using SIMCA (SIMCA-P + 1.3 − Umetrics) software. The validity of the models was assessed by statistical parameters R2 (correlation coefficient) and Q2 (cross-validation correlation coefficient). Average of each variable were first Log2 transformed and Two tailed student’s T-tests were carried out on metabolites to determined statistical differences between input variable and cut out point at p < 0.05.
Conclusion
Cell cultures respond to nutrient fluctuations through a myriad of strategies. Under N deficiency, production of photosynthetic pigments is severely stunted. NO3 addition lead to chlorophyll pigments and amino acid accumulation while NO3 limitation had very low amino acid content but was very effective at increasing neutral lipid content. Multi-variate analysis showed that more changes were observable in non-polar extracts when microalgae are exposed to nutrient deficiency.
A basic fingerprint of the biochemical changes using NMR technology coupled with multi-variate analysis proved the capability of NMR as a time-saving and efficient data mining analytical tool as biochemical changes in both polar and non-polar could be captured simultaneously. More studies need to be carried out in order to determine the correct proportions of nutrient content in which to stimulate production of favourable metabolites without compromising productivity. Metabolite activity are important indicators in pathway alterations which would come in handy when determining biomarkers or designing nutrient regimes for economical output.
Acknowledgements
The authors acknowledge the financial support by the Ministry of Higher Education and Universiti Malaysia Terengganu for the conduct of the research under the Fundamental Research Grant Scheme (Vot 59264).
Author Contributions
N.L., T.S. and A.A. designed the study; K.Y. performed and collected the data; Y.L., S.S. and K.Y. analysed the data; N.L., K.Y. and S.S. drafted the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ramanna L, Guldhe A, Rawat I, Bux F. The optimization of biomass and lipid yields of Chlorella sorokiniana when using wastewater supplemented with different nitrogen sources. Bioresource Technology. 2014;168:127–135. doi: 10.1016/j.biortech.2014.03.064. [DOI] [PubMed] [Google Scholar]
- 2.Sasso S, Pohnert G, Lohr M, Mittag M, Hertweck C. Microalgae in the postgenomic era: a blooming reservoir for new natural products. FEMS microbiology reviews. 2012;36:761–785. doi: 10.1111/j.1574-6976.2011.00304.x. [DOI] [PubMed] [Google Scholar]
- 3.Kumari P, Kumar M, Reddy CRK, Jha B. Nitrate and Phosphate Regimes Induced Lipidomic and Biochemical Changes in the Intertidal Macroalga Ulva lactuca (Ulvophyceae, Chlorophyta) Plant and Cell Physiology. 2014;55:52–63. doi: 10.1093/pcp/pct156. [DOI] [PubMed] [Google Scholar]
- 4.Adams, C. Studies on nitrogen and silicon deficiency in microalgal lipid production. (Utah State University, 2013).
- 5.Powell N, Shilton A, Chisti Y, Pratt S. Towards a luxury uptake process via microalgae–defining the polyphosphate dynamics. Water Research. 2009;43:4207–4213. doi: 10.1016/j.watres.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 6.Dean AP, Sigee DC, Estrada B, Pittman JK. Using FTIR spectroscopy for rapid determination of lipid accumulation in response to nitrogen limitation in freshwater microalgae. Bioresource Technology. 2010;101:4499–4507. doi: 10.1016/j.biortech.2010.01.065. [DOI] [PubMed] [Google Scholar]
- 7.Sanz-Luque, E., Chamizo-Ampudia, A., Llamas, A., Galvan, A. & Fernandez, E. Understanding nitrate assimilation and its regulation in microalgae. Frontiers in plant science6 (2015). [DOI] [PMC free article] [PubMed]
- 8.Cuellar-Bermudez SP, et al. Extraction and purification of high-value metabolites from microalgae: essential lipids, astaxanthin and phycobiliproteins. Microbial Biotechnology. 2015;8:190–209. doi: 10.1111/1751-7915.12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.El-Sheek, M. & Rady, A. Effect of phosphorus starvation on growth, photosynthesis and some metabolic processes in the unicellular green alga Chlorella kessleri. Phyton (1995).
