Abstract
Fowl cholera caused by Pasteurella multocida has always been a disease of global importance for poultry production. The aim of this study was to obtain more information about the epidemiology of avian P. multocida infection in southwest China and the genetic characteristics of clinical isolates. P. multocida isolates were characterized by biochemical and molecular-biological methods. The distributions of the capsular serogroups, the phenotypic antimicrobial resistance profiles, lipopolysaccharide (LPS) genotyping and the presence of 19 virulence genes were investigated in 45 isolates of P. multocida that were associated with clinical disease in poultry. The genetic diversity of P. multocida strains was performed by 16S rRNA and rpoB gene sequence analysis as well as multilocus sequence typing (MLST). The results showed that most (80.0%) of the P. multocida isolates in this study represented special P. multocida subspecies, and 71.1% of the isolates showed multiple-drug resistance. 45 isolates belonged to capsular types: A (100%) and two LPS genotypes: L1 (95.6%) and L3 (4.4%). MLST revealed two new alleles (pmi77 and gdh57) and one new sequence type (ST342). ST129 types dominated in 45 P. multocida isolates. Isolates belonging to ST129 were with the genes ompH+plpB+ptfA+tonB, whereas ST342 included isolates with fur+hgbA+tonB genes. Population genetic analysis and the MLST results revealed that at least one new ST genotype was present in the avian P. multocida in China. These findings provide novel insights into the epidemiological characteristics of avian P. multocida isolates in southwest China.
Keywords: epidemiology, MLST, Pasteurella multocida, poultry
Fowl cholera is an acute, fatal septicemic disease of various domestic and wild bird species, which are responsible for significant economic loss in poultry industries around the world [18, 28, 32]. Pasteurella multocida (P. multocida) has been reported to be the cause of fowl cholera outbreaks [43]. P. multocida can also lead to atrophic rhinitis in swine, hemorrhagic septicemia in bovine and buffaloes, fowl cholera, snuffles in rabbits and infected finger bone in humans [44]. P. multocida includes three subspecies: P. multocida subspecies multocida, P. multocida subspecies septica, and P. multocida subspecies gallicida [26]. Although P. multocida subspecies multocida is the most common cause of fowl cholera, the P. multocida subspecies septica and gallicida can also cause fowl cholera disease [26].
The pathogenicity of P. multocida is associated with the different virulence factors. A number of virulence factors have been identified to date and include fimbriae, adherence and colonization factors (ptfA, hsf-1, fimA, pfhA and tadD), iron-regulated and acquisition proteins (tonB, hgbA, hgbB, tbpA and fur), extracellular enzymes such as neuraminidase (nanB and nanH), superoxide dismutase (sodA and sodC), hyaluronidase (pmHAS), dermonecrotoxin (toxA) and a variety of outer-membrane proteins (OMPs) such as protectins (ompA, ompH and plpB) [31, 38]. These virulence factors are believed to play an important role in the pathogenesis of P. multocida.
P. multocida is genetically diverse on the basis of numerous studies [5, 13, 24]. Since different subspecies, serogroups and molecular characterization of P. multocida have different epidemiological significances in the fowl cholera disease, in order to effectively prevent this disease, it is important to identify the biological characteristics of P. multocida clinical isolates that causes infections. P. multocida are grouped into 5 capsular serogroups (serogroups A, B, D, E and F) and 16 somatic serotypes (serotypes 1 to 16) based on capsule and LPS, respectively [4, 15]. More recently, Harper et al. [14] simplified the typing of LPS antigens (L1-L8) and developed a multiplex PCR targeting the genes encoding the LPS structures (LPS-mPCR). Gene technologies such as 16S rRNA, restriction endonuclease analysis (REA), ribotyping, random amplification of polymorphic DNA (RAPD)-PCR, pulsed-field gel electrophoresis (PFGE) and multilocus sequence type analysis (MLST), have pushed forward studies of molecular epidemiology and genetic diversity of P. multocida [24, 35, 43]. MLST is considered the appropriate bacterial typing techniques for global and long-term studies [13, 36] and the results can be easily compared among different laboratories for long-term and global surveillance of bacterial strains [36, 43]. MLST is effective in identifying cases or outbreaks related to P. multocida infection, as well as identifying the source and dissemination events [33].
Although antimicrobials remain main tools for prevention and therapy of P. multocida infections, the increased incidence of multidrug-resistant pathogenic bacteria has been widely reported in last several decades [12, 17, 38]. Antibiotic resistance and the multidrug resistant phenotypes in pathogenic bacteria from food-producing animals are recognized as public health problems, which has drawn increasing concern in the world [38]. Control of P. multocida in poultry could be achieved with a vaccine-based approach. However, the protective efficacy of P. multocida vaccines is variable, particularly against heterologous strains [23]. Hence, more information about different types of avian P. multocida is needed in order to identify potential vaccine candidates.
The aim of this study was to understand the genetic characteristics of P. multocida associated with clinical cases of fowl cholera disease in southwest China. The investigation focused on isolates collected from China during a narrow time interval of two years and comparison was made to other strains by MLST. The data can be used to develop P. multocida vaccines for poultry and to set up other control measures.
MATERIALS AND METHODS
Bacterial isolates and DNA preparation
From January 2016 to August 2017, 283 liver samples of diseased ducks, geese and chicken from different farms located in Rongchang, Yongchuan, Dazu, Neijiang and Longchang in southwest China were collected. An average of three samples comes from one farm. We define one isolated strain comes from one farm. The average distance of sampled farms is 28.62km in each region. The nearest two of the farms are 7 km apart nearly. The samples were plated on Luria-Bertani (LB, Oxoid, Thermo Fisher Scientific, Shanghai, China) agar supplemented with 5% defibrinated sheep blood. All plates were incubated at 37°C under appropriate air conditions for 24 hr. DNA of all clinical isolates were extracted by Lysis Buffer for Microorganism to Direct PCR (Takara, Dalian, China). Afterward, presumptive isolates of P. multocida were confirmed by using the specific primers Kmt1 stated in Townsend et al [39]. All isolates were freeze-dried and kept at −80°C.
