Abstract
Cannabigerol (CBG) is one of the major phytocannabinoids present in Cannabis sativa L. that is attracting pharmacological interest because it is non-psychotropic and is abundant in some industrial hemp varieties. The aim of this work was to investigate in parallel the binding properties of CBG to cannabinoid CB1 (CB1R) and CB2 (CB2R) receptors and the effects of the compound on agonist activation of those receptors and of CB1–CB2 heteroreceptor complexes. Using [3H]-CP-55940, CBG competed with low micromolar Ki values the binding to CB1R and CB2R. Homogeneous binding in living cells, which is only technically possible for the CB2R, provided a 152 nM Ki value. Also interesting, CBG competed the binding of [3H]-WIN-55,212-2 to CB2R but not to CB1R (Ki: 2.7 versus >30 μM). The phytocannabinoid modulated signaling mediated by receptors and receptor heteromers even at low concentrations of 0.1–1 μM. cAMP, pERK, β-arrestin recruitment and label-free assays in HEK-293T cells expressing the receptors and treated with endocannabinoids or selective agonists proved that CBG is a partial agonist of CB2R. The action on cells expressing heteromers was similar to that obtained in cells expressing the CB2R. The effect of CBG on CB1R was measurable but the underlying molecular mechanisms remain uncertain. The results indicate that CBG is indeed effective as regulator of endocannabinoid signaling.
Keywords: cannabinoid receptor, cannabigerol, G-protein-coupled receptor, phytocannabinoid, TR-FRET, partial agonist
Introduction
Cannabinoid compounds bind and activate cannabinoid CB1 (CB1R) and CB2 (CB2R) receptors, which belong to the superfamily of G-protein-coupled receptors. There are many ways to classify them, but the most used distinguishes between endogenous molecules (endocannabinoids), phytocannabinoids and synthetic cannabinoids. Endocannabinoids and one of the most studied phytocannabinoids, Δ9-tetrahydrocannabinol (Δ9-THC), are agonists with more or less CB1R/CB2R selectivity. Furthermore, synthetic cannabinoids mainly act (as agonists or antagonists) by binding to the orthosteric site of receptors (Mechoulam, 2016). Indeed, there is a limited number of molecules, either synthetic or phytocannabinoids, that behave as allosteric modulators of cannabinoid receptor function.
Anandamide and 2-arachidonoyl glycerol (2-AG) are the two main endocannabinoids, being synthesized from membrane lipids and having an alkyl-amide chemical structure. They are retrograde effectors being produced in the post-synaptic neuron to act in the pre-synaptic neuron where they regulate the release of neurotransmitters (Diana and Marty, 2004).
Phytocannabinoids are phenolic terpenes biosynthesized in nature nearly exclusively in the Cannabis sativa L. plant. In the Cannabis plant, all cannabinoids are biosynthesized in the acid form, mainly Δ9-THCA, CBDA, etc. CBGA is the first molecule formed in the biosynthetic pathway and the substrate of Δ9-tetrahydrocannabinol-synthase and CBD-synthase (Fellermeier and Zenk, 1998). The pharmacologic effects of Cannabis components, traditionally consumed through inhalation, are attributed to the decarboxylated neutral products of above mentioned acids: Δ9-THC, CBD, and CBG.
Synthetic cannabinoids are very different in chemical structure. For instance, they may be indoles like WIN-55,212-2, AM-1241 or JWH-018, or phenolic, phenols lacking the pyrene ring, like CP-55,940 or HU-308. All these compounds have been used in cannabinoid research and have helped to unveil pharmacological aspects of the endocannabinoid system. It should be noted that some of these compounds have recently arrived at the streets sold as legal highs, thus raising Public Health concerns (Adams et al., 2017; Weinstein et al., 2017).
The endocannabinoid system is constituted by the endogenous cannabinoids, the enzymes that produce and degrade them, and by the receptors that mediate their actions. Whereas endocannabinoids consist of molecules with aliphatic structure, AEA and 2-AG, the structure of natural cannabinoids, derived from C. sativa L., is fairly different [see (Lu and Mackie, 2016) and references therein]. Although it is well established that one of the main active components of the plant and one of the few that are psychoactive, namely Δ9-THC, acts via cannabinoid receptors, there is controversy on whether these receptors mediate the action of phytocannabinoids such as CBN, CBD or CBG. As happened the last years for CBD, a new research and revision of the cannabinoid receptor pharmacology must be done with the rest of phytocannabinoids as CBG. A further phenomenon that may be considered to understand the action of molecules from C. sativa L. and its extracts is the fact that cannabinoid receptors may form heteromers, namely CB1–CB2 heteroreceptors, which display particular functional properties (Callén et al., 2012). It should be noted that in CNS those heteromers are mainly expressed in pallidal neurons (Lanciego et al., 2011; Sierra et al., 2015) and in activated microglia (Navarro et al., 2018a).
Cannabigerol was isolated, characterized and synthetized by the same researchers than reported the structure of the main psychotropic agent of Cannabis, Δ9-THC (Gaoni and Mechoulan, 1964). Few years later in vivo assays showed that CBG was non-psychoactive (Grunfeld and Edery, 1969; Mechoulam et al., 1970). The lower concentration and the lack of psychoactivity was probably the cause that CBG was shadowed by Δ9-THC. In fact, CBG has attracted less attention than Δ9-THC and even than CBD, but nowadays is gaining interest among the scientific community. Some commercial hemp varieties have CBG and CBGA as main cannabinoids and, therefore, CBG is another of the phytocannabinoids to be considered by the unregulated market of hemp oils and derivatives. As recently pointed out, the increased therapeutic potential of C. sativa L. components requires a more in deep understanding of the pharmacology of phytocannabinoids other than Δ9-THC, namely CBD, CBG, CBN, Δ9-THCV, Δ8-THC, CBC and CBDV (Turner et al., 2017).
Preliminary results using membranes from mice brain or from CHO cells expressing the human CB2R led to postulate that CBG could be a partial agonist at both CB1R and CB2R with Ki values in the 300–500 nM range (Gauson et al., 2007; Pertwee, 2008). The first published data on the binding of CBG to human CB1R and CB2R were provided by (Rosenthaler et al., 2014) working with [3H]CP-55,940 as radioligand and with preparations from Sf9 cells co-expressing one receptor and the Gαi3β1γ2 protein. The Ki values obtained in competition assays are 897 and 153 nM for CB1R and CB2R, respectively. CBG may modulate the activity of transient receptor potential channels of ankyrin type-1; however, the EC50 values lie in the micromolar range (De Petrocellis et al., 2008). It has been reported that CBG binds to CB1R (Ki = 381 nM) from mouse brain membranes and CB2R (Ki = 2.6 μM) from CHO cells expressing the human receptor; CBG at high concentrations (10 μM) antagonized [35S]GTPγS binding in mouse brain membranes treated with AEA or CP-55940 (Cascio et al., 2010). Authors also reported CBG as α2-adrenoceptor agonist at nanomolar levels (EC50 = 0.2 nM), and being also able to antagonize [35S]GTPγS binding upon stimulation of the 5HT1A receptor by 1 μM 8-OH-DPAT (Cascio et al., 2010). Other findings indicate that CBG can act as (i) agonist/desensitizer of TRPA1 (EC50 = 700 nM), (ii) agonist of TRPV1 (EC50 = 1.3 μM) (iii) agonist of TRPV2 (EC50 = 1.7 μM), (iv) antagonist of TRPM8 channels (IC50 = 160 nM) and v) inhibitor of AEA cell uptake (Ki = 11.3 μM) (De Petrocellis et al., 2011). More recently, the PPARγ has been reported as target of the phytocannabinoid CBG (Ki = 11.7 μM) that at high concentrations, in the 10–25 μM range, may enhance the PPARγ transcriptional activity (Granja et al., 2012; Nadal et al., 2017). A recent review substantiates the complexity of the field and highlights that other players, GPR55 for instance, are also targeted by cannabinoids (Solymosi and Kofalvi, 2017).
The aim of this work was to characterize CBG pharmacology on the cannabinoid receptors using binding and measurement of different signal transduction mechanisms in living HEK-293T cells expressing human CB1R, CB2R, or CB1–CB2 heteroreceptor complexes. The results indicate that, in our experimental conditions, CBG mainly acts on CB2R and behaves as a partial agonist.
