ABSTRACT
Daptomycin is a last-resort membrane-targeting lipopeptide approved for the treatment of drug-resistant staphylococcal infections, such as bacteremia and implant-related infections. Although cases of resistance to this antibiotic are rare, increasing numbers of clinical, in vitro, and animal studies report treatment failure, notably against Staphylococcus aureus. The aim of this study was to identify the features of daptomycin and its target bacteria that lead to daptomycin treatment failure. We show that daptomycin bactericidal activity against S. aureus varies significantly with the growth state and strain, according to the membrane fatty acid composition. Daptomycin efficacy as an antibiotic relies on its ability to oligomerize within membranes and form pores that subsequently lead to cell death. Our findings ascertain that daptomycin interacts with tolerant bacteria and reaches its membrane target, regardless of its bactericidal activity. However, the final step of pore formation does not occur in cells that are daptomycin tolerant, strongly suggesting that it is incapable of oligomerization. Importantly, membrane fatty acid contents correlated with poor daptomycin bactericidal activity, which could be manipulated by fatty acid addition. In conclusion, daptomycin failure to treat S. aureus is not due to a lack of antibiotic-target interaction, but is driven by its capacity to form pores, which depends on membrane composition. Manipulation of membrane fluidity to restore S. aureus daptomycin bactericidal activity in vivo could open the way to novel antibiotic treatment strategies.
KEYWORDS: Staphylococcus aureus, bacterial membranes, biofilms, daptomycin, fatty acids, fluorescence, tolerance
INTRODUCTION
Staphylococcus aureus is a major opportunistic human pathogen that causes a wide range of severe clinical infections, including bacteremia and endocarditis, as well as infections related to prosthetic implants (1). These invasive diseases are often associated with biofilm formation (2, 3). The distinct properties of bacteria in biofilms, compared to their planktonic counterparts, render such infections particularly difficult to treat, resulting in severely restricted antimicrobial treatment options (4).
Daptomycin was approved by the U.S. Food and Drug Administration in 2003 for the treatment of severe Gram-positive infections, including those caused by methicillin-resistant S. aureus (MRSA) (5). This cyclic lipopeptide, which comprises a hydrophilic depsipeptide covalently linked to a decanoyl fatty acid chain (5, 6), generally displays rapid bactericidal activity by binding to the cytoplasmic membrane. Calcium ions (Ca2+) enhance daptomycin amphiphilicity by assembling the charged amino acids on one side of the molecule and exposing its lipophilic tail on the other side, leading to micelle formation as well. When in contact with bacterial membranes, daptomycin micelles undergo structural transitions, enabling the lipophilic tail to interact with the membrane, which provokes pore formation and membrane destabilization, followed by potassium ion leakage and subsequent cell death (7). Daptomycin aggregation in the membrane also interferes with membrane-associated processes that impair cell envelope synthesis (7–10).
Membrane-active antimicrobials, such as daptomycin, are believed to be bactericidal against slow-growing or dormant bacteria, as well as against biofilms (6, 11, 12). However, in clinical practice, daptomycin failure to treat S. aureus infections (5, 13–22) can reach 10% for all infection types (23). Interestingly, such daptomycin treatment failure does not seem to correlate exclusively with resistance-based genetic mutations (24, 25) that occur after prolonged treatment courses (26). Failure can also correspond to an adaptive event involving physiological changes of bacteria in the context of biofilm-associated infections, for example. Indeed, daptomycin showed decreased bactericidal activity against slow-growing and biofilm cells (termed tolerance; reference 27) without a concomitant increase in the MICs (20). In this context, we reasoned that, as the bacterial membrane is the daptomycin target, an investigation of membrane composition properties would give insight on the parameters affecting bactericidal activity. In particular, slow-growing and biofilm bacteria were recently shown to exhibit more rigid membranes than those of actively growing cells, which may be one reason for bacterial recalcitrance to daptomycin (28).
The bacterial membrane equilibrium between rigidity and fluidity is driven by multiple factors that impact membrane order (water, metal ions, proteins, carotenoids, staphyloxanthin, etc.) (29–33), but it is primarily related to lipid packing within the membrane, which is determined by the structure and composition of phospholipid fatty acids (29, 34). In the case of S. aureus, bacterial membranes mainly consist of straight-chain and branched-chain saturated fatty acids. Straight-chain fatty acids pack together to produce a bilayer with low permeability properties, while branched-chain iso or anteiso methyl species promote a more fluid membrane structure (34). Interestingly, the presence of exogenous fatty acids found in the growth environment (serum, fluids, etc.) can impact membrane composition and fluidity (35, 36).
Previous studies showed a correlation between daptomycin resistance and cell membrane alterations by comparing daptomycin-susceptible and resistant strains (obtained in vitro by submitting bacteria to serial daptomycin passages) (31, 37–40). However, none of these studies aimed to identify the factors depending on cell physiology (i.e., growth state) that are responsible for drug failure (41). Here, we investigate the impact of growth phase and membrane composition on daptomycin efficacy against S. aureus.
RESULTS
Daptomycin bactericidal activity varies as a function of S. aureus growth state and strains.
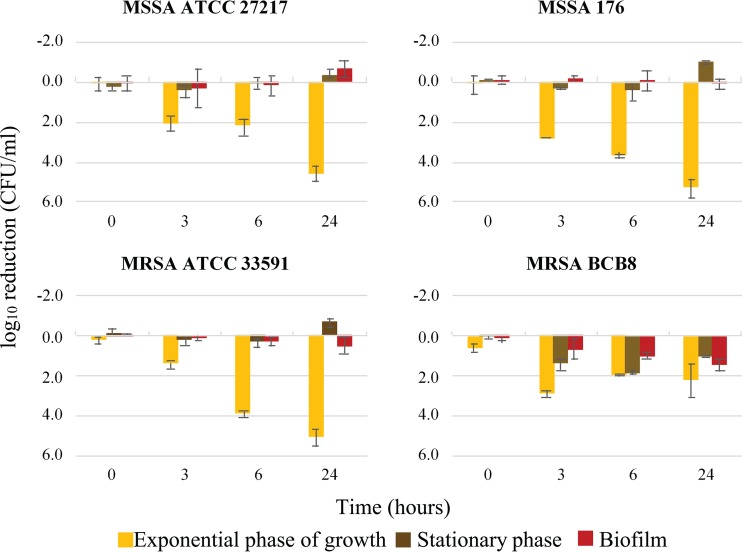
Daptomycin bactericidal activity was assessed against two methicillin-resistant strains (S. aureus strains MRSA ATCC 33591 and BCB8, with respective MIC values of 0.25 mg/liter and 0.125 mg/liter [42]), but also against two methicillin-susceptible strains (methicillin-susceptible S. aureus [MSSA] ATCC 27217 and 176, with respective MIC values of 0.25 mg/liter and 0.5 mg/liter; reference [41]). All four strains scored as daptomycin-susceptible in standard MIC tests (i.e., below 1 mg/liter, the breakpoint value according to EUCAST [61]) (42). These strains, depending on their growth phase, were then examined for daptomycin bactericidal activity using a therapeutic concentration (20 μg/ml) (Fig. 1).
FIG 1.
Daptomycin bactericidal activity on four S. aureus strains as a function of their growth state. Each graph represents daptomycin-induced reduction in bacterial counts over time, measured at 0, 3, 6, and 24 h for each strain. MSSA ATCC 27217 and MRSA ATCC 33591 are two collection strains. MSSA 176 and MRSA BCB8 are clinical strains isolated from bloodstream infections in patients. Error bars represent the analysis of variance (ANOVA) based on at least 4 independent experiments (P < 0.05).
