Abstract
Aim:
To develop a SULT1A1 multiplex ligation-dependent probe amplification assay and to investigate multi-ethnic copy number variant frequencies.
Methods:
A novel multiplex ligation-dependent probe amplification assay was developed and tested on 472 African–American, Asian, Caucasian, Hispanic and Ashkenazi Jewish individuals.
Results:
The frequencies of atypical total copy number (i.e., greater or less than two) were 38.7% for Hispanics, 38.9% for Ashkenazi Jewish, 43.2% for Caucasians, 53.6% for Asians and 64.1% for African–Americans. Heterozygous SULT1A1 deletion carriers (slow sulfators) were most common among Caucasians (8.4%), whereas African–Americans had the highest frequencies of three or more copies (rapid sulfators; 60.9%).
Conclusion:
Different ethnic and racial populations have varying degrees of SULT1A1-mediated sulfation activity, which warrants further research and that may have utility for drug response prediction among SULT1A1-metabolized medications.
Keywords: : copy number variation, multiplex ligation-dependent probe amplification (MLPA), pharmacogenetics, pharmacogenomics, SULT1A1
The detection of human genetic variation has evolved from karyotyping large chromosomal rearrangements to sequencing exomes and genomes to identify single nucleotide variants (SNVs) [1]. The ability to detect common SNVs has enabled association studies on genetic susceptibility to human diseases and other phenotypes; however, copy number variants (CNVs) have increasingly been implicated in many phenotypes and human adaptation to environmental variables based on their structural alteration of gene dosage [2,3]. It is estimated that up to 60% of the human genome contains structural variants, which typically range in size from 100 to 50 kb [4]. Structural variation also occurs among the polymorphic genes that encode drug metabolizing enzymes, including the cytochrome P450s (CYP2B6, CYP2D6), glutathione S-transferases (GSTT1, GSTM1) and sulfotransferases (SULT1A1, SULT2A1) [5–8].
Sulfation is an important step in metabolizing xenobiotics, drugs and hormones, which is carried out by members of the SULT superfamily. The homologous SULT1A subfamily includes SULT1A1, SULT1A2, SULT1A3 and SULT1A4, yet interrogating these genes is challenging due to SULT1A1 being 96% homologous with SULT1A2 and 93% with SULT1A3 and SULT1A4 (which are identical) [9]. SULT1A1 is involved in the metabolism and detoxification of carcinogens, steroid hormones (e.g., estrogen) and other common medications (e.g., tamoxifen) [10]; however, SULT1A1 activity is known to vary significantly between individuals [11,12]. This variability is partly explained by SULT1A1 promoter SNVs [13], as well as structural variation at chromosome 16p11.2 that encompasses the major SULT1A1 transcript (NM_001055.3) [5,10]. The major SULT1A1 transcript has eight exons (seven coding) and encodes a 295 amino acid protein expressed in many human tissues, whereas the minor SULT1A1 transcript has two alternatively spliced upstream noncoding exons and encodes a 219 amino acid protein that is largely expressed in brain tissues [14,15]. Recurrent SULT1A1 CNVs are highly correlated with enzyme activity, which translates to individuals with copy number losses (less than two copies) or gains (greater than two copies) being classified as ‘slow sulfators’ or ‘rapid sulfators’ respectively [10].
The frequencies of pharmacogenetic CNV alleles vary by ancestry, ethnicity and geography [7]; however, detecting these structural variants is challenging due to high sequence homology within many pharmacogene families. Using quantitative PCR (qPCR) for exons 2 and 3 and intron 2 of SULT1A1, previous studies have identified frequencies of one copy ranging from 0 to 20% in selected populations, and three copies or more ranging from 25 to 63% [6,10]. We aimed to extend these previous studies by investigating the frequencies of SULT1A1 CNV alleles using a novel multiplex ligation-dependent probe amplification (MLPA) assay across a large multi-ethnic cohort of almost 500 unrelated individuals from the greater New York metropolitan area.
Materials & methods
Study population
Peripheral blood samples from healthy donors who indicated their racial/ethnic background and gave informed consent for the use of their DNA for research were obtained from the New York Blood Center as previously described [7,16,17]. In addition, blood samples were obtained with informed consent from unrelated, healthy 100% Ashkenazi Jewish (AJ) individuals from the greater New York metropolitan area as previously described [7,17–20]. All personal identifiers were removed, and isolated DNA samples were tested anonymously. Genomic DNA was isolated using the Puregene® DNA Purification kit (Qiagen, CA, USA) according to the manufacturer's instructions. For the current study, five different populations were subjected to SULT1A1 copy number interrogation: African–American (n = 97), Asian (n = 100), Caucasian (n = 102), Hispanic (n = 98) and AJ (n = 99).