Materials and Methods
Reagents
ACEA, JWH133, and AEA were purchased from Tocris Bioscience (Bristol, United Kingdom), CBD and CBG analytical standard solutions were purchased from THCpharm (Frankfurt, DE). Concentrated (10 mM) stock solutions prepared in ethanol (CBG, ACEA, and AEA) or DMSO (JWH133 and CM-157) were stored at -20°C. In each experimental session, aliquots of concentrated solutions of compounds were thawed and conveniently diluted in the appropriate experimental solution. For non-radioactive binding assays, TLB was obtained from Cisbio Bioassays (LABMED; Codolet, France). The Tb derivative of O6-benzylguanine was synthesized by Cisbio Bioassays and is commercialized as SNAP-Lumi4-Tb (SSNPTBC; Cisbio Assays). The plasmid encoding for the SNAP-tagged human CB2R used for transient transfection was obtained from Cisbio Bioassays (PSNAP-CB2). CB2R agonist 3-[[4-[2-tert-butyl-1-(tetrahydropyran-4-ylmethyl)benzimidazol-5-yl]sulfonyl-2-pyridyl]oxy]propan-1-amine (CM-157) conjugated to a fluorescent probe was developed in collaboration with Cisbio Bioassays (Martínez-Pinilla et al., 2016).
Cannabinoid Isolation, Purification and Analysis
Cannabidiol was purified from dried leaves and inflorescences of the Cannabis variety SARA (CPVO file number: 20150098), CBG from the variety AIDA (CPVO file number: 20160167) following a previously described method (Nadal, 2016) that provides compounds with >95% purity. An Agilent liquid chromatography set-up (Model 1260, Pittsburgh, PA, United States) consisting of a binary pump, a vacuum degasser, a column oven, an autosampler and a diode array detector (DAD) equipped with a 150 mm length × 2.1 mm internal diameter, 2.7 μm pore size Poroshell 120 EC-C18 column was used for the quality control of the purified cannabinoids. The analysis was performed using water and acetonitrile both containing ammonium formate 50 mM as mobile phases. Flow-rate was 0.2 mL/min and the injection volume was 3 μL. Chromatographic peaks were recorded at 210 nm. All determinations were carried out at 35°C. All samples were analyzed in duplicate. The results of each cannabinoid purity, 96.04% for CBD and 99.9% for CBG, were calculated as weight (%) versus a commercial standard from THCpharm (CBD batch n° L01258-M-1.0; CBG batch n° L01260-M-1.0).
Radioligand Binding Assays
Cell Culture and Membrane Preparation
For radioligand binding experiments CHO cells, stably transfected with cDNA for human CB1 or CB2 cannabinoid receptors, were grown adherently and maintained in Ham’s F12 containing 10% fetal bovine serum, penicillin (100 U/mL), streptomycin (100 μg/mL) and geneticin (G418, 0.4 mg/mL) at 37°C in a humid atmosphere of 5% CO2. Membranes were prepared from cells washed with PBS and scraped off plates in ice-cold hypotonic buffer (5 mM Tris HCl, 2 mM EDTA, pH 7.4). The cell suspension was homogenized with a Polytron and then centrifuged for 30 min at 40,000 × g.
Saturation Binding Experiments
[3H]-CP-55940 saturation binding experiments (specific activity 169 Ci/mmol, Perkin Elmer) were performed incubating different concentrations of the radioligand (0.03 – 10 nM) in binding buffer (50 mM Tris-HCl, pH 7.4, 2.5 mM EDTA, 5 mM MgCl2 for CB1R or 50 mM Tris-HCl, pH 7.4, 1 mM EDTA, 5 mM MgCl2 for CB2R) using CHO membranes expressing the human versions of CB1R or CB2R (10 μg protein/sample) at 30°C. Non-specific binding was determined in the presence of 1 μM WIN-55,212-2. At the end of the incubation period (90 min for CB1R or 60 min for CB2R) bound and free radioactivity were separated in a cell harvester (Brandel Instruments) by filtering the assay mixture through Whatman GF/B glass fiber filters. The filter-bound radioactivity was counted in a 2810 TR liquid scintillation counter (Perkin Elmer).
[3H]-WIN-55,212-2 saturation binding experiments (specific activity 48 Ci/mmol, Perkin Elmer) were performed incubating different concentrations of the radioligand (0.5–100 nM for CB1R or 0.2–40 nM for CB2R) in binding buffer (50 mM Tris-HCl, pH 7.4, 1 mM EDTA, 5 mM MgCl2) with CB1R- or CB2R-containing CHO cell membranes (10 μg protein/sample) at 30°C. Non-specific binding was determined in the presence of 1 μM WIN-55,212-2. At the end of the incubation period (60 min) bound and free radioactivity were separated in a cell harvester (Brandel Instruments) by filtering the assay mixture through Whatman GF/B glass fiber filters. The filter-bound radioactivity was counted in a 2810 TR liquid scintillation counter (Perkin Elmer).
Competition Binding Experiments
[3H]-CP-55940 competition binding experiments were performed incubating 0.3 nM of radioligand and different concentrations of the tested compounds with membranes obtained from CHO cells expressing human CB1 or CB2 receptors (10 μg protein/sample) for 90 min (CB1R) or 60 min (CB2R) at 30°C. Non-specific binding was determined in the presence of 1 μM WIN-55,212-2. Bound and free radioactivity were separated by filtering the assay mixture as above indicated. The filter bound radioactivity was counted using a Packard Tri Carb 2810 TR scintillation counter (Perkin Elmer).
Competition binding experiments were also performed incubating 3 nM [3H]-WIN-55,212-2 and different concentrations of the tested compounds with membranes obtained from CHO cells transfected with human CB1 or CB2 receptors (10 μg protein/sample) for 60 min at 30°C. Non-specific binding was determined in the presence of 1 μM WIN-55,212-2. Bound and free radioactivity were separated by filtering the assay mixture as above indicated. The filter bound radioactivity was counted using a Packard Tri Carb 2810 TR scintillation counter (Perkin Elmer).
Homogeneous Binding Assays in Living Cells
Expression Vector
cDNAs for the human version of cannabinoid CB2R without their stop codon were obtained by PCR and subcloned to SNAP-containing vector (PSNAP; Cisbio Bioassays) using sense and antisense primers harboring unique restriction sites for HindIII and BamHI generating the SNAP tagged CB2R (CB2R-SNAP).
Cell Culture and Transfection
For HTRF assays, HEK-293T cells were used. HEK 293T (HEK-293T) cells were grown in DMEM supplemented with 2 mM L-glutamine, 1 mM sodium pyruvate, 100 units/mL penicillin/streptomycin, and 5% (v/v) FBS [all supplements were from Invitrogen, (Paisley, Scotland, United Kingdom)]. Cells were maintained at 37°C in a humidified atmosphere of 5% CO2 and were passaged, with enzyme-free cell dissociation buffer (13151-014, Gibco®, Thermo Fisher, Waltham, MA, United States), when they were 80–90% confluent, i.e., approximately twice a week. Cells were transiently transfected with the PEI (Polyethylenimine, Sigma, St. Louis, MO, United States) method as previously described (Medrano et al., 2017; Navarro et al., 2018b). Experiments were carried out in cells expressing SNAP-tagged CB2R in the presence or in the absence of CB1R.
Labeling of Cells Expressing SNAP-Tagged CB2R
Cell culture medium was removed from the 25-cm2 flask and 100 nM SNAP-Lumi4-Tb, previously diluted in 3 mL of TLB 1X, was added to the flask and incubated for 1 h at 37°C under 5% CO2 atmosphere in a cell incubator. Cells were then washed four times with 2 mL of TLB 1X to remove the excess of SNAP-Lumi4-Tb, detached with enzyme-free cell dissociation buffer, centrifuged 5 min at 1,500 rpm and collected in 1 mL of TLB 1X. Tag-lite-based binding assays were performed 24 h after transfection. Densities in the 2,500–3,000 cells/well range were used to carry out binding assays in white opaque 384-well plates.
Non-radioactive Competition Binding Assays
For competition binding assays, the fluorophore-conjugated CB2R ligand (labeled CM-157), unconjugated CM-157 and CBG were diluted in TLB 1X. HEK-293T cells transiently expressing Tb-labeled SNAP-CB2R with or without CB1R were incubated with 20 nM fluorophore-conjugated CB2R ligand, in the presence of increasing concentrations (0–10 μM range) of CBG or CM-157. Plates contained 10 μL of labeled cells, and 5 μL of TLB 1X or 5 μL of CBG or 5 μL CM-157 were added prior to the addition of 5 μL of the fluorescent ligand. Plates were then incubated for at least 2 h at room temperature before signal detection. Detailed description of the HTRF assay is found in Martínez-Pinilla et al. (2016).
