ABSTRACT
The in vitro activities of ceftazidime-avibactam and comparators against 9,149 isolates of Enterobacteriaceae and 2,038 isolates of Pseudomonas aeruginosa collected by 42 medical centers in nine countries in the Asia-Pacific region from 2012 to 2015 were determined as part of the International Network for Optimal Resistance Monitoring (INFORM) global surveillance program. Antimicrobial susceptibility testing was conducted by Clinical and Laboratory Standards Institute (CLSI) broth microdilution, and isolate subset analysis was performed on the basis of the resistant phenotypes and β-lactamase content. Ceftazidime-avibactam demonstrated potent in vitro activity (MIC, ≤8 μg/ml) against all Enterobacteriaceae tested (99.0% susceptible) and was the most active against isolates that were metallo-β-lactamase (MBL) negative (99.8% susceptible). Against P. aeruginosa, 92.6% of all isolates and 96.1% of MBL-negative isolates were susceptible to ceftazidime-avibactam (MIC, ≤8 μg/ml). The rates of susceptibility to ceftazidime-avibactam ranged from 97.0% (Philippines) to 100% (Hong Kong, South Korea) for Enterobacteriaceae and from 83.1% (Thailand) to 100% (Hong Kong) among P. aeruginosa isolates, with lower susceptibilities being observed in countries where MBLs were more frequently encountered (Philippines, Thailand). Ceftazidime-avibactam inhibited 97.2 to 100% of Enterobacteriaceae isolates, per country, that carried serine β-lactamases, including extended-spectrum β-lactamases, AmpC cephalosporinases, and carbapenemases (KPC, GES, OXA-48-like). It also inhibited 91.3% of P. aeruginosa isolates that were carbapenem nonsusceptible in which no acquired β-lactamase was detected. Among MBL-negative Enterobacteriaceae isolates that were ceftazidime nonsusceptible, meropenem nonsusceptible, colistin resistant, and multidrug resistant, ceftazidime-avibactam inhibited 96.1, 87.7, 100, and 98.8% of isolates, respectively, and among MBL-negative P. aeruginosa isolates that were ceftazidime nonsusceptible, meropenem nonsusceptible, colistin resistant, and multidrug resistant, ceftazidime-avibactam inhibited 79.6, 83.6, 83.3, and 68.2% of isolates, respectively. Overall, clinical isolates of Enterobacteriaceae and P. aeruginosa collected in nine Asia-Pacific countries from 2012 to 2015 were highly susceptible to ceftazidime-avibactam.
KEYWORDS: ceftazidime-avibactam, Asia-Pacific, surveillance, Gram negative, Enterobacteriaceae, Pseudomonas aeruginosa, INFORM
INTRODUCTION
Avibactam (AVI) is a non-β-lactam β-lactamase inhibitor that inhibits the activity of Ambler class A β-lactamases, including extended-spectrum β-lactamases (ESBLs; e.g., TEM-type, SHV-type, and CTX-M-type β-lactamases); KPC carbapenemases; AmpC cephalosporinases (Ambler class C β-lactamases); and some Ambler class D β-lactamases (e.g., OXA-48) (1–3). Avibactam in combination with ceftazidime (CAZ) has previously demonstrated potent in vitro activity against KPC-producing clinical isolates of Enterobacteriaceae, including isolates that also carry AmpC and ESBL enzymes and/or that have impaired permeability, as well as Pseudomonas aeruginosa (1–7). Metallo-β-lactamase-(MBL)-producing Enterobacteriaceae and Pseudomonas aeruginosa are not susceptible to ceftazidime-avibactam (1, 6).
To date, only a limited number of surveillance studies have been conducted in which data for isolates from the Asia-Pacific region have been specifically analyzed (8–10) and not grouped with data from countries outside the region. Previous surveillance studies have reported that the prevalence of β-lactam-resistant and multidrug-resistant (MDR) Enterobacteriaceae and nonfermentative Gram-negative bacilli (GNB) is higher in certain countries within the Asia-Pacific region than in other nations in that region and in other geographic locations (2, 4, 5, 8–12). β-Lactam resistance rates have been observed to be lower in Australia, New Zealand, and Japan and higher in China, India, Indonesia, Philippines, Taiwan, and Thailand (2, 4, 5, 8–12). To date, the majority of published studies have not included molecular characterization of β-lactamases specifically from the Asia-Pacific region or provided ceftazidime-avibactam susceptibility data for Gram-negative bacilli isolated from patients in many Asia-Pacific countries (8–10). The intent of the current study was to augment currently published phenotypic data by determining the in vitro susceptibilities to ceftazidime-avibactam and comparators of clinical isolates of Enterobacteriaceae and P. aeruginosa collected from hospitalized patients in Asia-Pacific countries over a recent 4-year time period (2012 to 2015), as well as to analyze the activity of ceftazidime-avibactam against antimicrobial-resistant and molecularly characterized β-lactamase-producing subsets at both the regional and country levels. These data were collected as part of the International Network for Optimal Resistance Monitoring (INFORM) global surveillance program. The INFORM global surveillance program was established in 2012 to benchmark and track the in vitro activity of ceftazidime-avibactam and comparative agents against clinical isolates of β-lactamase-producing Enterobacteriaceae and nonfermentative Gram-negative bacilli, including P. aeruginosa.
RESULTS
Of the 9,149 isolates of Enterobacteriaceae tested, 99.0% were susceptible to ceftazidime-avibactam (MIC90, 0.5 μg/ml); the percentages of susceptibility were lower for meropenem (98.4%), doripenem (98.3%), amikacin (97.5%), tigecycline (93.9%), piperacillin-tazobactam (86.9%), colistin (83.0%), and the other agents tested (<80%) (Table 1). Ceftazidime-avibactam MIC90 values for individual species or species groups within the Enterobacteriaceae family ranged from 0.12 μg/ml (Proteeae) to 1 μg/ml (Enterobacter spp.), with only minor variation (<2%) in the percent susceptibility to ceftazidime-avibactam, which ranged from 98.1% (Enterobacter spp.) to 99.9% (Escherichia coli), being observed (Table 1). Percent susceptibility to ceftazidime-avibactam was higher for Enterobacteriaceae isolates that did not carry MBLs (99.8% susceptible) than for MBL-positive isolates (1.4% susceptible) (Table 1). Accordingly, the percent susceptibilities to ceftazidime-avibactam for MBL-negative isolates of individual species or species groups of Enterobacteriaceae (99.5 to 100% susceptible) were marginally higher (<2%) than the percent susceptibilities for all isolates of individual species or species groups (98.1 to 99.9%) (Table 1). Percent susceptibility to ceftazidime-avibactam for Enterobacteriaceae isolates from each of the Asia-Pacific countries surveyed ranged from 99.1 to 100% (MIC90, 0.25 to 0.5 μg/ml) for all countries except China (98.8% susceptible; MIC90, 0.5 μg/ml), Thailand (98.7%; MIC90, 0.5 μg/ml), and Philippines (97.0%; MIC90, 0.5 μg/ml) (Fig. 1; see also Tables S2A to S10A in the supplemental material).
TABLE 1.
In vitro activities of ceftazidime-avibactam and comparator antimicrobial agents tested against 9,149 isolates of Enterobacteriaceae and 2,038 isolates of P. aeruginosa collected in the Asia-Pacific region as part of the INFORM global surveillance program from 2012 to 2015
| Organism, phenotype/genotype (no. of isolates)a | Antimicrobial agent | MIC (μg/ml)b |
% susceptiblec | ||
|---|---|---|---|---|---|
| 50% | 90% | Range | |||
| Enterobacteriaceae (9,149) | Ceftazidime-avibactam | 0.12 | 0.5 | ≤0.015 to >128 | 99.0 |
| Ceftazidime | 0.25 | 64 | ≤0.015 to >128 | 75.1 | |
| Cefepime | ≤0.12 | >16 | ≤0.12 to >16 | 77.8 | |
| Aztreonam | 0.12 | 64 | ≤0.015 to >128 | 73.9 | |
| Piperacillin-tazobactam | 2 | 64 | ≤0.25 to >128 | 86.9 | |
| Doripenem | 0.06 | 0.25 | ≤0.008 to >4 | 98.3 | |
| Imipenem | 0.25 | 2 | ≤0.03 to >8 | 85.1 | |
| Meropenem | 0.03 | 0.12 | ≤0.004 to >8 | 98.4 | |
| Amikacin | 2 | 8 | ≤0.25 to >32 | 97.5 | |
| Colistin (n = 4,140)d | 0.5 | >4 | ≤0.12 to >4 | 83.0 | |
| Tigecycline | 0.5 | 2 | ≤0.015 to >8 | 93.9 | |
| Levofloxacin | 0.12 | >4 | ≤0.03 to >4 | 74.6 | |
| Enterobacteriaceae, MBL negative (9,075) | Ceftazidime-avibactam | 0.12 | 0.5 | ≤0.015 to >128 | 99.8 |
| Ceftazidime | 0.25 | 64 | ≤0.015 to >128 | 75.7 | |
| Cefepime | ≤0.12 | >16 | ≤0.12 to >16 | 78.4 | |
| Aztreonam | 0.12 | 64 | ≤0.015 to >128 | 74.3 | |
| Piperacillin-tazobactam | 2 | 32 | ≤0.25 to >128 | 87.5 | |
| Doripenem | 0.06 | 0.25 | ≤0.008 to >4 | 99.1 | |
| Imipenem | 0.25 | 2 | ≤0.03 to >8 | 85.7 | |
| Meropenem | 0.03 | 0.12 | ≤0.004 to >8 | 99.1 | |
| Amikacin | 2 | 8 | ≤0.25 to >32 | 97.8 | |
| Colistin (n = 4,103)d | 0.5 | >4 | ≤0.12 to >4 | 82.9 | |
| Tigecycline | 0.5 | 2 | ≤0.015 to >8 | 94.0 | |
| Levofloxacin | 0.12 | >4 | ≤0.03 to >4 | 74.9 | |
| Escherichia coli (3,140) | Ceftazidime-avibactam | 0.12 | 0.25 | ≤0.015 to >128 | 99.9 |
| Ceftazidime | 0.25 | 32 | ≤0.015 to >128 | 72.6 | |
| Cefepime | ≤0.12 | >16 | ≤0.12 to >16 | 69.4 | |
| Aztreonam | 0.12 | 64 | ≤0.015 to >128 | 68.5 | |
| Piperacillin-tazobactam | 2 | 16 | ≤0.25 to >128 | 92.1 | |
| Doripenem | 0.03 | 0.06 | ≤0.008 to >4 | 99.6 | |
| Imipenem | 0.25 | 0.25 | ≤0.03 to >8 | 99.1 | |
| Meropenem | 0.03 | 0.06 | ≤0.004 to >8 | 99.4 | |
| Amikacin | 2 | 8 | ≤0.25 to >32 | 98.2 | |
| Colistin (n = 1,344)d | 0.5 | 1 | ≤0.12 to >4 | 99.1 | |
| Tigecycline | 0.25 | 0.5 | ≤0.015 to 8 | 99.9 | |
| Levofloxacin | 0.5 | >4 | ≤0.03 to >4 | 55.8 | |
| Escherichia coli, MBL negative (3,137) | Ceftazidime-avibactam | 0.12 | 0.25 | ≤0.015 to >128 | >99.9 |
| Ceftazidime | 0.25 | 32 | ≤0.015 to >128 | 72.6 | |
| Cefepime | ≤0.12 | >16 | ≤0.12 to >16 | 69.5 | |
| Aztreonam | 0.12 | 64 | ≤0.015 to >128 | 68.5 | |
| Piperacillin-tazobactam | 2 | 16 | ≤0.25 to >128 | 92.2 | |
| Doripenem | 0.03 | 0.06 | ≤0.008 to >4 | 99.7 | |
| Imipenem | 0.25 | 0.25 | ≤0.03 to >8 | 99.2 | |
| Meropenem | 0.03 | 0.06 | ≤0.004 to >8 | 99.5 | |
| Amikacin | 2 | 8 | ≤0.25 to >32 | 98.3 | |
| Colistin (n = 1,343)d | 0.5 | 1 | ≤0.12 to >4 | 99.1 | |
| Tigecycline | 0.25 | 0.5 | ≤0.015 to 8 | 99.9 | |
| Levofloxacin | 0.5 | >4 | ≤0.03 to >4 | 55.8 | |
| Klebsiella pneumoniae (2,538) | Ceftazidime-avibactam | 0.12 | 0.5 | ≤0.015 to >128 | 98.3 |
| Ceftazidime | 0.25 | 128 | ≤0.015 to >128 | 69.4 | |
| Cefepime | ≤0.12 | >16 | ≤0.12 to >16 | 71.7 | |
| Aztreonam | 0.12 | 128 | ≤0.015 to >128 | 69.6 | |
| Piperacillin-tazobactam | 4 | >128 | ≤0.25 to >128 | 79.4 | |
| Doripenem | 0.06 | 0.12 | 0.015 to >4 | 96.8 | |
| Imipenem | 0.25 | 1 | ≤0.03 to >8 | 94.3 | |
| Meropenem | 0.03 | 0.12 | ≤0.004 to >8 | 96.7 | |
| Amikacin | 1 | 4 | ≤0.25 to >32 | 96.3 | |
| Colistin (n = 1,288)d | 1 | 1 | 0.12 to >4 | 97.9 | |
| Tigecycline | 0.5 | 2 | ≤0.015 to >8 | 96.3 | |
| Levofloxacin | 0.12 | >4 | ≤0.03 to >4 | 78.6 | |
| Klebsiella pneumoniae, MBL negative (2,501) | Ceftazidime-avibactam | 0.12 | 0.5 | ≤0.015 to >128 | 99.7 |
| Ceftazidime | 0.25 | 128 | ≤0.015 to >128 | 70.4 | |
| Cefepime | ≤0.12 | >16 | ≤0.12 to >16 | 72.7 | |
| Aztreonam | 0.06 | 128 | ≤0.015 to >128 | 70.4 | |
| Piperacillin-tazobactam | 4 | >128 | ≤0.25 to >128 | 80.5 | |
| Doripenem | 0.06 | 0.12 | 0.015 to >4 | 98.2 | |
| Imipenem | 0.25 | 1 | ≤0.03 to >8 | 95.5 | |
| Meropenem | 0.03 | 0.06 | ≤0.004 to >8 | 98.0 | |
| Amikacin | 1 | 4 | ≤0.25 to >32 | 96.8 | |
| Colistin (n = 1,266)d | 1 | 1 | 0.12 to >4 | 98.0 | |
| Tigecycline | 0.5 | 2 | ≤0.015 to >8 | 96.3 | |
| Levofloxacin | 0.12 | >4 | ≤0.03 to >4 | 79.2 | |
| Klebsiella oxytoca (432) | Ceftazidime-avibactam | 0.12 | 0.25 | ≤0.015 to 128 | 98.8 |
| Ceftazidime | 0.12 | 8 | ≤0.015 to >128 | 89.8 | |
| Cefepime | ≤0.12 | 2 | ≤0.12 to >16 | 92.1 | |
| Aztreonam | 0.25 | 32 | ≤0.015 to >128 | 82.9 | |
| Piperacillin-tazobactam | 2 | 64 | ≤0.25 to >128 | 89.1 | |
| Doripenem | 0.06 | 0.12 | 0.015 to >4 | 98.8 | |
| Imipenem | 0.25 | 0.5 | 0.06 to >8 | 98.2 | |
| Meropenem | 0.03 | 0.06 | 0.015 to >8 | 98.8 | |
| Amikacin | 1 | 4 | 0.5 to >32 | 98.6 | |
| Colistin (n = 187)d | 0.5 | 1 | 0.25 to 4 | 98.9 | |
| Tigecycline | 0.25 | 1 | 0.06 to 4 | 99.5 | |
| Levofloxacin | 0.06 | 1 | ≤0.03 to >4 | 94.0 | |
| Klebsiella oxytoca, MBL negative (428) | Ceftazidime-avibactam | 0.12 | 0.25 | ≤0.015 to 16 | 99.8 |
| Ceftazidime | 0.12 | 4 | ≤0.015 to >128 | 90.7 | |
| Cefepime | ≤0.12 | 2 | ≤0.12 to >16 | 92.8 | |
| Aztreonam | 0.25 | 32 | ≤0.015 to >128 | 83.4 | |
| Piperacillin-tazobactam | 2 | 32 | ≤0.25 to >128 | 89.7 | |
| Doripenem | 0.06 | 0.06 | 0.015 to 2 | 99.8 | |
| Imipenem | 0.25 | 0.5 | 0.06 to 8 | 99.1 | |
| Meropenem | 0.03 | 0.06 | 0.015 to 8 | 99.8 | |
| Amikacin | 1 | 4 | 0.5 to >32 | 99.1 | |
| Colistin (n = 186)d | 0.5 | 1 | 0.25 to 4 | 98.9 | |
| Tigecycline | 0.25 | 1 | 0.06 to 4 | 99.5 | |
| Levofloxacin | 0.06 | 1 | ≤0.03 to >4 | 94.4 | |
| Enterobacter spp.e (1,088) | Ceftazidime-avibactam | 0.25 | 1 | ≤0.015 to >128 | 98.1 |
| Ceftazidime | 0.5 | 128 | ≤0.015 to >128 | 66.0 | |
| Cefepime | ≤0.12 | 8 | ≤0.12 to >16 | 84.8 | |
| Aztreonam | 0.12 | 64 | ≤0.015 to >128 | 67.6 | |
| Piperacillin-tazobactam | 4 | 128 | ≤0.25 to >128 | 76.3 | |
| Doripenem | 0.06 | 0.25 | 0.015 to >4 | 97.7 | |
| Imipenem | 1 | 2 | ≤0.03 to >8 | 80.7 | |
| Meropenem | 0.06 | 0.12 | 0.008 to >8 | 97.8 | |
| Amikacin | 1 | 4 | ≤0.25 to >32 | 98.2 | |
| Colistin (n = 481)d | 0.5 | >4 | ≤0.12 to >4 | 86.7 | |
| Tigecycline | 0.5 | 1 | 0.06 to 8 | 97.2 | |
| Levofloxacin | 0.06 | 2 | ≤0.03 to >4 | 90.7 | |
| Enterobacter spp., MBL negative (1,069) | Ceftazidime-avibactam | 0.25 | 1 | ≤0.015 to 64 | 99.7 |
| Ceftazidime | 0.5 | 128 | ≤0.015 to >128 | 67.2 | |
| Cefepime | ≤0.12 | 8 | ≤0.12 to >16 | 86.3 | |
| Aztreonam | 0.12 | 64 | ≤0.015 to >128 | 68.2 | |
| Piperacillin-tazobactam | 4 | 128 | ≤0.25 to >128 | 77.3 | |
| Doripenem | 0.06 | 0.25 | 0.015 to >4 | 99.4 | |
| Imipenem | 1 | 2 | ≤0.03 to >8 | 82.1 | |
| Meropenem | 0.06 | 0.12 | 0.008 to >8 | 99.4 | |
| Amikacin | 1 | 4 | ≤0.25 to >32 | 98.5 | |
| Colistin (n = 471)d | 0.5 | >4 | ≤0.12 to >4 | 86.8 | |
| Tigecycline | 0.5 | 1 | 0.06 to 8 | 97.8 | |
| Levofloxacin | 0.06 | 2 | ≤0.03 to >4 | 91.9 | |
| Citrobacter spp.f (558) | Ceftazidime-avibactam | 0.12 | 0.5 | ≤0.015 to >128 | 98.4 |
| Ceftazidime | 0.5 | 128 | 0.06 to >128 | 73.5 | |
| Cefepime | ≤0.12 | 8 | ≤0.12 to >16 | 88.0 | |
| Aztreonam | 0.12 | 64 | ≤0.015 to >128 | 73.8 | |
| Piperacillin-tazobactam | 4 | 64 | ≤0.25 to >128 | 81.9 | |
| Doripenem | 0.06 | 0.12 | ≤0.008 to >4 | 98.4 | |
| Imipenem | 0.25 | 1 | 0.06 to >8 | 91.6 | |
| Meropenem | 0.03 | 0.06 | 0.008 to >8 | 98.6 | |
| Amikacin | 2 | 4 | ≤0.25 to >32 | 97.3 | |
| Colistin (n = 229)d | 0.5 | 1 | ≤0.12 to >4 | 99.6 | |
| Tigecycline | 0.5 | 1 | 0.06 to 4 | 99.6 | |
| Levofloxacin | 0.06 | 4 | ≤0.03 to >4 | 88.7 | |
| Citrobacter spp., MBL negative (552) | Ceftazidime-avibactam | 0.12 | 0.5 | ≤0.015 to >128 | 99.5 |
| Ceftazidime | 0.25 | 128 | 0.06 to >128 | 74.3 | |
| Cefepime | ≤0.12 | 4 | ≤0.12 to >16 | 89.0 | |
| Aztreonam | 0.12 | 64 | ≤0.015 to >128 | 74.3 | |
| Piperacillin-tazobactam | 4 | 64 | ≤0.25 to >128 | 82.6 | |
| Doripenem | 0.06 | 0.12 | ≤0.008 to >4 | 99.3 | |
| Imipenem | 0.25 | 1 | 0.06 to >8 | 92.4 | |
| Meropenem | 0.03 | 0.06 | 0.008 to >8 | 99.3 | |
| Amikacin | 2 | 4 | ≤0.25 to >32 | 97.5 | |
| Colistin (n = 227)d | 0.5 | 1 | ≤0.06 to >4 | 99.6 | |
| Tigecycline | 0.25 | 1 | 0.06 to 4 | 99.6 | |
| Levofloxacin | 0.06 | 4 | ≤0.03 to >4 | 89.1 | |
| Proteeaeg (1,190) | Ceftazidime-avibactam | 0.06 | 0.12 | ≤0.015 to 64 | 99.5 |
| Ceftazidime | 0.06 | 1 | ≤0.015 to >128 | 94.8 | |
| Cefepime | ≤0.12 | 0.5 | ≤0.12 to >16 | 94.0 | |
| Aztreonam | ≤0.015 | 0.25 | ≤0.015 to 128 | 96.8 | |
| Piperacillin-tazobactam | 0.5 | 1 | ≤0.25 to >128 | 99.0 | |
| Doripenem | 0.25 | 0.5 | 0.03 to >4 | 98.5 | |
| Imipenem | 2 | 4 | 0.06 to >8 | 24.3 | |
| Meropenem | 0.06 | 0.12 | ≤0.004 to >8 | 99.5 | |
| Amikacin | 4 | 8 | ≤0.25 to >32 | 97.1 | |
| Colistin (n = 521)d | >4 | >4 | 0.25 to >4 | 0.4 | |
| Tigecycline | 2 | 4 | 0.03 to >8 | 65.0 | |
| Levofloxacin | 0.12 | >4 | ≤0.03 to >4 | 83.8 | |
| Proteeae, MBL negative (1,186) | Ceftazidime-avibactam | 0.06 | 0.12 | ≤0.015 to 32 | 99.8 |
| Ceftazidime | 0.06 | 0.5 | ≤0.015 to >128 | 95.1 | |
| Cefepime | ≤0.12 | 0.5 | ≤0.12 to >16 | 94.3 | |
| Aztreonam | ≤0.12 | 0.25 | ≤0.015 to 128 | 96.9 | |
| Piperacillin-tazobactam | 0.5 | 1 | ≤0.25 to >128 | 99.2 | |
| Doripenem | 0.25 | 0.5 | 0.03 to >4 | 98.8 | |
| Imipenem | 2 | 4 | 0.06 to >8 | 24.4 | |
| Meropenem | 0.06 | 0.12 | ≤0.004 to >8 | 99.7 | |
| Amikacin | 4 | 8 | ≤0.25 to >32 | 97.2 | |
| Colistin (n = 520)d | >4 | >4 | 0.25 to >4 | 0.4 | |
| Tigecycline | 2 | 4 | 0.03 to >8 | 64.9 | |
| Levofloxacin | 0.12 | >4 | ≤0.03 to >4 | 84.0 | |
| Other Enterobacteriaceaeh (203) | Ceftazidime-avibactam | 0.12 | 0.5 | ≤0.015 to >128 | 99.5 |
| Ceftazidime | 0.25 | 1 | ≤0.015 to >128 | 93.6 | |
| Cefepime | ≤0.12 | 1 | ≤0.12 to >16 | 92.6 | |
| Aztreonam | 0.12 | 2 | ≤0.015 to >128 | 92.6 | |
| Piperacillin-tazobactam | 2 | 8 | ≤0.25 to >128 | 94.1 | |
| Doripenem | 0.12 | 0.25 | 0.015 to >4 | 99.0 | |
| Imipenem | 0.5 | 2 | 0.06 to >8 | 88.7 | |
| Meropenem | 0.06 | 0.12 | 0.03 to >8 | 99.0 | |
| Amikacin | 2 | 4 | ≤0.25 to 32 | 99.5 | |
| Colistin (n = 90)d | >4 | >4 | 0.25 to >4 | 10.0 | |
| Tigecycline | 1 | 2 | 0.25 to >8 | 96.6 | |
| Levofloxacin | 0.12 | 2 | ≤0.03 to >4 | 93.1 | |
| Other Enterobacteriaceae, MBL negative (202) | Ceftazidime-avibactam | 0.12 | 0.5 | ≤0.015 to 1 | 100 |
| Ceftazidime | 0.25 | 1 | ≤0.015 to >128 | 94.1 | |
| Cefepime | ≤0.12 | 1 | ≤0.12 to >16 | 93.1 | |
| Aztreonam | 0.12 | 2 | ≤0.015 to >128 | 92.6 | |
| Piperacillin-tazobactam | 2 | 8 | ≤0.25 to >128 | 94.1 | |
| Doripenem | 0.12 | 0.25 | 0.015 to >4 | 99.5 | |
| Imipenem | 0.5 | 2 | 0.06 to >8 | 89.1 | |
| Meropenem | 0.06 | 0.12 | 0.03 to >8 | 99.5 | |
| Amikacin | 2 | 4 | ≤0.25 to 32 | 99.5 | |
| Colistin (n = 90)d | >4 | >4 | 0.25 to >4 | 10.0 | |
| Tigecycline | 1 | 2 | 0.25 to >8 | 96.5 | |
| Levofloxacin | 0.12 | 2 | ≤0.03 to 4 | 93.6 | |
| Pseudomonas aeruginosa (2,038) | Ceftazidime-avibactam | 2 | 8 | 0.03 to >128 | 92.6 |
| Ceftazidime | 2 | 64 | 0.12 to >128 | 77.9 | |
| Cefepime | 4 | >16 | ≤0.12 to >16 | 80.6 | |
| Aztreonam | 8 | 32 | 0.06 to >128 | 64.5 | |
| Piperacillin-tazobactam | 8 | >128 | ≤0.25 to >128 | 71.0 | |
| Doripenem | 0.5 | >4 | 0.03 to >4 | 78.7 | |
| Imipenem | 2 | >8 | 0.06 to >8 | 67.9 | |
| Meropenem | 0.5 | >8 | 0.015 to >8 | 77.6 | |
| Amikacin | 4 | 8 | ≤0.25 to >32 | 94.4 | |
| Colistin (n = 1,278)d | 2 | 2 | ≤0.12 to >8 | 93.5 | |
| Levofloxacin | 0.5 | >4 | ≤0.03 to >4 | 77.1 | |
| Pseudomonas aeruginosa, MBL negative (1,964) | Ceftazidime-avibactam | 2 | 8 | 0.03 to >128 | 96.1 |
| Ceftazidime | 2 | 64 | 0.12 to >128 | 80.8 | |
| Cefepime | 4 | 16 | ≤0.12 to >16 | 83.5 | |
| Aztreonam | 8 | 32 | 0.06 to >128 | 65.8 | |
| Piperacillin-tazobactam | 8 | >128 | ≤0.25 to >128 | 73.4 | |
| Doripenem | 0.5 | >4 | 0.03 to >4 | 81.7 | |
| Imipenem | 2 | >8 | 0.06 to >8 | 70.4 | |
| Meropenem | 0.5 | 8 | 0.015 to >8 | 80.5 | |
| Amikacin | 4 | 8 | ≤0.25 to >32 | 96.8 | |
| Colistin (n = 1,225)d | 2 | 2 | ≤0.12 to >8 | 93.5 | |
| Levofloxacin | 0.5 | >4 | ≤0.03 to >4 | 79.9 | |
MBL negative, no gene encoding a metallo-β-lactamase was detected by PCR.
