Abstract
This Perspective provides examples of current and future applications of deep learning in pharmacogenomics, including: identification of novel regulatory variants located in noncoding domains of the genome and their function as applied to pharmacoepigenomics; patient stratification from medical records; and the mechanistic prediction of drug response, targets and their interactions. Deep learning encapsulates a family of machine learning algorithms that has transformed many important subfields of artificial intelligence over the last decade, and has demonstrated breakthrough performance improvements on a wide range of tasks in biomedicine. We anticipate that in the future, deep learning will be widely used to predict personalized drug response and optimize medication selection and dosing, using knowledge extracted from large and complex molecular, epidemiological, clinical and demographic datasets.
Keywords: : adverse events, artificial intelligence, deep learning, drug discovery, drug–drug interaction, drug–gene interaction, noncoding regulatory variation, patient stratification, pharmacogenomics
Machine learning is a fundamental concept of artificial intelligence (AI), and is a key component of the ongoing big data revolution that is transforming biomedicine and healthcare [1–3]. Unlike many ‘expert system’-based methods in medicine that rely on sets of predefined rules about the domain, machine learning algorithms learn these rules from data, benefiting directly from the detail contained in large, complex and heterogeneous datasets [4]. Deep learning is one of the most successful types of machine learning techniques that has transformed many important subfields of AI over the last decade. Examples include data modeling and analytics, computer vision, speech recognition and natural language processing (NLP). Deep learning demonstrated breakthrough performance improvements over pre-existing techniques on a wide range of complex tasks across multiple biomedical research domains spanning from basic clinical to translational [5].
The deep learning methods landscape encompasses a variety of biologically inspired models that can be applied directly to raw data, automatically learn useful features and make a prediction without a need to form a hypothesis [5]. While biomedical applications of deep learning are still emerging, they have already shown promising advances over the prior state-of-the-art in several tasks [6–8]. We anticipate deep learning algorithms to have a substantial impact on pharmacogenomics, pharmaceutical discovery and more generally, on personalized clinical decision support in the near future.
Pharmacogenomics focuses on the identification of genetic variants that are correlated with drug effects in populations, cohorts and individual patients. It has traditionally straddled the intersection of genomics and pharmacology, with the greatest impact on clinical practice in oncology [9], psychiatry [10], neurology [11] and cardiology [12]. Pharmacogenomics offers promise for applications such as medication optimization for patients based on genotype in diagnostic testing, value as a companion diagnostic and drug discovery and development. However, physicians, caregivers, patients and pharmaceutical and biotechnology companies have all been slow to adopt pharmacogenomics, despite recommendations by the US FDA [13]. Recently, however, pharmaceutical companies that are faced with rising costs and resource investments required for drug development have begun to recognize the potential of genomics for drug discovery, and to a lesser extent, for stratification of participants in clinical trials to mitigate adverse events (AEs) and increase efficacy [14,15]. In addition, the adoption of pharmacogenomic testing for optimization of medication selection in psychiatry has shown promise for the clinical utility [10,16].
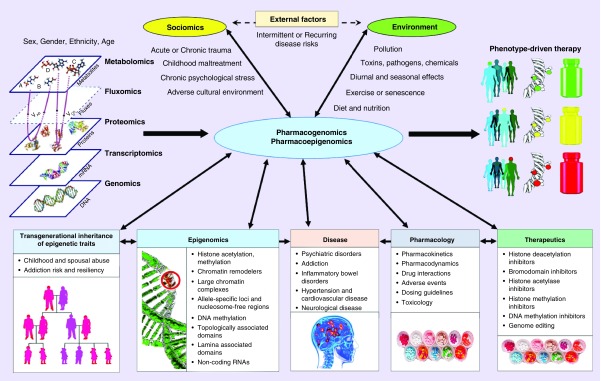
Historically, the exome has provided a rich source of single variants for genotyping in pharmacogenomics, with a focus on genes that encode ADME (absorption, distribution, metabolism, excretion), including DMET (drug metabolizing enzymes and transporters) proteins. More recently, the noncoding regulatory genome is proving to be the next domain for the discovery of new pharmacogenomic variants that will provide clinical utility [17]. This new research lies at the heart of the new field of ‘Pharmacoepigenomics’, which is the corresponding emerging subdomain of pharmacogenomics that focuses on studying the role of the epigenome in drug response [18]. However, these new variant discovery methods and their potential corresponding drug development processes can be very time consuming, where large trials are needed to assess clinical efficacy, toxicity and safety [19]. Moreover, the continuing growth of other types of collected data that can improve phenotype-driven therapy via pharmacogenomics also poses a number of challenges for accurate treatment response and outcome prediction (Figure 1).