Signal was detected using an EnVision microplate reader (PerkinElmer, Waltham, MA, United States) equipped with a FRET optic module allowing donor excitation at 337 nm and signal collection at both 665 and 620 nm. A frequency of 10 flashes/well was selected for the xenon flash lamp excitation. The signal was collected at both 665 and 620 nm using the following time-resolved settings: delay, 150 μs; integration time, 500 μs. HTRF® ratios were obtained by dividing the acceptor (665 nm) by the donor (620 nm) signals and multiplying by 10,000. The 10,000-multiplying factor is used solely for the purpose of easier data handling.
Functional Assays
Cell Culture and Transient Transfection
HEK-293T cells were grown in DMEM medium (Gibco, Paisley, Scotland, United Kingdom) supplemented with 2 mM L-glutamine, 100 U/mL penicillin/streptomycin, MEM Non-Essential Amino Acids Solution (1/100) and 5% (v/v) heat inactivated Foetal Bovine Serum (FBS) (Invitrogen, Paisley, Scotland, United Kingdom). Cells were maintained in a humid atmosphere of 5% CO2 at 37°C. Cells were transiently transfected with the PEI (Polyethylenimine, Sigma, St. Louis, MO, United States) method as previously described (Medrano et al., 2017; Navarro et al., 2018b) and used for functional assays 48 h later (unless otherwise stated).
cAMP Determination
Signaling experiments have been performed as previously described (Navarro et al., 2010, 2016, 2018b; Hinz et al., 2018). Two hours before initiating the experiment, HEK-293T cell-culture medium was replaced by serum-starved DMEM medium. Then, cells were detached, resuspended in growing medium containing 50 μM zardaverine and placed in 384-well microplates (2,500 cells/well). Cells were pretreated (15 min) with CBG -or vehicle- and stimulated with agonists (15 min) before adding 0.5 μM forskolin or vehicle. Readings were performed after 15 min incubation at 25°C. HTRF energy transfer measures were performed using the Lance Ultra cAMP kit (PerkinElmer, Waltham, MA, United States). Fluorescence at 665 nm was analyzed in a PHERAstar Flagship microplate reader equipped with an HTRF optical module (BMG Lab Technologies, Offenburg, Germany).
ERK Phosphorylation Assays
To determine ERK1/2 phosphorylation, 50,000 HEK-293T cells/well were plated in transparent Deltalab 96-well microplates and kept at the incubator for 24 h. 2 to 4 h before the experiment, the medium was substituted by serum-starved DMEM medium. Then, cells were pre-treated at 25°C for 10 min with vehicle or CBG in serum-starved DMEM medium and stimulated for an additional 7 min with the specific agonists. Cells were then washed twice with cold PBS before addition of lysis buffer (20 min treatment). 10 μL of each supernatant were placed in white ProxiPlate 384-well microplates and ERK 1/2 phosphorylation was determined using AlphaScreen®SureFire® kit (Perkin Elmer) following the instructions of the supplier and using an EnSpire® Multimode Plate Reader (PerkinElmer, Waltham, MA, United States).
Dynamic Mass Redistribution Assays (DMR)
Cell mass redistribution induced upon receptor activation was detected by illuminating the underside of a biosensor with polychromatic light and measuring the changes in the wavelength of the reflected monochromatic light. The magnitude of this wavelength shift (in picometers) is directly proportional to the amount of DMR. HEK-293T cells were seeded in 384-well sensor microplates to obtain 70–80% confluent monolayers constituted by approximately 10,000 cells per well. Previous to the assay, cells were washed twice with assay buffer (HBSS with 20 mM HEPES, pH 7.15) and incubated for 2 h with assay-buffer containing 0.1% DMSO (24°C, 30 μL/well). Hereafter, the sensor plate was scanned and a baseline optical signature was recorded for 10 min before adding 10 μL of CBG for 30 min followed by the addition of 10 μL of specific agonists; all test compounds were dissolved in assay buffer. The cell signaling signature was determined using an EnSpire® Multimode Plate Reader (PerkinElmer, Waltham, MA, United States) by a label-free technology. Then, DMR responses were monitored for at least 5,000 s. Results were analyzed using EnSpire Workstation Software v 4.10.
β-Arrestin 2 Recruitment
Arrestin recruitment was determined as previously described (Medrano et al., 2017; Navarro et al., 2018b). Briefly, BRET experiments were performed in HEK-293T cells 48 h after transfection with the cDNA corresponding to the CB2R-YFP or CB1R-YFP and 1 μg cDNA corresponding to β-arrestin 2-Rluc. Cells (20 μg protein) were distributed in 96-well microplates (Corning 3600, white plates with white bottom) and were incubated with CBG for 15 min and stimulated with the agonist for 10 min prior the addition of 5 μM coelenterazine H (Molecular Probes, Eugene, OR, United States). After 1 min of adding coelenterazine H, BRET between β-arrestin 2-Rluc and receptor-YFP was determined and quantified. The readings were collected using a Mithras LB 940 (Berthold Technologies, Bad Wildbad, Germany) that allows the integration of the signals detected in the short-wavelength filter at 485 nm and the long-wavelength filter at 530 nm. To quantify protein-RLuc expression luminescence readings were also performed 10 min of adding 5 μM coelenterazine H.
Data Handling and Statistical Analysis
Affinity values (Ki) were calculated from the IC50 obtained in competition radioligand binding assays according to the Cheng and Prusoff equation: Ki = IC50/(1 + [C]/KD), where [C] is the free concentration of the radioligand and KD its dissociation constant (Cheng, 2001).
Data from homogeneous binding assays were analyzed using Prism 6 (GraphPad Software, Inc., San Diego, CA, United States). Ki values were determined according to the Cheng and Prusoff equation with KD = 21 nM for CM-157 (Cheng, 2001). Signal-to-background (S/B ratio) calculations were performed by dividing the mean of the maximum value (μmax) by that of the minimum value (μmin) obtained from the sigmoid fits.
The data are shown as the mean ± SEM. Statistical analysis was performed with SPSS 18.0 software. The test of Kolmogorov–Smirnov with the correction of Lilliefors was used to evaluate normal distribution and the test of Levene to evaluate the homogeneity of variance. Significance was analyzed by one-way ANOVA, followed by Bonferroni’s multiple comparison post hoc test. Significant differences were considered when p < 0.05.
Results
Saturation and Competition Radioligand-Based Assays in Membranes Expressing CB1R or CB2R
The effect of CBG on radioligand binding to CB1R or CB2R was first tested using the classical radioligand-binding assay in membranes isolated from CHO cells expressing human CB1R or CB2R and incubated with radioligands: [3H]-CP-55940 or [3H]-WIN-55,212-2. Data obtained from binding isotherms using increasing [3H]-CP-55940 or [3H]-WIN-55,212-2 concentrations lead to a monophasic saturation curve. Saturation curves, receptor density (Bmax values) and affinity (KD values) are shown in Figures 1A–D. The affinity of the two radioligands was in the nanomolar range for both CB1R and CB2R. KD for [3H]-CP-55940 to CB1R and CB2R was similar with values around 0.3 nM. KD values for WIN-55,212-2 were 9.4 and 3.2 nM for CB1R and CB2R, respectively (Figures 1C,D). Overall the results agree with previously reported data (McPartland et al., 2007; Merighi et al., 2010).
FIGURE 1.
Radioligand binding assays to CB1R and CB2R. (A–D) Saturation curves of either [3H]-CP-55940 or [3H]-WIN-55,212-2 binding on membranes from CHO cells stably expressing human CB1R (A,C) or CB2R (B,D). (E,F) Competition curves for WIN-55,212-2 in radioligand-based assays using either [3H]-CP-55940 (E) or [3H]-WIN-55,212-2 (F) binding on membranes from CHO cells stably expressing human CB1R or CB2R. Data are expressed as the mean ± SEM of five independent experiments performed in duplicate. KD (obtained from saturation isotherms) are shown in Table 1.
Competition binding assays of WIN-55,212-2 showed similar Ki values using the two radioligands to CB1R and CB2R and agreed with the KD values for [3H]-WIN-55,212-2 binding (Table 1 and Figures 1E,F). Table 1 reports the affinity values of CBG. Ki values of CBG obtained using [3H]-CP-55940 as radioligand were in the low micromolar range in both CB1R and CB2R. The affinity value of CBG obtained using [3H]-WIN-55,212-2 for CB2R was 2.7 μM, about twofold higher than that obtained using [3H]-CP-55940. Using [3H]-WIN-55,212-2 in competition binding experiments on CB1R, CBG was not able to displace the radioligand (Figures 2A,B). In summary, CBG displayed Ki values in the low micromolar range when competing for the binding to the CB2R. Surprisingly, significant competition in the binding to the CB1R was only observed when using [3H]-CP-55940 as radioligand.
Table 1.
