Abstract
The crystal and molecular structure of 2′-O-Me(CGCGCG)2 has been determined at 1.19 Å resolution, at 100 K, using synchrotron radiation. The structure in space group P3212 is a half-turn right-handed helix that includes two 2-methyl-2,4-pentanediol (MPD) molecules bound in the minor groove. The structure deviates from A-form RNA. The duplex is overwound with an average value of 9.7 bp per turn, characterised as having a C3′-endo sugar pucker, very low base pair rise and high helical twist and inclination angles. The structure includes 65 ordered water molecules. Only a single row of water molecules is observed in the minor groove due to the presence of hydrophobic 2′-O-methyl groups. As many as five magnesium ions are located in the structure. Two are in the major groove and interact with O6 and N7 of guanosine and N4 of cytidine residues through their hydration spheres. This work provides the first example of molecular interactions of nucleic acids with MPD, which was used as a precipitant, cryo-solvent and resolution enhancing agent. The two MPD molecules intrude into the hydration network in the minor groove, each forming hydrogen bonds between their secondary hydroxyl group and exo-amino functions of guanosine residues. Comparison of the 2′-O-Me(CGCGCG)2 structure in the P3212 and P6122 crystals delineates stability of the water network within the minor groove to dehydration by MPD and is of interest for evaluating factors governing small molecule binding to RNA. Intrusion of MPD into the minor groove of 2′-O-Me(CGCGCG)2 is discussed with respect to RNA dehydration, a prerequisite of Z-RNA formation.
INTRODUCTION
Double-stranded regions, predominant in RNA structure, exist mostly in the right-handed helical A-form with a geometry close to the average parameters determined from fibre diffraction (1,2). Inspection of high resolution X-ray RNA structures currently available in the Nucleic Acids Database (NDB) (3) reveals not only conformational details of helices and other domains of considerable complexity but also allows a detailed interpretation of RNA hydration schemes (4) and magnesium binding sites (5–7). It is now generally accepted that hydration (8,9) and metal-binding sites (5,10,11) are integral parts of RNA structure and are of crucial importance for the physical properties of RNA and a wide variety of biological functions.
One of the most intriguing features of some RNAs, directly related to the phenomena of RNA hydration and metal binding, is their tendency to form left-handed double helices, termed Z-RNA. Poly[r(CG)] (12) and shorter duplexes like (CGCGCG)2 (13) are able to form Z-RNA under conditions of high salt, such as 6 M NaClO4 or 2.7 M MgCl2. Salts have a high impact on the polyanionic structure of nucleic acids and their water shells (2,14). In the case of helicity reversal of right-handed (CG)n duplexes, salt at high concentrations is necessary to screen repulsion between electronegative phoshates in the left-handed form (15,16) and interact with the water shell around RNA leading to its dehydration (13). A similar effect can also be induced under conditions of high pressure (17,18) or by addition of alcohols, e.g. ethanol, which, due to its dehydrating properties, decreases the threshold of salt concentration necessary to induce helicity reversal (16,19). The mode of action of alcohols on a molecular level is unknown.
In DNA, the Z-form has been observed in vivo, where it is stabilised by negative supercoiling of DNA upstream of the moving RNA polymerase (20). The biological role of Z-RNA (21,22) has long been in question, due to non-physiological environmental conditions necessary to induce formation of left-handed double helical tracts in vitro. A turning point in this respect may be the recent report on Z-RNA induction and stabilisation by human dsRNA adenosine deaminase, an important enzyme involved in hypermutation of RNA viruses (23).
Despite a wealth of accumulated physical data, including the preliminary X-ray structure of (CGBrCGBr)2 (24) and NMR structures of (CGCGCG)2 (25) in the Z-form, the mechanism of RNA helicity reversal still awaits explanation. It also remains to be seen why right-handed RNA duplexes containing alternating CG base pairs undergo this transition much more easily than those containing UA base pairs (17). Clearly, elucidation of the factors governing the tendency to form Z-RNA requires more data on the structure of right-handed RNA and its hydration, the latter being highly influenced by environmental conditions.
Recently we reported low salt solution structures of (CGCGCG)2 (PDB no. 1PBM) and 2′-O-Me(CGCGCG)2 (PDB no. 1PBL) duplexes (26). These short right-handed duplexes revealed structures with large deviations from canonical A-RNA. To our surprise the structure of (CGCGCG)2 was little changed upon 2′-O-methylation. In parallel, the crystal structure of the 2′-O-Me(CGCGCG)2 duplex was solved in space group P6122 at 1.30 Å resolution, at 277 K (NDB no. ARFS26) (27). The structure was similar to the model obtained in solution and revealed a characteristic hydration pattern consisting of regular clusters of water molecules in the major groove and a single row of water molecules in the minor groove. Although these observations have raised some interest (28), it remains to be seen whether structural similarities between 2′-O-MeRNA and RNA will be found for other sequences.
A knowledge of the structure and hydration of this small subclass of right-handed RNA helices has given us new insights in our ongoing studies of salt- and alcohol-induced helicity reversal in RNA duplexes. In contrast to (CG)n RNA duplexes, 2′-O-Me(CGCGCG)2 does not undergo reversal to Z-RNA even under conditions of high pressure and remains in the right-handed form (29).
2-Methyl-2,4-pentanediol (MPD), due to its mild dehydrating properties, is often used as a precipitating agent in the crystallisation of biomolecules. It is also commonly used as a cryo-protectant in crystallography. Several examples are known of MPD binding to proteins in an orderly manner recognisable in electron density maps (30). To date no such examples are available among the coordinates for nucleic acids structures deposited in the NDB.
Here we present the X-ray structure of 2′-O-Me(CGCGCG)2 at 1.19 Å resolution, a 2′-O-modified RNA molecule with stereoselectively bound (S)-MPD, measured at 100 K in space group P3212. Detailed analysis of the hydration scheme in the cryo-cooled structure led to identification of five bound magnesium ions and provides the first example of detailed molecular interactions of nucleic acids with MPD. The intrusion of MPD into the hydration shell of right-handed (CG)n RNA will be discussed in view of known MPD properties to promote helical transitions and condensation of DNA duplexes in solution (31). We suggest that the 2′-O-Me(CGCGCG)2 cryo-structure might be taken as a model of a molecule arrested, due to 2′-O-methylation, in the right-handed state and incapable of Z-RNA formation. We hope that the results presented here lead to a deeper understanding of the RNA hydration shell, modes of water-mediated binding of small molecules (inhibitors) to RNA and factors governing RNA dehydration.
