ABSTRACT
Sequence type 36 (ST36) Klebsiella pneumoniae is distributed worldwide. We found an ST36 K. pneumoniae clinical isolate that was carbapenem resistant, carried blaKPC-2, had mucoid regulator gene rmpA, and exhibited high virulence. The finding suggests the emergence of carbapenem-resistant hypervirulent K. pneumoniae of ST36, and surveillance of carbapenem-resistant hypervirulent K. pneumoniae is required.
KEYWORDS: carbapenem resistance, hypervirulence, Klebsiella pneumoniae
TEXT
Hypervirulent Klebsiella pneumoniae (hvKP) is a particular threat because it can cause severe infections in apparently healthy people, with high mortality rates (1). hvKP is usually susceptible to carbapenems, and its infections have been successfully treated using carbapenems (1, 2). However, some carbapenem-resistant K. pneumoniae strains of the widely distributed sequence type 11 (ST11) became hypervirulent by acquiring a pLVPK-like virulence plasmid (3). The combination of carbapenem resistance and hypervirulence significantly compromises the options of antimicrobial agents for treating the life-threatening infections caused by the carbapenem-resistant hvKP and therefore represents a major urgent challenge for clinical treatment, infection control, and public health (4). Carbapenem-resistant hvKP is not restricted to ST11 (5, 6), as such strains can emerge either from carbapenem-resistant strains by acquiring the virulence plasmid or from hvKP by acquiring carbapenem resistance. Here, we report the acquisition of carbapenem resistance in a non-ST11 hvKP strain.
Strain WCHKP13F2 was recovered in the intensive care unit of a hospital in Sichuan province in 2015 from the blood of a male patient in his late 40s with severe burns. The patient developed a health care-associated bloodstream infection, with rigors, high fever (temperature >39°C), headache, and shock, that was caused by strain WCHKP13F2. He received the combination of imipenem and amikacin plus surgical debridement. However, he had a poor outcome and was discharged with unresolved critical illness on his own will. The strain was identified as K. pneumoniae by Vitek II (bioMérieux, Marcy-l'Étoile, France) and was resistant to aztreonam (MIC, 128 μg/ml), ceftazidime (MIC, 16 μg/ml), gentamicin (MIC, 32 μg/ml), imipenem (MIC, 32 μg/ml), meropenem (MIC, 16 μg/ml), piperacillin-tazobactam (MIC, 256/4 μg/ml), sulfisoxazole (MIC, >512 μg/ml), and tigecycline (MIC, 4 μg/ml); intermediate to levofloxacin (MIC, 4 μg/ml); and susceptible to amikacin (MIC, 1 μg/ml) and colistin (MIC, 1 μg/ml), as determined by the microdilution method of the Clinical and Laboratory Standards Institute (CLSI) (7). The strain was mucoid on the agar plate. A string test was performed by stretching bacterial colonies using an inoculation loop with a hypervirulent ST23:K1 K. pneumoniae strain WCHKP030925 used as a control (1). Strain WCHKP13F2 formed a 2-mm viscous string, which is below the >5 mm that defines hypermucous conditions.