The clinical isolate MRSA BCB8 responded differently to daptomycin than did the other tested strains, as it was more sensitive to daptomycin in stationary phase and in biofilm (Fig. 1).
The three other strains, independently of their resistance to methicillin (MSSA or MRSA), showed accrued susceptibility during exponential growth (Fig. 1). In strong contrast, daptomycin had a primarily bacteriostatic effect on cells maintained in the stationary phase or included in biofilms (Fig. 1).
These results show that daptomycin bactericidal activity against S. aureus varies with the growth state and according to the strain.
Daptomycin is available to interact with tolerant S. aureus cells.
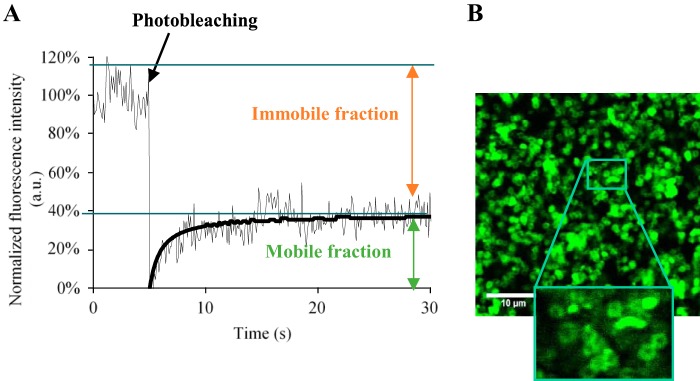
We investigated whether failure of daptomycin to eradicate S. aureus stationary-phase and biofilm cells was simply due to a loss of antibiotic binding in comparison to exponential-phase cells. We performed image-based fluorescence recovery after photobleaching (FRAP) to measure daptomycin diffusion and its availability to interact with planktonic cells, either during exponential growth or in stationary phase, and compared the results to those already obtained with biofilm cells in reference 42. The experiments were performed on two representative S. aureus strains, either tolerant (MSSA ATCC 27217 collection strain) or not tolerant to daptomycin (MRSA BCB8 clinical isolate). Fluorescence recovery curves were equivalent, regardless of the bacterial growth phase. Stationary-phase S. aureus-bound fluorescently labeled boron-dipyrromethene (BODIPY-FL)-labeled daptomycin (BODIPY-FL-daptomycin) was ∼60% ± 5% (versus 20% BODIPY-daptomycin bound to S. aureus biofilms [42]) over the 30-s observation period (Fig. 2A). The local diffusion coefficient at equilibrium (D), determined using the Braga mathematical model (41) was 11.0 ± 1.2 μm2/s, and was independent of the growth phase (compared to 7.1 ± 0.6 μm2/s in biofilms). As a control, BODIPY-FL alone did not associate with bacteria, and fluorescence was fully recovered after photobleaching (100% freely diffusing molecules); its local diffusion coefficient was 180 ± 60 μm2/s (41). Daptomycin appeared to be homogenously localized at the surface of stationary-phase S. aureus bacteria, as imaged by fluorescence confocal laser scanning microscopy (CLSM) (Fig. 2B). Together, these results demonstrate that daptomycin interacts with S. aureus independently of the cell state, suggesting that the drug is cell-associated, but that it is inactive against stationary-phase and biofilm cells.
FIG 2.
Daptomycin interacts with exponentially growing and stationary-phase cells, and 40% of free molecules are still bioavailable. (A) Typical fluorescence recovery curve obtained for BODIPY-FL-daptomycin within exponentially growing and stationary-phase MSSA ATCC 27217 or MRSA BCB8 cells. The normalized fluorescence intensity is represented as a function of the time. Experiments were performed on at least 5 independent samples under each condition. (B) Typical image acquired for stationary-phase cells deposited 5 min prior to observation on a coverslip, in the presence of BODIPY-FL-daptomycin. The inset shows that BODIPY-FL-daptomycin appears to be homogenously localized at the surface of bacterial cells.
Daptomycin reaches its cytoplasmic membrane target in tolerant S. aureus cells.
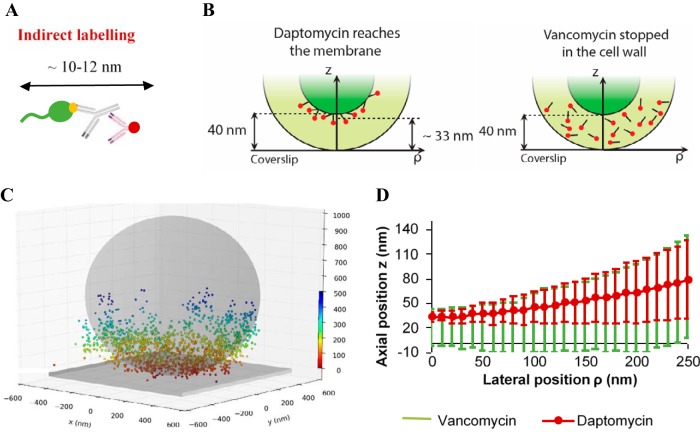
We asked whether cell-bound daptomycin actually reaches its target, the cytoplasmic membrane, in the case of poor or absent antibacterial activity. Fluorescence CLSM did not provide sufficient resolution of the thin cytoplasmic membrane to discriminate between wall or membrane location of daptomycin (200-nm resolution, versus ∼40-nm thickness for the cell wall and 10 to 15 nm for the cell membrane [43–45]). We therefore performed direct optical nanoscopy with axially localized detection (DONALD) to determine antibiotic three-dimensional (3D) localization with a nanometric precision (10 nm in the lateral and 15 to 20 nm in the axial directions [46]) in S. aureus strain ATCC 27217 in stationary phase. For this experiment, the antibiotic was fluorescently labeled by means of a 10- to 12-nm antibody-based construction (shown in Fig. 3A; see Materials and Methods). It should be noted that this antibody-based labeling construction targets the cyclic decapeptide core of daptomycin, such that the lipid tail is left free to interact with the lipid bilayer and is still bioactive. Consequently, antibiotic molecules inserted into the membrane would be expected to be localized at ∼30 nm from the coverslip surface, when considering the 10- to 12-nm fluorescent label together with the cell wall thickness (daptomycin; Fig. 3B). In contrast, antibiotic molecules targeting the cell wall would be expected to be localized randomly inside the cell wall, and hence distributed over the ∼40 nm thickness (vancomycin; Fig. 3B). Data processing of DONALD acquisitions (see Methods) allowed us to determine the 3D distribution of the fluorescently labeled daptomycin in the bacterium (Fig. 3C). More precisely, the mean height of fluorophores from the coverslip surface (axial position) is represented as a function of the bacterial fluorophore lateral position in Fig. 3D. These results showed that the labeled daptomycin was localized around 33 nm (measurements distributed between 28 and 38 nm) above the substratum, corresponding to a membrane localization in stationary-phase cells (Fig. 3B and D).
FIG 3.
Daptomycin reaches the cytoplasmic membrane of S. aureus stationary-phase cells. (A) Antibody-based construction used to label daptomycin. In green, daptomycin; in yellow, the BODIPY-FL grafted on the cyclic decapeptide core of daptomycin; in gray, the anti-BODIPY-FL rabbit antibody; and in red, the AF647-fragment of goat antibody. (B) The two cases considered here are either localization at the cell membrane (daptomycin) or within the cell wall (vancomycin). (C) A typical 3D DONALD image acquired for stationary-phase cells. The colored scale bar represents the z position (nm). The gray sphere and plane represent the expected shape of the bacterium and the coverslip surface, respectively. (D) Daptomycin (red) and vancomycin (green) localization curves represent the axial positions (z) of the fluorescently labeled antibiotics as a function of their lateral positions (ρ). Error bars represent ± twice the standard deviation of the fluorescent probe distribution. It must be noted that total internal reflection fluorescence (TIRF) depth detection (∼300 nm) limits observations to the bottom side of each bacterium deposited on the substratum.
