ABSTRACT
A collection of 126 pigs was screened for carriage of colistin-resistant Enterobacteriaceae in a farm in Minas Gerais, Brazil. Out of this collection, eight colistin-resistant Escherichia coli isolates were recovered, including one from Minas Gerais State producing a new MCR-3 variant (MCR-3.12). Analysis of the lipopolysaccharide revealed that MCR-3.12 had a function similar to that of MCR-1 and MCR-2 as a result of the addition of a phosphoethanolamine group to the lipid A moiety. Genetic analysis showed that the mcr-3.12 gene was carried by an IncA/C2 plasmid and was embedded in an original genetic environment. This study reports the occurrence of the MCR-3-like determinant in South America and is the first to demonstrate the functionality of this group of enzymes as a phosphoethanolamine transferase.
KEYWORDS: MCR-3, polymyxins, plasmid, swine, mcr
INTRODUCTION
The increasing occurrence of colistin-resistant Enterobacteriaceae is of great concern since colistin represents one of the last-resort treatments for infections caused by carbapenem-resistant Enterobacteriaceae (CRE). In addition to chromosomally encoded resistance mechanisms corresponding to mutations or deletions in genes involved in the biosynthesis of the lipopolysaccharide (LPS), the acquisition of resistance through horizontal gene transfer has recently been described (1). Five different plasmid-mediated colistin resistance genes have been identified so far in Enterobacteriaceae, including mcr-1, mcr-2, mcr-3, mcr-4, and mcr-5 (2–6). They code for enzymes that modify the lipid A moiety of the LPS of Gram-negative bacteria and consequently confer resistance to polymyxin B and colistin (1). To date, only MCR-1 and MCR-2 have been shown to function as phosphoethanolamine transferases (7). The mcr-1 and mcr-2 genes likely originate from Moraxella species (8), with Moraxella pluranimalium being the progenitor of mcr-2 (9), Aeromonas spp. being the progenitor of mcr-3-like genes (4), and Shewanella spp. being the progenitor of mcr-4-like genes (5). The origin of the newly discovered mcr-5 gene remains unknown (6). The high prevalence of MCR-1-producing Escherichia coli isolates in food-producing animals and, therefore, the high rate of occurrence of colistin-resistant isolates may be explained by the constant use of colistin in veterinary medicine, in particular, for the treatment of livestock (poultry, swine, and cattle) (1). To date, six mcr-3 variants have been reported since the discovery of mcr-3.1 in June 2017, identified from an Escherichia coli isolate from a healthy pig in China (4) and in a Salmonella isolate from human infections in Denmark (10). The mcr-3.2 variant was identified in E. coli isolates from cattle in Spain (11). The mcr-3.3 to -3.9 variants were identified in Aeromonas spp. (12–15), and the mcr-3.10 variant was identified in E. coli isolates from ducks in China (15). Finally, the mcr-3.11 gene was from an E. coli isolate recovered from a chicken in China (unpublished; GenBank accession number MG489958.1). Even if Aeromonas spp. were described to be the progenitor of the mcr-3 gene, this gene might also be found as an acquired determinant in that species (13).
Here we report on a novel mcr-3 variant detected in an E. coli isolate recovered from a pig with postweaning diarrhea that had previously been treated with colistin in Brazil.
RESULTS
Characterization of a new mcr-3 variant and susceptibility testing.