- 10.Pollesello P, et al. Lipid extracts from different algal species:1H and13C-NMR spectroscopic studies as a new tool to screen differences in the composition of fatty acids, sterols and carotenoids. J Appl Phycol. 1992;4:315–322. doi: 10.1007/BF02185788. [DOI] [Google Scholar]
- 11.Markou G, Nerantzis E. Microalgae for high-value compounds and biofuels production: A review with focus on cultivation under stress conditions. Biotechnology Advances. 2013;31:1532–1542. doi: 10.1016/j.biotechadv.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 12.Kang CD, An JY, Park TH, Sim SJ. Astaxanthin biosynthesis from simultaneous N and P uptake by the green alga Haematococcus pluvialis in primary-treated wastewater. Biochemical Engineering Journal. 2006;31:234–238. doi: 10.1016/j.bej.2006.08.002. [DOI] [Google Scholar]
- 13.Stacey GN, Day JG. Putting cells to sleep for future science. Nat Biotech. 2014;32:320–322. doi: 10.1038/nbt.2869. [DOI] [PubMed] [Google Scholar]
- 14.Singh S, Kate BN, Banerjee UC. Bioactive Compounds from Cyanobacteria and Microalgae: An Overview. Critical Reviews in Biotechnology. 2005;25:73–95. doi: 10.1080/07388550500248498. [DOI] [PubMed] [Google Scholar]
- 15.Beckonert O, et al. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nature protocols. 2007;2:2692–2703. doi: 10.1038/nprot.2007.376. [DOI] [PubMed] [Google Scholar]
- 16.Rosenberg JN, et al. Comparative Analyses of Three Chlorella Species in Response to Light and Sugar Reveal Distinctive Lipid Accumulation Patterns in the Microalga C. sorokiniana. PLoS ONE. 2014;9:e92460. doi: 10.1371/journal.pone.0092460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mahrous EA, Farag MA. Two dimensional NMR spectroscopic approaches for exploring plant metabolome: a review. Journal of advanced research. 2015;6:3–15. doi: 10.1016/j.jare.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hegewald E, Wolf M, Keller A, Friedl T, Krienitz L. ITS2 sequence-structure phylogeny in the Scenedesmaceae with special reference to Coelastrum (Chlorophyta, Chlorophyceae), including the new genera Comasiella and Pectinodesmus. Phycologia. 2010;49:325–335. doi: 10.2216/09-61.1. [DOI] [Google Scholar]
- 19.Tuchman N, Schollett M, Rier S, Geddes P. Differential Heterotrophic Utilization of Organic Compounds by Diatoms and Bacteria under Light and Dark Conditions. Hydrobiologia. 2006;561:167–177. doi: 10.1007/s10750-005-1612-4. [DOI] [Google Scholar]
- 20.Patey MD, et al. Determination of nitrate and phosphate in seawater at nanomolar concentrations. TrAC Trends in Analytical Chemistry. 2008;27:169–182. doi: 10.1016/j.trac.2007.12.006. [DOI] [Google Scholar]
- 21.Cuellar-Bermudez SP, et al. Nutrients utilization and contaminants removal. A review of two approaches of algae and cyanobacteria in wastewater. Algal Research. 2017;24:438–449. doi: 10.1016/j.algal.2016.08.018. [DOI] [Google Scholar]
- 22.Delrue F, Álvarez-Díaz PD, Fon-Sing S, Fleury G, Sassi J-F. The environmental biorefinery: Using microalgae to remediate wastewater, a win-win paradigm. Energies. 2016;9:132. doi: 10.3390/en9030132. [DOI] [Google Scholar]
- 23.Bui TVL, Ross IL, Jakob G, Hankamer B. Impact of Procedural Steps and Cryopreservation Agents in the Cryopreservation of Chlorophyte Microalgae. PLoS ONE. 2013;8:e78668. doi: 10.1371/journal.pone.0078668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prathima Devi M, Venkata Subhash G, Venkata Mohan S. Heterotrophic cultivation of mixed microalgae for lipid accumulation and wastewater treatment during sequential growth and starvation phases: Effect of nutrient supplementation. Renewable Energy. 2012;43:276–283. doi: 10.1016/j.renene.2011.11.021. [DOI] [Google Scholar]
- 25.Pérez-Pérez M, Couso I, Heredia-Martínez L, Crespo J. Monitoring Autophagy in the Model Green Microalga Chlamydomonas reinhardtii. Cells. 2017;6:36. doi: 10.3390/cells6040036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pancha I, et al. Nitrogen stress triggered biochemical and morphological changes in the microalgae Scenedesmus sp. CCNM 1077. Bioresource Technology. 2014;156:146–154. doi: 10.1016/j.biortech.2014.01.025. [DOI] [PubMed] [Google Scholar]
- 27.Mahmud I, Kousik C, Hassell R, Chowdhury K, Boroujerdi AF. NMR Spectroscopy Identifies Metabolites translocated from powdery mildew resistant rootstocks to susceptible watermelon scions. Journal of Agricultural and Food Chemistry. 2015;63:8083–8091. doi: 10.1021/acs.jafc.5b02108. [DOI] [PubMed] [Google Scholar]
- 28.Khozin-Goldberg, I. In The Physiology of Microalgae 413–484 (Springer, 2016).