Copy number analysis by MLPA
MLPA was performed using the SULT1A1 MLPA kit (P344-B1; MRC-Holland, Amsterdam, The Netherlands) according to the manufacturer's instructions. In brief, all DNA samples were diluted with TE buffer and denatured for 5 min at 98°C. After cooling to 25°C, probemix and MLPA buffer were added to each sample, mixed and incubated for 1 min at 95°C followed by 16-h hybridization at 60°C. The ligation reaction was performed at 54°C by adding 32 μl of ligase-65 mix followed by heating for 5 min at 98°C. PCR buffer, water and MLPA ligation reactions were maintained in a thermocycler at 60°C while polymerase mix was added to each tube. Exon-specific probes with universal tagged primers underwent PCR, which consisted of 35 amplification cycles (95°C for 30 s, 60°C for 30 s and 72°C for 60 s), followed by a 20 min incubation at 72°C. Amplified products were separated by capillary gel electrophoresis on an ABI 3730XL DNA Analyzer (Thermo Fisher Scientific, CA, USA) and analyzed using Coffalyser software (MRC-Holland). After quality control and data normalization to autosome reference probes, copy number was determined according to the following peak ratio ranges: zero copies: 0 <0.15; one copy: 0.40 <0.75; two copies: 0.85 <1.15; three copies: 1.25 <1.6; four copies: 1.6 <1.9; greater than four copies: >1.9. These ratios were determined by testing control Coriell DNA samples previously reported to have 1, 2, 3 or 4 copies of SULT1A1 by qPCR (NA17244, NA17245, NA17224 and NA17204, respectively) [10]. The frequencies were compared between the different ethnicities with the likelihood-ratio χ2 test.
Copy number analysis by qPCR
MLPA analysis of SULT1A1 copy number was validated by commercially available TaqMan® real-time qPCR Copy Number Assays that interrogated exon 1 and intron 2 of the minor transcript variant 5 (NM_177536.2) and the 3′UTR region of both major and minor SULT1A1 transcripts (Hs00736771_cn, Hs04461762_cn, Hs04461427_cn; Thermo Fisher Scientific) as per the manufacturer's instructions. In brief, FAM-labeled SULT1A1 TaqMan minor groove binding probes and unlabeled PCR primers were individually run in a duplex qPCR with a VIC-labeled RNase P TaqMan Copy Number Reference Assay (catalog number: 4403326; Thermo Fisher Scientific). Quadruplicate experiments were each performed in 10 μl reactions containing ∼10 ng of DNA, 1X TaqMan Genotyping Master mix, 0.5 μl each of TaqMan Copy Number and Reference Assays in 384 well plates. Covered plates were run in a 7900HT Fast Real-Time PCR System (Applied Biosystems, CA, USA) and the amplification consisted of a denaturation step at 95°C for 10 min followed by 40 amplification cycles (95°C for 15 s and 60°C for 60 s). Data were captured using absolute quantitation with a manual CT threshold and autobaseline, followed by analysis using CopyCaller™ v1.0 Software (Applied Biosystems) where the number of copies of target sequence was determined by relative quantitation with the comparative CT (ΔΔCT) method. This method measures the CT difference (ΔCT) between target and reference sequences, and then compares the ΔCT values of test samples to a calibrator sample known to have two copies of the target sequence. The copy number of the target was calculated to be two times the relative quantity.
SULT1A1 Sanger sequencing
Selected samples were subjected to Sanger sequencing of an 1807 bp fragment amplified by the SequalPrep Long PCR kit (Thermo Fisher Scientific) using forward (5′-GAGGAGTTGGCTCTGCAGGGTTTC-3′) and reverse (5′-CCTCTCCAAACAGGTCAAGT-3′) primers targeted to exon 8 of the major SULT1A1 transcript. The amplification consisted of an initial denaturation step at 94°C for 2 min followed by ten cycles (94°C for 10 s, 58°C for 30 s and 68°C for 3 min) plus 20 cycles in which the extension step had a 20 s increase per cycle. PCR products were visualized by 1% agarose gel electrophoresis, and amplicons were sequenced with an ABI 3730XL DNA Analyzer (Applied Biosystems).