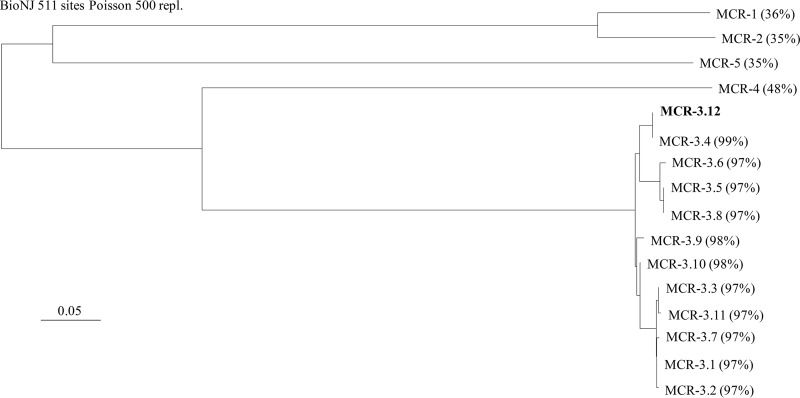
Out of the 126 pig samples, 8 samples were found to contain colistin-resistant E. coli isolates. All the animals had received a treatment including colistin for 15 days after the weaning period. Out of the 8 colistin-resistant E. coli isolates, only a single isolate (I112) was positive by PCR for the mcr-3 gene. The other colistin-resistant E. coli isolates remained negative for other mcr-like genes. Sequencing of the PCR products revealed that the mcr-3-like gene corresponded to a new variant, named mcr-3.12 (GenBank accession number MG564491), encoding the MCR-3.12 enzyme, which shared 97% amino acid sequence identity with the original MCR-3 variant and between 97% and 99% amino acid sequence identity with the other MCR-3-like variants (Fig. 1). Isolate I112 showed resistance to broad-spectrum cephalosporins, tetracycline, chloramphenicol, florfenicol, nalidixic acid, sulfonamides, sulfamethoxazole-trimethoprim, and kanamycin. It was found to be positive by the Rapid Polymyxin NP test and showed a colistin MIC of 4 μg/ml, determined using the broth microdilution method. Multilocus sequence typing (MLST) analysis showed that isolate I112 belonged to sequence type 64 (ST641) and to phylogroup A. Analysis with the SerotypeFinder program (version 1.1) indicated that it belonged to the O160:H25 serotype. Phylogenetic analysis of the known mcr-3 gene showed a significant diversity among the variants. Three major subgroups could be identified, including (i) MCR-3.5, MCR-3.6, and MCR-3.8, (ii) MCR-3.4 and MCR-3.11, and (iii) MCR-3.1, MCR-3.2, MCR-3.3, MCR-3.7, and MCR-3.11, respectively. Both the MCR-3.9 and MCR-3.10 enzymes were found to be close to the MCR-3.12 and MCR-3.1 variants (Fig. 1).
FIG 1.
Phylogenetic tree obtained for all the identified MCR-like enzymes, including all MCR-3 variants, by the distance method using the neighbor-joining algorithm (SeaView, version 4, software). Branch lengths are drawn to scale and are proportional to the number of amino acid substitutions with 500 bootstrap replications. The distance along the vertical axis has no significance. The percent amino acid sequence identity shared between the MCR-3.12 variant and the other MCR-like enzymes is indicated in parentheses.
MCR-3 is a phosphoethanolamine transferase conferring resistance to colistin.
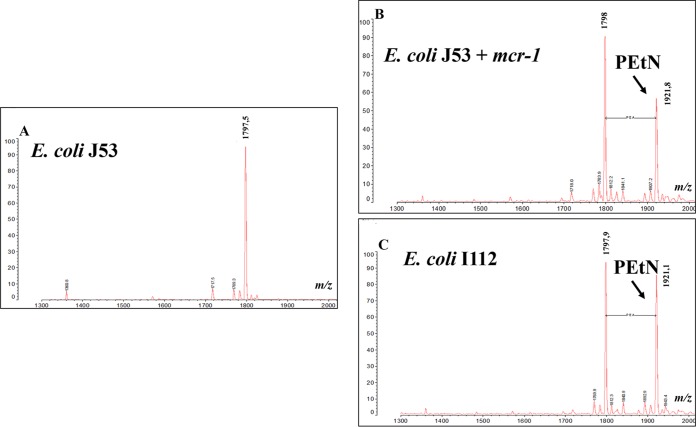
Mass spectrometry analysis of the LPS showed that unlike the negative control, strain J53, showing a single m/z 1798 peak corresponding to the bis-phosphorylated hexa-acylated lipid A, the MCR-1 and MCR-3 producers showed an identical additional peak at m/z 1921 (change in m/z, m/z 123), corresponding to addition of a phosphoethanolamine (PEtN) group to lipid A, as was previously described (7, 16) (Fig. 2). Induction of the pBADb-mcr-3-like plasmid allowed a colistin MIC of 4 μg/ml to be obtained, whereas the noninduced clone presented a colistin MIC of 0.03 μg/ml, showing that the production of MCR-3.12 conferred a 130-fold increase in the colistin MIC. Altogether, these results show the phosphoethanolamine transferase activity of the MCR-3.12 enzyme and its impact on colistin susceptibility.
FIG 2.
Mass spectrometry analysis of lipid A from strain E. coli J53 (A), its transconjugant carrying the mcr-1 gene (B), and clinical isolate I112 expressing the mcr-3.12 gene (C). The addition of a PEtN group is indicated by an arrow.
Plasmid analysis.
