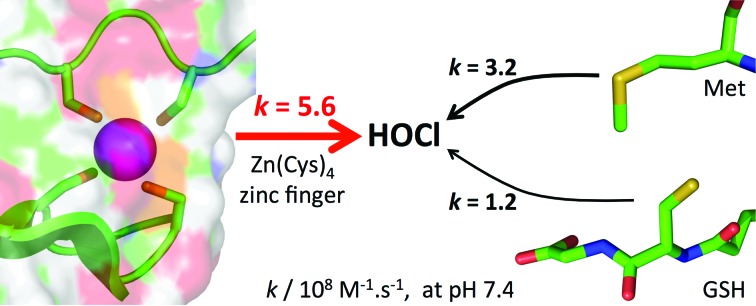
 Reaction rate constants of HOCl with zinc-bound cysteines are determined, demonstrating that zinc fingers are potent targets for HOCl and may serve as HOCl sensors.
Reaction rate constants of HOCl with zinc-bound cysteines are determined, demonstrating that zinc fingers are potent targets for HOCl and may serve as HOCl sensors.
Abstract
Hypochlorous acid (HOCl) is one of the strongest oxidants produced in mammals to kill invading microorganisms. The bacterial response to HOCl involves proteins that are able to sense HOCl using methionine, free cysteines or zinc-bound cysteines of zinc finger sites. Although the reactivity of methionine or free cysteine with HOCl is well documented at the molecular level, this is not the case for zinc-bound cysteines. We present here a study that aims at filling this gap. Using a model peptide of the Zn(Cys)4 zinc finger site of the chaperone Hsp33, a protein involved in the defence against HOCl in bacteria, we show that HOCl oxidation of this model leads to the formation of two disulfides. A detailed mechanistic and kinetic study of this reaction, relying on stopped-flow measurements and competitive oxidation with methionine, reveals very high rate constants: the absolute second-order rate constants for the reaction of the model zinc finger with HOCl and its conjugated base ClO– are (9.3 ± 0.8) × 108 M–1 s–1 and (1.2 ± 0.2) × 104 M–1 s–1, the former approaching the diffusion limit. Revised values of the second-order rate constants for the reaction of methionine with HOCl and ClO– were also determined to be (5.5 ± 0.8) × 108 M–1 s–1 and (7 ± 5) × 102 M–1 s–1, respectively. At physiological pH, the zinc finger site reacts faster with HOCl than methionine and glutathione or cysteine. This study demonstrates that zinc fingers are potent targets for HOCl and confirms that they may serve as HOCl sensors as proposed for Hsp33.
Introduction
Hypochlorous acid (HOCl) is one of the strongest oxidants produced by living organisms, especially to kill invading microorganisms.1,2 This is achieved within neutrophils, which are the most abundant white blood cells of the mammalian innate immune system.3 They internalize bacterial invaders into phagosomes, which contain a high concentration of hydrolytic enzymes and where a huge amount of reactive oxygen species is produced in order to kill microbes. Among reactive oxygen species, HOCl is considered as the main species responsible for oxidative killing.3 Myeloperoxidases catalyse its production from chloride ions and H2O2.4 The main targets of HOCl are proteins, but DNA and cell membranes can be damaged as well.5 Concerning proteins, at physiological pH, the fastest reactions occur with the sulfur-containing amino acids cysteine and methionine (k > 107 M–1 s–1).6–9 The reaction of HOCl with cysteine yields mainly disulfides, sulfinates or sulfonates whereas the reaction with methionine yields sulfoxides. Much slower are the reactions with aromatic side chains of tyrosine, tryptophan or histidine and with amino groups of lysine (k ∼ 102 to 105 M–1 s–1), which yield chlorinated aromatics and chloramines, respectively.10 HOCl-mediated protein oxidation is associated with severe protein aggregation, which is not the case with other oxidants.5 The bacterial arsenal to combat HOCl involves both low-molecular weight thiols, such as glutathione, and proteins.5 They scavenge HOCl, repair oxidative damage or prevent aggregation. These proteins can be regulated at the transcriptional or post-transcriptional level. HOCl-responsive transcription factors make use of cysteines11,12 or methionine,13 the most reactive amino acids, to sense HOCl. Among defence proteins is the holdase chaperone Hsp33, which prevents aggregation of key proteins.14–16 The active site of Hsp33 is constituted by a Zn(Cys)4 zinc finger site. Zinc fingers generally refer to small protein domains where a set of four cysteine or histidine side chains bind a zinc ion in a tetrahedral geometry with three types of arrangements: (Cys)4, (Cys)3(His) or (Cys)2(His)2. Zinc fingers can adopt various folds and have different functions: they can be pure structural elements, mediate protein/DNA, protein/RNA, or protein/protein interactions, act as redox switches, or catalyze alkyl transfer reactions.17–25 The four cysteines of the zinc finger site of Hsp33 are arranged in a CXC motif and a CXXC one.26 Activation of Hsp33 is caused by rapid HOCl oxidation of the zinc finger leading to the formation of two disulfides, one in the CXC motif, the other in the CXXC one, and the partial unfolding of Hsp33 that allows interaction with target proteins.14,27 In vitro, Hsp33 can also be activated by combination of H2O2 oxidation and elevated temperature.16 It is striking to note that Hsp33 uses zinc-bound cysteines to sense HOCl since it has been demonstrated that cysteines of zinc finger sites are less reactive than free cysteines toward other oxidants like H2O2 or O2.28 Whereas the reactivity of free cysteine and methionine toward HOCl is well documented,5,7–10 literature is scarce on molecular information concerning zinc-bound cysteines, especially regarding kinetics. Rare studies on proteins relate to Hsp33,14 metallothioneins,29,30 alcohol dehydrogenase29,31,32 or pro-matrilysin MMP-7.33 However, with proteins, the precise determination of the reaction rate constant of zinc-bound cysteines with HOCl is hampered by the presence of several other amino acids, like methionine or additional cysteines, that contribute to the reactivity of the whole protein. Herein, using a model peptide, we describe the detailed reactivity toward HOCl of an isolated zinc finger site mimicking that of Hsp33.
Results
Experimental design
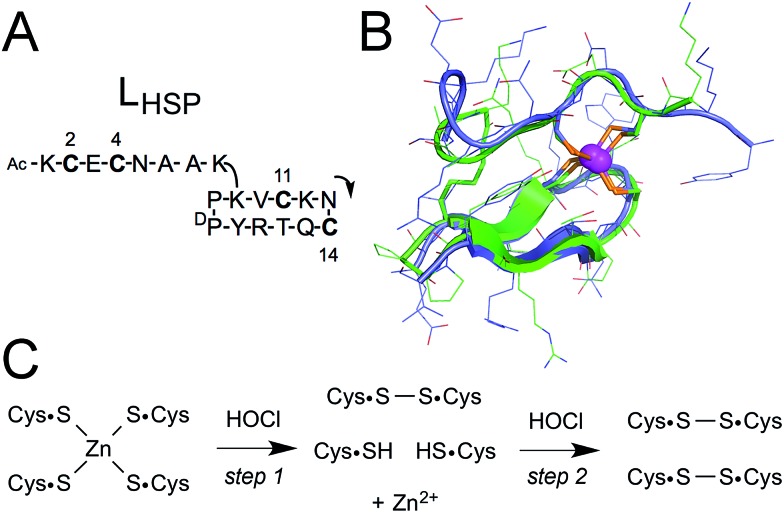
We have described previously a model peptide for the zinc finger site of Hsp33.34 This peptide, namely LHSP, is based on a cyclic peptide bearing a linear tail35 and is designed to match the amino acid sequence and the structure of Hsp33 (Fig. 1A).34 Both the cycle and the tail comprise a pair of cysteines (a CXXC motif in the cycle and a CXC motif in the linear tail). A structural analysis in solution by NMR has revealed that the zinc complex Zn·LHSP adopts the same fold as Hsp33 around the Zn2+ ion (Fig. 1B) and a kinetic study has shown that Zn·LHSP reacts with H2O2 at the same rate as Hsp33, thereby showing that Zn·LHSP is an excellent model to study the reactivity of the zinc finger site of Hsp33.34 Therefore, it provides a unique opportunity to assess the reactivity of an isolated zinc finger site without the interference of other reactive amino acids.
Fig. 1. (A) Amino acid sequence of LHSP. The arrow indicates the N-to-C direction in the cycle. The numbering of the four cysteines is also shown. (B) Superimposition of the NMR solution structure of the Zn·LHSP complex (green)34 and the crystallographic structure of the ZF site of Thermotoga maritima Hsp33 (blue, pdb ; 1VQ0).36 (C) Reaction scheme for the oxidation of the Zn(Cys)4 site by HOCl. Note that Zn2+ can also be coordinated to the two remaining cysteines after the first step.
In order to fully characterize the reaction between Zn·LHSP and HOCl, we first investigated by various techniques the stoichiometry of the reaction and the nature of the products. Then, a detailed kinetic study was carried out to determine the rate constants by a combination of stopped-flow measurements at high pH and competitive oxidation with methionine at physiological pH.
Identification of oxidation products
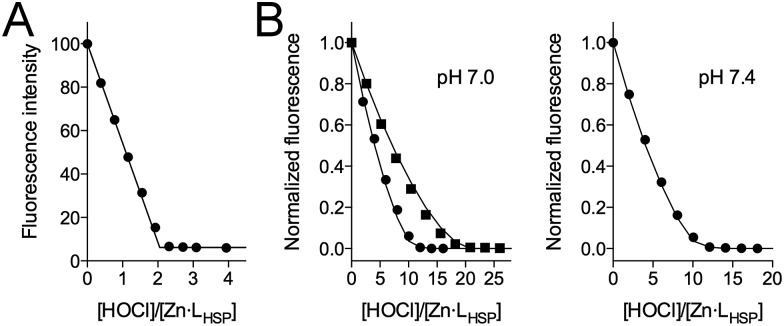
The titration of Zn·LHSP (20–40 μM) by HOCl in a phosphate buffer (pH 7.0) was first monitored by electronic absorption and tyrosine fluorescence. Upon addition of HOCl, the Cys·S– → Zn2+ ligand to metal charge transfer (LMCT) bands in the 200–250 nm region34,37 disappear until the end-point of the titration is reached at 2.0 equivalents of added HOCl (see Fig. S1 of ESI†). Similarly, the tyrosine fluorescence band at 305 nm (λex = 280 nm) decreases up to 2.0 equiv. HOCl (Fig. 2A). This indicates the oxidation of the cysteines by HOCl – leading to disulfides (see below) which are known quenchers of tyrosine fluorescence – and the release of Zn2+ as observed previously for H2O2.28,34 However, the reaction with HOCl is much faster than with H2O2 as it is completed within the mixing time, i.e. within 10 s in comparison with several tens of minutes for H2O2.28,38
Fig. 2. Evolution of tyrosine fluorescence (λex = 280 nm) upon titrating Zn·LHSP by HOCl in a phosphate buffer (20 mM) in the absence (A) and presence (B) of methionine. (A) [Zn·LHSP] = 40 μM at pH 7.0. (B) [Zn·LHSP] = 20 μM, [Met] = 190 μM (circles) and 380 μM (squares) at pH 7.0 (left) and [Met] = 190 μM (circles) at pH 7.4 (right). The solid lines in (B) correspond to the simulations of the fluorescence decrease with kapp = 1.75 × kMet (see ESI† for details).
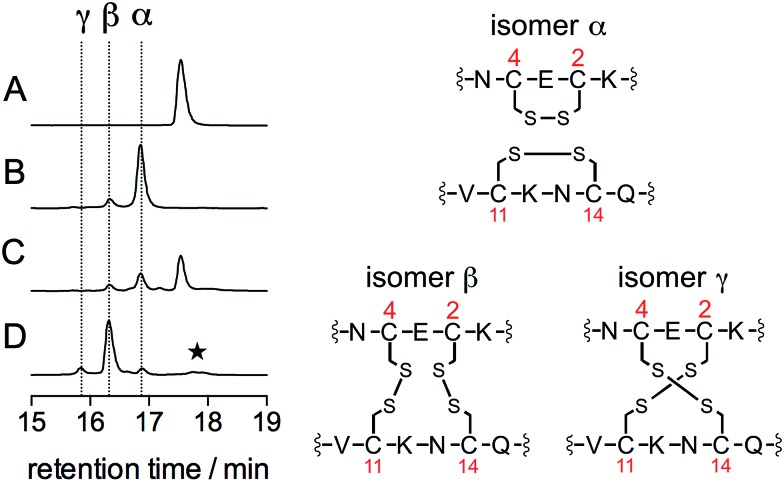
The oxidation products were identified by a combination of HPLC and mass spectrometry analyses. Fig. 3 shows the chromatograms recorded before (A) and after (B) addition of 3 equiv. of HOCl to a solution of Zn·LHSP in a phosphate buffer (20 mM, pH 7.0). Chromatogram (A) displays a single peak at 17.6 min corresponding to free LHSP (the Zn2+ ion is lost during elution due to the acidic gradient used, which contains 0.1% TFA). After reaction with HOCl, this peak has disappeared and two new peaks, α and β, were detected at 16.8 min (85%) and 16.3 min (15%), respectively. Mass analysis reveals a loss of 4 mass units compared to free LHSP for both peaks. The oxidation can be fully reversed by tris(carboxyethyl)phosphine, a well-known disulfide reductant. This indicates that α and β correspond to two out of the three possible bis-disulfide forms of LHSP. For comparison, as shown on chromatogram (D), oxidation of Zn·LHSP by excess H2O2 yields products α and β together with the third bis-disulfide γ (15.8 min) and also dimers of LHSP comprising a total of four disulfides at ca. 17.8 min.28 The relative proportion (α : β : γ) upon H2O2 oxidation is (8 : 72 : 12). It is noteworthy that the respective proportions of the bis-disulfide isomers vary markedly between HOCl and H2O2 oxidation but this will be discussed elsewhere. The three bis-disulfide isomers α, β and γ were prepared by oxidation of Zn·LHSP with either H2O2 or HOCl, isolated by HPLC and identified by LC/MS after digestion with both trypsin and GluC (Table 1). Indeed, each of the four cysteines of LHSP is flanked by a pair of amino acids prone to hydrolysis with either trypsin (K, R) or GluC (E). This allows isolation of each disulfide upon digestion and facile identification. Isomer α, the major bis-disulfide product obtained by HOCl oxidation, comprises one disulfide in the tail within the CXC motif and the other in the cycle within the CXXC motif, whereas β and γ feature disulfides that bridge the tail and the cycle (Fig. 3). Noteworthy, the disulfide pattern observed for α corresponds to that of the active form of Hsp33.14,27
Fig. 3. HPLC monitoring of the reaction of Zn·LHSP (35 μM) in a phosphate buffer (50 mM, pH 7.0, 298 K) with HOCl or H2O2: chromatograms (A)–(D) correspond to unreacted Zn·LHSP (A), Zn·LHSP + 3.0 eq. HOCl (B), Zn·LHSP + 1.0 eq. HOCl (C) and Zn·LHSP + 3000 eq. H2O2 (D). The star on chromatogram (D) denotes LHSP dimers with intermolecular disulfides.28 The disulfide patterns of oxidation products α, β and γ are shown on the right as identified by endopeptidase digestion and ESI-MS analyses.
Table 1. Summary of ESI-MS analysis after trypsin and GluC digestion of the three LHSP bis-disulfide isomers.
| Fragment | Mass/Da | Observed m/z |
||
| Isomer α (tR = 16.8 min) | Isomer β (tR = 16.3 min) | Isomer γ (tR = 15.8 min) | ||
| YDPPK | 503.27 | 504.2 (+), 252.5 (2+) | 504.2 (+), 252.5 (2+) | 504.2 (+), 252.5 (2+) |
| C 2EC4NAAK (1 S–S) | 735.27 | 736.3 (+), 368.6 (2+) | — | — |
| VC11K + NC14QTR (1 S–S) | 966.44 | 967.5 (+), 484.2 (2+) | — | — |
| AcKC2E + NC14QTR (1 S–S) | 1038.42 | — | 1039.4 (+), 520.1 (2+) | — |
| C 2E + NC14QTR (1 S–S) | 868.31 | — | 869.3 (+), 435.1 (2+) | — |
| C 4NAAK + VC11K (1 S–S) | 851.40 | — | 852.4 (+), 426.6 (2+) | — |
| AcKC2E + VC11K (1 S–S) | 766.33 | — | — | 767.3 (+), 384.1 (2+) |
| C 2E + VC11K (1 S–S) | 596.23 | — | — | 597.2 (+), 299.1 (+) |
| C 4NAAK + NC14QTR (1 S–S) | 1123.49 | — | — | 562.7 (2+) |
| AcKC2EC4NAAK + C11K + NC14QTR (2 S–S) | 1871.81 | — | 936.9 (2+), 625.1 (3+) | 937.4 (2+), 625.1 (3+) |
| AcKC2EC4NAAK + VC11K + NC14QTR (2 S–S) | 1701.71 | — | 851.9 (2+), 568.4 (3+) | 851.9 (2+), 568.5 (3+) |
| AcKC2EC4NAAK + C11KNC14QTR (2 S–S) | 1853.80 | — | 927.9 (2+), 619.2 (3+) | — |
| C 2EC4NAAK + C11KNC14QTR (2 S–S) | 1683.69 | — | 842.8 (2+), 562.5 (3+) | — |
Kinetic study: stopped-flow measurements at high pH
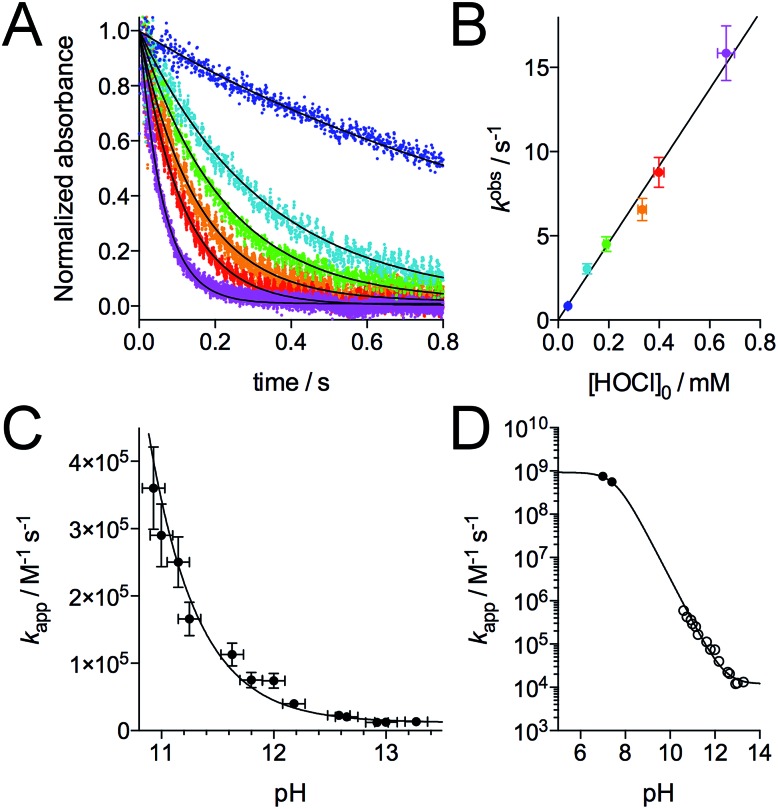
Regarding the mechanism of the reaction, the formation of the two disulfides may be sequential, as observed for H2O2,28 or simultaneous if two molecules of HOCl react simultaneously with the zinc finger. As noted above, the reaction between Zn·LHSP and HOCl is very fast. In order to get a deeper insight into its kinetics and its mechanism, we monitored the reaction using a stopped-flow apparatus, following the disappearance of the LMCT band at 230 nm. However, at pH 7.0, the reaction is completed within the 3 ms mixing time of the apparatus. Nevertheless, both HOCl and its conjugate base hypochlorite (ClO–, pKa(HOCl/ClO–) = 7.54)39 can react with the zinc finger but ClO– is generally far less reactive than HOCl.7,8,40–42 Therefore, we studied the reaction at higher pH to slow it down. Fig. 4A shows the decay of the LMCT band observed at pH 12.6 for various concentrations of HOCl under pseudo-first-order conditions (at least a 10-fold excess of oxidant relative to Zn·LHSP (2–10 μM), see ESI† for details). All kinetic traces could be fitted to a single exponential and the amplitude of the absorbance decay is in agreement with the oxidation of the four cysteines (ESI†). The observed first-order rate constant kobs shows a linear dependence on the total concentration of oxidant [HOCl]0 = [HOCl] + [ClO–] (Fig. 4B and ESI†). This indicates that the rate-determining step is second-order, first-order in complex and first-order in oxidant (eqn (1)) and that the formation of the two disulfides is sequential.
| Rate = kapp[HOCl]0[Zn·LHSP] | 1 |
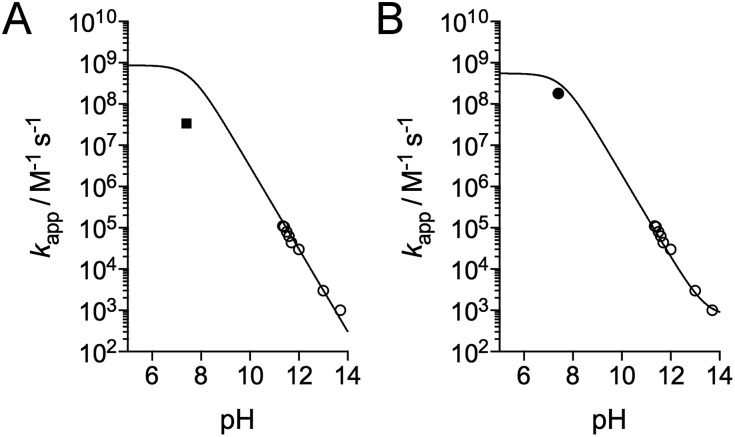
Fig. 4. Kinetics of the reaction of Zn·LHSP with HOCl. (A) Stopped-flow absorbance monitoring (λ = 230 nm) of the reaction at pH 12.6 (298 K) with various concentration of HOCl (monoexponential fits are shown as solid lines). (B) Plot of the observed first order rate constant kobs derived from monoexponential fits of kinetic traces against the concentration of HOCl. The slope of the linear regression yielded kapp = (2.3 ± 0.4) × 104 M–1 s–1 at pH 12.6. (C) pH-Dependence of kapp in the high-pH region. (D) pH-Dependence of kapp between pH 5 and 14 showing kapp values determined by stopped-flow measurements (open circles) and by competitive titrations of Zn·LHSP and methionine by HOCl (black circles). The solid lines in (C) and (D) correspond to the fit of the pH-dependence of kapp using eqn (4). The fit over the whole pH range yielded k(Zn·LHSP + HOCl) = (9.3 ± 0.8) × 108 M–1 s–1 and k(Zn·LHSP + ClO–) = (1.2 ± 0.2) × 104 M–1 s–1.
The major peaks observed on the HPLC chromatogram recorded after oxidation of Zn·LHSP by default HOCl (only one equivalent) correspond to unreacted LHSP and the bis-disulfides isomers α and β (Fig. 3, chromatogram C). This means that the mono-disulfide forms of LHSP are not significantly accumulated during the reaction. The attack of HOCl by the Zn(Cys)4 site, which leads to the formation of the first disulfide, is the rate-determining step (step 1 in Fig. 1C), the formation of the second disulfide (step 2) being at least 5 times faster. A similar trend was observed previously for the H2O2 oxidation of Zn·LHSP with reaction rate constants of 0.11 M–1 s–1 and 0.8 M–1 s–1 for the formation of the first and second disulfide, respectively.28 The apparent second-order rate constant of the rate-determining step kapp was determined by stopped-flow measurements at various pH values over the range 10.8–13.3.‡ § Fig. 4C shows the pH-dependence of kapp. Indeed, Zn·LHSP can react with both HOCl and ClO– to yield the mono-disulfide intermediate. Therefore, the rate of consumption of Zn·LHSP is given by eqn (2), where k(Zn·LHSP + HOCl) and k(Zn·LHSP + ClO–) are the second-order rate constants for the reaction of Zn·LHSP with HOCl and ClO–, respectively.¶
| Rate = k(Zn·LHSP + HOCl)[HOCl][Zn·LHSP] + k(Zn·LHSP + ClO–)[ClO–][Zn·LHSP] | 2 |
The rate can also be expressed as a function of [HOCl]0, the total concentration of oxidant used for stopped-flow measurements under pseudo-first-order conditions by eqn (3), where Ka is the ionization constant of HOCl.
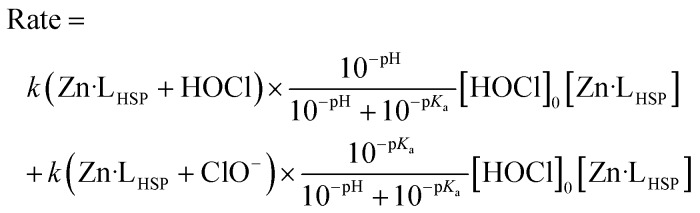 |
3 |
Finally, combining eqn (1) and (3) gives eqn (4) linking kapp and pH.
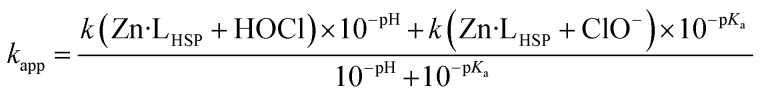 |
4 |
Eqn (4) was used to fit the pH-dependence of the experimental kapp values. An excellent fit was obtained (Fig. 4C, solid line), which yielded k(Zn·LHSP + HOCl) = (9.2 ± 0.9) × 108 M–1 s–1 and k(Zn·LHSP + ClO–) = (1.2 ± 0.2) × 104 M–1 s–1. As mentioned above, the reaction between Zn·LHSP and HOCl at physiological pH is too fast to be monitored directly. In order to confirm the values of rate constants derived from high-pH stopped-flow measurements, we decided to perform competition experiments between Zn·LHSP and methionine at physiological pH. However, there is some discrepancy in the literature concerning the rate constants of the reaction between methionine and HOCl/ClO–.7,9 Therefore, we have reassessed these rate constants.
Reassessment of the oxidation rate constant of methionine by HOCl
The oxidation of methionine (Met) by HOCl yields the sulfoxide MetO.7 As for Zn·LHSP, both HOCl and its conjugated base ClO– can react with Met to give MetO and the respective second-order rate constants for these reactions are k(Met + HOCl) and k(Met + ClO–). Armesto et al. have measured the rate constants for this reaction at 298 K.7 Again, the reaction is too fast to be monitored directly around pH 7, even by stopped-flow techniques, and stopped-flow measurements were performed at high pH. Apparent second-order rate constants were obtained at various pH values above 11. Armesto et al. fitted the pH-dependence of the apparent rate constants to eqn (5), where Ka is the ionization constant of HOCl, assuming that k(Met + ClO–) is negligible (Fig. 5A, open circles and solid line). The fit yielded k(Met + HOCl) = (8.7 ± 0.2) × 108 M–1 s–1.7Eqn (5) allows calculating kapp(Met) = 5.1 × 108 M–1 s–1 at pH 7.4 from the high-pH values.
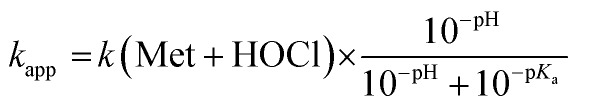 |
5 |
Fig. 5. pH-Dependence of the apparent rate constant for the reaction of methionine with HOCl. (A) Data reported by Armesto et al.7 (open circles) and Storkey et al.9 (square). The solid line corresponds to the fit proposed by Armesto et al. with eqn (5) and k(Met + HOCl) = 8.7 × 108 M–1 s–1.7 (B) Data reported by Armesto et al. (open circles) and re-evaluated apparent constant at pH 7.4 determined in this work (black circle). The solid line corresponds to the fit to eqn (9), which yielded k(Met + HOCl) = (5.5 ± 0.8) × 108 M–1 s–1 and k(Met + ClO–) = 700 ± 500 M–1 s–1.
Recently, Storkey et al. reported the apparent second-order rate constant for the oxidation of Met by HOCl at pH 7.4 and 295 K.9 First, they determined the apparent oxidation rate constant of FmocMet, kapp(FmocMet), by competition with a dithiol (3,3′-dithiodipropionic acid) to be (1.5 ± 0.2) × 108 M–1 s–1. This value was double-checked by competition with SCN–. Then, they performed a competition with Met using a method relying on HPLC separation with detection of the Fmoc group fluorescence. The apparent second-order rate constant kapp(Met) for the oxidation of Met at pH 7.4 was (0.34 ± 0.05) × 108 M–1 s–1, which is 15 times smaller than Armesto's value (Fig. 5A). In addition, they report an apparent second-order rate constant for N-acetylmethionine of (1.7 ± 0.2) × 108 M–1 s–1, which is very close to that of FmocMet, in contrast to that of Met. Since no strong influence is expected from substituents on the amino group, the Met value appears doubtful and this prompted us to reassess the apparent second-order rate constant for Met at pH 7.4. As proposed by Storkey et al.,9 we performed competition studies with FmocMet using HPLC and fluorescence monitoring.
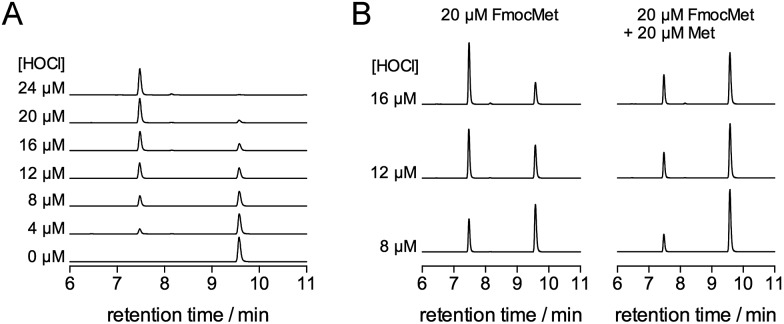
Fig. 6A shows the HPLC monitoring of the oxidation of FmocMet (20 μM) by HOCl at pH 7.4. Upon HOCl addition, the peak at 9.6 min corresponding to FmocMet disappears with concomitant appearance of a peak at 7.4 min corresponding to the sulfoxide product FmocMetO. Fig. 6B compares oxidations of FmocMet (20 μM) performed in the absence and presence of Met (20 μM). The chromatograms show that the presence of one equivalent of Met decreases the yield of FmocMetO by ca. 50%, suggesting similar reaction rate constants for FmocMet and Met.
Fig. 6. (A) Oxidation of FmocMet (20 μM) by HOCl at pH 7.4 (phosphate buffer 10 mM, 298 K) monitored by HPLC. FmocMet elutes at 9.6 min and its oxidation product FmocMetO at 7.5 min. (B) Competition between FmocMet and Met at pH 7.4 monitored by HPLC. The left and right panels compare the chromatograms obtained after oxidation of FmocMet (20 μM) by HOCl in the absence (left) and presence (right) of Met (20 μM). Oxidations were performed at 298 K in a phosphate buffer (10 mM).
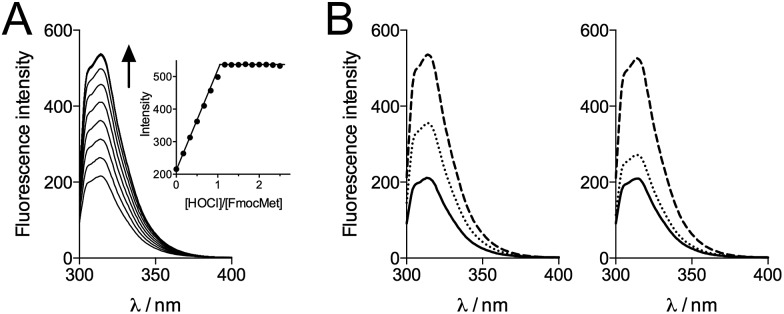
In order to get more precise data, we monitored the oxidation of FmocMet by fluorescence spectroscopy. Indeed, the Fmoc moiety is fluorescent when excited at 265 nm. Titration of FmocMet by HOCl at pH 7.4 (298 K) shows that the fluorescence emission increases linearly up to one equivalent HOCl and remains constant above one equivalent (Fig. 7A). This indicates that the sulfoxide FmocMetO emits more than FmocMet and that fluorescence can be used to monitor the reaction.
Fig. 7. (A) Fluorescence monitoring (λex = 265 nm) of the oxidation of FmocMet (20 μM) by HOCl at pH 7.4 in a phosphate buffer (10 mM, 298 K). The inset shows the intensity of the emission at 314 nm against added HOCl equivalents. (B) Competitive oxidation of FmocMet and Met by HOCl at pH 7.4 monitored by fluorescence (λex = 265 nm) in a phosphate buffer (10 mM, 298 K): fluorescence spectra of solutions of FmocMet (20 μM) in the absence (left panel) and presence (right panel) of Met (20 μM). Solid, dotted and dashed lines correspond to spectra recorded before addition of HOCl, after addition of HOCl (10 μM) and after addition of excess HOCl, respectively.
We thus performed competition experiments between FmocMet and Met. For this purpose, solutions containing FmocMet and Met or only FmocMet in various concentrations were oxidized at pH 7.4 in a phosphate buffer by identical but variable amounts of HOCl. The fluorescence spectra were recorded (i) before and (ii) after addition of HOCl, and (iii) after addition of excess HOCl for complete oxidation of FmocMet (Fig. 7B). For each solution, the yield of FmocMetO can be calculated from such a set of three spectra. Analysis of the concentration-dependent inhibition of FmocMetO formation by Met, as described by Storkey et al.9 (see ESI† for details), gave kapp(Met)/kapp(FmocMet) = 1.3 ± 0.1. Therefore, our competition experiments lead to a new experimental value for kapp(Met) at pH 7.4, (1.9 ± 0.3) × 108 M–1 s–1, which is ca. one order of magnitude higher than that proposed by Storkey et al. and more in line with the values they determined for FmocMet and N-acetylmethionine. This value (plotted as a black circle in Fig. 5B) is also in better agreement with the high-pH values of Armesto et al.7 (open circle) as shown by the fit of the pH-dependence of kapp(Met) fitted over the pH range 5–14 using eqn (6) (solid line). This fit yielded revised values of the second-order rate constants for the reaction of Met with HOCl and ClO–: k(Met + HOCl) = (5.5 ± 0.8) × 108 M–1 s–1 and k(Met + ClO–) = 700 ± 500 M–1 s–1.
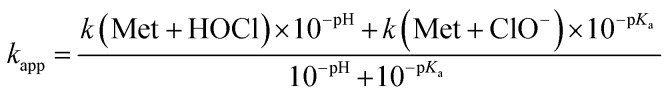 |
6 |
The value of kapp(Met) at pH 7.4 can be refined using eqn (6) and values determined for k(Met + HOCl) and k(Met + ClO–): kapp(Met) = 3.2 × 108 M–1 s–1. Similarly, at pH 7.0, kapp(Met) = 4.3 × 108 M–1 s–1.
Competition between Zn·LHSP and methionine at physiological pH
In order to confirm the rate constants of the reaction between Zn·LHSP and HOCl/ClO–, competitive titrations of Zn·LHSP and methionine by HOCl were conducted at pH 7.0 and 7.4 with tyrosine fluorescence monitoring (Fig. 2B). Simulations of the titrations yielded a value of 1.75 for the ratio kapp(Zn·LHSP)/kapp(Met) at both pH (see ESI† for simulation details). Therefore, values of 7.5 × 108 M–1 s–1 and 5.6 × 108 M–1 s–1 can be calculated at pH 7.0 and 7.4, respectively, for the apparent second-order rate constant of the reaction between Zn·LHSP and HOCl. These values are displayed in Fig. 4D (black circles) together with the stopped-flow-derived values at high pH (open circles). Fitting the two sets of kapp values together to eqn (4) (Fig. 4D, solid line) shows a remarkable agreement between them and yields values for k(Zn·LHSP + HOCl) = (9.3 ± 0.8) × 108 M–1 s–1 and k(Zn·LHSP + ClO–) = (1.2 ± 0.2) × 104 M–1 s–1 that are identical to those derived from high-pH values only, within error margin.