Affinity values of CB compounds obtained from radioligand binding assays.
| [3H]-CP-55940 competition binding experiments | [3H]-WIN-55,212-2 competition binding experiments | |||
|---|---|---|---|---|
| CB1 – KD (nM) | CB2 – KD (nM) | CB1 – KD (nM) | CB2 – KD (nM) | |
| 0.29 ± 0.02 | 0.32 ± 0.02 | 9.43 ± 0.83 | 3.16 ± 0.24 | |
| CB1 – Ki (nM) | CB2 – Ki (nM) | CB1 – Ki (nM) | CB2 – Ki (nM) | |
| WIN-55,212-2 | 8.08 ± 0.65 | 3.22 ± 0.31 | 9.86 ± 0.84 | 3.48 ± 0.27 |
| CBG | 1,045 ± 74 | 1,225 ± 85 | >30,000 | 2,656 ± 130 |
| CBD | 1,690 ± 110 | 1,714 ± 70 | >30,000 | 4,019 ± 342 |
KD values were obtained from saturation isotherms and Ki from data in competition assays using the indicated radiolabelled compounds ([3H]-CP-55940 or [3H]-WIN-55,212-2).
FIGURE 2.
Competition by CBG of agonist binding to CB1R and/or CB2R. (A,B) Competition curves for CBG in radioligand-based assays using either [3H]-CP-55940 (A) or [3H]-WIN-55,212-2 (B) binding on membranes from CHO cells stably expressing human CB1R or CB2R. (C) Scheme of the HTRF-based competitive binding assay. The GPCR of interest with the SNAP-tagged enzyme fused to its N-terminal domain is expressed at the cell surface. SNAP is a commercially available tag consisting of circa 180 amino acids, that can be labeled with fluorophores or other probes in a covalent fashion. The GPCR–SNAP-tagged cells are subsequently labeled with a Tb-containing probe (SNAP-Lumi4-Tb) through a covalent bond between the Tb and the reactive side of the SNAP enzyme. The Tb acts as FRET donor of an acceptor covalently linked to a selective CB2 receptor ligand. Thus, upon binding of a fluorophore-conjugated ligand (FRET acceptor) on the donor-labeled SNAP-tagged/GPCR fusion protein, an HTRF signal from the sensitized acceptor can be detected since the energy transfer can occur only when the donor and the acceptor are in close proximity. In competition binding assays using CM-157, the unlabelled specific ligand competes for receptor binding site with the fluorophore-conjugated ligand, leading to a decrease in the HTRF signal detected. (D–G) HEK-293T were transiently transfected with 1 μg cDNA for SNAP-CB2R in the absence (D,E) or presence of 0.5 μg cDNA for CB1R (F,G). Competition curves of specific binding of 20 nM fluorophore-conjugated CM-157 using CM-157 (0–10 μM) (D,F) or of CBG (0–10 μM) (E,G) as competitors are shown. Data represent the mean ± SEM of five experiments in triplicates.
CBG Binds to the Orthosteric Site of Cannabinoid CB2R at Nanomolar Concentrations
Competition experiments were performed using 20 nM of a fluorophore-conjugated selective CB2R agonist (CM-157) and a homogeneous non-radioactive method performed in living cells expressing SNAP-CB2R (details in Martínez-Pinilla et al., 2016; Figure 2C). Unfortunately, the equivalent fluorophore-conjugated selective CB1R ligand is not available to perform HTRF assays in SNAP-CB1R-expressing living cells. Competition assays were performed in HEK-293T cells expressing Lumi4-Tb-labeled CB2R fused to the SNAP protein and incubated with a fixed amount of the fluorophore-conjugated agonist and different CBG concentrations. As observed in Figure 2, both the unlabelled selective agonist (CM-157) and CBG decreased the binding to SNAP-CB2R in monophasic fashion and with Ki values in the nanomolar range (16 nM for CM-157 and of 152 nM for CBG; Figures 2D,E). The Ki obtained for CM-157 matches with previously reported dissociation constant KD values (Martínez-Pinilla et al., 2016). These results indicate that CBG can significantly bind to the orthosteric site of cannabinoid CB2R at nanomolar concentrations.
Similar experiments were carried out in HEK-293T cells expressing SNAP-CB2R fusion protein and a similar amount of CB1R, i.e., in cells that express CB2R in a CB1–CB2 receptor heteromer context. In the presence of cannabinoid CB1R the Ki for CM-157 was 19 nM (Figure 2F) and Ki for CBG was reduced (56 nM, Figure 2G). These results indicate that in cells expressing both cannabinoid receptors, CB1 and CB2, CBG shows higher affinity for cannabinoid CB2R.
CBG Effects on Cannabinoid Receptor-Agonist-Induced Effects
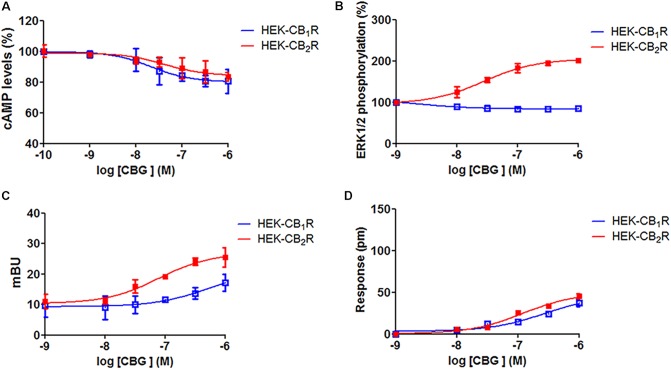
Previous reports Gauson et al. (2007), Cascio et al. (2010) suggest that CBG may be a partial agonist of cannabinoid receptors. To investigate this possibility, HEK-293T cells expressing CB1R or CB2R were treated with increasing concentrations of CBG (1 nM to 10 μM) and cAMP, MAPK, β-arrestin recruitment and dynamic mass cell redistribution (DMR) assays were developed. Interestingly, it was observed that in cells expressing CB1R (Figure 3, blue curves), CBG induced a small decrease in forskolin induced cAMP levels and a small increase in β-arrestin recruitment (Figures 3A,C), while having no significant action on MAPK phosphorylation assay (Figure 3B). Consequently, CBG in label-free assays induced a slight effect in the DMR signal (Figure 3D) that is consistent with a G protein-dependent action on cAMP levels; label-free signal is based on optical detection of DMR following receptor activation and mainly reflects G-protein-coupling (Kebig et al., 2009; Schröder et al., 2009; Hamamoto et al., 2015). On the other hand, in HEK-293T cells expressing CB2R (Figure 3, red curves), the action on forskolin-induced cAMP levels and on the DMR signal was small and similar to that exerted in CB1R-expressing cells (Figure 3A). On the contrary, the activation of the MAP kinase pathway was notable (Figure 3B). Also noteworthy was the CBG-induced β-arrestin recruitment (Figure 3C). Taken together these data suggest that CBG is a poor agonist of CB1R, whereas it acts as a partial agonist in some of the signaling pathways analyzed in cells expressing CB2R.
FIGURE 3.
Cannabigerol action in cells expressing CB1R or CB2R. HEK-293T cells were transfected with 0.75 μg cDNA for CB1R (red line) or 1 μg cDNA for CB2R (blue line). Dose–effect curves for cAMP production are expressed as % of levels obtained by 0.5 μM forskolin treatment (A). Dose-effect curves for ERK1/2 phosphorylation are expressed as % respect to basal levels (B). Dose-effect curves for β-arrestin recruitment (C) and label-free (D) assays are expressed, respectively, in mBRET units and pm. In β-arrestin-2 recruitment assays cells were transfected with 1 μg cDNA for β-arrestin-Rluc and either 0.75 μg cDNA for CB1R-YFP or 1 μg cDNA for CB2R-YFP. Data are the mean ± SEM of a representative experiment in triplicates (n = 6).
