Abstract
Identity elements play essential roles in the recognition of tRNAs by their cognate aminoacyl-tRNA synthetase. An operational RNA code relates amino acids to specific sequences and structural features of tRNA acceptor stems. In this study, a series of tRNATrp variants was prepared by in vitro transcription and their efficiencies of aminoacylation by tryptophan (kcat/Km) were measured with the aid of Bacillus subtilis and human tryptophanyl-tRNA synthetases (TrpRS). The identity elements in the operational RNA code of human tRNATrp were found to be: major element, discriminator base A73; minor elements, G1/C72 and U5/G68. From the cross-species aminoacylation assays, we conclude that the identity elements in tRNATrp from B.subtilis and human all contribute to species-specific aminoacylation by TrpRS. Analyses of 22 TrpRS sequences covering three taxonomic domains (bacteria, eukarya and archaea) reveal that the sequences are divided into two evolutionarily distant groups. The same partition is also observed in the analyses of tRNATrp acceptor stem sequences. Our data suggest that the two TrpRS groups may reflect co-adaptations needed to accommodate changes in the operational RNA code for tryptophan.
INTRODUCTION
The accuracy of protein synthesis is essential to the survival of all organisms. Aminoacyl-tRNA synthetases (aaRS) catalyze the esterification of tRNA by their cognate amino acids. The fidelity of protein synthesis thus depends on the accurate recognition by each synthetase of its substrate tRNA and amino acid (1). The aminoacyl-tRNA synthetase family can be divided into two groups based primarily on the shared structural motifs in their catalytic domains (2). In recent years, the identification of nucleotide determinants, specific functional groups and subtle tRNA structural features in each of the 20 aminoacylation systems has revealed the mechanism of specific tRNA selection by the synthetases (3,4). Moreover, the observation of at least 11 tRNA synthetases that charge minihelices based on the sequences of the acceptor stems of their cognate tRNAs (5–8) provides strong evidence for an early ‘operational RNA code’ (9) based on acceptor stems that may have preceded the genetic code. The operational RNA code is typically comprised of 1–3 bp and the N73 ‘discriminator’ base (10). Strong evidence supports the notion that N73 plays a key role in the operational RNA code for many amino acids (11,12).
Although the ‘discriminator’ base is essential in synthetase recognition, the identity of N73 is not always conserved across taxa. For example, in the glycyl system, A73 is conserved in all tRNAGly sequences from archaea and eukarya, while the tRNAGly sequences from bacteria conserve a U at this position (13). Meanwhile, the enzymes from Escherichia coli and human do not cross-acylate their respective tRNAs (14). Moreover, an in vitro study indicated that species-specific aminoacylation could be switched by a single nucleotide exchange at position 73 (15). Similarly, the conservation of discriminator base identities was found in the tRNAs of lysine, proline and histidine (13). All these facts imply that the operational RNA code in archaea and eukarya is distinct from that in bacteria, and this difference is the main reason for the failure of cross-species tRNA recognition.
The identity elements of Bacillus subtilis tRNATrp were characterized previously by both in vivo and in vitro assays (12). The identity elements recognized by B.subtilis tryptophanyl-tRNA synthetase (TrpRS) were established to be: major elements, the discriminator base G73 and anticodon; minor elements, A1/U72, G5/C68 and A9, in decreasing importance. The non-anticodon identity elements are mainly located in the acceptor stem of the B.subtilis tRNATrp sequence.
Through sequence alignment of tRNAs for Trp from different sources (12), 12 bases that are uniquely common in bacterial sequences, but unshared by archaeal and eukaryotic tRNATrp, were selected. These 12 bases, according to the sequence of tRNATrp from B.subtilis, included A1/U72, G5/C68 and the discriminator base G73 in the acceptor stem, A9 and U11/A24 in the D-arm and G29/C41 and U31/A39 in the anticodon stem. The counterparts in tRNATrp from human were G1/C72, U5/G68, A73, G9, C11/G24, U29/A41 and A31/U39. In order to investigate the species-specific differences in tRNATrp recognition, all these different bases were studied by site-directed mutagenesis. Two enzymes were chosen in our study, one is B.subtilis TrpRS, a representative synthetase from bacteria, the other is human TrpRS, a typical eukaryotic enzyme. Because of the repeatedly manifested importance of the operational RNA code in the recognition of tRNAs by aaRS (15–23), different bases in the acceptor stem were examined in detail to identify the cross-species barrier in the aminoacylation by TrpRS from human and B.subtilis. Finally, sequences of TrpRS and tRNATrp covering three taxa were analyzed to investigate the aminoacylation systems in evolution. The results of this study contribute to our understanding of the relationship between the evolution of tRNA synthetases and the corresponding changes in tRNA acceptor stem recognition.
MATERIALS AND METHODS
Plasmids and reagents
Plasmid pKSW1 (24), which directs overexpression of B.subtilis TrpRS, was a kind gift from Dr Xue Hong. The high level expression vector pET24a(+)-HTrpRS was constructed in our laboratory (25). Restriction endonucleases, T4 DNA ligase and RiboMAX Large Scale RNA Production System-T7 were from Promega (Madison, WI). l-[5-3H]Tryptophan and DEAE Sepharose CL-6B were from Amersham Pharmacia (Piscataway, NJ). l-Tryptophan was from Gibco BRL (Gaithersburg, MD). Ni–NTA–agarose was from Qiagen (Chatsworth, CA). Bovine tRNA was from Sigma (St Louis, MO).