MIC50 and MIC90 were not calculated for <10 isolates.
Percent susceptibility was determined according to CLSI 2016 breakpoints, with the exception of that to ceftazidime-avibactam and tigecycline, where U.S. FDA breakpoints were applied. Because CLSI colistin breakpoints are not available for the Enterobacteriaceae, susceptibility for this organism group was determined using the EUCAST 2016 breakpoints for colistin.
Values are for colistin tested without 0.002% polysorbate 80; data are for isolates collected in 2014 and 2015 only.
The Enterobacter spp. included Enterobacter aerogenes (n = 439), Enterobacter asburiae (n = 89), Enterobacter cloacae (n = 525), Enterobacter kobei (n = 29), and Enterobacter ludwigii (n = 6).
The Citrobacter spp. included Citrobacter amalonaticus (n = 10), Citrobacter braakii (n = 23), Citrobacter farmeri (n = 3), Citrobacter freundii (n = 274), Citrobacter koseri (n = 246), Citrobacter sedlakii (n = 1), and a Citrobacter isolate whose species was not determined (n = 1).
The Proteeae included Morganella morganii (n = 253), Proteus mirabilis (n = 566), Proteus penneri (n = 14), Proteus vulgaris (n = 274), Providencia alcalifaciens (n = 2), Providencia rettgeri (n = 50), and Providencia stuartii (n = 31).
Other Enterobacteriaceae included Klebsiella ozaenae (n = 1), Kluyvera ascorbata (n = 1), Raoultella ornithinolytica (n = 12), Raoultella planticola (n = 2), Serratia liquefaciens (n = 5), Serratia marcescens (n = 181), and Serratia rubidaea (n = 1).
FIG 1.
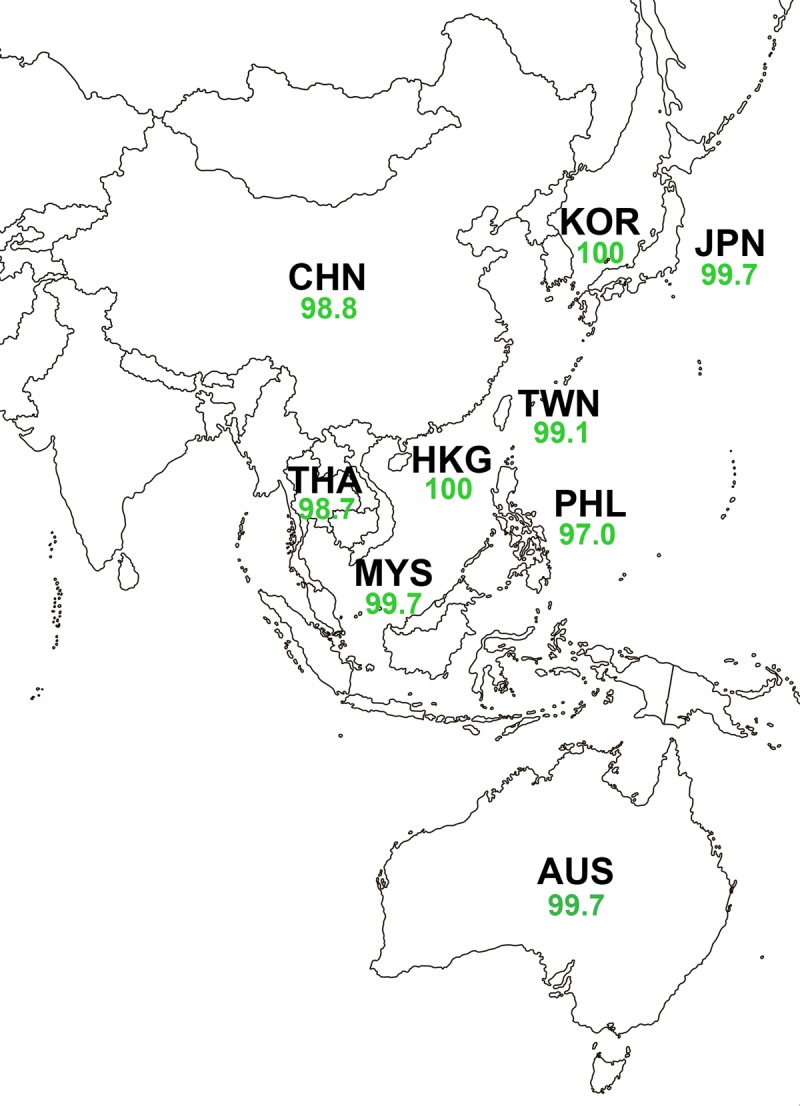
Percent susceptibility to ceftazidime-avibactam for isolates of Enterobacteriaceae collected from 2012 to 2015, by Asia-Pacific country. Ceftazidime-avibactam susceptible was defined as an MIC of ≤8 μg/ml; ceftazidime-avibactam resistant was defined as an MIC of ≥16 μg/ml. The green font indicates that >90% of isolates were ceftazidime-avibactam susceptible. Country abbreviations are as follows: AUS, Australia; CHN, China; HKG, Hong Kong; JPN, Japan; KOR, South Korea; MYS, Malaysia; PHL, Philippines; TWN, Taiwan; THA, Thailand. No isolates were obtained from patients in mainland China in 2014 or 2015 or patients in Hong Kong in 2015 due to export restrictions.
Table 2 depicts the in vitro activity of ceftazidime-avibactam and the comparator agents against subsets of Enterobacteriaceae isolates that were molecularly characterized for β-lactamase content. Ceftazidime-avibactam inhibited 99.6%, 99.3%, 98.9%, 97.7%, and 97.2% of ESBL-positive, AmpC-positive, ESBL-positive and AmpC-positive, original-spectrum β-lactamase (OSBL)-positive, and KPC-positive isolates, respectively, as well as all GES-positive (class A carbapenemase) and OXA-48-like-positive (class D carbapenemase) isolates. The MIC90 values for ceftazidime-avibactam against these subsets of β-lactamase-positive Enterobacteriaceae ranged from 0.5 μg/ml (OSBL-positive and ESBL-positive isolates) to 4 μg/ml (KPC-positive isolates). The percentages of isolates susceptible to doripenem (93.2 to 99.0%) and meropenem (96.2 to 98.5%) were equivalent to or slightly lower than the percentages of isolates susceptible to ceftazidime-avibactam observed for ESBL-positive, AmpC-positive, ESBL-positive and AmpC-positive, and OSBL-positive isolates. As anticipated, ceftazidime-avibactam was poorly active against isolates carrying MBLs (MIC90, >128 μg/ml; 1.4% susceptible); only tigecycline (MIC90, 4 μg/ml; 89.2% susceptible) and colistin (MIC90, >4 μg/ml; 86.5% susceptible) retained in vitro activity against the majority of MBL-positive isolates. The distribution of serine β-lactamases and MBLs among molecularly characterized isolates from each country in the Asia-Pacific region is summarized in the supplemental material (Tables S2B to S10B; Fig. S1A to D). CTX-M-type ESBLs accounted for 85.2% of all ESBLs, with CTX-M-15 (41.4%) being the most common ESBL identified (Fig. S1B). DHA-1 (55.3%) and CMY-2 (37.1%) accounted for the majority of all AmpC enzymes identified (Fig. S1C). ESBLs and AmpC β-lactamases were found in all countries in the region but differed in prevalence across the different countries (Fig. S1B and C). Carbapenemases were found in all countries in the Asia-Pacific region except Hong Kong. NDM (35.2%) and KPC (30.4%) were the most common carbapenemases identified in the region. The majority of KPCs were KPC-2 (97.4%), and NDM-1 (81.8%) was the most common NDM type. Considerable differences in the types and prevalence of individual carbapenemases were observed across the countries. The in vitro activity of ceftazidime-avibactam against Enterobacteriaceae in each country was related to the proportion of MBL-positive isolates (Tables S2A to S10A).
TABLE 2.
In vitro activities of ceftazidime-avibactam and comparator antimicrobial agents tested against 3,020 isolates of β-lactamase-positive Enterobacteriaceae (n = 2,388) and P. aeruginosa (n = 632) collected in the Asia-Pacific region as part of the INFORM global surveillance program from 2012 to 2015
| Organism, genotype (no. of isolates)a | Antimicrobial agent | MIC (μg/ml)b |
% susceptiblec | ||
|---|---|---|---|---|---|
| 50% | 90% | Range | |||
| Enterobacteriaceae (2,388) | |||||
| OSBL positive (44) | Ceftazidime-avibactam | 0.25 | 0.5 | 0.03 to >128 | 97.7 |
| Ceftazidime | 2 | 16 | 0.06 to >128 | 68.2 | |
| Cefepime | 0.5 | 8 | ≤0.12 to >16 | 81.8 | |
| Aztreonam | 1 | 64 | ≤0.015 to >128 | 75.0 | |
| Piperacillin-tazobactam | 4 | >128 | ≤0.25 to >128 | 61.4 | |
| Doripenem | 0.06 | 0.25 | 0.015 to >4 | 93.2 | |
| Imipenem | 0.25 | 2 | 0.12 to 4 | 72.7 | |
| Meropenem | 0.03 | 0.25 | 0.015 to 8 | 97.7 | |
| Amikacin | 1 | 4 | ≤0.25 to >32 | 95.5 | |
| Colistin (n = 20)d | 1 | 1 | 0.25 to >4 | 95.0 | |
| Tigecycline | 0.5 | 1 | 0.06 to 4 | 97.7 | |
| Levofloxacin | 0.5 | >4 | 0.06 to >4 | 56.8 | |
| SHV, spectrum undefined (1)e | Ceftazidime-avibactam | — | — | 0.25 | 100 |
| Ceftazidime | — | — | 128 | 0 | |
| Cefepime | — | — | ≤0.12 | 100 | |
| Aztreonam | — | — | 64 | 0 | |
| Piperacillin-tazobactam | — | — | 64 | 0 | |
| Doripenem | — | — | 0.12 | 100 | |
| Imipenem | — | — | 2 | 0 | |
| Meropenem | — | — | 0.12 | 100 | |
| Amikacin | — | — | 1 | 100 | |
| Colistin (n = 0)d | — | — | NDf | ND | |
| Tigecycline | — | — | 1 | 100 | |
| Levofloxacin | — | — | 1 | 100 | |
| ESBL positive (1,593)g | Ceftazidime-avibactam | 0.25 | 0.5 | ≤0.015 to >128 | 99.6 |
| Ceftazidime | 32 | >128 | 0.03 to >128 | 24.7 | |
| Cefepime | >16 | >16 | ≤0.12 to >16 | 8.5 | |
| Aztreonam | 64 | >128 | ≤0.015 to >128 | 11.5 | |
| Piperacillin-tazobactam | 8 | >128 | ≤0.25 to >128 | 73.4 | |
| Doripenem | 0.06 | 0.12 | 0.015 to >4 | 99.0 | |
| Imipenem | 0.25 | 0.5 | ≤0.03 to >8 | 96.9 | |
| Meropenem | 0.03 | 0.06 | 0.008 to >8 | 98.5 | |
| Amikacin | 4 | 16 | ≤0.25 to >32 | 95.1 | |
| Colistin (n = 729)d | 0.5 | 1 | ≤0.12 to >4 | 96.2 | |
| Tigecycline | 0.5 | 2 | 0.06 to 8 | 96.2 | |
| Levofloxacin | >4 | >4 | ≤0.03 to >4 | 34.5 | |
| AmpC positive (446)h | Ceftazidime-avibactam | 0.25 | 1 | ≤0.015 to 64 | 99.3 |
| Ceftazidime | 32 | >128 | 0.03 to >128 | 33.9 | |
| Cefepime | 0.25 | 4 | ≤0.12 to >16 | 87.0 | |
| Aztreonam | 8 | 64 | ≤0.015 to >128 | 46.0 | |
| Piperacillin-tazobactam | 8 | >128 | ≤0.25 to >128 | 65.5 | |
| Doripenem | 0.12 | 0.5 | 0.015 to >4 | 97.5 | |
| Imipenem | 1 | 2 | 0.12 to >8 | 54.3 | |
| Meropenem | 0.06 | 0.25 | ≤0.004 to >8 | 98.4 | |
| Amikacin | 2 | 4 | ≤0.25 to >32 | 96.2 | |
| Colistin (n = 167)d | 0.5 | >4 | 0.25 to >4 | 88.0 | |
| Tigecycline | 0.5 | 2 | 0.06 to 8 | 94.2 | |
| Levofloxacin | 0.5 | >4 | ≤0.03 to >4 | 69.7 | |
| ESBL positive + AmpC positive (186)i | Ceftazidime-avibactam | 0.5 | 2 | ≤0.015 to 128 | 98.9 |
| Ceftazidime | 128 | >128 | 0.12 to >128 | 3.8 | |
| Cefepime | >16 | >16 | ≤0.12 to >16 | 17.7 | |
| Aztreonam | 64 | >128 | 0.03 to >128 | 3.8 | |
| Piperacillin-tazobactam | 32 | >128 | 0.5 to >128 | 44.6 | |
| Doripenem | 0.12 | 0.5 | 0.03 to >4 | 97.3 | |
| Imipenem | 1 | 2 | 0.06 to >8 | 74.7 | |
| Meropenem | 0.06 | 0.25 | 0.015 to >8 | 96.2 | |
| Amikacin | 4 | >32 | ≤0.25 to >32 | 77.4 | |
| Colistin (n = 66)d | 1 | 1 | 0.25 to 4 | 98.5 | |
| Tigecycline | 0.5 | 2 | 0.06 to 8 | 97.3 | |
| Levofloxacin | >4 | >4 | ≤0.03 to >4 | 28.5 | |
| KPC positive (36)j | Ceftazidime-avibactam | 1 | 4 | 0.06 to 16 | 97.2 |
| Ceftazidime | 128 | >128 | 0.12 to >128 | 5.6 | |
| Cefepime | >16 | >16 | ≤0.12 to >16 | 2.8 | |
| Aztreonam | >128 | >128 | 0.06 to >128 | 2.8 | |
| Piperacillin-tazobactam | >128 | >128 | 0.5 to >128 | 2.8 | |
| Doripenem | >4 | >4 | 0.06 to >4 | 8.3 | |
| Imipenem | >8 | >8 | 2 to >8 | 0 | |
| Meropenem | >8 | >8 | 0.06 to >8 | 2.8 | |
| Amikacin | 4 | >32 | ≤0.25 to >32 | 77.8 | |
| Colistin (n = 9)d | — | — | 0.25 to 1 | 100 | |
| Tigecycline | 1 | 2 | 0.06 to 4 | 91.7 | |
| Levofloxacin | >4 | >4 | 0.12 to >4 | 22.2 | |
| GES carbapenemase positive (2)k | Ceftazidime-avibactam | — | — | 0.06 to 1 | 100 |
| Ceftazidime | — | — | 4 to >128 | 50.0 | |
| Cefepime | — | — | 2 to 16 | 50.0 | |
| Aztreonam | — | — | 4 to >128 | 50.0 | |
| Piperacillin-tazobactam | — | — | 1 to >128 | 50.0 | |
| Doripenem | — | — | 0.06 to 4 | 50.0 | |
| Imipenem | — | — | 0.5 to 4 | 50.0 | |
| Meropenem | — | — | 0.06 to >8 | 50.0 | |
| Amikacin | — | — | 2 to 2 | 100 | |
| Colistin (n = 2)d | — | — | 0.5 to 0.5 | 100 | |
| Tigecycline | — | — | 0.05 to 1 | 100 | |
| Levofloxacin | — | — | 0.5 to 2 | 100 | |
| OXA-48-like positive (6)l | Ceftazidime-avibactam | — | — | 0.25 to 2 | 100 |
| Ceftazidime | — | — | 0.5 to >128 | 16.7 | |
| Cefepime | — | — | ≤0.12 to >16 | 16.7 | |
| Aztreonam | — | — | 0.12 to >128 | 16.7 | |
| Piperacillin-tazobactam | — | — | 128 to >128 | 0 | |
| Doripenem | — | — | 0.25 to >4 | 83.3 | |
| Imipenem | — | — | 1 to >8 | 33.3 | |
| Meropenem | — | — | 1 to >8 | 33.3 | |
| Amikacin | — | — | 1 to 8 | 100 | |
| Colistin (n = 6)d | — | — | 0.25 to >4 | 83.3 | |
| Tigecycline | — | — | 0.12 to 0.5 | 100 | |
| Levofloxacin | — | — | >4 to >4 | 0 | |
| MBL positive (74)m | Ceftazidime-avibactam | >128 | >128 | 2 to >128 | 1.4 |
| Ceftazidime | >128 | >128 | 32 to >128 | 0 | |
| Cefepime | >16 | >16 | 1 to >16 | 2.7 | |
| Aztreonam | 64 | >128 | ≤0.015 to >128 | 25.7 | |
| Piperacillin-tazobactam | >128 | >128 | 0.5 to >128 | 16.2 | |
| Doripenem | >4 | >4 | 1 to >4 | 4.1 | |
| Imipenem | >8 | >8 | 0.5 to >8 | 6.8 | |
| Meropenem | >8 | >8 | 0.5 to >8 | 8.1 | |
| Amikacin | 4 | >32 | 0.5 to >32 | 66.2 | |
| Colistin (n = 37)d | 1 | >4 | 0.25 to >4 | 86.5 | |
| Tigecycline | 1 | 4 | 0.06 to 8 | 89.2 | |
| Levofloxacin | >4 | >4 | 0.06 to >4 | 36.5 | |
| P. aeruginosa (632) | |||||
| OSBL positive (3)n | Ceftazidime-avibactam | — | — | 8 to 32 | 66.7 |
| Ceftazidime | — | — | 16 to >128 | 0 | |
| Cefepime | — | — | 16 to >16 | 0 | |
| Aztreonam | — | — | 16 to 128 | 0 | |
| Piperacillin-tazobactam | — | — | >128 to >128 | 0 | |
| Doripenem | — | — | >4 to >4 | 0 | |
| Imipenem | — | — | >8 to >8 | 0 | |
| Meropenem | — | — | >8 to >8 | 0 | |
| Amikacin | — | — | >32 to >32 | 0 | |
| Colistin (n = 1)d | — | — | 1 | 100 | |
| Levofloxacin | — | — | 4 to >4 | 0 | |
| ESBL positive (19)n,o | Ceftazidime-avibactam | 64 | >128 | 4 to >128 | 21.1 |
| Ceftazidime | >128 | >128 | 128 to >128 | 0 | |
| Cefepime | >16 | >16 | >16 to >16 | 0 | |
| Aztreonam | >128 | >128 | 16 to >128 | 0 | |
| Piperacillin-tazobactam | 128 | >128 | 16 to >128 | 5.3 | |
| Doripenem | >4 | >4 | 4 to >4 | 0 | |
| Imipenem | >8 | >8 | 4 to >8 | 0 | |
| Meropenem | >8 | >8 | 4 to >8 | 0 | |
| Amikacin | 32 | >32 | 8 to >32 | 36.8 | |
| Colistin (n = 13)d | 2 | 2 | 1 to 2 | 100 | |
| Levofloxacin | >4 | >4 | >4 to >4 | 0 | |
| GES carbapenemase positive (3)n | Ceftazidime-avibactam | — | — | 4 to 8 | 100 |
| Ceftazidime | — | — | 8 to >128 | 33.3 | |
| Cefepime | — | — | 16 to >16 | 0 | |
| Aztreonam | — | — | 8 to 128 | 33.3 | |
| Piperacillin-tazobactam | — | — | 64 to >128 | 0 | |
| Doripenem | — | — | >4 to >4 | 0 | |
| Imipenem | — | — | 2 to >8 | 33.3 | |
| Meropenem | — | — | 4 to >8 | 0 | |
| Amikacin | — | — | 8 to >32 | 33.3 | |
| Colistin (n = 2)d | — | — | 2 to 2 | 100 | |
| Levofloxacin | — | — | >4 to >4 | 0 | |
| GES, spectrum undefined (3)n,p | Ceftazidime-avibactam | — | — | 8 to 128 | 33.3 |
| Ceftazidime | — | — | 64 to >128 | 0 | |
| Cefepime | — | — | 16 to >16 | 0 | |
| Aztreonam | — | — | 16 to 32 | 0 | |
| Piperacillin-tazobactam | — | — | 16 to >128 | 33.3 | |
| Doripenem | — | — | >4 to >4 | 0 | |
| Imipenem | — | — | 4 to >8 | 0 | |
| Meropenem | — | — | 8 to >8 | 0 | |
| Amikacin | — | — | 32 to >32 | 0 | |
| Colistin (n = 3)d | — | — | 1 to 2 | 100 | |
| Levofloxacin | — | — | >4 to >4 | 0 | |
| KPC positive (1)n | Ceftazidime-avibactam | — | — | 4 | 100 |
| Ceftazidime | — | — | 64 | 0 | |
| Cefepime | — | — | >16 | 0 | |
| Aztreonam | — | — | >128 | 0 | |
| Piperacillin-tazobactam | — | — | >128 | 0 | |
| Doripenem | — | — | >4 | 0 | |
| Imipenem | — | — | >8 | 0 | |
| Meropenem | — | — | >8 | 0 | |
| Amikacin | — | — | 4 | 100 | |
| Colistin (n = 0)d | — | — | NDf | ND | |
| Levofloxacin | — | — | 0.12 | 100 | |
| MBL positive (74)n,q | Ceftazidime-avibactam | 128 | >128 | 8 to >128 | 1.4 |
| Ceftazidime | >128 | >128 | 8 to >128 | 1.4 | |
| Cefepime | >16 | >16 | 8 to >16 | 2.7 | |
| Aztreonam | 16 | >128 | 4 to >128 | 28.4 | |
| Piperacillin-tazobactam | 128 | >128 | 8 to >128 | 6.8 | |