Results
SULT1A1 MLPA design
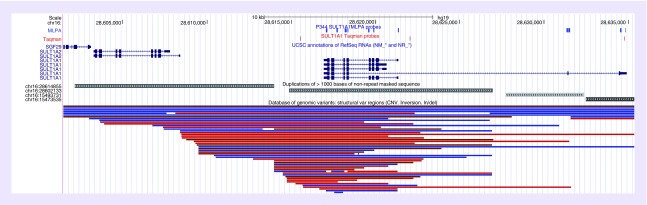
MLPA probes consist of bidirectional hybridizing oligonucleotides that are ligated together when they hybridize to directly adjacent target DNA sequences. The length of the hybridizing oligonucleotides, GC content, and melting temperature facilitate stable binding during MLPA hybridization and efficient PCR amplification. MLPA probe design is dependent on the availability of a unique target sequence to prevent unintended probe binding and amplification of off-target homologous sequences. However, probe specificity can often be enabled by single nucleotide differences between target DNA and homologous sequences. For the SULT1A1 MLPA assay, probes were designed to interrogate seven of eight exons of the major SULT1A1 transcript (NM_001055.3), two probes for each of the two upstream exons of the minor SULT1A1 transcript (NM_177536.2), and 11 autosomal control probes. It was not possible to design specific MLPA probes for exon 2 of the major SULT1A1 transcript due to the high sequence homology between this exon and other SULT1 genes. The locations of the 11 total SULT1A1 MLPA probes with respect to transcripts and coding regions are illustrated in Figure 1.
Figure 1. . The SULT1A1 gene region on chromosome 16p11.2, highlighting the location of SULT1A1 transcript variants, multiplex ligation-dependent probe amplification and quantitative PCR probes, segmental duplications, and copy number variants (CNVs) catalogued in the Database of Genomic Variants (DGV; http://dgv.tcag.ca).
Multi-ethnic SULT1A1 MLPA Testing
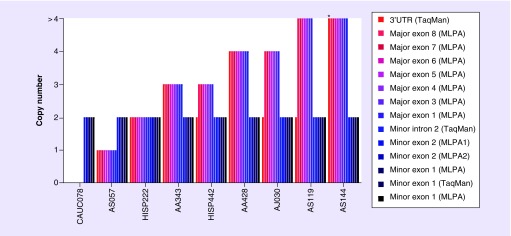
A total of 496 healthy unrelated individuals were subjected to SULT1A1 MLPA testing; however, 24 samples were excluded due to inadequate DNA quantity or quality. As such, data from 472 samples (95.1%) were analyzed, which included African–American (n = 92), Asian (n = 97), Caucasian (n = 95), Hispanic (n = 93) and AJ (n = 95) individuals. All populations tested harbored SULT1A1 CNV alleles, and the frequencies of individuals with an atypical total copy number (i.e., any copy number other than two) were 38.7% for Hispanics, 38.9% for AJ, 43.2% for Caucasians, 53.6% for Asians, and 64.1% for African–Americans (Table 1). Of note, all SULT1A1 CNVs were only observed in the major transcript, whereas the upstream exons 1 and 2 of the minor SULT1A1 transcript were consistently two copies in all tested individuals (Figure 2).
Table 1. . Multi-ethnic SULT1A1 copy number frequencies.
| Gene | Total copies | African–American (n = 92) | Asian (n = 97) | Caucasian (n = 95) | Hispanic (n = 93) | Ashkenazi Jewish (n = 95) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Freq. | 95% CI | Freq. | 95% CI | Freq. | 95% CI | Freq. | 95% CI | Freq. | 95% CI | ||
| SULT1A1 | 0 | – | – | – | – | 0.011† | 0.000–0.031 | – | – | – | – |
| 1 | 0.033 | 0.000–0.069 | 0.031 | 0.000–0.065 | 0.074 | 0.021–0.126 | 0.011 | 0.000–0.032 | 0.063 | 0.014–0.112 | |
| 2 | 0.359 | 0.261–0.457 | 0.464 | 0.365–0.563 | 0.568 | 0.469–0.668 | 0.613 | 0.514–0.712 | 0.611 | 0.512–0.709 | |
| 3 | 0.391‡ | 0.292–0.491 | 0.330 | 0.236–0.423 | 0.274 | 0.184–0.363 | 0.280 | 0.188–0.371 | 0.284 | 0.194–0.375 | |
| 4 | 0.141† | 0.070–0.212 | 0.072 | 0.021–0.124 | 0.053 | 0.008–0.098 | 0.086 | 0.029–0.143 | 0.042 | 0.002–0.082 | |
| >4 | 0.076 | 0.022–0.130 | 0.103‡ | 0.043–0.164 | 0.021 | 0.000–0.050 | 0.011 | 0.000–0.032 | – | – | |
| 0 + 1 (SS) | 0.033 | 0.000–0.069 | 0.031 | 0.000–0.065 | 0.084† | 0.028–0.140 | 0.011 | 0.000–0.032 | 0.063 | 0.014–0.112 | |
| 2 (NS) | 0.359‡ | 0.261–0.457 | 0.464 | 0.365–0.563 | 0.568 | 0.469–0.668 | 0.613 | 0.514–0.712 | 0.611 | 0.512–0.709 | |
| 3 + 4 + >4 (RS) | 0.609‡ | 0.509–0.708 | 0.505 | 0.406–0.605 | 0.347 | 0.252–0.443 | 0.376 | 0.278–0.475 | 0.326† | 0.232–0.421 | |
†p < 0.05.
