Abstract
This study reports a novel adenovirus that was found circulating in pigeons in China. Nucleotide homology analysis of the hexon gene showed a nucleotide similarity of 79.0 and 70.9% with PiAd-2 variant A and PiAd-1, respectively. Phylogenetic analysis suggested that the identified virus, together with PiAd-2 variant, constitutes a monophyletic group (proposed as Pigeon Aviadenovirus B) in the genus Aviadenovirus. The present study contributes to the understanding of the epidemiology, ecology, and taxonomy of adenoviruses in pigeons.
Keywords: aviadenovirus, novel adenovirus, pigeon
Until recently, five genera within the family Adenoviridae were known, namely Atadenovirus, Aviadenovirus, Ichtadenovirus, Mastadenovirus and Siadenovirus (https://talk.ictvonline.org/taxonomy/). Reportedly, adenoviruses (AdVs) can infect a large number of vertebrate species [2, 9]. Birds are considered common hosts for AdVs, and this fact is mirrored by a large number of so-called fowl AdVs (FAdVs), described in chicken already several decades ago. Moreover, birds can be infected with genetically diverse adenoviruses of the family Adenoviridae [1, 8], which are classified into three different genera, namely Atadenovirus (e.g., Duck atadenovirus A and Psittacine atadenovirus A) [16, 19], Siadenovirus (e.g., Turkey siadenovirus A) [4], and Aviadenovirus, which includes twelve species, some of which are major bird adenoviruses (https://talk.ictvonline.org/taxonomy/).
The viral genome of adenoviruses consists of linear double-stranded DNA varying between 26 and 45 kb, depending on the species [11, 12, 17, 18]. The major structural proteins are the hexon protein and the fiber protein, which is non-covalently linked to the penton base, forming a structure called a penton. The hexon protein is the major capsid protein of the non-enveloped icosahedral virion, on which type-, group-, and subgroup-specific determinants are located [7, 15].
Adenoviral infection in pigeons was firstly described in Belgium in 1984 and has since been observed worldwide [20]. De Herdt et al. (1995) described two adenovirus-associated disease courses in specific pigeons. Type 1 adenovirus (also known as the classic adenovirus, pigeon adenovirus 1, or PiAd-1) has striking similarities with young pigeon disease syndrome (YPDS), as mainly young pigeons are affected, showing diarrhea, vomiting, and weight loss for approximately one week. Type 2 adenovirus (also known as pigeon adenovirus 2 or PiAd-2) affects pigeons of all ages and is characterized by sudden death and extensive hepatic necrosis [3, 20].
In this study, we examined a commercial pigeon flock located in Fujian, which had a disease characterized by anorexia, diarrhea, and ataxia. The rate of illness was usually high (up to 35%), and mortality rates were at nearly 10%. Samples from each farm were collected; samples of the liver, spleen, and pancreases were taken from the diseased pigeons for diagnosis. The samples that were suspected to have bacterial infection (Escherichia coli or Salmonella anatum spp.) were cultured and detected as described by Liu et al. [10]; however, no bacterial infection was found.
The rest of the samples were homogenized in phosphate-buffered saline (10%, w/v) containing antibiotics (10,000 U/ml of penicillin and 10 mg/ml of streptomycin). The suspension was then clarified by centrifugation at 8,000 g at 4°C for 20 min and stored at −80°C until use. The supernatants were used for DNA/RNA extraction using the EasyPure Viral DNA/RNA Kit (TransGen Biotech, Beijing, China), according to the manufacturer’s instructions. The Premix TaqTM (Ex TaqTM Version 2.0 plus dye) and TaKaRa PrimeScript one-step RT-PCR Kit Ver.2 (TaKaRa, Dalian, China) were used for PCR and RT-PCR, respectively. Pigeon herpesvirus (PiHV) and pigeon circovirus (PiCV) were detected by PCR as described by Freick et al. [5]; avian paramyxovirus type 1 (APMV-1) was detected by RT-PCR as described by Mase et al. [13]; and avian influenza virus (AIV) was detected by RT-PCR as described by Hoffmann et al. [6]. These classical endemic and emerging pathogens were excluded as the causative agents of the disease.
The only positive results were observed using the HexF1/ HexR1 (Table 1) primers, established by Mase et al. (2009) for targeting the hexon gene of avian adenoviruses [14]. Other primers, which were previously used for detecting adenoviral infections in pigeons (such as PiAd-2, PiAd-2 variant A, and PiAd-2 variant B; Table 1) [14, 18] and which target the hexon gene, were also used for excluding co-infections with other pigeon adenoviruses.