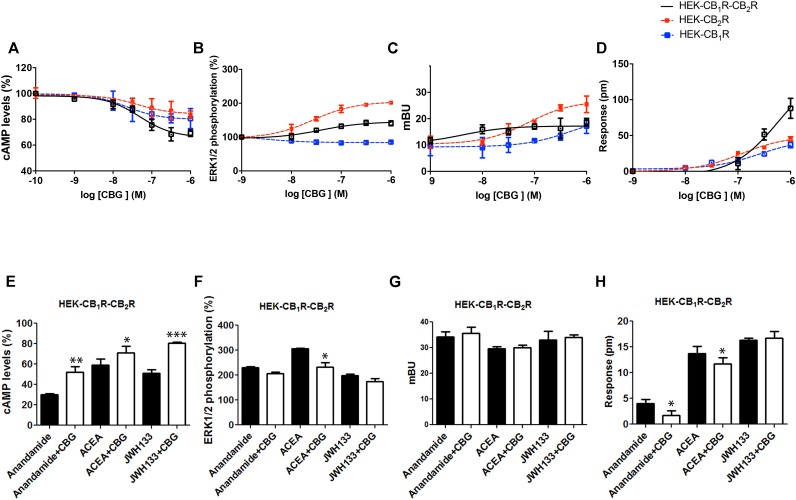
To further examine the CBG effect over CB1R, HEK-293T cells expressing CB1R were treated with the endocannabinoid agonist, AEA, or with ACEA in the presence or in the absence of 100 nM or 1 μM CBG. In forskolin-induced cAMP assays we found that 100 nM or 1 μM CBG pretreatment induced a significant decrease in both, AEA and ACEA induced effects (Figure 4A). In contrast, CBG (100 nM or 1 μM) was unable to modify the agonist-induced MAPK phosphorylation and β-arrestin recruitment (Figures 4B,C). In label-free DMR assays the results were similar to those obtained in cAMP determination assays, i.e., CBG reduced the effect of the agonists (Figure 4D).
FIGURE 4.
Effect of CBG on the action of CB1R and CB2R agonists. (A–D) HEK-293T cells were transfected with 0.75 μg cDNA for CB1R and treated with 100 nM AEA or a selective CB1R ligand (100 nM ACEA) in the absence (black bars) or presence of 100 nM (white bars) or 1 μM (gray bars) CBG. (E–H) HEK-293T cells were transfected with 1 μg cDNA for CB2R and treated with 100 nM AEA or a selective CB2R ligand (100 nM JWH133) in the absence (black bars) or presence of 100 nM (white bars) or 1 μM (gray bars) CBG. cAMP production (A,E) is expressed as % of levels obtained by 0.5 μM forskolin. ERK1/2 phosphorylation data are expressed as % respect to basal levels (B,F). In β-arrestin-2 recruitment assays cells were transfected with 1 μg cDNA for β-arrestin-Rluc and either 0.75 μg cDNA for CB1R-YFP or 1 μg cDNA for CB2R-YFP. Data for β-arrestin recruitment (C,G) and label-free (D,H) assays are expressed, respectively, in mBRET units and pm. Data represent the mean ± SEM of six different experiments performed with six replicates. One-way ANOVA and Bonferroni’s multiple comparison post hoc test were used for statistical analysis (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; versus treatment with AEA, ACEA, or JWH133 alone).
Cannabigerol (100 nM or 1 μM) was also tested in HEK-293T cells expressing CB2R and using AEA and a receptor selective agonist, JWH133. Pretreatment with CBG reduced the effects of AEA and JWH133 in experiments of forskolin-induced cAMP levels, ERK1/2 phosphorylation and in label-free DMR read-outs (Figure 4). In contrast, CBG did not affect the recruitment of β-arrestin induced by agonists (Figure 4G). This last result may be due to the low sensitivity of the assay as β-arrestin recruitment BRET signal was virtually negligible. Energy transfer techniques completely depend on the correct orientation of the fusion proteins and the reduced signal may be due to poor recruitment of β-arrestin and/or to a high distance between BRET donor/acceptor in the putative β-arrestin-Rluc/CB2R-YFP complex. Thus, CBG in cells activated by endocannabinoids or by selective agonists behaves as a partial agonist of the CB2R.
CBG Effect in HEK-293T Cells Expressing CB1R and CB2R
Experiments were finally performed in cells co-expressing the two cannabinoid receptors, which are able to form heteromeric complexes. A CB1–CB2 receptor heteromer print consists of a negative cross-talk observed in Akt phosphorylation and neurite outgrowth; i.e., activation of one receptor reduces the signaling originated upon partner receptor activation (Callén et al., 2012). To characterize the CBG effect, experiments were performed in HEK-293T cells expressing the two cannabinoid receptors. Dose-effect curves were provided for cAMP level and ERK1/2 phosphorylation determination, and for label-free DMR signal and β-arresting recruitment. Interestingly, the effect on cAMP level determination and DMR assays was additive (Figure 5), i.e., the presence of CBG blunted the negative cross-talk in these signaling pathways. However, the negative cross-talk was still evident in both ERK1/2 phosphorylation and β-arrestin recruitment experiments (Figures 5B,C).
FIGURE 5.
Effect of CBG in cells expressing CB1 and CB2 receptors. (A–D) Effect of CBG in HEK-293T cells transfected with 0.75 μg cDNA for CB1R and 1 μg cDNA for CB2R (A,B,D) or 1 μg cDNA for β-arrestin-Rluc, 0.75 μg cDNA for CB1R and 1 μg cDNA for CB2R-YFP (C). Dose–effect curves for cAMP production are expressed as % of levels obtained by 0.5 μM forskolin treatment (A). Dose-effect curves for ERK1/2 phosphorylation are expressed as % respect of basal levels (B). Dose-effect curves for β-arrestin recruitment (C) and label-free (D) assays are expressed, respectively, in mBRET units and pm. Dotted lines (red and blue) are the same than those shown in Figure 3 and serve as a reference for differential effects in cells coexpressing both receptors. Data are the mean ± SEM of a representative experiment in triplicates (n = 6). (E–H) HEK-293T cells transfected with 0.75 μg cDNA for CB1R and 1 μg cDNA for CB2R (E,F,H) or 1 μg cDNA for β-arrestin-Rluc, 0.75 μg cDNA for CB1R and 1 μg cDNA for CB2R-YFP (G) were treated with 100 nM AEA or a selective CB2R ligand (100 nM JWH133) in the absence (black bars) or presence (white bars) of 100 nM CBG. cAMP production (E) is expressed as % of levels obtained by 0.5 μM forskolin. ERK1/2 phosphorylation data are expressed as % respect of basal levels (F). Data for β-arrestin recruitment (G) and label-free (H) assays are expressed, respectively, in mBRET units and pm. Data represent the mean ± SEM of six different experiments performed with three replicates. One-way ANOVA and Bonferroni’s multiple comparison post hoc test were used for statistical analysis (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; versus treatment with AEA, ACEA, or JWH133 alone).
Finally, the effect of 100 nM CBG (100 nM) on AEA, ACEA and/or JWH133 actions was investigated in cells co-expressing CB1R and CB2R. CBG pretreatment led to significant effects, always reducing the effect of the agonists, in cAMP-related assays (Figure 5E). However, the effect in the other assay types was negligible except for the negative modulation of the ACEA effect on ERK1/2 phosphorylation and DMR, and of the AEA effect on DMR read-outs (Figures 5F–H). Therefore, CBG either blunted the cAMP-dependent signaling or did not significantly alter the negative cross-talk when other CB1/CB2-mediated signaling read-outs were determined (see Figures 5B,C). It should be noted that cross-talk at the intracellular signaling level, cannot be ruled out to partly explain some of the findings (Bayewitch et al., 1995; Wartmann et al., 1995; Mcguinness et al., 2009; Peters and Scott, 2009; Van Der Lee et al., 2009).
Discussion
The aim of this paper was to comparatively address CBG pharmacology and effects on CB1 and CB2 receptors, and on CB1–CB2 heteroreceptor complexes. The binding experiments using radiolabelled- and non-radiolabelled-based approaches have provided relevant results. The results on CB2R are clear an indicate that CBG acts as a competitive partial agonist ligand. There is, however, an interesting observation as the Ki values for competing both [3H]-CP-55940 and [3H]-WIN-55,212-2 are in the low micromolar range (Table 1), whereas displaying a value of 152 nM in HTRF-based assays. As pointed out in previous reports, the conditions of the approach using a fluorescent-conjugated CM-157 allows identification of different states of the receptor. Irrespective of the molecular mechanism, the marked differences in affinity constants suggest different ways to accommodate the ligand within the orthosteric center. To our knowledge this is the first report performed in parallel binding assays using three different ligands that reportedly bind to the orthosteric center of the CB2R ([3H]-CP-55940, [3H]-WIN-55,212-2 and fluorescence-conjugated-CM-157). In summary, the most reasonable assumption is that CBG binds to the orthosteric center of CB2R but with marked differences in affinity depending on the assay. It should be noted that differences in affinity may result from the fact that HTRF binding is performed in living cells whereas radioligand binding assays are performed in isolated membranes. The already existing data concerning CBG affinity for CB1 and CB2 receptors, all performed using [3H]-CP-55940 also indicate that the affinity may vary depending on the context of the receptor, by inter alia the constraints of the membrane, heteromerization or interaction with G-proteins. Comparing our results with similar data using [3H]-CP-55940, the affinity is higher for receptors expressed in HEK-293 cells or in brain membranes (Gauson et al., 2007; Pertwee, 2008; Pollastro et al., 2011) that in receptors expressed in CHO cells (Table 1). In competition assays of radioligand binding to CB1R or to CB2R, affinity for CBG is similar to that previously published (Gauson et al., 2007; Pertwee, 2008), except in the case of Sf9 cells (Ki: 897 and 153 nM for, respectively, CB1R and CB2R). This piece of data would indicate conformational changes induced by third molecules that affect the binding of the radioligand and/or of CBG. In fact, Sf9 are insect cells that do not express the cognate Gi protein and, therefore, Gαi3β1γ2 was heterologously expressed to perform the binding assays that led to different affinities for CBG (897 and 153 nM for, respectively, CB1R and CB2R) (Rosenthaler et al., 2014).