Enzyme purification and activity assays
The purification procedure of human TrpRS was carried out as described previously with Ni–NTA–agarose (25). The purified product was human TrpRS with a histidine tag at the C-terminus. Bacillus subtilis TrpRS was purified to homogeneity as described previously (24). Human enzyme was assayed at 30°C in an aminoacylation mixture containing 4 mM ATP, 0.8 mM dithiothreitol (DTT),1 µCi l-[5-3H]tryptophan, 8 mM MgCl2, 80 mM Tris–HCl pH 7.5, 0.02 µM in vitro transcribed tRNATrp, in a total volume of 50 µl. Bacillus subtilis enzyme was assayed at 22°C in an aminoacylation mixture containing 4 mM ATP, 1 mM DTT, 1 µCi l-[5-3H]tryptophan, 40 mM magnesium acetate, 140 mM Tris–HCl pH 7.8, 0.02 µM in vitro transcribed tRNATrp, in a total volume of 50 µl (25,26). The kinetic constants were derived from Lineweaver–Burk plots.
RNA preparation
All the tRNA variants were prepared by in vitro transcription (25). Mutagenesis of tRNAs was accomplished as described previously (27). All the desired mutations were verified by sequencing. The transcripts were purified by 10% PAGE under denaturing conditions. Before aminoacylation assay, transcripts were heated at 70°C for 2 min and slowly cooled to room temperature.
Sequence analyses
Multiple sequence alignments were performed using the ClustalX program (28). TrpRS sequences used for alignments were obtained from the aaRS database at http://www.pozman.edu.pl/aars (29). The sequences from 22 different species across three taxa were as follows: Archaeoglobus fulgidus (O28579); Methanococcus jannaschii (Q58810); Pyrococcus abyssi (CAB50601); Mycobacterium leprae (Q49901); Aquifex aeolicus (O67115); B.subtilis (P21656); Borrelia burgdorferi (O51038); Chlamydia muridarum (AAF39670); Chlamydophila pneumoniae AR39 (AAF38841); E.coli (P00954); Haemophilus influenzae (P43835); Neisseria meningitidis (AAF41828); Rickettsia prowazekii (Q9ZD76); Streptomyces coelicolor (CAB84910); Thermotoga maritima (Q9WYW2); Ureaplasma urealyticum (AAF30582); Bos taurus (P17248); Drosophila melanogaster (AAF20166); Homo sapiens (P23381); Mus musculus (P32921); Oryctolagus cuniculus (P23612); Saccharomyces cerevisiae (Q12109). In addition, 10 tRNATrp acceptor stem sequences were aligned using the same program (28). These sequences, obtained from a tRNA sequences database at http://www.uni-bayreuth.de/departments/biochemie/trna/ (13), were as follows: A.fulgidus (DW0340); U.urealyticum; E.coli (DW1660); B.subtilis (DW1540); H.sapiens; H.influenzae (DW2000); R.prowazekii (DW1870); S.cerevisiae (DW7630) (12); Halo volcanii (12).
To construct the TrpRS phylogenetic tree, all gaps and insertions were deleted manually from the aligned results, the remaining 244 residues were used. The tRNATrp evolutionary tree was constructed using the acceptor stem sequences N1–N7 and N66–N73. CCA sequence at the 3′-end of the acceptor stem was not included in the calculations because it is strictly conserved in all tRNATrp. The programs DNADIST, DNAPARS, PROTDIST, PROTPARS, KITSCH and CONSENSE in the PHILIP program package were used (30). Bootstrap analyses were performed with 100 replicates made by SEQBOOT (30).
RESULTS
tRNA molecules prepared by in vitro transcription do not contain the modified nucleotides found in naturally occurring tRNAs. Despite this fact, most tRNA transcripts are efficient substrates for their cognate synthetase (31–33). In the B.subtilis TrpRS system (12), modified bases do not appear to contribute significantly to aminoacylation by B.subtilis TrpRS. Bovine and human tRNA sequences are generally very closely related or identical (13). As purified native human tRNATrp was not available, unmodified human tRNATrp transcript was aminoacylated by human TrpRS and the rate was similar to the rate of bovine tRNATrp present in a mixture of native bovine tRNA (data not shown). Therefore, using in vitro transcribed tRNA as substrate, the aminoacylation activities of B.subtilis and human TrpRS could be evaluated.