As a control, we performed experiments under the same conditions with vancomycin, a cell-wall-targeting antibiotic that was labeled with the same antibody-based construction. In this last case, labeled vancomycin was distributed between 0 and 40 nm above the coverslip (Fig. 3D), characterizing a random localization of vancomycin within the bacterial cell wall.
Daptomycin induces low rates of membrane damage and potassium ion release in tolerant S. aureus cells.
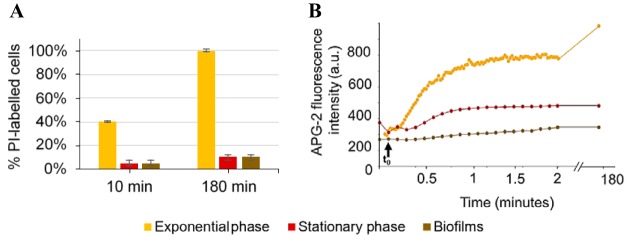
The pore-forming ability of daptomycin was evaluated using propidium iodide (PI), an indicator of membrane permeabilization. Due to the large size of this cationic fluorescent dye, PI is excluded from cells with intact cytoplasmic membranes, but it can penetrate membrane-damaged bacteria if pores of sufficient size are created (10). We observed that PI was able to penetrate ∼40% of exponentially growing cells 10 min after daptomycin treatment. In contrast, only few cells were PI labeled (∼5%) after treatment of stationary phase and biofilms, indicative of less membrane damage (Fig. 4A; see also Fig. S1 in the supplemental material). At 3 h post-daptomycin treatment, essentially all exponentially growing cells were PI labeled (100%), whereas stationary-phase and biofilm cells were predominantly unlabeled (10%) and thus viable.
FIG 4.
PI uptake and potassium ion release in response to daptomycin treatment. Significant membrane damage only occurs within exponentially growing S. aureus cells. (A) Percentage of PI-labeled bacterial cells 10 min and 180 min after daptomycin addition. Error bars represent the standard deviation, based on at least 5 independent acquired images. (B) APG-2 fluorescence intensity evolution over time after daptomycin addition. Bacteria were incubated for 10 min with APG-2 (a membrane-impermeant K+ ion marker) to check the fluorescence intensity signal. t0 corresponds to daptomycin addition (indicated by black arrow).
Daptomycin-induced release of potassium ions was evaluated using the membrane-impermeant K+ fluorescent probe Asante potassium green 2 (APG-2) (Fig. 4B). Within 1 min after daptomycin addition to exponentially growing bacteria the fluorescence signal increased from a baseline of 200 arbitrary units (AU) to 700 AU, revealing a rapid release of K+ ions into the surrounding medium. In contrast, fluorescence intensity measured within stationary-phase cells was comparable before and after antibiotic treatment. For biofilms, a slight increase in fluorescence signal was observed after daptomycin addition, before reaching a plateau within 1 min (400 AU). Three hours after daptomycin incubation with bacterial cells in the presence of APG-2, extracellular fluorescence intensity continued to increase for exponentially growing cells, whereas it remained stable for stationary-phase and biofilm bacteria.
Membrane fatty acid composition is a determinant of daptomycin bactericidal activity.
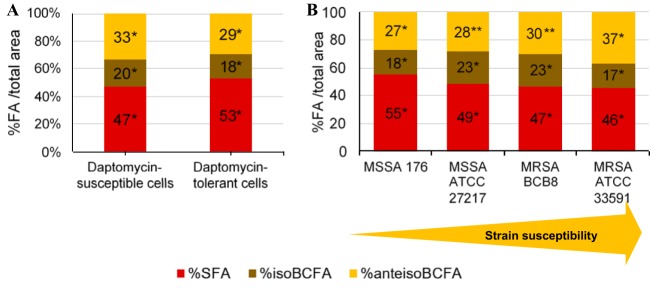
We considered that membrane modifications, in particular those affecting membrane order, could impact daptomycin bactericidal activity. We thus investigated S. aureus fatty acid composition as a possible determinant of membrane fluidity on all strains (MSSA 176 and 27217 and MRSA 33591 and BCB8; Fig. 1) and during different growth phases (see Fig. S5 in the supplemental material). The data were compiled with daptomycin bactericidal activity results (Fig. 5A) using variance analyses, to determine whether daptomycin bactericidal activity correlated with the nature of fatty acids constituting bacterial membranes. Significantly, poor daptomycin bactericidal activity correlated with higher percentages of saturated fatty acids (SFA) and a concomitant decrease iso branched-chain fatty acids (isoBCFA) and anteiso branched-chain fatty acids (anteisoBCFA) (P < 0.05) (Fig. 5A). We further correlated the susceptibility of S. aureus strains to daptomycin (regardless of growth phase) to their membrane fatty acid content (Fig. 5B). Variance analysis showed that strains against which daptomycin displayed lower bactericidal activity (i.e., the tested MSSA isolates) had more SFA-rich membranes than did the susceptible strains (MRSA isolates), which had higher anteiso BCFA contents (P < 0.05). These results suggest that high proportions of SFA may be a determining factor in poor efficiency of daptomycin against S. aureus.
FIG 5.
Fatty acid composition of S. aureus membranes is a determinant of daptomycin bactericidal activity. (A) Percentages in total area of each class of fatty acids present in the membrane whether or not bacteria were tolerant. An asterisk (*) represents a statistically significant difference between fatty acid percentages, regardless of whether bacterial cells are susceptible or tolerant to daptomycin (P < 0.05). (B) Correlation between the fatty acid percentages and the susceptibility of the strains (defined below). The results shown here consider the susceptibilities of the four strains independently of the phase of growth. Each bar represents the percentage values for each type of fatty acid. *, SFA and iso BCFA percentages were statistically different between the four strains (P < 0.05); **, anteiso BCFA percentages were statistically not different between the two strains (P < 0.05). For both (panels A and B), the results shown consider all the data obtained, independently of the strain and the growth phase. In order to classify the different experiments, we considered that bacteria were susceptible to daptomycin when the log reduction in bacterial counts was positive (a decrease in bacterial populations), and, a contrario, that bacteria were tolerant when the log reduction in bacterial counts was negative (an increase in bacterial counts) or zero.
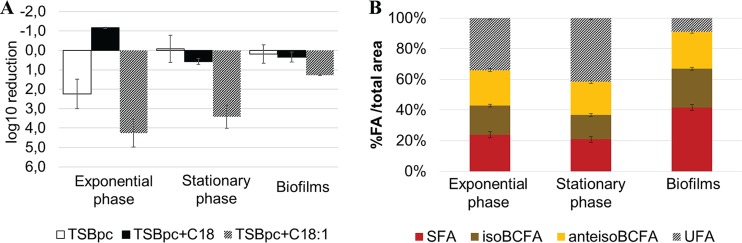
To further confirm the impact of bacterial membrane order on daptomycin inefficacy, we manipulated the membrane fatty acid composition by adding different exogenous fatty acids to culture medium, namely, either stearic acid (C18), a long-chain SFA that rigidifies membranes, or oleic acid (C18:1), a long-chain unsaturated fatty acid that fluidifies membranes (28, 34). Bacterial growth was simultaneously determined in the presence of either C18 or C18:1 without daptomycin, showing that no fatty acid-induced toxicity occurred in the tested conditions (see Fig. S3 and S4 in the supplemental material). While C18 incorporation led to a decreased or equal daptomycin efficacy in all growth phases, C18:1 incorporation led to a significant increase in daptomycin bactericidal activity (Fig. 6A). Indeed, a 4.1-log10 reduction in bacterial cell counts was observed at 6 h in exponentially growing cells, compared to a 2-log10 reduction in the nonsupplemented culture, (Fig. 6A). Interestingly, while daptomycin was inefficient in killing stationary-phase cultures in tryptic soy broth (TSB) enriched with proteins and calcium (TSBpc) (Fig. 6A), C18:1 addition led to a 3.3-log10 reduction in bacterial counts after 6 h of treatment. Biofilm bacteria were also sensitized to daptomycin by C18:1 addition, but to a lesser extent (1.1-log10 reduction in bacterial counts; Fig. 6A). Fatty acid profiles determined for C18:1-supplemented bacterial cultures (Fig. 6B) demonstrated that stationary-phase cells incorporated oleic acid, which is partly elongated to C20:1, to reach 34% (C18:1) and 42% (C20:1) unsaturated fatty acids (UFA) of bacterial fatty acids, while biofilm cells only incorporated 9%. This result is in agreement with the lower impact of oleic acid addition on daptomycin bactericidal activity in biofilms compared to that in exponential and stationary-phase cells (Fig. 6A).