‡p < 0.01.
n: Number of individuals; NS: Normal sulfators; RS: Rapid sulfators; SS: Slow sulfators.
Figure 2. . Representative cases tested by Multiplex ligation-dependent probe amplification and quantitative PCR illustrating differences in copy number consistency across probes, and localization of the SULT1A1 copy number variation alleles to the major and minor transcripts.
The probe loci that interrogated the 5′ exons and introns of the minor transcript are located upstream of the major transcript (see Figure 1 for genomic locations). The asterisk (*) indicates a sample with five and six copies by qPCR, which was interpreted by MLPA to be >4 copies.
MLPA: Multiplex ligation-dependent probe amplification; qPCR: Quantitative PCR.
SULT1A1 copy number loss
Heterozygous SULT1A1 deletion carriers (i.e., one total copy) were observed in all tested populations, which ranged in frequency from 1.1% in Hispanics to 8.4% in Caucasians (Table 1 & Figure 2). As such, the highest frequency of ‘slow sulfators’ was observed in the Caucasian population (∼1 in 10; p < 0.05), including one individual that harbored a homozygous SULT1A1 deletion.
SULT1A1 copy number gains
The multi-ethnic frequencies of SULT1A1 copy number gains (greater than two copies) are summarized in Table 1 and representative cases are illustrated in Figure 2. All tested populations harbored SULT1A1 copy number gains, which had significantly different frequencies across the five racial/ethnic groups (p < 0.001). The frequencies of three total SULT1A1 copies ranged from 27.4% in Caucasians to 39.1% in African–Americans, and the frequencies of four or more total copies ranged from 4.2% in the AJ to 21.7% in African–Americans. Taken together, African–Americans had the highest frequency of ‘rapid sulfators’ (60.9%; p < 0.01), which translated to ∼1 in 2 individuals having greater than two copies of SULT1A1.
SULT1A1 MLPA confirmation by qPCR & Sanger sequencing
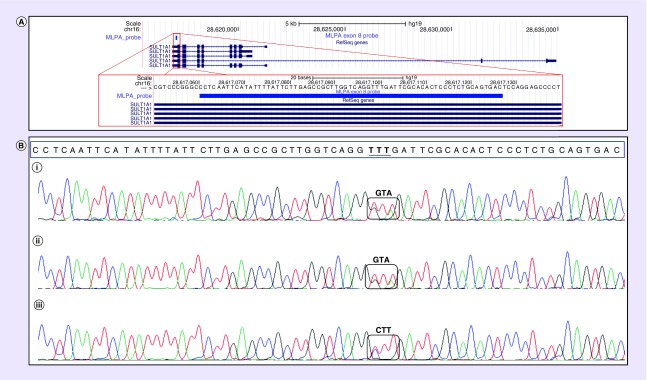
A subset of samples (n = 48; 10.2%) were also tested by TaqMan qPCR copy number assays targeted to the SULT1A1 locus (Figures 1 & 2). All samples tested by qPCR were consistent with MLPA results from the major and minor transcripts, which confirmed that SULT1A1 CNVs were restricted to the major transcript (Figure 1 &Supplementary Table 1). Inconsistent MLPA results were observed in three independent samples at exon 8 of the major SULT1A1 transcript, which prompted subsequent Sanger sequencing across this exon in these samples. As illustrated in Figure 3, Sanger sequencing determined that the discrepancy in exon 8 MLPA copy number was due to SNV polymorphisms (rs377290662, rs114793911) underlying the hybridization sequence close to the ligation site of one exon 8 MLPA probe.
Figure 3. . Sanger sequencing results for exon 8 in three samples with discordant multiplex ligation-dependent probe amplification results.
(A) The red box is a schematic representation of exon 10 and the multiplex ligation-dependent probe amplification probe location. (B) The multiplex ligation-dependent probe amplification probe sequence is within the blue box and identified single nucleotide variants that likely interfered with probe hybridization are denoted within black boxes.
