Abstract
Aim:
Staphylococcus aureus is a major cause of severe hospital-acquired infections, and biofilm formation is an important part of staphylococcal pathogenesis. Therefore, developing new antimicrobial agents against both planktonic cells and biofilm of S. aureus is a major challenge.
Results:
Three 1,3,4-oxadiazole derivatives exhibited antimicrobial activity against seven S. aureus strains in vitro, with minimum inhibitory concentrations ranging from 4 to 32 μg/ml. At 4 × minimum inhibitory concentration, all compounds killed cells within 24 h, demonstrating bactericidal activity. In addition to their effects against planktonic cells, these compounds prevented biofilm formation in a dose-dependent manner, with inhibitory concentrations for biofilm formation ranging from 8 to 32 μg/ml. Interestingly, higher concentrations of these compounds were effective against mature biofilms and all compounds downregulated the transcription of the biofilm-related gene spa.
Conclusion:
We report three new 1,3,4-oxadiazole derivatives that have bactericidal activity and could provide as alternatives to combat S. aureus.
Keywords: : 1,3,4-oxadiazole derivatives; antibiotic resistance; biofilm; planktonic cells; Staphylococcus aureus
Staphylococcus aureus is involved in various infections, such as sepsis, bacteremia, endocarditis, pneumonia and device-related infections [1,2]. Drug-resistant strains of the bacteria, such as methicillin-resistant S. aureus (MRSA), are associated with over 5400 deaths per year in the USA [3], and drug resistance continues to evolve. The continued emergence of antibiotic resistance, especially against β-lactam antibiotics, such as methicillin, has posed a challenge for treating staphylococcal infections, thus resulting in a need for continued drug discovery and development.
Exacerbating the drug-tolerance problem is the ability of pathogens to form biofilms that protect the bacteria from host defenses and prevent the delivery of some antibiotics [4]. Within biofilms, S. aureus adopts a phenotype that confers intrinsic resistance to antibiotics, resulting in persistent and chronic bacterial infections [5–7]. This biofilm formation is closely correlated with protein A, fibronectin-binding proteins and polysaccharide intercellular adhesion, that are the products of spa, fnb and ica, respectively [8–10]. The regulation of these genes is particularly important for biofilm-mediated infections. Notably, the expression levels of the above genes are regulated by a network of interacting regulators, including the staphylococcal accessory regulator sar [7,8,11,12], that has been confirmed to promote biofilm formation [13,14].
Drug recalcitrance by biofilms and the continued development of resistance and tolerance drive an urgent need to identify new drugs against both planktonic and biofilm MRSA cells. To this end, we investigated the antimicrobial potential of 1,3,4-oxadiazoles, which comprise a class of heterocyclic compounds with a wide range of biological activities, including antiviral, anti-inflammatory, antitubercular, hypoglycemic, antineoplastic, fungicidal and antibacterial properties [15–19]. Moreover, 1,3,4-oxadiazole heterocycles are excellent bioisosteres of amides and esters and can contribute in enhancing biological activity by participating in hydrogen-bonding interactions [20]. Accordingly, compounds featuring the 1,3,4-oxadiazole nucleus have demonstrated antimicrobial activity against S. aureus [21,22], that is attributed in part to the presence of a toxophoric -N=C-O- linkage group, which may react with the nucleophilic centers of the microbial cells [23].
In the course of our search for new antimicrobial agents, we identified three novel compounds featuring 1,3,4-oxadiazole that demonstrated potent antimicrobial activity against S. aureus. In this work, we evaluated the antimicrobial efficacy of these oxadiazole derivatives against S. aureus planktonic cells and biofilms, and evaluated the impact of these compounds on the transcription of biofilm-related genes.
Materials & methods
Bacterial strains & nematodes
The bacterial strains listed in Table 1 were selected from an existing strain collection maintained in our laboratory and grown at 37°C in tryptic soy broth (BD Diagnostics, NJ, USA). The Caenorhabditis elegans glp-4(bn2);sek-1(km4) strain was maintained on a lawn of Escherichia coli strain HB101 on 10 cm plates at 15°C [24].
Table 1. . Antibacterial activities of compounds against staphylococcal strains.
| Strains | Antibacterial activity of compounds (MIC, μg/ml) | |||||
|---|---|---|---|---|---|---|
| Name | Type | OZE-I | OZE-II | OZE-III | Vancomycin | Oxacillin |
| Newman | MSSA | 16 | 16 | 32 | 2 | ≤0.0625 |
| MW2 | MRSA | 8 | 8 | 16 | 4 | 64 |
| ATCC 29213 | MSSA | 8 | 8 | 16 | 2 | 0.5 |
| ATCC 33591 | MRSA | 4 | 4 | 16 | 2 | >64 |
| USA100 | MSSA | 4 | 8 | 16 | 2 | ≤0.0625 |
| USA300 | MRSA | 4 | 4 | 8 | 4 | >64 |
| USA400 | MRSA | 8 | 8 | 16 | 2 | >64 |
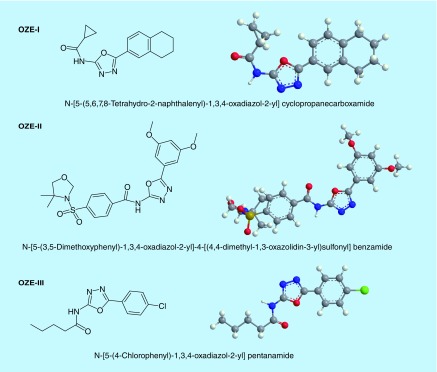
MIC: Minimum inhibitory concentration; MRSA: Methicillin-resistant Staphylococcus aureus; MSSA: Methicillin-susceptible S. aureus; OZE-I: N-(5-(5,6,7,8-tetrahydronaphthalen-2-yl)-1,3,4-oxadiazol-2-yl) cyclopropanecarboxamide; OZE-II: N-(5-(3,5-dimethoxyphenyl)-1,3,4-oxadiazol-2-yl)-4-((4,4-dimethyloxazolidin-3-yl) sulfonyl) benzamide; OZE-III: N-(5-(4-chlorophenyl)-1,3,4-oxadiazol-2-yl) pentanamide.
Caenorhabditis elegans-MRSA liquid infection
Three oxadiazole derivatives obtained from Life Chemicals, Inc. (ON, Canada) were dissolved in DMSO to make working stock solutions. As reported in our earlier study [25,26], the standard 384-well plates with the investigated compounds or 1% DMSO were adopted to detect their antistaphylococcal activity. Briefly, 15 adult glp-4(bn2);sek-1(km4) worms were transferred to each well that was previously seeded by S. aureus MW2. The transfer of nematodes was performed using a COPAS large particle sorter (Union Biometrica, MA, USA). The worms stained with the vital dye Sytox Orange (Life Technologies, CA, USA) were imaged using an Image-Xpress Micro automated microscope (Molecular Devices, CA, USA).