FIG 6.
Incorporation of unsaturated fatty acids into S. aureus ATCC 27217 bacterial membranes increases daptomycin bactericidal activity, whereas incorporation of stearic acid (C18) decreases the antibiotic activity. (A) Log reduction in bacterial counts after 6 h of daptomycin treatment compared with the initial bacterial concentration. TSBpc+C18 corresponds to stearic acid-supplemented medium, and TSBpc+C18:1 corresponds to oleic acid-supplemented medium. (B) Proportions of S. aureus ATCC 27217 fatty acids with added oleic acid in medium. Error bars represent the standard deviation based on six experiments.
DISCUSSION
Daptomycin is a last-resort antibiotic used to treat infections due to multiantibiotic-resistant bacteria (5). However, daptomycin efficacy in S. aureus is diminished in stationary-phase and biofilm cells (12, 47). The physical basis for conditional S. aureus daptomycin tolerance has remained an unsolved question. Our main findings identify the step at which daptomycin bactericidal activity fails and provide a means of restoring its efficacy.
Previous studies suggested that daptomycin failure against stationary-phase and biofilm cells was due to poor access to its membrane target and consequent lack of availability (6, 48). Our results disprove this hypothesis. As reported previously, daptomycin diffuses throughout the biofilm structure and interacts with bacteria (42). In addition, DONALD experiments revealed here that daptomycin intercalates into the cell membrane of tolerant cells. Thus, limited antibiotic access to its biological target does not explain daptomycin failure to kill bacteria. To our knowledge, this is the first report in which an antibiotic is localized with such precision in live S. aureus cells. Previous studies of antibiotic localization were performed on larger bacteria, e.g., Bacillus subtilis (2 to 4 μm long and 0.5 to 2 μm wide) or giant unilamellar vesicles (49, 50).
Our results also suggest that pore formation at the membrane site, required for daptomycin activity, fails to occur in stationary-phase and biofilm cells; no membrane damage or potassium ion leakage was observed in these growth states. Taken together, these results demonstrate that daptomycin reaches its membrane target in tolerant bacteria, and that oligomerization for pore formation seems to be the crucial missing step.
Stationary and biofilm S. aureus cell membranes were previously shown to produce less-mobile lipids (increased saturated fatty acid content) than did exponentially growing cells (28), leading us to hypothesize that membrane order could be one determining factor for daptomycin capacity to form pores. Furthermore, adding free exogenous fatty acids is an effective means of altering membrane composition, and thus manipulating membrane lipid packing (28, 35, 51, 52). As reported in the literature, incorporation of unsaturated fatty acids generates kinks that confer greater mobility to membrane phospholipids (6). Consistently, we found that oleic acid supplementation increased daptomycin bactericidal activity against tolerant S. aureus cells, whereas stearic acid addition had the inverse effect and decreased daptomycin efficacy. Restoration of daptomycin efficacy thus correlated with cell capacity to incorporate oleic acid and was thereby impacted by membrane order.
Additional effects of oleic acid might also be considered in view of a recent study showing that daptomycin efficacy is attenuated in S. aureus agr mutants (53). Authors showed that an S. aureus agr mutant forms vesicles that are shed from membranes, which sequester daptomycin to lower its effective concentration (53). Interestingly, oleic acid reportedly stimulates agr gene transcription (54). It is therefore tempting to speculate that blebbing would not occur when agr expression is high, thus leaving cells daptomycin sensitive.
Interestingly, fatty acid supplementation led to an opposite effect in a recent study on Enterococcus faecalis, i.e., daptomycin failed to kill bacteria in the presence of unsaturated fatty acids or lipid-containing fluids, including bile. Those effects did not correlate with membrane fluidity (52). These results might suggest that species-specific differences would account for different responses. Alternatively, cotreatment with fatty acids and daptomycin could lead to titration of the antibiotic into micelles. In our study, daptomycin was added to fatty-acid-loaded but washed cells, which might account for the observed differences.
In summary, results of this study show that daptomycin reaches its membrane target in S. aureus regardless of growth state, and that the key parameter for daptomycin bactericidal activity is its capacity to form pores, which varies with membrane fatty acid composition. Daptomycin (in)efficacy against S. aureus can be manipulated by fatty acid addition, which could open the way to novel strategies of antibiotic treatment, such as encapsulation for drug deliveries (55) or the use of bitherapies, including daptomycin and membrane-fluidizing agents.
MATERIALS AND METHODS
Bacterial strains.
Four S. aureus strains were tested in the present study; two were collection strains (methicillin-susceptible S. aureus [MSSA] ATCC 27217 and methicillin-resistant S. aureus [MRSA] ATCC 33591) and two others were isolated from patients with S. aureus bloodstream infections (MSSA 176 and MRSA BCB8). All strains were kept at −80°C in tryptic soy broth (TSB) (bioMérieux, France) containing 20% (vol/vol) glycerol. The frozen cells were subcultured once in TSB (overnight culture) to constitute the stock cultures, from which aliquots were kept at −20°C. Bacterial growth and experiments were both conducted at 37°C.
Antibiotic and culture medium.
Daptomycin was purchased from Sigma (France), and the fluorescently labeled antibiotic BODIPY-FL-labeled daptomycin (BODIPY-FL-daptomycin) (BODIPY-FL is fluorescently labeled boron-dipyrromethene) was kindly provided by Cubist Pharmaceuticals (Massachusetts, USA). According to the manufacturer's instructions, the stock solutions were prepared by diluting daptomycin and BODIPY-FL-daptomycin in dimethyl sulfoxide (1 mg/ml), which was then kept at −20°C. Before the solutions were used, they were diluted in TSB enriched with proteins (bovine serum albumin [BSA], 36 g/liter; Sigma, France) and calcium (CaCl2 · 2H2O, 50 mg/liter; Sigma, France) to mimic in vivo physiological levels. It has been determined that, under these conditions, the final concentration of dimethyl sulfoxide was noncytotoxic for the bacteria. A clinically meaningful concentration of 20 μg/ml for daptomycin was used in this study.
When indicated, an exogenous fatty acid was added to the culture medium, namely, either stearic acid (C18, +99% puriss [the products have a certificate of purity; Larodan], a saturated fatty acid) or oleic acid (C18:1cis9, +99% puriss [Larodan], a monounsaturated fatty acid). Stock solutions (100 mM) of exogenous fatty acids were prepared in dimethyl sulfoxide (DMSO) (final concentrations of DMSO [0.1%] did not induce bacterial toxicity). Final concentrations of fatty acids were 0.25 mM in the growth medium.
Determination of daptomycin bactericidal activity against S. aureus.