Discussion
The paucity of available SULT1A1 copy number assays prompted our development of a multiplexed MLPA assay that simultaneously interrogates 11 loci across the major and minor transcripts of the SULT1A1 gene. This novel MLPA assay was validated by screening almost 500 human DNA samples from five different racial and ethnic populations (African–American, Asian, Caucasian, Hispanic and AJ), including additional confirmatory testing by qPCR and Sanger sequencing. Despite the structural complexity of the SULT1A1 gene region, these results together indicated that this multiplexed assay is robust at detecting SULT1A1 copy number, with atypical total copy numbers detected in all populations, ranging from 38.7% in Hispanics to 64.1% in African–Americans.
SULT1A1 belongs to a family of Phase II detoxification enzymes and plays a key role in catalyzing the sulfate conjugation of several xenobiotics, steroid hormones, neurotransmitters and medications (e.g., tamoxifen, fulvestrant and toremefine). Functional genetic studies have identified SNVs in the SULT1A1 coding and regulatory regions that are associated with transcription, translation and enzyme activity [13,21–23]. Of note, the SULT1A1 c.638G>A (SULT1A1*2) allele has been associated with adverse endpoints among breast cancer patients treated with tamoxifen [24,25], and other regulatory variants have been implicated in SULT1A1 activity [13]. However, SULT1A1 CNV alleles are strongly associated with SULT1A1 enzymatic activity and explain more of the observed in vitro variability than SULT1A1 SNVs [13,26]. Individuals with less than two copies of SULT1A1 (slow sulfators) have lower enzyme activity and those with greater than two copies (rapid sulfators) have increased enzyme activity [10,27].
The SULT1A1 CNV alleles had significantly different frequencies across the five racial/ethnic groups, which were consistent with previously reported data from other populations assessed by targeted qPCR [6,10,27]. ‘Slow sulfators’ were detected in all tested populations, which ranged in frequency from 1.1% in Hispanics to 8.4% in Caucasians (∼1 in 10), including one rare individual that harbored a homozygous SULT1A1 deletion. In addition, SULT1A1 ‘rapid sulfators’ were detected in all tested populations, which ranged in frequency from 32.6% in the AJ to 60.9% in African–Americans (∼1 in 2). Given the high frequencies of SULT1A1 duplication/triplication alleles in the African–American population, future clinical SULT1A1 pharmacogenetic studies are warranted in patient cohorts of African descent.
The significance of structural variation in human disease and phenotypic diversity has increasingly become recognized, and several genomic studies have generated catalogs of CNVs to facilitate a better understanding of their clinical relevance [28,29]. Some of the common mechanisms of CNV formation include nonallelic homologous recombination, microhomology-mediated break-induced repair and nonhomologous end joining [30]. The SULT1A1 locus at chromosome 16p11.2 is encompassed by several homologous segmental duplications, including two ∼11 kb repeats in direct orientation that share ∼95% homology and which underlie the major SULT1A2 and SULT1A1 transcripts. These repeats most likely mediate SULT1A1 structural variation by acting as substrates for nonallelic homologous recombination during human meiosis. This is further evidenced by the regional CNVs that are cataloged in the DGV (Figure 1), which are primarily localized between these two segmental duplications. The CNV sizes in the DGV are consistent with the SULT1A1 CNV alleles detected in our study, as copy number gains and losses were only detected by MLPA and qPCR across the major SULT1A1 transcript and not including the upstream exons of the minor transcript (Figure 1 & Supplementary Data).
Of note, three independent African–American samples had an inconsistent exon 8 MLPA result compared with all other tested loci across the SULT1A1 gene, suggesting a small single exon deletion. However, Sanger sequencing of this region across these samples identified two polymorphisms (rs377290662, rs114793911) close to the ligation site of one of the exon 8 probes. Although all of the SULT1A1 probes were designed to avoid known sequence variants, these two variants were not in dbSNP at the time of probe design. Polymorphisms that interfere with MLPA probe hybridization can result in allele dropout, as previously reported for other PCR-based molecular techniques [31–34]. However, given the increasing identification and prevalence of rare sequence variants in the human genome [35,36], allele dropout from individual MLPA targeted regions will never be completely avoidable. These data underscore the importance of not over-interpreting single loci deletions by MLPA and the need to confirm these inconsistent results by an orthogonal method. Furthermore, the two variants in exon 8 are the discriminating nucleotides between SULT1A1 and the other three members of the SULT1A subfamily, which could indicate the possibility of gene conversion between subfamily members.