The results from binding to the CB1R are not very robust and more difficult to interpret. Unfortunately, there are no ligands available to perform HTRF binding to SNAP-CB1R-expressing living cells, whereas the data from competition assays using [3H]-CP-55940 or [3H]-WIN-55,212-2 were contradictory. On the one hand, the Ki for binding to the CB1R using [3H]-CP-55940 was in the low micromolar range, as it occurred with data from radioligand binding to the CB2R. However, CBG was unable to compete [3H]-WIN-55,212-2 binding to the CB1R. Taking into account that recognition sites for CP-55940 and WIN-55,212-2 are not identical in the CB1R, one possibility is that CBG binds to the orthosteric center but displaying different equilibrium binding parameters depending on the radioligand. It was early observed that Lys192 in the CB1R third transmembrane domain (TM3) was crucial for binding of CP-55940 and AEA but not for WIN-55,212-2 (Bonner et al., 1996; Chin et al., 1998). Later, in silico models pointed to an hydrophobic pocket for CP-55940 binding that involved residues in different transmembrane domains (not only in TM3) and in the second extracellular loop (Shim et al., 2003). Those models showed that WIN-55,212-2 not only binds to the hydrophobic pocket described for CP-55940 but to another hydrophobic region involving residues in TM2 and TM3 (Shim and Howlett, 2006). The structure of CBG is more similar to CP-55940 than to WIN-55,212,2, bearing an OH in the A ring that may interact with the TM3 Lys192 residue. In brief, CBG binds to the orthosteric center of CB1R as indicated by the fact that CBG affects CP-55940 binding without affecting the binding of [3H]-WIN-55,212-2. In other words, CBG was able to distinguish between two subregions of the CB1R orthosteric center. We therefore suggest that pharmacological studies concerning the CB1R should be run in parallel using radiolabelled CP-55940 and WIN-55,212-2. Interestingly CP-55940 and WIN-55,212-2 are able to fix the CB1R in two different conformations (Georgieva et al., 2008) and, therefore, CBG would affect more the conformation and signaling arising from occupation of the CP-55940 binding site. Other possibilities cannot be ruled out and, in this respect, we assayed CBD in competition assays and obtained similar results than those obtained using CBG (Table 1). Accordingly, CBG could act on CB1R (but not on CB2R) as non-competitive (allosteric) modulator, as described for CBD (Laprairie et al., 2015).
When one compound binds to the orthosteric center and affects several signaling pathways with different potency as in the case of CBG in cells expressing CB1R, the phenomenon is known as functional selectivity or biased agonism. In cells expressing CB1R, CBG effect is skewed toward the Gi-mediated signaling pathway. This is in agreement with our finding of significant effect in label-free assays; often DMR signals correlate with effect on cAMP levels in the case of receptors coupled to Gi or Gs proteins (Grundmann and Kostenis, 2015a,b; Hamamoto et al., 2015). It is, however, intriguing that CBG was unable to displace the binding of [3H]-WIN-55,212-2 to the CB1R. Therefore, an action of CBG on a particular state of the receptor, which, in the case of CB2R may be disclosed by HTRF binding in living cells (Martínez-Pinilla et al., 2016), cannot be ruled out. Taking together all results, an allosteric action of CBG on the CB1R would not explain why it is able to engage Gi –mediating signaling. Another possibility, which was suggested for AM630, a previously considered CB2R antagonist (Bolognini et al., 2012), is that CBG is a protean agonist displaying biased agonism.
Data from CB2R-mediated functional assays were easier to interpret. First of all, the efficacy was lower compared to selective synthetic agonists and endocannabinoids. Also, CBG led to biased agonism as the effect on cAMP levels was small while being quite marked in ERK phosphorylation and β-arrestin recruitment. Therefore, CBG acted as a partial agonist and, as such, it was able to reduce the effects of other cannabinoid agonists. At 1 μM the effect of CBG on receptor activation by other agonists was similar to that exerted by 100 nM (Figure 4) thus suggesting that the effective affinity in living cells is that obtained in HTRF non-radioactive-based assays.
Due to the complex pharmacology of cannabinoids this research was undertaken to investigate whether CBG could be exerting a differential action on the CB1–CB2 receptor heteromers. Previous data have shown that the interplay between the two receptors in an heteromeric context is also complex. Whereas Callén et al. (2012) showed a negative cross-talk in a heterologous expression system, the allosteric interaction in the CB1–CB2 heteroreceptor complex is synergistic in primary cultures of activated microglia activated with LPS and interferon gamma and in primary cultures of microglia from a transgenic model of Alzheimer’s disease (Navarro et al., 2018a). Dose-effect experiments here undertaken in the HEK-293T-based heterologous expression system showed that CBG treatment in the absence of any other agonist, led to additive/synergistic effects on cAMP and label-free read-outs. In contrast, in ERK phosphorylation and β-arrestin recruitment, we found the negative cross-talk already described for this heteromer when full agonists are used to activate the receptors (Callén et al., 2012). These results suggest that partial agonism on the CB2R is regulated by the presence of CB1R; however, more complex alternative scenarios cannot be ruled out as CBG may act on the orthosteric site of the CB2R protomer and as protean agonist of the CB1R protomer. In cells expressing the two receptors, the overall effect of 100 nM CBG on agonist-induced activation is more consistent with acting on CB2R than on CB1R. In fact, the results in co-expressing cells, which likely express heteromers, are similar to those encountered in CB2R-expressing cells. In summary, CBG significantly modulates CB2R- or CB1R/CB2R-mediated endocannabinoid action, while the effects are weak in CB1R-expressing cells. Our findings demonstrating the action of CBG on the cannabinoid receptors are in complete agreement and may explain the in vitro results, reporting the protection of macrophages against oxidative stress (Giacoppo et al., 2017), and the beneficial in vivo effects in a model of inflammatory bowel disease (Borrelli et al., 2013). In the first of these two studies CBG-mediated protection is blocked by AM630, a selective CB2R ligand, whereas the CB1R antagonist, SR141716A, had no effect on CBG action (Giacoppo et al., 2017). The second study reported that CBG may both reduce the histological and molecular changes of experimental colitis and nitrite release from macrophages after LPS stimulation; again these effects were seemingly mediated by CB2R (Borrelli et al., 2013). These results can be explained by our findings; CBG acting as a partial agonist and exerting actions via CB2R in macrophages (Giacoppo et al., 2017) or “antagonizing” the effects of endogenous or synthetic cannabinoids, as in LPS-stimulated macrophages (Borrelli et al., 2013). In conclusion, the results presented in this study reveal that the non-psychotropic phytocannabinoid, CBG, may exert beneficial actions with therapeutic potential via cannabinoid receptors.
Author Contributions
XN and RF had the original idea, designed and coordinated actions in the different participating institutions, and wrote the initial manuscript. GN performed non-radiolabelled-based homogeneous binding assays, participated in the signaling experiments, and significantly contributed to manuscript preparation. IR-R participated in the signaling experiments and in writing methods. RR-S actively participated in data analysis and parameter calculation. EC supervised data analysis, provided pharmacological expertise, and insight into data interpretation. FV performed the radioligand binding experiments. KV and PB performed the radioligand binding data analysis and interpretation. SC selected the Cannabis varieties and supervised the production of the vegetal raw material used for the isolation and purification of cannabinoids. VSM performed the isolation and purification of cannabinoids. CS-CC and CF-V performed the analytical quality control to the purified cannabinoids. All co-authors critically revised, contributed to the editing, and approved the manuscript.
Conflict of Interest Statement
Authors declare that this research was undertaken in collaboration with Phytoplant Research S.L. Co-authors working in the Spanish and Italian public institutions do not receive honoraria from the company and do not have any participation in the company (stock shares or similar).