Table 1. Primers used in this study.
| Primers | Sequences (5′→3′) | Size of amplicons (bp) | Reference |
|---|---|---|---|
| HexF1 | GAYRGYHGGRTNBTGGAYATGGG | 800 | Mase et al., 2009 [14] |
| HexR1 | TACTTATCNACRGCYTGRTTCCA | ||
| PiAdV-2 Hex-3-F | GTAACATGAGCGTGCTGTTTG | 643 | Teske et al., 2017 [18] |
| PiAdV-2 Hex-3-R | CTGAGAAACGAAACCCGAATTG | ||
| PiAdV-2 A 2404+ | CTGACACTAATGATACGGAG | 330 | |
| PiAdV-2 2734- | GGGTCCAGTTCGAAGTTGATYA | ||
| PiAdV-2 B 2404+ | CGGACACTGGAAGTAGCACC | 330 | |
| PiAdV-2 2734- | GGGTCCAGTTCGAAGTTGATYA | ||
| NPiAdV-HexF1a) | TGAAACATGGCTGCGCTCACT | 1,064 | This study |
| HexR1a) | TACTTATCNACRGCYTGRTTCCA | ||
| NPiAdV-HexF2a) | ATCCGGCATGAACGTGGTAGTAGA | 1,895 | |
| NPiAdV-HexR2a) | CTTAGACTGCGTTGCCTGT | ||
a) Primer pairs (NPiAdV-HexF1 and HexR1 and NPiAdV-HexF2 and NPiAdV-HexR2) were used to amplify the complete hexon gene of the pigeon adenovirus strain FJ2017.
The complete hexon gene of the identified pigeon adenovirus, designated as strain FJ2017, was amplified using the primers listed in Table 1. PCR was carried out in a PCR system (Eppendorf AG, Hamburg, Germany). The PCR mixture (50 µl) contained 25 µl Premix Taq™ (Ex Taq™ Version 2.0 plus dye; TaKaRa), 1 µl forward primer and reverse primer (20 µM each), 2 µl template DNA, and 21 µl distilled water. The PCR cycle consisted of 1 cycle at 94°C for 5 min; 35 cycles of 94°C for 50 sec (denaturation), 53°C for 35 sec (annealing), and 72°C for 120 sec (extension); and 1 cycle at 72°C for 10 min (final extension). The PCR products were subjected to electrophoresis on 1.0% agarose gels for analysis. To confirm the presence of adenoviruses in samples, all PCR amplicons with the expected product size were subjected to DNA sequencing. PCR products were purified using QIAquick Gel Extraction Kit (Qiagen, Hilden, Germany) and then cloned into a pMD18-T vector (TaKaRa), according to the manufacturer’s instructions. The vector was used to transform the competent Escherichia coli strain DH5α (TaKaRa). After identification, positive transformants were submitted to a company (Sangon, Shanghai, China) for nucleotide sequence determination. For each PCR product, three colonies were selected for Sanger sequencing in both directions.
After sequence-independent amplification, sequencing and BLAST search (https://blast.ncbi.nlm.nih.gov/Blast.cgi) were performed. The sequences were assembled; the length of the hexon gene was 2,832 base pairs (bp), and it encoded 943 amino acids. The obtained sequences were submitted to the GenBank under the accession number MF576429.
In the present study, the hexon gene from previously known 18 aviadenoviruses (including 12 species in the genus and the unclassified PiAd-2 variant A identified in pigeons), 1 siadenovirus (Turkey adenovirus A), and 3 atadenoviruses (Psittacine adenovirus 3 and duck adenoviruses) retrieved from GenBank were used for further analyses. Nucleotide and amino acid identities were analyzed using Lasergene software v10.0 (DNAStar, Madison, WI, U.S.A.), by the ClustalW method.
In terms of sequence identity, the identified PiAd strain FJ2017 showed nucleotide and amino acid similarities of 79.0 and 88.6% with PiAd-2 variant A, respectively (GenBank No. KX121164) [18]. It should be noted that no classic PiAd-2 hexon complete gene sequences could be obtained from GenBank and was therefore not used for analysis in this study. In addition, the PiAd strain FJ2017 showed nucleotide and amino acid similarities of 70.3 and 80.4% with PiAd-1, respectively (GenBank No. NC024474) [12]. The lower nucleotide identities (51.8–68.1%) and amino acid identities (49.3–76.0%) shared by the strain FJ2017 and other selected bird adenoviruses are shown in Table 2.