Mating-out assays with E. coli J53 and Klebsiella pneumoniae CIP53153 as the recipients, but also with Aeromonas punctata CIP102629 as the recipient, were successful, highlighting the broad host range of the plasmid carrying the mcr-3.12 variant. In contrast, no transconjugant was obtained using Pseudomonas aeruginosa PAO1 as the recipient. Conjugation followed by PCR showed that mcr-3.12 was located on a conjugative plasmid named p112. That latter plasmid carries genes for resistance to tetracyclines, sulfonamides, chloramphenicol, and florfenicol. PCR-based replicon typing (PBRT) analysis showed that plasmid p112 belongs to the IncA/C2 incompatibility group. Kieser extraction followed by gel electrophoresis identified its size to be ca. 140 kb. The MICs of colistin for the E. coli and K. pneumoniae transconjugants were 4 and 8 μg/ml, respectively, and therefore, the transconjugants were categorized as resistant according to the EUCAST breakpoint (the original MICs for the bacterial hosts were 0.25 and 0.12 μg/ml, respectively) (http://www.eucast.org). Interestingly, the MIC of colistin for the A. punctata transconjugant was 16 μg/ml (original MIC, 0.12 μg/ml), indicating a very significant impact of MCR-3.12 on colistin susceptibility in that species.
Bioinformatic analysis and genetic context of the mcr-3.12 gene.
Whole-genome sequencing of E. coli I112 identified a series of resistance determinants, including genes encoding resistance to β-lactams (the blaTEM-1B and blaCTX-M-8 genes), aminoglycosides (aph[3′]-Ia, strA, and strB), tetracyclines (tetA), phenicols (catA1 and floR), sulfonamides (sul2), and trimethoprim (dfr18). The mcr-3-like gene was found in association with a gene encoding a diacylglycerol kinase dgkA-like gene sharing 98% nucleotide sequence identity with the dgkA gene identified in association with the first mcr-3 gene described on plasmid pWJ1 (4).
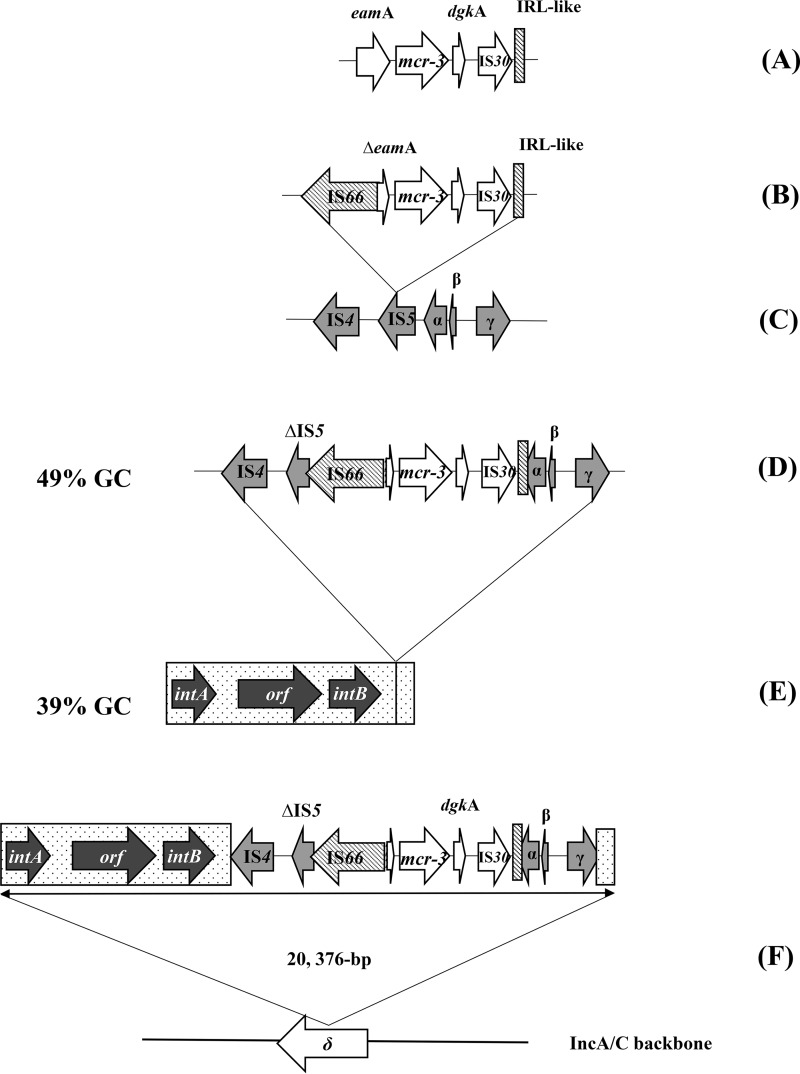
The mcr-3.12 gene was located between two insertion sequences belonging to the IS66 and IS30 families, respectively (Fig. 3). Interestingly, an inverted repeat left (IRL)-like sequence of IS66 was detected 90 bp after the end of the inverted repeat right (IRR) sequence of the IS30-like sequence and was found to share 100% nucleotide sequence identity with the first 24 nucleotides (nt) of the IRL-like sequence of IS66 (Fig. 3). This IRL-like sequence downstream of the IS30-like sequence could form a putative transposon with IS66.