Antimicrobial susceptibility testing
The minimum inhibitory concentrations of tested compounds against S. aureus strains were determined as described in Clinical and Laboratory Standards Institute protocols [27]. Briefly, the S. aureus strains were cultured in Müller–Hinton (MH) broth (Becton Dickinson and Company, NJ, USA). Twofold serial dilutions of compounds (50 μl) were mixed with 50 μl of bacterial culture (1 × 106 colony-forming unit/ml) and incubated at 35°C for 16–20 h in 96-well plates (Corning, NY, USA). The MIC was determined as the lowest concentration of the sample in which no visible bacterial growth was observed after incubation. Subsequently, 10 μl of culture from the MIC assay was plated on Müller–Hinton agar and incubated overnight at 35°C. The lowest concentration at which colonies were not observed was regarded as the minimum bactericidal concentration (MBC).
Hemolysis assay
The hemolytic activity of compounds was determined in triplicate according to previous protocol [28]. Fifty microliters of 2% human erythrocytes (Rockland Immunochemicals, PA, USA) in phosphate-buffered saline (PBS) was added to 50 μl of serially diluted compounds and incubated at 37°C for 1 h. The plate was then centrifuged at 3724 × g for 5 min. Fifty microliters of the supernatant was transferred to a new plate, and the absorbance was measured at 540 nm.
Cytotoxicity assay
Cytotoxicity assays were performed as described by the method of Dzoyem et al. [29] with some modifications. Briefly, HepG2 cells (ATCC HB 8065; ATCC, VA, USA) were cultivated in Dulbecco’s modified eagle medium (DMEM; Gibco, NY, USA) containing 10% fetal bovine serum (Gibco) and 1% penicillin/streptomycin (Gibco) at 37°C in CO2-humidified chamber. HepG2 cells were harvested and resuspended in DMEM and 100 μl of cells were added to each well of 96-well plates (Corning) at a final concentration of 5 × 104 cells. Test compounds were serially diluted in serum and antibiotic-free DMEM and added to the wells and incubated as described earlier for 24 h. For the last 4 h of the 24 h incubation period, 10 μl of 2-(4-iodophenyl)-3-(4-nitrophenyl)-5-(2, 4-disulfophenyl)-2H-tetrazolium (WST-1) solution (Roche, Mannheim, Germany) was added to each well. WST-1 reduction was measured at 450 nm using Vmax microplate reader (Molecular Device). This assay was performed in triplicate, and survival was calculated by comparing to cells treated with the vehicle control (DMSO).
In vitro time-kill studies
The time-kill assay was conducted in cation-adjusted MH broth with logarithmically growing bacterial cultures and compounds at concentrations equivalent to four-times the MIC. At periodic intervals, samples were serially diluted and plated onto tryptic soy agar (BD Biosciences) plates for colony counts as described previously [30]. The experiments were performed in triplicate.
Bacterial cell membrane permeabilization assay
The permeability of bacterial membranes was detected by measuring Sytox Green (Life Technologies) uptake by bacterial cells in 96-well plates, as described in an earlier study [3]. In brief, overnight cultures of bacteria were harvested by centrifugation at 3724 × g for 5 min, and then the pellets were washed twice and suspended in PBS to an absorbance of 0.5 at 600 nm. Sytox Green was added to the cells at a final concentration of 5 μM and incubated in the dark for 30 min. Fifty microliters of serial compound dilutions in PBS were added to 50 μl bacterial suspension. The fluorescence intensity of Sytox Green (excitation wavelength 485 nm; emission wavelength 530 nm) was read every 5 min for 1 h at room temperature. The experiments were performed in triplicate.
Prevention of S. aureus static biofilm formation
Biofilm formation was detected in triplicate in the presence of compounds or DMSO by the crystal violet method with a slight modification [31]. Briefly, in 96-well microtiter plates, 50 μl of compounds in MH broth with 2% glucose were mixed with 50 μl of bacterial suspension (OD600 = 0.03) and incubated at 37°C for 48 h without shaking. After aspirating the planktonic cells, the residual adhesive cells in each well were washed with water for three-times and stained with 125 μl of 0.1% CV solution for 10 min at room temperature. Subsequently, the plates were washed twice with water to remove excess dye and then air dried for 10 min. Two hundred microliters of ethanol (95%) were added to each well and the plates were incubated at room temperature for 20 min. The intensity of CV at 595 nm was measured by the SpectraMax M2Multi-mode Microplate Reader (Molecular Devices).
Viability assays of biofilms cells
As reported previously [31], the plates were filled with 100 μl of 106 colony-forming unit/ml bacterial suspensions in tryptic soy broth containing 1% glucose and incubated at 37°C for 24 h under static conditions. Subsequently, the medium was discarded and the wells were rinsed twice with PBS to remove any unbound cells. After culturing with 100 μl different concentrations of compounds at 37°C for 24 h, the plates were washed to remove the supernatant. Each well was rinsed with 100 μl of PBS containing 25 μg methylthiazoltetrazolium (MTT; Sigma-Aldrich, MO, USA). The formazan crystals, generated from the reduction of MTT by (respiratory) reductases of living staphylococcal cells, were dissolved in DMSO and the absorbance at 570 nm was detected. Another way to measure the viability of staphylococcal cells was based on the viable cell count method of Saising et al. [7].
Real-time Reverse Transcription-Polymerase Chain Reaction
To evaluate the effects of compounds on bacterial gene expression, overnight culture of S. aureus MW2 was grown to OD600 0.5–0.6 in brain heart infusion broth [32]. Then, the cells were exposed to the compounds (at sublethal concentrations) or 1% DMSO for 3 h. RNA was prepared as recommended by the manufacturer (Qiagen, Inc., CA, USA) and its concentration was determined by a nanodrop. Verso cDNA synthesis kit (Thermo Fisher Scientific, MA, USA) was used to reverse-transcribe RNA into cDNA, and the included RT Enhancer in this kit prevented genomic DNA carryover, eliminating the need for separate DNase I treatment. Reverse transcription-polymerase chain reactions (RT-PCR) were performed as recommended by the manufacturer (Bio-Rad, CA, USA) using the primers in Supplementary Table 1 and the iCycler iQ real-time detection system (Bio-Rad). According to the Mitchell et al. [32], the relative expression ratios were calculated as follows: n-fold expression = 2-ΔΔCt, ΔΔCt = ΔCt (drug-treated)/ΔCt (untreated), where ΔCt represents the difference between the cycle threshold (Ct) of the gene studied and the Ct of housekeeping gyrA gene (internal control). Student’s t test was used for analyzing those PCR results.