Twelve different bases across acceptor stem, D-arm and anticodon stem
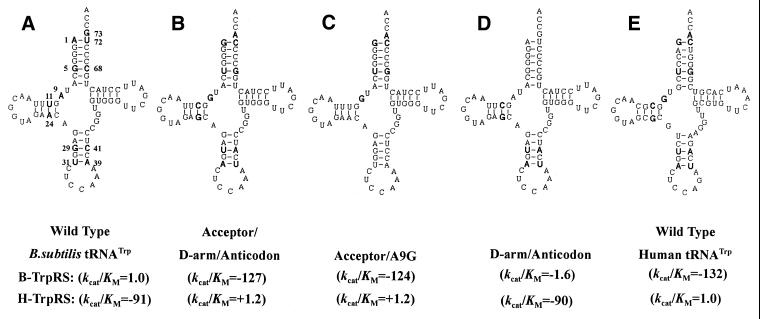
In order to determine which part of tRNATrp contributes to the species-specific aminoacylation, three multiple mutations were generated and assayed using both B.subtilis and human TrpRS. Figure 1 shows the secondary structures of the two wild-types (Fig. 1A and E) and three mutants of tRNATrp (Fig. 1B–D). Aminoacylation results (decrease in kcat/Km relative to that of the wild-type transcript) are also shown in Figure 1. The individual kinetic parameters measured for the wild-type tRNA were as follows: human tRNATrp aminoacylated by human TrpRS, kcat 1.0 s–1, Km 1.1 µM; B.subtilis tRNATrp aminoacylated by B.subtilis TrpRS, kcat 0.69 s–1, Km 0.56 µM. As in the ProRS system (17), human TrpRS weakly catalyzes the aminoacylation reaction of B.subtilis tRNATrp, with a 91-fold decrease in kcat/Km relative to that of the human transcript. Human tRNATrp is also a poor substrate for B.subtilis TrpRS, significant loss of aminoacylation efficiency (–132-fold) being observed in the assay.
Figure 1.
Secondary structure of wild-type B.subtilis tRNATrp and human tRNATrp and the three mutants studied. Mutated elements and their counterparts in wild-type tRNATrp are in a larger and bold font. kcat/Km is given relative to that of a wild-type tRNATrp transcript aminoacylated by its cognate TrpRS, which was set at 1.0. In these assays, the tRNATrp concentration ranged from 0.1 to 20 µM and the enzyme concentration was 25, 50 or 100 nM. B-TrpRS and H-TrpRS refer to B.subtilis and human TrpRS, respectively.
As shown in Figure 1B, a multiple mutation at all 12 different positions was made in B.subtilis tRNATrp; these nucleotides were changed to the corresponding nucleotides present in human tRNATrp. The resultant mutant shows a sharp decrease in aminoacylation efficiency by B.subtilis TrpRS as compared to wild-type B.subtilis tRNATrp transcript (–127-fold). At the same time, this mutant exhibits a strong positive effect on kcat/Km value (from –91-fold to +1.2-fold) by human TrpRS. On the basis of these results, two additional mutants were generated and assayed in order to find the exact part of tRNATrp contributing to the species-specific aminoacylation. One mutant included multiple changes in the acceptor stem and at A9G (Fig. 1C); the other had six substitutions in the D-arm and anticodon stem (Fig. 1D). Aminoacylation assays by both bacterial and eukaryotic enzymes show that the acceptor/A9G mutant has almost the same kinetic parameters as the acceptor/D-arm/anticodon mutant (–124-fold), indicating the minor effects of the substitutions in the D-arm and anticodon stem. Subsequent examination of the D-arm/anticodon mutant further confirmed this notion. In aminoacylation by B.subtilis TrpRS, only a minor negative effect (–1.6-fold as compared to wild-type B.subtilis tRNATrp) was observed for the D-arm/anticodon mutant. Meanwhile, it was not surprising that the D-arm/anticodon mutant was a poor substrate for human TrpRS (–90-fold), just like the wild-type B.subtilis tRNATrp (Fig. 2D).
Figure 2.
Secondary structure of wild-type B.subtilis tRNATrp and human tRNATrp and the six mutants studied. Mutated elements in the operational RNA code and their counterparts in wild-type tRNATrp are in a larger and bold font. kcat/Km is given relative to that of a wild-type tRNATrp transcript aminoacylated by its cognate TrpRS, which was set at 1.0. In these assays, the tRNATrp concentrations ranged from 0.1 to 20 µM and the enzyme concentration was 25, 50 or 100 nM. B-TrpRS and H-TrpRS refer to B.subtilis and human TrpRS, respectively.
Based on these data, we propose that the elements contributing to species-specific aminoacylation are mainly located in the acceptor stem.
Cross-species aminoacylation by human TrpRS
On the basis of wild-type B.subtilis tRNATrp (WTB, Fig. 2A), three mutants named MBH1, MBH2 and MBH3 were made in order to determine the exact role of these five elements in species-specific aminoacylation (Fig. 2B–D). Likewise, three other mutants, MHB1, MHB2 and MHB3 (Fig. 2F–H), were generated on the basis of wild-type human tRNATrp (WTH, Fig. 2E). Results are summarized in Figure 2 and detailed values are given in Table 1.
Table 1. Effect of single and multiple nucleotide mutations on aminoacylation of tRNATrp transcripts by B.subtilis and human TrpRS.
| Variant | B.subtilis TrpRS | Human TrpRS | ||
| |
kcat/KM (relative) |
Change in efficiency (x-fold) |
kcat/Km (relative) |
Change in efficiency (x-fold) |
| WTB | 1.0 | 1.0 | 0.011 | –91 |
| MBH1 | 0.0056 | –179 | 0.30 | –3.3 |
| MBH2 | 0.0055 | –181 | 0.48 | –2.1 |
| MBH3 | 0.0023 | –435 | 0.75 | –1.3 |
| WTH | 0.0076 | –132 | 1.0 | 1.0 |
| MHB1 | 0.075 | –13 | 0.10 | –10 |
| MHB2 | 0.11 | –9.1 | 0.091 | –11 |
| MHB3 | 0.14 | –7.1 | 0.064 | –16 |
The specificity constant (kcat/Km) for aminoacylation of tRNATrp variant transcripts was derived from a Lineweaver–Burk plot. kcat/Km is given relative to that of a wild-type tRNATrp transcript aminoacylated by its cognate TrpRS, which was set at 1.0. Concentrations of tRNATrp ranging from 0.1 to 20 µM were used and the TrpRS concentration was 25, 50 or 100 nM.