MATERIALS AND METHODS
Oligoribonucleotide crystals
The hexamer 2′-O-Me(CGCGCG) was prepared by phosphoramidite chemistry (32). Duplex crystals were grown at 20°C by hanging drop/vapour diffusion. After an extensive search, two forms of monocrystals were obtained under the following conditions: 5 mg/ml RNA in 50 mM HEPES buffer pH 7.5, 15 mM MgCl2, 1 mM spermine tetrahydrochloride and 30–40% (R,S) MPD (Sigma) as precipitating agent. The structure related to one form, P6122, was solved earlier (27). In this work we deal with a second form crystallised in the P3212 space group and subjected to cryo-protection with MPD.
Crystallographic data collection and processing
All the X-ray diffraction data were collected from a single crystal that prior to measurement had been immersed for ∼2 min in 100% (R,S) MPD. The data were collected at 100 K using synchrotron radiation from the BW7A wiggler line of the DORIS storage ring at the EMBL outstation at DESY, Hamburg, using an 18 cm MarResearch imaging plate scanner. Three data sets were collected, at long, medium and short exposures, to record intensities at high, medium and low resolution. The intensities were integrated and scaled using the DENZO/SCALEPACK program suite (33). Outliers were rejected based on the χ2 test implemented in SCALEPACK. The post-refinement option was used to refine the cell parameters. The X-ray data are summarised in Table 1.
Table 1. Summary of X-ray data collection.
| Beam line at DORIS | BW7A |
| Temperature | 100 K |
| Maximum resolution (Å) | 1.19 |
| Wavelength (Å) | 0.900 |
| Number of images (three passes) | 167 |
| Oscillation range | 0.5–3.0° |
| Rmergea | 0.049 |
| Raw measurements used | 150 605 |
| Unique reflections | 8715 |
| Per cent completeness (20–1.2 Å) | 95.8 |
| Per cent completeness in high resolution bin | 94 (1.21–1.19 Å) |
| Per cent greater than 2σ | 81.5 |
| Per cent greater than 2σ in high resolution bin | 49.7 |
| I/σ in highest resolution bin | 2.1 |
| Space group | P3212 |
| Unit cell parameters: | |
| a = b (Å) | 24.81 |
| c (Å) | 77.37 |
aRmerge = Σ|Ii – <I>|/Σ<I>, where Ii is an individual intensity measurement and <I> is the average intensity for this reflection with summation over all the data.
Structure solution and refinement
The structure was solved by molecular replacement as implemented in the program AMORE (34) from the CCP4 program suite (35). The solution was obtained using as the starting model the refined structure of 2′-O-Me(CGCGCG)2 solved previously (27) at 277 K in space group P6122 (NDB no. ARFS26). The rotation function did not give any outstanding peaks but the translation function in space group P3212 gave a number of related peaks at a correlation coefficient of ∼0.45, compared to 0.25 for other peaks. After rigid body refinement the correlation coefficient was 0.75, against a background of ∼0.5. The model was positioned in the unit cell according to the highest peak to reveal favourable intermolecular contacts. (3Fo – 2Fc) and (Fo – Fc) difference maps were inspected to reveal good overall agreement with the model, although considerable deviations were observed for the terminal base pairs.
The structure was refined by stereochemically restrained least squares minimisation as implemented in the program SHELXL (36). The integrated diffraction intensities between 20 and 1.19 Å were used in the refinement, rather than the structure factor amplitudes. The geometric restraints were derived from the standard dictionary used in the CCP4 program suite (35). Planarity restraints were imposed on the guanine and cytosine rings, as well as restraints on bond lengths and bond angles. Cycles of least squares refinement were interspersed with rounds of manual rebuilding based on (3Fo – 2Fc) and (Fo – Fc) maps, using the program TURBO-FRODO (37). Initially only isotropic temperature factors were refined, but in the later stages of refinement the B factors were refined anisotropically. Solvent molecules were inserted automatically using the program ARP_WARP (38) and inspected individually for agreement with the electron density maps. In the final stage of refinement occupancy factors were refined for water and magnesium ions, except for one magnesium lying on the two-fold axis and bridging two RNA molecules. Two MPD molecules were built into the electron density as the (S) enantiomorph, which seems to predominate at both sites, although some weak features in the electron density could be interpreted as the other enantiomorph or possibly another orientation of the MPD molecules. Refinement was terminated when it was felt that no further significant improvement in the model could be achieved. Refinement was performed using the conjugate gradient algorithm. After it was completed, one additional cycle of minimisation was executed using the full-matrix least-squares method in order to obtain direct estimates of errors in atomic positions and temperature factors. To decrease the size of the computation the model was divided into three blocks, two containing one RNA strand each and one block for the remaining atoms. All restraints and shift damping were removed in that cycle. Because of the unrestrained mode of this refinement cycle the resulting model was not used. Helical parameters were calculated using program CURVES 5.11 (39).
RESULTS
The refined model and crystal packing
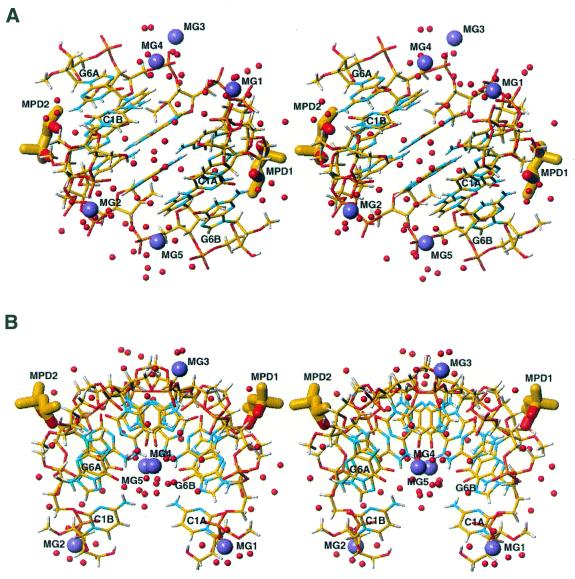
The model of the 2′-O-methylated RNA duplex is complete, forming a half-turn right-handed helix. It also includes 65 water molecules and five Mg2+ ions in partially occupied sites, but recognisable by their characteristic coordination. The model also includes two (S)-enantiomeric MPD molecules. The final R factor (Σ||Fo| – |Fc||/Σ|Fo|) for the atomic model was 0.155 for all data. The final data/parameter (d/p) ratio value was 2.8 for the refined model with anisotropic temperature factors. When restraints are taken into account the d/p value effectively becomes higher. The overall estimated standard deviation (e.s.d.) for atomic positions is 0.096 Å. For the RNA atoms it is 0.063 Å (oxygen, 0.055 Å; carbon, 0.073 Å; nitrogen, 0.050 Å; phosphorus, 0.023 Å), for the water molecules 0.140 Å, for MPD 0.240 Å (oxygen, 0.180 Å; carbon, 0.252 Å) and for the magnesium ions 0.100 Å.