Subspecies identification for clinical isolates of P. multocida
Three subspecies (subsp. multocida, subsp. septica and subsp. gallicida) of all isolates of P. multocida were identified by standard biochemical procedures, 16S rRNA and rpoB gene sequences. All isolates of P. multocida were characterized biochemically by using standard biochemical procedures, including dulcitol, sorbitol, trehalose, arabinose, xylose, and indole reactions. The 16S rRNA gene and rpoB gene from all clinical isolates of the P. multocida were amplified by polyermase chain reaction (PCR) using the specific primer pairs [6, 20]. Afterwards, amplification products were sequenced by Sangon Biotech Co., Ltd (Shanghai, China). The nucleotide sequences of the 16s rRNA and rpoB gene were aligned by using the L-INS-I algorithm implemented in MAFFT [19]. Phylogenetic trees based on 16S rRNA and rpoB genes were constructed with the neighbor-joining (NJ) algorithm implemented in MEGA 7.0 software [22]. Conserved regions were determined using the Gblocks program [37]. The best-fitting nucleotide substitution model with the lowest BIC score was determined using MEGA7.0. NJ trees were built under the Jukes-Cantor model, and the robustness of the tree topology was assessed with 1,000 bootstrap replicates.
Antimicrobial susceptibility and antimicrobial resistance genes (ARGs)
All determinations of antimicrobial minimum inhibitory concentrations (MIC) for the P. multocida strains were performed by the broth microdilution method on Mueller-Hinton broth (Oxoid, Thermo Fisher Scientific) in a 96-well microplate, according to the Clinical and Laboratory Standards Institute (CLSI) [7, 8]. Thirteen antimicrobial agents (Sangon Biotech, Shanghai, China) were tested in 2-fold dilution series. Breakpoints of streptomycin was those defined by the European Committee on Antimicrobial Susceptibility Testing (EUCAST, 2012) [41]. For the other antimicrobials, the breakpoint values were taken from the CLSI guidelines. Reference strains Escherichia coli ATCC 25922 and Staphylococcus aureus ATCC 29213 served as quality control strains. As defined as the multidrug-resistance (MDR) is an acquired non-susceptibility to at least one agent in three or more antimicrobial categories [38]. Seven pairs of oligonucleotide primers (Table 1) to be used for the detection of specific resistance genes were designed by using the primer 5.0 software (Premier Biosoft International, Palo Alto, CA, U.S.A.). Amplification products of ARG were purified and sequenced by Sangon Shanghai Co., Ltd.
Table 1. Primers used for the identification of resistance gene.
| Target genes | Name | Sequnence (5'-3') | Product (bp) |
|---|---|---|---|
| floR | floR-1 | ATTTATCTCCCTGTCGTTCC | 982 |
| floR-2 | TCCCGACAATGCTGACTAT | ||
| tetA | tetA-1 | TTTCGCTTGCCGCATTTGG | 329 |
| tetA-2 | ATAGATCGCCGTGAAGAGGAGG | ||
| tetB | tetB-1 | GTTATCTTTGCTCCTTGGC | 835 |
| tetB-2 | CACCTTGCTGATGACTCTTT | ||
| tetH | tetH-1 | CAGAAAACCCATCTTGCT | 904 |
| tetH-2 | CCATAACAGACCATCCCA | ||
| strA | strA-1 | GTTCACAGCCTATCGGTTG | 453 |
| strA-2 | GTCCAATCGCAGATAGAAGG | ||
| sul2 | sul2-1 | CCGTCTCGCTCGACAGTTA | 509 |
| sul2-2 | CTCGTGTGTGCGGATGAAGT | ||
| TEM | TEM-1 | TCGTGTCGCCCTTATTCCC | 778 |
| TEM-2 | CTGACTCCCCGTCGTGTAGAT |
Reference sequence: floR: FM179941.1; tetA: ALBX01002091.1; tetB: FR872822.1; tetH: AJ514834.1; strA: NC_019381; sul2: CP003022.1; TEM: NZ_JQAH01000009.1.
Detection of capsule typing and virulence-associated genes (VAGs)
All isolates were screened by PCR analysis for the presence of capsule biosynthesis genes and the virulence-associated genes. PCR analysis of VAGs for capsule biosynthesis (capA, capB, capD, capE and capF), LPS-mPCR, iron-regulated and iron acquisition proteins (tonB, tbpA, fur, hgbA and hgbB), fimbriae and adhesins (fimA, ptfA, hsf-1, tadD, pfhA, nanH and nanB), superoxide dismutase (sodA and sodC), hyaluronidase (pmHAS), toxA and outer membrane proteins (ompH, ompA and plpB) was conducted as described previously [14, 31, 38].
MLST genotyping
All 45 P. multocida isolates were further analyzed using MLST, which was performed as described on the MLST database, utilizing partial sequences of seven genes for genotypic characterization. PCR amplification for seven housekeeping genes (adk, est, pmi, zwf, mdh, gdh and pgi) was carried out using primers and protocols available at the RIRDC MLST Database. (https://pubmlst.org/pmultocida/). This RIRDC scheme was developed by Sounthi Subaaharan and Pat Blackall [36]. Amplification products were sequenced (GeneCreate, Wuhan, China). The sequence of each locus was checked on the PubMLST RIRDC Database website for determination of the allele designations and ST of each isolate. The allele and ST of isolate were deposited in the PubMLST database.
Analysis of population diversity
The eBURST v3 software (http://spneumoniae.mlst.net/eburst/) was used to investigate the population diversity and relationship between MLST sequence types and to analyze clonal complexes (CC) based on the stringent group definition of six out of seven alleles. Phylogenetic and molecular evolutionary analyzes were conducted on the basis of the concatenation of seven MLST loci using MEGA 7.0. The UPGMA tree was built under the Kimura 2-parameter model, and the robustness of the tree topology was assessed with 1,000 bootstrap replicates. Population structure and ancestry of the major ST complexes from all available STs in the MLST database was inferred using the admixture model with independent allele frequencies implemented in STRUCTURE version 2.3.4. Sequence data (seven core genes) were formatted using xmfa2struct software (http://www.xavierdidelot.xtreemhost.com/clonalframe.htm). STRUCTURE software uses a Bayesian clustering framework and assumes that the observed data are derived from K ancestral populations (lineages) [10, 29]. Data statistical testing was performed with SPSS software (version 17.0; SPSS Inc., Chicago, IL, U.S.A.). Comparisons of proportions were made by χ2. Differences with P<0.05 were considered statistically significant.