Figure 1. . Pharmacogenomics and pharmacoepigenomics for phenotype-driven therapy.
Pharmacogenomics and pharmacoepigenomics will be facilitated by various data sources that include not only traditional ‘omics’ databases, but also: growing knowledge in epigenomics, including novel variant and their functional annotation; pharmacological data, including new therapeutics, drug interactions and dosing guidelines; and patient data, including clinical, socio-economic and information about the environment.
Partially adapted with permission from [20] (2012) under CC BY 3.0 license.
Extracting usable knowledge from large databases requires advanced computational methods that can find patterns, conduct prediction, detection and classification [2,3] along with visual data analytics [21,22]. Current approaches for knowledge extraction in pharmacogenomics include statistical methods [23,24], machine learning [24,25] and recently, deep learning. Therefore, new deep-learning-based predictive analytic methods are desirable to accelerate the discovery of new pharmacogenomic markers, forecast drug efficacy in stratified cohorts of patients to minimize potential adverse effects of drugs and to maximize the success of treatment.
In this Perspective, we provide examples of current and potential future applications of AI, and more specifically deep learning, in pharmacogenomics and pharmacoepigenomics to illustrate the utility and the future potential of these methods. It should be recognized that this Perspective provides an overview of selective pharmacogenomic applications, and is not intended to provide an authoritative and rigorous evaluation of the technical foundations of these methods. In addition, like other machine learning methods, deep learning techniques are prone to error if not grounded in computational, statistical and experimental expertise [6]. Indeed, proper controls and performance metrics are critical for the performance evaluation of such models [26]. For those seeking extensive reviews, there are a number of existing deep learning application-focused review articles in computational biology [6,7,27], pharmacology and drug discovery [28–32] and other areas of biomedicine [8]. The most successful applications of deep learning require expertise in both the methodology and in the subject matter under investigation, in this case, pharmacogenomics. Thus, we have two synergistic objectives – to increase awareness while stimulating dialogue among pharmacogenomics and pharmacology researchers about promising future applications of this powerful machine learning computational methodology.
Opportunities & challenges for deep learning applications
Methodological advantages of deep learning
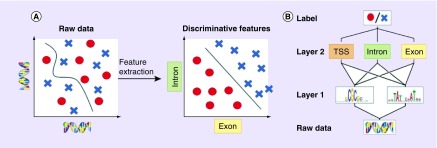
Conventional machine learning algorithms are typically limited in their ability to process raw data [5]. Their performance heavily depends on the extraction of relevant representations or features that require careful engineering and considerable domain expertise (Figure 2A). In the past, biomedical datasets have typically been limited by sample size, and since often many more features could be measured, the performance of conventional machine learning algorithms degraded when useful information was buried in an excess of extracted features. This posed a challenge for the determination and extraction of the optimal feature set for the problem under examination. Two related and widely-used solutions are used to overcome this limitation: dimensionality reduction methods that shrink the feature space to the set of most informative components [24]; and feature selection methods that identify a relatively small number of features that can accurately predict an outcome [25]. While many of these general-purpose methods already exist, they are not necessarily optimized for pharmacogenomic biomarker discovery. This and other related pharmacological research applications require careful experimental design and choice of validation techniques. Overall, limitations of conventional machine learning methods include the need for extensive human guidance, painstaking feature handcrafting, careful data preprocessing and the above-mentioned dimensionality reduction to achieve top performance.
Figure 2. . Feature extraction versus representation learning.
(A) Raw input data are often high-dimensional and related to the corresponding label in a complicated way, which is challenging for many conventional machine learning algorithms (left plot). Alternatively, higher-level features extracted using a deep model may be able to better discriminate between classes (right plot). (B) Deep networks use a hierarchical structure to learn increasingly abstract feature representations from the raw data recommendation.
Adapted with permission from [7] (2016) under CC BY 3.0 license.