Statistical analysis
Statistical analysis was performed using analysis of variance. Comparisons between means were carried out according to Student’s t test. Differences were considered significant at a level of p < 0.05.
Results
Identification of novel 1,3,4-oxadiazole derivatives & in vitro evaluation of antibacterial activity against staphylococcal strains
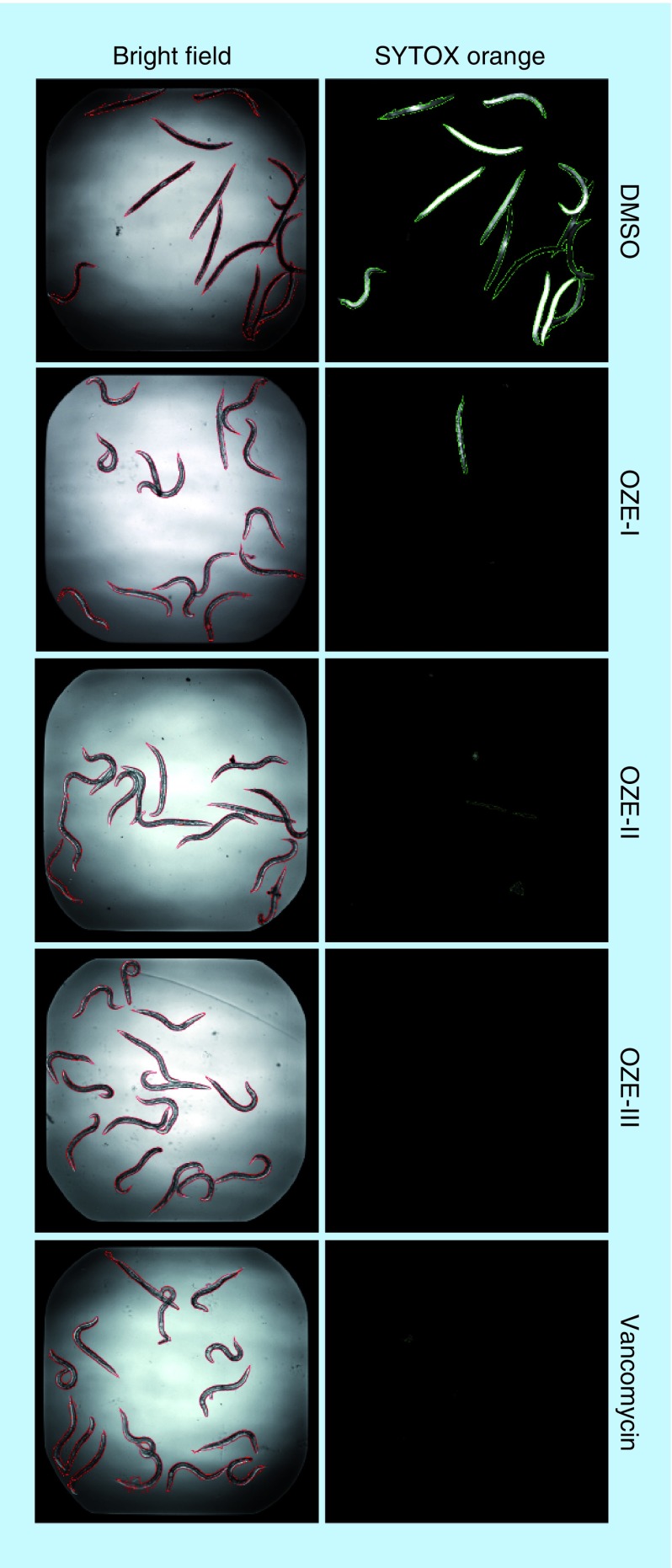
Previous work in our lab established a C. elegans-MRSA whole animal liquid infection model for high-throughput screening of chemical libraries to identify anti-infective compounds. Through our screening efforts, we identified three 1,3,4-oxadiazole derivatives, N-(5-(5,6,7,8-tetrahydronaphthalen-2-yl)-1,3,4-oxadiazol-2-yl) cyclopropanecarboxamide (OZE-I), N-(5-(3,5-dimethoxyphenyl)-1,3,4-oxadiazol-2-yl)-4-((4,4-dimethyloxazolidin-3-yl) sulfonyl) benzamide (OZE-II) and N-(5-(4-chlorophenyl)-1,3,4-oxadiazol-2-yl) pentanamide (OZE-III) (Figure 1). As compared with DMSO-treated worms, OZE-I, OZE-II and OZE-III were highly effective at prolonging the survival of worms infected with MRSA at 7.14 μg/ml (screening concentration) (Figure 2).
Figure 1. . Chemical structure of compounds.
The chemical structures of OZE-I {N-[5-(5,6,7,8-Tetrahydro-2-naphthalenyl)-1,3,4-oxadiazol-2-yl] cyclopropanecarboxamide; C16H17N3O2; MW: 283.32}, OZE-II {N-[5-(3,5-Dimethoxyphenyl)-1,3,4-oxadiazol-2-yl]-4-[(4,4-dimethyl-1,3-oxazolidin-3-yl)sulfonyl] benzamide; C22H24N4O7S; MW: 488.51} and OZE-III {N-[5-(4-Chlorophenyl)-1,3,4-oxadiazol-2-yl] pentanamide; C13H14ClN3O2; MW: 279.72}.
Figure 2. . Bright-field and Sytox Orange-stained images of methicillin-resistant Staphylococcus aureus-infected Caenorhabditis elegans with 1% DMSO, 7.14 μg/ml OZE-I, OZE-II, OZE-III or 10 μg/ml vancomycin.
Image analysis was carried out using CellProfiler; worms identified in the bright field are outlined in red, and dead worms identified in the Sytox Orange fluorescent image are in green.
For color figures please see online at https://www.future-science.com/doi/10.4155/fmc-2017-0159
To confirm the antistaphylococcal activity of the 1,3,4-oxadiazole derivatives identified in the screen, we tested their ability to inhibit bacterial growth using the broth microdilution assay. As shown in Table 1, all three oxadiazole derivatives demonstrated potent antibacterial activity against S. aureus. With MICs of 4–16 μg/ml, OZE-I demonstrated similar activity to OZE-II in inhibiting the growth of all tested strains, except of S. aureus USA100 that was inhibited by OZE-I and -II at the MIC of 4 and 8 μg/ml, respectively. Compared with these two compounds, relative higher MIC values were obtained for OZE-III and they ranged from 8 to 32 μg/ml. The antimicrobial activity of OZE-I, -II and -III was inclusive of inhibitory activity against MRSA strains, such as S. aureus MW2 and USA300. Additionally, the MBCs of OZE-I, OZE-II and OZE-III against S. aureus MW2 were observed at concentrations 16, 16 and 64 μg/ml, respectively, while for S. aureus USA300, the MBC values were 16, 8 and 8 μg/ml, respectively for OZE-I, -II and -III.