Determination of bacterial virulence
PM-N1701 (ST129), PM-R1602 (ST342) and PM-L1706 (ST8) strains were randomly selected to estimate the median lethal dose (LD50) by toxicity test in mice (TTM). All animal procedures were reviewed and approved by the Ethics Committee of Southwest University. Eighty SPF mice at 6 weeks of age (Chongqing Academy of Animal Science, Chongqing, China) were randomly assigned to eight groups of ten animals each. P. multocida was cultured on trypticase soy broth supplemented with 5% fetal calf serum and incubated on a shaking table at 220 rpm and temperature of 37°C for 12–16 hr. The number of viable P. multocida were enumerated by plate counts. Cultures were serially diluted 2-fold and diluted to 7 concentrations in sterile phosphate buffered saline (PBS). The mice in groups 1–7 were inoculated intraperitoneally with 0.2 ml per mouse of the cultures, and the mice in group 8 were injected with 0.2 ml of sterile PBS, as a control group. The LD50 was calculated following the method described by Reed-Muench [30].
RESULTS
Isolation and sub-species classification
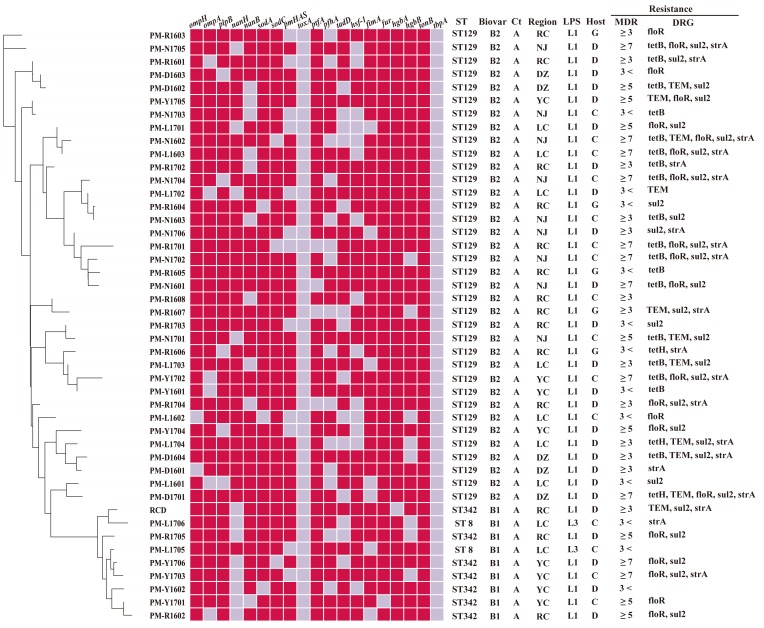
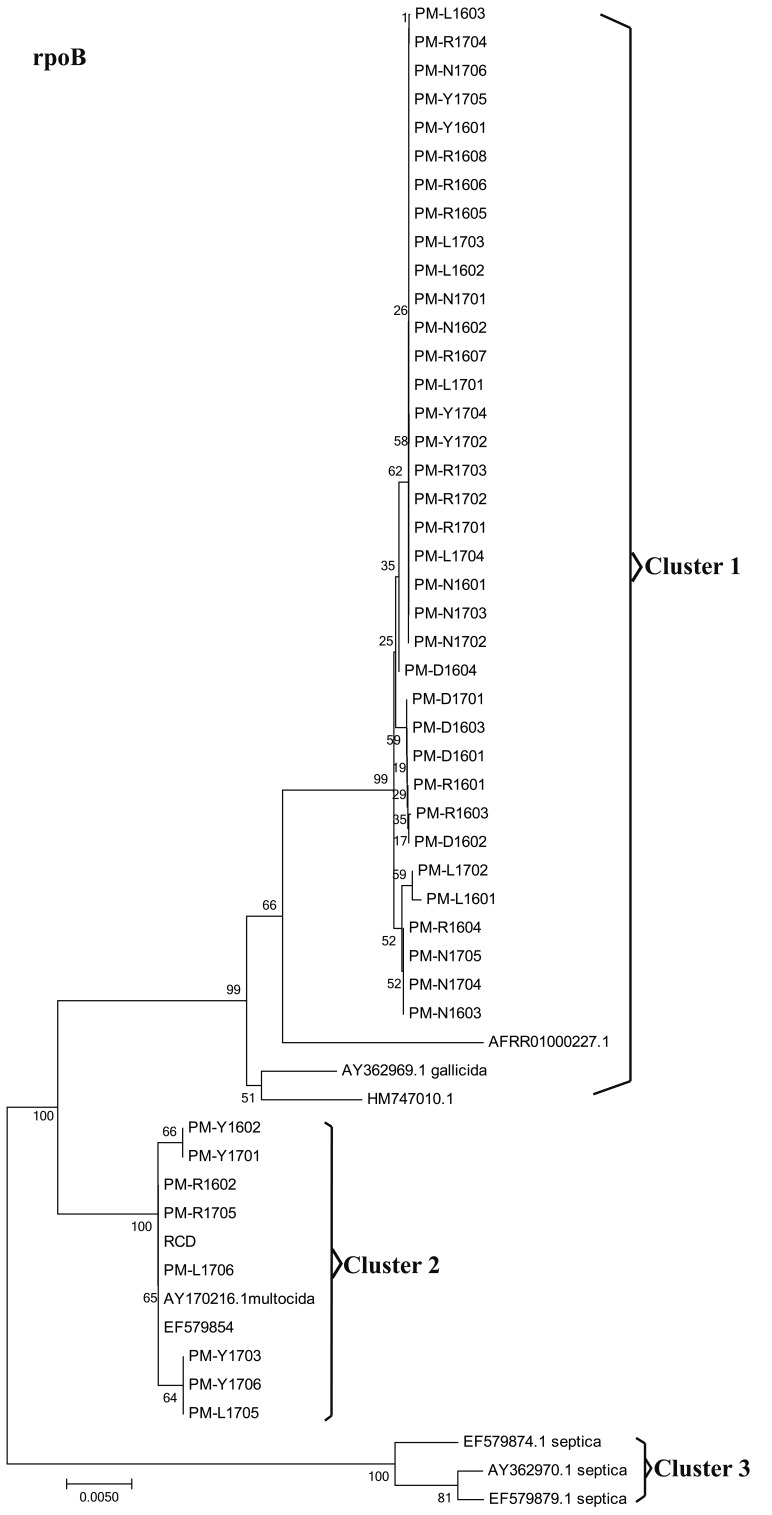
A total of 45 P. multocida strains were isolated from outbreaks of fowl cholera occurring in southwest China. Of the 45 P. multocida strains studied, all isolates were found to express capsule type A (Fig. 1). All strains tested positive in sorbitol and indole tests, and were negative in the dulcitol tests. However, the strains differed in their abilities to ferment some sugars (Table 2). This study noticed differences among strains in their abilities to ferment trehalose, arabinose and xylose, and these differences permitted us to classify 45 P. multocida strains into two distinct biochemical types (Table 2). Biovar 1 matched the typical subspecies multocida defined [11, 24]. However, Biovars 2 revealed some differences from the three typical subspecies of P. multocida in biochemical characterization. All isolates were characterized by rpoB and 16S rRNA sequencing for further verification at subspecies level of P. multocida based on the molecular method. According to the N-J phylogenetic tree (Fig. 2) based on 16S rRNA sequences, 45 P. multocida strains were located in cluster 1 along with the type strains of P. multocida ssp. Multocida CCUG17976T (accession no. AF294410) and P. multocida ssp. Gallicida CCUG17978T (accession no. AF294412). Four reference strains of P. multocida ssp. Septica were clustered in clusters 2. The subsp. Multocida and subsp. Gallicida of 45 P. multocida strains were not recognized in the 16S rRNA phylogeny, and therefore further distinguished them by rpoB gene sequence comparison. Within the rpoB gene phylogenetic tree (Fig. 3), three distinct clusters can be observed with tree topologies, which is not the case in the 16S rRNA tree. Cluster 2 included 9 isolates and P. multocida ssp. Multocida CCUG17976T strains. Cluster 1 within the rpoB gene phylogenetic tree contained 36 isolates and P. multocida ssp. Gallicida CCUG17978T strains. Four reference strains of P. multocida ssp. Spetica were clustered in cluster 3. Thus, the resolution of the 16S rRNA within the family P. multocida is generally lower than that of the rpoB gene sequence in this study, and the results obtained confirmed previous research [24]. The 16S rRNA and rpoB sequences of 45 P. multocida isolates were deposited in the GenBank database under accession numbers MG597069-MG597111, MG859656-MG859657 and MG602835-MG602877, MG813901-MG813902 respectively.