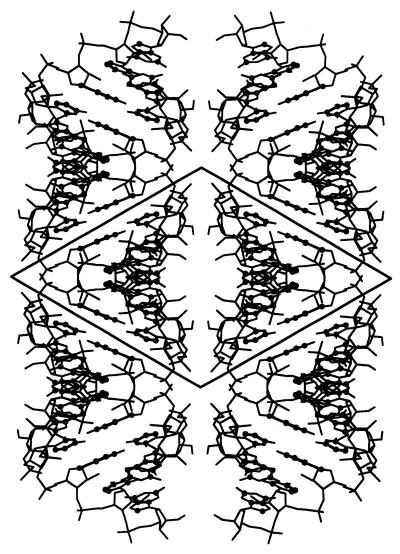
The crystal lattice consists of helical half-turns of hexamer duplexes base stacked head-to-tail, forming pseudo-infinite helices (Fig. 1), arranged in layers one molecule thick, perpendicular to the 3-fold screw axis. The neighbouring layers are at 120° to each other. The structure is similar to the P6122 crystal structure although the space group is different. The solvent content is 38% of the crystal volume, assuming a constant RNA density of 1.7 g/cm3 (40). The asymmetric unit in both crystal forms contains one duplex, but the volume of the asymmetric unit is smaller than in the hexagonal crystal by 14% and the solvent content is reduced by 20%.
Figure 1.
Crystal packing of 2′-O-Me(CGCGCG)2 in the P3212 form. The crystal lattice consists of pseudo-infinite columns of duplexes base stacked head-to-tail, arranged in layers one molecule thick, perpendicular to the 3-fold screw axis. The neighbouring layers are at 120° to each other.
Overall structure of 2′-O-Me(CGCGCG)2
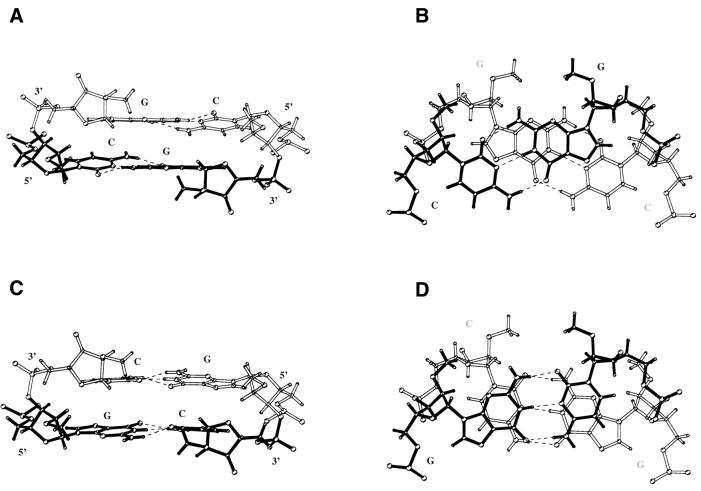
In the crystal lattice the self-complementary hexamer 2′-O-Me(CGCGCG) forms approximately one half-turn of a right- handed RNA helix (Fig. 2). The overall structure of 2′-O-Me(CGCGCG)2 deviates from canonical A-RNA and is similar to the structure found in the P6122 crystal form at 1.30 Å resolution, at 277 K (27), and to solution structures of (CGCGCG)2 and 2′-O-Me(CGCGCG)2 (26). The duplex is overwound and contains 9.7 bp per helical turn. The 2′-O-methyl groups point towards the minor groove. All sugars are in the stabilised C3′-endo pucker with PN values of 6–25°. All residues are characterised by high pucker amplitudes (average Φ = 45) and by anti glycosidic bond angles (Table 2). The γ torsion angle ranged from 30° to 52°, describing the (+) gauche conformations. Watson–Crick base pairing is observed throughout the duplex. Table 3 shows the helical parameters of the duplex structure calculated using the program CURVES 5.11 (39). The rise parameter within the core of the duplex is very small (1.9 Å) and its value rises to 2.7–3.0 Å at the ends of the duplex. Helical twist angles (36–39°) are higher than is typical for A-RNA helices; the inclination and propeller twist angles are also elevated. A characteristic stacking pattern was observed for 2′-O-Me(CGCGCG)2 similar to the P6122 crystal structure (Fig. 3).
Figure 2.
Two orthogonal stereo views of the molecular structure of the 2′-O-Me(CGCGCG)2 duplex in its P3212 form showing 65 water molecules (red spheres), five magnesium ions (violet spheres) and two MPD molecules (thick sticks) bound to the minor groove. The relationship between views (A) and (B) is a 90° rotation about a horizontal axis in the plane of the paper.
Table 2. Sugar, backbone and glycosidic torsion anglesa for the 2′-O-Me(CGCGCG)2 X-ray structure in the P3212 and P6122 space groups (33).
| Residue | α | β | γ | δ | ɛ | ζ | χ | ||||||||
| A strand |
strand B |
P3212 |
P6122 |
P3212 |
P6122 |
P3212 |
P6122 |
P3212 |
P6122 |
P3212 |
P6122 |
P3212 |
P6122 |
P3212 |
P6122 |
| C1A | – | – | – | – | 33 | 41 | 84 | 93 | 218 | 202 | 285 | 288 | 198 | 202 | |
| C1B | – | – | – | – | 43 | 52 | 95 | 76 | 210 | 219 | 280 | 280 | 200 | 193 | |
| G2A | 290 | 297 | 179 | 176 | 53 | 46 | 75 | 82 | 222 | 215 | 297 | 291 | 195 | 193 | |
| G2B | 294 | 291 | 178 | 177 | 46 | 51 | 84 | 93 | 220 | 203 | 282 | 287 | 196 | 195 | |
| C3A | 294 | 277 | 173 | 172 | 43 | 61 | 77 | 68 | 210 | 219 | 293 | 290 | 199 | 197 | |
| C3B | 278 | 292 | 173 | 170 | 56 | 49 | 79 | 72 | 208 | 217 | 291 | 290 | 197 | 201 | |
| G4A | 287 | 288 | 180 | 177 | 49 | 55 | 79 | 75 | 214 | 215 | 288 | 285 | 198 | 200 | |
| G4B | 291 | 293 | 179 | 176 | 47 | 54 | 79 | 76 | 213 | 215 | 290 | 290 | 196 | 198 | |
| C5A | 293 | 293 | 172 | 173 | 51 | 54 | 73 | 72 | 212 | 207 | 295 | 296 | 193 | 196 | |
| C5B | 295 | 293 | 172 | 171 | 49 | 52 | 80 | 73 | 205 | 211 | 293 | 291 | 192 | 197 | |
| G6A | 294 | 291 | 182 | 182 | 49 | 63 | 95 | 75 | – | – | – | – | 204 | 201 | |
| G6B | 296 | 296 | 181 | 174 | 52 | 63 | 81 | 72 | – | – | – | – | 203 | 199 | |
| Mean | 291 | 291 | 177 | 175 | 48 | 53 | 82 | 77 | 213 | 212 | 289 | 288 | 197 | 198 | |
| A-RNA | 294 | 186 | 49 | 95 | 202 | 294 | 202 |
aP α O5′ β C5′ γ C4′ δ C3′ ɛ O3′ ζ P.