- 29.Boroda A, Aizdaicher N, Odintsova N. The influence of ultra-low temperatures on marine microalgal cells. J Appl Phycol. 2014;26:387–397. doi: 10.1007/s10811-013-0093-5. [DOI] [Google Scholar]
- 30.Constantinou MA, et al. 1H NMR-based metabonomics for the diagnosis of inborn errors of metabolism in urine. Analytica Chimica Acta. 2005;542:169–177. doi: 10.1016/j.aca.2005.03.059. [DOI] [Google Scholar]
- 31.Day, J. G. & Harding, K. In Plant Cryopreservation: A Practical Guide (ed Barbara M. Reed) 95–116 (Springer New York, 2008).
- 32.Araujo GS, et al. Extraction of lipids from microalgae by ultrasound application: prospection of the optimal extraction method. Ultrasonics sonochemistry. 2013;20:95–98. doi: 10.1016/j.ultsonch.2012.07.027. [DOI] [PubMed] [Google Scholar]
- 33.Smolinska A, Blanchet L, Buydens LM, Wijmenga SS. NMR and pattern recognition methods in metabolomics: from data acquisition to biomarker discovery: a review. Analytica chimica acta. 2012;750:82–97. doi: 10.1016/j.aca.2012.05.049. [DOI] [PubMed] [Google Scholar]
- 34.Machado M, et al. Comprehensive metabolic reprograming in freshwater Nitzschia palea strains undergoing nitrogen starvation is likely associated with its ecological origin. Algal Research. 2016;18:116–126. doi: 10.1016/j.algal.2016.06.003. [DOI] [Google Scholar]
- 35.Chen F. High cell density culture of microalgae in heterotrophic growth. Trends in Biotechnology. 1996;14:421–426. doi: 10.1016/0167-7799(96)10060-3. [DOI] [Google Scholar]
- 36.Antti H, et al. Statistical experimental design and partial least squares regression analysis of biofluid metabonomic NMR and clinical chemistry data for screening of adverse drug effects. Chemometrics and intelligent laboratory systems. 2004;73:139–149. doi: 10.1016/j.chemolab.2003.11.013. [DOI] [Google Scholar]
- 37.Ramachandra T, Sajina K, Supriya G. Lipid composition in microalgal community under laboratory and outdoor conditions. Indian Journal of Science and Technology. 2011;4:1488–1494. [Google Scholar]
- 38.Rastoll MJ, et al. The development of a cryopreservation method suitable for a large cyanobacteria collection. J Appl Phycol. 2013;25:1483–1493. doi: 10.1007/s10811-013-0001-z. [DOI] [Google Scholar]
- 39.Grimi N, et al. Selective extraction from microalgae Nannochloropsis sp. using different methods of cell disruption. Bioresource Technology. 2014;153:254–259. doi: 10.1016/j.biortech.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 40.Sarpal AS, et al. Determination of lipid content of oleaginous microalgal biomass by NMR spectroscopic and GC–MS techniques. Analytical and bioanalytical chemistry. 2015;407:3799–3816. doi: 10.1007/s00216-015-8613-6. [DOI] [PubMed] [Google Scholar]
- 41.Sobolev AP, Brosio E, Gianferri R, Segre AL. Metabolic profile of lettuce leaves by high‐field NMR spectra. Magnetic Resonance in Chemistry. 2005;43:625–638. doi: 10.1002/mrc.1618. [DOI] [PubMed] [Google Scholar]
- 42.Gupta V, Ratha SK, Sood A, Chaudhary V, Prasanna R. New insights into the biodiversity and applications of cyanobacteria (blue-green algae)—Prospects and challenges. Algal Research. 2013;2:79–97. doi: 10.1016/j.algal.2013.01.006. [DOI] [Google Scholar]
- 43.Mimouni V, et al. The potential of microalgae for the production of bioactive molecules of pharmaceutical interest. Current pharmaceutical biotechnology. 2012;13:2733–2750. doi: 10.2174/138920112804724828. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y, et al. Free amino acids and small molecular acids profiling of marine microalga Isochrysis zhangjiangensis under nitrogen deficiency. Algal Research. 2016;13:207–217. doi: 10.1016/j.algal.2015.12.001. [DOI] [Google Scholar]