In conclusion, we describe the development of a novel multiplexed MLPA assay that simultaneously interrogates 11 loci across the SULT1A1 gene. This assay was validated by screening 472 African–American, Asian, Caucasian, Hispanic and AJ individuals, which detected multi-ethnic SULT1A1 CNV alleles that were consistent with copy numbers derived from orthogonal qPCR testing. Importantly, atypical total copy numbers (i.e., copy numbers other than two) were detected in all populations, with ∼1 in 10 Caucasians being ‘slow sulfators’ and ∼1 in 2 African–Americans being ‘rapid sulfators.’ In addition to highlighting the accuracy and specificity of this robust MLPA assay, these data indicate that different ethnic and racial populations have varying degrees of SULT1A1-mediated sulfation activity, which warrants further research and that may have utility for drug response prediction among SULT1A1-metabolized medications.
Future perspective
Structural variation in the human genome is increasingly becoming appreciated as an important contributor to some Mendelian diseases, common disease and pediatric neuropsychiatric disorder risk, and human drug response phenotypes. Pharmacogenomic structural variants (deletions, duplications, and other rearrangements) can be interrogated for some drug metabolism genes including CYP2B6, CYP2D6, GSTT1, GSTM1 and SULT1A1, and this has been enabled by molecular technologies that can quantitatively assess gene copy number. Importantly, common genotyping and sequencing techniques have historically not been able to concurrently measure copy number, which resulted in increased effort and burden to incorporate the additional testing when wanted or needed. MLPA provides a convenient and robust platform to measure the copy number at multiple loci in a single experiment; however, the ability to infer copy number from high-throughput sequencing data suggests that in the future we may be able to comprehensively interrogate targeted gene regions for nucleotide and copy number variants with only a single platform. But given the sequence homology between pharmacogenomic and other gene family members, the ongoing challenges related to short-read sequencing alignment will remain as necessary hurdles to overcome.
Executive summary.
SULT1A1 multiplex ligation-dependent probe amplification design
Probes were designed to interrogate seven of eight exons of the major SULT1A1 transcript (NM_001055.3), two probes for each of the two upstream exons of the minor SULT1A1 transcript (NM_177536.2) and 11 autosomal control probes.
Multi-ethnic SULT1A1 multiplex ligation-dependent probe amplification testing
A total of 472 African–American, Asian, Caucasian, Hispanic and Ashkenazi Jewish (AJ) individuals were subjected to SULT1A1 MLPA testing.
The frequencies of individuals with an atypical total copy number (i.e., any copy number other than two) were 38.7% for Hispanics, 38.9% for AJ, 43.2% for Caucasians, 53.6% for Asians and 64.1% for African–Americans.
All SULT1A1 copy number variant alleles were only observed in the major transcript, whereas the upstream exons 1 and 2 of the minor SULT1A1 transcript were consistently two copies in all tested individuals.
SULT1A1 copy number losses & gains
Caucasians had the highest frequency of ‘slow sulfators’ (8.4%), which translated to ∼1 in 10 individuals having less than two copies of SULT1A1.
African–Americans had the highest frequency of ‘rapid sulfators’ (60.9%), which translated to ∼1 in 2 individuals having greater than two copies of SULT1A1.
Selected samples were also tested by quantitative PCR and the results were consistent with multiplex ligation-dependent probe amplification, which confirmed that SULT1A1 copy number variants are restricted to the major transcript.
Conclusion & future direction
In addition to highlighting the accuracy and specificity of this robust multiplex ligation-dependent probe amplification assay, these data indicate that different ethnic and racial populations have varying degrees of SULT1A1 activity, which warrants further research and that may have utility for drug response prediction among SULT1A1-metabolized medications.
Supplementary Material
Acknowledgements
The SULT1A1 MLPA kit reagents used in this study were generously provided by MRC-Holland (Amsterdam, The Netherlands).