Table 3. Helical parametersa for 2′-O-Me(CGCGCG)2 X-ray structure in the P3212 and P6122 space groups (33).
| Base pair | x-Displacement (Å) | Inclination (°) | Propeller twist (°) | |||
| |
P3212 |
P6122 |
P3212 |
P6122 |
P3212 |
P6122 |
| C1A–G6B | –4.9 | –4.8 | 24 | 19 | –13 | –6 |
| G2A–C5B | –4.8 | –4.8 | 22 | 19 | –15 | –16 |
| C3A–G4B | –4.8 | –4.9 | 24 | 21 | –21 | –12 |
| G4A–C3B | –4.8 | –5.0 | 24 | 22 | –14 | –8 |
| C5A–G2B | –4.7 | –5.0 | 20 | 21 | –17 | –16 |
| G6A–C1B |
–4.7 |
–5.0 |
20 |
22 |
– 7 |
– 7 |
| Base step | Rise (Å) | Twist (°) | Roll (°) | |||
| |
P3212 |
P6122 |
P3212 |
P6122 |
P3212 |
P6122 |
| C1A–G2A | 2.7 | 3.2 | 38 | 40 | –4 | –4 |
| G2A–C3A | 2.0 | 2.2 | 36 | 34 | 4 | 6 |
| C3A–G4A | 1.9 | 2.2 | 39 | 34 | 6 | 4 |
| G4A–C5A | 1.9 | 2.2 | 36 | 38 | 6 | 4 |
| C5A–G6A | 3.0 | 2.9 | 36 | 37 | –6 | –4 |
aParameters were calculated with the program CURVES 5.11 (39).
Figure 3.
The pattern of inter-strand stacking within CG (A and B) and GC steps (C and D) of the 2′-O-Me(CGCGCG)2 duplex in the P3212 form. In CG steps inter-strand guanines are parallel while cytosines are not parallel; in GC steps this motif is reversed.
Hydration, magnesium and MPD binding
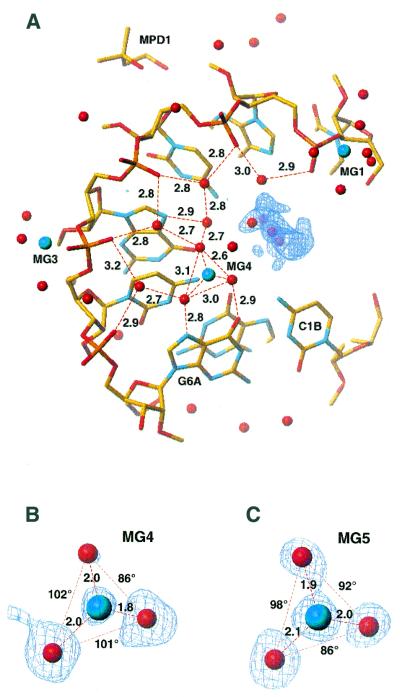
The duplex is hydrated by 65 water molecules clearly defined in the (3Fo – 2Fc) map. The intra-strand phosphate oxygens (O1P) are water bridged with W–O1P distances ranging from 2.8 to 3.3 Å. These and other water molecules localised in the major groove form fused, planar, six-membered rings and non-planar, five-membered rings for each RNA strand (Fig. 4), as previously observed in the P6122 crystal structure. The hydration network is completed by hydrogen bonds, with N7 and O6 of guanines acting as accepting sites (average distance 2.9 Å). The N4-H of cytosines are less strongly involved, with an average distance 3.6 Å. The same water molecules form weak hydrogen bonds with C5-H of cytosines (3.8 Å).
Figure 4.
Hydration of the 2′-O-Me(CGCGCG)2 duplex in the P3212 form within the major groove. Atoms lying in the further perspective are omitted for clarity (A). The pattern of fused 5- and 6-membered rings seen in the P6122 crystal (33) is observed. The (3Fo – 2Fc) electron density is shown for magnesium ions MG4 (B) and MG5 (of a second strand) (B), together with their three coordinating ordered water molecules; two of them are hydrogen bonded to N7 and O6 of guanine and the third to N4 of cytosine. The hydration pattern is disrupted near the 5′-ends by electron density [blue contours, (3Fo – 2Fc) map at 1 r.m.s. level], probably corresponding to partially disordered MPD.
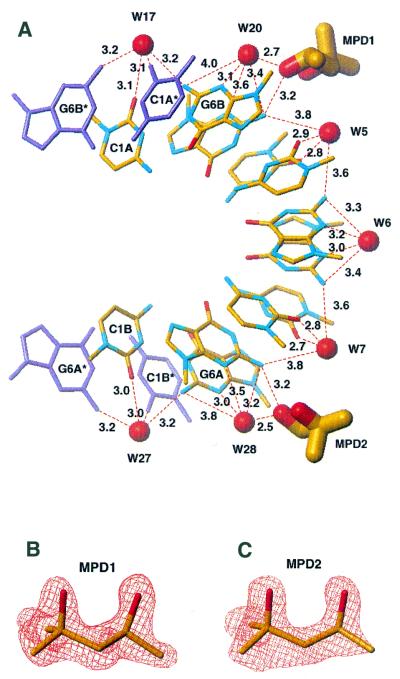
The minor groove, which is narrowed due to the presence of 2′-O-methyl groups, is hydrated by a single row of seven well-defined water molecules with distances ranging from 5.8 to 6.3 Å (Fig. 5). Each water is wedged between two inter-strand 2′-O-methyl groups at a distance of ∼2.5 Å from the nearest water oxygen and 3.5 Å from the nearest methyl hydrogen. The waters bridge the duplex strands by donating a pair of hydrogen bonds either to two self-parallel cytosines or to two self-parallel guanines. The waters hydrogen bonded to the carbonyl O2 of cytosines are at distances of 2.7–3.1 Å whilst the ones bridging the endo-cyclic N3 of guanines are bound less strongly (3.1–3.5 Å). The hydration pattern in the minor groove extends across the axially stacked RNA molecules, with single water sites, lying exactly on the 2-fold axes, bridging C1A and C1B with their symmetry-related equivalents (Fig. 5).
Figure 5.
Hydration and MPD binding within the minor groove of 2′-O-Me(CGCGCG)2 in its P3212 form. A single row of seven water molecules spans both duplex strands: each water bridges, by donating a pair of hydrogen bonds, either to two self-parallel cytosines or two self-parallel guanines. Two MPD molecules intrude into the minor groove. For each MPD molecule the oxygen of its secondary hydroxyl group is the hydrogen bond acceptor from the exo-NH2 group of the G2A (MPD1) or G2B (MPD2) guanine residue (A). ‘Omit’ (Fo – Fc) maps, contoured at the 3 r.m.s. level, are shown for the two MPD molecules (B and C). The maps were calculated after omitting MPD from the model and 10 cycles of least squares minimisation.