| Doripenem | >4 | >4 | 4 to >4 | 0 | |
| Imipenem | >8 | >8 | 4 to >8 | 0 | |
| Meropenem | >8 | >8 | 2 to >8 | 1.4 | |
| Amikacin | 32 | >32 | 4 to >32 | 28.4 | |
| Colistin (n = 53)d | 2 | 2 | 0.5 to 4 | 94.3 | |
| Levofloxacin | >4 | >4 | 0.25 to >4 | 4.1 | |
| No acquired β-lactamase detected (529)n | Ceftazidime-avibactam | 4 | 8 | 0.12 to >128 | 91.3 |
| Ceftazidime | 4 | 64 | 0.25 to >128 | 66.2 | |
| Cefepime | 8 | >16 | ≤0.12 to >16 | 66.5 | |
| Aztreonam | 16 | 64 | 0.25 to >128 | 45.4 | |
| Piperacillin-tazobactam | 16 | >128 | ≤0.25 to >128 | 52.2 | |
| Doripenem | 4 | >4 | 0.03 to >4 | 38.6 | |
| Imipenem | >8 | >8 | 0.5 to >8 | 6.6 | |
| Meropenem | 4 | >8 | ≤0.06 to >8 | 32.9 | |
| Amikacin | 4 | 16 | ≤0.25 to >32 | 94.0 | |
| Colistin (n = 299)d | 2 | 2 | 0.12 to 8 | 94.0 | |
| Levofloxacin | 2 | >4 | ≤0.03 to >4 | 58.4 | |
OSBL, original-spectrum β-lactamase (e.g., TEM-1, SHV-1, SHV-11); ESBL, extended-spectrum β-lactamase; MBL, metallo-β-lactamase.
—, MIC50 and MIC90 were not calculated for <10 isolates.
Percent susceptibility was determined according to CLSI 2016 breakpoints, with the exception of that to ceftazidime-avibactam and tigecycline, where U.S. FDA breakpoints were applied. Because CLSI colistin breakpoints are not available for the Enterobacteriaceae, susceptibility for this organism group was determined using the EUCAST 2016 breakpoints for colistin.
Values are for colistin tested without 0.002% polysorbate 80; data are for isolates collected in 2014 and 2015 only.
SHV, spectrum undefined, SHV-type β-lactamase for which the spectrum of activity (original spectrum or extended spectrum) has not been biochemically determined.
ND, not determined; MIC range and percent susceptible were not calculated for 0 isolates.
Includes isolates carrying the chromosomal ESBL common to K. oxytoca, inhibitor-resistant β-lactamases (SHV-type and/or TEM-type β-lactamases that are not inhibited by clavulanic acid), SHV-type and/or TEM-type β-lactamases with an undefined spectrum of activity, and/or OSBLs.
Includes isolates carrying the chromosomal AmpC common to Citrobacter spp., Enterobacter spp., Morganella morganii, Providencia spp., and Serratia spp. and isolates cocarrying SHV-type and/or TEM-type β-lactamases with an undefined spectrum of activity and/or OSBLs.
Includes isolates carrying the chromosomal β-lactamases common to Citrobacter spp., Enterobacter spp., and K. oxytoca and isolates cocarrying OSBLs.
Includes isolates carrying ESBLs, plasmidic and chromosomal AmpC β-lactamases, and/or OSBLs.
Isolates cocarry ESBLs.
Includes isolates carrying ESBLs, plasmidic AmpC β-lactamases, and/or OSBLs.
Includes isolates cocarrying ESBLs, plasmidic and chromosomal AmpC β-lactamases, OSBLs, and serine carbapenemases (IMP-4 and KPC-2, 2 isolates; NDM-1 and OXA-232, 5 isolates).
Assumed to carry the chromosomal AmpC common to P. aeruginosa.
Includes isolates carrying OSBLs.
Includes isolates carrying GES-24 (1 isolate) and GES-29 (2 isolates) β-lactamases, for which the spectrum of activity (ESBL or carbapenemase) has not been biochemically determined.
Includes isolates cocarrying ESBLs.
Table 3 shows the in vitro activity of ceftazidime-avibactam and the comparator agents against 2,277 ceftazidime-nonsusceptible isolates of Enterobacteriaceae (24.9% of all isolates). Overall, 96.1% of ceftazidime-nonsusceptible isolates were susceptible to ceftazidime-avibactam (MIC90, 1 μg/ml), with the MIC90s against individual species of Enterobacteriaceae ranging from 0.5 to 2 μg/ml (93.9 to 99.5% susceptible) for E. coli, Klebsiella pneumoniae, Enterobacter spp., and Citrobacter spp. Ceftazidime-nonsusceptible isolates of Proteeae (MIC90, 8 μg/ml; 90.3% susceptible) and Klebsiella oxytoca (MIC90, 16 μg/ml; 88.6% susceptible) were less susceptible to ceftazidime-avibactam. The percentage of isolates susceptible to ceftazidime-avibactam increased for all species when the activity against isolates that did not produce an MBL was evaluated. Across the Asia-Pacific region, the percentage of Enterobacteriaceae isolates that tested nonsusceptible to ceftazidime ranged from 9.0% (Australia) to 35.5% (Philippines) (Fig. S2). The percentage of ceftazidime-nonsusceptible isolates that were susceptible to ceftazidime-avibactam was >95% (MIC90, 0.5 to 2 μg/ml) in all countries except Philippines (MIC90, 2 μg/ml; 91.5% susceptible) (Tables S2A to S10A). The lower activity demonstrated by ceftazidime-avibactam in Philippines, as well as in Thailand and China, can be attributed to the greater numbers of isolates carrying MBLs, as 99.1%, 99.5%, and 98.4% of ceftazidime-nonsusceptible MBL-negative isolates from these countries, respectively, were susceptible to ceftazidime-avibactam (Tables S3A and B, S7A and B, and S10A and B; Fig. S1D).
TABLE 3.
In vitro activities of ceftazidime-avibactam and comparator antimicrobial agents tested against 2,277 isolates of ceftazidime-nonsusceptible Enterobacteriaceae and 451 isolates of ceftazidime-nonsusceptible P. aeruginosa collected in the Asia-Pacific region as part of the INFORM global surveillance program from 2012 to 2015
| Organism, phenotype (no. of isolates)a | Antimicrobial agent | MIC (μg/ml)b |
% susceptiblec | ||
|---|---|---|---|---|---|
| 50% | 90% | Range | |||
| Enterobacteriaceae (2,277) | Ceftazidime-avibactam | 0.25 | 1 | ≤0.015 to >128 | 96.1 |
| Ceftazidime | 64 | >128 | 8 to >128 | 0 | |
| Cefepime | 16 | >16 | ≤0.12 to >16 | 29.3 | |
| Aztreonam | 64 | >128 | ≤0.015 to >128 | 8.0 | |
| Piperacillin-tazobactam | 16 | >128 | ≤0.25 to >128 | 55.3 | |
| Doripenem | 0.06 | 0.25 | 0.015 to >4 | 93.9 | |
| Imipenem | 0.25 | 2 | ≤0.03 to >8 | 86.8 | |
| Meropenem | 0.06 | 0.25 | ≤0.004 to >8 | 93.6 | |
| Amikacin | 2 | 16 | ≤0.25 to >32 | 92.1 | |
| Colistin (n = 1,037)d | 0.5 | 1 | ≤0.12 to >4 | 92.4 | |
| Tigecycline | 0.5 | 2 | 0.06 to 8 | 95.2 | |
| Levofloxacin | >4 | >4 | ≤0.03 to >4 | 44.1 | |
| Enterobacteriaceae, MBL negative (2,203) | Ceftazidime-avibactam | 0.25 | 1 | ≤0.015 to >128 | 99.2 |
| Ceftazidime | 64 | >128 | 8 to >128 | 0 | |
| Cefepime | 16 | >16 | ≤0.12 to >16 | 30.1 | |
| Aztreonam | 64 | >128 | ≤0.015 to >128 | 7.4 | |
| Piperacillin-tazobactam | 16 | >128 | ≤0.25 to >128 | 56.6 | |
| Doripenem | 0.06 | 0.25 | 0.015 to >4 | 97.0 | |
| Imipenem | 0.25 | 2 | ≤0.03 to >8 | 89.5 | |
| Meropenem | 0.06 | 0.12 | ≤0.004 to >8 | 96.5 | |
| Amikacin | 2 | 16 | ≤0.25 to >32 | 92.9 | |
| Colistin (n = 1,000)d | 0.5 | 1 | ≤0.12 to >4 | 92.6 | |
| Tigecycline | 0.5 | 2 | 0.06 to 8 | 95.4 | |
| Levofloxacin | >4 | >4 | ≤0.03 to >4 | 44.4 | |
| Escherichia coli (862) | Ceftazidime-avibactam | 0.12 | 0.5 | ≤0.015 to >128 | 99.5 |
| Ceftazidime | 32 | 128 | 8 to >128 | 0 | |
| Cefepime | >16 | >16 | ≤0.12 to >16 | 18.8 | |
| Aztreonam | 64 | 128 | 0.5 to >128 | 6.0 | |
| Piperacillin-tazobactam | 4 | 128 | 0.5 to >128 | 81.2 | |
| Doripenem | 0.06 | 0.06 | 0.015 to >4 | 98.6 | |
| Imipenem | 0.25 | 0.5 | ≤0.03 to >8 | 96.9 | |
| Meropenem | 0.03 | 0.06 | ≤0.004 to >8 | 98.0 | |
| Amikacin | 4 | 16 | 0.5 to >32 | 95.1 | |
| Colistin (n = 362)d | 0.5 | 1 | ≤0.12 to 4 | 98.1 | |
| Tigecycline | 0.25 | 0.5 | 0.06 to 8 | 99.8 | |
| Levofloxacin | >4 | >4 | ≤0.03 to >4 | 22.7 | |
| Escherichia coli, MBL negative (859) | Ceftazidime-avibactam | 0.12 | 0.5 | ≤0.015 to >128 | 99.9 |
| Ceftazidime | 32 | 128 | 8 to >128 | 0 | |
| Cefepime | >16 | >16 | ≤0.12 to >16 | 18.9 | |
| Aztreonam | 64 | 128 | 0.5 to >128 | 6.1 | |
| Piperacillin-tazobactam | 4 | 128 | 0.5 to >128 | 81.5 | |
| Doripenem | 0.06 | 0.06 | 0.015 to >4 | 99.0 | |
| Imipenem | 0.25 | 0.5 | ≤0.03 to >8 | 97.2 | |
| Meropenem | 0.03 | 0.06 | ≤0.004 to >8 | 98.4 | |
| Amikacin | 4 | 16 | 0.5 to >32 | 95.2 | |
| Colistin (n = 361)d | 0.5 | 1 | ≤0.12 to 4 | 98.1 | |
| Tigecycline | 0.25 | 0.5 | 0.06 to 8 | 99.8 | |
| Levofloxacin | >4 | >4 | ≤0.03 to >4 | 22.8 | |
| Klebsiella pneumoniae (778) | Ceftazidime-avibactam | 0.5 | 2 | ≤0.015 to >128 | 94.3 |
| Ceftazidime | 128 | >128 | 8 to >128 | 0 | |
| Cefepime | >16 | >16 | ≤0.12 to >16 | 15.9 | |
| Aztreonam | 64 | >128 | 0.06 to >128 | 5.7 | |
| Piperacillin-tazobactam | 64 | >128 | 0.5 to >128 | 39.2 | |
| Doripenem | 0.12 | 2 | 0.015 to >4 | 89.9 | |
| Imipenem | 0.25 | 2 | ≤0.03 to >8 | 84.6 | |
| Meropenem | 0.06 | 2 | 0.015 to >8 | 89.2 | |
| Amikacin | 2 | 32 | ≤0.25 to >32 | 88.4 | |
| Colistin (n = 407)d | 1 | 1 | 0.25 to >4 | 95.1 | |
| Tigecycline | 1 | 2 | 0.06 to 8 | 93.6 | |
| Levofloxacin | >4 | >4 | ≤0.03 to >4 | 44.9 | |
| Klebsiella pneumoniae, MBL negative (741) | Ceftazidime-avibactam | 0.5 | 1 | ≤0.015 to >128 | 99.1 |
| Ceftazidime | 64 | >128 | 8 to >128 | 0 | |
| Cefepime | >16 | >16 | ≤0.12 to >16 | 16.7 | |
| Aztreonam | 64 | >128 | 0.5 to >128 | 5.1 | |
| Piperacillin-tazobactam | 32 | >128 | 0.5 to >128 | 40.6 | |
| Doripenem | 0.06 | 0.25 | 0.015 to >4 | 94.2 | |
| Imipenem | 0.25 | 2 | ≤0.03 to >8 | 88.3 | |
| Meropenem | 0.06 | 0.25 | 0.015 to >8 | 93.5 | |
| Amikacin | 2 | 32 | ≤0.25 to >32 | 89.9 | |
| Colistin (n = 385)d | 1 | 1 | 0.25 to >4 | 95.3 | |
| Tigecycline | 1 | 2 | 0.06 to 8 | 93.4 | |
| Levofloxacin | >4 | >4 | ≤0.03 to >4 | 44.9 | |
| Klebsiella oxytoca (44) | Ceftazidime-avibactam | 0.25 | 16 | ≤0.015 to 128 | 88.6 |
| Ceftazidime | 64 | >128 | 8 to >128 | 0 | |
| Cefepime | 2 | >16 | ≤0.12 to >16 | 50.0 | |
| Aztreonam | 32 | >128 | 1 to >128 | 2.3 | |
| Piperacillin-tazobactam | 4 | >128 | 1 to >128 | 65.9 | |
| Doripenem | 0.06 | 2 | 0.03 to >4 | 88.6 | |
| Imipenem | 0.25 | 4 | 0.12 to >8 | 84.1 | |
| Meropenem | 0.06 | 4 | 0.03 to >8 | 88.6 | |
| Amikacin | 2 | 32 | 0.5 to >32 | 88.6 | |
| Colistin (n = 22)d | 0.5 | 1 | 0.25 to 1 | 100 | |
| Tigecycline | 0.5 | 1 | 0.06 to 2 | 100 | |
| Levofloxacin | 0.5 | >4 | ≤0.03 to >4 | 70.5 | |
| Klebsiella oxytoca, MBL negative (40) | Ceftazidime-avibactam | 0.25 | 1 | ≤0.015 to 16 | 97.5 |
| Ceftazidime | 32 | >128 | 8 to >128 | 0 | |
| Cefepime | 2 | 16 | ≤0.12 to >16 | 52.5 | |
| Aztreonam | 32 | >128 | 8 to >128 | 0 | |
| Piperacillin-tazobactam | 4 | >128 | 1 to >128 | 70.0 | |
| Doripenem | 0.06 | 0.12 | 0.03 to 2 | 97.5 | |
| Imipenem | 0.25 | 1 | 0.12 to 8 | 92.5 | |
| Meropenem | 0.06 | 0.12 | 0.03 to 8 | 97.5 | |
| Amikacin | 2 | 8 | 0.5 to >32 | 92.5 | |
| Colistin (n = 21)d | 0.5 | 1 | 0.25 to 1 | 100 | |
| Tigecycline | 0.25 | 1 | 0.06 to 2 | 100 | |
| Levofloxacin | 0.5 | >4 | ≤0.03 to >4 | 72.5 | |
| Enterobacter spp.e (370) | Ceftazidime-avibactam | 0.5 | 2 | ≤0.015 to >128 | 94.3 |
| Ceftazidime | 64 | >128 | 8 to >128 | 0 | |
| Cefepime | 1 | >16 | ≤0.12 to >16 | 60.5 | |
| Aztreonam | 32 | 128 | 0.06 to >128 | 7.6 | |
| Piperacillin-tazobactam | 64 | >128 | 0.5 to >128 | 31.1 | |
| Doripenem | 0.12 | 0.5 | 0.03 to >4 | 93.8 | |
| Imipenem | 1 | 2 | ≤0.03 to >8 | 82.7 | |
| Meropenem | 0.12 | 0.25 | 0.015 to >8 | 93.8 | |
| Amikacin | 2 | 8 | 0.5 to >32 | 95.1 | |
| Colistin (n = 160)d | 0.5 | >4 | 0.25 to >4 | 87.5 | |
| Tigecycline | 0.5 | 2 | 0.06 to 8 | 93.8 | |
| Levofloxacin | 0.25 | >4 | ≤0.03 to >4 | 79.2 | |
| Enterobacter spp., MBL negative (351) | Ceftazidime-avibactam | 0.5 | 1 | ≤0.015 to 64 | 99.2 |
| Ceftazidime | 64 | >128 | 8 to >128 | 0 | |
| Cefepime | 1 | >16 | ≤0.12 to >16 | 63.5 | |
| Aztreonam | 32 | 128 | 0.5 to >128 | 6.3 | |
| Piperacillin-tazobactam | 64 | >128 | 0.5 to >128 | 31.6 | |
| Doripenem | 0.12 | 0.25 | 0.03 to >4 | 98.6 | |
| Imipenem | 1 | 2 | ≤0.03 to >8 | 87.2 | |
| Meropenem | 0.12 | 0.25 | 0.015 to >8 | 98.3 | |
| Amikacin | 2 | 8 | 0.5 to >32 | 96.0 | |
| Colistin (n = 150)d | 0.5 | 4 | 0.25 to >4 | 88.0 | |
| Tigecycline | 0.5 | 2 | 0.06 to 8 | 95.2 | |
| Levofloxacin | 0.12 | >4 | ≤0.03 to >4 | 82.1 | |
| Citrobacter spp.f (148) | Ceftazidime-avibactam | 0.5 | 2 | 0.03 to >128 | 93.9 |
| Ceftazidime | 128 | >128 | 8 to >128 | 0 | |
| Cefepime | 1 | >16 | ≤0.12 to >16 | 63.5 | |
| Aztreonam | 32 | 128 | 0.5 to >128 | 8.8 | |
| Piperacillin-tazobactam | 32 | >128 | 0.5 to >128 | 34.5 | |
| Doripenem | 0.06 | 0.25 | 0.015 to >4 | 93.9 | |
| Imipenem | 0.5 | 2 | 0.12 to >8 | 85.8 | |
| Meropenem | 0.06 | 0.25 | 0.015 to >8 | 94.6 | |
| Amikacin | 2 | 16 | 0.5 to >32 | 90.5 | |
| Colistin (n = 52)d | 0.5 | 1 | 0.25 to 2 | 100 | |
| Tigecycline | 0.5 | 1 | 0.12 to 4 | 99.3 | |
| Levofloxacin | 1 | >4 | ≤0.03 to >4 | 71.0 | |
| Citrobacter spp., MBL negative (142) | Ceftazidime-avibactam | 0.5 | 1 | 0.03 to >128 | 97.9 |
| Ceftazidime | 64 | >128 | 8 to >128 | 0 | |
| Cefepime | 1 | >16 | ≤0.12 to >16 | 66.2 | |
| Aztreonam | 32 | 128 | 0.5 to >128 | 7.8 | |
| Piperacillin-tazobactam | 32 | >128 | 0.5 to >128 | 35.2 | |
| Doripenem | 0.06 | 0.12 | 0.015 to >4 | 97.2 | |
| Imipenem | 0.5 | 2 | 0.12 to >8 | 88.7 | |
| Meropenem | 0.06 | 0.12 | 0.015 to >8 | 97.2 | |
| Amikacin | 2 | 16 | 0.5 to >32 | 90.9 | |
| Colistin (n = 50)d | 0.5 | 1 | 0.25 to 2 | 100 | |
| Tigecycline | 0.5 | 1 | 0.12 to 4 | 99.3 | |
| Levofloxacin | 1 | >4 | ≤0.03 to >4 | 71.8 | |
| Proteeaeg (62) | Ceftazidime-avibactam | 0.25 | 8 | ≤0.015 to 64 | 90.3 |
| Ceftazidime | 32 | >128 | 8 to >128 | 0 | |
| Cefepime | 1 | >16 | ≤0.12 to >16 | 61.3 | |
| Aztreonam | 2 | 32 | ≤0.015 to 128 | 66.1 | |
| Piperacillin-tazobactam | 1 | 32 | ≤0.25 to >128 | 83.9 | |
| Doripenem | 0.25 | 2 | 0.06 to >4 | 87.1 | |
| Imipenem | 4 | 8 | 0.5 to >8 | 9.7 | |
| Meropenem | 0.12 | 1 | 0.03 to >8 | 90.3 | |
| Amikacin | 4 | >32 | 0.5 to >32 | 82.3 | |
| Colistin (n = 26)d | >4 | >4 | >4 to >4 | 0 | |
| Tigecycline | 2 | 4 | 0.12 to 8 | 50.0 | |
| Levofloxacin | >4 | >4 | ≤0.03 to >4 | 33.9 | |
| Proteeae, MBL negative (58) | Ceftazidime-avibactam | 0.12 | 1 | ≤0.015 to 32 | 96.6 |
| Ceftazidime | 32 | >128 | 8 to >128 | 0 | |
| Cefepime | 1 | >16 | ≤0.12 to >16 | 65.5 | |
| Aztreonam | 2 | 32 | ≤0.015 to 128 | 65.5 | |
| Piperacillin-tazobactam | 1 | 32 | ≤0.25 to >128 | 87.9 | |
| Doripenem | 0.25 | 1 | 0.06 to >4 | 93.1 | |
| Imipenem | 4 | 8 | 0.5 to >8 | 10.3 | |
| Meropenem | 0.12 | 0.25 | 0.03 to >8 | 94.8 | |
| Amikacin | 4 | >32 | 0.5 to >32 | 84.5 | |
| Colistin (n = 25)d | >4 | >4 | >4 to >4 | 0 | |
| Tigecycline | 4 | 4 | 0.12 to 8 | 48.3 | |
| Levofloxacin | 4 | >4 | ≤0.03 to >4 | 34.5 | |
| Other Enterobacteriaceaeh (13) | Ceftazidime-avibactam | 0.5 | 1 | 0.12 to >128 | 92.3 |
| Ceftazidime | 16 | >128 | 8 to >128 | 0 | |
| Cefepime | 16 | >16 | 2 to >16 | 15.4 | |
| Aztreonam | 64 | >128 | 0.25 to >128 | 15.4 | |
| Piperacillin-tazobactam | 32 | >128 | 2 to >128 | 46.2 | |
| Doripenem | 0.12 | >4 | 0.06 to >4 | 84.6 | |
| Imipenem | 0.5 | >8 | 0.25 to >8 | 61.5 | |
| Meropenem | 0.06 | 8 | 0.06 to >8 | 84.6 | |
| Amikacin | 4 | 16 | 0.5 to 32 | 92.3 | |
| Colistin (n = 8)d | — | — | 0.25 to >4 | 25.0 | |
| Tigecycline | 1 | 4 | 0.25 to 8 | 84.6 | |
| Levofloxacin | 1 | >4 | 0.12 to >4 | 69.2 | |
| Other Enterobacteriaceae, MBL negative (12) | Ceftazidime-avibactam | 0.25 | 1 | 0.12 to 1 | 100 |
| Ceftazidime | 16 | 128 | 8 to >128 | 0 | |
| Cefepime | 8 | >16 | 2 to >16 | 16.7 | |
| Aztreonam | 128 | >128 | 2 to >128 | 8.3 | |
| Piperacillin-tazobactam | 64 | >128 | 2 to >128 | 41.7 | |
| Doripenem | 0.12 | 0.5 | 0.06 to >4 | 91.7 | |
| Imipenem | 0.5 | 2 | 0.25 to >8 | 66.7 | |
| Meropenem | 0.06 | 0.25 | 0.06 to >8 | 91.7 | |
| Amikacin | 4 | 16 | 0.5 to 32 | 91.7 | |
| Colistin (n = 8)d | — | — | 0.25 to >4 | 25.0 | |
| Tigecycline | 1 | 4 | 0.25 to 8 | 83.3 | |
| Levofloxacin | 1 | >4 | 0.12 to >4 | 75.0 | |
| Pseudomonas aeruginosa (451) | Ceftazidime-avibactam | 8 | 128 | 0.03 to >128 | 66.7 |
| Ceftazidime | 64 | >128 | 16 to >128 | 0.0 | |
| Cefepime | 16 | >16 | 1 to >16 | 21.7 | |
| Aztreonam | 32 | 128 | 1 to >128 | 13.1 | |
| Piperacillin-tazobactam | 128 | >128 | 2 to >128 | 7.1 | |
| Doripenem | 4 | 8 | 0.06 to >4 | 43.5 | |
| Imipenem | 8 | >8 | 0.25 to >8 | 39.5 | |
| Meropenem | 4 | >8 | 0.03 to >8 | 44.6 | |
| Amikacin | 4 | >32 | ≤0.25 to >32 | 78.7 | |
| Colistin (n = 301)d | 2 | 2 | 0.25 to >8 | 92.4 | |
| Levofloxacin | >4 | >4 | ≤0.03 to >4 | 43.7 | |
| Pseudomonas aeruginosa, MBL negative (378) | Ceftazidime-avibactam | 8 | 32 | 0.03 to >128 | 79.6 |
| Ceftazidime | 64 | >128 | 16 to >128 | 0 | |
| Cefepime | 16 | >16 | 1 to >16 | 25.7 | |
| Aztreonam | 32 | 128 | 1 to >128 | 10.3 | |
| Piperacillin-tazobactam | 128 | >128 | 2 to >128 | 7.4 | |
| Doripenem | 2 | 8 | 0.06 to >4 | 51.9 | |
| Imipenem | 4 | >8 | 0.25 to >8 | 47.1 | |
| Meropenem | 2 | >8 | 0.03 to >8 | 52.9 | |
| Amikacin | 4 | 32 | ≤0.25 to >32 | 88.4 | |
| Colistin (n = 248)d | 2 | 2 | 0.25 to >8 | 91.9 | |
| Levofloxacin | 2 | >4 | ≤0.03 to >4 | 51.3 | |
MBL negative, no gene encoding a metallo-β-lactamase was detected by PCR.
—, MIC50 and MIC90 were not calculated for <10 isolates.
Percent susceptibility was determined according to CLSI 2016 breakpoints, with the exception of that to ceftazidime-avibactam and tigecycline, where U.S. FDA breakpoints were applied. Because CLSI colistin breakpoints are not available for the Enterobacteriaceae, susceptibility for this organism group was determined using the EUCAST 2016 breakpoints for colistin.
Values are for colistin tested without 0.002% polysorbate 80; data are for isolates collected in 2014 and 2015 only.
The Enterobacter spp. included Enterobacter aerogenes (n = 147), Enterobacter asburiae (n = 26), Enterobacter cloacae (n = 185), Enterobacter kobei (n = 10), and Enterobacter ludwigii (n = 2).
The Citrobacter spp. included Citrobacter amalonaticus (n = 1), Citrobacter braakii (n = 10), Citrobacter freundii (n = 115), Citrobacter koseri (n = 21), and Citrobacter sedlakii (n = 1).
The Proteeae included Morganella morganii (n = 27), Proteus mirabilis (n = 22), Proteus vulgaris (n = 3), Providencia rettgeri (n = 6), and Providencia stuartii (n = 4).
Other Enterobacteriaceae included Raoultella ornithinolytica (n = 2) and Serratia marcescens (n = 11).
Ceftazidime-avibactam displayed reduced activity (47.7% susceptible) against 149 isolates of meropenem-nonsusceptible Enterobacteriaceae (Table 4). This result is again largely explained by the observation that 45.6% of meropenem-nonsusceptible isolates were MBL positive; the activity against MBL-negative meropenem-nonsusceptible isolates was vastly improved (87.7% susceptible). The MIC90 values for ceftazidime-avibactam against all meropenem-nonsusceptible isolates and isolates of individual species or species groups of Enterobacteriaceae were >128 μg/ml. However, MIC90 values against subsets of MBL-negative meropenem-nonsusceptible isolates were significantly decreased, yielding susceptibility percentages of 85.7 to 93.8% for E. coli, Enterobacter spp., and K. pneumoniae (the only species with >10 isolates tested). Meropenem-nonsusceptible isolates were the most susceptible to tigecycline (85.9%), colistin (85.1%), and amikacin (69.8%), with the rates of susceptibility to β-lactams other than ceftazidime-avibactam being ≤15%. The percentage of isolates that tested nonsusceptible to meropenem was relatively low among the isolates from the Asia-Pacific countries, ranging from 0.3 to 4.2% (Fig. S2). Ceftazidime-avibactam displayed high rates of activity (≥99%) against isolates collected in countries in which meropenem nonsusceptibility was conferred primarily by serine carbapenemases (KPC, OXA-48-like) or ESBL and/or AmpC β-lactamases presumably combined with additional resistance mechanisms (Tables S2A and B and S10A and B). In countries where MBLs were the major cause of nonsusceptibility to meropenem, the susceptibility of meropenem-nonsusceptible isolates to ceftazidime-avibactam was reduced.
TABLE 4.
In vitro activities of ceftazidime-avibactam and comparator antimicrobial agents tested against 149 isolates of meropenem-nonsusceptible Enterobacteriaceae and 457 isolates of meropenem-nonsusceptible P. aeruginosa collected in the Asia-Pacific region as part of the INFORM global surveillance program from 2012 to 2015
| Organism, phenotype (no. of isolates)a | Antimicrobial agent | MIC (μg/ml)b |
% susceptiblec | ||
|---|---|---|---|---|---|
| 50% | 90% | Range | |||
| Enterobacteriaceae (149) | Ceftazidime-avibactam | 32 | >128 | 0.06 to >128 | 47.7 |
| Ceftazidime | >128 | >128 | 0.5 to >128 | 2.7 | |
| Cefepime | >16 | >16 | ≤0.12 to >16 | 3.4 | |
| Aztreonam | 128 | >128 | ≤0.015 to >128 | 14.8 | |
| Piperacillin-tazobactam | >128 | >128 | 0.5 to >128 | 10.7 | |
| Doripenem | >4 | >4 | 0.03 to >4 | 10.1 | |
| Imipenem | >8 | >8 | 0.12 to >8 | 12.1 | |
| Meropenem | >8 | >8 | 2 to >8 | 0 | |
| Amikacin | 4 | >32 | ≤0.25 to >32 | 69.8 | |
| Colistin (n = 67)d | 1 | >4 | 0.25 to >4 | 85.1 | |
| Tigecycline | 1 | 4 | 0.06 to 8 | 85.9 | |
| Levofloxacin | >4 | >4 | 0.06 to >4 | 26.9 | |
| Enterobacteriaceae, MBL negative (81) | Ceftazidime-avibactam | 1 | 16 | 0.06 to >128 | 87.7 |
| Ceftazidime | 128 | >128 | 0.5 to >128 | 4.9 | |
| Cefepime | >16 | >16 | ≤0.12 to >16 | 3.7 | |
| Aztreonam | >128 | >128 | 0.12 to >128 | 6.2 | |
| Piperacillin-tazobactam | >128 | >128 | 0.5 to >128 | 7.4 | |
| Doripenem | >4 | >4 | 0.03 to >4 | 18.5 | |
| Imipenem | 8 | >8 | 0.12 to >8 | 18.5 | |
| Meropenem | 8 | >8 | 2 to >8 | 0 | |
| Amikacin | 4 | >32 | ≤0.25 to >32 | 75.3 | |
| Colistin (n = 30)d | 0.5 | 4 | 0.25 to >4 | 83.3 | |
| Tigecycline | 1 | 4 | 0.06 to 8 | 84.0 | |
| Levofloxacin | >4 | >4 | 0.25 to >4 | 19.8 | |
| Escherichia coli (19) | Ceftazidime-avibactam | 1 | >128 | 0.06 to >128 | 79.0 |
| Ceftazidime | 128 | >128 | 0.5 to >128 | 10.5 | |
| Cefepime | >16 | >16 | 2 to >16 | 5.3 | |
| Aztreonam | 128 | >128 | 1 to >128 | 10.5 | |
| Piperacillin-tazobactam | >128 | >128 | 1 to >128 | 15.8 | |
| Doripenem | 2 | >4 | 0.03 to >4 | 31.6 | |
| Imipenem | 4 | >8 | 0.12 to >8 | 26.3 | |
| Meropenem | 8 | >8 | 2 to >8 | 0 | |
| Amikacin | 4 | >32 | 1 to >32 | 84.2 | |
| Colistin (n = 3)d | — | — | 0.25 to 1 | 100 | |
| Tigecycline | 0.25 | 1 | 0.06 to 2 | 100 | |
| Levofloxacin | >4 | >4 | 1 to >4 | 10.5 | |
| Escherichia coli, MBL negative (16) | Ceftazidime-avibactam | 0.5 | 4 | 0.06 to >128 | 93.8 |
| Ceftazidime | 128 | >128 | 0.5 to >128 | 12.5 | |
| Cefepime | >16 | >16 | 2 to >16 | 6.3 | |
| Aztreonam | 128 | >128 | 1 to >128 | 12.5 | |
| Piperacillin-tazobactam | >128 | >128 | 1 to >128 | 18.8 | |
| Doripenem | 2 | >4 | 0.03 to >8 | 37.5 | |
| Imipenem | 4 | >8 | 0.12 to >8 | 31.3 | |
| Meropenem | 4 | >8 | 2 to >8 | 0 | |
| Amikacin | 4 | >32 | 1 to >32 | 87.5 | |
| Colistin (n = 2)d | 0.25 | 0.25 | 0.25 to 0.25 | 100 | |
| Tigecycline | 0.25 | 1 | 0.06 to 2 | 100 | |
| Levofloxacin | >4 | 8 | 1 to >8 | 12.5 | |
| Klebsiella pneumoniae (85) | Ceftazidime-avibactam | 4 | >128 | 0.12 to >128 | 51.8 |
| Ceftazidime | >128 | >128 | 0.5 to >128 | 1.2 | |
| Cefepime | >16 | >16 | ≤0.12 to >16 | 2.4 | |
| Aztreonam | 128 | >128 | 0.06 to >128 | 7.1 | |
| Piperacillin-tazobactam | >128 | >128 | 4 to >128 | 4.7 | |
| Doripenem | >4 | >4 | 0.12 to >4 | 8.2 | |
| Imipenem | >8 | >8 | 0.25 to >8 | 11.8 | |
| Meropenem | >8 | >8 | 2 to >8 | 0 | |
| Amikacin | 4 | >32 | ≤0.25 to >32 | 67.1 | |
| Colistin (n = 49)d | 1 | 4 | 0.25 to >4 | 87.8 | |
| Tigecycline | 1 | 4 | 0.06 to 8 | 89.4 | |
| Levofloxacin | >4 | >4 | 0.06 to >4 | 25.9 | |
| Klebsiella pneumoniae, MBL negative (49) | Ceftazidime-avibactam | 1 | 16 | 0.12 to >128 | 89.8 |
| Ceftazidime | >128 | >128 | 0.5 to >128 | 2.0 | |
| Cefepime | >16 | >16 | ≤0.12 to >16 | 4.1 | |
| Aztreonam | >128 | >128 | 0.12 to >128 | 2.0 | |
| Piperacillin-tazobactam | >128 | >128 | 16 to >128 | 2.0 | |
| Doripenem | >4 | >8 | 0.12 to >8 | 14.3 | |
| Imipenem | >8 | >8 | 0.25 to >8 | 14.3 | |
| Meropenem | >8 | >8 | 2 to >8 | 0 | |
| Amikacin | 4 | >32 | ≤0.25 to >32 | 73.5 | |
| Colistin (n = 27)d | 0.5 | 4 | 0.25 to >8 | 85.2 | |
| Tigecycline | 1 | 4 | 0.06 to 8 | 83.7 | |
| Levofloxacin | >4 | >8 | 0.5 to >8 | 14.3 | |
| Klebsiella oxytoca (5) | Ceftazidime-avibactam | — | — | 16 to 128 | 0 |
| Ceftazidime | — | — | 64 to >128 | 0 | |
| Cefepime | — | — | 2 to >16 | 20.0 | |
| Aztreonam | — | — | 1 to >128 | 20.0 | |
| Piperacillin-tazobactam | — | — | 4 to >128 | 20.0 | |
| Doripenem | — | — | 2 to >4 | 0 | |
| Imipenem | — | — | 2 to >8 | 0 | |
| Meropenem | — | — | 4 to >8 | 0 | |
| Amikacin | — | — | 0.5 to >32 | 60.0 | |
| Colistin (n = 1)d | — | — | 1 to 1 | 100 | |
| Tigecycline | — | — | 0.5 to 1 | 100 | |
| Levofloxacin | — | — | 1 to 4 | 60.0 | |
| Klebsiella oxytoca, MBL negative (1) | Ceftazidime-avibactam | — | — | 16 | 0 |
| Ceftazidime | — | — | 128 | 0 | |
| Cefepime | — | — | 4 | 0 | |
| Aztreonam | — | — | 64 | 0 | |
| Piperacillin-tazobactam | — | — | 128 | 0 | |
| Doripenem | — | — | 2 | 0 | |
| Imipenem | — | — | 8 | 0 | |
| Meropenem | — | — | 8 | 0 | |
| Amikacin | — | — | 8 | 100 | |
| Colistin (n = 0)d | — | — | NDe | ND | |
| Tigecycline | — | — | 1 | 100 | |
| Levofloxacin | — | — | 2 | 100 | |
| Enterobacter spp.f (24) | Ceftazidime-avibactam | >128 | >128 | 1 to >128 | 25.0 |
| Ceftazidime | >128 | >128 | 2 to >128 | 4.2 | |
| Cefepime | >16 | >16 | 1 to >16 | 4.2 | |
| Aztreonam | 128 | >128 | 0.06 to >128 | 25.0 | |
| Piperacillin-tazobactam | >128 | >128 | 4 to >128 | 12.5 | |
| Doripenem | >4 | >4 | 1 to >4 | 4.2 | |
| Imipenem | >8 | >8 | 0.5 to >8 | 4.2 | |
| Meropenem | >8 | >8 | 2 to >8 | 0 | |
| Amikacin | 4 | >32 | 0.5 to >32 | 75.0 | |
| Colistin (n = 10)d | 0.5 | >4 | 0.5 to >4 | 80.0 | |
| Tigecycline | 2 | 4 | 0.5 to 8 | 58.3 | |
| Levofloxacin | >4 | >4 | 0.06 to >4 | 25.0 | |
| Enterobacter spp., MBL negative (7) | Ceftazidime-avibactam | — | — | 1 to 16 | 85.7 |
| Ceftazidime | — | — | 2 to >128 | 14.3 | |
| Cefepime | — | — | 8 to >16 | 0 | |
| Aztreonam | — | — | 64 to >128 | 0 | |
| Piperacillin-tazobactam | — | — | 128 to >128 | 0 | |
| Doripenem | — | — | 1 to >4 | 14.3 | |
| Imipenem | — | — | 0.5 to >8 | 14.3 | |
| Meropenem | — | — | 2 to >8 | 0 | |
| Amikacin | — | — | 0.5 to >32 | 71.4 | |
| Colistin (n = 0)d | — | — | NDe | ND | |
| Tigecycline | — | — | 0.5 to 4 | 42.9 | |
| Levofloxacin | — | — | 0.25 to >4 | 42.9 | |
| Citrobacter spp.g (8) | Ceftazidime-avibactam | — | — | 0.25 to >128 | 37.5 |
| Ceftazidime | — | — | 16 to >128 | 0 | |
| Cefepime | — | — | >16 to >16 | 0 | |
| Aztreonam | — | — | 0.5 to >128 | 25.0 | |
| Piperacillin-tazobactam | — | — | 16 to >128 | 12.5 | |
| Doripenem | — | — | 2 to >4 | 0 | |
| Imipenem | — | — | 1 to >8 | 12.5 | |
| Meropenem | — | — | 2 to >8 | 0 | |
| Amikacin | — | — | 2 to >32 | 62.5 | |
| Colistin (n = 2)d | — | — | 1 to 1 | 100 | |
| Tigecycline | — | — | 0.5 to 1 | 100 | |
| Levofloxacin | — | — | 0.06 to >4 | 50.0 | |
| Citrobacter spp., MBL negative (4) | Ceftazidime-avibactam | — | — | 0.25 to 128 | 75.0 |
| Ceftazidime | — | — | 16 to >128 | 0 | |
| Cefepime | — | — | >16 to >16 | 0 | |
| Aztreonam | — | — | 64 to >128 | 0 | |
| Piperacillin-tazobactam | — | — | 64 to >128 | 0 | |
| Doripenem | — | — | 2 to >4 | 0 | |
| Imipenem | — | — | 1 to >8 | 25.0 | |
| Meropenem | — | — | 4 to >8 | 0 | |
| Amikacin | — | — | 4 to >32 | 50.0 | |
| Colistin (n = 0)d | — | — | ND | ND | |
| Tigecycline | — | — | 0.5 to 1 | 100 | |
| Levofloxacin | — | — | 1 to >4 | 25.0 | |
| Proteeaeh (6) | Ceftazidime-avibactam | — | — | 0.06 to 64 | 33.3 |
| Ceftazidime | — | — | 8 to >128 | 0 | |
| Cefepime | — | — | 4 to >16 | 0 | |
| Aztreonam | — | — | ≤0.015 to 32 | 66.7 | |
| Piperacillin-tazobactam | — | — | 0.5 to 128 | 50.0 | |
| Doripenem | — | — | 0.25 to >4 | 16.7 | |
| Imipenem | — | — | 1 to >8 | 16.7 | |
| Meropenem | — | — | 2 to >8 | 0 | |
| Amikacin | — | — | 1 to >32 | 50.0 | |
| Colistin (n = 2)d | — | — | >4 to >4 | 0 | |
| Tigecycline | — | — | 0.5 to 8 | 66.7 | |
| Levofloxacin | — | — | 0.12 to >4 | 33.3 | |
| Proteeae, MBL negative (3) | Ceftazidime-avibactam | — | — | 0.06 to 32 | 66.7 |
| Ceftazidime | — | — | 8 to 128 | 0 | |
| Cefepime | — | — | 8 to >16 | 0 | |
| Aztreonam | — | — | 0.5 to 32 | 66.7 | |
| Piperacillin-tazobactam | — | — | 0.5 to 32 | 66.7 | |
| Doripenem | — | — | 0.25 to >4 | 33.3 | |
| Imipenem | — | — | 1 to >8 | 33.3 | |
| Meropenem | — | — | 2 to >8 | 0 | |
| Amikacin | — | — | 1 to >32 | 66.7 | |
| Colistin (n = 1)d | — | — | >4 | 0 | |
| Tigecycline | — | — | 0.5 to 8 | 66.7 | |
| Levofloxacin | — | — | 2 to >4 | 33.3 | |
| Other Enterobacteriaceaei (2) | Ceftazidime-avibactam | — | — | 1 to >128 | 50.0 |
| Ceftazidime | — | — | 16 to >128 | 0 | |
| Cefepime | — | — | >16 to >16 | 0 | |
| Aztreonam | — | — | 0.25 to >128 | 50.0 | |
| Piperacillin-tazobactam | — | — | 4 to >128 | 50.0 | |
| Doripenem | — | — | >4 to >4 | 0 | |
| Imipenem | — | — | >8 to >8 | 0 | |
| Meropenem | — | — | 8 to >8 | 0 | |
| Amikacin | — | — | 0.5 to 1 | 100 | |
| Colistin (n = 0)d | — | — | NDe | ND | |
| Tigecycline | — | — | 0.5 to 1 | 100 | |
| Levofloxacin | — | — | 2 to >4 | 50.0 | |
| Other Enterobacteriaceae, MBL negative (1) | Ceftazidime-avibactam | — | — | 1 | 100 |
| Ceftazidime | — | — | 16 | 0 | |
| Cefepime | — | — | >16 | 0 | |
| Aztreonam | — | — | >128 | 0 | |
| Piperacillin-tazobactam | — | — | >128 | 0 | |
| Doripenem | — | — | >4 | 0 | |
| Imipenem | — | — | >8 | 0 | |
| Meropenem | — | — | >8 | 0 | |
| Amikacin | — | — | 0.5 | 100 | |
| Colistin (n = 0)d | — | — | NDe | ND | |
| Tigecycline | — | — | 1 | 100 | |
| Levofloxacin | — | — | 2 | 100 | |
| Pseudomonas aeruginosa (457) | Ceftazidime-avibactam | 8 | 128 | 0.25 to >128 | 70.5 |
| Ceftazidime | 16 | >128 | 0.25 to >128 | 45.3 | |
| Cefepime | 16 | >16 | 1 to >16 | 44.6 | |
| Aztreonam | 32 | 128 | 0.5 to >128 | 28.7 | |
| Piperacillin-tazobactam | 64 | >128 | 1 to >128 | 31.1 | |
| Doripenem | >4 | >4 | 0.5 to >4 | 9.6 | |
| Imipenem | >8 | >8 | 0.5 to >8 | 5.5 | |
| Meropenem | >8 | >8 | 4 to >8 | 0 | |
| Amikacin | 8 | >32 | ≤0.25 to >32 | 77.9 | |
| Colistin (n = 286)d | 2 | 2 | 0.25 to 8 | 96.2 | |
| Levofloxacin | >4 | >4 | 0.12 to >4 | 39.8 | |
| Pseudomonas aeruginosa, MBL negative (384) | Ceftazidime-avibactam | 4 | 32 | 0.25 to >128 | 83.6 |
| Ceftazidime | 8 | 128 | 0.25 to >128 | 53.7 | |
| Cefepime | 8 | >16 | 1 to >16 | 52.6 | |
| Aztreonam | 32 | 128 | 0.5 to >128 | 28.9 | |
| Piperacillin-tazobactam | 32 | >128 | 1 to >128 | 35.7 | |
| Doripenem | >4 | 4 | 0.5 to >4 | 11.5 | |
| Imipenem | >8 | >8 | 0.5 to >8 | 6.5 | |
| Meropenem | 8 | >8 | 4 to >8 | 0 | |
| Amikacin | 4 | 32 | ≤0.25 to >32 | 87.2 | |
| Colistin (n = 233)d | 2 | 2 | 0.25 to 8 | 96.6 | |
| Levofloxacin | 4 | >4 | 0.12 to >4 | 46.6 | |
MBL negative, no gene encoding a metallo-β-lactamase was detected by PCR.
—, MIC50 and MIC90 were not calculated for <10 isolates.
Percent susceptibility was determined according to CLSI 2016 breakpoints, with the exception of that to ceftazidime-avibactam and tigecycline, where U.S. FDA breakpoints were applied. Because CLSI colistin breakpoints are not available for the Enterobacteriaceae, susceptibility for this organism group was determined using the EUCAST 2016 breakpoints for colistin.
Values are for colistin tested without 0.002% polysorbate 80; data are for isolates collected in 2014 and 2015 only.
ND, not determined; MIC range and percent susceptible were not calculated for 0 isolates.
The Enterobacter spp. included Enterobacter aerogenes (n = 3), Enterobacter asburiae (n = 3), and Enterobacter cloacae (n = 18).
The Citrobacter spp. included Citrobacter braakii (n = 1), Citrobacter freundii (n = 4), and Citrobacter koseri (n = 3).
The Proteeae included Proteus mirabilis (n = 3), Proteus vulgaris (n = 1), Providencia rettgeri (n = 1), and Providencia stuartii (n = 1).
Other Enterobacteriaceae included Serratia marcescens (n = 2).
Table 5 shows the in vitro activity of ceftazidime-avibactam and the comparator agents against 106 colistin-resistant isolates of Enterobacteriaceae (excluding data for the Proteeae and Serratia spp., which are intrinsically resistant to colistin). The percentage of isolates that were resistant to colistin ranged from <1% in Japan, South Korea, and Taiwan to 2.1% in Hong Kong and 2.3% in Philippines (Fig. S2). Ceftazidime-avibactam inhibited 96.2% of isolates at the MIC breakpoint (MIC90, 1 μg/ml) and demonstrated activity similar to that of amikacin (96.2% susceptible) and tigecycline (98.1% susceptible). Ceftazidime-avibactam inhibited colistin-resistant isolates of Enterobacteriaceae from all countries (Tables S2A to S10A).
TABLE 5.
In vitro activities of ceftazidime-avibactam and comparator antimicrobial agents tested against 106 isolates of colistin-resistant Enterobacteriaceae and 6 isolates of colistin-resistant P. aeruginosa collected in the Asia-Pacific region as part of the INFORM global surveillance program from 2012 to 2015
| Organism, phenotype (no. of isolates)a | Antimicrobial agent | MIC (μg/ml)b |
% susceptiblec | ||
|---|---|---|---|---|---|
| 50% | 90% | Range | |||
| Enterobacteriaceaed (106) | Ceftazidime-avibactam | 0.25 | 1 | ≤0.015 to >128 | 96.2 |
| Ceftazidime | 2 | >128 | 0.06 to >128 | 55.7 | |
| Cefepime | ≤0.12 | >16 | ≤0.12 to >16 | 71.7 | |
| Aztreonam | 0.5 | 128 | ≤0.015 to >128 | 58.5 | |
| Piperacillin-tazobactam | 4 | >128 | 0.5 to >128 | 74.5 | |
| Doripenem | 0.06 | 0.5 | 0.03 to >4 | 93.4 | |
| Imipenem | 0.5 | 4 | 0.06 to >8 | 80.2 | |
| Meropenem | 0.06 | 0.25 | 0.015 to >8 | 92.5 | |
| Amikacin | 2 | 8 | 1 to >32 | 96.2 | |
| Colistin (n = 106)e | >4 | >4 | 4 to >4 | 0 | |
| Tigecycline | 0.5 | 2 | 0.12 to 8 | 98.1 | |
| Levofloxacin | 0.12 | >4 | ≤0.03 to >4 | 75.5 | |
| Enterobacteriaceae, MBL negatived (102) | Ceftazidime-avibactam | 0.25 | 1 | ≤0.015 to 4 | 100 |
| Ceftazidime | 1 | 128 | 0.06 to >128 | 57.8 | |
| Cefepime | ≤0.12 | >16 | ≤0.12 to >16 | 73.5 | |
| Aztreonam | 0.5 | 128 | ≤0.015 to >128 | 58.8 | |
| Piperacillin-tazobactam | 4 | >128 | 0.5 to >128 | 77.5 | |
| Doripenem | 0.06 | 0.25 | 0.03 to 4 | 97.1 | |
| Imipenem | 0.5 | 2 | 0.06 to 4 | 83.3 | |
| Meropenem | 0.06 | 0.25 | 0.015 to 8 | 96.1 | |
| Amikacin | 2 | 8 | 1 to >32 | 98.0 | |
| Colistin (n = 102)e | >4 | >4 | 4 to >4 | 0 | |
| Tigecycline | 0.5 | 1 | 0.12 to 8 | 98.0 | |
| Levofloxacin | 0.12 | >4 | ≤0.03 to >4 | 77.5 | |
| Escherichia coli (12) | Ceftazidime-avibactam | 0.12 | 0.25 | 0.03 to 0.5 | 100 |
| Ceftazidime | 16 | 64 | 0.12 to >128 | 41.7 | |
| Cefepime | 4 | >16 | ≤0.12 to >16 | 41.7 | |
| Aztreonam | 16 | 64 | 0.06 to 64 | 41.7 | |
| Piperacillin-tazobactam | 2 | 8 | 0.5 to >128 | 91.7 | |
| Doripenem | 0.06 | 0.12 | 0.03 to 0.12 | 100 | |
| Imipenem | 0.25 | 0.5 | 0.12 to 2 | 91.7 | |
| Meropenem | 0.03 | 0.06 | 0.03 to 0.12 | 100 | |
| Amikacin | 4 | 4 | 2 to 8 | 100 | |
| Colistin (n = 12)e | 4 | 4 | 4 to >4 | 0 | |
| Tigecycline | 0.25 | 1 | 0.12 to 2 | 100 | |
| Levofloxacin | 2 | >4 | ≤0.03 to >4 | 58.3 | |
| Klebsiella pneumoniae (27) | Ceftazidime-avibactam | 0.5 | 4 | 0.12 to >128 | 92.6 |
| Ceftazidime | 128 | >128 | 0.12 to >128 | 25.9 | |
| Cefepime | >16 | >16 | ≤0.12 to >16 | 33.3 | |
| Aztreonam | 128 | >128 | 0.06 to >128 | 29.6 | |
| Piperacillin-tazobactam | 64 | >128 | 0.5 to >128 | 44.4 | |
| Doripenem | 0.12 | >4 | 0.03 to >4 | 81.5 | |
| Imipenem | 0.5 | 4 | 0.06 to >8 | 74.1 | |
| Meropenem | 0.06 | 8 | 0.03 to >8 | 77.8 | |
| Amikacin | 2 | >32 | 1 to >32 | 85.2 | |
| Colistin (n = 27)e | >4 | >4 | 4 to >4 | 0 | |
| Tigecycline | 0.5 | 2 | 0.12 to 8 | 92.6 | |
| Levofloxacin | >4 | >4 | 0.06 to >4 | 37.0 | |
| Klebsiella pneumoniae, MBL negative (25) | Ceftazidime-avibactam | 0.5 | 2 | 0.12 to 4 | 100 |
| Ceftazidime | 128 | >128 | 0.12 to >128 | 28.0 | |
| Cefepime | >16 | >16 | ≤0.12 to >16 | 36.0 | |
| Aztreonam | 128 | >128 | 0.06 to >128 | 32.0 | |
| Piperacillin-tazobactam | 32 | >128 | 0.5 to >128 | 48.0 | |
| Doripenem | 0.12 | 4 | 0.03 to >4 | 88.0 | |
| Imipenem | 0.5 | 4 | 0.06 to 4 | 80.0 | |
| Meropenem | 0.06 | 2 | 0.03 to 8 | 84.0 | |
| Amikacin | 2 | 16 | 1 to >32 | 92.0 | |
| Colistin (n = 25)e | >4 | >4 | 4 to >4 | 0 | |
| Tigecycline | 0.5 | 2 | 0.12 to 8 | 92.0 | |
| Levofloxacin | >4 | >4 | 0.06 to >4 | 40.0 | |
| Klebsiella oxytoca (2) | Ceftazidime-avibactam | — | — | 0.12 to 0.12 | 100 |
| Ceftazidime | — | — | 0.12 to 0.12 | 100 | |
| Cefepime | — | — | ≤0.12 to ≤0.12 | 100 | |
| Aztreonam | — | — | 0.5 to 0.5 | 100 | |
| Piperacillin-tazobactam | — | — | 2 to 4 | 100 | |
| Doripenem | — | — | 0.06 to 0.06 | 100 | |
| Imipenem | — | — | 0.25 to 0.5 | 100 | |
| Meropenem | — | — | 0.06 to 0.06 | 100 | |
| Amikacin | — | — | 2 to 2 | 100 | |
| Colistin (n = 2)e | — | — | 4 to 4 | 0 | |
| Tigecycline | — | — | 0.5 to 0.5 | 100 | |
| Levofloxacin | — | — | 0.06 to 0.12 | 100 | |
| Enterobacter spp.f (64) | Ceftazidime-avibactam | 0.25 | 1 | 0.03 to >128 | 96.9 |
| Ceftazidime | 0.5 | 128 | 0.12 to >128 | 68.8 | |
| Cefepime | ≤0.12 | 2 | ≤0.12 to >16 | 92.2 | |
| Aztreonam | 0.12 | 32 | ≤0.015 to 64 | 71.9 | |
| Piperacillin-tazobactam | 4 | 64 | 0.5 to >128 | 82.8 | |
| Doripenem | 0.06 | 0.25 | 0.03 to >4 | 96.9 | |
| Imipenem | 1 | 4 | 0.25 to >8 | 81.3 | |
| Meropenem | 0.06 | 0.25 | 0.015 to >8 | 96.9 | |
| Amikacin | 2 | 2 | 1 to 16 | 100 | |
| Colistin (n = 64)e | >4 | >4 | 4 to >4 | 0 | |
| Tigecycline | 0.5 | 1 | 0.12 to 2 | 100 | |
| Levofloxacin | 0.06 | 1 | ≤0.03 to >4 | 93.8 | |
| Enterobacter spp., MBL negative (62) | Ceftazidime-avibactam | 0.25 | 0.5 | 0.03 to 2 | 100 |
| Ceftazidime | 0.5 | 128 | 0.12 to 128 | 71.0 | |
| Cefepime | ≤0.12 | 2 | ≤0.12 to >16 | 93.6 | |
| Aztreonam | 0.12 | 32 | ≤0.015 to 64 | 71.0 | |
| Piperacillin-tazobactam | 4 | 32 | 0.5 to >128 | 85.5 | |
| Doripenem | 0.06 | 0.25 | 0.03 to 0.5 | 100 | |
| Imipenem | 1 | 2 | 0.25 to 4 | 83.9 | |
| Meropenem | 0.06 | 0.12 | 0.015 to 0.5 | 100 | |
| Amikacin | 2 | 2 | 1 to 16 | 100 | |
| Colistin (n = 62)e | >4 | >4 | 4 to >4 | 0 | |
| Tigecycline | 0.5 | 1 | 0.12 to 2 | 100 | |
| Levofloxacin | 0.06 | 1 | ≤0.03 to >4 | 95.2 | |
| Citrobacter sp.g (1) | Ceftazidime-avibactam | — | — | ≤0.015 | 100 |
| Ceftazidime | — | — | 0.06 | 100 | |
| Cefepime | — | — | ≤0.12 | 100 | |
| Aztreonam | — | — | ≤0.015 | 100 | |
| Piperacillin-tazobactam | — | — | 0.5 | 100 | |
| Doripenem | — | — | 0.25 | 100 | |
| Imipenem | — | — | 4 | 0 | |
| Meropenem | — | — | 0.12 | 100 | |
| Amikacin | — | — | 2 | 100 | |
| Colistin (n = 1)e | — | — | >4 | 0 | |
| Tigecycline | — | — | 1 | 100 | |
| Levofloxacin | — | — | 0.12 | 100 | |
| Pseudomonas aeruginosa (6) | Ceftazidime-avibactam | — | — | 2 to 16 | 83.3 |
| Ceftazidime | — | — | 1 to 128 | 66.7 | |
| Cefepime | — | — | 2 to >16 | 66.7 | |
| Aztreonam | — | — | 8 to 64 | 33.3 | |
| Piperacillin-tazobactam | — | — | 4 to >128 | 66.7 | |
| Doripenem | — | — | 0.25 to >4 | 83.3 | |
| Imipenem | — | — | 1 to >8 | 66.7 | |
| Meropenem | — | — | 0.25 to 8 | 83.3 | |
| Amikacin | — | — | 2 to 16 | 100 | |
| Colistin (n = 6)e | — | — | 8 to >8 | 0 | |
| Levofloxacin | — | — | 0.5 to 4 | 83.3 | |
| Pseudomonas aeruginosa, MBL negative (6) | Ceftazidime-avibactam | — | — | 2 to 16 | 83.3 |
| Ceftazidime | — | — | 1 to 128 | 66.7 | |
| Cefepime | — | — | 2 to >16 | 66.7 | |
| Aztreonam | — | — | 8 to 64 | 33.3 | |
| Piperacillin-tazobactam | — | — | 4 to >128 | 66.7 | |
| Doripenem | — | — | 0.25 to >4 | 83.3 | |
| Imipenem | — | — | 1 to >8 | 66.7 | |
| Meropenem | — | — | 0.25 to 8 | 83.3 | |
| Amikacin | — | — | 2 to 16 | 100 | |
| Colistin (n = 6)e | — | — | 8 to >8 | 0 | |
| Levofloxacin | — | — | 0.5 to 4 | 83.3 | |
MBL negative, no gene encoding a metallo-β-lactamase was detected by PCR.
—, MIC50 and MIC90 were not calculated for <10 isolates.
Percent susceptibility was determined according to CLSI 2016 breakpoints, with the exception of that to ceftazidime-avibactam and tigecycline, where U.S. FDA breakpoints were applied. Because CLSI colistin breakpoints are not available for the Enterobacteriaceae, susceptibility for this organism group was determined using the EUCAST 2016 breakpoints for colistin.
Excludes isolates of Proteeae and Serratia spp., which are intrinsically resistant to colistin.
Values are for colistin tested without 0.002% polysorbate 80; data are for isolates collected in 2014 and 2015 only.
The Enterobacter spp. included Enterobacter asburiae (n = 28), Enterobacter cloacae (n = 33), and Enterobacter kobei (n = 3).
The Citrobacter sp. included Citrobacter freundii (n = 1).
Table 6 depicts the in vitro activity of ceftazidime-avibactam and the comparator agents against 1,193 isolates of MDR Enterobacteriaceae. The rate of susceptibility to ceftazidime-avibactam was 93.8% (MIC90, 2 μg/ml), which was similar to that for tigecycline (93.4% susceptible), and exceeded the rates of susceptibility to doripenem (89.4% susceptible), meropenem (88.8% susceptible), colistin (88.2% susceptible), amikacin (84.8% susceptible), imipenem (84.0% susceptible), and the other tested agents (<50% susceptible). The MIC90 values of ceftazidime-avibactam against MDR isolates varied from 0.5 to >128 μg/ml for different species of Enterobacteriaceae. Greater than 90% of E. coli, K. pneumoniae, and Proteeae isolates were susceptible to ceftazidime-avibactam (MIC90, 0.5 to 4 μg/ml). The reduced activity of ceftazidime-avibactam against K. oxytoca (83.3% susceptible; MIC90, 128 μg/ml), Enterobacter spp. (76.3% susceptible; MIC90, >128 μg/ml), and Citrobacter spp. (81.8% susceptible; MIC90, 128 μg/ml) could be attributed to the relatively high number of MBL-producing isolates among these species subsets. Again, the activity of ceftazidime-avibactam was much greater (as reflected by lower MIC values) against all and species-specific subsets of MDR MBL-negative isolates (98.8% susceptible; MIC90, 1 μg/ml). The rate of susceptibility to ceftazidime-avibactam was greater than or comparable to that of colistin, tigecycline, and carbapenems among MDR isolates of all species of Enterobacteriaceae.
TABLE 6.
In vitro activities of ceftazidime-avibactam and comparator antimicrobial agents tested against 1,193 isolates of MDR Enterobacteriaceae and 301 isolates of MDR P. aeruginosa collected in the Asia-Pacific region as part of the INFORM global surveillance program from 2012 to 2015
| Organism, genotype (no. of isolates)a | Antimicrobial agent | MIC (μg/ml)b |
% susceptiblec | ||
|---|---|---|---|---|---|
| 50% | 90% | Range | |||
| Enterobacteriaceae (1,193) | Ceftazidime-avibactam | 0.25 | 2 | ≤0.015 to >128 | 93.8 |
| Ceftazidime | 64 | >128 | 0.06 to >128 | 6.5 | |
| Cefepime | >16 | >16 | ≤0.12 to >16 | 5.1 | |
| Aztreonam | 64 | >128 | ≤0.015 to >128 | 2.6 | |
| Piperacillin-tazobactam | 32 | >128 | ≤0.25 to >128 | 46.9 | |
| Doripenem | 0.06 | 2 | 0.015 to >4 | 89.4 | |
| Imipenem | 0.25 | 4 | 0.06 to >8 | 84.0 | |
| Meropenem | 0.06 | 2 | 0.015 to >8 | 88.8 | |
| Amikacin | 4 | >32 | ≤0.25 to >32 | 84.8 | |
| Colistin (n = 509)d | 0.5 | 4 | 0.25 to >4 | 88.2 | |
| Tigecycline | 0.5 | 2 | 0.06 to 8 | 93.4 | |
| Levofloxacin | >4 | >4 | ≤0.03 to >4 | 11.8 | |
| Enterobacteriaceae, MBL negative (1,133) | Ceftazidime-avibactam | 0.25 | 1 | ≤0.015 to >128 | 98.8 |
| Ceftazidime | 64 | >128 | 0.06 to >128 | 6.8 | |
| Cefepime | >16 | >16 | ≤0.12 to >16 | 5.3 | |
| Aztreonam | 64 | >128 | ≤0.015 to >128 | 1.8 | |
| Piperacillin-tazobactam | 32 | >128 | ≤0.25 to >128 | 49.1 | |
| Doripenem | 0.06 | 0.5 | 0.015 to >4 | 94.1 | |
| Imipenem | 0.25 | 2 | 0.06 to >8 | 88.2 | |
| Meropenem | 0.06 | 0.25 | 0.015 to >8 | 93.3 | |
| Amikacin | 4 | >32 | ≤0.25 to >32 | 86.2 | |
| Colistin (n = 476)d | 0.5 | 4 | 0.25 to >4 | 88.2 | |
| Tigecycline | 0.5 | 2 | 0.06 to 8 | 93.7 | |
| Levofloxacin | >4 | >4 | ≤0.03 to >4 | 10.9 | |
| Escherichia coli (569) | Ceftazidime-avibactam | 0.25 | 0.5 | ≤0.015 to >128 | 99.3 |
| Ceftazidime | 32 | >128 | 0.5 to >128 | 7.0 | |
| Cefepime | >16 | >16 | 0.25 to >16 | 2.6 | |
| Aztreonam | 64 | >128 | 8 to >128 | 0 | |
| Piperacillin-tazobactam | 8 | >128 | 0.5 to >128 | 76.1 | |
| Doripenem | 0.06 | 0.12 | 0.015 to >4 | 97.9 | |
| Imipenem | 0.25 | 0.5 | 0.06 to >8 | 96.8 | |
| Meropenem | 0.03 | 0.12 | 0.015 to >8 | 97.2 | |
| Amikacin | 4 | 16 | 0.5 to >32 | 92.6 | |
| Colistin (n = 205)d | 0.5 | 1 | 0.25 to 4 | 97.6 | |
| Tigecycline | 0.25 | 0.5 | 0.06 to 4 | 99.8 | |
| Levofloxacin | >4 | >4 | 0.06 to >4 | 1.4 | |
| Escherichia coli, MBL negative (566) | Ceftazidime-avibactam | 0.25 | 0.5 | ≤0.015 to >128 | 99.8 |
| Ceftazidime | 32 | >128 | 0.5 to >128 | 7.1 | |
| Cefepime | >16 | >16 | 0.25 to >16 | 2.7 | |
| Aztreonam | 64 | >128 | 8 to >128 | 0 | |
| Piperacillin-tazobactam | 8 | >128 | 0.5 to >128 | 76.5 | |
| Doripenem | 0.06 | 0.12 | 0.015 to >4 | 98.4 | |
| Imipenem | 0.25 | 0.5 | 0.06 to >8 | 97.4 | |
| Meropenem | 0.03 | 0.12 | 0.015 to >8 | 97.7 | |
| Amikacin | 4 | 16 | 0.5 to >32 | 92.8 | |
| Colistin (n = 204)d | 0.5 | 1 | 0.25 to 4 | 97.6 | |
| Tigecycline | 0.25 | 0.5 | 0.06 to 4 | 99.8 | |
| Levofloxacin | >4 | >4 | 0.06 to >4 | 1.4 | |
| Klebsiella pneumoniae (453) | Ceftazidime-avibactam | 0.5 | 4 | 0.03 to >128 | 91.4 |
| Ceftazidime | 128 | >128 | 0.25 to >128 | 1.3 | |
| Cefepime | >16 | >16 | ≤0.12 to >16 | 3.8 | |
| Aztreonam | 128 | >128 | 0.12 to >128 | 0.9 | |
| Piperacillin-tazobactam | >128 | >128 | 0.5 to >128 | 18.1 | |
| Doripenem | 0.12 | >4 | 0.015 to >4 | 83.2 | |
| Imipenem | 0.5 | >8 | 0.06 to >8 | 78.2 | |
| Meropenem | 0.06 | >8 | 0.015 to >8 | 82.3 | |
| Amikacin | 4 | >32 | ≤0.25 to >32 | 81.0 | |
| Colistin (n = 234)d | 1 | 2 | 0.25 to >4 | 91.5 | |
| Tigecycline | 1 | 2 | 0.06 to 8 | 91.4 | |
| Levofloxacin | >4 | >4 | ≤0.03 to >4 | 20.3 | |
| Klebsiella pneumoniae, MBL negative (421) | Ceftazidime-avibactam | 0.5 | 2 | 0.03 to >128 | 98.3 |
| Ceftazidime | 128 | >128 | 0.25 to >128 | 1.4 | |
| Cefepime | >16 | >16 | ≤0.12 to >16 | 4.0 | |
| Aztreonam | 128 | >128 | 0.12 to >128 | 0.5 | |
| Piperacillin-tazobactam | >128 | >128 | 0.5 to >128 | 19.2 | |
| Doripenem | 0.12 | 2 | 0.015 to >4 | 89.6 | |
| Imipenem | 0.25 | 2 | 0.06 to >8 | 83.6 | |
| Meropenem | 0.06 | 2 | 0.015 to >8 | 88.6 | |
| Amikacin | 4 | >32 | ≤0.25 to >32 | 83.1 | |
| Colistin (n = 214)d | 0.5 | 2 | 0.25 to >4 | 91.6 | |
| Tigecycline | 1 | 2 | 0.06 to 8 | 91.0 | |
| Levofloxacin | >4 | >4 | ≤0.03 to >4 | 18.8 | |
| Klebsiella oxytoca (18) | Ceftazidime-avibactam | 0.5 | 128 | 0.25 to 128 | 83.3 |
| Ceftazidime | 64 | >128 | 1 to >128 | 38.9 | |
| Cefepime | 16 | >16 | 0.5 to >16 | 22.2 | |
| Aztreonam | 128 | >128 | 32 to >128 | 0 | |
| Piperacillin-tazobactam | >128 | >128 | 16 to >128 | 5.6 | |
| Doripenem | 0.12 | >4 | 0.03 to >4 | 83.3 | |
| Imipenem | 0.25 | >8 | 0.12 to >8 | 83.3 | |
| Meropenem | 0.06 | >8 | 0.03 to >8 | 83.3 | |
| Amikacin | 2 | >32 | 1 to >32 | 77.8 | |
| Colistin (n = 5)d | — | — | 0.25 to 1 | 100 | |
| Tigecycline | 0.5 | 1 | 0.25 to 1 | 100 | |
| Levofloxacin | 4 | >4 | 0.06 to >4 | 38.9 | |
| Klebsiella oxytoca, MBL negative (16) | Ceftazidime-avibactam | 0.5 | 4 | 0.25 to 16 | 93.8 |
| Ceftazidime | 16 | >128 | 1 to >128 | 43.8 | |
| Cefepime | 8 | >16 | 0.5 to >16 | 25.0 | |
| Aztreonam | 128 | >128 | 32 to >128 | 0 | |
| Piperacillin-tazobactam | >128 | >128 | 16 to >128 | 6.3 | |
| Doripenem | 0.12 | 0.12 | 0.03 to 2 | 93.8 | |
| Imipenem | 0.25 | 0.5 | 0.12 to 8 | 93.8 | |
| Meropenem | 0.06 | 0.12 | 0.03 to 8 | 93.8 | |
| Amikacin | 2 | >32 | 1 to >32 | 87.5 | |
| Colistin (n = 5)d | 0.25 | 1 | 0.25 to 1 | 100 | |
| Tigecycline | 0.5 | 1 | 0.25 to 1 | 100 | |
| Levofloxacin | >4 | >4 | 0.06 to >4 | 43.8 | |
| Enterobacter spp.e (76) | Ceftazidime-avibactam | 1 | >128 | 0.12 to >128 | 76.3 |
| Ceftazidime | >128 | >128 | 2 to >128 | 4.0 | |
| Cefepime | >16 | >16 | 0.5 to >16 | 14.5 | |
| Aztreonam | 128 | >128 | 0.06 to >128 | 6.6 | |
| Piperacillin-tazobactam | >128 | >128 | 4 to >128 | 13.2 | |
| Doripenem | 0.25 | >4 | 0.03 to >4 | 72.4 | |
| Imipenem | 1 | >8 | 0.25 to >8 | 64.5 | |
| Meropenem | 0.25 | >8 | 0.03 to >8 | 71.1 | |
| Amikacin | 2 | >32 | 0.5 to >32 | 81.6 | |
| Colistin (n = 34)d | 0.5 | >4 | 0.25 to >4 | 70.6 | |
| Tigecycline | 1 | 4 | 0.25 to 8 | 79.0 | |
| Levofloxacin | >4 | >4 | ≤0.03 to >4 | 27.6 | |
| Enterobacter spp., MBL negative (61) | Ceftazidime-avibactam | 1 | 8 | 0.12 to 64 | 95.1 |
| Ceftazidime | 128 | >128 | 2 to >128 | 4.9 | |
| Cefepime | >16 | >16 | 0.5 to >16 | 16.4 | |
| Aztreonam | 128 | >128 | 16 to >128 | 0 | |
| Piperacillin-tazobactam | >128 | >128 | 4 to >128 | 14.8 | |
| Doripenem | 0.12 | 1 | 0.03 to >4 | 90.2 | |
| Imipenem | 1 | 2 | 0.25 to >8 | 80.3 | |
| Meropenem | 0.12 | 2 | 0.03 to >8 | 88.5 | |
| Amikacin | 2 | >32 | 0.5 to >32 | 83.6 | |
| Colistin (n = 24)d | 1 | >4 | 0.25 to >4 | 66.7 | |
| Tigecycline | 1 | 4 | 0.25 to 8 | 83.6 | |
| Levofloxacin | >4 | >4 | ≤0.03 to >4 | 31.2 | |
| Citrobacter spp.f (33) | Ceftazidime-avibactam | 0.5 | 128 | 0.12 to >128 | 81.8 |
| Ceftazidime | 128 | >128 | 4 to >128 | 3.0 | |
| Cefepime | >16 | >16 | ≤0.12 to >16 | 15.2 | |
| Aztreonam | 64 | >128 | 0.5 to >128 | 9.1 | |
| Piperacillin-tazobactam | >128 | >128 | 4 to >128 | 3.0 | |
| Doripenem | 0.12 | >4 | 0.03 to >4 | 75.8 | |
| Imipenem | 1 | >8 | 0.12 to >8 | 72.7 | |
| Meropenem | 0.12 | >8 | 0.03 to >8 | 78.8 | |
| Amikacin | 8 | >32 | 1 to >32 | 63.6 | |
| Colistin (n = 6)d | — | — | 0.25 to 1 | 100 | |
| Tigecycline | 0.5 | 1 | 0.12 to 2 | 100 | |
| Levofloxacin | >4 | >4 | 0.06 to >4 | 18.2 | |
| Citrobacter spp., MBL negative (28) | Ceftazidime-avibactam | 0.5 | 4 | 0.12 to 128 | 96.4 |
| Ceftazidime | 128 | >128 | 4 to >128 | 3.6 | |
| Cefepime | >16 | >16 | ≤0.12 to >16 | 17.9 | |
| Aztreonam | 64 | >128 | 4 to >128 | 3.6 | |
| Piperacillin-tazobactam | >128 | >128 | 4 to >128 | 3.6 | |
| Doripenem | 0.12 | 4 | 0.03 to >4 | 85.7 | |
| Imipenem | 1 | 4 | 0.12 to >8 | 82.1 | |
| Meropenem | 0.06 | 8 | 0.03 to >8 | 85.7 | |
| Amikacin | 8 | >32 | 1 to >32 | 60.7 | |
| Colistin (n = 4)d | 0.25 | 0.5 | 0.25 to 0.5 | 100 | |
| Tigecycline | 0.5 | 2 | 0.12 to 2 | 100 | |
| Levofloxacin | >4 | >4 | 0.12 to >4 | 14.3 | |
| Proteeaeg (35) | Ceftazidime-avibactam | 0.12 | 4 | ≤0.015 to 64 | 91.4 |
| Ceftazidime | 4 | >128 | 0.06 to >128 | 54.3 | |
| Cefepime | >16 | >16 | ≤0.12 to >16 | 25.7 | |
| Aztreonam | 4 | 64 | ≤0.015 to 128 | 51.4 | |
| Piperacillin-tazobactam | 1 | 32 | ≤0.25 to 128 | 85.7 | |
| Doripenem | 0.25 | 2 | 0.06 to >4 | 88.6 | |
| Imipenem | 4 | 8 | 0.5 to >8 | 8.6 | |
| Meropenem | 0.12 | 2 | 0.03 to >8 | 88.6 | |
| Amikacin | >32 | >32 | 1 to >32 | 34.3 | |
| Colistin (n = 19)d | >4 | >4 | >4 to >4 | 0 | |
| Tigecycline | 4 | 8 | 0.5 to 8 | 42.9 | |
| Levofloxacin | >4 | >4 | 0.12 to >4 | 5.7 | |
| Proteeae, MBL negative (33) | Ceftazidime-avibactam | 0.12 | 1 | ≤0.015 to 32 | 97.0 |
| Ceftazidime | 4 | >128 | 0.06 to >128 | 57.6 | |
| Cefepime | >16 | >16 | ≤0.12 to >16 | 27.3 | |
| Aztreonam | 4 | 64 | ≤0.015 to 128 | 51.5 | |
| Piperacillin-tazobactam | 1 | 32 | ≤0.25 to 128 | 87.9 | |
| Doripenem | 0.25 | 0.5 | 0.06 to >4 | 93.9 | |
| Imipenem | 2 | 4 | 0.5 to >8 | 9.1 | |
| Meropenem | 0.12 | 0.25 | 0.03 to >8 | 93.9 | |
| Amikacin | >32 | >32 | 1 to >32 | 36.4 | |
| Colistin (n = 19)d | >4 | >4 | >4 to >4 | 0 | |
| Tigecycline | 4 | 8 | 0.5 to 8 | 42.4 | |
| Levofloxacin | >4 | >4 | 0.12 to >4 | 6.1 | |
| Other Enterobacteriaceaeh (9) | Ceftazidime-avibactam | — | — | 0.12 to >128 | 88.9 |
| Ceftazidime | — | — | 4 to >128 | 11.1 | |
| Cefepime | — | — | 4 to >16 | 0 | |
| Aztreonam | — | — | 0.25 to >128 | 11.1 | |
| Piperacillin-tazobactam | — | — | 4 to >128 | 33.3 | |
| Doripenem | — | — | 0.06 to >4 | 77.8 | |
| Imipenem | — | — | 0.25 to >8 | 66.7 | |
| Meropenem | — | — | 0.06 to >8 | 77.8 | |
| Amikacin | — | — | 0.5 to 16 | 100 | |
| Colistin (n = 6)d | — | — | >4 to >4 | 0 | |
| Tigecycline | — | — | 0.5 to 8 | 66.7 | |
| Levofloxacin | — | — | 0.12 to >4 | 55.6 | |
| Other Enterobacteriaceae, MBL negative (8) | Ceftazidime-avibactam | — | — | 0.12 to 1 | 100 |
| Ceftazidime | — | — | 4 to >128 | 12.5 | |
| Cefepime | — | — | 4 to >16 | 0 | |
| Aztreonam | — | — | 16 to >128 | 0 | |
| Piperacillin-tazobactam | — | — | 4 to >128 | 25.0 | |
| Doripenem | — | — | 0.06 to >4 | 87.5 | |
| Imipenem | — | — | 0.25 to >8 | 75.0 | |
| Meropenem | — | — | 0.06 to >8 | 87.5 | |
| Amikacin | — | — | 0.5 to 16 | 100 | |
| Colistin (n = 6)d | — | — | >4 to >4 | 0 | |
| Tigecycline | — | — | 0.5 to 8 | 62.5 | |
| Levofloxacin | — | — | 0.12 to >4 | 62.5 | |
| Pseudomonas aeruginosa (301) | Ceftazidime-avibactam | 8 | >128 | 1 to >128 | 52.8 |
| Ceftazidime | 64 | >128 | 1 to >128 | 8.3 | |
| Cefepime | >16 | >16 | 4 to >16 | 6.3 | |
| Aztreonam | 32 | >128 | 4 to >128 | 9.3 | |
| Piperacillin-tazobactam | >128 | >128 | 4 to >128 | 4.0 | |
| Doripenem | >4 | >4 | 0.06 to >4 | 19.3 | |
| Imipenem | >8 | >8 | 0.5 to >8 | 18.9 | |
| Meropenem | >8 | >8 | 0.12 to >8 | 19.6 | |
| Amikacin | 8 | >32 | 0.5 to >32 | 68.4 | |
| Colistin (n = 193)d | 2 | 2 | 0.25 to >8 | 94.8 | |
| Levofloxacin | >4 | >4 | 0.12 to >4 | 20.3 | |
| Pseudomonas aeruginosa, MBL negative (233) | Ceftazidime-avibactam | 8 | 64 | 1 to >128 | 68.2 |
| Ceftazidime | 64 | >128 | 1 to >128 | 10.7 | |
| Cefepime | >16 | >16 | 4 to >16 | 8.2 | |
| Aztreonam | 64 | >128 | 4 to >128 | 4.3 | |
| Piperacillin-tazobactam | >128 | >128 | 4 to >128 | 3.4 | |
| Doripenem | >4 | >4 | 0.06 to >4 | 24.9 | |
| Imipenem | >8 | >8 | 0.5 to >8 | 24.5 | |
| Meropenem | >8 | >8 | 0.12 to >8 | 25.3 | |
| Amikacin | 8 | >32 | 0.5 to >32 | 80.3 | |
| Colistin (n = 143)d | 2 | 2 | 0.25 to >8 | 95.1 | |
| Levofloxacin | >4 | >4 | 0.12 to >4 | 25.8 | |
MBL negative, no gene encoding a metallo-β-lactamase was detected by PCR.
—, MIC50 and MIC90 were not calculated for <10 isolates.
Percent susceptibility was determined according to CLSI 2016 breakpoints, with the exception of that to ceftazidime-avibactam and tigecycline, where U.S. FDA breakpoints were applied. Because CLSI colistin breakpoints are not available for the Enterobacteriaceae, susceptibility for this organism group was determined using the EUCAST 2016 breakpoints for colistin.
Values are for colistin tested without 0.002% polysorbate 80; data are for isolates collected in 2014 and 2015 only.
The Enterobacter spp. included Enterobacter aerogenes (n = 11), Enterobacter asburiae (n = 5), Enterobacter cloacae (n = 59), and Enterobacter kobei (n = 1).
The Citrobacter spp. included Citrobacter braakii (n = 2), Citrobacter freundii (n = 21), and Citrobacter koseri (n = 10).
The Proteeae included Morganella morganii (n = 5), Proteus mirabilis (n = 26), Proteus vulgaris (n = 1), and Providencia stuartii (n = 3).
Other Enterobacteriaceae included Serratia marcescens (n = 9).
MDR rates ranged from 2.7% (Australia) to 19.4% (Thailand) across the nine Asia-Pacific countries (Fig. 2). In eight countries, ≥93.5% of MDR isolates of Enterobacteriaceae remained susceptible to ceftazidime-avibactam (Tables 7 and S2A to S10A). The activity of ceftazidime-avibactam was lowest against MDR isolates collected in Philippines (83.8% susceptible) (Table 7). The reduced activity observed can be explained by the relatively high number of MBL-producing isolates, as the activity of ceftazidime-avibactam against subsets of MBL-negative MDR isolates (98.0% susceptible) was restored (Tables 7 and S7A). Ceftazidime-avibactam demonstrated activity comparable or superior to that of all other agents tested against MDR isolates collected from individual countries (Tables S2A to S10A).
FIG 2.
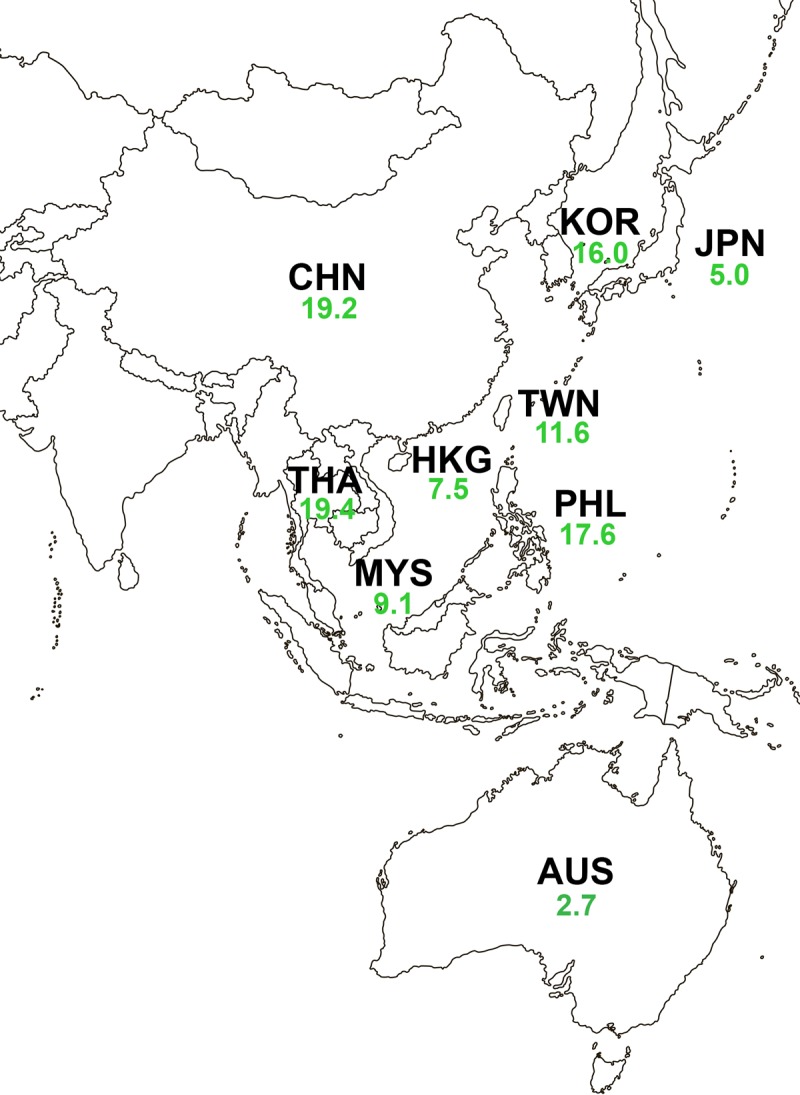
Percentage of isolates of Enterobacteriaceae collected from 2012 to 2015 that were multidrug resistant (MDR), by Asia-Pacific country. Multidrug resistance was defined as resistance by CLSI criteria to three or more sentinel antimicrobial agents from different chemical classes. The green font indicates that <20% of isolates were MDR. Country abbreviations are as follows: AUS, Australia; CHN, China; HKG, Hong Kong; JPN, Japan; KOR, South Korea; MYS, Malaysia; PHL, Philippines; TWN, Taiwan; THA, Thailand. No isolates were obtained from patients in mainland China in 2014 or 2015 or patients in Hong Kong in 2015 due to export restrictions.
TABLE 7.
In vitro activity of ceftazidime-avibactam tested against 1,193 isolates of MDR Enterobacteriaceae and 301 isolates of MDR P. aeruginosa stratified by Asia-Pacific region country of origin and collected as part of the INFORM global surveillance program from 2012 to 2015
| Organism, genotypea | Country (no. of isolates) | Ceftazidime-avibactam MIC (μg/ml)b |
% susceptiblec | ||
|---|---|---|---|---|---|
| 50% | 90% | Range | |||
| Enterobacteriaceae | Australia (36) | 0.25 | 1 | 0.06 to 32 | 97.2 |
| China (239) | 0.25 | 4 | ≤0.015 to >128 | 94.1 | |
| Hong Kong (11)d | 0.12 | 0.5 | 0.06 to 0.5 | 100 | |
| Japan (39)d | 0.25 | 0.5 | ≤0.015 to 8 | 100 | |
| Malaysia (59) | 0.25 | 1 | 0.03 to >128 | 98.3 | |
| Philippines (229) | 0.5 | >128 | 0.03 to >128 | 83.8 | |
| South Korea (190)d | 0.5 | 1 | ≤0.015 to 4 | 100 | |
| Taiwan (145) | 0.5 | 2 | 0.03 to >128 | 96.6 | |
| Thailand (245) | 0.25 | 2 | ≤0.015 to >128 | 93.5 | |
| Enterobacteriaceae, MBL negative | Australia (35) | 0.25 | 1 | 0.06 to 4 | 100 |
| China (231) | 0.25 | 4 | ≤0.015 to >128 | 97.4 | |
| Hong Kong (11) | 0.12 | 0.5 | 0.06 to 0.5 | 100 | |
| Japan (39) | 0.25 | 0.5 | ≤0.015 to 8 | 100 | |
| Malaysia (58) | 0.25 | 1 | 0.03 to 4 | 100 | |
| Philippines (196) | 0.25 | 1 | 0.03 to >128 | 98.0 | |
| South Korea (190) | 0.5 | 1 | ≤0.015 to 4 | 100 | |
| Taiwan (142) | 0.5 | 2 | 0.03 to 16 | 98.6 | |
| Thailand (231) | 0.25 | 1 | ≤0.015 to 64 | 99.1 | |
| Pseudomonas aeruginosa | Australia (17)d | 8 | 32 | 1 to 128 | 82.4 |
| China (30)d | 8 | 64 | 1 to 128 | 83.3 | |
| Hong Kong (3)d | — | — | 4 to 8 | 100 | |
| Japan (23) | 8 | 32 | 2 to >128 | 78.3 | |
| Malaysia (12) | 16 | >128 | 4 to >128 | 41.7 | |
| Philippines (73) | 32 | >128 | 2 to >128 | 37.0 | |
| South Korea (44) | 8 | 128 | 2 to >128 | 68.2 | |
| Taiwan (35)d | 8 | 32 | 2 to >128 | 65.7 | |
| Thailand (64) | 64 | >128 | 2 to >128 | 21.9 | |
| Pseudomonas aeruginosa, MBL negative | Australia (17) | 8 | 32 | 1 to 128 | 82.4 |
| China (30) | 8 | 64 | 1 to 128 | 83.3 | |
| Hong Kong (3) | — | — | 4 to 8 | 100 | |
| Japan (21) | 8 | 16 | 2 to 32 | 85.7 | |
| Malaysia (8) | — | — | 4 to >128 | 62.5 | |
| Philippines (36) | 8 | 32 | 2 to 32 | 75.0 | |
| South Korea (39) | 8 | 128 | 2 to 128 | 76.9 | |
| Taiwan (35) | 8 | 32 | 2 to >128 | 65.7 | |
| Thailand (44) | 16 | >128 | 2 to >128 | 31.8 | |
MBL negative, no gene encoding a metallo-β-lactamase was detected by PCR.
—, MIC50 and MIC90 were not calculated for <10 isolates.
Percent susceptibility for ceftazidime-avibactam was determined by applying U.S. FDA breakpoints.
All isolates were MBL negative.
Of the 2,038 isolates of P. aeruginosa tested from 2012 to 2015, 92.6% were susceptible to ceftazidime-avibactam (MIC, ≤8 μg/ml); the MIC90 for ceftazidime-avibactam against all P. aeruginosa was 8 μg/ml (Table 1). Percent susceptibilities to the other agents tested were lower than those to ceftazidime-avibactam, with the exception of amikacin (94.4% susceptible) and colistin (93.5% susceptible). The percent susceptible rate for ceftazidime-avibactam increased to 96.1% when only MBL-negative isolates of P. aeruginosa were considered. There were 632 isolates of carbapenem-nonsusceptible P. aeruginosa that were screened for β-lactamase genes. Of these, 74 isolates (11.7%) were found to carry genes encoding an MBL with or without additional acquired serine β-lactamases. In the majority of isolates (83.7%), no acquired β-lactamase was identified (Tables 2 and S2A to S10A), and these isolates were inferred to harbor alterations in OprD or efflux pump expression, likely combined with the hyperproduction of a chromosomal AmpC enzyme (13). VEB-type β-lactamases (80.6% of ESBLs) were the most common ESBLs identified and were found primarily in isolates from Thailand (Fig. S3A). VIM-type (58.0%) and IMP-type (35.8%) MBLs were the most common carbapenemases identified in the region. The majority of VIM-type enzymes detected were VIM-2 (80.9%), while the prevalence of the IMP-type enzymes was more varied, with IMP-26 (34.5%) and IMP-48 (20.7%) being the most common (Fig. S3B). In contrast to the Enterobacteriaceae, carbapenem nonsusceptibility in P. aeruginosa does not appear to be primarily mediated by the expression of carbapenemases (Fig. S3C). Ceftazidime-avibactam was not active against isolates carrying MBLs, as expected, but also demonstrated reduced activity against MBL-negative, ESBL-positive isolates (21.1% susceptible), while 91.3% of isolates with no acquired β-lactamase detected were susceptible to ceftazidime-avibactam (Table 2). The rate of susceptibility to ceftazidime-avibactam among all collected P. aeruginosa isolates ranged from 83.1% (Thailand) to 100% (Hong Kong) across the Asia-Pacific region, and the rate of susceptibility to ceftazidime-avibactam exceeded 90% for isolates from seven of the nine countries surveyed (Fig. 3).
FIG 3.
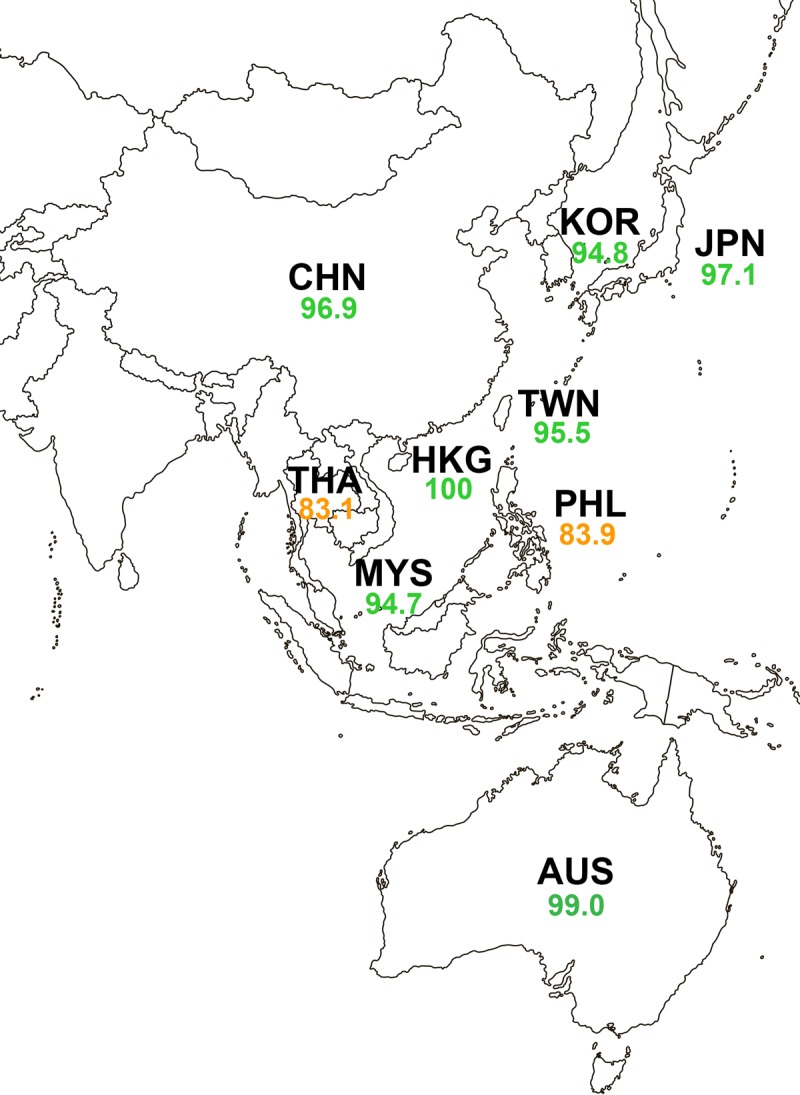
Percent susceptibility to ceftazidime-avibactam for isolates of P. aeruginosa collected from 2012 to 2015, by Asia-Pacific country. Ceftazidime-avibactam susceptible was defined as an MIC of ≤8 μg/ml; ceftazidime-avibactam resistant was defined as an MIC of ≥16 μg/ml. The green font indicates that >90% of isolates were ceftazidime-avibactam susceptible. The orange font indicates that 80 to 89.9% of isolates were ceftazidime-avibactam susceptible. Country abbreviations are as follows: AUS, Australia; CHN, China; HKG, Hong Kong; JPN, Japan; KOR, South Korea; MYS, Malaysia; PHL, Philippines; TWN, Taiwan; THA, Thailand. No isolates were obtained from patients in mainland China in 2014 or 2015 or patients in Hong Kong in 2015 due to export restrictions.
Ceftazidime-nonsusceptible P. aeruginosa isolates were found across the region, with percentages varying from 12.0% (Australia) to 32.2% (Philippines) (Fig. S4). A total of 66.7% of ceftazidime-nonsusceptible P. aeruginosa isolates were susceptible to ceftazidime-avibactam, with the percent susceptibility ranging from 40.7% (Thailand) to 100% (Hong Kong) (Tables 3 and S2A to S10A). All comparator agents other than colistin (92.4% susceptible) and amikacin (78.7% susceptible) demonstrated lower activities than ceftazidime-avibactam against this subset of isolates. The activity of ceftazidime-avibactam against the MBL-negative ceftazidime-nonsusceptible subset (79.6% susceptible) was improved (Table 3).
The percentage of P. aeruginosa isolates that were meropenem nonsusceptible was 22.4% and ranged from 8.7% (Australia) to 31.5% (Thailand) (Tables 4 and S2A to S10A; Fig. S4). Overall, 70.5% of meropenem-nonsusceptible isolates collected in the region remained susceptible to ceftazidime-avibactam, which was a percent susceptibility higher than that observed for all other agents tested except colistin (96.2% susceptible) and amikacin (77.9% susceptible). The rate of susceptibility of meropenem-nonsusceptible isolates to ceftazidime-avibactam was the lowest in Thailand (48.4% susceptible), Philippines (54.4% susceptible), and Malaysia (66.7% susceptible) and increased by 12.9 to 39.9% when isolates carrying MBLs were excluded (Tables S2A to S10A).
Only six isolates of P. aeruginosa collected in Asia-Pacific countries were resistant to colistin (0.3% of the total) (Table 5). No colistin-resistant isolates of P. aeruginosa that carried MBL genes were identified. Five of the six colistin-resistant isolates were susceptible to ceftazidime-avibactam. The single colistin- and ceftazidime-avibactam-resistant isolate was collected in Philippines (Table S7A).
A total of 301 MDR isolates of P. aeruginosa were identified (14.8% of all isolates), with the percentages of isolates testing as MDR varying from 5.7% (Australia) to 24.0% (Philippines) (Fig. 4). Of these, 52.8% remained susceptible to ceftazidime-avibactam; this percent susceptibility was higher than that observed to all other agents except colistin (94.8% susceptible) and amikacin (68.4% susceptible) (Table 6). Ceftazidime-avibactam was the least active against MDR isolates from Thailand (21.9% susceptible; MIC90, >128 μg/ml), Philippines (37.0% susceptible; MIC90, >128 μg/ml), and Malaysia (41.7% susceptible; MIC90, >128 μg/ml) and was the most active against MDR isolates from China (83.3% susceptible; MIC90, 64 μg/ml) and Australia (82.4% susceptible; MIC90, 32 μg/ml) (Tables 7 and S2A to S10A). Although the activity of ceftazidime-avibactam against MBL-negative isolates improved, MIC90 values remained above the susceptibility breakpoint (≤8 μg/ml) in all countries (Tables 7 and S2A to S10A). Nonetheless, ceftazidime-avibactam remained the second or third most active agent after colistin (and sometimes amikacin) against MDR isolates in each of the nine countries.
FIG 4.
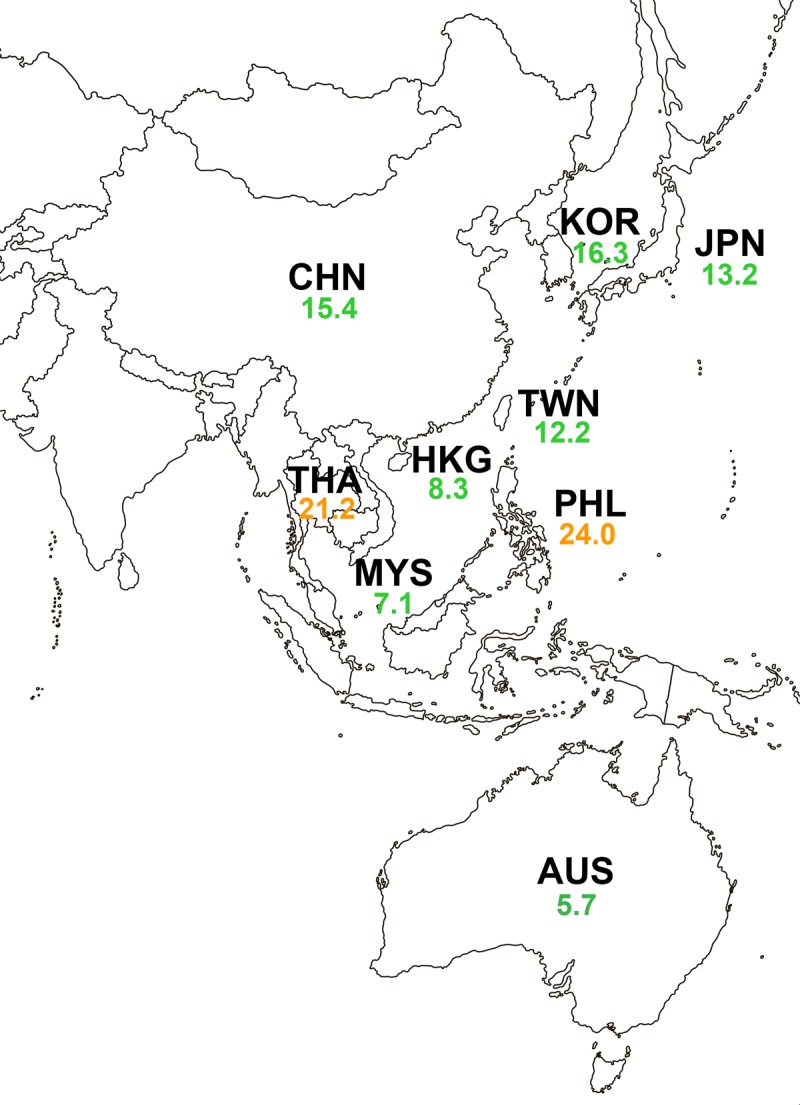
Percentage of isolates of P. aeruginosa collected from 2012 to 2015 that were multidrug resistant (MDR), by Asia-Pacific country. Multidrug resistance was defined as resistance by CLSI criteria to three or more sentinel antimicrobial agents from different chemical classes. The green font indicates that <20% of isolates were MDR. The orange font indicates that 20 to 29.9% of isolates were MDR. Country abbreviations are as follows: AUS, Australia; CHN, China; HKG, Hong Kong; JPN, Japan; KOR, South Korea; MYS, Malaysia; PHL, Philippines; TWN, Taiwan; THA, Thailand. No isolates were obtained from patients in mainland China in 2014 or 2015 or patients in Hong Kong in 2015 due to export restrictions.
DISCUSSION
Only two previous studies have specifically described the in vitro activity of ceftazidime-avibactam against clinical isolates of Enterobacteriaceae and P. aeruginosa from the Asia-Pacific region (2, 14). In the first study, Flamm et al. tested 137 urinary isolates of Enterobacteriaceae collected in Australia, China, India, South Korea, Malaysia, Singapore, Thailand, and Hong Kong in 2011 (14). They reported the ceftazidime-avibactam MIC90 to be 1 μg/ml, slightly higher than the MIC90 values (0.25 to 0.5 μg/ml) for urinary isolates of Enterobacteriaceae from the United States, Europe, and Latin America (14). In the current study, we found that 99.0% of Enterobacteriaceae isolates were susceptible to ceftazidime-avibactam (MIC, ≤8 μg/ml) with a MIC90 of 0.5 μg/ml and that ceftazidime-avibactam MIC90 values for individual species or species groups within the family Enterobacteriaceae ranged from 0.12 μg/ml (Proteeae) to 1 μg/ml (Enterobacter spp.). We observed only a minor variation (<2%) in the percent susceptibility to ceftazidime-avibactam, which ranged from 98.1% (Enterobacter spp.) to 99.9% (E. coli) (Table 1). It was observed that country-specific variances in the incidence of MBL-producing isolates must be considered when assessing the activity of ceftazidime-avibactam against clinical isolates of Enterobacteriaceae as well as P. aeruginosa. In the second study, Nichols et al. reported that 93.2% of P. aeruginosa isolates from nine Asia-Pacific countries (41 laboratories, 1,392 isolates) were susceptible to ceftazidime-avibactam and that nonsusceptibility to ceftazidime-avibactam could be explained by the possession of genes encoding MBLs in approximately one-half of the nonsusceptible isolates (2). Nichols et al. also found that 68.9% of ceftazidime-nonsusceptible isolates were susceptible to ceftazidime-avibactam, as were 71.7% of meropenem-nonsusceptible isolates (2). In the current study, which is an extension of the Nichols et al. study with one additional year of data, 92.6% of all P. aeruginosa isolates were susceptible to ceftazidime-avibactam with a MIC90 of 8 μg/ml (Table 1); 66.7% of ceftazidime-nonsusceptible isolates (range, 40.7% [Thailand] to 100% [Hong Kong]) and 70.5% of meropenem-nonsusceptible isolates (range, 48.4% [Thailand] to 100% [Hong Kong]) were also susceptible to ceftazidime-avibactam.
In studies in which the prevalence of specific β-lactamases was assessed, Kiratisin et al. reported that among 500 Enterobacteriaceae isolates collected from 20 centers in five Asia-Pacific countries in 2010, ESBL-positive rates were the highest in Thailand (45.2%) and Vietnam (55.1%) and that 97.2% of Enterobacteriaceae were susceptible to all carbapenems tested (9). A total of 625 isolates of P. aeruginosa were also tested, and the investigators reported that between 10.3% (New Zealand) and 46.7% (Vietnam) (mean, 29.8%) of P. aeruginosa isolates were nonsusceptible to at least one carbapenem (9). Mendes et al. studied isolates from 12 Asia-Pacific countries and reported that ESBL-positive rates in E. coli and K. pneumoniae varied widely between countries (ranging from 10 to 15% in Australia and New Zealand to 47 to 91% in Taiwan, India, Indonesia, and Philippines) (10). P. aeruginosa isolates collected as part of the same study demonstrated rates of resistance of >20% to all antimicrobial agents tested, except for colistin (99% susceptible), amikacin (92% susceptible), gentamicin (85% susceptible), and tobramycin (85% susceptible) (10). Pfaller et al. studied isolates from 11 Asia-Pacific countries (29 laboratories) and reported the rates of the ESBL phenotype (MICs, ≥2 μg/ml for ceftazidime, ceftriaxone, or aztreonam), MDR, and carbapenem resistance among E. coli and K. pneumoniae isolates (8). For isolates from Australia and New Zealand combined, the rates of the ESBL and MDR phenotypes were 8.4% and 4.2%, respectively, among E. coli isolates and 8.5% and 2.4%, respectively, among K. pneumoniae isolates, with no carbapenem-resistant isolates being identified in either species (8). For isolates from China, the rates of the ESBL phenotype, MDR, and carbapenem resistance were 66.9%, 36.8%, and 1.1%, respectively, among E. coli isolates and 45.6%, 26.3%, and 12.9%, respectively, among K. pneumoniae isolates (8). For isolates from other Asian countries (Hong Kong, Indonesia, Malaysia, Philippines, Singapore, South Korea, Taiwan, and Thailand) combined, the rates of the ESBL and MDR phenotypes were 39.2% and 21.7%, respectively, for E. coli and 41.9% and 29.1%, respectively, for K. pneumoniae; no E. coli isolates and 4.2% of K. pneumoniae isolates were carbapenem resistant (8). Sheng et al. studied 699 genotypically characterized β-lactamase-positive isolates of Enterobacteriaceae collected in 2008 and 2009 from 11 countries (34 medical centers) in the Asia-Pacific region (15). They reported that CTX-M-type enzymes were the most common ESBLs found, comprising 90.3% of all ESBLs (CTX-M-15 accounted for 72.7% of CTX-M β-lactamases). CMY was the most frequently identified AmpC, accounting for 56.1% of detected AmpCs (CMY-2 was the most prevalent subtype), and NDM-type MBLs were the most common carbapenemases found, accounting for 82.3% of MBLs detected (15). Carbapenemases were almost exclusively found in Enterobacteriaceae from India, except for IMP-type carbapenemases, which were also detected in isolates from Philippines and Australia (15). IMP-positive isolates of Enterobacteriaceae and P. aeruginosa have previously been reported to be more common in the Asia-Pacific region than in other regions of the world (11, 16). Isolates containing plasmid-mediated AmpC β-lactamases, such as DHA and CMY, together with ESBLs or carbapenemases were also common in the Asia-Pacific region (15). Even though the INFORM study is not structured to determine the prevalence of β-lactamases causing specific resistance phenotypes, it is noteworthy that the current study observed β-lactamase distributions similar to those in the published reports described above.
In the current study, for the Enterobacteriaceae, generally only the presence of an MBL gene was associated with in vitro resistance to ceftazidime-avibactam (Table 2) (6, 7). Isolates of the Enterobacteriaceae nonsusceptible to carbapenems by other mechanisms, such as production of GES, KPC (3, 6, 7, 17), and OXA-48 (6, 17) carbapenemases (Table 2) or production of an ESBL or AmpC enzyme combined with reduced drug permeability (6), were susceptible to ceftazidime-avibactam. Intrinsic imipenem resistance among Proteeae species (18) also did not affect susceptibility to ceftazidime-avibactam (Table 1). The current study identified only a few isolates of Enterobacteriaceae (17 isolates; 0.2%) for which the reduced ceftazidime-avibactam susceptibility could not be explained by the presence of an MBL. The mechanism(s) of reduced susceptibility remains to be determined for those isolates but may be attributable to porin or penicillin-binding protein changes or the presence of avibactam-insensitive β-lactamases (e.g., other MBLs) that were not detected by the current molecular testing algorithm (16). None of the isolates with reduced susceptibility to ceftazidime-avibactam identified in this study carried KPC β-lactamases with amino acid substitutions in Ser130 or the Ω loop (Arg164 to Asp179), reported to confer resistance to ceftazidime-avibactam (19–22). Upregulated efflux of ceftazidime-avibactam is a less likely explanation, as efflux was not implicated in reduced susceptibility to ceftazidime-avibactam in a direct test of that mechanism (23).
We conclude that clinical isolates of Enterobacteriaceae (99.0% susceptible) and P. aeruginosa (92.6% susceptible) collected from nine Asia-Pacific countries from 2012 to 2015 were highly susceptible to ceftazidime-avibactam and that ceftazidime-avibactam was as active as and frequently more active than currently available antimicrobial agents of last resort (i.e., amikacin, colistin, and tigecycline). Ceftazidime-avibactam retained its potent in vitro activity against the majority of MBL-negative Enterobacteriaceae and P. aeruginosa isolates with ceftazidime-nonsusceptible, meropenem-nonsusceptible, colistin-resistant, and MDR phenotypes collected in the Asia-Pacific region from 2012 to 2015. We observed that the percent susceptibility to ceftazidime-avibactam for Enterobacteriaceae isolates from six of the nine Asia-Pacific countries surveyed ranged from 99.1 to 100% (MIC90, 0.25 to 0.5 μg/ml), while isolates from China (98.8% susceptible; MIC90, 0.5 μg/ml), Thailand (98.7% susceptible; MIC90, 0.5 μg/ml), and Philippines (97.0% susceptible; MIC90, 0.5 μg/ml) were slightly less susceptible. This finding correlated with Philippines, Thailand, and China having the highest percentages of isolates carrying MBLs (2.7, 1.2, and 0.8%, respectively), while the percentages of MBL-positive isolates from the other six countries ranged from none (no isolates in Hong Kong and South Korea) to 0.6% (Taiwan). We also noted that the percent susceptibility to ceftazidime-avibactam for P. aeruginosa isolates was associated with the percentage of isolates carrying MBLs. For the seven countries with percent susceptibilities to ceftazidime-avibactam of >94%, the percentage of MBL-positive isolates ranged from none (no isolates in Australia and Hong Kong) to 3.0% (Malaysia), while Philippines (where 83.9% of isolates were susceptible to ceftazidime-avibactam) and Thailand (where 83.1% of isolates were susceptible to ceftazidime-avibactam) had 13.2 and 6.6% of isolates carrying an MBL, respectively. The differences in antimicrobial susceptibilities observed across countries in a specific geographic region, such as the Asia-Pacific region, demonstrate the importance of performing regional surveillance.
MATERIALS AND METHODS
Clinical isolates of Enterobacteriaceae and P. aeruginosa.
The INFORM global surveillance program collected and confirmed the identities of 11,187 nonduplicate clinical isolates of Gram-negative bacilli (9,149 isolates of Enterobacteriaceae; 2,038 isolates of P. aeruginosa) from 42 medical center laboratories in nine Asia-Pacific countries from 2012 to 2015. Each participating medical center laboratory was requested to collect predefined numbers of selected bacterial pathogens isolated from patients with specific infection types on an annual basis (2, 5). Isolates were limited to one per patient, were determined by the participating laboratory algorithms to be clinically significant, and were collected irrespective of their antimicrobial susceptibility profile (2, 5). Isolates of the Enterobacteriaceae belonging to the tribe Proteeae, which includes the three genera Proteus, Providencia, and Morganella, were frequently analyzed together as a group. The demographic information associated with the 11,187 isolates is summarized in Table S1 in the supplemental material. Isolates were transported to International Health Management Associates, Inc. (IHMA; Schaumburg, IL, USA), which served as the central reference laboratory for the INFORM global surveillance study. At IHMA, the identity of each isolate was confirmed using a Bruker Biotyper matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry instrument (Bruker Daltonics, Billerica, MA, USA).
Antimicrobial susceptibility testing.
Clinical and Laboratory Standards Institute (CLSI)-defined broth microdilution antimicrobial susceptibility testing was performed using in-house-prepared (at IHMA), 96-well broth microdilution panels (18, 24). Avibactam was tested at a fixed concentration of 4 μg/ml in combination with doubling dilutions of ceftazidime (18). MICs were interpreted using 2016 CLSI breakpoints (18), with the following exceptions. Ceftazidime-avibactam MICs were interpreted using U.S. FDA MIC breakpoints for Enterobacteriaceae and P. aeruginosa (susceptible, ≤8 μg/ml; resistant, ≥16 μg/ml) (25), as CLSI MIC breakpoints were not published at the time of the study. U.S. FDA MIC interpretative breakpoints were also used for tigecycline (26). EUCAST MIC interpretative breakpoints were used for colistin when it was tested against Enterobacteriaceae (27), as CLSI criteria for colistin are also not available.
Isolates of Escherichia coli, Klebsiella pneumoniae, Klebsiella oxytoca, and Proteus mirabilis that tested with MICs of ≥2 μg/ml to ceftazidime or aztreonam were subjected to phenotypic combination testing with clavulanic acid to confirm the presence of an ESBL (18). An MDR phenotype was defined according to the criteria of Magiorakos et al. as resistance to sentinel agents from three or more antimicrobial agent classes (the sentinel agents are indicated in parentheses), including cephalosporins (cefepime), monobactams (aztreonam), β-lactam–β-lactamase inhibitor combinations (piperacillin-tazobactam), carbapenems (meropenem), fluoroquinolones (levofloxacin), aminoglycosides (amikacin), glycylcyclines (tigecycline), and polymyxins (colistin) (28).
Screening for β-lactamase genes.
All Enterobacteriaceae isolates demonstrating a positive ESBL confirmatory test, MICs to ceftazidime of ≥16 μg/ml, or MICs to doripenem, imipenem, or meropenem of ≥2 μg/ml were screened for β-lactamase content using a combination of the microarray-based Check-MDR CT101 kit (Check-Points, Wageningen, Netherlands) and published multiplex PCR assays (29). These assays detected genes encoding carbapenemases (KPC, GES, NDM, IMP, VIM, SPM, GIM, OXA-48), ESBLs (TEM, SHV, CTX-M, VEB, PER, GES), original-spectrum β-lactamases (OSBLs; TEM and SHV enzymes that do not contain substitutions at amino acid position 104, 164, or 238 in TEM or 146, 238, or 240 in SHV, which are associated with ESBL activity, e.g., TEM-1, SHV-1, and SHV-11), and plasmid-mediated AmpC β-lactamases (ACC, ACT, CMY, DHA, FOX, MIR, MOX), as previously described (29). All P. aeruginosa isolates testing with MICs to doripenem, imipenem, or meropenem of ≥4 μg/ml were screened for genes encoding the carbapenemases, ESBLs, and OSBLs listed above, plus OXA-24, as described previously (2). Enzyme variants were identified by amplification of full-length β-lactamase genes, followed by DNA sequencing and comparison of the sequences to those deposited in the National Center for Biotechnology Information database (www.ncbi.nlm.nih.gov) and on the Lahey Clinic website (www.lahey.org/studies).
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge the contributions of the study investigators, laboratory personnel, and all members of the AstraZeneca INFORM surveillance program.
This study was sponsored by AstraZeneca Pharmaceuticals LP, which also included compensation fees for manuscript preparation. AstraZeneca's rights to ceftazidime-avibactam were acquired by Pfizer in December 2016.
Medical writing and editorial support were provided by J.A.K., a consultant for IHMA and an employee of the University of Manitoba and Diagnostic Services Manitoba; K.M.K. and S.K.B., employees of IHMA; and B.L.M.D.J. and G.G.S., employees of Pfizer. All authors provided analysis input and have read and approved the final manuscript.
D.F.S. is an employee of IHMA. None of the IHMA authors or J.A.K. has a personal financial interest in the sponsor of this paper (AstraZeneca Pharmaceuticals). B.L.M.D.J. and G.G.S. were employees of and shareholders in AstraZeneca at the time of the study.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.02569-17.
REFERENCES
- 1.de Jonge BLM, Karlowsky JA, Kazmierczak KM, Biedenbach DJ, Sahm DF, Nichols WW. 2016. In vitro susceptibility to ceftazidime-avibactam of carbapenem-nonsusceptible Enterobacteriaceae isolates collected during the INFORM global surveillance study (2012 to 2014). Antimicrob Agents Chemother 60:3163–3169. doi: 10.1128/AAC.03042-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nichols WW, de Jonge BLM, Kazmierczak KM, Karlowsky JA, Sahm DF. 2016. In vitro susceptibility of global surveillance isolates of Pseudomonas aeruginosa to ceftazidime-avibactam (INFORM 2012 to 2014). Antimicrob Agents Chemother 60:4743–4749. doi: 10.1128/AAC.00220-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Papp-Wallace KM, Bajaksouzian S, Abdelhamed AM, Foster AN, Winkler ML, Gatta JA, Nichols WW, Testa R, Bonomo R, Jacobs MR. 2015. Activities of ceftazidime, ceftaroline and aztreonam alone and combined with avibactam against isogenic Escherichia coli strains expressing selected single β-lactamases. Diagn Microbiol Infect Dis 82:65–69. doi: 10.1016/j.diagmicrobio.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kazmierczak KM, Biedenbach DJ, Hackel M, Rabine S, de Jonge BLM, Bouchillon SK, Sahm DF, Bradford PA. 2016. Global dissemination of blaKPC into bacterial species beyond Klebsiella pneumoniae and in vitro susceptibility to ceftazidime-avibactam and aztreonam-avibactam. Antimicrob Agents Chemother 60:4490–4500. doi: 10.1128/AAC.00107-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karlowsky JA, Biedenbach DJ, Kazmierczak KM, Stone GG, Sahm DF. 2016. Activity of ceftazidime-avibactam against extended-spectrum- and AmpC β-lactamase-producing Enterobacteriaceae collected in the INFORM global surveillance study from 2012 to 2014. Antimicrob Agents Chemother 60:2849–2857. doi: 10.1128/AAC.02286-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Livermore DM, Mushtaq S, Warner M, Zhang J, Maharjan S, Doumith M, Woodford N. 2011. Activities of NXL104 combinations with ceftazidime and aztreonam against carbapenemase-producing Enterobacteriaceae. Antimicrob Agents Chemother 55:390–394. doi: 10.1128/AAC.00756-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li H, Estabrook M, Jacoby GA, Nichols WW, Testa RT, Bush K. 2015. In vitro susceptibility of characterized β-lactamase-producing strains tested with avibactam combinations. Antimicrob Agents Chemother 59:1789–1793. doi: 10.1128/AAC.04191-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pfaller MA, Flamm RK, Duncan LR, Mendes RE, Jones RN, Sader HS. 2017. Antimicrobial activity of tigecycline and cefoperazone/sulbactam tested against 18,386 Gram-negative organisms from Europe and the Asia-Pacific region (2013-2014). Diagn Microbiol Infect Dis 88:177–183. doi: 10.1016/j.diagmicrobio.2017.02.020. [DOI] [PubMed] [Google Scholar]
- 9.Kiratisin P, Chongthaleong A, Yen Tan T, Lagamayo E, Roberts S, Garcia J, Davies T. 2012. Comparative in vitro activity of carbapenems against major Gram-negative pathogens: results of Asia-Pacific surveillance from the COMPACT II study. Int J Antimicrob Agents 39:311–316. doi: 10.1016/j.ijantimicag.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 10.Mendes RE, Mendoza M, Banga Singh KK, Castanheira M, Bell JM, Turnidge JD, Lin SSF, Jones RN. 2013. Regional resistance surveillance program results for 12 Asia-Pacific nations (2011). Antimicrob Agents Chemother 57:5721–5726. doi: 10.1128/AAC.01121-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kazmierczak KM, Rabine S, Hackel M, McLaughlin RE, Biedenbach DJ, Bouchillon SK, Sahm DF, Bradford PA. 2016. Multi-year, multi-national survey of the incidence and global distribution of metallo-β-lactamase-producing Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob Agents Chemother 60:1067–1078. doi: 10.1128/AAC.02379-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biedenbach D, Bouchillon S, Hackel M, Hoban D, Kazmierczak K, Hawser S, Badal R. 2015. Dissemination of NDM metallo-β-lactamase genes among clinical isolates of Enterobacteriaceae collected during the SMART Global Surveillance Study from 2008 to 2012. Antimicrob Agents Chemother 59:826–830. doi: 10.1128/AAC.03938-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lister PD, Wolter DJ, Hanson ND. 2009. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev 22:582–610. doi: 10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flamm RK, Sader HS, Farrell DJ, Jones RN. 2014. Ceftazidime-avibactam and comparator agents tested against urinary tract isolates from a global surveillance program (2011). Diagn Microbiol Infect Dis 80:233–238. doi: 10.1016/j.diagmicrobio.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 15.Sheng W-H, Badal RE, Hsueh P-R. 2013. Distribution of extended-spectrum β-lactamases, AmpC β-lactamases, and carbapenemases among Enterobacteriaceae isolates causing intra-abdominal infections in the Asia-Pacific region: results of the Study for Monitoring Antimicrobial Resistance Trends (SMART). Antimicrob Agents Chemother 57:2981–2988. doi: 10.1128/AAC.00971-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Logan LK, Weinstein RA. 2017. The epidemiology of carbapenem-resistant Enterobacteriaceae: the impact and evolution of a global menace. J Infect Dis 215(Suppl 1):S28–S36. doi: 10.1093/infdis/jiw282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castanheira M, Mills JC, Costello SE, Jones RN, Sader HS. 2015. Ceftazidime-avibactam activity tested against Enterobacteriaceae isolates from U.S. hospitals (2011 to 2013) and characterization of β-lactamase-producing strains. Antimicrob Agents Chemother 59:3509–3517. doi: 10.1128/AAC.00163-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clinical and Laboratory Standards Institute. 2016. M100-S26. Performance standards for antimicrobial susceptibility testing, 26th informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 19.Papp-Wallace KM, Winkler ML, Taracila MA, Bonomo RA. 2015. Variants of β-lactamase KPC-2 that are resistant to inhibition by avibactam. Antimicrob Agents Chemother 59:3710–3717. doi: 10.1128/AAC.04406-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winkler ML, Papp-Wallace KM, Bonomo R. 2015. Activity of ceftazidime/avibactam against isogenic strains of Escherichia coli containing KPC and SHV β-lactamases with single amino acid substitutions in the Ω-loop. J Antimicrob Chemother 70:2279–2286. doi: 10.1093/jac/dkv094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Compain F, Arthur M. 2017. Impaired inhibition by avibactam and resistance to the ceftazidime-avibactam combination due to the D179Y substitution in the β-lactamase KPC-2. Antimicrob Agents Chemother 61:e00451-. doi: 10.1128/AAC.00451-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shields RK, Chen L, Cheng S, Chavada KD, Press EG, Synder A, Pandey R, Doi Y, Kreiswirth BN, Nguyen MH, Clancy CJ. 2017. Emergence of ceftazidime-avibactam resistance due to plasmid-borne blaKPC-3 mutations during treatment of carbapenem-resistant Klebsiella pneumoniae infections. Antimicrob Agents Chemother 61:e02097-16. doi: 10.1128/AAC.02097-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pagès J-M, Peslier S, Keating TA, Lavigne J-P, Nichols WW. 2016. The role of the outer membrane and porins in the susceptibility of β-lactamase-producing Enterobacteriaceae to ceftazidime-avibactam. Antimicrobial Agents Chemother 60:1349–1359. doi: 10.1128/AAC.01585-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clinical and Laboratory Standards Institute. 2015. M07-A10. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard, 10th ed Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 25.Allergan USA, Inc. 2018. Avycaz® (ceftazidime and avibactam) for injection, for intravenous use, prescribing information. Revised February. Allergan USA, Inc, Irvine, CA. [Google Scholar]
- 26.Pfizer Inc. 2016. Tygacil® (tigecycline) injection, powder, lyophilized, for solution, prescribing information. Revised June. Pfizer Inc, Philadelphia, PA. [Google Scholar]
- 27.European Committee on Antimicrobial Susceptibility Testing. 2016. Breakpoint tables for interpretation of MICs and zone diameters. Version 6.1 http://www.eucast.org.
- 28.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 29.Lob SH, Kazmierczak KM, Badal RE, Hackel MA, Bouchillon SK, Biedenbach DJ, Sahm DF. 2015. Trends in susceptibility of Escherichia coli from intra-abdominal infections to ertapenem and comparators in the United States according to data from the SMART Program, 2009 to 2013. Antimicrob Agents Chemother 59:3606–3610. doi: 10.1128/AAC.05186-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