Fig. 1.
Dendrogram representing relatedness among P. multocida strains isolated from poultry according to biological characteristics of isolates. Note: phylogenetic tree: Multiple genes combined to build phylogenetic tree base on 16S rRNA and rpoB gene. Red square: positive reaction; Gray squares: negative reaction; Ct: capsular type RC: Rongchang; NJ: Neijiang; DZ: Dazu; YC: Yongchuan; LC: Longchang. G: goose; D: duck; C: chicken.
Table 2. Differential properties and identity of 45 clinical isolates of Pasteurella.
| Bt1 (n=9) |
Bt2 (n=36) |
P. multocida ssp. Multocidaa) |
P. multocida ssp. septicaa) |
P. multocida ssp. gallicidaa) |
|
|---|---|---|---|---|---|
| Dulcitol | - | - | - | - | + |
| Sorbitol | + | + | + | - | + |
| Trehalose | + | - | + | + | - |
| Arabinose | - | + | - | - | + |
| Xylose | + | - | + | + | - |
| Indole | + | + | + | + | + |
Fig. 2.
Neighbor joining phylogenetic analysis of 16S rRNA gene sequences.
Fig. 3.
Neighbor joining phylogenetic analysis of partial rpoB sequences
Antimicrobial susceptibility
Forty five P. multocida isolates resistance rates to Amoxicillin, Sulfamethazine, Tetracycline, Doxycycline, Florfenicol and Streptomycin were 82.2, 71.1, 53.3, 48.9, 46.7 and 44.4%, respectively. In addition, 71.1% of isolates were MDR. Of the 24 tetracycline-resistant isolates, 18 isolates harbored tetB and 3 isolates harbored tetH. The remaining 3 isolates were negative for all the tet genes tested in this study. Of the 37 amoxicillin-resistant isolates, 11 isolates harbored the TEM gene. All isolates that showed resistance to florfenicol were positive for the floR gene, and all streptomycin-resistant isolates also were positive for the strA gene in this study. Of the 32 sulfamethazine-resistant isolates, 30 harbored the sul2 gene.
Virulence genes and toxicity test
The relative frequencies of the VAGs are presented in Fig. 1. Among the 45 avian P. multocida strains, the 19 VAG regions ranged in prevalence from 0% (toxA and tbpA) to 100% (tonB). More than 90% of isolates harbored the tonB (100.0%), fur (97.8%), hgbA (97.8%), ompH (95.6%), sodA (93.3%), sodC (93.3%) and ptfA genes (91.1%). The tbpA and toxA gene were not detected in any of the 45 clinical strains studied. The great majority of VAGs were equally distributed in each source animal (ducks, chicken and geese). The results of toxicity test showed that PM-N1701 (LD50=53.3) and strain PM-R1602 (LD50=65) have similar virulence (Table 4).
Table 4. Determination of the LD50 of PM-N1701, PM-R1602 and PM-L1706 strains.
| Strains | Dose (CFU/0.2 ml) and total dead | LD 50 (CFU) | ||||||
|---|---|---|---|---|---|---|---|---|
| 400 | 200 | 100 | 50 | 25 | 12 | 6 | ||
| PM-N1701 | 34/34 | 24/26 | 16/21 | 9/19 | 4/21 | 1/27 | 0/36 | 53.3 |
| PM-R1602 | 31/31 | 21/23 | 13/19 | 7/18 | 2/21 | 0/29 | 0/39 | 65 |
| PM-L1706 | 3/11 | 1/18 | 0/27 | 0/37 | 0/47 | - | - | ND |
ND: Not detected.