Five magnesium ions were observed in the structure (Fig. 2), three interacting with phosphate residues and two in the major groove (Fig. 4). All three phosphate-bound magnesium ions form inner- or outer-sphere complexes, each magnesium bridging two phosphates from symmetry-related molecules in a lateral, column-to-column direction. MG1 forms an inner- sphere complex with phosphate G2A and an outer-sphere complex with phosphate C3A of the neighbouring molecule. MG2 forms an inner-sphere complex with phosphate G2B and an outer-sphere complex with phosphate C3B. MG3 bridges two duplexes through an inner-sphere complex with C5A and C5B phosphate oxygens (O2P). The coordination sphere of these magnesium ions is completed by water molecules.
The two magnesium ions located in the major groove form outer-sphere complexes spaning steps C5A–G6A (MG4) and C5B–G6B (MG5) (Fig. 4). The magnesium is recognisable by its well-defined coordination distances and angles to its three visible ligands. Each magnesium ion coordinates three well-ordered water molecules arrayed to the first RNA hydration shell; two of them are hydrogen bonded to N7 and O6 of guanine, and the third to N4 of cytosine bases. The magnesium ion is positioned in the second hydration sphere of RNA and the remaining three water ligands would lie in the third hydration sphere, which is disordered in the crystal structure.
Two MPD molecules are located in the minor groove (Fig. 5). Their sites are clearly defined in the (3Fo – 2Fc) map with occupancies of 0.7 for both (average B = 30 Å2 for MPD1 and 37 Å2 for MPD2). For each MPD molecule the oxygen of the secondary hydroxyl group is the hydrogen bond acceptor from the exo-NH2 group of the G2A (MPD1) or G2B (MPD2) guanine residue (average hydrogen bond donor–acceptor distance 3.2 Å). The MPD molecule is in its compact form, being stabilised by an intramolecular hydrogen bond with the secondary hydroxyl group as the donor.
Inspection of the (3Fo – 2Fc) map where the 5′-ends of the duplex are close to each other reveals the presence of electron density which might correspond to a low occupancy MPD site, but no satisfactory fit could be obtained for a complete, ordered molecule (Fig. 4).
DISCUSSION
Although the P3212 crystal of 2′-O-Me(CGCGCG)2 was obtained under similar conditions to the P6122 form (27), namely in the presence of 30–40% (R,S)-MPD, the crystal was then transferred to anhydrous (R,S)-MPD prior to diffraction measurement, to enhance diffraction, i.e. in a similar way to that developed in DNA crystallography (41,42). The high crystal quality, cryogenics and the use of synchrotron radiation allowed refinement of the model of the P3212 structure and its hydration with exceptional accuracy. This presented a unique opportunity to investigate how high concentrations of MPD influence the structure in comparison to the X-ray structure in the P6122 space group. As indicated above, there are some differences in conformational parameters of the 2′-O-methylated RNA duplex structures in the two crystal forms (Tables 2 and 3). However, the main novelty in the current P3212 structure concerns hydration, magnesium binding and the presence of MPD molecules stereoselectively bound to RNA.
Major groove hydration and magnesium binding
The P3212 crystal contains 20% less water than the P6122 space group, making the RNA more condensed. The duplex structure is hydrated in a regular way with approximately 11 ordered water sites per base pair step. This value is lower than the average (15–20) experimentally determined for nucleic acid structures (2) and the value estimated for CG base pairs in A-DNA (19 sites) (43) and (CG)12 RNA (22 sites) (44,45).
Most of the water molecules are located near the phosphate groups and in the major groove, forming a regular pattern. The intra-strand phosphate oxygens (O1P) are water bridged, as in other crystal structures of oligonucleotide duplexes in the A-form (8,46); the concept of economy of hydration (46), formerly put forward to explain why more compact A-DNA structures need less hydration than B-DNA, finds a wider meaning when considering hydration of 2′-O-MeRNA versus RNA. The phosphate–water bridges form one edge of a sheet of water clusters consisting of fused five- and six-membered rings, each including one phosphate oxygen and four or five waters, respectively (Fig. 4). The five-membered rings are non-planar while the hexameric rings are nearly planar. Such clusters were first seen in the 2′-O-Me(CGCGCG)2 structure in the P6122 crystal (27). The water network is stabilised by hydrogen bonds donated to O6 and N7 of guanines as primary base sites, a feature seen in other RNA structures (4,8,47). The remaining water molecules within the same hydration ring are less strongly bound, with only distant interactions (3.6–3.8 Å) between N4-H of cytosines and two of the remaining waters. The same cytosine residues form weak hydrogen bonds of C5-H type with waters (3.8 Å) in a similar manner to other CG pairs in RNA duplexes (4).
To our knowledge, the crystal structure of 2′-O-Me(CGCGCG)2 in the P3212 crystal has the highest ratio of magnesium ions per base pair for a short RNA duplex crystal structure. All five magnesium ions are embedded in the hydration network in a unified architecture allowing water- and magnesium-mediated backbone–base contacts (Fig. 4) in a manner observed in several other RNA duplexes (5,7,11) and RNA–DNA chimeric duplexes (48). The magnesium ions play two different roles: interacting with the phosphates and with the bases in the major groove. The strongest of the inter-phosphate interactions (via MG3) is an inner-sphere complex with phosphate ligands (C5A and C5B) from neighbouring duplexes. MG3 facilitates specific intermolecular contacts that compensate for the lack in 2′-O-Me(CGCGCG)2 of a 2′-hydroxyl function, which in several RNA crystal structures plays a role in water-mediated RNA–RNA interactions (49). It also compensates for the repulsive phosphate–phosphate interactions between two closely spaced duplexes. The other type of interaction involves mixed inner- and outer-sphere complexes neutralising charges on the phosphates. Formation of such magnesium bridges alters the typical pattern of intra-strand phosphate hydration in A-helices and has the effect on hydration statistics explained above.
Two magnesium ions are located in the major groove near the 3′-ends of the duplex, spanning CG step residues through outer-sphere complexes. The hydration of the magnesium ions is such that the geometry of their visible ligands fits well into the scheme of the first hydration sphere of the RNA molecule (Fig. 4). As in other RNA duplexes (7) and RNA–DNA chimeric duplexes (48), guanine O6 and N7 sites are primary Mg2+ acceptors, in this case contributing two waters to its coordination sphere. The CG step, comprising C5A–G6A and C5B–G6B residues, is additionally stabilised by a third water of the magnesium coordination sphere accepting the hydrogen bond of the cytosine N4-H site. Although this interaction is much weaker, it plays an important role in positioning the magnesium within the characteristic hydration scheme of the major groove.