FIG 3.
Proposed model of the chronology of the acquisition of the mcr-3.12 gene into the IncA/C2 plasmid. The genes eamA and dgkA encode a metabolite transporter and a diacylglycerol kinase, respectively. intA and intB represent putative integrases; α, β, and γ are ORFs encoding a reverse transcriptase, a transcriptional regulator, and a diguanylate cyclase, respectively; δ corresponds to the ORF encoding a DNA methyltransferase located on the IncA/C2 plasmid backbone.
Further analysis showed that this putative transposon was embedded in a longer structure that was inserted between nucleotides 1049 and 1050 of a DNA methyltransferase gene located on the IncA/C2 backbone. This structure was 20,376 bp long and is represented in Fig. 3F. It could be defined into three different regions: (i) a 5′ region characterized by a 7,666-bp region with a GC content of 39% containing three putative open reading frames (ORFs), including two encoding putative site-specific integrases, (ii) the putative transposon containing the mcr-3 variant and three ORFs (α, β, and γ) presenting a GC content of 49%, and (iii) a 3′ region of 526 bp with a GC content similar to that of the first 7,666-bp region (Fig. 3F). ORFs α, β, and γ encoded a reverse transcriptase, a transcriptional regulator, and a diguanylate cyclase, respectively. Their products showed strong amino acid sequence identity (98%) with the sequences of putative proteins from Aeromonas dhakensis.
DISCUSSION
We report here the identification of a novel variant of the mcr-3 gene, detected in an E. coli isolate recovered from a pig in Brazil. Interestingly, previous studies also described MCR-3 producers recovered from animal samples (11, 13), suggesting the same link between animal and colistin resistance that has been established for the mcr-1 gene. The pigs screened in this study had previously been treated with colistin for 15 days after the weaning period. This suggests the possible selection of the colistin-resistant strain during this period, as we showed in our previous study describing a high prevalence of MCR-1 producers in a pig farm in Portugal where animals had received colistin (17). There have been many reports of MCR producers in Brazil, with MCR-1 being the only variant systematically identified. These isolates were a single Salmonella enterica serotype Typhimurium isolate that was recovered from retail meat (18) and E. coli isolates recovered from chicken meat (19), from migratory penguins (20), on public beaches (21), or from patients with bloodstream infections (22, 23). Also, KPC-2-producing E. coli isolates (24) and KPC-2-producing Klebsiella pneumoniae isolates belonging to ST392 and ST437 (25, 26) were identified. A quite extensive study identified a series of 59 MCR-1-producing E. coli isolates recovered from humans, chicken, chicken meat, bovine, turkey, swine, and penguin (27). However, we might speculate that most studies have been designed to detect only the mcr-1 gene so far, and few have investigated the occurrence of the most recently identified other variants.
Isolate I112 carried a novel mcr-3 variant named mcr-3.12. It belonged to ST641, isolates of which corresponding to isolates recovered from pigs in Germany in 2016 were previously found to carry the mcr-1 gene (28). It belongs to phylogroup A of E. coli and therefore corresponds to a commensal strain. Sequence alignment analysis showed that mcr-3.12 shares 99% nucleotide sequence identity with the sequence from an Aeromonas veronii isolate. This suggests that this new variant may have originated from that particular species or may have widely disseminated as an acquired resistance trait within that species. It is noteworthy that we showed here that the IncA/C2-type plasmid bearing the mcr-3.12 gene could replicate in Aeromonas spp. We may therefore speculate that such a plasmid type might have been involved in the original spread of mcr-3-like genes from their progenitors to other bacterial species, including members of the family Enterobacteriaceae.
The results of induction experiments and analysis of the lipid A of the isolate strongly indicate that the MCR-3 enzyme confers colistin resistance in the same way that the MCR-1 and MCR-2 enzymes do by adding a phosphoethanolamine group to the lipid A moiety, although this enzyme shared only 45 and 47% amino acid sequence identity with the amino acid sequences of MCR-1 and MCR-2, respectively. The fact that MCR-1, -2, and-3 share similar functions was previously hypothesized through in silico protein structure analysis (4).
The mcr-3 gene was previously described on IncHI2 and IncX4 plasmids, which are commonly found in association with the mcr-1 and mcr-2 genes. Here, we have described the first IncA/C2 plasmid carrying a plasmid-mediated colistin resistance determinant. This plasmid backbone is commonly identified to be a support for many different antibiotic resistance genes. Here, the determinants tetA, sul2, and floR, encoding resistance to tetracycline, sulfonamides, and phenicols, respectively, were also detected on the same plasmid. The broad host range of this plasmid was demonstrated by evidencing its ability to replicate not only in E. coli and K. pneumoniae but also in A. punctata.