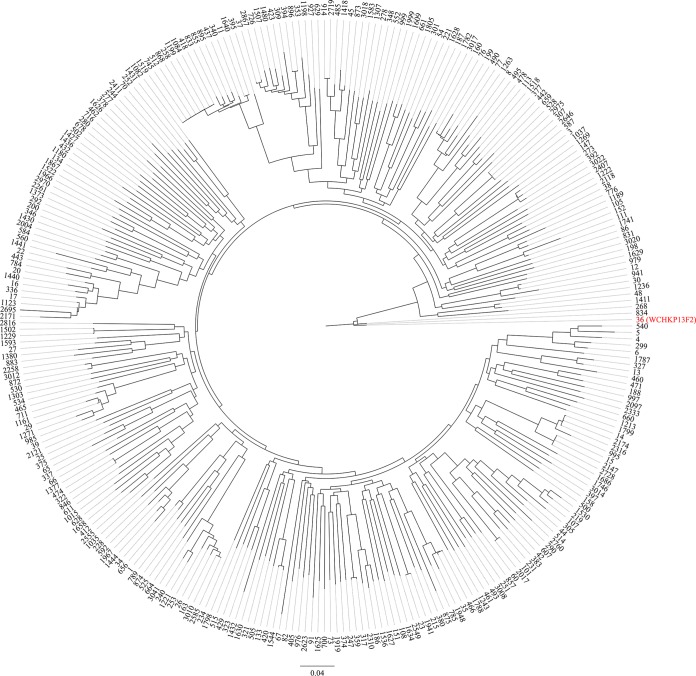
The strain was subjected to whole-genome sequencing using both HiSeq X10 sequencer (Illumina, San Diego, CA), with the 150-bp paired-end protocol and 200× coverage, and the long-read MinION sequencer (Nanopore, Oxford, United Kingdom). The de novo hybrid assembly of both short and long reads was performed using Unicycler under the conservative mode for increased accuracy (8). The complete circular contigs generated were then corrected using Plion with Illumina reads for several rounds until no change was detected (9). The genome size of strain WCHKP13F2 was 5,749,649 bp, including a 5.38-Mb chromosome and two plasmids. Annotation was carried out using Prokka (10). Strain WCHKP13F2 belonged to ST36 (gapA-infB-mdh-pgi-phoE-rpoB-tonB allele number 2-1-2-1-7-1-7) and the capsular type K62, as determined using the assembled contigs to query the multilocus sequence typing (MLST) and wzi allele databases (http://bigsdb.pasteur.fr/klebsiella/klebsiella.html). To study the phylogenetics of ST36 K. pneumoniae with those of other STs, all assembled K. pneumoniae genomes (n = 3,423; accessed by 27 March 2018) were retrieved from GenBank and were checked for quality and contaminations using Kraken with the MiniKraken 8-GB database (11). Assemblies with >200 contigs were discarded. MLST was performed using the script (https://github.com/tseemann/mlst). For each known ST, the complete genome or a draft one with a higher N50 value was selected as the representative, whereas the assemblies of unknown STs were excluded. A total of 299 assemblies with distinct STs were included and were annotated using Prokka (10), followed by identification of the core genome using Roary (12). A phylogenetic tree was inferred based on 86,396 SNP sites from 2,839 genes presented in all assemblies using RAxML with a GTRGAMMA model (13) (Fig. 1). ST36 is closely related to ST268 (2-1-2-1-7-1-81) and ST834 (2-1-2-1-7-1-25) with only one allele (tonB) variance among the three STs (Fig. 1).
FIG 1.
Core genome-based phylogenetic tree of K. pneumoniae belonging to different STs. ST36 is indicated in red. ST36 is closely related to ST268 and ST834 but is distinct from other STs.