MLST
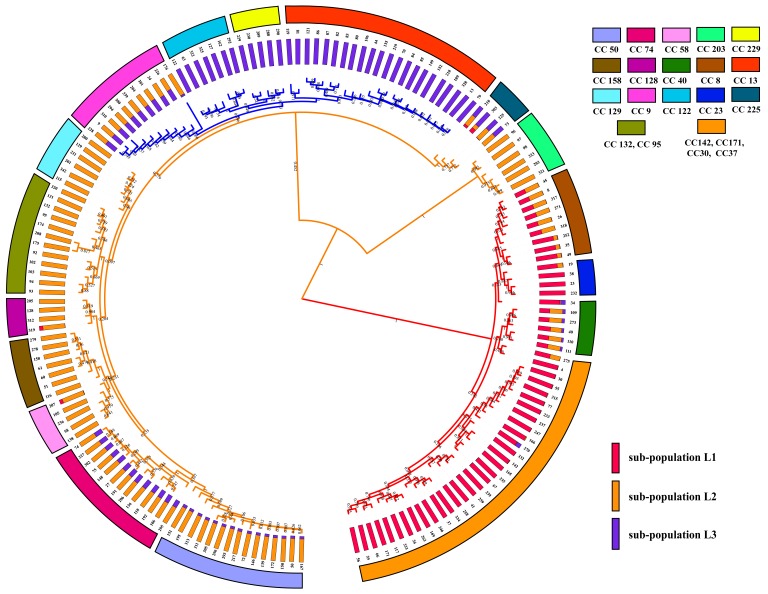
In this study, MLST analysis revealed three different STs (ST129, ST8 and ST342) (Fig. 1). The ST342 (adk21-est33-pmi77-zwf2-mdh17-gdh57-pgi20) was identified for the first time in this study. ST342 has two new allele pmi77 and gdh57, which have a G/A mutation at position 58 of pmi26 locus and a C/T mutation at position 378 of gdh20 locus, respectively. According to the rpoB-base tree and MLST scheme, we can observe that 36 isolates of special P. multocida are ST129 genotype. Seven ST342 and two ST8 strains were detected among 45 isolates. All ST342 and ST8 strains were from P. multocida subsp. multocida. In Fig. 1, we can observe that ST129 (88.9%) was more statistically significant than ST342 (0%) for the nanH gene. To analyze the genetic characteristics of 45 P. multocida isolates and previously identified P. multocida strains globally, the DNA sequences (n=168) of representative ST complexes of P. multocida were downloaded from the P. multocida MLST website, and the ST phylogenetic tree was built using MEGA 7.0 (Fig. 4). According to the ST phylogenetic tree (Fig. 4) and the eBURST v3 software, the ST129 and newly discovered ST342 are clustered in the CC129 clonal complex, of which ST129 was defined as the primary founder of the CC129 clonal complex. ST8 was clustered into CC8 clonal complex. The mixture model of STRUCTURE was applied to the sequence dataset of 168 STs. Multiple runs (iterations=10) with K values from 2 to 7 showed maximal posterior probability at K=3. The 168 STs fell into three distinct sub-populations (lineage: L1, L2 and L3) according to the results of group structure analysis (Fig. 4). There was no admixture of ancestral sources between ST129 and ST342. They are classified as lineage L2 and tended to be highly homogenous.
Fig. 4.
Diagrams denoting population structure. Note: UPGMA tree of the 168 STs (21 representative clonal complexes) based on the concatenated sequences of 7 loci and the three lineages (L1 to L3) were separated with 100% bootstrap value support. Proportions of ancestral subpopulations of the 168 STs and different colors represented distinct assuming subpopulations corresponding to lineage L1 to L3.
DISCUSSION
Pasteurellosis is one of the most common diseases of some wild and domestic animals worldwide. The cases of P. multocida infected animals have been widely reported in recent years in China [38, 43]. However, the distribution and prevalence of serotypes and pathotypes of P. multocida isolates may vary considerably from different regions and times. Molecular characterization and genetic features of P. multocida isolates from poultry are seldom reported in southwest China. In this study, although 36 arabinose-positive strains from the rpoB-base tree cluster I were grouped closely to each other with strains of P. multocida subsp. gallicida CCUG17978T strains, we do not think that these strains were subsp. gallicida strains because of their aberrant phenotype (dulcitol-negative) (Table 2). However, on the basis of previous reports [11, 21, 25], arabinose-positive isolates were detected only among P. multocida subsp. gallicida strains. Thus, these strains isolated from poultry may be some special P. multocida strains. Muhldorfer et al. [24] and Stahel et al. [34] identified some P. multocida subsp. gallicida strains by the rpoB gene. However, there are differences between the results of rpoB-base tree and biochemical test in this study. Varga et al. [40] thought that discrimination between P. multocida subsp. multocida and P. multocida subsp. gallicida strains based on the phylogenetic analysis of the rpoB and thdF genes is problematic due to very limited information in GenBank about the effective rpoB gene sequences for P. multocida subsp. gallicida and subsp. multocida strains of typical biochemical characteristics. Therefore, the ability of the rpoB gene to distinguish between P. multocida subsp. multocida and P. multocida subsp. gallicida strains needs further study, and our results also provide some sample data for the future study of the rpoB gene.
It was reported [31, 38] that a number of VAGs are correlated with the pathogenic mechanisms of P. multocida. Among these important virulence genes, the iron acquisition genes (tonB, hgbA, hgbB, tbpA and fur) can help pathogenic bacteria to obtain a variety of heme iron source from the host infected. In this study, the detection rates of tonB, hgbA, hgbB and fur genes were over 82.2%, of which the detection rate of tonB gene was 100%. This result is in line with the theoretical expectation and similar results have also been reported [31]. However, tbpA gene was not detected for all 45 isolates in the present study. Many previous studies [1, 9] have shown that the tbpA encoding gene is closely related to the ruminant strains and Shirzad et al. [31] also observed that a high frequency of the tbpA gene was only found in the ruminant isolates. They speculated [1, 9, 31] that the tbpA encoding gene could be an epidemiological marker in these ruminant isolates, and our results of the tbpA gene in avian isolates reinforce this idea. The pfhA gene, a filamentous hemagglutinin, plays a major role in the initial colonization of the upper respiratory tract. 66.7% of the poultry isolates were positive for the pfhA gene in the study, and a similar frequency of pfhA gene was also found by Furian et al. [12] in Brazil. Ewers et al. [9] and Shirzad et al. [31] suggested that the frequency of pfhA gene varies greatly among the strains of P. multocida. Shirzad et al. [31] observed that the frequency of pfhA gene was higher among poultry isolates (46.66%) in comparison with other isolates and similar results were reported by Furian et al [12]. As the number of samples expanded, the pfhA gene appeared more frequently in poultry isolates. Therefore, the pfhA gene may be an important virulence factor for poultry isolates.