The mcr-3.12 gene is located in a putative transposon including the IS66 upstream sequence of the mcr-3 gene and an IS30-like downstream sequence. Interestingly, a 24-nt region found 90 bp downstream of the IS30-like sequence was found to be identical to the IRL of IS66. Further experiments will be conducted to confirm whether IS66 could have played a role in the acquisition of this phosphoethanolamine transferase gene by a mechanism similar to the one-handed transposition that has been described for ISEcp1 in the mobilization of blaCTX-M-15 (29).
The genetic context of the putative mcr-3 transposon is complex, and the chronology of acquisition of this structure by the IncA/C2 plasmid can hardly be explained. One hypothesis is summarized in Fig. 3. IS66 might have been involved in the original mobilization of the mcr-3.12 gene from Aeromonas spp. (Fig. 3A to D). Then, a second mobilization event involving an unknown mechanism between the genetic structure containing the putative integrases (Fig. 3E) and the mcr-3-carrying structure may have occurred, forming a 20,376-bp integron-like genetic complex. Finally, this whole structure may have been mobilized and inserted between nt 1049 and nt 1050 of a DNA methyltransferase gene located on an IncA/C2 plasmid backbone (Fig. 3F). The resulting resistance plasmid is, in the end, one of those responsible for the spread of mcr genes among the Enterobacteriaceae.
MATERIALS AND METHODS
Bacterial isolate and susceptibility testing.
Screening of colistin-resistant isolates from 126 different pigs in 10 swine herds from different sites in the state of Minas Gerais in Brazil was performed. All pigs presented with postweaning diarrhea. The isolates were initially tested for colistin resistance using agar dilution methods. All colonies growing on plates supplemented with >2 μg/ml of colistin were confirmed by the commercialized Rapid Polymyxin NP test (ELITech Microbiology, France) (30), and MICs were determined by the broth microdilution method using cation-adjusted Mueller-Hinton (MH) broth. Antimicrobial susceptibility testing for determination of susceptibility to other antibiotic families was performed according to the standard disk diffusion method on MH agar plates following CLSI recommendations (31).
Whole-genome sequencing and molecular analysis.
PCR screening for mcr genes was performed using primers designed to detect all known variants of MCR-3. Primers MCR-3allF (5′-GCA TTT ATG CTG AAC TGG CG-3′) and MCR-3allR (5′-AGC GGC TTT CTG CTG CAA AC-3′) were used, and the corresponding amplicons were subsequently sequenced (Microsynth, Balgach, Switzerland). Whole genomic DNA of the MCR-3-positive isolate was extracted by use of a Sigma-Aldrich GenElute bacterial genomic DNA kit. Genomic libraries were assessed using a Nextera XT library preparation kit (Illumina Inc., San Diego, CA), and sequencing was performed using an Illumina MiniSeq system with 300-bp paired-end reads and a coverage of 50 times. The generated FastQ data were compiled and analyzed using the CLC Genomic Workbench (version 7.5.1; CLC Bio, Aarhus, Denmark). Reads were de novo assembled with automatic bubble and word size, and contigs with a minimum contig length of 800 nucleotides were generated using the mapping mode map reads back to contigs.
The resulting contigs were uploaded into the Center for Genomic Epidemiology server (http://www.genomicepidemiology.org/). The plasmid replicon type, multilocus sequence type, serotype, and antimicrobial resistance determinants were determined using the PlasmidFinder (version 1.3), MLST (version 1.8), SerotypeFinder (version 1.1), and ResFinder (version 3.0) programs, respectively (32–34). Phylogroup analysis was performed by using the method described by Clermont et al. (35). Sequence alignments and construction of phylogenetic trees were performed with the SeaView alignment tool (version 4; Prabi, La Doua, Lyon, France) (36).
Plasmid analysis was performed using the Kieser extraction method (37) followed by gel electrophoresis in order to estimate the size of the plasmid containing the mcr-3 gene using E. coli strain 50192 harboring four plasmids of 154, 66, 48, and 7 kb, respectively, as the plasmid size marker. Determination of the incompatibility group was confirmed by PCR-based replicon typing (PBRT) (38).