Human TrpRS weakly aminoacylates the wild-type B.subtilis tRNATrp transcript, with a 91-fold decrease in kcat/Km relative to that of the unmodified human transcript (Fig. 2A). The relatively low efficiency of cross-acylation may be due to the large differences between B.subtilis and human tRNATrp in the acceptor stem (different N73 discriminator base and 6 of 7 bp). A single mutation of the acceptor stem at position 73 (MBH1) was made because it was shown previously to be important for B.subtilis tRNATrp aminoacylation by its cognate synthetase (12). This mutation resulted in a drastic increase in aminoacylation efficiency from –91- to –3.3-fold as compared with the wild-type B.subtilis tRNATrp (Fig. 2A and B). Then, the transcripts of MBH2 and MBH3 were subjected to aminoacylation assay, which showed that the A1G/U72C and G5U/C68G mutations further improved aminoacylation activity (from –3.3- to –2.1- and –1.3-fold).
The mutants based on human tRNATrp were transcribed in vitro and assayed with human TrpRS. The single mutant MHB1 showed a large decrease in aminoacylation efficiency (–10-fold, Fig. 2F) as compared to the wild-type human tRNATrp. The double and multiple mutants (MHB2 and MHB3) showed minor additional negative effects (from –10- to –11- and –16-fold) in the recognition of human tRNATrp.
Cross-species aminoacylation by B.subtilis TrpRS
As reported previously (12), B.subtilis TrpRS weakly aminoacylates tRNATrp from eukaryotic sources. Human tRNATrp is also a poor substrate of B.subtilis TrpRS, with a 132-fold decrease in aminoacylation efficiency as compared to wild-type B.subtilis tRNATrp. A single mutation A73G greatly increased the kcat/Km value of human tRNATrp by 10 times (from –132- to –13-fold). Additional mutations G1A/C72U and U5G/G68C also had minor positive effects on B.subtilis TrpRS recognition (from –13- to –9.1- and –7.1-fold, Fig. 2F–H).
A set of single mutations described in a previous work (12) established G73 in B.subtilis tRNATrp as the major identity element and A1/U72, G5/C68 as minor elements. In our present study, these elements were examined again by the combination of single and multiple mutations. The substitution of G73A resulted in a sharp decrease in kcat/Km relative to B.subtilis tRNATrp (–179-fold, Fig. 2B). The changes at A1/U72 and G5/C68 had minor additional effects on B.subtilis TrpRS recognition (from –179- to –181- and –435-fold, Fig. 2B–D).
Sequence analyses of TrpRS
Previous genetic and biochemical studies have identified many genes of TrpRS from all three taxa. From a comparison of these different sequences, we observed great similarities among the sequences from the same taxon and very low sequence identities across taxa. For instance, the sequence similarity between B.subtilis and Bacillus stearothermophilus TrpRS is 76%, the sequence identity between human and M.musculus TrpRS is even higher (89%), but the sequence similarity between the B.subtilis and human enzymes is only 11%. To gain further insight into the evolution of TrpRS, we searched a well-established aaRS sequence database and the obtained TrpRS sequences were subjected to computer analysis. Twenty-two sequences were used for the analysis in this work, including 13 from bacteria, three from archaea and six from eukarya.
The TrpRS sequences can be aligned so that 14 positions are completely conserved in their amino acid residue identities (Fig. 3, marked by asterisks). The crystal structure of B.stearothermophilus TrpRS has been determined (34), thus a framework to locate these conserved residues in the structure is available. Conserved residues are located not only in the HIGH and KMSKS class-defining motifs (residues G18, K193, M194, S195 and S197 in the B.subtilis enzyme) but also in other regions (residues G8, D42, G70, A132, P143, D147, R156, A159 and K270 in the B.subtilis enzyme), indicating that TrpRSs from B.subtilis and human evolved from a common ancestral gene in spite of their low overall sequence similarity.
Figure 3.
Alignment of 22 tryptophanyl-tRNA synthetases. Amino acids identical for all the sequences are marked by asterisks. Gaps in the sequences are indicated by – and numbers represent residues that were omitted from the alignment. Sequences are colored automatically on the basis of an alignment consensus, which is calculated using the colprot.par file (28). Abbreviations are as follows: A-Af, A.fulgidus; A-Mj, M.jannaschii; A-Pa, P.abyssi; B-Ml, M.leprae; B-Aa, A.aeolicus; B-Bs, B.subtilis; B-Bb, B.burgdorferi; B-Cm, C.muridarum; B-Cp, C.pneumoniae AR39; B-Ec, E.coli; B-Hi, H.influenzae; B-Nm, N.meningitidis; B-Rp, R.prowazekii; B-Sc, S.coelicolor; B-Tm, T.maritima; B-Uu, U.urealyticum; E-Bt, B.taurus; E-Dm, D.melanogaster; E-Hs, H.sapiens; E-Mm, M.musculus; E-Oc, O.cuniculus; E-Sc, S.cerevisiae.