In contrast, deep learning methods model data by learning high-level representations with multilayer computational models such as artificial neural networks (ANNs) [5]. While classic feed-forward ANNs might serve as drop-in replacement for other machine learning models and require input preprocessing and feature extraction, deep learning architectures, such as convolutional neural networks (CNNs), allow the algorithm to automatically learn features from raw and noisy data. Deep neural networks rely on algorithms that optimize feature engineering processes to provide the classifier with relevant information that maximizes its performance with respect to the final task. Such deep learning models can be thought of as automated ‘feature learning’ or ‘feature detection’, which facilitates learning of hierarchical, increasingly abstract representations of high-dimensional heterogeneous data [5], also known as ‘representation learning’ (Figure 2B). Some common deep learning methods include deep feed-forward ANNs, CNNs, recurrent neural networks (RNNs), stacked autoencoders, deep belief networks and deep reinforcement learning techniques [5–7,27]. In biomedicine, these models are capable of unguided extraction of highly complex, nonlinear dependencies from raw data such as raw sequence data [8].
Recent applications of deep learning in biomedicine have already demonstrated their superior performance compared with other machine learning approaches in a number of biomedical problems [8], including those in image analysis [33–36], genomics [7,27], as well as drug discovery and repurposing [30,31]. This great success of deep learning models in many tasks is thought to be enabled by the explosive growth of volume of raw data along with significant progress in computing, including the use of powerful graphical processing units that are specifically well-suited for the optimization of deep learning models.
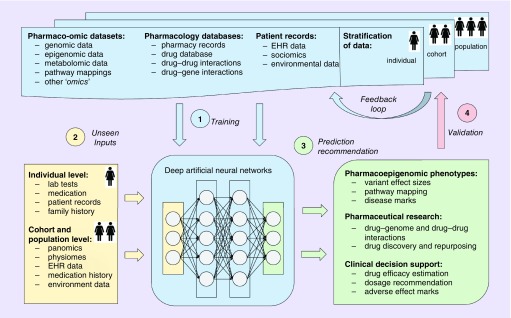
Figure 3 shows an idealized collective example of deep learning applications in pharmacogenomics. First, deep neural networks are trained on various existing datasets and/or their combinations. Depending on the type of data and a task in hand, prediction outcomes for a dataset can be known (supervised learning), partially known (semisupervised learning) or not known at all (unsupervised learning). Due to the flexibility of architectures, neural networks are capable of multimodal learning, in other words, jointly learning from a number of different datasets and data types without explicit definition of common features [37]. For example, Chaudhary et al. [38] trained a deep autoencoder model jointly on RNA-seq, miRNA-seq and methylation data from The Cancer Genome Atlas to predict subgroups of hepatocellular carcinoma patients. Moreover, deep networks can be used in a multitask learning regime by learning multiple objectives simultaneously and providing a number of outputs such as prediction of the regulatory function of a sequence, pathway mapping, disease and ADE mark identification, drug efficacy and dosage recommendation [31].
Figure 3. . An idealized example of deep learning applications to a number of common problems in pharmacogenomics, including prediction of pharmacoepigenomic phenotypes, novel regulatory variants and their function and stratified clinical decision support.
(A) First, deep neural networks are trained on various existing datasets and/or their combinations with known outcomes. (B) To perform prediction, unseen data of the same format as training samples are given to trained networks as inputs. (C) At the prediction step, a model produces outcome(s) that it was trained to predict during the training phase, for example, personalized drug response, probability of adverse events or novel pharmacogenomic variants and their pathway mapping. (D) Validation of the predicted responses occurs when true outcomes become available on various population stratification levels. For example, it can be individual patient treatment response, clinical trial results or new pharmaceutical study results. Data with observed true outcomes can again be used as a training set via a feedback loop, as shown.
EHR: Electronic health records.
Challenges & limitations of deep learning
Deep learning is a very fast-paced research domain and although its potential utility is impressive, there are several challenges associated with the practical usage of these models. Typically, CNNs are complex, heavily parameterized models, with little theoretical guarantees of performance or proven ways to construct effective problem-specific architectures. Due to their complexity, time-consuming training process and high representational power, such models should be used in cases when sufficient volumes of input data are available and conventional machine learning fails to provide an effective, simpler solution. For biological and clinical applications, adoption of deep learning is slower due to a few reasons, including skepticism arising from the data and hardware requirements, as well as the ‘black-box’ nature of these algorithms, which specifically challenges the notion of model interpretability [5,8]. Below we discuss some of these challenges in more detail.