Hemolytic activity & cytotoxicity
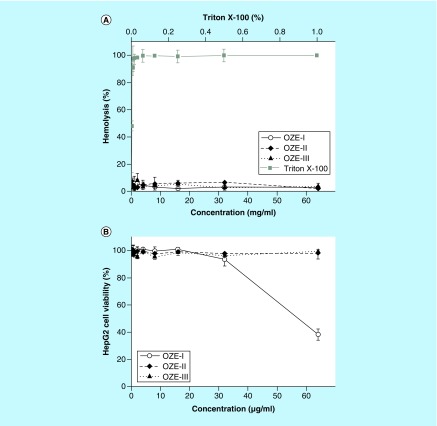
In order to evaluate the toxicity of these compounds, we tested the hemolytic activity of OZE-I, -II and -III using the human erythrocytes lysis assay and their cytotoxicity against human HepG2 cells (Figure 3). Compared with the highly hemolytic triton X-100 (used as positive control), none of these oxadiazole derivatives resulted in noteworthy hemolysis (Figure 3A). Moreover, as depicted in Figure 3B, OZE-II and -III demonstrated no cytotoxicity (LD50 >64 mg/ml), while cells treated with OZE-I exhibited slight toxic effect on HepG2 cells with a LD50 >32 μg/ml.
Figure 3. . Hemolytic activity and cytotoxicity of OZE-I, -II and OZE-III.
(A) Human erythrocytes were treated with serial dilutions of Triton X-100 (0.002–001%) or compounds (0.125–164 μg/ml). (B) Cytotoxicity of compounds was evaluated using HepG2 cells. The viability of HepG2 cells was measured after treatment with serially diluted concentrations (0.125–164 μg/ml) of each compound.
Time-killing studies
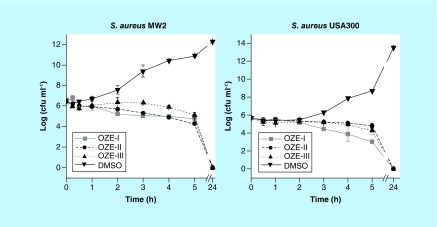
The antimicrobial activity of the oxadiazole derivatives on planktonic cells of S. aureus MW2 and USA300 were further measured in experiments assessing the rate of killing by using concentrations at 4 × MIC. As depicted in Figure 4, all three compounds inhibited the growth of S. aureus strains MW2 and USA300. OZE-I, -II and -III showed gradual killing, as demonstrated by decreasing cell counts of S. aureus MW2 after 5 h by 1.7 log10 (p < 0.001), 2.2 log10 (p < 0.001) and 1.4 log10 (p = 0.0018), respectively (Figure 4A). In the case of S. aureus USA300, OZE-I exhibited a remarkable viable bacterial reduction with a significant decrease in bacterial killing of 2.7 log10 after a period of 5 h (p = 0.0024, Figure 4B), while OZE-II and -III had limited activity against this strain. It is particularly interesting, however, that no colonies were detected in any of the three investigational compounds after 24 h interaction with S. aureus MW2 or USA300, indicating all the three oxadiazole derivatives were lethal to S. aureus after prolonged exposure.
Figure 4. . Growth curve of Staphylococcus aureus MW2 and USA300 in the presence of OZE-I, OZE-II and OZE-III at the concentrations of 4× minimum inhibitory concentration.
1,3,4-oxadiazole derivatives inhibit biofilm formation
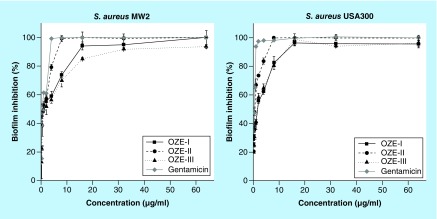
To determine if our investigational compounds prevent biofilm formation, we used a quantitative microtiter plate method [33]. The bacteria were cultured in microtiter plates for 24 h in the presence of the three individual compounds (0.125–164 μg/ml). Subsequently, the formed biofilms were stained with crystal violet. As shown in Figure 5, the compounds effectively inhibited biofilm formation in a dose-dependent manner. The concentrations of OZE-I and -III required to prevent biofilm formation of S. aureus USA300 were 16 μg/ml, whereas OZE-II was more effective and required a concentration of 8 μg/ml. These results indicated that these three oxadiazole derivatives exert inhibitory effects on biofilm formation.
Figure 5. . Biofilm formation of Staphylococcus aureus MW2 and USA300 in the presence of OZE-I, -II and -III.
Growth and biofilm formation occurred in microtiter plates. Shown are the turbidity (OD600) of the cell culture (A) and the inhibition rate of biofilm formation (B).
Effect of compounds on mature biofilms
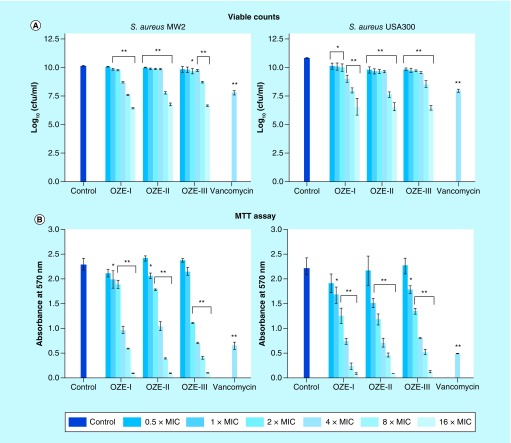
In addition to the staining assay above, we studied the effect of oxadiazole derivatives on cell viability in 24 h mature biofilm, which is extremely robust against antibiotics. Serial concentrations of compounds were added to 1-day-old biofilm, and the mixtures were incubated for 24 h at 37°C. As described in Figure 6A, OZE-I, -II and -III caused dose-dependent reduction of biofilm. Among them, only OZE-II was able to result in significant reduction of biofilms at all tested doses (0.5–16 × MIC), as compared with the control (p < 0.01). The minimal dose of OZE-I that causes a significant biofilm loss of S. aureus MW2 was 1 × MIC, while OZE-III did not exhibit notable ability until 2 × MIC. Remarkably, all compounds performed well against mature biofilms of S. aureus USA300, even at the low concentrations of 0.5 × MIC.
Figure 6. . Effects of OZE-I, -II and -III on 24-h mature biofilms of Staphylococcus aureus MW2 and USA300.
The viability of bacterial cells in 24-h biofilms was detected by viable count or methylthiazoltetrazolium assay. Error bars represent standard deviation.
*p < 0.05 and **p < 0.01, compared with the results of the control. Control, strain treated with 1% DMSO.