Footnotes
Supplementary data
To view the supplementary data that accompany this paper please visit the journal website at: https://www.futuremedicine.com/doi/suppl/10.2217/pgs-2018-0047
Financial & competing interests disclosure
This research was supported in part by the National Institute of General Medical Sciences (NIGMS) of the NIH, through Grant K23 GM104401 (SA Scott). R Vijzelaar and L Stolk are paid employees of MRC-Holland, Amsterdam, The Netherlands, and MR Botton and SA Scott. are paid employees of Sema4, Stamford, CT, USA. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Open access
This work is licensed under the Attribution-NonCommercial-NoDerivatives 4.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
References
- 1.Stankiewicz P, Lupski JR. Structural variation in the human genome and its role in disease. Annu. Rev. Med. 2010;61:437–455. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- 2.Stranger BE, Forrest MS, Dunning M, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315(5813):848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaunt TR, Rodriguez S, Guthrie PA, Day IN. An expectation-maximization program for determining allelic spectrum from CNV data (CoNVEM): insights into population allelic architecture and its mutational history. Hum. Mutat. 2010;31(4):414–420. doi: 10.1002/humu.21199. [DOI] [PubMed] [Google Scholar]
- 4.Escaramis G, Docampo E, Rabionet R. A decade of structural variants: description, history and methods to detect structural variation. Brief. Funct. Genomics. 2015;14(5):305–314. doi: 10.1093/bfgp/elv014. [DOI] [PubMed] [Google Scholar]
- 5.Gaedigk A, Gaedigk R, Leeder JS. UGT2B17 and SULT1A1 gene copy number variation (CNV) detection by LabChip microfluidic technology. Clin. Chem. Lab. Med. 2010;48(5):627–633. doi: 10.1515/CCLM.2010.128. [DOI] [PubMed] [Google Scholar]
- 6.Schulze J, Johansson M, Thorngren JO, Garle M, Rane A, Ekstrom L. SULT2A1 gene copy number variation is associated with urinary excretion rate of steroid sulfates. Front. Endocrinol. (Lausanne) 2013;4:88. doi: 10.3389/fendo.2013.00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martis S, Mei H, Vijzelaar R, Edelmann L, Desnick RJ, Scott SA. Multi-ethnic cytochrome-P450 copy number profiling: novel pharmacogenetic alleles and mechanism of copy number variation formation. Pharmacogenomics J. 2013;13(6):558–566. doi: 10.1038/tpj.2012.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gjerde J, Hauglid M, Breilid H, et al. Effects of CYP2D6 and SULT1A1 genotypes including SULT1A1 gene copy number on tamoxifen metabolism. Ann. Oncol. 2008;19(1):56–61. doi: 10.1093/annonc/mdm434. [DOI] [PubMed] [Google Scholar]
- 9.Hebbring SJ, Moyer AM, Weinshilboum RM. Sulfotransferase gene copy number variation: pharmacogenetics and function. Cytogenet. Genome Res. 2008;123(1–4):205–210. doi: 10.1159/000184710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hebbring SJ, Adjei AA, Baer JL, et al. Human SULT1A1 gene: copy number differences and functional implications. Hum. Mol. Genet. 2007;16(5):463–470. doi: 10.1093/hmg/ddl468. [DOI] [PubMed] [Google Scholar]
- 11.Price RA, Spielman RS, Lucena AL, Van Loon JA, Maidak BL, Weinshilboum RM. Genetic polymorphism for human platelet thermostable phenol sulfotransferase (TS PST) activity. Genetics. 1989;122(4):905–914. doi: 10.1093/genetics/122.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Loon J, Weinshilboum RM. Human platelet phenol sulfotransferase: familial variation in thermal stability of the TS form. Biochem. Genet. 1984;22(11–12):997–1014. doi: 10.1007/BF00499627. [DOI] [PubMed] [Google Scholar]
- 13.Ning B, Nowell S, Sweeney C, et al. Common genetic polymorphisms in the 5′-flanking region of the SULT1A1 gene: haplotypes and their association with platelet enzymatic activity. Pharmacogenet. Genomics. 2005;15(7):465–473. doi: 10.1097/01.fpc.0000166823.74378.79. [DOI] [PubMed] [Google Scholar]
- 14.Hildebrandt M, Adjei A, Weinshilboum R, et al. Very important pharmacogene summary: sulfotransferase 1A1. Pharmacogenet. Genomics. 2009;19(6):404–406. doi: 10.1097/FPC.0b013e32832e042e. [DOI] [PubMed] [Google Scholar]
- 15.GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45(6):580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scott SA, Jaremko M, Lubitz SA, Kornreich R, Halperin JL, Desnick RJ. CYP2C9*8 is prevalent among African–Americans: implications for pharmacogenetic dosing. Pharmacogenomics. 2009;10(8):1243–1255. doi: 10.2217/pgs.09.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scott SA, Khasawneh R, Peter I, Kornreich R, Desnick RJ. Combined CYP2C9, VKORC1 and CYP4F2 frequencies among racial and ethnic groups. Pharmacogenomics. 2010;11(6):781–791. doi: 10.2217/pgs.10.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scott SA, Edelmann L, Kornreich R, Erazo M, Desnick RJ. CYP2C9, CYP2C19 and CYP2D6 allele frequencies in the Ashkenazi Jewish population. Pharmacogenomics. 2007;8(7):721–730. doi: 10.2217/14622416.8.7.721. [DOI] [PubMed] [Google Scholar]