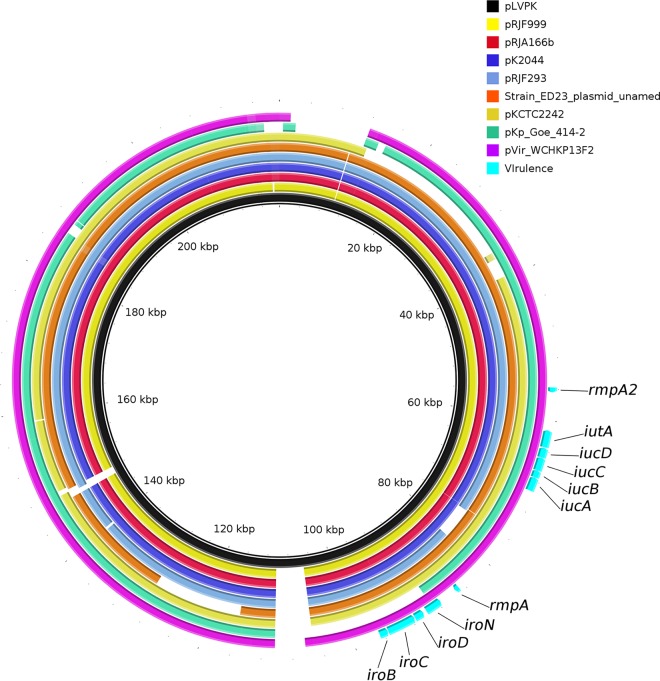
It is known that ST36 K. pneumoniae is an hvKP that causes bacteremia (2) and has worldwide distribution (14–16). Virulence genes were identified using the database available at http://bigsdb.pasteur.fr/klebsiella/klebsiella.html. Strain WCHKP13F2 had a number of virulence factors, including the regulator of mucoid phenotype (rmpA), aerobactin (iucABCD, iutA), colibactin (clbA-R), salmochelin (iroN, iroBCDN), yersiniabactin (fyuA, irp2, and ybtAEPQTUX) (17), and the type 3 fimbriae-encoding system (mrkABCDFHIJ) (18). Strain WCHKP13F2 also had a truncated rmpA2 (another regulator of mucoid phenotype) gene, which was due to a mutation introducing a stop codon. The rmpA, iroBCDN, and iucABCD genes were carried by a 208,166-bp IncHI1/IncFIB plasmid, designated pVir-KP13F2, which was highly similar (97% coverage and 99% identity) to the known virulence plasmid pLVPK (GenBank accession no. AY378100) (Fig. 2). pLVPK (IncHI1/IncFIB) is 219,385 bp and possesses rmpA, rmpA2, iucABCD, iutA, and iroBCDN, while the recently identified IncHI1/IncFIB virulence plasmid pVir-CR-HvKP4 (178,154 bp) in ST11 carbapenem-resistant hvKP carries rmpA2, iucABCD, and iutA but not rmpA or iroBCDN (3).
FIG 2.
Alignment of pVir-KP13F2 and plasmids encoding hypervirulence. pLVPK is used as a reference. The alignment is a pairwise BLASTn alignment performed using BLAST Ring Image Generator (BRIG). Accession numbers for the plasmids are AP006726 (pK2044), AY378100 (pLVPK), CP016815 (unnamed plasmid of ED23), CP014011 (pRJF999), CP014009 (pRJF293), CP018338 (pKp_Goe_414-2), CP019049 (pRJA166b), CP002911 (pKCTC2242), and MF943217 (pVir-KP13F2). The locations of virulence genes rmpA, rmpA2, iucABCD, iutA, and iroBCDN are indicated.
Wax moth (Galleria mellonella) larvae weighing ∼250 to 350 mg (purchased from Tianjin Huiyude Biotech Company, Tianjin, China) were used to assess the virulence of the strain. Overnight culture of strain WCHKP13F2 was washed with phosphate-buffered saline (PBS) and then adjusted with PBS to concentrations of 1 × 104 CFU/ml, 1 × 105 CFU/ml, 1 × 106 CFU/ml, and 1 × 107 CFU/ml. All experiments were performed in triplicate. An aliquot of 10 μl of each inoculum was injected into the hemocoel of 16 larvae via the last left proleg using 25-μl Hamilton syringes (19). The larvae were then incubated at 37°C in plastic containers. We counted the number of live larvae every 12 h for 3 days. Two carbapenem-resistant K. pneumoniae clinical isolates, WCHKP10 and WCHKP13F4, both of which carried blaKPC-2 and belonged to ST11, were used as the controls of low virulence. Carbapenem-susceptible hypermucous ST23:K1 K. pneumoniae strain WCHKP030925 was used as the control of hypervirulence. At an inoculum of 1 × 106 CFU/ml at 72 h after infection, survival of G. mellonella was 0% with strains WCHKP13F2 and WCHKP030925 and 62.5% and 93.8% with WCHKP10 and WCHKP13F4, respectively (Fig. 3; see Table S1 in the supplemental material). This suggests that strain WCHKP13F2 was truly hypervirulent.
FIG 3.