Data requirements
While most successful deep learning applications relied on large labeled data, many biological and clinical datasets until recently were limited by amount of available labeled samples compared with the big data analytics applications such as natural image processing and NLP. Over time, as the number of samples increases and the number of relevant high-quality labeled datasets expands, the wealth of pertinent pharmacogenomics data that can be used for analytics will begin to resemble a big data challenge on par with contemporary applications in other domains. The rich variety of heterogeneous data types in pharmacogenomics can improve the utilization of highly flexible deep networks that can deal with sparse, high-dimensional, multimodal data. The real power of deep learning in a domain such as pharmacogenomics will be realized when it is combined with a prior domain knowledge [8], such as gene networks or pathways; a relevant example being used for the prediction of the pharmacological properties of drugs using transcriptomic data combined with pathway information [39]. Multimodal, multitask and transfer learning are often used to alleviate data limitations to some extent. Transfer learning approaches include training a deep network on a large existing dataset, and then using this preinitialized model to learn from a smaller dataset, which typically leads to improved performance [8]. When training data is not (fully) labeled, various semisupervised techniques can be employed [8,34,40]. Data quality is another important concern in deep learning applications. Although deep learning models can be trained directly on raw data, low quality datasets may require additional preprocessing and cleaning. Publicly sharing the preprocessing code (e.g., Basset [41]) and cleaned data (e.g., MoleculeNet [42]) is important to expedite further research and practical applications.
Overfitting in deep learning
A trained machine learning model may represent some attributes of the dataset that do not accurately reflect their underlying relationships. This problem may be magnified as the size of the feature or parameter set is increased relative to the size of the input sample set. Such models exhibit poor predictive performance, as they over-represent minor variations in the training data. Overfitting is an issue of trade-off between generalization and approximation of the training data in a model. A model can underfit high-dimensional, heterogeneous dataset with complex hidden patterns if the model's representational power is low, which is often the case, for example, for linear models. Although overfitting is a common issue in machine learning, it is more likely to affect complex models, especially when there are not enough samples in the training set, learning is performed for too long or where training examples are rare, causing the learner to follow the patterns in training data too closely. In the case of deep learning, overfitting is often a threat due to the high representational power of a model, which leads to the ability to ‘memorize’ the whole training set very efficiently. Thus, deep learning methods require careful choice of model architecture and hyperparameters. Although there are specific methods to prevent overfitting [5,7], in general, the trained model should exhibit robust performance on test data using relevant properties of the trained data. For more detailed description of overfitting and model selection, see [43].
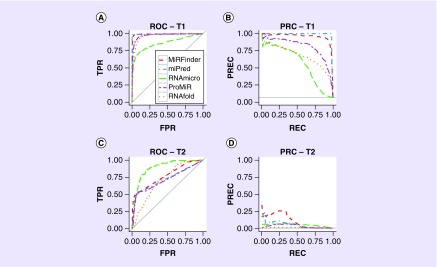
Preventing overfitting also requires a very careful design of the model evaluation scheme, including usage of cross-validation techniques, normalization by subjects, etc. Validation metrics may include mean absolute error or root-mean-square error (sample standard deviation of the differences between predicted and observed outcomes) for regression; accuracy; precision (also known as positive predictive value – the fraction of retrieved instances that are relevant); recall (sensitivity – the fraction of relevant instances that are retrieved); precision-recall curve and area under the precision-recall curve; receiver operating characteristic and area under the receiver operating characteristic curve (AUC); and mean average precision, for ranking [26,44]. Although some of these may seem intuitive, correct determination requires great care, and is often fraught with sources of error that are not easily understood, except in the context of the problem under study. For example, while the AUC plot is a common visual method for classification performance evaluation, it is not the most informative when classes are represented largely by a different number of samples in the dataset [44], which is a common situation in pharmacogenomics; see Figure 4. One test of the quality of the trained machine learning model is its ability to faithfully generalize into varying test sets that constitute different manifestations of the same problem.
Figure 4. . A re-evaluation of a previously published study confirms the advantages of the precision-recall curve plot over the receiver operating characteristic plot in an imbalanced binary classification task.
ROC and PRC plots show the performances of six different tools, MiRFinder (red), miPred (blue), RNAmicro (green), ProMiR (purple) and RNAfold (orange) for microRNA (miRNA) gene discovery from MiRFinder study [45]. The re-analysis used two independent test sets, T1 and T2. The four plots are for (A) ROC on T1, (B) PRC on T1, (C) ROC on T2 and (D) PRC on T2.
PRC: Precision-recall curve; ROC: Receiver operating characteristic.
Adapted with permission from [44] © Saito, Rehmsmeier (2015) published by PLoS under CC BY 4.0 license.