MIC: Minimum inhibitory concentration.
To confirm the above results, we also utilized an indirect MTT method to test cell viability. The MTT assay is based on the activity of MTT-reducing enzymes (frequently NADH-dependent dehydrogenases) as an indication of live cells. As expected, increasing concentrations (1 × to 16 × MIC) of all three compounds led to a decrease in formazan production. At all tested concentrations >0.5 × MIC, OZE-1, -II and -III resulted in a dose-dependent decrease, which is consistent with the viable count method results and the formazan production in the presence of subinhibitory concentrations of our investigational compounds was comparable to that of the control cultures without compounds (Figure 6B).
Influence on transcription of S. aureus sarA, icaA, spa, fnbA & fnbB
To evaluate the effects of OZE-I, -II and -III on the integrity of the bacterial cell membrane, bacteria were exposed to compounds at different concentrations (1–64 μg/ml) and the uptake of Sytox Green over a period of 60 min was measured. Cells treated with all the investigated compounds showed no uptake of the dye even at a high concentration of 64 μg/ml (Supplementary Figure 1), indicating that activity of OZE-I, -II and -III was not associated with disruption of the physical integrity of the cell membrane.
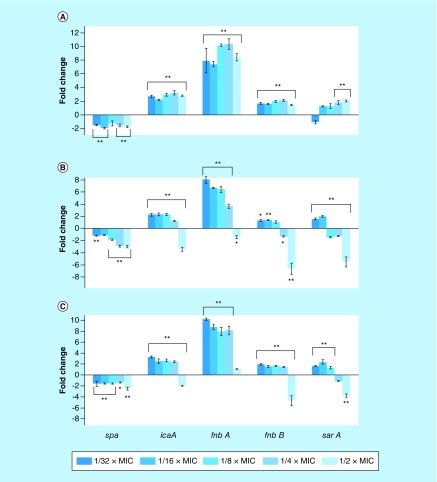
Next, we set to investigate the hypothesis if, similar to other antistaphylococcal compounds, these investigational compounds could alter the transcription of sarA, icaA, spa, fnbA and fnbB. Therefore, we detected the transcription levels of sarA, icaA, spa, fnbA and fnbB by using real-time RT-PCR. As expected, our results showed that OZE-I, -II and -III exerted significant and dose-dependent effects in the transcription levels of these genes (Figure 7). More specifically, the transcriptional levels of spa were significantly decreased by 1.75-, 2.96- and 2.43-fold (p < 0.001), when exposed with 1/2 × MIC of OZE-I, -II and -III, respectively. Interestingly, the transcription levels of icaA, fnbA and fnbB were markedly upregulated after exposure to all subinhibitory concentrations of OZE-I (p < 0.001). The expression of gene regulator sarA was increased along with the increasing concentrations of OZE-I, whereas the increasing sub-MICs of OZE-II and -III led to gradual reduction of sarA, icaA, fnbA and fnbB. Particularly, 1/2 × MIC of OZE-II and -III had the strongest capacity to suppress the tested genes. For instance, OZE-II and -III at 1/2 × MIC downregulated sarA transcription by, respectively, 5.5- and 3.75-fold (p < 0.001); the levels of transcription of fnbB were significantly decreased by 6.6- and 4.7-fold (p < 0.001), respectively, following the addition of 1/2 × MIC of OZE-II and -III.
Figure 7. . Relative expression levels of spa, icaA, fnbA, fnbB and sarA in Staphylococcus aureus MW2 after culture with subinhibitory concentrations of OZE-I, -II and -III.
The bar graph is a mean of three independent experiments. Error bars represent standard deviation.
*p < 0.05 and **p < 0.01, compared with the results of the corresponding control. Control, strain treated with 1% DMSO.
MIC: Minimum inhibitory concentration.
Discussion
Staphylococcus aureus is associated with numerous acute and chronic infections and these infections remain problematic to treat, partly due to the emergence of multiresistant strains. An alternative strategy for overcoming antibiotic resistance and treating S. aureus infections is to explore innovative heterocyclic agents with a novel mode of action. In this context, oxadiazole and its derivatives, have been playing a crucial role in these drug discovery efforts. The introduction of 1,3,4-oxadiazole ring to the inhibitors can change their polarity, flexibility, as well as metabolic stability [20,34]. As the acceptor of hydrogen bonds formation, meanwhile, the 1,3,4-oxadiazole scaffold has the potential to be an isosteric substituent for amide or ester groups, so as to interfere protein or lipid biosynthesis in pathogens. Accordingly, we identified and characterized the antimicrobial properties of three novel 1,3,4-oxadiazole derivatives against S. aureus.
When developing new antimicrobial agents, an initial and crucial aspect to take into consideration is their safety. All OZE agents were able to prolong survival of C. elegans infected with MRSA strain MW2, underlying the nonlethal properties of all investigated compounds. To gain further insight into their safety, we tested the toxicity of three compounds using mammalian cells. Compared with triton X-100, a known hemolytic agent, OZE-I, -II and -III were well tolerated by human erythrocytes and did not show hemolytic activity even at the maximum tested concentration of 64 μg/ml. As expected, OZE-II and -III had no detectable toxicity, whereas OZE-I was found to be toxic at the maximum tested concentration of 64 μg/ml. Considering its nontoxicity within the range of MICs, OZE-I was retained for use in the succeeding experiments.
OZE-I, -II and -III were not only effective in prolonging the survival of MRSA-infected C. elegans, but also exhibited antimicrobial activity against S. aureus strains in vitro. To our knowledge, the antimicrobial activity of OZE-I, -II and -III had not been elucidated, but studies with other types of 1,3,4-oxadiazole derivatives have shown that the MIC values against S. aureus were ≥62.5 μg/ml [22,35]. Our study demonstrates the antibacterial activities of OZE-I, -II and -III, building upon this family of compounds. Additionally, the MBC values revealed that all the tested compounds were bactericidal, which offers several distinct advantages over bacteriostatic agents [36,37]. Time-killing curves further pointed out that OZE-I, -II and -III at 4 × MIC were slowly bactericidal against S. aureus MW2 and USA300, demonstrating around 2 log10 decrease in the viable cell counts within 5 h and no colonies within 24 h.
While many compounds are effective against planktonic cells, this does not necessarily represent that state of cells during an infection. During the infection process, S. aureus and other bacteria, including S. aureus, establish biofilms in the infected tissues. To exploit the antibiofilm potentials of OZE agents, we detected two main phases of biofilms in this work: early biofilm formation and mature biofilms. We discovered that OZE-I, -II and -III were able to prevent biofilm formation and kill some cells in mature biofilms. The cell counts of mature biofilm declined significantly, but some colonies were still detected after exposure to the three investigational compounds for 24 h. Accordingly, OZE-I, -II and -III were more effective in killing planktonic cells and preventing biofilm formation, than in eradicating the robust and protected cells in the established biofilms. Similarly, Saising et al. [7] found gallidermin performed excellent on inhibiting biofilm formation, but its significant effect on old biofilms of S. aureus was only seen at 4 × MIC. From this respect, all three OZE agents behaved like many other antibiotics that displayed good abilities against planktonic cells and biofilm inhibition but comparatively low activity on mature biofilms.