Table 2. Nucleotide similarity between hexon genes of strain FJ2017 with other adenoviruses.
| Genus | Species | GenBank No. | Length of hexon (bp) | Nucleotide identity (%) | Amino acids identity (%) |
|---|---|---|---|---|---|
| Aviadenovirus | Duck aviadenovirus B | KR135164 | 2,814 | 65.1 | 72.7 |
| NC024486 | 2,823 | 65.3 | 71.7 | ||
| Falcon aviadenovirus A | AY683541 | 2,807 | 65.8 | 73.2 | |
| Fowl aviadenovirus A | KX247012 | 2,829 | 68.1 | 76.0 | |
| MF168407 | 2,829 | 67.9 | 75.9 | ||
| Fowl aviadenovirus B | KC493646 | 2,862 | 66.2 | 74.3 | |
| Fowl aviadenovirus C | KY436520 | 2,814 | 66.7 | 73.7 | |
| KY436522 | 2,814 | 66.7 | 73.7 | ||
| Fowl aviadenovirus D | KU746335 | 2,853 | 66.6 | 74.5 | |
| KY426992 | 2,853 | 66.5 | 74.2 | ||
| Fowl aviadenovirus E | KX258422 | 2,844 | 67.4 | 74.9 | |
| KY968968 | 2,844 | 67.6 | 74.9 | ||
| Goose aviadenovirus A | NC017979 | 2,802 | 65.8 | 72.8 | |
| Pigeon aviadenovirus A | NC024474 | 2,855 | 70.3 | 80.4 | |
| Turkey aviadenovirus B | NC014564 | 2,823 | 66.5 | 73.8 | |
| Turkey aviadenovirus C | NC022612 | 2,907 | 67.2 | 74.1 | |
| Turkey aviadenovirus D | NC022613 | 2,829 | 67.2 | 75.2 | |
| Atadenovirus | Duck atadenovirus A | KJ452173 | 2,733 | 52.5 | 49.5 |
| NC001813 | 2,733 | 52.5 | 49.5 | ||
| Psittacine atadenovirus A | NC025962 | 2,742 | 51.8 | 49.3 | |
| Siadenovirus | Turkey siadenovirus A | AC000016 | 2,721 | 52.0 | 53.1 |
| Unclassified | PiAd-2 variant A | KX121164 | 2,826 | 79.0 | 88.6 |
| This study | Aviadenovirus variant | MF576429 | 2,832 | / | / |
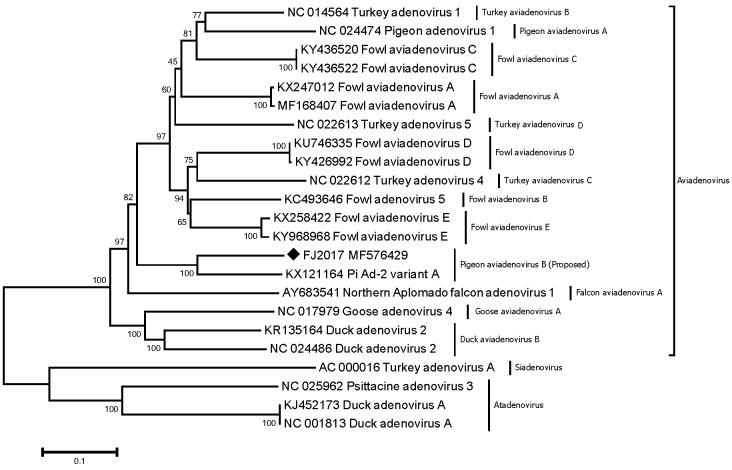
Phylogenetic trees based on the hexon amino acid sequences, deduced from the complete hexon gene, were constructed using the neighbor-joining method implemented in MEGA 6. Bootstrap analysis was performed with 1,000 replications. The phylogenetic tree (Fig. 1) showed that the PiAd strain FJ2017, together with PiAd-2 variant A, forms a distinct clade, separate from the 12 species previously classified in the genus Aviadenovirus. These data suggest that the PiAd strain FJ2017, together with PiAd-2 variant A, can be classified into a novel cluster (herein proposed as pigeon aviadenovirus B, as opposed to pigeon aviadenovirus A) in the genus Aviadenovirus, family Adenoviridae.
Fig. 1.
Phylogenetic tree constructed based on the hexon proteins of bird adenoviruses of the family Adenoviridae. The tree was generated by MEGA 6.0 software, using the neighbor-joining method (bootstrap=1,000). The scale bar represents the number of nucleotide substitutions per site. The pigeon adenovirus FJ2017 strain in this study is indicated with a black diamond (♦). Reference sequences obtained from GenBank are indicated by strain name and accession number.