- 19.Scott SA, Edelmann L, Kornreich R, Desnick RJ. Warfarin pharmacogenetics: CYP2C9 and VKORC1 genotypes predict different sensitivity and resistance frequencies in the Ashkenazi and Sephardi Jewish populations. Am. J. Hum. Genet. 2008;82(2):495–500. doi: 10.1016/j.ajhg.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scott SA, Martis S, Peter I, Kasai Y, Kornreich R, Desnick RJ. Identification of CYP2C19*4B: pharmacogenetic implications for drug metabolism including clopidogrel responsiveness. Pharmacogenomics J. 2012;12(4):297–305. doi: 10.1038/tpj.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raftogianis RB, Wood TC, Otterness DM, Van Loon JA, Weinshilboum RM. Phenol sulfotransferase pharmacogenetics in humans: association of common SULT1A1 alleles with TS PST phenotype. Biochem. Biophys. Res. Commun. 1997;239(1):298–304. doi: 10.1006/bbrc.1997.7466. [DOI] [PubMed] [Google Scholar]
- 22.Raftogianis RB, Wood TC, Weinshilboum RM. Human phenol sulfotransferases SULT1A2 and SULT1A1: genetic polymorphisms, allozyme properties, and human liver genotype-phenotype correlations. Biochem. Pharmacol. 1999;58(4):605–616. doi: 10.1016/s0006-2952(99)00145-8. [DOI] [PubMed] [Google Scholar]
- 23.Yu X, Dhakal IB, Beggs M, et al. Functional genetic variants in the 3′-untranslated region of sulfotransferase isoform 1A1 (SULT1A1) and their effect on enzymatic activity. Toxicol. Sci. 2010;118(2):391–403. doi: 10.1093/toxsci/kfq296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nowell S, Sweeney C, Winters M, et al. Association between sulfotransferase 1A1 genotype and survival of breast cancer patients receiving tamoxifen therapy. J. Natl Cancer Inst. 2002;94(21):1635–1640. doi: 10.1093/jnci/94.21.1635. [DOI] [PubMed] [Google Scholar]
- 25.Nowell SA, Ahn J, Rae JM, et al. Association of genetic variation in tamoxifen-metabolizing enzymes with overall survival and recurrence of disease in breast cancer patients. Breast Cancer Res. Treat. 2005;91(3):249–258. doi: 10.1007/s10549-004-7751-x. [DOI] [PubMed] [Google Scholar]
- 26.Moyer AM, Suman VJ, Weinshilboum RM, et al. SULT1A1, CYP2C19 and disease-free survival in early breast cancer patients receiving tamoxifen. Pharmacogenomics. 2011;12(11):1535–1543. doi: 10.2217/pgs.11.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu X, Kubota T, Dhakal I, et al. Copy number variation in sulfotransferase isoform 1A1 (SULT1A1) is significantly associated with enzymatic activity in Japanese subjects. Pharmgenomics Pers. Med. 2013;6:19–24. doi: 10.2147/PGPM.S36579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johansson AC, Feuk L. Characterization of copy number-stable regions in the human genome. Hum. Mutat. 2011;32(8):947–955. doi: 10.1002/humu.21524. [DOI] [PubMed] [Google Scholar]
- 29.Sudmant PH, Rausch T, Gardner EJ, et al. An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526(7571):75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat. Rev. Genet. 2009;10(8):551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blais J, Lavoie SB, Giroux S, et al. Risk of Misdiagnosis due to allele dropout and false-positive PCR artifacts in molecular diagnostics: analysis of 30,769 genotypes. J. Mol. Diagn. 2015;17(5):505–514. doi: 10.1016/j.jmoldx.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 32.Hahn S, Garvin AM, Di Naro E, Holzgreve W. Allele drop-out can occur in alleles differing by a single nucleotide and is not alleviated by preamplification or minor template increments. Genet. Test. 1998;2(4):351–355. doi: 10.1089/gte.1998.2.351. [DOI] [PubMed] [Google Scholar]
- 33.Wang C, Schroeder KB, Rosenberg NA. A maximum-likelihood method to correct for allelic dropout in microsatellite data with no replicate genotypes. Genetics. 2012;192(2):651–669. doi: 10.1534/genetics.112.139519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mullins FM, Dietz L, Lay M, et al. Identification of an intronic single nucleotide polymorphism leading to allele dropout during validation of a CDH1 sequencing assay: implications for designing polymerase chain reaction-based assays. Genet. Med. 2007;9(11):752–760. doi: 10.1097/gim.0b013e318159a369. [DOI] [PubMed] [Google Scholar]
- 35.Genomes Project Consortium. Abecasis GR, Altshuler D, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tennessen JA, Bigham AW, O'Connor TD, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337(6090):64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.