Conjugation experiments were performed using the azide-resistant strain E. coli J53. In addition, conjugations were also performed with temocillin-resistant Pseudomonas aeruginosa strain PAO1, azide-resistant Klebsiella pneumoniae strain CIP53153, and azide-resistant Aeromonas punctata strain CIP102629 as the recipient strains to test the plasmid carrying the mcr-3.12 variant for a broad host range. Both donor and recipient strains were cultured in exponential phase and then mixed on solid LB agar using filters at a 1:10 donor/recipient ratio. After 5 h of incubation, the filters were resuspended in 0.85% NaCl and the bacterial mixture was plated onto agar plates supplemented with colistin (1 μg/ml) and sodium azide (100 μg/ml) for E. coli or with temocillin (50 μg/ml) and sodium azide (100 μg/ml) for P. aeruginosa. Since the plasmid bearing the mcr-3.12 gene conferred resistance to tetracycline, conjugations using K. pneumoniae and A. punctata as the recipients were attempted using tetracycline (100 μg/ml) and sodium azide (100 μg/ml) as selective molecules. The susceptibility of all transconjugants to antibiotics was confirmed by use of the antibiogram followed by PCR for the mcr-3-like gene.
Analysis of the LPS modification.
The LPS of E. coli J53 (unmodified lipid A), TCAf24 (J53 mcr-1 transconjugant), and I112 (MCR-3-like producer) were analyzed by mass spectrometry (MS). Lipid A was obtained by hydrolysis of 3 mg of lyophilized bacteria in 120 μl of isobutyric acid and 1 M ammonium hydroxide (5:3; vol/vol), heating for 1 h at 100°C, and cooling at 4°C before centrifugation, as previously described (39). The supernatant was then diluted with water and lyophilized before it was washed with methanol. The insoluble lipid A obtained was finally extracted in a chloroform-methanol-water (3:1:0.25, vol/vol/vol) mixture. Matrix-assisted laser desorption ionization-MS analysis was performed using a PerSeptive Voyager STR (PE Biosystems, France) time of flight mass spectrometer in linear negative-ion mode. Dihydroxybenzoic acid (DHB) at 10 mg/ml in 0.1 M citric acid in chloroform-methanol-water (3:1.5:0.25, vol/vol/vol) was used as the matrix.
Cloning and overexpression of the mcr-3.12 gene.
The new mcr-3 variant was cloned into the arabinose-inducible pBADb vector in order to determine the impact of the expression of the MCR-3.12 phosphoethanolamine transferase on colistin susceptibility. Induction of the pBADb vector was performed using MH broth supplemented with 1% l-arabinose as previously described (8).
ACKNOWLEDGMENTS
This work has been funded by the University of Fribourg; by the Swiss National Science Foundation (project FNS-407240_177381); and by grants from the ANIWHA ERA-NET project, Switzerland; OFSP, Bern, Switzerland (grant no. 16009294); and the Novartis Foundation for Medical-Biological Research.
REFERENCES
- 1.Poirel L, Jayol A, Nordmann P. 2017. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev 30:557–596. doi: 10.1128/CMR.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu JH, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 3.Xavier BB, Lammens C, Ruhal R, Kumar-Singh S, Butaye P, Goossens H, Malhotra-Kumar S. 2016. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill 21(27):pii=30280 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=30280. [DOI] [PubMed] [Google Scholar]
- 4.Yin W, Li H, Shen Y, Liu Z, Wang S, Shen Z, Zhang R, Walsh TR, Shen J, Wang Y. 2017. Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. mBio 8(3):e00543-17. doi: 10.1128/mBio.00543-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carattoli A, Villa L, Feudi C, Curcio L, Orsini S, Luppi A, Pezzotti G, Magistrali CF. 2017. Novel plasmid-mediated colistin resistance mcr-4 gene in Salmonella and Escherichia coli, Italy 2013, Spain and Belgium, 2015 to 2016. Euro Surveill 22(31):pii=30589 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=30589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borowiak M, Fischer J, Hammerl JA, Hendriksen RS, Szabo I, Malorny B. 2017. Identification of a novel transposon-associated phosphoethanolamine transferase gene, mcr-5, conferring colistin resistance in d-tartrate fermenting Salmonella enterica subsp. enterica serovar Paratyphi B. J Antimicrob Chemother 72:3317–3324. doi: 10.1093/jac/dkx327. [DOI] [PubMed] [Google Scholar]