A re-examination of electron density in the P6122 crystal structure, where only two magnesium ions were localised (27) forming salt bridges, revealed two small peaks in the second hydration shell of the major groove at locations corresponding to the Mg2+ ions in the cryogenic structure and in similar configuration with the water ligands. This is consistent with observations made in DNA crystallography at near-atomic resolution that immersion of crystals in concentrated MPD results in better localisation and visualisation of metals (41,42) and therefore a deeper understanding of their role in DNA structure.
Minor groove hydration and MPD binding
The 2′-O-methyl groups in the 2′-O-Me(CGCGCG)2 structure point towards the minor groove, leading not only to its narrowing but also to increased hydrophobicity. The average distance between the inter-strand 2′-O-methyl carbons atoms is only 7.2 Å. This means that the space between 2′-O-methyl groups across the minor groove is only ∼3.2 Å, after taking into account the van der Waals radii.
Therefore, only a single row of seven water molecules is observed in the minor groove of 2′-O-Me(CGCGCG)2 (Fig. 5), instead of two rows of water spanning free 2′-hydroxy groups in A-RNA duplexes, e.g. (CCCCGGGG)2 (8). The seven water molecules are not in contact with each other and, similar to the P6122 crystal, none have access to RNA 2′-oxygens as hydrogen bond acceptors. Water molecules act as strong hydrogen bond donors spanning both oligoribonucleotide strands. Each water bridges, via a pair of hydrogen bonds, either two self-parallel cytosines or two self-parallel guanines. Hydrogen bonds from water to the O2 carbonyl function of cytosines are stronger than those to the endo-cyclic N3 of guanines. Such a pattern of water-mediated inter-base contacts in the minor groove offers an explanation as to why the bases in (CG)n alternating base pairs maintain their uncommon cross-strand parallel arrangement. This distinct architecture of hydration in the minor groove in this crystal structure and in the similar P6122 space group (27) strongly suggests that the single row of water molecules within the minor groove is necessary to maintain the right-handed architecture in the (CG)n family of RNA duplexes.
The 2′-O-Me(CGCGCG)2 structure in the P3212 space group includes two MPD molecules bound in the minor groove. The (S) enantiomorph of MPD was primarily selected upon binding from the (R,S)-MPD mixture used during crystallography experiments. This is the first example of molecular interactions of a nucleic acid structure with MPD that was used as a precipitant, cryo-solvent and resolution enhancing agent. We find this surprising, since soaking crystals in concentrated MPD solution (42) has been common in recent years in the DNA field. It remains to be seen whether the increase in hydrophobicity of the minor groove due to the 2′-O-methyl groups is of importance in attracting MPD from bulk solution, as the methyl groups at the distal end of the MPD molecules point towards the 2′-O-methyl groups of the duplex.
Both the MPD molecules are located near the duplex 5′-ends. The MPD molecules intrude into the hydration layer of the minor groove (Fig. 5) without displacing any of the ordered water molecules observed in both the P3212 and P6122 crystals. This makes the overall scheme of hydrogen bonded and van der Waals interactions very dense within the minor groove. For each MPD molecule, the oxygen of the secondary hydroxyl group is the hydrogen bond acceptor from the exo-NH2 group of the G2A (MPD1) or G2B (MPD2) residue. These interactions, with an average hydrogen bond donor–acceptor distance of 3.2 Å, are very well matched to the minor groove geometry and, most probably, are of importance for selection of the (S) enantiomorph upon binding from the (R,S)-MPD used. It should be pointed out that this mode of MPD intrusion cancels involvement of waters in the minor groove as weak acceptors from exo-NH2 groups of guanines, a feature observed in the P6122 crystal structure (27).
The stereoselective binding of the (S) enantiomorph of MPD within the hydration network of the 2′-O-Me(CGCGCG)2 structure gives an insight into the way alcohols and small ligands containing hydroxy groups interact with the primary hydration shells of RNA. This should be of interest in designing small molecules (inhibitors) with water-mediated binding affinities for the minor groove of nucleic acid duplexes (50). The mode of MPD binding could also be relevant in elucidation of MPD involvement in oligonucleotide bending of A-DNA duplexes (51).
Does the binding of MPD to 2′-O-Me(CGCGCG)2 in the crystal structure help in evaluation of the dehydration processes that precede Z-RNA formation? It has been reported that MPD acts as a dehydrating agent inducing a helicity reversal transition in DNA duplexes containing alternating CG base pairs (31). Interestingly, the resulting CD spectra of d(CGCGCG)2 were similar to that of (CGCGCG)2, resembling spectra of Z-RNA reported earlier (13), rather than Z-DNA (15). As noted above, 2′-O-Me(CGCGCG)2 does not undergo reversal to Z-RNA (29), in contrast to (CG)n RNA duplexes. This points primarily to the influence of 2′-O-methylation, which due to conjunction of both electronegative 2′-substitution and steric effects stabilises the pucker of sugar residues in their high C3′-endo conformation (26,52), thus making the sugar transitions necessary for Z-RNA formation energetically unfavourable. Nonetheless, the hydration pattern of these non-canonical RNA duplexes (53) might also be of importance.
We suggest that the 2′-O-Me(CGCGCG)2 cryo-structure might be considered as a model of the (CG)n RNA molecule arrested, due to 2′-O-methylation, in the right-handed state and incapable of Z-RNA formation. Thus, the intrusion of MPD into the hydration shell of 2′-O-Me(CGCGCG)2 indicates the manner of RNA dehydration that precedes Z-RNA formation. Besides the two (S)-MPD molecules clearly shown in the minor groove, we have also observed electron density close to the duplex 5′-ends. The density remained uninterpreted, being too disordered for clear interpretation, but it probably corresponds to a disordered MPD molecule(s), partial fitting of MPD into the density being possible (Fig. 5).
Although comparison of the 2′-O-Me(CGCGCG)2 structure in the P6122 and P3212 crystals delineates the stability of the water shell in the minor groove under dehydrating conditions, the intrusion of MPD alters the water network, i.e. the waters in the minor groove no longer function as weak acceptors from exo-NH2 groups of guanines, as previously observed (27). One might speculate whether release of the exo-amino group of guanine residues from bonding to the minor groove water network has a role in promoting change in the guanosine residues to a syn conformation.
Results presented here shed new light on the structure and hydration shell within the (CG)n family of RNA duplexes and might be of interest to all studying 2′-O-methylated RNA in a broader sense (28,54). Stereoselective binding of MPD to the minor groove of 2′-O-MeRNA, observed for the first time, delineates conceivable modes of water-mediated binding of alcohols or other hydroxy group-containing small molecules to RNA (50) and points to the factors governing RNA dehydration as a prerequisite of Z-RNA formation.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Richard Lavery for kindly supplying us with the program CURVES 5.11, Mariusz Popenda for discussions and Mieczyslawa Kluge for technical assistance. This work was supported in part by a grant from the State Committee for Scientific Research, Republic of Poland (7T09A09720). The generous support of the Poznañ Supercomputing and Networking Centre is acknowledged.