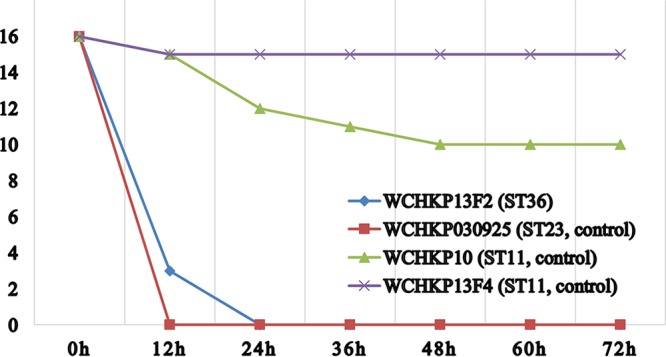
Survival of G. mellonella after infection by carbapenem-resistant strains. The effect of 1 × 106 CFU/ml of each hvKP isolate on survival of G. mellonella is shown, whereas those of other inoculums are shown in Table S1 in the supplemental material. WCHKP10 and WCHKP13F4 were two blaKPC-2-carrying carbapenem-resistant K. pneumoniae clinical isolates of ST11 and were used as controls of low virulence. WCHKP030925 was a carbapenem-susceptible clinical isolate of ST23:K1 and was used as the control of hypervirulence.
Antimicrobial resistance genes were predicted using ResFinder (Center for Genomic Epidemiology, http://genomicepidemiology.org/). Strain WCHKP13F2 had the carbapenemase gene blaKPC-2, which was carried by a 166,034-bp IncFII plasmid, designated pKPC-KP12F2. Conjugation was performed in broth using the azide-resistant Escherichia coli strain J53 as the recipient. Transconjugants were selected on agar plates containing 2 μg/ml meropenem and 150 μg/ml azide, and the presence of blaKPC-2 in transconjugants was examined by PCR. This suggests that pKPC-KP12F2 was a self-transmissible plasmid. Similar to its multidrug resistance phenotype, strain WCHKP13F2 had multiple genes mediating resistance to aminoglycosides [aac(6′)-Ib-cr, aac(3)-IId, aadA16, aph(3′)-Ia, strA, strB], β-lactams (blaTEM-1, blaSHV-11), fosfomycin (fosA), macrolides [mph(A)], quinolones [aac(6′)-Ib-cr, oqxA, oqxB, qnrS1, qnrB49], rifampin (arr3), sulfonamides (sul1, sul2), tetracycline [tet(A)], and trimethoprim (dfrA27). blaSHV-11, fosA, oqxA, and oqxB were located in the chromosome, whereas all other resistance genes were carried by pKPC-KP12F2.
The findings in the present study together with those from several recent reports (3, 5, 6) suggest that carbapenem-resistant hvKP isolates are diverse in clonal background and have emerged independently. As both carbapenem resistance and hypervirulence are encoded by plasmids, it is possible that carbapenem-susceptible strains may acquire plasmids carrying virulence genes or nonhypervirulent strains may acquire plasmids encoding carbapenem resistance to form carbapenem-resistant hvKP. Surveillance of carbapenem-resistant hvKP is urgently required to generate essential information for preventing their spread.
Accession number(s).
Complete sequences of the chromosome of strain WCHKP13F2, plasmid pVir-KP13F2, and pKPC-KP12F2 have been deposited in the DDBJ/EMBL/GenBank databases under accession numbers CP028389 to CP028391.
Supplementary Material
ACKNOWLEDGMENTS
The work was supported by a grant from the National Natural Science Foundation of China (project nos. 81572030 and 81772233).
The funders had no role in study design, data collection, or interpretation or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02644-17.