To investigate the prevalence of this novel adenovirus in pigeons, viral nucleic acids were extracted from all 35 samples using the EasyPure Viral DNA/RNA Kit (TransGen). The individual samples were collected in Fujian, from January 2015 to December 2017. No positive results were obtained using the pigeon adenovirus detection primers listed in Table 1. Two samples with a similar syndrome were collected from the same commercial pigeon flock; in these, the PiAd strain FJ2017 was identified, but they also tested positive using NPiAdV-HexF1/HexR1 (NPiAdV-HexF1/HexR1). The expected length of the amplified fragment was 1,064 bp. The PCR amplicons were T-A cloned and sequenced by Sanger method, described earlier. Sequence analysis showed that the two amplicons shared 100% nucleotide identity with the PiAd strain FJ2017, indicating that a single virus strain was prevalent in the pigeons in this area.
Moreover, positive samples were submitted to viral isolation by using 9-day-SPF chicken embryos, but no deaths or pathological changes were observed in SPF chicken embryos after five passages. These data show that attempts to isolate the virus were unsuccessful, which means that it cannot be concluded that the adenovirus described herein is also the causative agent of the disease. Further studies, including propagation assays and pathogenicity tests of the newfound virus as well as of other possible pathogens, are needed to determine the causative agent of the disease.
In summary, in our study, we identified a novel adenovirus that circulates among pigeons. To the best of our knowledge, this is the first report on this adenovirus in pigeons in China. The present study contributes to the understanding of the epidemiology, ecology, and taxonomy of the diverse adenoviruses in pigeons.
CONFLICTS OF INTEREST
The authors declare that they have no conflicts of interest.
Acknowledgments
This work was funded by the Natural Science Foundation of China (31602068), China Agriculture Research System (CARS-42), Fujian Academy of Agriculture Science Innovative Research Team Project (STIT2017-3-10) and Young Talent Program Project (YC2015-12), and the Fujian Public Welfare Project (2018R1023-5).
REFERENCES
- 1.Changjing L., Haiying L., Dongdong W., Jingjing W., Youming W., Shouchun W., Jida L., Ping L., Jianlin W., Shouzhen X., Shangjin C., Yi Z., Yanbo Y.2016. Characterization of fowl adenoviruses isolated between 2007 and 2014 in China. Vet. Microbiol. 197: 62–67. doi: 10.1016/j.vetmic.2016.11.005 [DOI] [PubMed] [Google Scholar]
- 2.Dadáková E., Chrudimský T., Brožová K., Modrý D., Celer V., Hrazdilová K.2017. New adenoviruses from new primate hosts - growing diversity reveals taxonomic weak points. Mol. Phylogenet. Evol. 107: 305–307. doi: 10.1016/j.ympev.2016.11.013 [DOI] [PubMed] [Google Scholar]
- 3.De Herdt P., Ducatelle R., Lepoudre C., Charlier G., Nauwynck H.1995. An epidemic of fatal hepatic necrosis of viral origin in racing pigeons (Columba livia). Avian Pathol. 24: 475–483. doi: 10.1080/03079459508419087 [DOI] [PubMed] [Google Scholar]
- 4.Dhama K., Gowthaman V., Karthik K., Tiwari R., Sachan S., Kumar M. A., Palanivelu M., Malik Y. S., Singh R. K., Munir M.2017. Haemorrhagic enteritis of turkeys - current knowledge. Vet. Q. 37: 31–42. doi: 10.1080/01652176.2016.1277281 [DOI] [PubMed] [Google Scholar]
- 5.Freick M., Müller H., Raue R.2008. Rapid detection of pigeon herpesvirus, fowl adenovirus and pigeon circovirus in young racing pigeons by multiplex PCR. J. Virol. Methods 148: 226–231. doi: 10.1016/j.jviromet.2007.11.003 [DOI] [PubMed] [Google Scholar]
- 6.Hoffmann E., Stech J., Guan Y., Webster R. G., Perez D. R.2001. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 146: 2275–2289. doi: 10.1007/s007050170002 [DOI] [PubMed] [Google Scholar]