- 7.Sun J, Xu Y, Gao R, Lin J, Wei W, Srinivas S, Li D, Yang RS, Li XP, Liao XP, Liu YH, Feng Y. 2017. Deciphering MCR-2 colistin resistance. mBio 8(3):e00625-17. doi: 10.1128/mBio.00625-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kieffer N, Nordmann P, Poirel L. 2017. Moraxella species as potential sources of MCR-like polymyxin resistance determinants. Antimicrob Agents Chemother 61:e00129-17. doi: 10.1128/AAC.00129-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poirel L, Kieffer N, Fernandez-Garayzabal JF, Vela AI, Larpin Y, Nordmann P. 2017. MCR-2-mediated plasmid-borne polymyxin resistance most likely originates from Moraxella pluranimalium. J Antimicrob Chemother 72:2947–2949. doi: 10.1093/jac/dkx225. [DOI] [PubMed] [Google Scholar]
- 10.Litrup E, Kiil K, Hammerum AM, Roer L, Nielsen EM, Torpdahl M. 2017. Plasmid-borne colistin resistance gene mcr-3 in Salmonella isolates from human infections, Denmark, 2009-17. Euro Surveill 22(31):pii=30587 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=30587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hernández M, Iglesias MR, Rodríguez-Lázaro D, Gallardo A, Quijada N, Miguela-Villoldo P, Campos MJ, Píriz S, López-Orozco G, de Frutos C, Sáez JL, Ugarte-Ruiz M, Domínguez L, Quesada A. 2017. Co-occurrence of colistin-resistance genes mcr-1 and mcr-3 among multidrug-resistant Escherichia coli isolated from cattle, Spain, September 2015. Euro Surveill 22(31):pii=30586 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=30586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu L, Feng Y, Zhang X, McNally A, Zong Z. 2017. New variant of mcr-3 in an extensively drug-resistant Escherichia coli clinical isolate carrying mcr-1 and blaNDM-5. Antimicrob Agents Chemother 61:e01757-17. doi: 10.1128/AAC.01757-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ling Z, Yin W, Li H, Zhang Q, Wang X, Wang Z, Ke Y, Wang Y, Shen J. 2017. Chromosome-mediated mcr-3 variants in Aeromonas veronii from chicken meat. Antimicrob Agents Chemother 61:e01272-17. doi: 10.1128/AAC.01272-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eichhorn I, Feudi C, Wang Y, Kaspar H, Feßler AT, Lübke-Becker A, Michael GB, Shen J, Schwarz S. 31 January 2018. Identification of novel variants of the colistin resistance gene mcr-3 in Aeromonas spp. from the national resistance monitoring programme GERM-Vet and from diagnostic submissions. J Antimicrob Chemother. doi: 10.1093/jac/dkx538. [DOI] [PubMed] [Google Scholar]
- 15.Wang X, Zhai W, Li J, Liu D, Zhang Q, Shen Z, Wang S, Wang Y. 2018. Presence of an mcr-3 variant in Aeromonas caviae, Proteus mirabilis, and Escherichia coli from one domestic duck. Antimicrob Agents Chemother 62:e02106-17. doi: 10.1128/AAC.02106-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu YY, Chandler CE, Leung LM, McElheny CL, Mettus RT, Shanks RMQ, Liu JH, Goodlett DR, Ernst RK, Doi Y. 2017. Structural modification of lipopolysaccharide conferred by mcr-1 in Gram-negative ESKAPE pathogens. Antimicrob Agents Chemother 61:e00580-17. doi: 10.1128/AAC.00580-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kieffer N, Aires-de-Sousa M, Nordmann P, Poirel L. 2017. High rate of MCR-1-producing Escherichia coli and Klebsiella pneumoniae among pigs, Portugal. Emerg Infect Dis 23:2023–2029. doi: 10.3201/eid2312.170883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rau RB, de Lima-Morales D, Wink PL, Ribeiro AR, Martins AF, Barth AL. 2018. Emergence of mcr-1 producing Salmonella enterica serovar Typhimurium from retail meat: first detection in Brazil. Foodborne Pathog Dis 15:58–59. doi: 10.1089/fpd.2017.2346. [DOI] [PubMed] [Google Scholar]
- 19.Monte DF, Mem A, Fernandes MR, Cerdeira L, Esposito F, Galvão JA, Franco BDGM, Lincopan N, Landgraf M. 2017. Chicken meat as a reservoir of colistin-resistant Escherichia coli strains carrying mcr-1 genes in South America. Antimicrob Agents Chemother 61:e02718-16. doi: 10.1128/AAC.02718-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sellera FP, Fernandes MR, Sartori L, Carvalho MP, Esposito F, Nascimento CL, Dutra GH, Mamizuka EM, Pérez-Chaparro PJ, McCulloch JA, Lincopan N. 2017. Escherichia coli carrying IncX4 plasmid-mediated mcr-1 and blaCTX-M genes in infected migratory Magellanic penguins (Spheniscus magellanicus). J Antimicrob Chemother 72:1255–1256. doi: 10.1093/jac/dkw543. [DOI] [PubMed] [Google Scholar]