As OZE agents could decrease the planktonic cells and biofilms, we hypothesized that OZE-I, -II and -III could affect the transcription of biofilm-related genes, such as sarA, icaA, spa, fnbA and fnbB. SarA, a major global regulator of a diverse range of virulence determinants in S. aureus, is essential for biofilm formation in S. aureus [7,38,39]; icaA encodes polysaccharide intercellular adhesin or polymeric N-acetyl-glucosamine [39]; spa (encoding the surface protein A) is able to induce cell aggregation and biofilm formation [9]; fnbA and fnbB encode fibronectin-binding proteins FnbA and FnbB, which promote biofilm accumulation [40]. We found that spa was consistently downregulated upon treatment with all three investigational compounds at different sub-MICs. As an essential component of the biofilm, staphylococcal protein A (encoded by spa) is known for its ability to bind to IgG and von Willebrand factor [4,41,42]. Moreover, Merino et al. [9] reported that deletion of spa significantly reduced the capacity of S. aureus to colonize subcutaneously implanted catheters. In this respect, we reason that the influence of subinhibitory concentrations of OZE-I, -II and -III on the biofilm development may, at least partially, depend on inhibition of spa. In addition to the significant downregulation of spa, the transcription levels of protein-encoded genes icaA, fnbA and fnbB were decreased by OZE-II and -III in a dose-dependent manner.
Thus, based on the findings discussed in the previous paragraph, subinhibitory concentrations of antimicrobial agents may affect gene regulators by interfering with the translation of regulatory gene products [43–45]. Reportedly, the expression levels of fnbA and fnbB could be positively controlled and regulated by sarA [11] and sarA may act as a master regulator controlling the transcription of icaA, fnbA and fnbB, thereby affecting the biosynthesis of polysaccharide intercellular adhesin/polymeric N-acetyl-glucosamine and mediating fibronectin-binding proteins [10,46]. In agreement with this point, we observed that the global regulator sarA was impacted by OZE-II and -III in a dose-dependent manner. Published studies also demonstrated that down regulation of sarA might contribute to limit biofilm formation [13,39,47]. It is reasonable to assume that the influence of subinhibitory concentrations of OZE-II and -III on the biofilm development may depend on their induced inhibition of the global regulator sar. However, future studies should also include the study of biofilm cells.
Conclusion
We report three novel 1,3,4-oxadiazole derivatives that prolonged the survival of MRSA-infected C. elegans and exhibited in vitro antimicrobial and bactericidal activity against S. aureus. In addition to their efficacy against planktonic cells, OZE-I, -II and OZE-III inhibited biofilm formation and killed cells on established MRSA biofilms. This effect on biofilms appears to be mediated, at least in part, by the down regulation of genes sarA, icaA, spa, fnbA and fnbB. These compounds, with little hemolytic activity and low toxicity against mammalian cells, should be evaluated further as alternatives to combat biofilm and drug-resistant S. aureus strains.
Future perspective
We report three novel 1,3,4-oxadiazole derivatives OZE-I, -II and -III that were effective against planktonic cells and biofilms, providing promising candidates for treating staphylococcal infections. In vivo antistaphylococcal activities of these three compounds should be conducted, followed by evaluating their efficacy against the virulence factors.
Summary points.
Three 1,3,4-oxadiazole derivatives, N-(5-(5,6,7,8-tetrahydronaphthalen-2-yl)-1,3,4-oxadiazol-2-yl) cyclopropanecarboxamide (OZE-I), N-(5-(3,5-dimethoxyphenyl)-1,3,4-oxadiazol-2-yl)-4-((4,4-dimethyloxazolidin-3-yl) sulfonyl) benzamide (OZE-II) and N-(5-(4-chlorophenyl)-1,3,4-oxadiazol-2-yl) pentanamide (OZE-III) identified from Caenorhabditis elegans-methicillin-resistant Staphylococcus aureus infection model exhibited antimicrobial activity against seven S. aureus strains in vitro and prolonged nematode survival.
OZE-I and OZE-II showed the minimum inhibitory concentration (MIC) values against seven tested isolates ranging from 4 to 16 μg/ml, while the MIC to OZE-III exhibited an MIC range of 8 to 32 μg/ml.
All three compounds demonstrated no hemolytic activity and cytotoxicity, except OZE-I, while exhibited slight toxic effect on HepG2 cells with a LD50 >32 mg/ml.
During the time-killing course, all three oxadiazole derivatives could kill S. aureus strains MW2 and USA300 within 24 h at 4 × MIC.
In addition to inhibiting the planktonic cells of S. aureus, the three investigational compounds were effective at inhibiting biofilm formation and reducing mature biofilms in a dose-dependent manner.
All 1,3,4-oxadiazole derivatives at sub-MICs inhibited the transcription of the biofilm-related gene spa.
OZE-I, OZE-II and OZE-III could be the promising antistaphylococcal candidates against both planktonic cells and biofilms.
Supplementary Material
Footnotes
Supplementary data
To view the supplementary data that accompany this paper please visit the journal website at: https://www.future-science.com/doi/suppl/10.4155/fmc-2017-0159
Financial & competing interests disclosure
This study was supported by NIH grant P01 AI083214 to E Mylonakis. Z Zheng was supported by the China Scholarship Council through Chinese Government Graduate Student Overseas Study Program. Q Liu was supported by Shanghai General Hospital Characteristic Discipline Construction Fund and Shanghai Jiao Tong University K C Wong Medical Fellowship Fund. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Tong SY, Davis JS, Eichenberger E, Holland TL, Fowler VG. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015;28(3):603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benfield T, Espersen F, Frimodt-Møller N, et al. Increasing incidence but decreasing in-hospital mortality of adult Staphylococcus aureus bacteraemia between 1981 and 2000. Clin. Microbiol. Infect. 2007;13(3):257–263. doi: 10.1111/j.1469-0691.2006.01589.x. [DOI] [PubMed] [Google Scholar]
- 3.Rajamuthiah R, Jayamani E, Conery AL, et al. A defensin from the model beetle Tribolium castaneum acts synergistically with Telavancin and Daptomycin against multidrug resistant Staphylococcus aureus . PloS ONE. 2015;10(6):e0128576. doi: 10.1371/journal.pone.0128576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beenken KE, Dunman PM, McAleese F, et al. Global gene expression in Staphylococcus aureus biofilms. J. Bacteriol. 2004;186(14):4665–4684. doi: 10.1128/JB.186.14.4665-4684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stewart PS. Mechanisms of antibiotic resistance in bacterial biofilms. Int. J. Med. Microbiol. 2002;292(2):107–113. doi: 10.1078/1438-4221-00196. [DOI] [PubMed] [Google Scholar]
- 6.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284(5418):1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 7.Saising J, Dube L, Ziebandt AK, Voravuthikunchai SP, Nega M, Gotz F. Activity of gallidermin on Staphylococcus aureus and Staphylococcus epidermidis biofilms. Antimicrob. Agents Chemother. 2012;56(11):5804–5810. doi: 10.1128/AAC.01296-12. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Demonstrates that gallidermin prevents biofilm formation of Staphylococcus aureus and Staphylococcus epidermidis, but shows limited activity on cells within mature biofilms.