REFERENCES
- 1.Shon AS, Russo TA. 2012. Hypervirulent Klebsiella pneumoniae: the next superbug? Future Microbiol 7:669–671. doi: 10.2217/fmb.12.43. [DOI] [PubMed] [Google Scholar]
- 2.Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, Jenney A, Connor TR, Hsu LY, Severin J, Brisse S, Cao H, Wilksch J, Gorrie C, Schultz MB, Edwards DJ, Nguyen KV, Nguyen TV, Dao TT, Mensink M, Minh VL, Nhu NT, Schultsz C, Kuntaman K, Newton PN, Moore CE, Strugnell RA, Thomson NR. 2015. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci U S A 112:E3574–E3581. doi: 10.1073/pnas.1501049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gu D, Dong N, Zheng Z, Lin D, Huang M, Wang L, Chan EW-C, Shu L, Yu J, Zhang R, Chen S. 2018. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis 18:37–46. doi: 10.1016/S1473-3099(17)30489-9. [DOI] [PubMed] [Google Scholar]
- 4.Chen L, Kreiswirth BN. 2018. Convergence of carbapenem-resistance and hypervirulence in Klebsiella pneumoniae. Lancet Infect Dis 18:2–3. doi: 10.1016/S1473-3099(17)30517-0. [DOI] [PubMed] [Google Scholar]
- 5.Yao B, Xiao X, Wang F, Zhou L, Zhang X, Zhang J. 2015. Clinical and molecular characteristics of multi-clone carbapenem-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in a tertiary hospital in Beijing, China. Int J Infect Dis 37:107–112. doi: 10.1016/j.ijid.2015.06.023. [DOI] [PubMed] [Google Scholar]
- 6.Zhang R, Lin D, Chan EW, Gu D, Chen GX, Chen S. 2015. Emergence of carbapenem-resistant serotype K1 hypervirulent Klebsiella pneumoniae strains in China. Antimicrob Agents Chemother 60:709–711. doi: 10.1128/AAC.02173-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute. 2017. Performance standards for antimicrobial susceptibility testing; 23rd informational supplement. CLSI M100-S27. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 8.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 11.Wood DE, Salzberg SL. 2014. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol 15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MT, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen YT, Lai YC, Tan MC, Hsieh LY, Wang JT, Shiau YR, Wang HY, Lin AC, Lai JF, Huang IW, Lauderdale TL. 2017. Prevalence and characteristics of pks genotoxin gene cluster-positive clinical Klebsiella pneumoniae isolates in Taiwan. Sci Rep 7:43120. doi: 10.1038/srep43120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Papagiannitsis CC, Izdebski R, Baraniak A, Fiett J, Herda M, Hrabak J, Derde LP, Bonten MJ, Carmeli Y, Goossens H, Hryniewicz W, Brun-Buisson C, Gniadkowski M, MOSAR WP2, WP3 and WP5 Study Groups . 2015. Survey of metallo-β-lactamase-producing Enterobacteriaceae colonizing patients in European ICUs and rehabilitation units, 2008-11. J Antimicrob Chemother 70:1981–1988. doi: 10.1093/jac/dkv055. [DOI] [PubMed] [Google Scholar]
- 16.Alouache S, Estepa V, Messai Y, Ruiz E, Torres C, Bakour R. 2014. Characterization of ESBLs and associated quinolone resistance in Escherichia coli and Klebsiella pneumoniae isolates from an urban wastewater treatment plant in Algeria. Microb Drug Resist 20:30–38. doi: 10.1089/mdr.2012.0264. [DOI] [PubMed] [Google Scholar]
- 17.Carniel E. 1999. The Yersinia high-pathogenicity island. Int Microbiol 2:161–167. [PubMed] [Google Scholar]
- 18.Johnson JG, Clegg S. 2010. Role of MrkJ, a phosphodiesterase, in type 3 fimbrial expression and biofilm formation in Klebsiella pneumoniae. J Bacteriol 192:3944–3950. doi: 10.1128/JB.00304-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peleg AY, Jara S, Monga D, Eliopoulos GM, Moellering RC Jr, Mylonakis E. 2009. Galleria mellonella as a model system to study Acinetobacter baumannii pathogenesis and therapeutics. Antimicrob Agents Chemother 53:2605–2609. doi: 10.1128/AAC.01533-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.