Daptomycin bactericidal activity was assessed against the four S. aureus strains in the exponential phase of growth, stationary phase, and 24-h biofilms.
(i) Planktonic cells.
After two subcultures in TSB (one 8-h culture followed by an overnight culture), stationary-phase cells were harvested directly using the overnight culture, whereas exponentially growing cells were obtained with a third subculture in TSB enriched with proteins and calcium (TSBpc) with shaking at 100 rpm for 3 h. Bacterial suspensions were washed twice in sterile NaCl (150 mM) and resuspended in TSBpc. Optical density was measured at 600 nm to control the initial bacterial concentration. The bacterial solution was then divided equally into cell culture flasks where daptomycin solution was added. Bacterial growth was also controlled with a sample without daptomycin, corresponding to the control experiment. Viable culturable bacteria were then counted at regular interval times of 0 (when antibiotics were added), 3, 6, and 24 h after injection of antibiotics. For each time, 1.5 ml of bacterial culture was collected and centrifuged for 10 min at 7,000 × g in order to eliminate the excess of antibiotic. The bacterial pellet was dispersed in 1.5 ml of sterile NaCl (150 mM), centrifuged again, and dispersed under the same conditions. Successive decimal dilutions were then realized. For each dilution, six drops (10 μl) were deposited on tryptic soy agar (TSA) plates (bioMérieux, France) and incubated at 37°C for 24 h. CFU were counted and averaged for each dilution at each time. The detection limit of viable culturable cells here was 100 CFU/ml.
(ii) Biofilms.
For the preparation of S. aureus biofilms, 250 μl of an overnight subculture was added to 96-well microplates (μClear; Greiner Bio-One, France). After a 1.5-h adhesion period at 37°C, the wells were rinsed with a NaCl solution (150 mM) to eliminate nonadherent cells, refilled with sterile TSBpc, and then incubated for 24 h at 37°C to allow biofilm growth. To assess daptomycin bactericidal activity, the 24-h biofilms were rinsed with a sterile NaCl solution (150 mM) before adding daptomycin solution diluted in TSBpc, as described previously (42). Viable culturable bacteria were then enumerated at regular interval times of 0 (when daptomycin was added), 3, 6, and 24 h after the antibiotic injection. For each time, biofilms were scraped off, homogenized in a sterile NaCl solution, and centrifuged twice. Serial decimal dilutions were then realized and plated on TSA plates, and CFU were counted as for planktonic bacteria.
Antibiotic diffusion and availability inside biofilms using FRAP experiments.
Image-based fluorescence recovery after photobleaching (FRAP) measurements were performed on MSSA ATCC 27217 planktonic cells. Bacterial cultures were either grown to the exponential phase of growth or the stationary phase, as described previously. Then, 250 μl of these cultures was deposited on 96-well polystyrene microtiter plates 5 min prior to observations. All time-resolved measurements were obtained using a Leica TCS SP5 confocal laser scanning microscope (Leica Microsystems, France), implemented at the Centre de Photonique Biomédicale (CPBM; Orsay, France). The microscope is equipped with a 63× high numerical aperture (1.4) oil immersion objective and coupled with continuous lasers. BODIPY-FL-daptomycin was excited at 488 nm, and fluorescence emission was recorded between 500 and 600 nm. As described previously in references 41, 42, 56, FRAP experiments were conducted as follows. BODIPY-FL-daptomycin molecules were excited with a very intense light source in a user-defined region (in our case, a bacterium) to irreversibly photobleach their fluorescence. Fluorescence recovery was then probed over time at a low light power in the same photobleached region. The time course of fluorescence intensity recovery was analyzed with mathematical models, giving us the quantitative mobility of the fluorescent molecules and allowing us to determine the diffusion coefficients. For all FRAP experiments, the fluorescence intensity image size was fixed to 512 by 128 pixels with an 80-nm pixel size and recorded using a 16-bit resolution. The line scan rate was fixed at 1,400 Hz, corresponding to a total time between frames of 265 ms. The full widths at half maximum in xy and z (along the optical axis) of the bleached profile were 0.8 μm and 14 μm, respectively, allowing us to neglect diffusion along the axial/vertical axis and thus to consider only two-dimensional diffusion. Each FRAP experiment started with the acquisition of 50 images at 7% of laser maximum intensity (7 μW), followed by a 200-ms single bleached spot at 100% laser intensity. A series of 450 single-section images was then collected, with the laser power attenuated to its initial value (7% of the bleach intensity). The first image was recorded 365 ms after the beginning of bleaching.
DONALD.
The DONALD technique, described in detail in reference 46, relies on the localization of individual fluorescent molecules and requires them to be fluorescent for only a fraction of the time, so that they can be localized separately. To achieve this temporal separation, a reversible blinking (i.e., oscillation between bright and dark states) of the fluorescent molecules is induced by the addition of chemicals in the imaging buffer to influence their photophysical properties. One limitation, however, is that the mechanisms responsible for this chemically induced blinking have only been identified for a few dyes (57). Among them, Alexa Fluor (AF) 647, together with its imaging buffer (an enzymatic oxygen scavenging system to prevent from photobleaching), have widely been used to perform direct stochastic optical reconstruction microscopy (dSTORM) imaging (58, 59). Thus, to allow the antibiotic localization in our experiments, we designed a specific fluorescent labeling construction in order to detect AF647 fluorescence.
(i) Daptomycin labeling and buffer.
The labeling consisted of grafting a primary rabbit antibody (8 nm) that specifically targets BODIPY-FL and quenches its fluorescence. Then, a secondary goat anti-rabbit antibody fragment (5 nm) labeled with AF647 was used to target the rabbit antibody. The advantage of using such antibody-based labeling is that their rigid structure allows ascertainment of a precise size and orientation of the whole structure (60). The fluorophore detected and imaged here was Alexa Fluor 647, which was excited at 637 nm. To select only the fluorescence emission of this dye, a quadruple-band 446-523-600-677 (Semrock) filter was used. Acquisitions were performed using a thiol-based buffer (dSTORM super resolution buffer smart kit; Abbelight). We ensured that bacteria stayed alive during the time of acquisition by using PI to label damaged-membrane cells.
(ii) Localization, data processing, and statistical analysis.
Using total internal reflection fluorescence (TIRF) excitation to reduce background noise, most of the fluorophores were induced in a dark state until a sufficient density was obtained (typically 1 molecule per bacterium per frame). Images were acquired with 50-ms exposure time and 150 electron-multiplying charged coupled device (EMCCD) gain. The frames were then processed using a home-written Python code to detect the lateral position and the absolute depth of each spot. This detection relies on the direct optical nanoscopy with axially localized detection (DONALD) technique, which couples both single-molecule localization microscopy (SMLM) and supercritical angle fluorescence (SAF), using a Nikon Eclipse Ti-E microscope (46). SAF detection is based on the comparison of the far-field emission and the part of the near-field emission that is coupled into propagative waves at the sample/glass coverslip interface due to the refractive index mismatch. Since the near-field amplitude decays exponentially before it reaches the interface, an intensity measurement yields the absolute axial positions (z) of the fluorophores.
In order to determine the position of the antibiotic in bacteria, the data were processed as follows. First, the center (x0, y0) of each bacterium was measured. Then the lateral position (ρ) of each fluorophore (x, y, z) was calculated by . The localization results were pooled over approximately 80 bacteria for a total of 60,000 localizations. The results were then integrated radially and divided into ρ slices of 10-nm width, and the mean depth (<z>) of each ρ slice was displayed, along with its uncertainty with its distribution with (±2 standard deviations) which was obtained by subtracting the known localization precision variance from the measured variance. The value ρ gives the position of the fluorophore from the coverslip (ρ = 0 corresponds to the contact point between the bacterium and the coverslip). The advantage of measuring fluorophore heights at the contact point with the coverslip is that no assumptions of the size (i.e., the radius) or the shape (whether they flatten out near the contact point or not) of bacteria are required. It is worth noting that this does not immediately yield the position of the antibiotic, since the labeling process creates an antibody structure in which the fluorophore is 10 to 12 nm away from the antibiotic, which was taken into account in the analysis of our results.