- 45.Stringer KA, et al. Metabolic consequences of sepsis-induced acute lung injury revealed by plasma 1H-nuclear magnetic resonance quantitative metabolomics and computational analysis. American Journal of Physiology-Lung Cellular and Molecular Physiology. 2010;300:L4–L11. doi: 10.1152/ajplung.00231.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y, et al. A comparative study: the impact of different lipid extraction methods on current microalgal lipid research. Microb Cell Fact. 2014;13:14. doi: 10.1186/1475-2859-13-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Widjaja A, Chien C-C, Ju Y-H. Study of increasing lipid production from fresh water microalgae Chlorella vulgaris. Journal of the Taiwan Institute of Chemical Engineers. 2009;40:13–20. doi: 10.1016/j.jtice.2008.07.007. [DOI] [Google Scholar]
- 48.Shannon E, Abu-Ghannam N. Antibacterial derivatives of marine algae: An overview of pharmacological mechanisms and applications. Marine Drugs. 2016;14:81. doi: 10.3390/md14040081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Y, et al. A novel approach to transforming a non-targeted metabolic profiling method to a pseudo-targeted method using the retention time locking gas chromatography/mass spectrometry-selected ions monitoring. Journal of Chromatography A. 2012;1255:228–236. doi: 10.1016/j.chroma.2012.01.076. [DOI] [PubMed] [Google Scholar]
- 50.Menéndez M, Herrera-Silveira J, Comín FA. Effect of nitrogen and phosphorus supply on growth, chlorophyll content and tissue composition of the macroalga Chaetomorpha linum (OF Mull), Kutz, in a Mediterranean Coastal Lagoon. Scientia Marina, 2002, vol. 66, num. 2002;4:355–364. [Google Scholar]
- 51.Slama I, Abdelly C, Bouchereau A, Flowers T, Savouré A. Diversity, distribution and roles of osmoprotective compounds accumulated in halophytes under abiotic stress. Annals of Botany. 2015;115:433–447. doi: 10.1093/aob/mcu239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xin L, Hong-ying H, Ke G, Ying-xue S. Effects of different nitrogen and phosphorus concentrations on the growth, nutrient uptake, and lipid accumulation of a freshwater microalga Scenedesmus sp. Bioresource Technology. 2010;101:5494–5500. doi: 10.1016/j.biortech.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 53.Wikfors GH. Altering growth and gross chemical composition of two microalgal molluscan food species by varying nitrate and phosphate. Aquaculture. 1986;59:1–14. doi: 10.1016/0044-8486(86)90073-6. [DOI] [Google Scholar]
- 54.Arnold AA, et al. Identification of lipid and saccharide constituents of whole microalgal cells by 13C solid-state NMR. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2015;1848:369–377. doi: 10.1016/j.bbamem.2014.07.017. [DOI] [PubMed] [Google Scholar]
- 55.Juneja A, Ceballos RM, Murthy GS. Effects of environmental factors and nutrient availability on the biochemical composition of algae for biofuels production: a review. Energies. 2013;6:4607–4638. doi: 10.3390/en6094607. [DOI] [Google Scholar]
- 56.Aléman-Nava GS, et al. Two-stage cultivation of Nannochloropsis oculata for lipid production using reversible alkaline flocculation. Bioresource Technology. 2017;226:18–23. doi: 10.1016/j.biortech.2016.11.121. [DOI] [PubMed] [Google Scholar]
- 57.Parsaeimehr A, Mancera-Andrade EI, Robledo-Padilla F, Iqbal HMN, Parra-Saldivar R. A chemical approach to manipulate the algal growth, lipid content and high-value alpha-linolenic acid for biodiesel production. Algal Research. 2017;26:312–322. doi: 10.1016/j.algal.2017.08.016. [DOI] [Google Scholar]
- 58.Cuellar-Bermudez SP, et al. Effects of light intensity and carbon dioxide on lipids and fatty acids produced by Synechocystis sp. PCC6803 during continuous flow. Algal Research. 2015;12:10–16. doi: 10.1016/j.algal.2015.07.018. [DOI] [Google Scholar]
- 59.Ritchie R. Universal chlorophyll equations for estimating chlorophylls a, b, c, and d and total chlorophylls in natural assemblages of photosynthetic organisms using acetone, methanol, or ethanol solvents. Photosynthetica. 2008;46:115–126. doi: 10.1007/s11099-008-0019-7. [DOI] [Google Scholar]
- 60.Taylor R, Fletcher RL. Cryopreservation of eukaryotic algae – a review of methodologies. J Appl Phycol. 1998;10:481–501. doi: 10.1023/A:1008094622412. [DOI] [Google Scholar]