The alignment revealed that the 22 sequences could be sorted into two groups on the basis of an N-terminal extension pattern. The separation of these two groups has a perfect coincidence to the taxonomic origins of TrpRS. As shown in Figure 3, the sequences from archaea and eukarya are sorted into one group (the AE group) and the bacterial sequences are sorted into another group (the B group). The AE group has an extension of 55–152 amino acids located at the N-terminus. We also noted that the amino acid residues in many positions show great similarity in a group-specific manner. For instance, in the predicted acceptor binding loop, K110 (in the B.subtilis enzyme) is completely conserved in the B group, whereas G268 and F269 (in the human enzyme) are conserved in the AE group. Furthermore, the AE group shows a shortened pattern in the predicted acceptor binding loop. The large differences between the two groups in this small region may be due to the large differences between the two groups at the tRNATrp acceptor stem (see below), especially the N73 discriminator base. Besides the class-defining motifs (HIGH and the KMSKS loop), the 22 sequences show great similarity in the regions of the ‘specificity helix’, the ‘bottom of the AA pocket’ and the ‘putative C35 binding pocket’.
Partitioning of the 22 TrpRS sequences into two groups on the basis of the N-terminal extension pattern was further supported by phylogenetic analysis. After all gaps and insertions were deleted, the remaining 244 residues were used to compute a distance matrix of these sequences, from which the phylogenetic tree was constructed. As shown in Figure 5B, the two main branches are well separated and this result was supported by high bootstrap confidence values.
Figure 5.
Phylogenetic trees of 10 tRNATrp acceptor stem sequences (A), 22 TrpRSs (B) and the standard model for arrangement of the three domains of life (C) (adapted from ref. 42 with the kind permission of the author). The distance matrix used to construct the tree was computed using the Dayoff PAM matrix (30). Numbers show bootstrap values (percent) with 100 replicates. Abbreviations used in this figure are as in Figures 3 and 4.
The bipartite sorting of the sequences is consistent with taxonomic classification of the organisms from which the sequences were isolated. The prokaryotic group strictly consists of sequences from bacteria and the eukaryotic group contains sequences from eukarya and archaea.
Sequence analyses of tRNATrp
The alignment of 10 tRNATrp acceptor stem sequences also resulted in the partitioning of these sequences into two groups (Fig. 4). One group consists of sequences from bacteria and the other contains sequences from archaea and eukarya. The A73 discriminator base and G1/C72 are conserved in the eukaryotic group, whereas the prokaryotic group conserves G73 and A1/U72 at the same positions.
Figure 4.
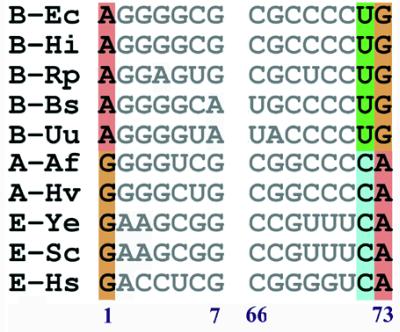
Alignment of 10 tRNATrp acceptor stem sequences. Numbers under the sequences indicate the positions in tRNA molecules. Abbreviations are as follows: B-Bs, B.subtilis; B-Ec, E.coli; B-Hi, H.influenzae,; B-Rp, R.prowazekii; B-Uu, U.urealyticum; A-Hv, H.volcanii; A-Af, A.fulgidus; E-Hs, H.sapiens; E-Sc, S.cerevisiae; E-Ye, yeast.
The phylogenetic tree of 10 tRNATrp acceptor stem sequences (Fig. 5A) was constructed to study the co-evolution of the enzymes and their cognate tRNATrp. From Figure 5A we can see that the two main branches are well separated and the sequences in each branch strictly derived from bacteria or from archaea and eukarya.
DISCUSSION
A previous sequence analysis of tRNATrp from different sources revealed that 12 bases were uniquely common to bacterial sequences, but unshared by the sequences from archaea and eukarya (12). These 12 bases are the best candidates as contributing to species-specific aminoacylation. Among them, discriminator base G73 was identified as a major identity element recognized by B.subtilis TrpRS and A1/U72, G5/C68 and A9 as minor elements (12). The counterparts in human tRNATrp were examined in this work. From aminoacylation assays of the in vitro transcripts, the identity elements recognized by human TrpRS were established to be: major element, discriminator base A73; minor elements, G1/C72 and U5/G68. Although the anticodon trinucleotide was considered a major identity element in the recognition of B.subtilis enzyme (12), it was not included in this study because it is common to all tRNATrp sequences and cannot contribute to species-specific aminoacylation. In order to determine which part of tRNATrp contributes to its species-specific aminoacylation, three multiple mutations were generated and assayed using both B.subtilis and human TrpRS. The aminoacylation efficiencies of these mutants are shown in Figure 1. If the mutant has B.subtilis tRNATrp identity elements G73, A1/U72 and G5/C68, then it is a good substrate for B.subtilis TrpRS and a poor substrate for human enzyme. On the other hand, if the mutant has human tRNATrp elements, its aminoacylation efficiency will be high for human enzyme and relatively low for B.subtilis TrpRS. From the experimental data we propose that the acceptor stem of the cloverleaf structure contributes most to species-specific aminoacylation of tRNATrp. With this in mind, a series of tRNATrp mutants were prepared to identify the exact roles of those species-specific elements in the operational RNA code.