Discussion
In this article, we describe for the first time the reactivity toward HOCl of an isolated zinc finger reproducing the Zn(Cys)4 site of Hsp33. We have shown that the zinc-bound cysteines are the primary targets of HOCl in this model and determined unambiguously the second-order reaction rate constant of the Zn(Cys)4 site. The role of HOCl as bactericidal agent is now well established but the understanding of how living organisms combat HOCl is still in its infancy. Hsp33 was one of the first proteins identified in bacterial defence against HOCl and amazingly, this protein uses its Zn(Cys)4 site to sense HOCl. The complete characterization of the reaction between a protein and HOCl (or ClO–) is generally not straightforward. The targets of HOCl may be multiple in a protein as several functional groups can react very rapidly with this oxidant in contrast to other less potent oxidants like H2O2 or O2 that react almost exclusively with the thiol function of cysteine. Therefore, careful identification of the products is required in order to clearly identify the reacting groups prior to determination of rate constants. This has been stressed for a small molecule like cystine (cysteine disulfide)43 and this is even more dramatic for a protein. Additionally, proteins contain several identical amino acids in their sequence providing several target sites for HOCl. This increases the number of possible products. Nevertheless, evaluations of rate constants for the reaction between HOCl and zinc-bound cysteines of zinc fingers or related sites in proteins have been reported in the literature. They concern yeast alcohol dehydrogenase31,32 and pro-matrilysin MMP-7.33 Although correlations between protein activity, cysteine oxidation, Zn2+ release and oxidant concentration were observed, the rate constants could not be unambiguously attributed to the oxidation of the active site zinc-bound cysteines, as pointed out in these studies. For example, in addition to its active Zn(Cys)2(His) site, yeast alcohol dehydrogenase features four zinc-bound cysteines arranged in a structural Zn(Cys)4 site, two free cysteines and eight methionines, which are other possible targets of HOCl.44 Our strategy relies on the use of a model peptide that includes only the zinc-bound cysteines but no methionine or free cysteines that would react at similar rate and preclude specific determination of the reaction rate constant of the zinc finger cysteines. The lysine or tyrosine side-chains of LHSP, which are important for the solubility of the model and its proper folding, respectively, react at much lower rate than the Zn(Cys)4 site thereby allowing the precise determination of the rate constant of the zinc finger.10 In addition, the compact design of our model based on a branched cyclic peptide makes it a robust molecule that remains stable and soluble even at very high pH. This allows stopped-flow measurements at pH above 10.5, which would not be possible with proteins that are generally not stable, soluble or correctly folded at such elevated pH.
The rate constants for HOCl oxidation of cysteine or methionine derivatives, including those for Zn·LHSP and Met determined in this study, are summarized in Table 2. All of them lie in the 107 to 109 M–1 s–1 range. Of note, the absolute rate constant of the reaction between Zn·LHSP and HOCl, k(Zn·LHSP + HOCl), approaches the diffusion limit. At pH 7.4, the apparent second-order rate constant for the reaction of HOCl with Zn·LHSP exceeds those of cysteine and methionine derivatives.9 Interestingly, Fliss et al. observed that HOCl can mobilize zinc in cells and they proposed that zinc might be released from metallothioneins, as happens in vitro.29,30,45,46 The rate constant at physiological pH reported here for zinc-bound cysteines shows that they could compete with the most reactive amino acid side chains and support the hypothesis of Fliss et al. As cysteine and methionine are used to sense HOCl in defence proteins, this confirms as well that the zinc finger site of Hsp33 can act as a HOCl sensor.
Table 2. Absolute and apparent (at pH 7.4) rate constants for the reaction of HOCl and ClO– with various sulfur-containing molecules (X).
| X | k(X + HOCl)/108 M–1 s–1 | k(X + ClO–)/103 M–1 s–1 | k app at pH 7.4/108 M–1 s–1 |
| Zn·LHSP a | 9.3 ± 0.8 | 12 ± 2 | 5.6 ± 0.5 |
| Cysteine b | — | — | 3.6 ± 0.5 |
| N-Acetylcysteine b | — | — | 0.29 ± 0.04 |
| Glutathione b | — | — | 1.2 ± 0.2 |
| Methionine a | 5.5 ± 0.8 | 0.7 ± 0.2 | 3.2 ± 0.3 |
| N-Acetylmethionine b | — | — | 1.7 ± 0.2 |
aThis work, measured at 298 K.
b Ref. 9, measured 295 K.
It is noteworthy that the zinc-bound cysteines of Zn·LHSP react faster with HOCl than free cysteines, which is surprising since Zn2+ exhibits a protective effect on cysteines regarding H2O2 or O2 oxidation.28 Indeed, the use of the zinc-bound cysteines to sense HOCl combines three advantages: (i) zinc, by lowering the reactivity of cysteines toward H2O2 and O2 and increasing their reactivity toward HOCl, enhances the selectivity of the detection of HOCl over H2O2 or O2 compared to free thiols, thereby approaching the perfect selectivity displayed by methionine, which is resistant to H2O2 or O2 in contrast to HOCl, (ii) large structural rearrangement is allowed upon oxidation, which can be obtained with cysteines when a disulfide is formed between remote cysteines47 but not with methionine, and (iii) disulfides can be easily reduced in contrast to methionine sulfoxide, which requires specialized reducing enzymes, allowing oxidation to be reversed easily. Altogether, this makes zinc fingers ideal redox switches for HOCl sensing, which is used by Hsp33.
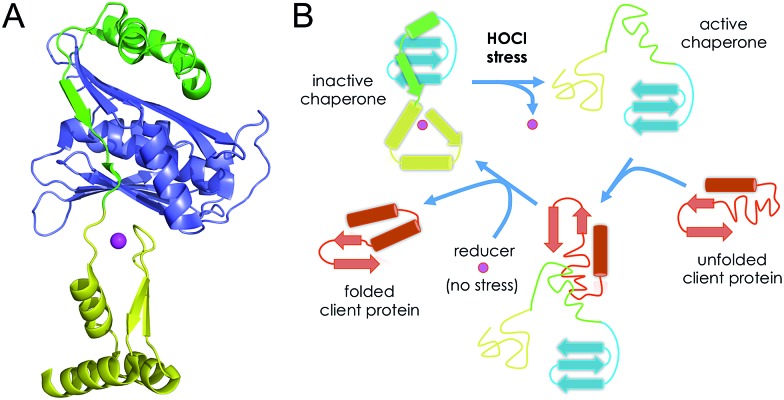
Hsp33 is composed of three domains: a C-terminal domain, which contains the Zn(Cys)4 zinc finger, a N-terminal domain and a metastable linker region. In its reduced and zinc-bound state, the three domains are well folded (Fig. 8A) and the linker region and the C-terminal domain wrap around the N-terminal domain.48 In its active form, in which the four cysteines of the zinc finger site are oxidized, both the C-terminal domain and the linker region are unfolded.16,49 Jakob et al. have identified the disordered linker region as the binding site for client proteins that need to be protected from unfolding.50 They proposed that the C-terminal zinc-binding domain primarily serves as a switch to control the folding of the linker region (Fig. 8B). In non-stress conditions, the cysteines are reduced and bound to zinc and Hsp33 is inactive with the folded C-terminal domain contacting the N-terminal domain, which locks the linker region in its inactive folded state. In HOCl stress conditions, the cysteines of the Zn(Cys)4 site are oxidized, the C-terminal domain unfolds thereby allowing the linker to unfold as well and bind client proteins. The data presented here support this mechanism. The higher rate constant measured for Zn·LHSP compared to methionine points to oxidation of the Zn(Cys)4 site prior to oxidation of methionine, which are at least partially buried in the case of the protein and, thus, probably less reactive than free methionine. Therefore, our data predict that the Zn(Cys)4 site should be the first target for HOCl in Hsp33. This is confirmed by mass spectrometry analysis of the HOCl-oxidized protein, which shows a predominant peak corresponding to the bis-disulfide form of the protein.14 Hence, the first event in the activation mechanism should be the rapid HOCl oxidation of the cysteine of the Zn(Cys)4 site into disulfides. The C-terminal domain has a low hydrophobic amino acid content and its helices contain several amino acids with β-branched side chains, which is unfavourable with respect to helix stabilization. The first cysteine pair (CXC motif) is located close to the linker region in the protein sequence and the other (CXXC motif) in the middle of C-terminal domain. In the reduced form of Hsp33, the zinc ion bridges the two pairs of cysteines and constrains the C-terminal domain into its folded state.51 Jakob et al. have shown that the disulfides in active Hsp33 are formed into each pair of cysteines,27 one in the CXC motif and the other in the CXXC motif, as observed with HOCl oxidation of Zn·LHSP. Hence, HOCl breaks the link between the two pairs of cysteines and elicits unfolding of the C-terminal domain, which unlocks the metastable linker region. When normal non-stress conditions are restored, disulfides are reduced and zinc binds to the cysteines forcing the C-terminal domain to fold: Hsp33 recovers its inactive fully folded form.
Fig. 8. (A) Structure of reduced zinc-loaded Hsp33 highlighting the C-terminal zinc finger domain (yellow), the N-terminal domain (in blue) and the linker region (in green). The zinc ion is shown in magenta. (B) Mechanism proposed by Jakob et al. for the HOCl activation of Hsp33.
H2O2 oxidation of Hsp33 at low temperature (30 °C) yields an inactive form in contrast to HOCl. Jakob et al. proposed that this inactive form is a partially oxidized protein with an oxidized CXXC motif and a reduced CXC one and they proposed that oxidation of the CXC is essential to unfold the linker region.14,16 However, in such a half-oxidized protein, it is not clear to us why the linker remains folded since, in the absence of a bridge between the two cysteine pairs, the C-terminal domain should unfold and unlock the linker region. We have observed that H2O2 oxidation of Zn·LHSP yields a disulfide pattern different from HOCl, with disulfides bridging the CXC and CXXC motifs. Our model suggests an alternative inactive form of Hsp33 that would be fully oxidized but would present disulfide bridges between the two pairs of cysteines, thereby preventing unfolding of the C-terminal domain. Therefore, insights are still to be gained to fully understand the mechanism of Hsp33 activation at the molecular level and the difference between HOCl and H2O2 oxidation with respect to Zn·LHSP or Hsp33 deserves to be investigated.
Another interesting point concerning Hsp33 is its high conservation in prokaryotes but absence from higher eukaryotes. Recently, Get3, another redox-regulated chaperone, was shown to protect eukaryotic cells against oxidative protein unfolding, as does Hsp33 in bacteria.52 Of particular note, Get3 features a zinc finger site and loses zinc upon oxidation. This suggests a sensing mechanism similar to Hsp33 with possible use of the zinc finger site to detect and signal HOCl.
Conclusion
In this article, we have characterized the products and the kinetics of the reaction of Zn·LHSP, a model Zn(Cys)4 zinc finger peptide, with hypochlorous acid. This reaction yields two disulfides. The absolute second-order rate constants of the reaction of Zn·LHSP with HOCl and its conjugated base ClO– are (9.3 ± 0.8) × 108 M–1 s–1 and (1.2 ± 0.2) × 104 M–1 s–1, respectively, the former approaching the diffusion limit. At physiological pH, the apparent rate constant is superior to that of reaction of methionine or free cysteines, the most reactive amino acids known so far. The study presented here constitutes the first unequivocal determination of the reaction rate constants of HOCl with zinc-bound cysteines. It establishes on strong molecular and kinetic bases that zinc fingers can be targets for HOCl and that some of them can be used as redox switches to sense HOCl as proposed for Hsp33. In addition, we provide a clarification regarding the discrepancy found in the literature concerning the rate constant of the reaction between methionine and HOCl. Work is now in progress to determine if other zinc fingers react at similar rate with HOCl.
Supplementary Material
Acknowledgments
We acknowledge the Agence Nationale de la Recherche (ANR-06-JCJC-0018) and Labex ARCANE (ANR-11-LABX-0003-01) for financial support.
Footnotes
†Electronic supplementary information (ESI) available: Experimental details concerning absorption, fluorescence and HPLC monitoring of the HOCl oxidation of Zn·LHSP, identification of oxidation products, stopped-flow experiments, re-assessment of rate constants for methionine oxidation and simulation of competitive HOCl titrations of methionine and Zn·LHSP. See DOI: 10.1039/c6sc00974c
‡The stability of the model peptide at high pH was checked by HPLC and circular dichroism over several hours.
§Below pH 10.8, the reaction is too fast to be monitored by stopped-flow methods.
¶The pKa of the first protonation of the Zn(Cys)4 core is 4.1,53 which is ca. 3 units below the lowest investigated pH. Therefore, the protonated forms of Zn·LHSP were not considered in the equations.
References
- Pullar J. M., Vissers M. C. M., Winterbourn C. C. IUBMB Life. 2000;50:259–266. doi: 10.1080/713803731. [DOI] [PubMed] [Google Scholar]
- Dahl J.-U., Gray M. J., Jakob U. J. Mol. Biol. 2015;427:1549–1563. doi: 10.1016/j.jmb.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterbourn C. C., Kettle A. J. Antioxid. Redox Signaling. 2013;18:642–660. doi: 10.1089/ars.2012.4827. [DOI] [PubMed] [Google Scholar]
- Klebanoff S. J. J. Leukocyte Biol. 2005;77:598–625. doi: 10.1189/jlb.1204697. [DOI] [PubMed] [Google Scholar]
- Gray M. J., Wholey W.-Y., Jakob U. Annu. Rev. Microbiol. 2013;67:141–160. doi: 10.1146/annurev-micro-102912-142520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterbourn C. C., in Encyclopedia of Radicals in Chemistry, Biology and Materials, John Wiley & Sons, Ltd, 2012. [Google Scholar]
- Armesto X. L., Canle M., Fernandez M. I., Garcia M. V., Santaballa J. A. Tetrahedron. 2000;56:1103–1109. [Google Scholar]
- Nagy P., Ashby M. T. J. Am. Chem. Soc. 2007;129:14082–14091. doi: 10.1021/ja0737218. [DOI] [PubMed] [Google Scholar]
- Storkey C., Davies M. J., Pattison D. I. Free Radical Biol. Med. 2014;73:60–66. doi: 10.1016/j.freeradbiomed.2014.04.024. [DOI] [PubMed] [Google Scholar]
- Pattison D. I., Davies M. J. Chem. Res. Toxicol. 2001;14:1453–1464. doi: 10.1021/tx0155451. [DOI] [PubMed] [Google Scholar]
- Parker B. W., Schwessinger E. A., Jakob U., Gray M. J. J. Biol. Chem. 2013;288:32574–32584. doi: 10.1074/jbc.M113.503516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M. J., Li Y., Leichert L. I.-O., Xu Z., Jakob U. Antioxid. Redox Signaling. 2015;23:747–754. doi: 10.1089/ars.2015.6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drazic A., Miura H., Peschek J., Le Y., Bach N. C., Kriehuber T., Winter J. Proc. Natl. Acad. Sci. U. S. A. 2013;110:9493–9498. doi: 10.1073/pnas.1300578110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter J., Ilbert M., Graf P. C. F., Oezcelik D., Jakob U. Cell. 2008;135:691–701. doi: 10.1016/j.cell.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wholey W.-Y., Jakob U. Mol. Microbiol. 2012;83:981–991. doi: 10.1111/j.1365-2958.2012.07982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilbert M., Horst J., Ahrens S., Winter J., Graf P. C. F., Lilie H., Jakob U. Nat. Struct. Mol. Biol. 2007;14:556–563. doi: 10.1038/nsmb1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laity J. H., Lee B. M., Wright P. E. Curr. Opin. Struct. Biol. 2001;11:39–46. doi: 10.1016/s0959-440x(00)00167-6. [DOI] [PubMed] [Google Scholar]
- Matthews J. M., Sunde M. IUBMB Life. 2002;54:351–355. doi: 10.1080/15216540216035. [DOI] [PubMed] [Google Scholar]
- Krishna S. S., Majumdar I., Grishin N. V. Nucleic Acids Res. 2003;31:532–550. doi: 10.1093/nar/gkg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maret W., Li Y. Chem. Rev. 2009;109:4682–4707. doi: 10.1021/cr800556u. [DOI] [PubMed] [Google Scholar]
- Andreini C., Bertini I., Cavallaro G. PLoS One. 2011;6:e26325. doi: 10.1371/journal.pone.0026325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalek J. L., Besold A. N., Michel S. L. J. Dalton Trans. 2011;40:12619–12632. doi: 10.1039/c1dt11071c. [DOI] [PubMed] [Google Scholar]
- Burdach J., O'Connell M. R., Mackay J. P., Crossley M. Trends Biochem. Sci. 2012;37:199–205. doi: 10.1016/j.tibs.2012.02.001. [DOI] [PubMed] [Google Scholar]
- Lee S. J., Michel S. L. J. Acc. Chem. Res. 2014;47:2643–2650. doi: 10.1021/ar500182d. [DOI] [PubMed] [Google Scholar]
- Pace N. J., Weerapana E. Biomolecules. 2014;4:419–434. doi: 10.3390/biom4020419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob U., Eser M., Bardwell J. C. A. J. Biol. Chem. 2000;275:38302–38310. doi: 10.1074/jbc.M005957200. [DOI] [PubMed] [Google Scholar]
- Barbirz S., Jakob U., Glocker M. O. J. Biol. Chem. 2000;275:18759–18766. doi: 10.1074/jbc.M001089200. [DOI] [PubMed] [Google Scholar]
- Bourlès E., Isaac M., Lebrun C., Latour J.-M., Sénèque O. Chem.–Eur. J. 2011;17:13762–13772. doi: 10.1002/chem.201101913. [DOI] [PubMed] [Google Scholar]
- Fliss H., Menard M. Arch. Biochem. Biophys. 1991;287:175–179. doi: 10.1016/0003-9861(91)90403-6. [DOI] [PubMed] [Google Scholar]
- Fliss H., Menard M. Arch. Biochem. Biophys. 1992;293:195–199. doi: 10.1016/0003-9861(92)90384-9. [DOI] [PubMed] [Google Scholar]
- Crow J. P., Beckman J. S., McCord J. M. Biochemistry. 1995;34:3544–3552. doi: 10.1021/bi00011a008. [DOI] [PubMed] [Google Scholar]
- Cook N. L., Pattison D. I., Davies M. J. Free Radical Biol. Med. 2012;53:2072–2080. doi: 10.1016/j.freeradbiomed.2012.09.033. [DOI] [PubMed] [Google Scholar]
- Fu X. Y., Kassim S. Y., Parks W. C., Heinecke J. W. J. Biol. Chem. 2001;276:41279–41287. doi: 10.1074/jbc.M106958200. [DOI] [PubMed] [Google Scholar]
- Sénèque O., Bourlès E., Lebrun V., Bonnet E., Dumy P., Latour J.-M. Angew. Chem., Int. Ed. 2008;47:6888–6891. doi: 10.1002/anie.200800677. [DOI] [PubMed] [Google Scholar]
- Jacques A., Mettra B., Lebrun V., Latour J.-M., Sénèque O. Chem.–Eur. J. 2013;19:3921–3931. doi: 10.1002/chem.201204167. [DOI] [PubMed] [Google Scholar]
- Jaroszewski L., Schwarzenbacher R., McMullan D., Abdubek P., Agarwalla S., Ambing E., Axelrod H., Biorac T., Canaves J. M., Chiu H. J., Deacon A. M., DiDonato M., Elsliger M. A., Godzik A., Grittini C., Grzechnik S. K., Hale J., Hampton E., Han G. W., Haugen J., Hornsby M., Klock H. E., Koesema E., Kreusch A., Kuhn P., Lesley S. A., Miller M. D., Moy K., Nigoghossian E., Paulsen J., Quijano K., Reyes R., Rife C., Spraggon G., Stevens R. C., van den Bedem H., Velasquez J., Vincent J., White A., Wolf G., Xu Q. P., Hodgson K. O., Wooley J., Wilson I. A. Proteins: Struct., Funct., Bioinf. 2005;61:669–673. doi: 10.1002/prot.20542. [DOI] [PubMed] [Google Scholar]
- Sénèque O., Bonnet E., Joumas F. L., Latour J.-M. Chem.–Eur. J. 2009;15:4798–4810. doi: 10.1002/chem.200900147. [DOI] [PubMed] [Google Scholar]
- Isaac M., Latour J.-M., Sénèque O. Chem. Sci. 2012;3:3409–3420. [Google Scholar]
- Morris J. C. J. Phys. Chem. 1966;70:3798–3805. [Google Scholar]
- Kumar K., Margerum D. W. Inorg. Chem. 1987;26:2706–2711. [Google Scholar]
- Fogelman K. D., Walker D. M., Margerum D. W. Inorg. Chem. 1989;28:986–993. [Google Scholar]
- Gerritsen C. M., Margerum D. W. Inorg. Chem. 1990;29:2757–2762. [Google Scholar]
- Nagy P., Ashby M. T. Chem. Res. Toxicol. 2005;18:919–923. doi: 10.1021/tx050079r. [DOI] [PubMed] [Google Scholar]
- Raj S. B., Ramaswamy S., Plapp B. V. Biochemistry. 2014;53:5791–5803. doi: 10.1021/bi5006442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliss H., Menard M., Desai M. Can. J. Physiol. Pharmacol. 1991;69:1686–1691. doi: 10.1139/y91-250. [DOI] [PubMed] [Google Scholar]
- Tatsumi T., Fliss H. J. Mol. Cell. Cardiol. 1994;26:471–479. doi: 10.1006/jmcc.1994.1058. [DOI] [PubMed] [Google Scholar]
- Luebke J. L., Giedroc D. P. Biochemistry. 2015;54:3235–3249. doi: 10.1021/acs.biochem.5b00085. [DOI] [PubMed] [Google Scholar]
- Janda I., Devedjiev Y., Derewenda U., Dauter Z., Bielnicki J., Cooper D. R., Graf P. C. F., Joachimiak A., Jakob U., Derewenda Z. S. Structure. 2004;12:1901–1907. doi: 10.1016/j.str.2004.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremers C. M., Reichmann D., Hausmann J., Ilbert M., Jakob U. J. Biol. Chem. 2010;285:11243–11251. doi: 10.1074/jbc.M109.084350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichmann D., Xu Y., Cremers C. M., Ilbert M., Mittelman R., Fitzgerald M. C., Jakob U. Cell. 2012;148:947–957. doi: 10.1016/j.cell.2012.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won H. S., Low L. Y., De Guzman R., Martinez-Yamout M., Jakob U., Dyson H. J. J. Mol. Biol. 2004;341:893–899. doi: 10.1016/j.jmb.2004.06.046. [DOI] [PubMed] [Google Scholar]
- Voth W., Schick M., Gates S., Li S., Vilardi F., Gostimskaya I., Southworth D. R., Schwappach B., Jakob U. Mol. Cell. 2014;56:116–127. doi: 10.1016/j.molcel.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sénèque O., Latour J.-M. J. Am. Chem. Soc. 2010;132:17760–17774. doi: 10.1021/ja104992h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.