- 7.Junnu S., Lertwatcharasarakul P., Jala S., Phattanakunanan S., Moonjit P., Songserm T.2014. Developing an indirect ELISA based on recombinant hexon protein for serological detection of inclusion body hepatitis in chickens. J. Vet. Med. Sci. 76: 289–293. doi: 10.1292/jvms.13-0196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaján G. L., Kecskeméti S., Harrach B., Benkő M.2013. Molecular typing of fowl adenoviruses, isolated in Hungary recently, reveals high diversity. Vet. Microbiol. 167: 357–363. doi: 10.1016/j.vetmic.2013.09.025 [DOI] [PubMed] [Google Scholar]
- 9.Lakatos B., Hornyák Á., Demeter Z., Forgách P., Kennedy F., Rusvai M.2017. Detection of a putative novel adenovirus by PCR amplification, sequencing and phylogenetic characterisation of two gene fragments from formalin-fixed paraffin-embedded tissues of a cat diagnosed with disseminated adenovirus disease. Acta Vet. Hung. 65: 574–584. doi: 10.1556/004.2017.056 [DOI] [PubMed] [Google Scholar]
- 10.Liu R., Chen C., Huang Y., Cheng L., Lu R., Fu G., Shi S., Chen H., Wan C., Fu Q., Lin J.2018. Microbiological identification and analysis of waterfowl livers collected from backyard farms in Southern China. J. Vet. Med. Sci. 80: (in press) doi: 10.1292/jvms.17-0452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marek A., Kosiol C., Harrach B., Kaján G. L., Schlötterer C., Hess M.2013. The first whole genome sequence of a Fowl adenovirus B strain enables interspecies comparisons within the genus Aviadenovirus. Vet. Microbiol. 166: 250–256. doi: 10.1016/j.vetmic.2013.05.017 [DOI] [PubMed] [Google Scholar]
- 12.Marek A., Kaján G. L., Kosiol C., Harrach B., Schlötterer C., Hess M.2014. Complete genome sequences of pigeon adenovirus 1 and duck adenovirus 2 extend the number of species within the genus Aviadenovirus. Virology 462−463: 107–114. doi: 10.1016/j.virol.2014.04.033 [DOI] [PubMed] [Google Scholar]
- 13.Mase M., Kanehira K.2015. Phylogenetic analysis of avian paramyxovirus serotype-1 in pigeons in Japan. J. Vet. Med. Sci. 77: 919–923. doi: 10.1292/jvms.14-0684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mase M., Mitake H., Inoue T., Imada T.2009. Identification of group I-III avian adenovirus by PCR coupled with direct sequencing of the hexon gene. J. Vet. Med. Sci. 71: 1239–1242. doi: 10.1292/jvms.71.1239 [DOI] [PubMed] [Google Scholar]
- 15.Pacesa M., Hendrickx R., Bieri M., Flatt J. W., Greber U. F., Hemmi S.2017. Small-size recombinant adenoviral hexon protein fragments for the production of virus-type specific antibodies. Virol. J. 14: 158. doi: 10.1186/s12985-017-0822-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schybli M., Sigrist B., Hess M., van Leerdam B., Hoop R. K., Vögtlin A.2014. Development of a new real-time polymerase chain reaction assay to detect Duck adenovirus A DNA and application to samples from Swiss poultry flocks. J. Vet. Diagn. Invest. 26: 189–194. doi: 10.1177/1040638714523426 [DOI] [PubMed] [Google Scholar]
- 17.Su X. N., Liu J. J., Zhou Q. F., Zhang X. H., Zhao L. C., Xie Q. M., Chen W. G., Chen F.2017. Isolation and genetic characterization of a novel adeno-associated virus from Muscovy ducks in China. Poult. Sci. 96: 3867–3871. doi: 10.3382/ps/pex235 [DOI] [PubMed] [Google Scholar]
- 18.Teske L., Rubbenstroth D., Meixner M., Liere K., Bartels H., Rautenschlein S.2017. Identification of a novel aviadenovirus, designated pigeon adenovirus 2 in domestic pigeons (Columba livia). Virus Res. 227: 15–22. doi: 10.1016/j.virusres.2016.09.024 [DOI] [PubMed] [Google Scholar]
- 19.To K. K., Tse H., Chan W. M., Choi G. K., Zhang A. J., Sridhar S., Wong S. C., Chan J. F., Chan A. S., Woo P. C., Lau S. K., Lo J. Y., Chan K. H., Cheng V. C., Yuen K. Y.2014. A novel psittacine adenovirus identified during an outbreak of avian chlamydiosis and human psittacosis: zoonosis associated with virus-bacterium coinfection in birds. PLoS Negl. Trop. Dis. 8: e3318. doi: 10.1371/journal.pntd.0003318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vereecken M., de Herdt P., Ducatelle R.1998. Adenovirus infections in pigeons: A review. Avian Pathol. 27: 333–338. doi: 10.1080/03079459808419348 [DOI] [PubMed] [Google Scholar]