AUTHOR CORRECTION
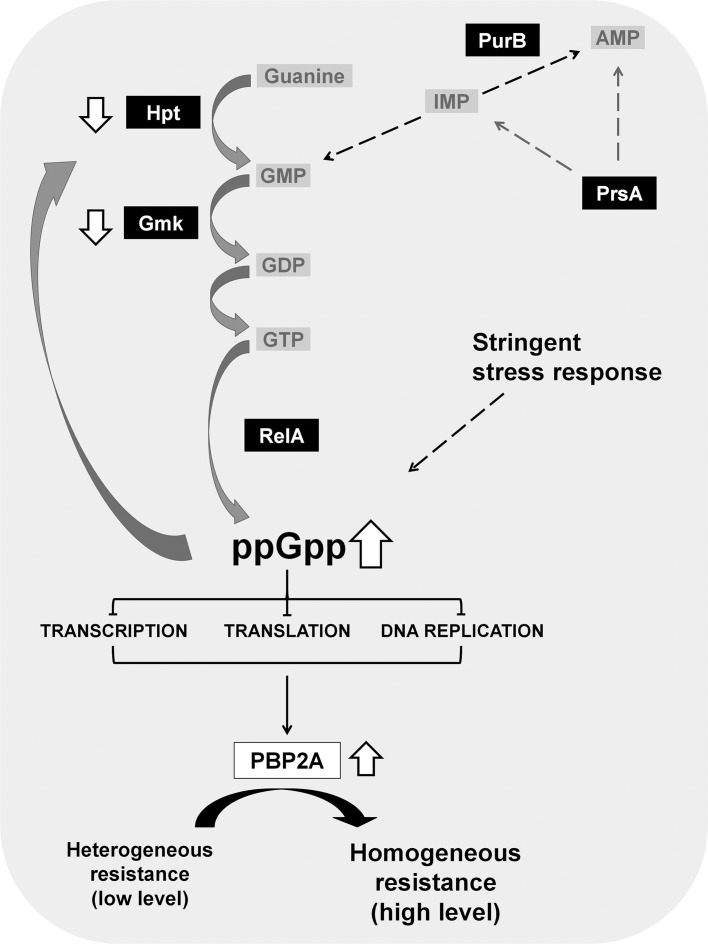
Volume 62, no. 6, e00206-18, 2018, https://doi.org/10.1128/AAC.00206-18. The authors note that (i) the gene that was affected in this study, encoding the ribose-phosphate pyrophosphokinase, is associated with different gene designations (prs or prsA) in genome annotations of different Staphylococcus aureus reference strains (as indicated in Table 1) and (ii) two genes (locus_tags SACOL0544 and SACOL1897) encoding different products are annotated as prsA in the genome of reference S. aureus COL strain (CP000046). This nomenclature issue has led to an erroneous description of the gene role in the Discussion section and in Figure 1 as linked to the foldase protein PrsA studied in reference 13 (Jousselin et al., 2015). Therefore, the reader should be aware of the following points:
1. Whenever prsA or PrsA are mentioned in the article, it may also read prs or Prs, respectively.
2. Page 6 of the PDF, 4th paragraph, the sentence “The chaperone PrsA was recently identified as a new auxiliary factor of oxacillin resistance in MRSA, affecting presumably the posttranscriptional maturation of penicillin-binding protein 2A (PBP2A), possibly at the stage of export and/or folding of newly synthesized PBP2A (13)” should be replaced with “Another target of amino acid replacements in distinct clones (RU-CAMP-28-3, RU-CAMP-28-5, and RU-CAMP-29-2) was the ribose-phosphate pyrophosphokinase (PrsA), which is involved in the direct or indirect production of purine/pyrimidine precursors.”
3. Figure 1 should appear as shown below: