 Chemical labeling coupled with LC-MS enables the sensitive and simultaneous detection of the oxidative products of 5-methylcytosine. With this method, we can determine 5-methylcytosine, 5-hydroxymethylcytosine, 5-formylcytosine and 5-carboxylcytosine in RNA of mammals.
Chemical labeling coupled with LC-MS enables the sensitive and simultaneous detection of the oxidative products of 5-methylcytosine. With this method, we can determine 5-methylcytosine, 5-hydroxymethylcytosine, 5-formylcytosine and 5-carboxylcytosine in RNA of mammals.
Abstract
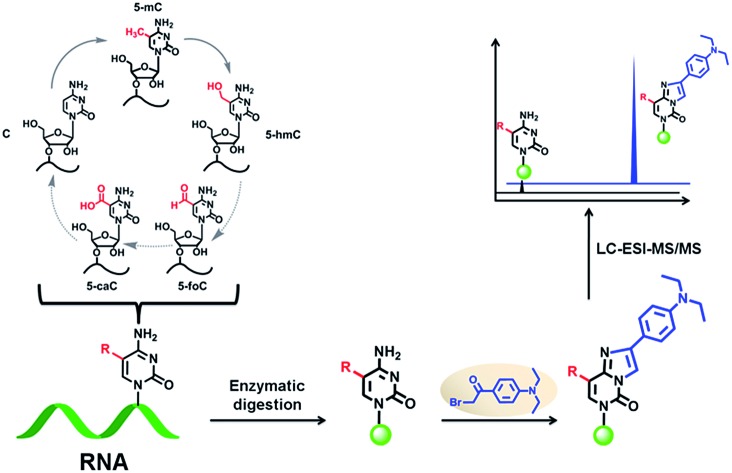
Similar to the reversible epigenetic modifications on DNA, dynamic RNA modifications were recently considered to constitute another realm for biological regulation in the form of “RNA epigenetics”. 5-Methylcytosine (5-mC) has long been known to be present in RNA from all three kingdoms of life. However, the functions of 5-mC in RNA have not been fully understood, especially for the RNA demethylation mechanism. The discovery of 5-hydroxymethylcytosine (5-hmC) in RNA together with our recently reported 5-formylcytosine (5-foC) in RNA indicated that 5-mC in RNA may undergo the same cytosine oxidation demethylation pathway with generating intermediates 5-hmC, 5-foC, and 5-carboxylcytosine (5-caC) by ten–eleven translocation (Tet) proteins as that in DNA. However, endogenous 5-caC in RNA has not been observed so far. In the current study, we established a method using chemical labeling coupled with liquid chromatography-mass spectrometry analysis for the sensitive and simultaneous determination of the oxidative products of 5-mC. Our results demonstrated that the detection sensitivities of 5-mC, 5-hmC, 5-foC and 5-caC in RNA increased by 70–313 folds upon 2-bromo-1-(4-diethylaminophenyl)-ethanone (BDEPE) labeling. Using this method, we discovered the existence of 5-caC in the RNA of mammals. In addition, we found the 5-mC occurs in all RNA species including mRNA, 28S rRNA, 18S rRNA and small RNA (<200 nt). However, 5-hmC, 5-foC and 5-caC mainly occur in mRNA, and barely detected in other types of RNA. Furthermore, we found that the content of 5-hmC in the RNA of human colorectal carcinoma (CRC) and hepatocellular carcinoma (HCC) tissues significantly decreased compared to tumor adjacent normal tissues, suggesting that 5-hmC in RNA may play certain functional roles in the regulation of cancer development and formation.
Introduction
Naturally occurring RNA molecules contain various chemically modified nucleosides.1 Up to date, more than 140 structurally distinct post-transcriptionally modified nucleosides have been identified in RNA.1 Similar to the reversible epigenetic marks on DNA, some RNA modifications were recently discovered to be dynamic and might have critical regulatory and/or signalling roles inside cells, which constitutes another realm for biological regulation in the form of “RNA epigenetics”.2–4
5-Methylcytosine (5-mC) has long been known to be present in the RNA from all three kingdoms of life.5,6 A recent study showed that 5-mC is widespread in both the coding and noncoding RNA of mammals, among which more than 8000 5-mC sites were identified in mRNA, implicating that 5-mC in RNA could be critical for the control and regulation of gene transcription and protein translation.7,8 However, the precise cellular functions of 5-mC in RNA have not been fully understood, especially for the RNA demethylation mechanism. On the other side, the functional role of 5-mC in DNA as an epigenetic mark is well established.9–14 DNA demethylation could be achieved through a consecutive oxidation of 5-mC by ten–eleven translocation (Tet) proteins with the generation of three intermediates, 5-hydroxymethylcytosine (5-hmC), 5-formylcytosine (5-foC), and 5-carboxylcytosine (5-caC).15–18 Due to the similarity of the chemical structure between DNA and RNA, DNA and RNA may share the same cytosine demethylation mechanism.
Indeed, Fu et al.19 recently reported that Tet proteins can also oxidize 5-mC to generate 5-hmC in RNA, which is similar to Tet proteins oxidizing 5-mC to 5-hmC in DNA. And our group further confirmed the existence of 5-foC in the RNA of mammals.20 These studies indicated that 5-mC in RNA may undergo the same cytosine demethylation pathway with generating the intermediates of 5-hmC, 5-foC, and 5-caC by Tet proteins as that in DNA. However, endogenous 5-caC in RNA has not been observed so far. To establish the above presumed RNA demethylation pathway, the identification of endogenous 5-caC in RNA is indispensable. However, the identification of these modified nucleosides is usually challenging due to their extremely low in vivo content as well as interference from the highly abundant normal nucleosides. The reported occurring frequency of 5-hmC and 5-foC in RNA is 1 to 20 per 106 cytosines.19,20 5-caC may be present at a lower levels than those of 5-hmC and 5-foC in the RNA of mammals. However, even though the content of modified nucleosides is extremely low, they may be biologically significant if they occur in specific gene regulatory elements, such as 5-hmC, 5-foC, 5-caC, N6-methyladenine, and N1-methyladenosine in DNA.2,21–24
In the current study, we established a method using chemical labeling coupled with liquid chromatography-electrospray ionization-tandem mass spectrometry analysis (LC-ESI-MS/MS) for the sensitive and simultaneous determination of all of the oxidation products of 5-mC in a possible RNA demethylation pathway. We first evaluated the detection sensitivities of labeled products with different labeling reagents carrying the same reactive group of the bromoacetonyl moiety. Our results demonstrated that the detection sensitivities of 5-mC, 5-hmC, 5-foC and 5-caC in RNA increased by 70–313 folds upon 2-bromo-1-(4-diethylaminophenyl)-ethanone (BDEPE) labeling. Using the developed analytical method, we were able to identify endogenous 5-caC in the RNA of mammals. In addition, we found that 5-mC occurs in all RNA species; however, 5-hmC, 5-foC and 5-caC mainly occur in mRNA, and barely detected in other types of RNA. Furthermore, we found that the content of 5-hmC in RNA of human colorectal carcinoma (CRC) and hepatocellular carcinoma (HCC) tissues significantly decreased compared to tumor adjacent normal tissues.
Experimental section
Chemicals and reagents
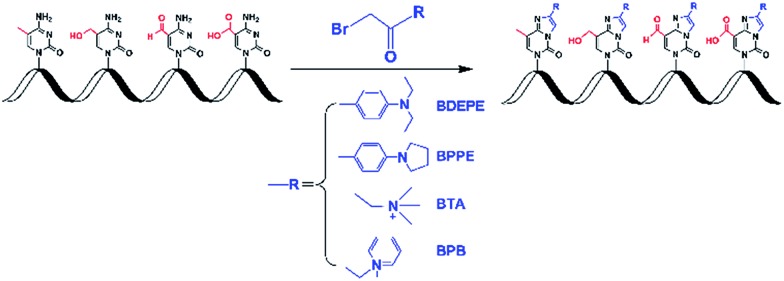
Cytidine (C), guanosine (G), adenosine (A), uridine (U), 5-methylcytidine (5-mC), and phosphodiesterase I were purchased from Sigma-Aldrich (Beijing, China). 5-Hydroxymethylcytidine (5-hmC), 5-formylcytidine (5-foC), and 5-carboxylcytidine (5-caC) were purchased from Berry & Associates (Dexter, MI). 2-Bromo-1-(4-diethylaminophenyl)-ethanone (BDEPE) and 2-bromo-1-(4-1-pyrrolidinylphenyl)-ethanone (BPPE) were purchased from Alfa Aesar (Ward Hill, MA). 3-Bromoacetonyltrimethylammonium bromide (BTA) and ω-bromoacetonylpyridinium bromide (BPB) were synthesized according to our previously described method.25,26 S1 nuclease and alkaline phosphatase (CIAP) were from Takara Biotechnology Co., Ltd. (Dalian, China).
Chromatographic grade methanol and acetonitrile (ACN) were purchased from Tedia Co. Inc. (Fairfield, OH). Formic acid and triethylamine (TEA) were purchased from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China). The water used throughout the study was purified on a Milli-Q apparatus (Millipore, Bedford, MA). Stock solutions of 5-mC, 5-hmC, 5-foC, and 5-caC were prepared in water at a concentration of 5 mM. BDEPE and BPPE were prepared in ACN at a concentration of 40 mM. BTA and BPB were prepared in ACN at a concentration of 20 mM.
Biological and clinical samples
Human embryonic kidney (293T) cells were obtained from the China Center for Type Culture Collection and maintained in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum, 100 U mL–1 penicillin, and 100 μg mL–1 streptomycin at 37 °C in a 5% CO2 atmosphere. Total RNA was extracted using E.Z.N.A.® HP Total RNA Kit (Omega Bio-Tek Inc., Norcross, GA, USA) according to the manufacture's recommended procedure.
Male Sprague-Dawley rat (4 weeks old) was obtained from the Center for Animal Experiment/ABSL-3 Laboratory of Wuhan University and sacrificed to collect tissues and stored under –80 °C. Liver tissue RNA was extracted using E.Z.N.A.® Tissue RNA Kit (Omega Bio-Tek Inc., Norcross, GA, USA).
A total of 40 formalin-fixed, paraffin-embedded (FFPE) tissue samples from colorectal carcinoma (CRC) patients, including 20 pairs of CRC tissues and matched tumor adjacent normal tissues, and a total of 16 fresh tissue samples from hepatocellular carcinoma (HCC) patients, including 8 pairs of HCC tissues and matched tumor adjacent normal tissues, were collected from Hubei Cancer Hospital. FFPE tissue RNA and fresh tissue RNA were extracted using the E.Z.N.A.® FFPE RNA Kit and E.Z.N.A.® Tissue RNA Kit (Omega Bio-Tek Inc., Norcross, GA), respectively. mRNA was further isolated from total RNA using the Promega PolyATtract® mRNA Isolation System (Madison, WI). Small RNA (<200 nt) was purified using the E.Z.N.A.® MiRNA Kit (Omega Bio-Tek Inc., Norcross, GA). 28S rRNA and 18S rRNA were purified using agarose electrophoresis, and the detailed isolation procedure can be found in the ESI.† An approval was granted by the Hubei Cancer Hospital Ethics Committee and met the declaration of Helsinki. All of the experiments were performed in accordance with Hubei Cancer Hospital Ethics Committee's guidelines and regulations.
Enzymatic digestion of RNA
Certain amounts of RNA (in 16 μL of H2O) were firstly denatured by heating at 95 °C for 5 min and then chilling on ice for 2 min. After adding 1/10 volume (2 μL) of S1 nuclease buffer (30 mM CH3COONa, pH 4.6, 280 mM NaCl, 1 mM ZnSO4) and 360 units (2 μL) of S1 nuclease, the mixture (20 μL) was then incubated at 37 °C for 2 h. To the resulting solution 10 μL of alkaline phosphatase buffer (50 mM Tris–HCl, 10 mM MgCl2, pH 9.0), 0.01 units (5 μL) of venom phosphodiesterase I, 30 units (1 μL) of alkaline phosphatase, and 64 μL of H2O were subsequently added. Then, incubation was continued at 37 °C for an additional 2 h followed by adding 100 μL of sterilized water and extraction with an equal volume of chloroform twice. The resulting aqueous layer was collected and further passed through a solid phase extraction (SPE) cartridge filled with 50 mg of sorbent of graphitized carbon black (Weltech Co., Ltd., Wuhan, China) to remove the salts. The elution was then dried with nitrogen gas at 37 °C for subsequent chemical labeling.
Chemical labeling
In this study, four different labeling reagents harboring the same reactive group of bromoacetonyl moiety (Fig. 1, and Table S1 in ESI†) were used to label 5-mC, 5-hmC, 5-foC, and 5-caC. Chemical labeling conditions, including TEA content, reaction temperature and time, and concentration of the labeling reagents were optimized to achieve the best labeling performance. The reactions were performed in 200 μL of ACN with 1 μM of 5-mC.
Fig. 1. Chemical labeling of the cytosine modifications.
The labeling products were examined on a Shimadzu LC-15C HPLC system (Tokyo, Japan) equipped with two LC-15C pumps, a CTO-15C thermostated column compartment, a SPD-15C UV/vis detector, and a RF-10A fluorescence detector (FLD). The UV/vis detector was connected with FLD in series. The reaction mixture was detected using a UV/vis detector with a wavelength of 260 nm, and the derivatives of 5-mC, 5-hmC, 5-foC, and 5-caC were detected by FLD with an excitation wavelength of 305 nm and an emission wavelength of 370 nm. A Hisep C18-T column (250 mm × 4.6 mm i.d., 5 μm, Weltech Co., Ltd., Wuhan, China) was used for the separation. The column temperature was set at 35 °C. Water containing 0.1% formic acid (v/v, solvent A) and acetonitrile (solvent B) were employed as mobile phase with a flow rate of 0.8 mL min–1. A gradient of 5% B for 4 min and 5–45% B for 40 min was used.
Analysis of labeled products of 5-mC, 5-hmC, 5-foC, and 5-caC by LC-ESI-MS/MS
Analysis of the labeled products of 5-mC, 5-hmC, 5-foC, and 5-caC was performed on the LC-ESI-MS/MS system consisting of an AB 3200 QTRAP mass spectrometer (Applied Biosystems, Foster City, CA, USA) with an ESI source (Turbo Ionspray) and a Shimadzu LC-20AD HPLC (Tokyo, Japan). Data acquisition and processing were performed using AB SCIEX Analyst 1.5 Software (Applied Biosystems, Foster City, CA, USA). The HPLC separation was performed on a Hisep C18-T column (150 mm × 2.1 mm i.d., 5 μm, Weltech Co., Ltd., Wuhan, China) at 35 °C. Water containing 0.05% formic acid (v/v, solvent A) and ACN (solvent B) was employed as the mobile phase. A gradient of 5–65% B for 40 min was used. The flow rate of the mobile phase was set at 0.2 mL min–1. The mass spectrometry detection was performed under positive ESI mode. The nucleosides and labeled products were monitored using the multiple reaction monitoring (MRM) mode. The MRM parameters were optimized to achieve maximal detection sensitivity (Table S2, ESI†).
Simultaneous analysis of 5-mC, 5-hmC, 5-foC, and 5-caC in the RNA of mammals
The chemical labeling of all these cytosine modifications from the digested RNA of mammals by BDEPE was performed under the optimized conditions. Briefly, the nucleoside mixture of the digested RNA was passed through an SPE cartridge filled with 50 mg of sorbent of graphitized carbon black (Weltech Co., Ltd., Wuhan, China) to remove the salts. The elution was then dried with nitrogen gas at 37 °C followed by BDEPE labeling, which was performed in 200 μL of ACN with 2 mM of BDEPE and 4 mM of TEA. Then, the resulting solution was incubated at 60 °C for 4 h with shaking at 1500 rpm to label 5-mC, 5-hmC, 5-foC, and 5-caC. The labeled products were dried with nitrogen gas at 37 °C and then reconstituted in 100 μL of water containing 5% ACN (v/v) followed by analysis using LC-ESI-MS/MS.
High resolution mass spectrometry experiments were also performed on the LC-QTOF-MS system consisting of a MicrOTOF-Q orthogonal-accelerated TOF mass spectrometer (Bruker Daltonics, Bremen, Germany) with an ESI source (Turbo Ionspray) and a Shimadzu LC-20AB binary pump HPLC (Tokyo, Japan). Data acquisition and processing were performed using Bruker Daltonics Control 3.4 and Bruker Daltonics Data analysis 4.0 software. The HPLC separation was performed on a Hisep C18-T column (150 mm × 2.1 mm i.d., 5 μm, Weltech Co., Ltd., Wuhan, China) at 35 °C. Water containing 0.05% formic acid (v/v, solvent A) and ACN (solvent B) was employed as mobile phase. A gradient of 5–65% B for 40 min was used. The flow rate of the mobile phase was set at 0.2 mL min–1.
Statistical analysis
The statistical data were processed with SPSS 19.0 software (SPSS Inc.). The paired t-test was performed to evaluate the differences of cytosine modifications in RNA between tumor tissues and tumor adjacent normal tissues. All p values were two-sided, and generally, p values <0.05 were considered to have statistical significance.
Results and discussion
Strategy for the sensitive identification and quantification of oxidation products of 5-mC in RNA
5-mC is one of the most important modifications in RNA molecules with potential functions on the control and regulation of gene transcription and protein translation.8 The mechanism of DNA cytosine demethylation in mammals has been well established.22,27 But how RNA cytosine demethylation is achieved is still unclear. Recent studies reported the discovery of 5-hmC and 5-foC in RNA in mammalian cells and found that Tet proteins also possess the activity of catalyzing the formation of 5-hmC from 5-mC in RNA, indicating that RNA may undergo the same Tet-mediated oxidative demethylation pathway as that in DNA.19,20 However, the further oxidation product of 5-mC in RNA (i.e. 5-caC) has not been identified in mammals, which therefore makes the hypothesis deficient.
In this respect, sensitive and accurate identification and quantification of the oxidation products of 5-mC in RNA will certainly facilitate the establishment of the RNA demethylation pathway. However, direct identification and quantification of these cytosine modifications in RNA has not been realized, which may be due to their extremely low in vivo abundances. Our recent study demonstrated that the N3 and N4 positions of cytosine could react with bromoacetonyl groups.28 Therefore, here we systemically evaluated the chemical labeling of cytosine modifications of 5-mC, 5-hmC, 5-foC, and 5-caC in RNA by different reagents bearing a bromoacetonyl moiety (Fig. 1). With the appropriate chemical labeling, the ionization efficiencies can be dramatically enhanced, which therefore can increase the detection sensitivities of these cytosine modifications. Along this line, we were able to identify and quantify all of the oxidation products of 5-mC in the RNA of mammals.
Identification of labeled products of 5-mC, 5-hmC, 5-foC, and 5-caC
We examined the labeled products of 5-mC, 5-hmC, 5-foC, and 5-caC with four different labeling reagents, including BDEPE, BPPE, BTA, and BPB, by LC-ESI-MS/MS. As expected, the four cytosine modifications successfully reacted with the labeling reagents and formed the desired derivatives. Typically, the labeled products were formed via the reaction of the bromoacetonyl group with the N3 and N4 positions of cytosine.
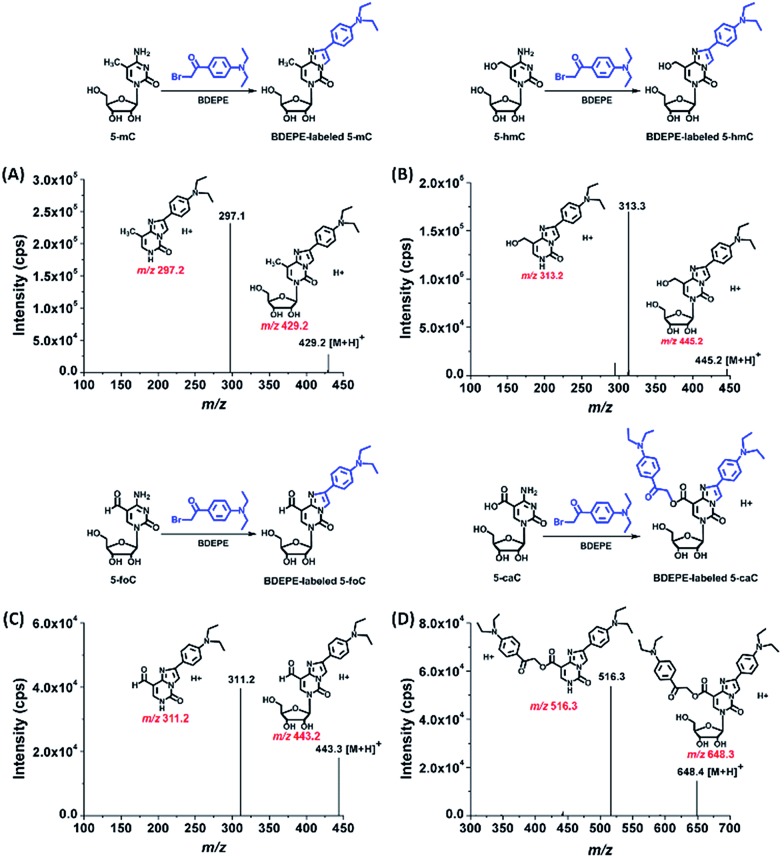
The product ion spectrum showed that m/z 429.2 and 297.1, m/z 445.2 and 313.2, m/z 443.2 and 311.2, m/z 648.3 and 516.3, which represent the parent ions of the 5-mC, 5-hmC, 5-foC, and 5-caC derivatives and their product ions, were observed after BDEPE labeling (Fig. 2). Similarly, the expected labeled products were also obtained via BPPE, BTA, and BPB labeling (Fig. S1–S3, ESI†). It is worth noting that the carboxyl group at the fifth position of 5-caC also reacts with the labeling reagents of BDEPE, BTA and BPB (Fig. 2D, S2D and S3D in ESI†). However, BPPE mainly reacts with the carboxyl group of 5-caC (Fig. S1D, ESI†). We reason that it is more favorable for the bromoacetonyl group of BPPE to form the ester with the carboxyl group of 5-mC since BPPE contains a pyrrolidyl group that increases the steric hindrance in the cyclization reaction between the bromoacetonyl group and the N3 and N4 positions of cytosine.
Fig. 2. Product ions spectra of BDEPE labeled 5-mC (A), 5-hmC (B), 5-foC (C), and 5-caC (D).
Improvement of detection sensitivity and LC separation upon chemical labeling
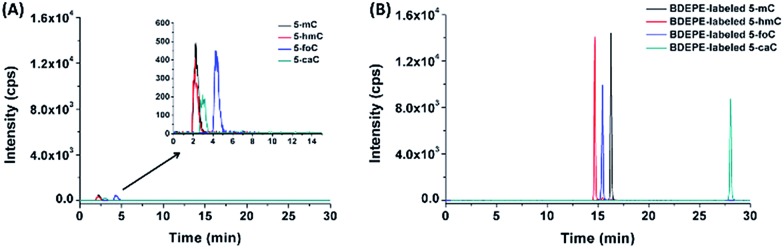
The main purpose for chemical labeling is to improve the LC separation and detection sensitivities of 5-mC, 5-hmC, 5-foC, and 5-caC during LC-ESI-MS/MS analysis. To obtain the best detection, we optimized the labeling conditions, including TEA concentration, reaction temperature and time, and the concentration of the labeling reagents. The detailed optimization conditions for the four labeling reagents are shown in Fig. S4–S7 and listed in Table S1 in ESI.† Then we investigated the LC separation and detection sensitivities of 5-mC, 5-hmC, 5-foC, and 5-caC upon labeling under their own optimized reaction conditions. The extracted ion chromatograms show that the retentions of native 5-mC, 5-hmC, 5-foC and 5-caC were relatively weak and they coeluted on the reversed-phase chromatographic column even under optimized separation conditions (Fig. 3A). However, after labeling using BDEPE and BPPE, the retention and separation resolution dramatically increased (Fig. 3B and S8A in ESI†), which can be attributed to the increased hydrophobicity of these labeled products. However, as for labeling using BTA or BPB, that harbors quaternary ammonium or pyridinium groups, the retentions of these derivatives did not increase on the reversed-phase chromatographic column (Fig. S8B and S8C in ESI†). The weak retentions of the BTA and BPB labeled products were due to the weak interaction between the positively charged derivatives and reversed-phase chromatographic column. In addition, the excess amounts of labeling reagents could be readily removed using a six-way valve through switching to a waste line after (for BDEPE and BPPE) or before (for BTA and BPB) the labeled products elute out, thus avoiding contamination of the mass spectrometer (Fig. S9, ESI†).
Fig. 3. Extracted ion chromatograms of 5-mC, 5-hmC, 5-foC, and 5-caC before (A) and after (B) labeling using BDEPE under the optimized conditions. The amount of 5-mC, 5-hmC, 5-foC, and 5-caC were 200 fmol.
Compared to the native forms of 5-mC, 5-hmC, 5-foC, and 5-caC, chemical labeling could also dramatically increase their detection sensitivities. The limits of detection (LODs) of 5-mC, 5-hmC, 5-foC, and 5-caC with and without chemical labeling are shown in Table 1. We found that these cytosine modifications labeled by BDEPE offered the best detection sensitivities with typically decreased LODs by 70 to 313 folds (Table 1). We reason that the differentially increased detection sensitivities using the four labeling reagents could be attributed to the differences of chemical properties of the derivatives as well as the labeling efficiency. Generally, the increased hydrophobicity of these labeled products results in a longer retention time in elution with a higher ratio of organic solvent. Thus, the analytes could be ionized more effectively in ESI owing to higher spraying and desolvation efficiency under higher ACN content. The BDEPE and BPPE labeled products showed longer retention times than the BTA and BPB labeled products, therefore the overall detection sensitivities of these cytosine modifications labeled by BDEPE and BPPE were better than those labeled by BTA and BPB. In addition, the labeling efficiencies obtained for BDEPE were the best among the four labeling reagents (Table S3, ESI†), which also contributed to the high detection sensitivities offered by BDEPE. The relative low labeling efficiencies for BTA and BPB could be due to the positive charge near the bromoacetonyl group that decreases the electrophilicity of the bromoacetonyl group. Taken together, here we chose BDEPE as the labeling reagent for the following experiments.
Table 1. Limits of detection of 5-mC, 5-hmC, 5-foC, and 5-caC with and without chemical labeling.
| LODs (fmol) |
||||
| 5-mC | 5-hmC | 5-foC | 5-caC | |
| Without labeling | 4.20 | 9.40 | 14.30 | 25.01 |
| BDEPE labeling | 0.06 | 0.07 | 0.10 | 0.08 |
| BPPE labeling | 0.43 | 0.69 | 0.79 | 1.21 |
| BTA labeling | 2.20 | 4.72 | 6.22 | 0.53 |
| BPB labeling | 5.25 | 2.72 | 16.59 | 4.41 |
Method validation
With BDEPE as the labeling reagent, the calibration curves of 5-mC, 5-hmC, 5-foC, and 5-caC were constructed by plotting the mean peak area ratios of 5-mC/103 G, 5-hmC/106 G, 5-foC/106 G, or 5-caC/107 G versus the mean molar ratios of 5-mC/103 G, 5-hmC/106 G, 5-foC/106 G, or 5-caC/107 G based on data obtained from triplicate measurements using BDEPE labeling coupled with LC-ESI-MS/MS analysis. The results showed that good linearities within the range of 0.2–50 5-mC/103 G, 0.2–100 5-hmC/106 G, 0.2–50 5-foC/106 G, and 0.2–50 5-caC/107 G were obtained with the coefficients of determination (R2) being greater than 0.99 (Table S4, ESI†). The accuracy and reproducibility of the developed method were evaluated with the REs and RSDs being less than 16.0% and 15.2% (Tables S5–S8, ESI†), respectively, demonstrating that good accuracy and reproducibility were achieved.
Determination of the oxidation products of 5-methylcytosine in the RNA of mammals
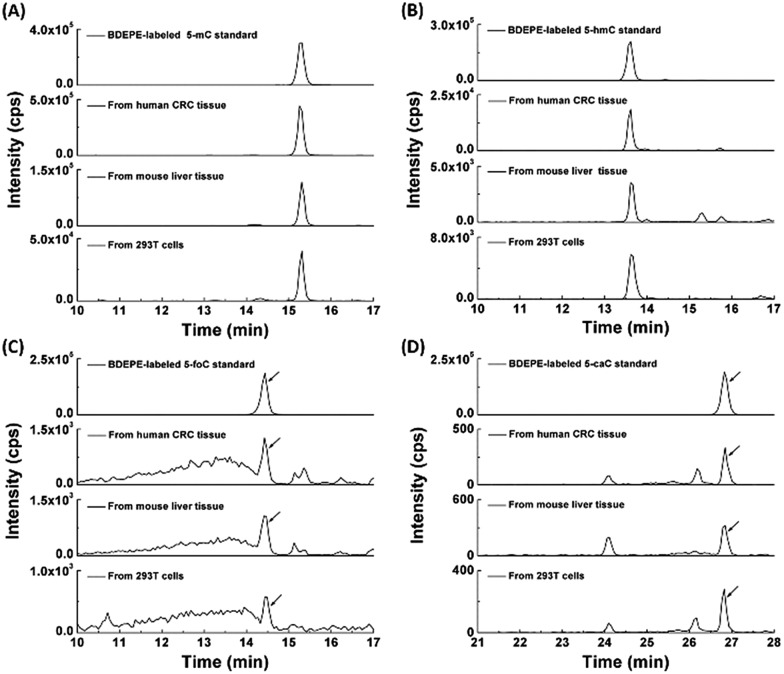
With the established method, we explored the oxidation products of 5-mC in the RNA of mammals, including 293T cells, mouse liver tissue and human CRC tissue. Shown in Fig. 4 are the typical extracted ion chromatograms of BDEPE labeled cytosine modification standards as well as the detected oxidation products of 5-mC in the RNA of mammals. The results demonstrated that all of these four cytosine modifications of 5-mC, 5-hmC, 5-foC, and 5-caC can be detected (Fig. 4). It is worth noting that 5-caC also can be distinctly observed in these samples even with much lower intensities than 5-mC and 5-hmC.
Fig. 4. Determination of 5-mC (A), 5-hmC (B), 5-foC (C), and 5-caC (D) in the RNA of mammals using BDEPE labeling coupled with LC-ESI-MS/MS analysis. Extracted ion chromatograms of the BDEPE labeled standards, BDEPE labeled cytosine modifications in the RNA from human CRC tissue, mouse liver tissue and 293T cells are shown in each panel.
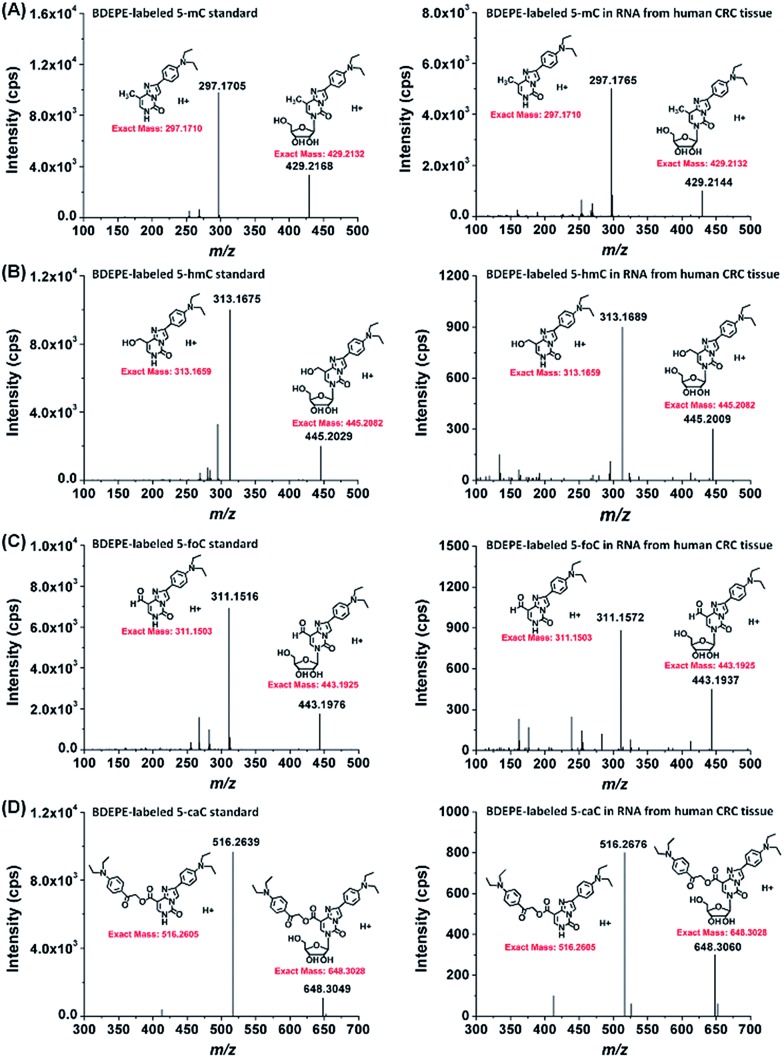
To further confirm these cytosine modifications in RNA, we also employed high resolution mass spectrometry to examine the BDEPE labeled products. The results showed that the molecular weight of the ions in the spectra of the BDEPE labeled standards were identical to the theoretical values, further demonstrating the successful labeling (Fig. 5, left panel). In addition, the high resolution MS spectra of the BDEPE labeled products from the RNA of human CRC tissue were similar to the BDEPE labeled standards and the fragment ions were identical to the theoretical values (Fig. 5, right panel), supporting the expected existence of these cytosine modifications in the RNA of mammals.
Fig. 5. Confirmation of the cytosine modifications in the RNA of mammals using high-resolution mass spectrometry. (A) Product-ion spectra of BDEPE labeled 5-mC standard (left) and 5-mC from the RNA of human CRC tissue. (B) Product-ion spectra of BDEPE labeled 5-hmC standard (left) and 5-hmC from the RNA of human CRC tissue. (C) Product-ion spectra of BDEPE labeled 5-foC standard (left) and 5-foC from the RNA of human CRC tissue. (D) Product-ion spectra of BDEPE labeled 5-caC standard (left) and 5-caC from the RNA of human CRC tissue.
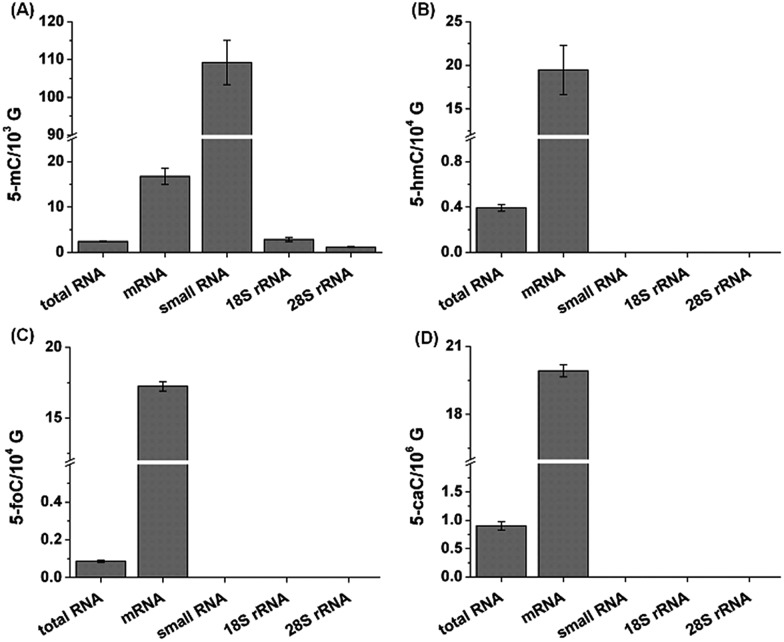
We then further explored these cytosine modifications in different RNA species. In this respect, we isolated different types of RNA, including mRNA, 28S rRNA, 18S rRNA and small RNA (<200 nt) from the total RNA of mouse liver tissue. The detailed isolation procedure of rRNA and small RNA can be found in the text in the ESI.† The purified 28S rRNA, 18S rRNA, small RNA (<200 nt) were examined via agarose and polyacrylamide gel electrophoresis (Fig. S10 in ESI†). And the purity of the isolated mRNA was evaluated using the N6,N6-dimethyladenosine according to previous methods (Fig. S11 in ESI†).29 Our results showed that 5-mC occurs in all RNA species with more abundant in mRNA and small RNA (Fig. 6). However, 5-hmC, 5-foC and 5-caC mainly occur in mRNA, and are barely detected in other types of RNA, such as 28S rRNA, 18S rRNA and small RNA (Fig. 6). The results are consistent with the recently just published paper stating that 5-hmC mainly exists in mRNA.30
Fig. 6. Determination of 5-mC (A), 5-hmC (B), 5-foC (C), and 5-caC (D) in different RNA species from mouse liver tissue. 10 μg total RNA and 1 μg other RNA species were used for quantification.
Collectively, the proved existence of 5-caC in RNA, together with previously identified 5-hmC and 5-foC, suggested the possible oxidative demethylation of 5-mC in the RNA of mammals. Therefore, 5-mC in RNA may potentially undergo demethylation through a similar pathway as Tet-mediated DNA demethylation. The further oxidative products of 5-hmC in DNA using Tet, i.e. 5-foC and 5-caC, could be readily recognized by thymine–DNA glycosylase, and the subsequent base excision repair machinery could result in the restoration of unmethylated cytosine. As for the 5-hmC in RNA, however, it is still not straightforward for this process. A recent study showed that 5-mC in mRNA can decrease translation and 5-hmC can favor the translation.30 Since the half-life of RNA is normally short, therefore, 5-hmC, 5-foC and 5-caC in mRNA may also be alternatively removed with degradation of mRNA once they fulfil their regulation functions. However, further investigation is required to elucidate the mechanism.
Content change of 5-mC, 5-hmC, 5-foC, and 5-caC in the RNA of human CRC and HCC tissues
CRC and HCC are common human cancers and the leading causes of cancer deaths worldwide.31 Our previous study demonstrated the significant decrease of the oxidation products of 5-mC in the DNA of CRC and HCC tissues compared to adjacent normal tissues.13,28 With the developed sensitive analytical method in the current study, we were able to identify and quantify all of the oxidation products of 5-mC in RNA. Therefore, here we further explored the correlation of the oxidation products of 5-mC in RNA with human CRC and HCC.
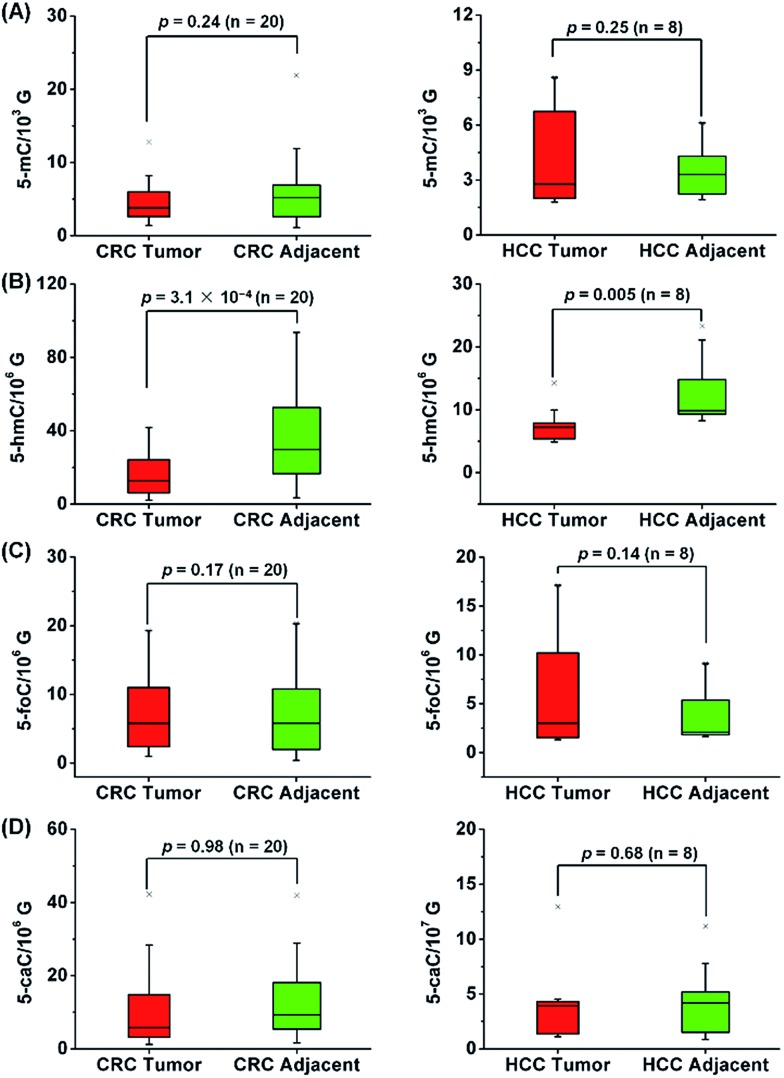
A total of 40 tissue samples derived from 20 CRC patients and 16 tissue samples from 8 HCC patients were analyzed. The mean content of 5-mC, 5-hmC, 5-foC and 5-caC in CRC tissues were 5.1 ± 0.7/103 G, 17.9 ± 2.9/106 G, 9.8 ± 2.3/106 G, 7.3 ± 1.2/107 G, respectively; and the mean content of these modifications in tumor adjacent normal tissues were 6.5 ± 1.2/103 G, 38.2 ± 5.6/106 G, 13.7 ± 2.6/106 G, 7.3 ± 1.3/107 G, respectively (Fig. 7, and Table S9 in ESI†). As for the HCC tissues, the mean content of 5-mC, 5-hmC, 5-foC and 5-caC were 4.5 ± 0.9/103 G, 7.9 ± 1.0/106 G, 6.8 ± 2.0/106 G, 4.2 ± 1.3/107 G, respectively; and the mean content of these modifications in tumor adjacent normal tissues were 3.7 ± 0.6/103 G, 12.8 ± 2.0/106 G, 4.2 ± 1.0/106 G, 4.9 ± 1.2/107 G, respectively (Fig. 7, and Table S10 in ESI†). The statistical results suggested the significant depletion of 5-hmC in human CRC (p = 3.1 × 10–4) and HCC (p = 0.005) tissues compared to tumor adjacent normal tissues; however, 5-mC, 5-foC, and 5-caC showed no significant difference between CRC or HCC tissues and tumor adjacent normal tissues (Fig. 7, and Tables S9 and S10 in ESI†).
Fig. 7. Quantification and statistical analysis of 5-mC (A), 5-hmC (B), 5-foC (C), and 5-caC (D) in the total RNA of human CRC tissues and tumor adjacent normal tissues (left panel) or in the total RNA of human HCC tissues and tumor adjacent normal tissues (right panel).
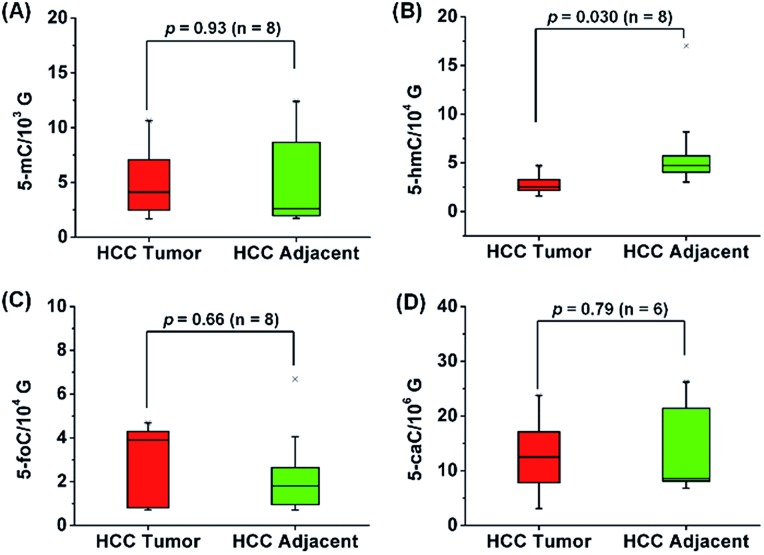
In addition, we further explored the content change of 5-hmC, 5-foC and 5-caC in the mRNA of HCC tissues since 5-hmC, 5-foC and 5-caC predominantly occur in mRNA. The results showed that 5-hmC significantly decreased in the mRNA of HCC tissues compared to tumor adjacent tissues (p = 0.03), but 5-foC and 5-caC showed no significant changes between HCC tissues and tumor adjacent tissues (Fig. 8, and Table S11 in ESI†).
Fig. 8. Quantification and statistical analysis of 5-mC (A), 5-hmC (B), 5-foC (C), and 5-caC (D) in the mRNA of human HCC tissues and tumor adjacent normal tissues.
The recent discovery of dynamic RNA modifications raised the possible regulatory roles of RNA modifications in biological processes.32,33 In the current study, we found that 5-hmC in RNA dramatically decreased in CRC and HCC tissues, which is consistent with previous reports of the significant decrease of 5-hmC in the DNA of CRC and HCC tissues.13,28 As the oxidation of 5-mC forms 5-hmC in RNA, similar to that of 5-mC to 5-hmC in DNA, the results demonstrated the depletion of 5-hmC in RNA may also have biological significance on epigenetic regulation in human tumors, which, however, still needs further investigation to fully understand their biological and pathological functions.
Conclusions
In the current study, we established a highly sensitive method using chemical labeling coupled with LC-ESI-MS/MS analysis for the simultaneous analysis of the oxidation products of 5-mC in the RNA of mammals. Upon BDEPE labeling, the detection sensitivities increased by 70 to 313 folds. Using the developed method, we were able to identify endogenous 5-caC in RNA. The discovery of endogenous 5-caC in RNA together with previously reported 5-hmC and 5-foC in RNA indicated that RNA may undergo the same cytosine oxidative demethylation as that in DNA. The generation of the intermediates of 5-mC oxidation products may present a signal mediating RNA degradation. In addition, we found that 5-hmC, 5-foC and 5-caC mainly occur in mRNA, and 5-hmC in RNA showed significant depletion in tumor tissues compared to tumor adjacent normal tissues. The marked depletion of 5-hmC in RNA in tumor tissues may have profound effects on epigenetic regulation in the development and formation of cancers.
Supplementary Material
Acknowledgments
The authors thank the financial support from the National Basic Research Program of China (973 Program) (2013CB910702, 2012CB720601), and the National Natural Science Foundation of China (21522507, 21475098).
Footnotes
†Electronic supplementary information (ESI) available. See DOI: 10.1039/c6sc01589a
References
- Machnicka M. A., Milanowska K., Osman Oglou O., Purta E., Kurkowska M., Olchowik A., Januszewski W., Kalinowski S., Dunin-Horkawicz S., Rother K. M., Helm M., Bujnicki J. M., Grosjean H. Nucleic Acids Res. 2013;41:D262–D267. doi: 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L., Song C. X., He C., Zhang Y. Annu. Rev. Biochem. 2014;83:585–614. doi: 10.1146/annurev-biochem-060713-035513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C. Nat. Chem. Biol. 2010;6:863–865. doi: 10.1038/nchembio.482. [DOI] [PubMed] [Google Scholar]
- Shen F., Huang W., Huang J. T., Xiong J., Yang Y., Wu K., Jia G. F., Chen J., Feng Y. Q., Yuan B. F., Liu S. M. J. Clin. Endocrinol. Metab. 2015;100:E148–E154. doi: 10.1210/jc.2014-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn D. B. Biochim. Biophys. Acta. 1960;38:176–178. doi: 10.1016/0006-3002(60)91219-1. [DOI] [PubMed] [Google Scholar]
- Saletore Y., Meyer K., Korlach J., Vilfan I. D., Jaffrey S., Mason C. E. Genome Biol. 2012;13:175. doi: 10.1186/gb-2012-13-10-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amort T., Souliere M. F., Wille A., Jia X. Y., Fiegl H., Worle H., Micura R., Lusser A. RNA Biol. 2013;10:1003–1009. doi: 10.4161/rna.24454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires J. E., Patel H. R., Nousch M., Sibbritt T., Humphreys D. T., Parker B. J., Suter C. M., Preiss T. Nucleic Acids Res. 2012;40:5023–5033. doi: 10.1093/nar/gks144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P. A. Nat. Rev. Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Yuan B. F. Adv. Clin. Chem. 2014;67:151–187. doi: 10.1016/bs.acc.2014.09.003. [DOI] [PubMed] [Google Scholar]
- Law J. A., Jacobsen S. E. Nat. Rev. Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Xiong J., Jiang H. P., Zheng S. J., Feng Y. Q., Yuan B. F. Anal. Chem. 2014;86:7764–7772. doi: 10.1021/ac5016886. [DOI] [PubMed] [Google Scholar]
- Chen M. L., Shen F., Huang W., Qi J. H., Wang Y., Feng Y. Q., Liu S. M., Yuan B. F. Clin. Chem. 2013;59:824–832. doi: 10.1373/clinchem.2012.193938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Lu Z. H. Chin. Chem. Lett. 2014;25:1559–1564. [Google Scholar]
- Kriaucionis S., Heintz N. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M., Koh K. P., Shen Y., Pastor W. A., Bandukwala H., Brudno Y., Agarwal S., Iyer L. M., Liu D. R., Aravind L., Rao A. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S., Shen L., Dai Q., Wu S. C., Collins L. B., Swenberg J. A., He C., Zhang Y. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y. F., Li B. Z., Li Z., Liu P., Wang Y., Tang Q., Ding J., Jia Y., Chen Z., Li L., Sun Y., Li X., Dai Q., Song C. X., Zhang K., He C., Xu G. L. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L., Guerrero C. R., Zhong N., Amato N. J., Liu Y., Liu S., Cai Q., Ji D., Jin S. G., Niedernhofer L. J., Pfeifer G. P., Xu G. L., Wang Y. J. Am. Chem. Soc. 2014;136:11582–11585. doi: 10.1021/ja505305z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H. Y., Xiong J., Qi B. L., Feng Y. Q., Yuan B. F. Chem. Commun. 2016;52:737–740. doi: 10.1039/c5cc07354e. [DOI] [PubMed] [Google Scholar]
- Ito S., Shen L., Dai Q., Wu S. C., Collins L. B., Swenberg J. A., He C., Zhang Y. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Zhang Y. Cell. 2014;156:45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Huang H., Liu D., Cheng Y., Liu X., Zhang W., Yin R., Zhang D., Zhang P., Liu J., Li C., Liu B., Luo Y., Zhu Y., Zhang N., He S., He C., Wang H., Chen D. Cell. 2015;161:893–906. doi: 10.1016/j.cell.2015.04.018. [DOI] [PubMed] [Google Scholar]
- Koziol M. J., Bradshaw C. R., Allen G. E., Costa A. S., Frezza C., Gurdon J. B. Nat. Struct. Mol. Biol. 2016;23:24–30. doi: 10.1038/nsmb.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M.-L., Fu X.-M., Liu J.-Q., Ye T.-T., Hou S.-Y., Huang Y.-Q., Yuan B.-F., Wu Y., Feng Y.-Q. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2012;905:67–74. doi: 10.1016/j.jchromb.2012.08.005. [DOI] [PubMed] [Google Scholar]
- Huang Y.-Q., Liu J.-Q., Gong H., Yang J., Li Y., Feng Y.-Q. Analyst. 2011;136:1515–1522. doi: 10.1039/c0an00736f. [DOI] [PubMed] [Google Scholar]
- Yuan B. F., Feng Y. Q. TrAC, Trends Anal. Chem. 2014;54:24–35. [Google Scholar]
- Tang Y., Zheng S. J., Qi C. B., Feng Y. Q., Yuan B. F. Anal. Chem. 2015;87:3445–3452. doi: 10.1021/ac504786r. [DOI] [PubMed] [Google Scholar]
- Li X., Zhu P., Ma S., Song J., Bai J., Sun F., Yi C. Nat. Chem. Biol. 2015;11:592–597. doi: 10.1038/nchembio.1836. [DOI] [PubMed] [Google Scholar]
- Delatte B., Wang F., Ngoc L. V., Collignon E., Bonvin E., Deplus R., Calonne E., Hassabi B., Putmans P., Awe S., Wetzel C., Kreher J., Soin R., Creppe C., Limbach P. A., Gueydan C., Kruys V., Brehm A., Minakhina S., Defrance M., Steward R., Fuks F. Science. 2016;351:282–285. doi: 10.1126/science.aac5253. [DOI] [PubMed] [Google Scholar]
- Brenner H., Kloor M., Pox C. P. Lancet. 2014;383:1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- Lee M., Kim B., Kim V. N. Cell. 2014;158:980–987. doi: 10.1016/j.cell.2014.08.005. [DOI] [PubMed] [Google Scholar]
- Fu Y., Dominissini D., Rechavi G., He C. Nat. Rev. Genet. 2014;15:293–306. doi: 10.1038/nrg3724. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.