PDB no. 1I7J; NDB no. AR0031
References
- 1.Arnott S., Hukins,D.W.L. and Dover,S.D. (1972) Optimised parameters for RNA double-helices. Biochem. Biophys. Res. Commun., 48, 1392–1399. [DOI] [PubMed] [Google Scholar]
- 2.Saenger W. (1984) Principles of Nucleic Acids Structure. Springer, Berlin, Germany.
- 3.Berman H.M., Zardecki,C. and Westbrook,J. (1998) The Nucleic Acid Database: A resource for nucleic acid science. Acta Crystallogr., 54D, 1095–1104. [DOI] [PubMed] [Google Scholar]
- 4.Auffinger P. and Westhof,E. (1998) Hydration of RNA base pairs. J. Biomol. Struct. Dyn., 16, 693–707. [DOI] [PubMed] [Google Scholar]
- 5.Cate J.H., Gooding,A.R., Podell,E., Zhou,K., Golden,B.L., Kundrot,C.E., Cech,T.R. and Doudna,J.A. (1996) Crystal structure of a group I ribozyme domain: principles of RNA packing. Science, 273, 1678–1685. [DOI] [PubMed] [Google Scholar]
- 6.Cate J.H., Hanna,R.L. and Doudna,J.A. (1997) A magnesium ion core at the heart of a ribozyme domain. Nature Struct. Biol., 4, 553–558. [DOI] [PubMed] [Google Scholar]
- 7.Correl C.C, Freeborn,B., Moore,P.B. and Steitz,T.A. (1997) Metals, motifs and recognition in the crystal structure of a 5S rRNA domain. Cell, 91, 705–712. [DOI] [PubMed] [Google Scholar]
- 8.Egli M., Portmann,S. and Usman,N. (1996) RNA hydration: a detailed look. Biochemistry, 35, 8489–8494. [DOI] [PubMed] [Google Scholar]
- 9.Westhof E. (1988) Water: an integral part of nucleic acid structure. Annu. Rev. Biophys. Biophys. Chem., 17, 125–144. [DOI] [PubMed] [Google Scholar]
- 10.Draper D.E. and Misra,V.K. (1998) RNA shows its metal. Nature Struct. Biol., 5, 927–930. [DOI] [PubMed] [Google Scholar]
- 11.Misra V.K and Draper,D.E. (1999) On the role of magnesium ions in RNA stability. Biopolymers, 48, 113–135. [DOI] [PubMed] [Google Scholar]
- 12.Hall K., Cruz,P., Tinoco,I.Jr, Jovin,T.M. and van de Sande,J.H. (1984) ‘Z-RNA’. A left-handed RNA double helix. Nature, 311, 584–586. [DOI] [PubMed] [Google Scholar]
- 13.Adamiak R.W., Galat,A. and Skalski,B. (1985) Salt- and solvent-dependent conformational transitions of ribo-CGCGCG duplex. Biochim. Biophys. Acta, 825, 345–352. [Google Scholar]
- 14.McFail-Isom L., Shui,X. and Williams,L.D. (1998) Divalent cations stabilize unstacked conformations of DNA and RNA by interacting with base pi systems. Biochemistry, 37, 17105–17111. [DOI] [PubMed] [Google Scholar]
- 15.Pohl F.M. and Jovin,T.M. (1972) Salt-induced co-operative conformational change of a synthetic DNA: equilibrium and kinetic studies with poly(dG-dC) J. Mol. Biol., 67, 375–396. [DOI] [PubMed] [Google Scholar]
- 16.Rich A., Nordheim,A. and Wang,A.H. (1984) The chemistry and biology of left-handed Z-DNA. Annu. Rev. Biochem., 53, 791–846. [DOI] [PubMed] [Google Scholar]
- 17.Krzyzaniak A., Barciszewski,J., Fürste,J.P., Bald,R., Erdmann,V.A., Salanski,P. and Jurczak,J. (1994) A-Z RNA conformational changes effected by high pressure. Int. J. Biol. Macromol., 16, 159–162. [DOI] [PubMed] [Google Scholar]
- 18.Barciszewski J., Jurczak,J., Porowski,S., Specht,T. and Erdmann,V.A. (1999) The role of water structure in conformational changes of nucleic acids in ambient and high-pressure conditions. Eur. J. Biochem., 260, 293–307. [DOI] [PubMed] [Google Scholar]
- 19.Tinoco I.Jr, Cruz,P., Davis,P., Hall,K., Hardin,C.C., Mathies,R.A., Puglisi,J.D., Trulson,M.A., Johnson,W.C.Jr and Neilson,T. (1986) Z-RNA: a left-handed double helix. In van Knippenberg,P.H. and Hilbers,C.W. (eds), Structure and Dynamics of RNA, NATO ASI Series A. Plenum Press, New York, NY, Vol. 110, pp. 55–69.
- 20.Liu L.F. and Wang,J.C. (1987) Supercoiling of the DNA template during transcription. Proc. Natl Acad. Sci. USA, 84, 7024–7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zarling D.A., Calhoun,C.J., Hardin,C.C. and Zarling,A.H. (1987) Cytoplasmic Z-RNA. Proc. Natl Acad. Sci. USA, 84, 6117–6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gagna C.E., Chen,J.H., Kuo,H.R. and Lambert,W.C. (1998) Localisation of left-handed Z-RNA in the outer cortical secondary fiber cells of the adult dog crystalline lens. Scanning, 20, 255–256. [PubMed] [Google Scholar]
- 23.Brown B.A., Lowenhoupt,K., Wilbert,C.M., Hanlone,E.B. and Rich,A. (2000) The Zα domain of the editing enzyme dsRNA adenosine deaminase binds left-handed Z-RNA as well as Z-DNA. Proc. Natl Acad. Sci. USA, 97, 13532–13536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakamura Y., Fujii,S., Urata,H., Uesugi,S., Ikehara,M. and Tomita,K. (1985) Crystal structure of left-handed RNA tetramer, r(C-br8G)2. Nucleic Acids Symp. Ser., 16, 29–32. [PubMed] [Google Scholar]
- 25.Davis P.W., Adamiak,R.W. and Tinoco,I.Jr (1990) Z-RNA: the solution NMR structure of r(CGCGCG). Biopolymers, 29, 109–122. [DOI] [PubMed] [Google Scholar]
- 26.Popenda M., Biala,E., Milecki,J. and Adamiak,R.W. (1997) Solution structure of RNA duplexes containing alternating CG base pairs: NMR study of r(CGCGCG)2 and 2′-O-Me(CGCGCG)2 under low salt conditions. Nucleic Acids Res., 25, 4589–4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adamiak D.A., Milecki,J., Popenda,M., Adamiak,R.W., Dauter,Z. and Rypniewski,W.R. (1997) Crystal structure of 2′-O-Me(CGCGCG)2, an RNA duplex at 1.30 Å resolution. Hydration pattern of 2′-O-methylated RNA. Nucleic Acids Res., 25, 4599–4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poole A., Penny,D. and Sjoberg,B.M. (2000) Methyl-RNA: an evolutionary bridge between RNA and DNA? Chem. Biol., 7, 207–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krzyzaniak A., Salanski,P., Adamiak,R.W., Jurczak,J. and Barciszewski,J. (1996) Structure and function of nucleic acids under high pressure. In Hayashi,R. and Balny,C. (eds), High Pressure Bioscience and Biotechnology. Elsevier Science, Amsterdam, The Netherlands, pp. 189–194.