Abbreviations
- Δ8-THC
Δ8-tetrahydrocannabinol
- Δ9-THC
Δ9-tetrahydrocannabinol
- Δ9-THCA
Δ9- tetrahy drocannabinolic acid
- Δ9-THCV
Δ9-tetrahydrocannabivarin
- 2-AG
2-arachidonoyl glicerol
- AEA
anandamide
- CB1R
cannabinoid receptor 1
- CB2R
cannabinoid receptor 2
- CBC
cannabichromene
- CBD
cannabidiol
- CBDA
cannabidiolic acid
- CBDV
cannabidivarin
- CBG
cannabigerol
- CBGA
cannabigerolic acid
- CBN
cannabinol
- CNS
central nervous system
- DMR
dynamic mass redistribution
- HEK
human embryonic kidney
- HTRF
homogeneous time-resolved fluorescence
- SNAP
protein used as a tag; it contains circa 180 amino acids and may be covalently labeled with different probes
- Tb
terbium
- TLB
Tag-lite labeling medium
Footnotes
Funding. This work was partially supported by grants from the Spanish Ministry of Economy and Competitiveness (Ref. No. BFU2015-64405-R and SAF2017-84117-R; they may include FEDER funds) and by a grant 201413-30 from: Fundació la Marató de TV3.
References
- Adams A. J., Banister S. D., Irizarry L., Trecki J., Schwartz M., Gerona R. (2017). Zombie” outbreak caused by the synthetic cannabinoid AMB-FUBINACA in New York. N. Engl. J. Med. 376 235–242. 10.1056/NEJMoa1610300 [DOI] [PubMed] [Google Scholar]
- Bayewitch M., Avidor-Reiss T., Levy R., Barg J., Mechoulam R., Vogel Z. (1995). The peripheral cannabinoid receptor: adenylate cyclase inhibition and G protein coupling. FEBS Lett. 375 143–147. 10.1016/0014-5793(95)01207-U [DOI] [PubMed] [Google Scholar]
- Bolognini D., Grazia A., Parolaro D., Pertwee R. G. (2012). AM630 behaves as a protean ligand at the human cannabinoid CB2 receptor. Br. J. Pharmacol. 165 2561–2574. 10.1111/j.1476-5381.2011.01503.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner I., Song Z. H., Bonner T. I. (1996). A lysine residue of the cannabinoid receptor is critical for receptor recognition by several agonists but not WIN55212-2. Mol. Pharmacol. 49 891–896. [PubMed] [Google Scholar]
- Borrelli F., Fasolino I., Romano B., Capasso R., Maiello F., Coppola D., et al. (2013). Beneficial effect of the non-psychotropic plant cannabinoid cannabigerol on experimental inflammatory bowel disease. Biochem. Pharmacol. 85 1306–1316. 10.1016/j.bcp.2013.01.017 [DOI] [PubMed] [Google Scholar]
- Callén L., Moreno E., Barroso-Chinea P., Moreno-Delgado D., Cortés A., Mallol J., et al. (2012). Cannabinoid receptors CB1 and CB2 form functional heteromers in brain. J. Biol. Chem. 287 20851–20865. 10.1074/jbc.M111.335273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascio M. G., Gauson L. A., Stevenson L. A., Ross R. A., Pertwee R. G. (2010). Evidence that the plant cannabinoid cannabigerol is a highly potent α 2-adrenoceptor agonist and moderately potent 5HT 1A receptor antagonist. Br. J. Pharmacol. 159 129–141. 10.1111/j.1476-5381.2009.00515.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H. C. (2001). The power issue: determination of KB or Ki from IC50 - A closer look at the Cheng-Prusoff equation, the Schild plot and related power equations. J. Pharmacol. Toxicol. Methods 46 61–71. 10.1016/S1056-8719(02)00166-1 [DOI] [PubMed] [Google Scholar]
- Chin C. N., Lucas-Lenard J., Abadji V., Kendall D. A. (1998). Ligand binding and modulation of cyclic AMP levels depend on the chemical nature of residue 192 of the human cannabinoid receptor 1. J. Neurochem. 70 366–373. [DOI] [PubMed] [Google Scholar]
- De Petrocellis L., Ligresti A., Moriello A. S., Allarà M., Bisogno T., Petrosino S., et al. (2011). Effects of cannabinoids and cannabinoid-enriched Cannabis extracts on TRP channels and endocannabinoid metabolic enzymes. Br. J. Pharmacol. 163 1479–1494. 10.1111/j.1476-5381.2010.01166.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Petrocellis L., Vellani V., Schiano-Moriello A., Marini P., Magherini P. C., Orlando P., et al. (2008). Plant-derived cannabinoids modulate the activity of transient receptor potential channels of ankyrin type-1 and melastatin type-8. J. Pharmacol. Exp. Ther. 325 1007–1015. 10.1124/jpet.107.134809 [DOI] [PubMed] [Google Scholar]
- Diana M. A., Marty A. (2004). Endocannabinoid-mediated short-term synaptic plasticity: depolarization-induced suppression of inhibition (DSI) and depolarization- induced suppression of excitation (DSE). Br. J. Pharmacol. 142 9–19. 10.1038/sj.bjp.0705726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellermeier M., Zenk M. H. (1998). Prenylation of olivetolate by a hemp transferase yields cannabigerolic acid, the precursor of tetrahydrocannabinol. FEBS Lett. 427 283–285. 10.1016/S0014-5793(98)00450-5 [DOI] [PubMed] [Google Scholar]
- Gaoni Y., Mechoulan R. (1964). Structure synthesis of cannabigerol new hashish constituent. Proc. Chem. Soc. Lond. 82 2189–2192. [Google Scholar]
- Gauson L. A., Stevenson L. A., Thomas A., Baillie G. L., Ross R. A., Pertwee R. G. (2007). “Cannabigerol behaves as a partial agonist at both CB1 and CB2 receptors,” in Proceedings of the 17th Annual Symposium on the Cannabinoids, (Burlington, VT: International Cannabinoid Research Society; ), 206. [Google Scholar]
- Georgieva T., Devanathan S., Stropova D., Park C. K., Salamon Z., Tollin G., et al. (2008). Unique agonist-bound cannabinoid CB1 receptor conformations indicate agonist specificity in signaling. Eur. J. Pharmacol. 581 19–29. 10.1016/j.ejphar.2007.11.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacoppo S., Gugliandolo A., Trubiani O., Pollastro F., Grassi G., Bramanti P., et al. (2017). Cannabinoid CB2 receptors are involved in the protection of RAW264.7 macrophages against the oxidative stress: An in vitro study. Eur. J. Histochem. 61 1–13. 10.4081/ejh.2017.2749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granja A. G., Carrillo-Salinas F., Pagani A., Gómez-Cañas M., Negri R., Navarrete C., et al. (2012). A cannabigerol quinone alleviates neuroinflammation in a chronic model of multiple sclerosis. J. Neuroimmune Pharmacol. 7 1002–1016. 10.1007/s11481-012-9399-3 [DOI] [PubMed] [Google Scholar]
- Grundmann M., Kostenis E. (2015a). Holistic methods for the analysis of cNMP effects. Handb. Exp. Pharmacol. 238 339–357. 10.1007/164_2015_42 [DOI] [PubMed] [Google Scholar]
- Grundmann M., Kostenis E. (2015b). Label-free biosensor assays in GPCR screening. Methods Mol. Biol. 1272 199–213. 10.1007/978-1-4939-2336-6_14 [DOI] [PubMed] [Google Scholar]
- Grunfeld Y., Edery H. (1969). Psychopharmacological activity of the active constituents of hashish and some related cannabinoids. Psychopharmacologia 14 200–210. 10.1007/BF00404218 [DOI] [PubMed] [Google Scholar]
- Hamamoto A., Kobayashi Y., Saito Y. (2015). Identification of amino acids that are selectively involved in Gi/o activation by rat melanin-concentrating hormone receptor 1. Cell. Signal. 27 818–827. 10.1016/j.cellsig.2015.01.008 [DOI] [PubMed] [Google Scholar]
- Hinz S., Navarro G., Borroto-Escuela D., Seibt B. F., Ammon C., de Filippo E., et al. (2018). Adenosine A2A receptor ligand recognition and signaling is blocked by A2B receptors. Oncotarget 9 13593–13611. 10.18632/oncotarget.24423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebig A., Kostenis E., Mohr K., Mohr-Andrä M. (2009). An optical dynamic mass redistribution assay reveals biased signaling of dualsteric GPCR activators. J. Recept. Signal Transduct. 29 140–145. 10.1080/10799890903047437 [DOI] [PubMed] [Google Scholar]
- Lanciego J. L., Barroso-Chinea P., Rico A. J., Conte-Perales L., Callén L., Roda E., et al. (2011). Expression of the mRNA coding the cannabinoid receptor 2 in the pallidal complex of Macaca fascicularis. J. Psychopharmacol. 25 97–104. 10.1177/0269881110367732 [DOI] [PubMed] [Google Scholar]