Pore formation and potassium ion release assays.
Exponentially growing and stationary-phase MSSA ATCC 27217 bacteria (250-μl portions of each), cultured as described previously, were deposited in 96-well microtiter plates 5 min before observation to allow sedimentation and further microscopy observations. The 24-h biofilms, grown as described previously, were rinsed with sterile NaCl (150 mM) and refilled with TSBpc, Fluorescence imaging was then performed using the same confocal microscope.
(i) Pore formation.
After daptomycin was added in each well to a final concentration of 20 μg/ml, 30 μM propidium iodide (PI; Sigma) was added as a membrane damage dye, indicative of pore formation (10). The fluorescent dye was excited at 543 nm, and fluorescence emission was collected between 640 and 750 nm. Transmission and fluorescence images were acquired 10 min and 3 h after daptomycin injection in the same wells.
(ii) Potassium ions release by bacterial cells.
To measure daptomycin-induced potassium ions release by bacterial cells, Asante potassium green (APG-2 tetramethylammonium [TMA+] salt; Abcam), a membrane-impermeant dye that fluoresces upon extracellular K+ ions chelation was used. The dye was excited at 488 nm, and its fluorescence emission was recorded between 500 and 600 nm. After addition of APG-2 to a final concentration of 2 μM in each bacterial sample, the fluorescence intensity was first controlled over 5 min without antibiotic. Fluorescence intensity images were then recorded each second over 100 s to observe the fluorescence intensity evolution over time. Daptomycin was injected ∼5 s after the beginning of the time-lapse acquisition.
Membrane fatty acid analysis.
Bacteria were grown to exponential or stationary growth phases and biofilms as described previously. Planktonic suspensions were washed twice in sterile NaCl (150 mM) or, when exogenous fatty acids were added in the medium, in 0.1% Triton X-100. Entire biofilms were rinsed twice in situ with 150 mM NaCl or Triton before analysis. Extraction and methylation of fatty acids were carried out directly on bacterial pellets or entire biofilms, as described in reference 28. Briefly, fatty acids of bacterial pellets or scratched biofilm cells were directly saponified and esterified by methanolic NaOH and methanolic HCl (1 ml NaOH 3.75 mol/l in 50% vol/vol methanol solution for 30 min at 100°C; addition of 2 ml HCl 3.25 mol/l in 45% vol/vol methanol solution for 10 min at 80°C). Fatty acid methyl esters were extracted with a diethyl ether/cyclohexane solution (1:1 vol/vol), and the organic phase was washed with a dilute base (NaOH 0.3 mol/l). Analytical gas chromatography of fatty acid methyl esters was carried out on a 6890 HP system (Agilent Technologies, Santa Clara, CA) equipped with a 0.25-μm BPX70 capillary column (25 m, 0.22-mm inside diameter [i.d.]) (SGE, Ring Wood, Victoria, Australia) and a flame-ionization detector. Column temperature was set at 100°C for 1 min and then increased to 170°C at the rate of 2°C/min. Data were acquired using a HPCORE ChemStation system (Agilent Technologies, Santa Clara, CA) and expressed as a percentage of the total area. Fatty acids were identified using fatty acid methyl ester standards and grouped into three classes, as follows: saturated fatty acids (SFA), iso branched-chain fatty acids (BCFA), and anteiso BCFA. Results are the average of at least six profiles (two injections of three extractions from independent cultures) for each condition.
Statistics.
For daptomycin bactericidal activity results and fatty acid compositions of bacterial membranes, analyses of variance (ANOVA) were performed using Statgraphics software (Manugistic, Rockville, MD). Evaluated factors were considered statistically significant when P values associated with the Fischer test were below 0.05.
Supplementary Material
ACKNOWLEDGMENTS
We thank Cynthia Arnassalom Apalama for performing membrane fatty acid analysis and the Centre de Photonique Biomédicale (CPBM) of the Centre Laser de l'Université Paris-Sud (CLUPS/LUMAT FR2764, Orsay, France) for the confocal microscope and L2 microbiology facilities.
Alexandra Gruss acknowledges funding from the Fondation pour la Recherche Médicale (DBF20161136769) for her involvement in this work.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.00023-18.
REFERENCES
- 1.Lowy FD. 1998. Staphylococcus aureus infections. N Engl J Med 339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 2.Davies D. 2003. Understanding biofilm resistance to antibacterial agents. Nat Rev Drug Discov 2:114–122. doi: 10.1038/nrd1008. [DOI] [PubMed] [Google Scholar]
- 3.Lebeaux D, Ghigo J-M, Beloin C. 2014. Biofilm-related infections: bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol Mol Biol Rev 78:510–543. doi: 10.1128/MMBR.00013-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shopsin B, Kaveri SV, Bayry J. 2016. Tackling difficult Staphylococcus aureus infections: antibodies show the way. Cell Host Microbe 20:555–557. doi: 10.1016/j.chom.2016.10.018. [DOI] [PubMed] [Google Scholar]
- 5.Humphries RM, Pollett S, Sakoulas G. 2013. a current perspective on daptomycin for the clinical microbiologist. Clin Microbiol Rev 26:759–780. doi: 10.1128/CMR.00030-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mingeot-Leclercq M-P, Décout J-L. 2016. Bacterial lipid membranes as promising targets to fight antimicrobial resistance, molecular foundations and illustration through the renewal of aminoglycoside antibiotics and emergence of amphiphilic aminoglycosides. Med Chem Comm 7:586–611. doi: 10.1039/C5MD00503E. [DOI] [Google Scholar]
- 7.Straus SK, Hancock REW. 2006. Mode of action of the new antibiotic for Gram-positive pathogens daptomycin: comparison with cationic antimicrobial peptides and lipopeptides. Biochim Biophys Acta 1758:1215–1223. doi: 10.1016/j.bbamem.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 8.Muthaiyan A, Silverman JA, Jayaswal RK, Wilkinson BJ. 2008. Transcriptional profiling reveals that daptomycin induces the Staphylococcus aureus cell wall stress stimulon and genes responsive to membrane depolarization. Antimicrob Agents Chemother 52:980–990. doi: 10.1128/AAC.01121-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mengin-Lecreulx D, Allen NE, Hobbs JN, Heijenoort J van. 1990. Inhibition of peptidoglycan biosynthesis in Bacillus megaterium by daptomycin. FEMS Microbiol Lett 69:245–248. doi: 10.1111/j.1574-6968.1990.tb04238.x. [DOI] [PubMed] [Google Scholar]