Interpretability of deep learning models
As applications of deep learning models in biomedicine emerge, the question of interpreting a model's outputs receives more attention from the domain experts and practitioners [8]. Unfortunately, the opinion that deep learning models are uninterpretable ‘black boxes’ still prevails outside of machine learning community. However, multiple different methods for deep learning model interpretation have been developed in recent years, including perturbation and back-propagation techniques to evaluate example-specific importance scores, exaggeration of hidden representations, activation maximization and other methods [6,8]. In addition, recent studies distinguish between interpreting a model and interpreting its decision, arguing that although interpreting a complex model is hard, often users only want an explanation for the prediction made by the model for a given example [46]. While most applications that motivated model interpretability techniques come from computer vision problems, there is a growing body of research that considers these methods in the life science and biomedical research context.
For example, DeepSEA [47] and DeepBind [48], deep learning-based algorithmic frameworks for predicting the chromatin effects of sequence alterations introduced individual virtual mutations in the input sequence to evaluate the change of the corresponding output (see also Table 2). A similar approach was used by Umarov et al. [49], where the sequence within each sliding window was substituted with a random sequence to estimate its effect on the result. In order to assess the change in predicted sequence accessibility, the Basset framework [41] implemented insertions of known protein-binding motifs into the centers of input sequences. DeepLIFT [50] allows computing feature importance scores based on explaining the difference of the output from some ‘reference’ output in terms of differences of the inputs from their ‘reference’ inputs. Lanchantin et al. [51] applied activation maximization to genomic sequence data to demonstrate patterns learned by an individual neuron in a network. Deming et al. [52] applied the attention mechanism to models trained on genomic sequence. Attention mechanisms provide insight into the model's mechanism of prediction by revealing which portions of the input are used by different outputs. While the interpretability of deep neural networks does not match that of most Bayesian models, recent developments in this area make it possible to interpret deep learning models, as well as many other commonly used machine learning algorithms such as support vector machines with nonlinear kernels or ensemble methods such as Random Forests [8]. For more detailed discussion of interpretability, we refer the reader to Ching et al. [8].
Table 2. . Examples of open-source deep learning software applications for the discovery of epigenomic regulatory interactions and variant annotation.
| Software | Source code | Description | Ref. |
|---|---|---|---|
| DeepSEA | http://deepsea.princeton.edu | Predicts the noncoding variant effects de novo from sequence by directly learning a regulatory sequence code from large-scale chromatin profile data, enabling prediction of chromatin effects of sequence alterations with single-nucleotide sensitivity | [47] |
| DeepBind | http://tools.genes.toronto.edu/deepbind/ | Predicts potential TFBSs and RBP binding sites; both in vitro and in vivo, outperforming 26 previously tested algorithms | [48] |
| deepnet-rbp | https://github.com/thucombio/deepnet-rbp | Predicts RBP binding sites taking (predicted) RNA tertiary structural information into account | [53] |
| Basset | www.github.com/davek44/Basset | Predicts DNA accessibility, simultaneously learning the relevant sequence motifs and the regulatory logic with which they are combined to determine cell-specific DNA accessibility. Predictions for the change in accessibility between variant alleles are greater for GWAS in SNPs that are likely to be causal relative to nearby SNPs in linkage disequilibrium with them | [41] |
| DanQ | https://github.com/uci-cbcl/DanQ | Uses the same features and data as the DeepSEA framework, outperforming DeepSEA for 97.6% of the targets | [54] |
| DeepChrome | https://github.com/QData/DeepChrome | Predicts gene expression from histone modification signals and enables the visualization of the combinatorial interactions among histone modifications via a novel optimization-based technique that generates feature pattern maps from the learned deep model | [55] |
| TFImpute | https://bitbucket.org/feeldead/tfimpute | Predicts cell-specific TF binding for TF-cell line combinations using an MTL setting to use information across TFs and cell lines | [56] |