- Laprairie R. B., Bagher A. M., Kelly M. E. M., Denovan-Wright E. M. (2015). Cannabidiol is a negative allosteric modulator of the cannabinoid CB1 receptor. Br. J. Pharmacol. 172 4790–4805. 10.1111/bph.13250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H.-C. C., Mackie K. (2016). an introduction to the endogenous cannabinoid system. Biol. Psychiatry 79 516–525. 10.1016/j.biopsych.2015.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Pinilla E., Rabal O., Reyes-Resina I., Zamarbide M., Navarro G., Sanchez-Arias J. A., et al. (2016). Two affinity sites of the cannabinoid subtype 2 receptor identified by a novel homogeneous binding assay. J. Pharmacol. Exp. Ther. 358 580–587. 10.1124/jpet.116.234948 [DOI] [PubMed] [Google Scholar]
- Mcguinness D., Malikzay A., Visconti R., Lin K., Bayne M., Monsma F., et al. (2009). Characterizing cannabinoid CB2 receptor ligands using DiscoveRx PathHunterTM β-arrestin assay. J. Biomol. Screen. 14 49–58. 10.1177/1087057108327329 [DOI] [PubMed] [Google Scholar]
- McPartland J. M., Glass M., Pertwee R. G. (2007). Meta-analysis of cannabinoid ligand binding affinity and receptor distribution: Interspecies differences. Br. J. Pharmacol. 152 583–593. 10.1038/sj.bjp.0707399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechoulam R. (2016). Cannabis - The Israeli perspective. J. Basic Clin. Physiol. Pharmacol. 27 181–187. 10.1515/jbcpp-2015-0091 [DOI] [PubMed] [Google Scholar]
- Mechoulam R., Shani A., Edery H., Grunfeld Y. (1970). Chemical basis of hashish activity. Science 169 611–612. 10.1126/science.169.3945.611 [DOI] [PubMed] [Google Scholar]
- Medrano M., Aguinaga D., Reyes-Resina I., Canela E. I., Mallol J., Navarro G., et al. (2017). Orexin A/Hypocretin modulates leptin receptor-mediated signaling by allosteric modulations mediated by the Ghrelin GHS-R1A receptor in hypothalamic neurons. Mol. Neurobiol. 55 4718–4730. 10.1007/s12035-017-0670-8 [DOI] [PubMed] [Google Scholar]
- Merighi S., Simioni C., Gessi S., Varani K., Borea P. A. (2010). Binding thermodynamics at the human cannabinoid CB1 and CB2 receptors. Biochem. Pharmacol. 79 471–477. 10.1016/j.bcp.2009.09.009 [DOI] [PubMed] [Google Scholar]
- Nadal X. (2016). Methods of purifying cannabinoids, compositions and kits thereof. U.S. Patent No 9765000 Washington, DC: U.S. Patent and Trademark Office. [Google Scholar]
- Nadal X., del Río C., Casano S., Palomares B., Ferreiro-Vera C., Navarrete C., et al. (2017). Tetrahydrocannabinolic acid is a potent PPARγ agonist with neuroprotective activity. Br. J. Pharmacol. 174 4263–4276. 10.1111/bph.14019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro G., Borroto-Escuela D., Angelats E., Etayo Í., Reyes-Resina I., Pulido-Salgado M., et al. (2018a). Receptor-heteromer mediated regulation of endocannabinoid signaling in activated microglia. Role of CB1 and CB2 receptors and relevance for Alzheimer’s disease and levodopa-induced dyskinesia. Brain Behav. Immun. 67 139–151. 10.1016/j.bbi.2017.08.015 [DOI] [PubMed] [Google Scholar]
- Navarro G., Cordomí A., Brugarolas M., Moreno E., Aguinaga D., Pérez-Benito L., et al. (2018b). Cross-communication between Gi and Gs in a G-protein-coupled receptor heterotetramer guided by a receptor C-terminal domain. BMC Biol. 16:24. 10.1186/s12915-018-0491-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro G., Cordomí A., Zelman-Femiak M., Brugarolas M., Moreno E., Aguinaga D., et al. (2016). Quaternary structure of a G-protein-coupled receptor heterotetramer in complex with Gi and Gs. BMC Biol. 14:26. 10.1186/s12915-016-0247-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro G., Ferré S., Cordomi A., Moreno E., Mallol J., Casadó V., et al. (2010). Interactions between intracellular domains as key determinants of the quaternary structure and function of receptor heteromers. J. Biol. Chem. 285 27346–27359. 10.1074/jbc.M110.115634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertwee R. G. (2008). The diverse CB 1 and CB 2 receptor pharmacology of three plant cannabinoids: Δ 9-tetrahydrocannabinol, cannabidiol and Δ 9-tetrahydrocannabivarin. Br. J. Pharmacol. 153 199–215. 10.1038/sj.bjp.0707442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters M. F., Scott C. W. (2009). Evaluating cellular impedance assays for detection of GPCR pleiotropic signaling and functional selectivity. J. Biomol. Screen. 14 246–255. 10.1177/1087057108330115 [DOI] [PubMed] [Google Scholar]
- Pollastro F., Taglialatela-Scafati O., Allarà M., Muñoz E., Di Marzo V., De Petrocellis L., et al. (2011). Bioactive prenylogous cannabinoid from fiber hemp (Cannabis sativa). J. Nat. Prod. 74 2019–2022. 10.1021/np200500p [DOI] [PubMed] [Google Scholar]
- Rosenthaler S., Pöhn B., Kolmanz C., Nguyen Huu C., Krewenka C., Huber A., et al. (2014). Differences in receptor binding affinity of several phytocannabinoids do not explain their effects on neural cell cultures. Neurotoxicol. Teratol. 46 49–56. 10.1016/j.ntt.2014.09.003 [DOI] [PubMed] [Google Scholar]
- Schröder R., Merten N., Mathiesen J. M., Martini L., Kruljac-Letunic A., Krop F., et al. (2009). The C-terminal tail of CRTH2 is a key molecular determinant that constrains Gαi and downstream signaling cascade activation. J. Biol. Chem. 284 1324–1336. 10.1074/jbc.M806867200 [DOI] [PubMed] [Google Scholar]
- Shim J. Y., Howlett A. C. (2006). WIN55212-2 docking to the CB 1 cannabinoid receptor and multiple pathways for conformational induction. J. Chem. Inf. Model. 46 1286–1300. 10.1021/ci0504824 [DOI] [PubMed] [Google Scholar]
- Shim J. Y., Welsh W. J., Howlett A. C. (2003). Homology model of the CB1 cannabinoid receptor: sites critical for nonclassical cannabinoid agonist interaction. Biopolym. Pept. Sci. Sect. 71 169–189. 10.1002/bip.10424 [DOI] [PubMed] [Google Scholar]
- Sierra S., Luquin N., Rico A. J., Gómez-Bautista V., Roda E., Dopeso-Reyes I. G., et al. (2015). Detection of cannabinoid receptors CB1 and CB2 within basal ganglia output neurons in macaques: changes following experimental parkinsonism. Brain Struct. Funct. 220 2721–2738. 10.1007/s00429-014-0823-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solymosi K., Kofalvi A. (2017). Cannabis: a treasure trove or pandora’s box? Mini Rev. Med. Chem. 17 1223–1291. 10.2174/1389557516666161004162133 [DOI] [PubMed] [Google Scholar]
- Turner S. E., Williams C. M., Iversen L., Whalley B. J. (2017). Molecular pharmacology of phytocannabinoids. Prog. Chem. Org. Nat. Prod. 103 61–101. 10.1007/978-3-319-45541-9_3 [DOI] [PubMed] [Google Scholar]
- Van Der Lee M. M. C., Blomenröhr M., Van Der Doelen A. A., Wat J. W. Y., Smits N., Hanson B. J., et al. (2009). Pharmacological characterization of receptor redistribution and β-arrestin recruitment assays for the cannabinoid receptor 1. J. Biomol. Screen. 14 811–823. 10.1177/1087057109337937 [DOI] [PubMed] [Google Scholar]
- Wartmann M., Campbell D., Subramanian A., Burstein S. H., Davis R. J. (1995). The MAP kinase signal transduction pathway is activated by the endogenous cannabinoid anandamide. FEBS Lett. 359 133–136. 10.1016/0014-5793(95)00027-7 [DOI] [PubMed] [Google Scholar]
- Weinstein A. M., Rosca P., Fattore L., London E. D. (2017). Synthetic cathinone and cannabinoid designer drugs pose a major risk for public health. Front. Psychiatry 8:156. 10.3389/fpsyt.2017.00156 [DOI] [PMC free article] [PubMed] [Google Scholar]