As shown in Figure 2B, single base mutation of the G73 discriminator base to A resulted in a sharp decrease in aminoacylation efficiency by B.subtilis TrpRS and a drastic increase in aminoacylation efficiency by human TrpRS. A similar pattern was observed for another single base mutation of the A73 discriminator base to G (Fig. 2F) in human tRNATrp. This mutation also resulted in a significant loss of aminoacylation efficiency by human TrpRS and a large increase in aminoacylation efficiency by B.subtilis TrpRS. The positive or negative effects of N1/N72 and N5/N68 mutations on aminoacylation efficiency by both the B.subtilis and human enzymes seemed to be minor. This fact may partly explain why G73 is always conserved in tRNATrp sequences from bacteria, whereas A is conserved at this position in the archaeal and eukaryotic sequences of tRNATrp. From these data, we can see that species-specific aminoacylation can be altered by a single mutation of the N73 discriminator base. The N1/N72 and N5/N68 base pairs also contribute to species-specific aminoacylation, but to a lesser extent. The G73 (A73) discriminator base is thus established to be the major species-specific aminoacylation element, with A1/U72 and G5/C68 (G1/C72, U5/G68) as minor elements.
A species-specific operational RNA code was recently revealed in the GlyRS (15,35) and LysRS (36) systems. In the GlyRS system, the conserved N73 discriminator base is U in all tRNAGly from bacteria and A for those from archaea and eukarya. Therefore, the partitioning of enzymes is consistent with conservation of N73, which is an important recognition element for both mammalian and bacterial GlyRS. There is a small difference in the LysRS system. Although A73 is conserved in bacteria and G73 is uniquely common to archaea and eukarya, G73 is not a critical element for the human enzyme. In contrast to the species-specific operational RNA code, specific bases in the acceptor stem have been identified as major identity determinants across taxa. For example, the G3/U70 wobble base pair of tRNAAla is crucial for tRNA recognition by the cognate AlaRS from E.coli, human and yeast (5,37–40).
From our present studies, B.subtilis TrpRS seems to be more stringent in the selection of substrate tRNATrp than human TrpRS. For example, MBH1 and MHB1 are both single substitution mutants, but the single mutation G73A (MBH1) has an 18 times larger effect on B.subtilis enzyme recognition than MHB1 on human TrpRS (–179- versus –10-fold, Table 1). At the same time, MBH1 can be aminoacylated at a relatively higher level by human TrpRS as compared to the aminoacylation of MHB1 by B.subtilis enzyme (–3.3- versus –13-fold, Table 1). This fact suggests that TrpRS has become more tolerant to changes in tRNATrp during evolution.
As shown in Figure 3, the class-defining HIGH and KMSKS motifs were highly conserved. Unlike other tRNAs, tRNATrp has only one anticodon, CCA, and C35 is strictly conserved in all tRNATrp. On the basis of this fact, we proposed that all the TrpRS sequences must share a conserved region for binding of the anticodon. The ClustalX multiple sequence alignment confirmed this notion. In the putative C35 binding pocket, K270 (in B.subtilis TrpRS) is completely conserved in the 22 sequences across three taxa. The high sequence similarity in the bottom of the AA pocket indicates that all TrpRS sequences share a common structure for binding of their cognate amino acid, tryptophan. The segment called the ‘specificity helix’ contains a high proportion of side chains that interact specifically with the substrate tryptophan and it is also highly conserved in these sequences.
These 22 TrpRS sequences are sorted into two groups on the basis of a specific N-terminal extension pattern. Sequences from archaea and eukarya with high similarity were sorted into the AE group and sequences from bacteria were sorted into the B group. Similar to the TrpRS system, 21 GlyRS sequences could also be sorted into two groups, one containing synthetases primarily from archaea and eukarya and the other from bacteria (41). The ProRS system is another example of such partition (17). In our present analyses, 14 residues of TrpRS are strictly conserved in the 22 sequences examined, indicating that in this case the two groups (the AE and B groups) were derived from a common ancestor. The partition was further supported by phylogenetic analysis of TrpRS sequences. In the phylogenetic tree of TrpRS, the sequences from bacteria and those from archaea and eukarya are well separated and the partition was supported by high bootstrap confidence values. This result is consistent with phylogenetic analysis of aaRS (42). In that report, 17 aaRS were grouped into three models: the standard, simple import and anomalous models. TrpRS, LeuRS, AspRS and GluRS belong to the standard model. In the standard model, the archaea are shown to be nearer to the eukarya than bacteria and mitochondrial sequences are more similar to those in bacteria (Fig. 5C). Ten tRNATrp acceptor stem sequences in the phylogenetic tree were also sorted into an AE and a B group. According to this partition, tRNATrp were found to be another example of the standard model. The similar partitions of TrpRS and tRNATrp sequences in the alignment and phylogenetic analyses indicate the co-evolutionary relationship of TrpRS and the acceptor stem of tRNATrp.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Dr Xue Hong (Hongkong University of Science and Technology) for her helpful advice and technical support. We also thank Prof. Youshang Zhang for his critical review of the manuscript. This work was supported by the Natural Science Foundation of China (grant 39730120), the Chinese Academy of Sciences (grant KSCX 2-2-04) and a grant from the Shanghai Institutes of Life Sciences.
References
- 1.Schimmel P. (1987) Aminoacyl-tRNA synthetases: general scheme of structure-functional relationships in the peptides and recognition of transfer RNAs. Annu. Rev. Biochem., 56, 125––158.. [DOI] [PubMed] [Google Scholar]
- 2.Eriani G., Delarue,M., Poch,O., Gangloff,J. and Moras,D. (1990) Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature, 347, 203––206.. [DOI] [PubMed] [Google Scholar]
- 3.McClain W.H. (1993) Rules that govern tRNA identity in protein synthesis. J. Mol. Biol., 234, 257––280.. [DOI] [PubMed] [Google Scholar]
- 4.Saks M.E., Sampson,J.R. and Abelson,J.N. (1994) The transfer RNA identity problem: a search for rules. Science, 263, 191––197.. [DOI] [PubMed] [Google Scholar]
- 5.Francklyn C. and Schimmel,P. (1989) Aminoacylation of RNA minihelices with alanine. Nature, 337, 478––481.. [DOI] [PubMed] [Google Scholar]
- 6.Francklyn C. and Schimmel,P. (1990) Enzymatic aminoacylation of an eight-base-pair microhelix with histidine. Proc. Natl Acad. Sci. USA, 87, 8655––8659.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Francklyn C., Shi,J.P. and Schimmel,P. (1992) Overlapping nucleotide determinants for specific aminoacylation of RNA microhelices. Science, 255, 1121––1125.. [DOI] [PubMed] [Google Scholar]
- 8.Frugier M., Florentz,C. and Giegé,R. (1992) Anticodon-independent aminoacylation of an RNA minihelix with valine. Proc. Natl Acad. Sci. USA, 89, 3990––3994.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schimmel P., Giegé,R., Moras,D. and Yokoyama,S. (1993) An operational RNA code for amino acids and possible relationship to genetic code. Proc. Natl Acad. Sci. USA, 90, 8763––8768.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crothers D.M., Seno,T. and Söll,D. (1972) Is there a discriminator site in transfer RNA? Proc. Natl Acad. Sci. USA, 69, 3063––3067.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hou Y.M. (1997) Discriminating among the discriminator bases of tRNAs. Chem. Biol., 4, 93––96.. [DOI] [PubMed] [Google Scholar]
- 12.Xue H., Shen,W., Giegé,R. and Wong,J.T. (1993) Identity elements of tRNATrp. J. Biol. Chem., 268, 9316––9322.. [PubMed] [Google Scholar]
- 13.Sprinzl M., Horn,C., Brown,M., Ioudovitch,A. and Steinberg,S. (1998) Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res., 26, 148––153.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shiba K., Schimmel,P., Motegi,H. and Noda,T. (1994) Human glycyl-tRNA synthetase. Wide divergence of primary structure from bacterial counterpart and species-specific aminoacylation. J. Biol. Chem., 269, 30049––30055.. [PubMed] [Google Scholar]
- 15.Hipps D., Shiba,K., Henderson,B. and Schimmel,P. (1995) Operational RNA code for amino acids: species-specific aminoacylation of minihelices switched by a single nucleotide. Proc. Natl Acad. Sci. USA, 92, 5550––5552.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ribas D.P.L. and Schimmel,P. (2001) Operational RNA code for amino acids in relation to genetic code in evolution. J. Biol. Chem., 276, 6881––6884.. [DOI] [PubMed] [Google Scholar]
- 17.Stehlin C., Burke,B., Yang,F., Liu,H., Shiba,K. and Musier-Forsyth,K. (1998) Species-specific differences in the operational RNA code for aminoacylation of tRNAPro. Biochemistry, 37, 8605––8613.. [DOI] [PubMed] [Google Scholar]
- 18.Rodin S.N. and Ohno,S. (1997) Four primordial modes of tRNA-synthetase recognition, determined by the (G,C) operational code. Proc. Natl Acad. Sci. USA, 94, 5183––5188.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agou F., Quevillon,S., Kerjan,P., Latreille,M.T. and Mirande,M. (1996) Functional replacement of hamster lysyl-tRNA synthetase by the yeast enzyme requires cognate amino acid sequences for proper tRNA recognition. Biochemistry, 35, 15322––15331.. [DOI] [PubMed] [Google Scholar]
- 20.Auld D.S. and Schimmel,P. (1996) Single sequence of a helix-loop peptide confers functional anticodon recognition on two tRNA synthetases. EMBO J., 15, 1142––1148.. [PMC free article] [PubMed] [Google Scholar]
- 21.Schimmel P. and Ripmaster,T. (1995) Modular design of components of the operational RNA code for alanine in evolution. Trends Biochem. Sci., 20, 333––334.. [DOI] [PubMed] [Google Scholar]
- 22.Schimmel P. (1995) An operational RNA code for amino acids and variations in critical nucleotide sequences in evolution. J. Mol. Evol., 40, 531––536.. [DOI] [PubMed] [Google Scholar]
- 23.Shiba K., Suzuki,N., Shigesada,K., Namba,Y., Schimmel,P. and Noda,T. (1994) Human cytoplasmic isoleucyl-tRNA synthetase: selective divergence of the anticodon-binding domain and acquisition of a new structural unit. Proc. Natl Acad. Sci. USA, 91, 7435––7439.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wen S., Chow,K.C. and Wong,J.T. (1990) High-level expression of Bacillus subtilis tryptophanyl-tRNA synthetase in Escherichia coli. Biochem. Cell Biol., 68, 492––495.. [DOI] [PubMed] [Google Scholar]
- 25.Xu F., Jia,J., Jin,Y.X. and Wang,D.B. (2001) High-level expression and single-step purification of human tryptophanyl-tRNA synthetase. Protein Expr. Purif., in press. [DOI] [PubMed] [Google Scholar]
- 26.Xu F., Chen,L., Jin,Y.X. and Wang,D.B. (2000) Immobilization of tryptophanyl-tRNA synthetase. Acta Biochim. Biophys. Sin., 32, 74––76.. [PubMed] [Google Scholar]
- 27.Chen L., Jin,Y.X. and Wang,D.B. (2000) Mitochondrial tRNATrp imply a eubacterial origin. Acta Biochim. Biophys. Sin., 32, 100––104.. [PubMed] [Google Scholar]
- 28.Thompson J.D., Gibson,T.J., Plewniak,F., Jeanmougin,F. and Higgins,D.G. (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res., 24, 4876––4882.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szymanski M., Deniziak,M.A. and Barciszewski,J. (2001) Aminoacyl-tRNA synthetases database. Nucleic Acids Res., 29, 288––290.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Felsenstein J. (1993) PHYLIP, Phylogeny Inference Package, version 3.5c. University of Washington, Seattle, WA.
- 31.Jahn M., Rogers,M.J. and Söll,D. (1991) Anticodon and acceptor stem nucleotides in tRNA(Gln) are major recognition elements for E. coli glutaminyl-tRNA synthetase. Nature, 352, 258––260.. [DOI] [PubMed] [Google Scholar]
- 32.Pütz J., Puglisi,J.D., Florentz,C. and Giegé,R. (1991) Identity elements for specific aminoacylation of yeast tRNA(Asp) by cognate aspartyl-tRNA synthetase. Science, 252, 1696––1699.. [DOI] [PubMed] [Google Scholar]
- 33.Sampson J.R. and Uhlenbeck,O.C. (1988) Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc. Natl Acad. Sci. USA, 85, 1033––1037.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doublie S., Bricogne,G., Gilmore,C. and Carter,C.W.Jr (1995) Tryptophanyl-tRNA synthetase crystal structure reveals an unexpected homology to tyrosyl-tRNA synthetase. Structure, 3, 17––31.. [DOI] [PubMed] [Google Scholar]
- 35.Nameki N., Tamura,K., Asahara,H. and Hasegawa,T. (1997) Recognition of tRNA(Gly) by three widely diverged glycyl-tRNA synthetases. J. Mol. Biol., 268, 640––647.. [DOI] [PubMed] [Google Scholar]
- 36.Shiba K., Stello,T., Motegi,H., Noda,T., Musier-Forsyth,K. and Schimmel,P. (1997) Human lysyl-tRNA synthetase accepts nucleotide 73 variants and rescues Escherichia coli double-defective mutant. J. Biol. Chem., 272, 22809––22816.. [DOI] [PubMed] [Google Scholar]
- 37.Hou Y.M. and Schimmel,P. (1988) A simple structural feature is a major determinant of the identity of a transfer RNA. Nature, 333, 140––145.. [DOI] [PubMed] [Google Scholar]
- 38.Shi J.P., Francklyn,C., Hill,K. and Schimmel,P. (1990) A nucleotide that enhances the charging of RNA minihelix sequence variants with alanine. Biochemistry, 29, 3621––3626.. [DOI] [PubMed] [Google Scholar]
- 39.Park S.J., Hou,Y.M. and Schimmel,P. (1989) A single base pair affects binding and catalytic parameters in the molecular recognition of a transfer RNA. Biochemistry, 28, 2740––2746.. [DOI] [PubMed] [Google Scholar]
- 40.McClain W.H. and Foss,K. (1988) Changing the acceptor identity of a transfer RNA by altering nucleotides in a “variable pocket”. Science, 241, 1804––1807.. [DOI] [PubMed] [Google Scholar]
- 41.Shiba K., Motegi,H. and Schimmel,P. (1997) Maintaining genetic code through adaptations of tRNA synthetases to taxonomic domains. Trends Biochem. Sci., 22, 453––437.. [DOI] [PubMed] [Google Scholar]
- 42.Doolittle R.F. and Handy,J. (1998) Evolutionary anomalies among the aminoacyl-tRNA synthetases. Curr. Opin. Genet. Dev., 8, 630––636.. [DOI] [PubMed] [Google Scholar]