- 10.Müller A, Wenzel M, Strahl H, Grein F, Saaki TNV, Kohl B, Siersma T, Bandow JE, Sahl H-G, Schneider T, Hamoen LW. 2016. Daptomycin inhibits cell envelope synthesis by interfering with fluid membrane microdomains. Proc Natl Acad Sci U S A 113:E7077–E7086. doi: 10.1073/pnas.1611173113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mascio CTM, Alder JD, Silverman JA. 2007. Bactericidal action of daptomycin against stationary-phase and nondividing Staphylococcus aureus cells. Antimicrob Agents Chemother 51:4255–4260. doi: 10.1128/AAC.00824-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lamp KC, Rybak MJ, Bailey EM, Kaatz GW. 1992. In vitro pharmacodynamic effects of concentration, pH, and growth phase on serum bactericidal activities of daptomycin and vancomycin. Antimicrob Agents Chemother 36:2709–2714. doi: 10.1128/AAC.36.12.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fowler VG, Boucher HW, Corey GR, Abrutyn E, Karchmer AW, Rupp ME, Levine DP, Chambers HF, Tally FP, Vigliani GA, Cabell CH, Link AS, DeMeyer I, Filler SG, Zervos M, Cook P, Parsonnet J, Bernstein JM, Price CS, Forrest GN, Fätkenheuer G, Gareca M, Rehm SJ, Brodt HR, Tice A, Cosgrove SE. 2006. Daptomycin versus standard therapy for bacteremia and endocarditis caused by Staphylococcus aureus. N Engl J Med 355:653–665. doi: 10.1056/NEJMoa053783. [DOI] [PubMed] [Google Scholar]
- 14.Mariani PG, Sader HS, Jones RN. 2006. Development of decreased susceptibility to daptomycin and vancomycin in a Staphylococcus aureus strain during prolonged therapy. J Antimicrob Chemother 58:481–483. doi: 10.1093/jac/dkl256. [DOI] [PubMed] [Google Scholar]
- 15.Julian K, Kosowska-Shick K, Whitener C, Roos M, Labischinski H, Rubio A, Parent L, Ednie L, Koeth L, Bogdanovich T, Appelbaum PC. 2007. Characterization of a daptomycin-nonsusceptible vancomycin-intermediate Staphylococcus aureus strain in a patient with endocarditis. Antimicrob Agents Chemother 51:3445–3448. doi: 10.1128/AAC.00559-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamp KC, Friedrich LV, Mendez-Vigo L, Russo R. 2007. Clinical experience with daptomycin for the treatment of patients with osteomyelitis. Am J Med 120(10 Suppl 1):S13–S20. doi: 10.1016/j.amjmed.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 17.Sharma M, Riederer K, Chase P, Khatib R. 2008. High rate of decreasing daptomycin susceptibility during the treatment of persistent Staphylococcus aureus bacteremia. Eur J Clin Microbiol Infect Dis 27:433–437. doi: 10.1007/s10096-007-0455-5. [DOI] [PubMed] [Google Scholar]
- 18.Levy DT, Steed ME, Rybak MJ, Guo Y, Gialanella P, Hanau L, Muggia V, Ostrowsky B. 2012. Successful treatment of a left ventricular assist device infection with daptomycin non-susceptible methicillin-resistant Staphylococcus aureus: case report and review of the literature. Transpl Infect Dis Off J Transplant Soc 14:E89–E96. doi: 10.1111/j.1399-3062.2012.00775.x. [DOI] [PubMed] [Google Scholar]
- 19.Moise PA, Amodio-Groton M, Rashid M, Lamp KC, Hoffman-Roberts HL, Sakoulas G, Yoon MJ, Schweitzer S, Rastogi A. 2013. Multicenter evaluation of the clinical outcomes of daptomycin with and without concomitant β-lactams in patients with Staphylococcus aureus bacteremia and mild to moderate renal impairment. Antimicrob Agents Chemother 57:1192–1200. doi: 10.1128/AAC.02192-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anthony SJ. 2008. Daptomycin failures in prosthetic joint infections and bone. Infect Dis Clin Prac 16:113. doi: 10.1097/IPC.0b013e3181660bf4. [DOI] [Google Scholar]
- 21.Stefani S, Campanile F, Santagati M, Mezzatesta ML, Cafiso V, Pacini G. 2015. Insights and clinical perspectives of daptomycin resistance in Staphylococcus aureus: a review of the available evidence. Int J Antimicrob Agents 46:278–289. doi: 10.1016/j.ijantimicag.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 22.Seaton RA, Menichetti F, Dalekos G, Beiras-Fernandez A, Nacinovich F, Pathan R, Hamed K. 2015. evaluation of effectiveness and safety of high-dose daptomycin: results from patients included in the European cubicin® outcomes registry and experience. Adv Ther 32:1192–1205. doi: 10.1007/s12325-015-0267-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gonzalez-Ruiz A, Beiras-Fernandez A, Lehmkuhl H, Seaton RA, Loeffler J, Chaves RL. 2011. Clinical experience with daptomycin in Europe: the first 2.5 years. J Antimicrob Chemother 66:912–919. doi: 10.1093/jac/dkq528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones T, Yeaman MR, Sakoulas G, Yang S-J, Proctor RA, Sahl H-G, Schrenzel J, Xiong YQ, Bayer AS. 2008. Failures in clinical treatment of Staphylococcus aureus infection with daptomycin are associated with alterations in surface charge, membrane phospholipid asymmetry, and drug binding. Antimicrob Agents Chemother 52:269–278. doi: 10.1128/AAC.00719-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skiest DJ. 2006. Treatment failure resulting from resistance of Staphylococcus aureus to daptomycin. J Clin Microbiol 44:655–656. doi: 10.1128/JCM.44.2.655-656.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Müller A, Grein F, Otto A, Gries K, Orlov D, Zarubaev V, Girard M, Sher X, Shamova O, Roemer T, François P, Becher D, Schneider T, Sahl HG. 2017. Differential daptomycin resistance development in Staphylococcus aureus strains with active and mutated gra regulatory systems. Int J Med Microbiol 308:335–348. doi: 10.1016/j.ijmm.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 27.Stewart PS. 2015. Antimicrobial tolerance in biofilms. Microbiol Spectr 3. doi: 10.1128/microbiolspec.MB-0010-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dubois-Brissonnet F, Trotier E, Briandet R. 2016. The biofilm lifestyle involves an increase in bacterial membrane saturated fatty acids. Front Microbiol 7:1673. doi: 10.3389/fmicb.2016.01673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Denich TJ, Beaudette LA, Lee H, Trevors JT. 2003. Effect of selected environmental and physico-chemical factors on bacterial cytoplasmic membranes. J Microbiol Methods 52:149–182. doi: 10.1016/S0167-7012(02)00155-0. [DOI] [PubMed] [Google Scholar]
- 30.Mishra NN, Yang S-J, Sawa A, Rubio A, Nast CC, Yeaman MR, Bayer AS. 2009. Analysis of cell membrane characteristics of in vitro-selected daptomycin-resistant strains of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 53:2312–2318. doi: 10.1128/AAC.01682-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mishra NN, Rubio A, Nast CC, Bayer AS. 2012. Differential adaptations of methicillin-resistant Staphylococcus aureus to serial in vitro passage in daptomycin: evolution of daptomycin resistance and role of membrane carotenoid content and fluidity. Int J Microbiol 2012:683450. doi: 10.1155/2012/683450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mishra NN, Liu GY, Yeaman MR, Nast CC, Proctor RA, McKinnell J, Bayer AS. 2011. Carotenoid-related alteration of cell membrane fluidity impacts Staphylococcus aureus susceptibility to host defense peptides. Antimicrob Agents Chemother 55:526–531. doi: 10.1128/AAC.00680-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mishra NN, Bayer AS. 2013. Correlation of cell membrane lipid profiles with daptomycin resistance in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 57:1082–1085. doi: 10.1128/AAC.02182-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y-M, Rock CO. 2008. Membrane lipid homeostasis in bacteria. Nat Rev Microbiol 6:222–233. doi: 10.1038/nrmicro1839. [DOI] [PubMed] [Google Scholar]