In order to further characterize these isolates, LPS-mPCR, capsular typing and MLST was used to analyze genetic diversity and molecular epidemiology of the isolated strains in southwest China. LPS-mPCR and capsular typing results indicated that 45 avian P. multocida strains were A:1 and A:3. However, MLST revealed that 45 avian P. multicida isolates were three ST types (ST129, ST8 and ST342), which demonstrated that MLST typing can obtain higher resolution in P. multocida typing compared with traditional typing (LPS and capsular). Thirty six avian P. multocida isolates were identified as ST129 genotypes (83.7%) in this study. Wang et al. [43] reported that 40 avian P. multocida isolated from the Jiangsu province of China were all identified as ST129, and Sun et al. [42] also reported a high detection rate (91%) of avian P. multocida ST129 genotype in China. This suggested that the ST129 may be the major genotype of avian P. multocida strain in China. The ST129 genotype was initially identified among bovine isolates in Sri Lanka [16], whereafter it was also identified in poultry in Zimbabwe [2], pigs and other animals in world [2]. As a result, some researchers think that the ST129 genotype of P. multocida can adapt to a wide variety of animal hosts [2, 16, 43]. It is noteworthy that the toxicity of PM-N1701 (ST129) strain (LD50=53.3) is strong by the TTM test. Therefore, we cannot ignore the potentially hostile of ST129 P. multocida strains with host diversity and high virulence found in China southwest to other animal species. The new ST342 strain from CC129 clonal complex also exhibits similar virulence (LD50=65) compared to the ST129 strain, so possible hazards from the ST342 strain in poultry cannot be underestimated. Interestingly, during virulence genotyping, we also found that the some genes displayed a certain level of “genotype-preference” [27]. For instance, isolates belonging to ST129 were with the genes ompH+plpB+ptfA+tonB, whereas ST342 included isolates with fur+hgbA+tonB genes. The detection rate of nanH genes in ST129 was significantly higher than their detection in ST342 and ST8 genotypes (P <0.05). In addition to the ST129 and ST342 strains found, two ST8 strains were found in this study. ST8 strains are rarely reported in China, and their isolation rate is low. Sun et al. [42] reported that only one ST8 strain was identified in 22 P. multocida isolates isolated from poultry in China. However, ST8 P. multocida strains has caused serious fowl cholera disease in Australia [3, 33]. Therefore, the potential hazard of ST8 P. multocida strain cannot be ignored. According to the sub-structuring analysis (Fig. 4), ST8 P. multocida strain is derived from the mixture of sub-population L1 and sub-population L2. However, ST129 and ST342 of P. multocida isolates were only observed in the sub-population L2 in the present study. This indicates that ST genotype strains from sub-population L1 exist in the avian P. multocida strains in China. The clonal complex of CC129, CC132, CC128, CC158, CC58, CC74, CC50 and CC95 clustered together (Fig. 4), which may suggest that these clonal complexes are closely related.
Antibiotic resistance has become a global problem. In this study, the prevalence of P. multocida isolate resistance to conventional veterinary drugs including florfenicol, doxycycline, tetracycline and sulfamethazine was found to be in excess of 46.7% (Table 3) and 71.1% of P. multocida isolates were resistant to at least three antimicrobials (Fig. 1). In addition, our results showed that the multi-drug resistance rate of ST129 and ST342 isolates reached 72.2 and 85.7%, respectively. Of which the percentage of ST129 and ST342 isolates resistant to at least five antimicrobial agents were 38.9 and 71.4%, respectively. This suggested that the resistance rate of over five antibiotics of ST342 isolates was significantly higher than that of ST129 isolates in this study. Tang et al. [38] revealing that P. multocida is exhibiting a rapid increase in the rate of resistance to a large number of antimicrobial agents. Therefore, it is especially important to use vaccines to prevent and control P. multocida strains.
Table 3. MICs for 13 antimicrobial agents against 45 strains of P. multocida.
| Antimicrobial | No. of isolates showing an MIC of agent (µg/ml) | Breakpoint (µg/ml) |
Resistance (%) |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ≤0.03 | 0.06 | 0.12 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | 256 | 512 | ≥1,024 | |||
| Amoxicillin | 2 | 1 | 2 | 3 | 10 | 6 | 7 | 6 | 5 | 3 | 32 | 82.2 | ||||||
| Ciprofloxacin | 8 | 2 | 18 | 5 | 10 | 2 | 4a) | 0.0 | ||||||||||
| Florfenicol | 6 | 8 | 10 | 13 | 2 | 3 | 3 | 8 | 46.7 | |||||||||
| Tetracycline | 8 | 11 | 2 | 9 | 6 | 3 | 3 | 1 | 2 | 16 | 53.3 | |||||||
| Tilmicosin | 5 | 2 | 4 | 13 | 3 | 8 | 5 | 2 | 3 | 32 | 22.2 | |||||||
| Doxycycline | 3 | 7 | 4 | 9 | 10 | 3 | 7 | 2 | 16b) | 48.9 | ||||||||
| Amikacin | 3 | 5 | 2 | 17 | 5 | 2 | 7 | 3 | 1 | 64 | 24.4 | |||||||
| Sulfamethazine | 3 | 3 | 7 | 21 | 11 | 512 | 71.1 | |||||||||||
| Spectinomycin | 17 | 2 | 9 | 8 | 5 | 1 | 3 | 128 | 6.7 | |||||||||
| Lincomycin | 1 | 5 | 13 | 9 | 5 | 2 | 10 | 4 | 37.8 | |||||||||
| Erythromycin | 17 | 5 | 7 | 6 | 3 | 5 | 2 | 8 | 22.2 | |||||||||
| Gentamicin | 7 | 3 | 10 | 16 | 2 | 1 | 5 | 1 | 16 | 20.0 | ||||||||
| Streptomycin | 1 | 9 | 15 | 13 | 7 | 512c) | 44.4 | |||||||||||
a) The value is based on Tang et al. standards [37]. b) The value is based on CLSI-2017 standards. c) The value is based on EUCAST standards.
The investigation disclosed epidemiological information of avian P. multocida infection in southwest China. In this study, the A:1 serotype strains dominated in 45 avian P. multocida isolates. Thirty six arabinose-positive isolates were detected as ST129 genotype. Seven ST342 strains and two ST8 strains had typical phenotypic characteristics of subsp. multocida. We found that three sequence types of P. multocida associated with pathogenic of poultry from China, of which ST342 strain were first discovered. In addition, our results showed that virulence-associated gene and antibiotic multidrug resistance profile is likely to be associated with ST genotype. This study will help in tracking evolution of P. multocida epidemic strains and provide the strain information to develop P. multocida vaccines for poultry.