- 21.Fernandes MR, Sellera FP, Esposito F, Sabino CP, Cerdeira L, Lincopan N. 2017. Colistin-resistant mcr-1-positive Escherichia coli on public beaches, an infectious threat emerging in recreational waters. Antimicrob Agents Chemother 61:e00234-17. doi: 10.1128/AAC.00234-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rossi F, Girardello R, Morais C, Cury AP, Martins LF, da Silva AM, Abdala E, Setubal JC, da Silva Duarte AJ. 2017. Plasmid-mediated mcr-1 in carbapenem-susceptible Escherichia coli ST156 causing a blood infection: an unnoticeable spread of colistin resistance in Brazil? Clinics (Sao Paulo) 72:642–644. doi: 10.6061/clinics/2017(10)09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rocha IV, Andrade CADN, Campos TL, Rezende AM, Leal NC, Vidal CFL, Xavier DE. 2017. Ciprofloxacin-resistant and extended-spectrum β-lactamase-producing Escherichia coli ST410 strain carrying the mcr-1 gene associated with bloodstream infection. Int J Antimicrob Agents 49:655–656. doi: 10.1016/j.ijantimicag.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 24.Conceição-Neto OC, Aires CAM, Pereira NF, da Silva LHJ, Picão RC, Siqueira BN, Albano RM, Asensi MD, Carvalho-Assef APD. 2017. Detection of the plasmid-mediated mcr-1 gene in clinical KPC-2-producing Escherichia coli isolates in Brazil. Int J Antimicrob Agents 50:282–284. doi: 10.1016/j.ijantimicag.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 25.Aires CAM, da Conceição-Neto OC, Tavares e Oliveira TR, Dias CF, Montezzi LF, Picão RC, Albano RM, Asensi MD, Carvalho-Assef APD. 2017. Emergence of the plasmid-mediated mcr-1 gene in clinical KPC-2-producing Klebsiella pneumoniae sequence type 392 in Brazil. Antimicrob Agents Chemother 61:e00317-17. doi: 10.1128/AAC.00317-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dalmolin TV, Martins AF, Zavascki AP, de Lima-Morales D, Barth AL. 2018. Acquisition of the mcr-1 gene by a high-risk clone of KPC-2-producing Klebsiella pneumoniae ST437/CC258, Brazil. Diagn Microbiol Infect Dis 90:132–133. doi: 10.1016/j.diagmicrobio.2017.09.016. [DOI] [PubMed] [Google Scholar]
- 27.Esposito F, Fernandes MR, Lopes R, Muñoz M, Sabino CP, Cunha MP, Silva KC, Cayô R, Martins WMBS, Moreno AM, Knöbl T, Gales AC, Lincopan N. 2017. Detection of colistin-resistant MCR-1-positive Escherichia coli by use of assays based on inhibition by EDTA and zeta potential. J Clin Microbiol 55:3454–3465. doi: 10.1128/JCM.00835-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pulss S, Semmler T, Prenger-Berninghoff E, Bauerfeind R, Ewers C. 2017. First report of an Escherichia coli strain from swine carrying an OXA-181 carbapenemase and the colistin resistance determinant MCR-1. Int J Antimicrob Agents 50:232–236. doi: 10.1016/j.ijantimicag.2017.03.014. [DOI] [PubMed] [Google Scholar]
- 29.Poirel L, Lartigue MF, Decousser JW, Nordmann P. 2005. ISEcp1B-mediated transposition of blaCTX-M in Escherichia coli. Antimicrob Agents Chemother 49:447–450. doi: 10.1128/AAC.49.1.447-450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poirel L, Larpin Y, Dobias J, Stephan R, Decousser JW, Madec JY, Nordmann P. 2018. Rapid Polymyxin NP test for the detection of polymyxin resistance mediated by the mcr-1/mcr-2 genes. Diagn Microbiol Infect Dis 90:7–10. doi: 10.1016/j.diagmicrobio.2017.09.012. [DOI] [PubMed] [Google Scholar]
- 31.Clinical and Laboratory Standards Institute. 2016. Performance standards for antimicrobial susceptibility testing; 26th informational supplement (M100–S26). Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 32.Joensen KG, Tetzschner AM, Iguchi A, Aarestrup FM, Scheutz F. 2015. Rapid and easy in silico serotyping of Escherichia coli using whole genome sequencing (WGS) data. J Clin Microbiol 53:2410–2426. doi: 10.1128/JCM.00008-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, Møller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol 66:4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 37.Kieser T. 1984. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid 12:19–36. doi: 10.1016/0147-619X(84)90063-5. [DOI] [PubMed] [Google Scholar]
- 38.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 39.Breton A, Novikov A, Martin R, Tissieres P, Caroff M. 2017. Structural and biological characteristics of different forms of V. filiformis lipid A: use of MS to highlight structural discrepancies. J Lipid Res 58:543–552. doi: 10.1194/jlr.M072900. [DOI] [PMC free article] [PubMed] [Google Scholar]