- 35.Morvan C, Halpern D, Kénanian G, Hays C, Anba-Mondoloni J, Brinster S, Kennedy S, Trieu-Cuot P, Poyart C, Lamberet G, Gloux K, Gruss A. 2016. Environmental fatty acids enable emergence of infectious Staphylococcus aureus resistant to FASII-targeted antimicrobials. Nat Commun 7:12944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lopez MS, Tan IS, Yan D, Kang J, McCreary M, Modrusan Z, Austin CD, Xu M, Brown EJ. 2017. Host-derived fatty acids activate type VII secretion in Staphylococcus aureus. Proc Natl Acad Sci U S A 114:11223–11228. doi: 10.1073/pnas.1700627114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bayer AS, Schneider T, Sahl H-G. 2013. Mechanisms of daptomycin resistance in Staphylococcus aureus: role of the cell membrane and cell wall. Ann N Y Acad Sci 1277:139–158. doi: 10.1111/j.1749-6632.2012.06819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li S, Yin Y, Chen H, Wang Q, Wang X, Wang H. 2017. Fitness cost of daptomycin-resistant Staphylococcus aureus obtained from in vitro daptomycin selection pressure. Front Microbiol 8:2199. doi: 10.3389/fmicb.2017.02199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Camargo ILBDC, Neoh H-M, Cui L, Hiramatsu K. 2008. Serial daptomycin selection generates daptomycin-nonsusceptible Staphylococcus aureus strains with a heterogeneous vancomycin-intermediate phenotype. Antimicrob Agents Chemother 52:4289–4299. doi: 10.1128/AAC.00417-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roch M, Gagetti P, Davis J, Ceriana P, Errecalde L, Corso A, Rosato AE. 2017. Daptomycin resistance in clinical MRSA strains is associated with a high biological fitness cost. Front Microbiol 8:2303. doi: 10.3389/fmicb.2017.02303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Daddi Oubekka S, Briandet R, Fontaine-Aupart M-P, Steenkeste K. 2012. Correlative time-resolved fluorescence microscopy to assess antibiotic diffusion-reaction in biofilms. Antimicrob Agents Chemother 56:3349–3358. doi: 10.1128/AAC.00216-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boudjemaa R, Briandet R, Revest M, Jacqueline C, Caillon J, Fontaine-Aupart M-P, Steenkeste K. 2016. New insight into daptomycin bioavailability and localization in Staphylococcus aureus biofilms by dynamic fluorescence imaging. Antimicrob Agents Chemother 60:4983–4990. doi: 10.1128/AAC.00735-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cui L, Ma X, Sato K, Okuma K, Tenover FC, Mamizuka EM, Gemmell CG, Kim M-N, Ploy M-C, Solh NE, Ferraz V, Hiramatsu K. 2003. Cell wall thickening is a common feature of vancomycin resistance in Staphylococcus aureus. J Clin Microbiol 41:5–14. doi: 10.1128/JCM.41.1.5-14.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cui L, Tominaga E, Neoh H, Hiramatsu K. 2006. Correlation between reduced daptomycin susceptibility and vancomycin resistance in vancomycin-intermediate Staphylococcus aureus. Antimicrob Agents Chemother 50:1079–1082. doi: 10.1128/AAC.50.3.1079-1082.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matias VRF, Beveridge TJ. 2006. Native cell wall organization shown by cryo-electron microscopy confirms the existence of a periplasmic space in Staphylococcus aureus. J Bacteriol 188:1011–1021. doi: 10.1128/JB.188.3.1011-1021.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bourg N, Mayet C, Dupuis G, Barroca T, Bon P, Lécart S, Fort E, Lévêque-Fort S. 2015. Direct optical nanoscopy with axially localized detection. Nat Photonics 9:587–593. doi: 10.1038/nphoton.2015.132. [DOI] [Google Scholar]
- 47.Sauermann R, Rothenburger M, Graninger W, Joukhadar C. 2008. Daptomycin: a review 4 years after first approval. Pharmacology 81:79–91. doi: 10.1159/000109868. [DOI] [PubMed] [Google Scholar]
- 48.Oldfield E, Feng X. 2014. Resistance-resistant antibiotics. Trends Pharmacol Sci 35:664–674. doi: 10.1016/j.tips.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pogliano J, Pogliano N, Silverman JA. 2012. Daptomycin-mediated reorganization of membrane architecture causes mislocalization of essential cell division proteins. J Bacteriol 194:4494–4504. doi: 10.1128/JB.00011-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen Y-F, Sun T-L, Sun Y, Huang HW. 2014. Interaction of daptomycin with lipid bilayers: a lipid extracting effect. Biochemistry (Mosc) 53:5384–5392. doi: 10.1021/bi500779g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brinster S, Lamberet G, Staels B, Trieu-Cuot P, Gruss A, Poyart C. 2009. Type II fatty acid synthesis is not a suitable antibiotic target for Gram-positive pathogens. Nature 458:83–86. doi: 10.1038/nature07772. [DOI] [PubMed] [Google Scholar]
- 52.Saito HE, Harp JR, Fozo EM. 2014. Incorporation of exogenous fatty acids protects Enterococcus faecalis from membrane-damaging agents. Appl Environ Microbiol 80:6527–6538. doi: 10.1128/AEM.02044-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pader V, Hakim S, Painter KL, Wigneshweraraj S, Clarke TB, Edwards AM. 2016. Staphylococcus aureus inactivates daptomycin by releasing membrane phospholipids. Nat Microbiol 2:16194. doi: 10.1038/nmicrobiol.2016.194. [DOI] [PubMed] [Google Scholar]
- 54.Kenny JG, Ward D, Josefsson E, Jonsson I-M, Hinds J, Rees HH, Lindsay JA, Tarkowski A, Horsburgh MJ. 2009. The Staphylococcus aureus response to unsaturated long chain free fatty acids: survival mechanisms and virulence implications. PLoS One 4:e4344. doi: 10.1371/journal.pone.0004344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reghal A, Potel G, Caillon J, Jacqueline C, Asehoune K, Gonnet M, Perrier T. August 2015. Nanocapsules lipidiques comprenant un antibiotique et leurs utilisations en therapie. Patent WO2015118496A4.
- 56.Daddi Oubekka S, Briandet R, Waharte F, Fontaine-Aupart M-P, Steenkeste K. 2011. Image-based fluorescence recovery after photobleaching (FRAP) to dissect vancomycin diffusion-reaction processes in Staphylococcus aureus, paper 80871I In Ramanujam N, Popp J (ed), Proc SPIE 8087, Clinical and Biomedical Spectroscopy and Imaging II, vol. 8087 of Proceedings of SPIE-OSA Biomedical Optics (Optical Society of America, 2011). [Google Scholar]
- 57.Olivier N, Keller D, Rajan VS, Gönczy P, Manley S. 2013. Simple buffers for 3D STORM microscopy. Biomed Opt Express 4:885–899. doi: 10.1364/BOE.4.000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dempsey GT, Vaughan JC, Chen KH, Bates M, Zhuang X. 2011. Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nat Methods 8:1027–1036. doi: 10.1038/nmeth.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin Y, Long JJ, Huang F, Duim WC, Kirschbaum S, Zhang Y, Schroeder LK, Rebane AA, Velasco MGM, Virrueta A, Moonan DW, Jiao J, Hernandez SY, Zhang Y, Bewersdorf J. 2015. Quantifying and optimizing single-molecule switching nanoscopy at high speeds. PLoS One 10(5):e0128135. doi: 10.1371/journal.pone.0128135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van de Linde S, Löschberger A, Klein T, Heidbreder M, Wolter S, Heilemann M, Sauer M. 2011. Direct stochastic optical reconstruction microscopy with standard fluorescent probes. Nat Protoc 6:991–1009. doi: 10.1038/nprot.2011.336. [DOI] [PubMed] [Google Scholar]
- 61.EUCAST 2016. Breakpoint tables for interpretation of MICs and zone diameters, version 6.0. The European Committee on Antimicrobial Susceptibility Testing. http://www.eucast.org/clinical_breakpoints/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.