Acknowledgments
We thank the team of the curators of the PubMLST system for importing novel alleles, profiles and isolates at https://pubmlst.org/pmultocida/. This work was supported by the Fundamental Research Funds for the Central Universities (number: XDJK2017D083). We would like to acknowledge Nicholas K. H. Ostan (PhD candidate in University of Toronto) for assistance with review of the paper.
REFERENCES
- 1.Atashpaz S., Shayegh J., Hejazi M. S.2009. Rapid virulence typing of Pasteurella multocida by multiplex PCR. Res. Vet. Sci. 87: 355–357. doi: 10.1016/j.rvsc.2009.04.004 [DOI] [PubMed] [Google Scholar]
- 2.Bisgaard M., Petersen A., Christensen H.2013. Multilocus sequence analysis of Pasteurella multocida demonstrates a type species under development. Microbiology 159: 580–590. doi: 10.1099/mic.0.063461-0 [DOI] [PubMed] [Google Scholar]
- 3.Blackall P. J., Fegan N., Chew G. T., Hampson D. J.1998. Population structure and diversity of avian isolates of Pasteurella multocida from Australia. Microbiology 144: 279–289. doi: 10.1099/00221287-144-2-279 [DOI] [PubMed] [Google Scholar]
- 4.Carter G. R.1952. The type specific capsular antigen of Pasteurella multocida. Can. J. Med. Sci. 30: 48–53. [DOI] [PubMed] [Google Scholar]
- 5.Davies R. L.2004. Genetic diversity among Pasteurella multocida strains of avian, bovine, ovine and porcine origin from England and Wales by comparative sequence analysis of the 16S rRNA gene. Microbiology 150: 4199–4210. doi: 10.1099/mic.0.27409-0 [DOI] [PubMed] [Google Scholar]
- 6.Davies R. L., Paster B. J., Dewhirst F. E.1996. Phylogenetic relationships and diversity within the Pasteurella haemolytica complex based on 16S rRNA sequence comparison and outer membrane protein and lipopolysaccharide analysis. Int. J. Syst. Bacteriol. 46: 736–744. doi: 10.1099/00207713-46-3-736 [DOI] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute (CLSI) 2013. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals; approved standard-fourth edition. CLSI document Vet01-A4. Wayne. [Google Scholar]
- 8.Clinical and Laboratory Standards Institute (CLSI) 2017. Performance Standards for Antimicrobial Susceptibility Testing. 27th ed. CLSI supplement M100.Wayne. [Google Scholar]
- 9.Ewers C., Lübke-Becker A., Bethe A., Kiebling S., Filter M., Wieler L. H.2006. Virulence genotype of Pasteurella multocida strains isolated from different hosts with various disease status. Vet. Microbiol. 114: 304–317. doi: 10.1016/j.vetmic.2005.12.012 [DOI] [PubMed] [Google Scholar]
- 10.Falush D., Stephens M., Pritchard J. K.2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164: 1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fegan N., Blackall P. J., Pahoff J. L.1995. Phenotypic characterisation of Pasteurella multocida isolates from Australian poultry. Vet. Microbiol. 47: 281–286. doi: 10.1016/0378-1135(95)00119-0 [DOI] [PubMed] [Google Scholar]
- 12.Furian T. Q., Borges K. A., Laviniki V., Rocha S. L., de Almeida C. N., do Nascimento V. P., Salle C. T., Moraes H. L.2016. Virulence genes and antimicrobial resistance of Pasteurella multocida isolated from poultry and swine. Braz. J. Microbiol. 47: 210–216. doi: 10.1016/j.bjm.2015.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.García-Alvarez A., Vela A. I., San Martín E., Chaves F., Fernández-Garayzábal J. F., Lucas D., Cid D.2017. Characterization of Pasteurella multocida associated with ovine pneumonia using multi-locus sequence typing (MLST) and virulence-associated gene profile analysis and comparison with porcine isolates. Vet. Microbiol. 204: 180–187. doi: 10.1016/j.vetmic.2017.04.015 [DOI] [PubMed] [Google Scholar]
- 14.Harper M., John M., Turni C., Edmunds M., St Michael F., Adler B., Blackall P. J., Cox A. D., Boyce J. D.2015. Development of a rapid multiplex PCR assay to genotype Pasteurella multocida strains by use of the lipopolysaccharide outer core biosynthesis locus. J. Clin. Microbiol. 53: 477–485. doi: 10.1128/JCM.02824-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heddleston K. L., Gallagher J. E., Rebers P. A.1972. Fowl cholera: gel diffusion precipitin test for serotyping Pasteruella multocida from avian species. Avian Dis. 16: 925–936. doi: 10.2307/1588773 [DOI] [PubMed] [Google Scholar]
- 16.Hotchkiss E. J., Hodgson J. C., Lainson F. A., Zadoks R. N.2011. Multilocus sequence typing of a global collection of Pasteurella multocida isolates from cattle and other host species demonstrates niche association. BMC Microbiol. 11: 115. doi: 10.1186/1471-2180-11-115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jamali H., Rezagholipour M., Fallah S., Dadrasnia A., Chelliah S., Velappan R. D., Wei K. S. C., Ismail S.2014. Prevalence, characterization and antibiotic resistance of Pasteurella multocida isolated from bovine respiratory infection. Vet. J. 202: 381–383. doi: 10.1016/j.tvjl.2014.07.024 [DOI] [PubMed] [Google Scholar]
- 18.Kardos G., Kiss I.2005. Molecular epidemiology investigation of outbreaks of fowl cholera in geographically related poultry flocks. J. Clin. Microbiol. 43: 2959–2961. doi: 10.1128/JCM.43.6.2959-2961.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katoh K., Standley D. M.2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30: 772–780. doi: 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korczak B., Christensen H., Emler S., Frey J., Kuhnert P.2004. Phylogeny of the family Pasteurellaceae based on rpoB sequences. Int. J. Syst. Evol. Microbiol. 54: 1393–1399. doi: 10.1099/ijs.0.03043-0 [DOI] [PubMed] [Google Scholar]
- 21.Kuhnert P., Korczak B. M.2006. Prediction of whole-genome DNA-DNA similarity, determination of G+C content and phylogenetic analysis within the family Pasteurellaceae by multilocus sequence analysis (MLSA). Microbiology 152: 2537–2548. doi: 10.1099/mic.0.28991-0 [DOI] [PubMed] [Google Scholar]