- 8.O’Neill E, Pozzi C, Houston P, et al. A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J. Bacteriol. 2008;190(11):3835–3850. doi: 10.1128/JB.00167-08. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Reports a novel methicillin-resistant S. aureus biofilm phenotype promoted by the fibronectin-binding proteins, FnBPA and FnBPB.
- 9.Merino N, Toledo-Arana A, Vergara-Irigaray M, et al. Protein A-mediated multicellular behavior in Staphylococcus aureus . J. Bacteriol. 2009;191(3):832–843. doi: 10.1128/JB.01222-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cramton SE, Gerke C, Schnell NF, Nichols WW, Götz F. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infection Immunity. 1999;67(10):5427–5433. doi: 10.1128/iai.67.10.5427-5433.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chien Y-T, Manna AC, Projan SJ, Cheung AL. SarA, a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for sar-dependent gene regulation. J. Biol. Chem. 1999;274(52):37169–37176. doi: 10.1074/jbc.274.52.37169. [DOI] [PubMed] [Google Scholar]
- 12.Mack D, Becker P, Chatterjee I, et al. Mechanisms of biofilm formation in Staphylococcus epidermidis and Staphylococcus aureus: functional molecules, regulatory circuits, and adaptive responses. Int. J. Med. Microbiol. 2004;294(2):203–212. doi: 10.1016/j.ijmm.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 13.Beenken KE, Blevins JS, Smeltzer MS. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infection Immunity. 2003;71(7):4206–4211. doi: 10.1128/IAI.71.7.4206-4211.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valle J, Toledo-Arana A, Berasain C, et al. SarA and not σB is essential for biofilm development by Staphylococcus aureus . Mol. Microbiol. 2003;48(4):1075–1087. doi: 10.1046/j.1365-2958.2003.03493.x. [DOI] [PubMed] [Google Scholar]
- 15.Desai N, Bhatt N, Somani H, Trivedi A. Synthesis, antimicrobial and cytotoxic activities of some novel thiazole clubbed 1, 3, 4-oxadiazoles. Eur. J. Med. Chem. 2013;67:54–59. doi: 10.1016/j.ejmech.2013.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Indicates that the novel thiazole clubbed 1, 3, 4-oxadiazoles with low cytotoxicity showed their antibacterial activities.
- 16.Barbuceanu S-F, Bancescu G, Cretu OD, Draghici C, Bancescu A, Radu-Popescu M. New heterocyclic compounds from 1, 3, 4-thiadiazole, 1, 3, 4-oxadiazole and 1, 2, 4-triazole class with potential antibacterial activity. Revista de Chimie (Bucharest), 2010;61(2):140–145. [Google Scholar]
- 17.Kumar GS, Rajendraprasad Y, Mallikarjuna B, Chandrashekar S, Kistayya C. Synthesis of some novel 2-substituted-5-[isopropylthiazole] clubbed 1, 2, 4-triazole and 1, 3, 4-oxadiazoles as potential antimicrobial and antitubercular agents. Eur. J. Med. Chem. 2010;45(5):2063–2074. doi: 10.1016/j.ejmech.2010.01.045. [DOI] [PubMed] [Google Scholar]
- 18.Mishra L, Said MK, Itokawa H, Takeya K. Antitumor and antimicrobial activities of Fe (II)/Fe (III) complexes derived from some heterocyclic compounds. Bioorganic Med. Chem. 1995;3(9):1241–1245. doi: 10.1016/0968-0896(95)00095-x. [DOI] [PubMed] [Google Scholar]
- 19.Tao J, Cao LH, Wang CF, Wang DZ. Synthesis of 1, 3, 4-oxadiazoles and 1, 3-thiazolidinones containing 1, 4, 5, 6-tetrahydro-6-pyridazinone. J. Chinese Chem. Soc. 2006;53(5):1193–1197. [Google Scholar]
- 20.Guimarães CRW, Boger DL, Jorgensen WL. Elucidation of fatty acid amide hydrolase inhibition by potent α-ketoheterocycle derivatives from Monte Carlo simulations. J. Am. Chem. Soc. 2005;127(49):17377–17384. doi: 10.1021/ja055438j. [DOI] [PubMed] [Google Scholar]
- 21.Adhikari V, Badiger V. Synthesis and biological activities of isoxazolo (5, 4-d) pyrimidinyloxymethyl-thiadiazoles,-oxadiazoles and-triazoles. ChemInform. 1988;19(47) [Google Scholar]
- 22.Şahin G, Palaska E, Ekizoğlu M, Özalp M. Synthesis and antimicrobial activity of some 1,3,4-oxadiazole derivatives. Il Farmaco. 2002;57(7):539–542. doi: 10.1016/s0014-827x(02)01245-4. [DOI] [PubMed] [Google Scholar]
- 23.Rigo B, Couturier D. Studies on pyrrolidinones. Synthesis of 5-(5-oxo-2-pyrrolidinyl)-1, 3, 5-oxadiazole-2-thione derivatives. J. Heterocyclic Chem. 1985;22(2):287–288. [Google Scholar]
- 24.Muhammed M, Coleman JJ, Mylonakis E. Caenorhabditis elegans: a nematode infection model for pathogenic fungi. Methods Mol. Biol. 2012;845:447–454. doi: 10.1007/978-1-61779-539-8_31. [DOI] [PubMed] [Google Scholar]
- 25.Rajamuthiah R, Fuchs BB, Jayamani E, et al. Whole animal automated platform for drug discovery against multi-drug resistant Staphylococcus aureus . PloS ONE. 2014;9(2):e89189. doi: 10.1371/journal.pone.0089189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim W, Conery AL, Rajamuthiah R, Fuchs BB, Ausubel FM, Mylonakis E. Identification of an antimicrobial agent effective against methicillin-resistant Staphylococcus aureus persisters using a fluorescence-based screening strategy. PLoS ONE. 2015;10(6):e0127640. doi: 10.1371/journal.pone.0127640. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Provides a fluorescence-based strategy for high-throughput screening of antimicrobial agents against methicillin-resistant S. aureus.