- 30.Freitag S. Le Trong,I., Klumb,L.A., Stayton,P.S. and Stenkamp,R.E. (1999) Atomic resolution structure of biotin-free Tyr43Phe streptavidin: what is in the binding site? Acta Crystallogr., 55D, 1118–1126. [DOI] [PubMed] [Google Scholar]
- 31.Vorlickova M., Subirana,J.A., Chladkova,J., Tejralova,I., HuynhDinh,T., Arnold.L. and Kypr,J. (1996) Comparison of the solution and crystal conformations of (G+C)-rich fragments of DNA. Biophys. J., 71, 1530–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Biala E., Milecki,J., Kowalewski,A., Popenda,M., Antkowiak,W.Z. and Adamiak,R.W. (1993) Z-RNA. The synthesis of 2′-O-[13C]methyl- and 5-methyl-analogs of ribo-CGCGCG. Acta Biochim. Pol., 40, 521–530. [PubMed] [Google Scholar]
- 33.Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–325. [DOI] [PubMed] [Google Scholar]
- 34.Navaza J. (1994) AMoRe: an automated package for molecular replacement. Acta Crystallogr., 50A, 157–163. [Google Scholar]
- 35.Collaborative Computational Project Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr., 50D, 760–763. [DOI] [PubMed] [Google Scholar]
- 36.Sheldrick G.M. and Schneider,T.R. (1997) SHELXL: high-resolution refinement. Methods Enzymol., 277, 319–343. [PubMed] [Google Scholar]
- 37.Roussel A. and Cambillau,C. (1991) Turbo Frodo. Silicon Graphics Geometry Partners Directory. Silicon Graphics, Mountain View, CA, p. 86.
- 38.Lamzin V.S. and Wilson,K.S. (1993) Automated refinement of protein models. Acta Crystallogr., 49D, 129–147. [DOI] [PubMed] [Google Scholar]
- 39.Lavery R. and Sklenar,H. (1988) Curves: the definition of generalised helicoidal parameters and of axis curvature for irregular nucleic acids. J. Biomol. Struct. Dyn., 6, 63–91. [DOI] [PubMed] [Google Scholar]
- 40.Feigin L.A. and Svergun,D.I. (1987) Structure Analysis by Small-angle X-ray and Neutron Scattering. Plenum Press, New York, NY, p. 120.
- 41.Fratini A.V., Kopka,M.L., Drew,H.R. and Dickerson,R.E. (1982) Reversible bending and helix geometry in a B-DNA dodecamer: CGCGAATTBrCGCG. J. Biol. Chem., 57, 14686–14707. [PubMed] [Google Scholar]
- 42.Shui X., McFail-Isom,L., Hu,G.G. and Williams,L.D. (1998) The B-DNA dodecamer at high resolution reveals a spine of water on sodium. Biochemistry, 37, 8341–8355. [DOI] [PubMed] [Google Scholar]
- 43.Feig M. and Pettitt,B.M. (1999) Modeling high-resolution hydration patterns in correlation with DNA sequence and conformation. J. Mol. Biol., 286, 1075–1095. [DOI] [PubMed] [Google Scholar]
- 44.Auffinger P. and Westhof,E. (2000) Water and ion binding around RNA and DNA (C,G) oligomers. J. Mol. Biol., 300, 1113–1131. [DOI] [PubMed] [Google Scholar]
- 45.Auffinger P. and Westhof,E. (2001) Water and ion binding around r(UpA)12 and d(TpA)12 oligomers—comparison with RNA and DNA (CpG)12 duplexes. J. Mol. Biol., 305, 1057–1072. [DOI] [PubMed] [Google Scholar]
- 46.Saenger W., Hunter,W.N. and Kennard,O. (1986) DNA conformation is determined by economics in the hydration of phosphate groups. Nature, 324, 385–388. [DOI] [PubMed] [Google Scholar]
- 47.Klosterman P.S., Shah,S.A. and Steitz,T.A. (1999) Crystal structures of two plasmid copy control related RNA duplexes: an 18 base pair duplex at 1.20 Å resolution and a 19 base pair duplex at 1.55 Å resolution. Biochemistry, 38, 14784–14792. [DOI] [PubMed] [Google Scholar]
- 48.Robinson H., Gao,Y.G., Sanishvili,R., Joachimiak,A. and Wang,A.H. (2000) Hexahydrated magnesium ions bind in the deep major groove and at the outer mouth of A-form nucleic acid duplexes. Nucleic Acids Res., 28, 1760–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schindelin H., Zhang M., Bald,R., Furste,J.P., Erdmann,V.A. and Heinemann,U. (1995) Crystal structure of an RNA dodecamer containing the Escherichia coli Shine-Dalgarno sequence. J. Mol. Biol., 249, 595–603. [DOI] [PubMed] [Google Scholar]
- 50.Hermann T. (2000) Strategies for the design of drugs targeting RNA and RNA-protein complexes. Angew. Chem. Int. Ed., 39, 1890–1905. [DOI] [PubMed] [Google Scholar]
- 51.Sprous D., Zacharias,W., Wood,Z.A. and Harvey,S.C. (1995) Dehydrating agents sharply reduce curvature in DNAs containing A tracts. Nucleic Acids Res., 23, 1816–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guschlbauer W. and Jankowski,K. (1980) Nucleoside conformation is determined by the electronegativity of the sugar substituent. Nucleic Acids Res., 8, 1421–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kulinska K., Kulinski,T., Lyubartsev,A., Laaksonen,A. and Adamiak,R.W. (2000) Spatial distribution functions as a tool in the analysis of ribonucleic acids hydration—molecular dynamics studies. Comput. Chem., 24, 451–457. [DOI] [PubMed] [Google Scholar]
- 54.Teplova M., Minasov,G., Tereshko,V., Inamati,G.B., Cook,P.D., Manoharan,M. and Egli,M. (1999) Crystal structure and improved antisense properties of 2′-O-(2-methoxyethyl)-RNA. Nature Struct. Biol., 6, 535–539. [DOI] [PubMed] [Google Scholar]