- 22.Kumar S., Stecher G., Tamura K.2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33: 1870–1874. doi: 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Massacci F. R., Magistrali C. F., Cucco L., Curcio L., Bano L., Mangili P., Scoccia E., Bisgaard M., Aalbæk B., Christensen H.2018. Characterization of Pasteurella multocida involved in rabbit infections. Vet. Microbiol. 213: 66–72. doi: 10.1016/j.vetmic.2017.11.023 [DOI] [PubMed] [Google Scholar]
- 24.Mühldorfer K., Schwarz S., Fickel J., Wibbelt G., Speck S.2011. Genetic diversity of Pasteurella species isolated from European vespertilionid bats. Vet. Microbiol. 149: 163–171. doi: 10.1016/j.vetmic.2010.10.002 [DOI] [PubMed] [Google Scholar]
- 25.Mutters R., Ihm P., Pohl S., Frederiksen W., Mannheim W.1985. Reclassification of the genus Pasteurella Trevisan 1887 on the basis of deoxyribonucleic acid homology, with proposals for the new species Pasteurella dagmatis, Pasteurella canis, Pasteurella stomatis, Pasteurella anatis, and Pasteurella langaa. Int. J. Syst. Bacteriol. 35: 309–322. doi: 10.1099/00207713-35-3-309 [DOI] [Google Scholar]
- 26.Peng Z., Liang W., Liu W., Chen H., Wu B.2017. Genome characterization of Pasteurella multocida subspecies septica and comparison with Pasteurella multocida subspecies multocida and gallicida. Arch. Microbiol. 199: 635–640. doi: 10.1007/s00203-017-1341-x [DOI] [PubMed] [Google Scholar]
- 27.Peng Z., Wang H., Liang W., Cheng Y., Tang X., Cheng H., Wu B.2017. A capsule/lipopolysaccharide/MLST genotype D/L6/ST11 of Pasteurella multocida is likely to be strongly associated with swine respiratory disease in China. Arch. Microbiol. 2: 1–12. [DOI] [PubMed] [Google Scholar]
- 28.Petersen K. D., Christensen J. P., Permin A., Bisgaard M.2001. Virulence of Pasteurella multocida subsp. multocida isolated from outbreaks of fowl cholera in wild birds for domestic poultry and game birds. Avian Pathol. 30: 27–31. doi: 10.1080/03079450020023168 [DOI] [PubMed] [Google Scholar]
- 29.Pritchard J. K., Stephens M., Donnelly P.2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reed L. J., Muench H.1938. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. 27. [Google Scholar]
- 31.Shirzad Aski H., Tabatabaei M.2016. Occurrence of virulence-associated genes in Pasteurella multocida isolates obtained from different hosts. Microb. Pathog. 96: 52–57. doi: 10.1016/j.micpath.2016.04.008 [DOI] [PubMed] [Google Scholar]
- 32.Singh R., Remington B., Blackall P., Turni C.2014. Epidemiology of fowl cholera in free range broilers. Avian Dis. 58: 124–128. doi: 10.1637/10656-090313-Reg.1 [DOI] [PubMed] [Google Scholar]
- 33.Singh R., Blackall P. J., Remington B., Turni C.2013. Studies on the presence and persistence of Pasteurella multocida serovars and genotypes in fowl cholera outbreaks. Avian Pathol. 42: 581–585. doi: 10.1080/03079457.2013.854861 [DOI] [PubMed] [Google Scholar]
- 34.Stahel A. B. J., Hoop R. K., Kuhnert P., Korczak B. M.2009. Phenotypic and genetic characterization of Pasteurella multocida and related isolates from rabbits in Switzerland. J. Vet. Diagn. Invest. 21: 793–802. doi: 10.1177/104063870902100605 [DOI] [PubMed] [Google Scholar]
- 35.Sthitmatee N., Kataoka Y., Sawada T.2010. Molecular epidemiology of Japanese avian Pasteurella multocida strains by the single-enzyme amplified fragment length polymorphism and pulsed-field gel electrophoresis. J. Vet. Med. Sci. 72: 1465–1470. doi: 10.1292/jvms.10-0181 [DOI] [PubMed] [Google Scholar]
- 36.Subaaharan S., Blackall L. L., Blackall P. J.2010. Development of a multi-locus sequence typing scheme for avian isolates of Pasteurella multocida. Vet. Microbiol. 141: 354–361. doi: 10.1016/j.vetmic.2010.01.017 [DOI] [PubMed] [Google Scholar]
- 37.Talavera G., Castresana J.2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56: 564–577. doi: 10.1080/10635150701472164 [DOI] [PubMed] [Google Scholar]
- 38.Tang X., Zhao Z., Hu J., Wu B., Cai X., He Q., Chen H.2009. Isolation, antimicrobial resistance, and virulence genes of Pasteurella multocida strains from swine in China. J. Clin. Microbiol. 47: 951–958. doi: 10.1128/JCM.02029-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Townsend K. M., Boyce J. D., Chung J. Y., Frost A. J., Adler B.2001. Genetic organization of Pasteurella multocida cap Loci and development of a multiplex capsular PCR typing system. J. Clin. Microbiol. 39: 924–929. doi: 10.1128/JCM.39.3.924-929.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Varga Z., Volokhov D. V., Stipkovits L., Thuma A., Sellyei B., Magyar T.2013. Characterization of Pasteurella multocida strains isolated from geese. Vet. Microbiol. 163: 149–156. doi: 10.1016/j.vetmic.2012.12.023 [DOI] [PubMed] [Google Scholar]
- 41.Version V. F.2012. European Committee on Antimicrobial Susceptibility Testing Breakpoint tables for interpretation of MICs and zone diameters.
- 42.Wang L., Sun J., Guo D., Cao P., Liu J., Liu C., Liu D., Qu L.2016. Identification of capsule serotype and genotype of Pasteurella multocida in some areas of China. Chin. J. Prev. Vet. Med. 38: 116–119. [Google Scholar]
- 43.Wang Y., Zhu J., Lu C., Wu B., Liu D., Hang W., Liu H., Liu X.2013. Evidence of circulation of an epidemic strain of Pasteurella multocida in Jiangsu, China by multi-locus sequence typing (MLST). Infect. Genet. Evol. 20: 34–38. doi: 10.1016/j.meegid.2013.07.027 [DOI] [PubMed] [Google Scholar]
- 44.Wilkie I. W., Harper M., Boyce J. D. , Adler B.2012. Pasteurella multocida: diseases and pathogenesis. Curr. Top. Microbiol. Immunol. 361: 1–22. [DOI] [PubMed] [Google Scholar]