- 27.Wayne P. Clinical and Laboratory Standards Institute: performance standards for antimicrobial susceptibility testing: twenty-fourth informational supplement, M100-S24. Clin. Lab. Stand. Inst. 2014;34(1):1–219. [Google Scholar]
- 28.Rosch JW, Boyd AR, Hinojosa E, et al. Statins protect against fulminant pneumococcal infection and cytolysin toxicity in a mouse model of sickle cell disease. J. Clin. Invest. 2010;120(2):627–635. doi: 10.1172/JCI39843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dzoyem JP, Nkuete AH, Kuete V, et al. Cytotoxicity and antimicrobial activity of the methanol extract and compounds from Polygonum limbatum . Planta Medica. 2012;78(08):787–792. doi: 10.1055/s-0031-1298431. [DOI] [PubMed] [Google Scholar]
- 30.Rajamuthiah R, Fuchs BB, Conery AL, et al. Repurposing salicylanilide anthelmintic drugs to combat drug resistant Staphylococcus aureus . PLoS ONE. 2015;10(4):e0124595. doi: 10.1371/journal.pone.0124595. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Shows that the salicylanilide anthelmintic drugs, niclosamide and oxyclozanide, are suitable candidates for treatment of staphylococcal infections.
- 31.Christensen GD, Simpson W, Younger J, et al. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 1985;22(6):996–1006. doi: 10.1128/jcm.22.6.996-1006.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitchell G, Lafrance M, Boulanger S, et al. Tomatidine acts in synergy with aminoglycoside antibiotics against multiresistant Staphylococcus aureus and prevents virulence gene expression. J. Antimicrob. Chemother. 2012;67(3):559–568. doi: 10.1093/jac/dkr510. [DOI] [PubMed] [Google Scholar]; •• Characterizes the antibacterial activity of tomatidine against S. aureus and examined the influence of tomatidine on the expression of virulence factors in S. aureus.
- 33.Berditsch M, Jager T, Strempel N, Schwartz T, Overhage J, Ulrich AS. Synergistic effect of membrane-active peptides polymyxin B and gramicidin S on multidrug-resistant strains and biofilms of Pseudomonas aeruginosa . Antimicrob. Agents Chemother. 2015;59(9):5288–5296. doi: 10.1128/AAC.00682-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rane RA, Gutte SD, Sahu NU. Synthesis and evaluation of novel 1, 3, 4-oxadiazole derivatives of marine bromopyrrole alkaloids as antimicrobial agent. Bioorgan. Med. Chem. Lett. 2012;22(20):6429–6432. doi: 10.1016/j.bmcl.2012.08.061. [DOI] [PubMed] [Google Scholar]
- 35.Joshi SD, Vagdevi HM, Vaidya VP, Gadaginamath GS. Synthesis of new 4-pyrrol-1-yl benzoic acid hydrazide analogs and some derived oxadiazole, triazole and pyrrole ring systems: a novel class of potential antibacterial and antitubercular agents. Eur. J. Med. Chem. 2008;43(9):1989–1996. doi: 10.1016/j.ejmech.2007.11.016. [DOI] [PubMed] [Google Scholar]
- 36.French G. Bactericidal agents in the treatment of MRSA infections – the potential role of daptomycin. J. Antimicrob. Chemother. 2006;58(6):1107–1117. doi: 10.1093/jac/dkl393. [DOI] [PubMed] [Google Scholar]
- 37.Mohammad H, Reddy PN, Monteleone D, et al. Antibacterial characterization of novel synthetic thiazole compounds against methicillin-resistant Staphylococcus pseudintermedius . PLoS ONE. 2015;10(6):e0130385. doi: 10.1371/journal.pone.0130385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheung AL, Zhang G. Global regulation of virulence determinants in Staphylococcus aureus by the SarA protein family. Front. Biosci. 2002;7:d1825–1842. doi: 10.2741/A882. [DOI] [PubMed] [Google Scholar]
- 39.O’Gara JP. Ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus . Fems. Microbiol. Lett. 2007;270(2):179–188. doi: 10.1111/j.1574-6968.2007.00688.x. [DOI] [PubMed] [Google Scholar]
- 40.O’Neill E, Humphreys H, O’Gara JP. Carriage of both the fnbA and fnbB genes and growth at 37 C promote FnBP-mediated biofilm development in meticillin-resistant Staphylococcus aureus clinical isolates. J. Med. Microbiol. 2009;58(4):399–402. doi: 10.1099/jmm.0.005504-0. [DOI] [PubMed] [Google Scholar]
- 41.Hartleib J, Köhler N, Dickinson RB, et al. Protein A is the von Willebrand factor binding protein on Staphylococcus aureus . Blood. 2000;96(6):2149–2156. [PubMed] [Google Scholar]
- 42.Jansson B, Uhlén M, Nygren P-Å. All individual domains of staphylococcal protein A show Fab binding. FEMS Immunol. Med. Microbiol. 1998;20(1):69–78. doi: 10.1111/j.1574-695X.1998.tb01112.x. [DOI] [PubMed] [Google Scholar]
- 43.Smith K, Gould KA, Ramage G, Gemmell CG, Hinds J, Lang S. Influence of tigecycline on expression of virulence factors in biofilm-associated cells of methicillin-resistant Staphylococcus aureus . Antimicrob. Agents Chemother. 2010;54(1):380–387. doi: 10.1128/AAC.00155-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herbert S, Barry P, Novick RP. Subinhibitory clindamycin differentially inhibits transcription of exoprotein genes in Staphylococcus aureus . Infect. Immun. 2001;69(5):2996–3003. doi: 10.1128/IAI.69.5.2996-3003.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stevens DL, Ma Y, Salmi DB, McIndoo E, Wallace RJ, Bryant AE. Impact of antibiotics on expression of virulence-associated exotoxin genes in methicillin-sensitive and methicillin-resistant Staphylococcus aureus . J. Infect. Dis. 2007;195(2):202–211. doi: 10.1086/510396. [DOI] [PubMed] [Google Scholar]
- 46.Ma Y, Xu Y, Yestrepsky BD, et al. Novel inhibitors of Staphylococcus aureus virulence gene expression and biofilm formation. PLoS ONE. 2012;7(10):e47255. doi: 10.1371/journal.pone.0047255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shenkman B, Rubinstein E, Cheung AL, et al. Adherence properties of Staphylococcus aureus under static and flow conditions: roles of agrand sar loci, platelets, and plasma ligands. Infect. Immun. 2001;69(7):4473–4478. doi: 10.1128/IAI.69.7.4473-4478.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.