| Rambutan | https://github.com/jmschrei/rambutan | Predicts Hi-C contacts at 1 kb resolution using nucleotide sequence and DNase I assay signal as inputs. Predicted contacts exhibit expected trends relative to histone modification ChIP-seq data, replication timing measurements and annotations of functional elements such as enhancers and promoters | [57] |
| CpGenie | https://github.com/gifford-lab/CpGenie/ | Produces allele-specific DNA methylation prediction with single-nucleotide sensitivity that enables accurate prediction of meQTLs. Contributes to the prediction of functional noncoding variants, including eQTLs and disease-associated mutations | [58] |
| DeepCpG | https://github.com/cangermueller/deepcpg | Identifies known and de novo sequence motifs that are predictive for DNA methylation levels or methylation variability, and to estimate the effect of single-nucleotide mutations | [59] |
| iDeep and iDeepS |
www.csbio.sjtu.edu.cn/bioinf/iDeep/ https://github.com/xypan1232/iDeepS |
Predicts RBP binding sites by multimodal learning from multiresource data, e.g., sequence, structure, domain-specific features and formats. Allows one to automatically capture the interpretable binding motifs for RBPs | [60] [61] |
| FactorNet | https://github.com/uci-cbcl/FactorNet | Predicts TFBS by leveraging a variety of features, including genomic sequences, genome annotations, gene expression and single-nucleotide resolution sequential signals, such as DNase I cleavage data | [62] |
| Basenji | https://github.com/calico/basenji | Predicts cell type-specific epigenetic and transcriptional profiles in large mammalian genomes from DNA sequence alone. Identifies promoters and distal regulatory elements, and synthesizes their content to make effective gene expression predictions. Model predictions for the influence of genomic variants on gene expression that align well to causal variants underlying eQTLs in human populations; and can be useful for generating mechanistic hypotheses to enable GWAS loci fine mapping | [63] |
| Concise | https://github.com/gagneurlab/concise | Predicts RBP binding sites using a spline transformation-based neural network module to model distances from regulatory sequences to genomic landmarks | [64] |
| DeepATAC | https://github.com/hiranumn/deepatac | Predicts binding locations from both DNA sequence and chromatin accessibility as measured by ATAC-seq, outperforming current approaches including DeepSEA | [65] |
ChIP-seq: Chromatin immunoprecipitation-sequencing; eQTL: Expression quantitative trait locus; GWAS: Genome-wide association study; meQTL: Methylation quantitative trait locus; MTL: Multitask learning; RBP: RNA binding protein; TFBS: Transcription factor binding site.
Identification of regulatory pharmacoepigenomic variants, drug target discovery & patient stratification
Inference of pharmacoepigenomic variants: the biology & machine learning
The noncoding human genome consists of a wealth of elements that regulate gene expression, and it accounts for the largest untapped potential source of drug targets and missing pharmacogenomic variation that has yet to be fully exploited [18]. Previous studies [66,67] demonstrate that psychotropic drugs such as valproic acid and lithium exert their impact in the human CNS through chromatin interaction pathways [67,68], regulating transcriptional networks that are constrained by the spatial and temporal dimensions of the 4D nucleome [18,69]. This led to the recognition that over 90% of the pharmacogenomic SNPs associated with drug efficacy and AEs in genome-wide association studies (GWAS) are located within enhancers, promoters and intron regions and impact their regulatory function. Coupled with a paucity of coding variation, this certainly contributes to drug response and comprises as yet uncharacterized pharmacogenomic variance requiring further exploration.
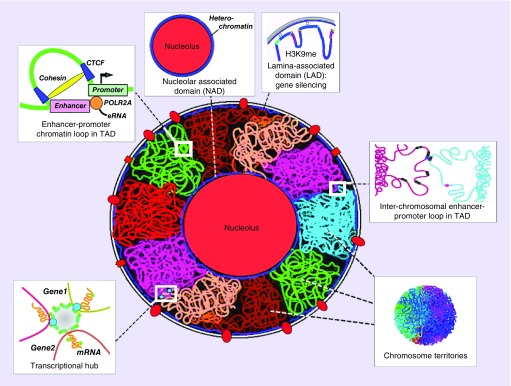
Results from the 4D nucleome program, funded by the US NIH, have led to the realization that significant molecular variation, which accounts for human differences in medication response and AEs are likely based on the intricate organization of the spatial genome [18,69]. Spatial and temporal morphological changes in the nucleus and nucleoli are associated with the underlying reorganization of the chromatin architecture in 3D and 4D (time dimension added) [70,71]. Cell-type-specific activation of topologically associated domains (TADs), which are defined areas within chromosome territories (chromosomes) and which provide a foundation for regulation of gene expression, while other nuclear zones of transcriptional regulation include repression (silencing) of gene expression within lamina-associated domains at the nuclear periphery (Figure 5) [72]. Nuclear pore complexes control the flow of molecular transport, splicing and are transcriptional hubs. Nucleoli are surrounded by heterochromatin and help to serve as an organizing scaffold for chromosome territories. Interchromosomal spatial contacts provide a mechanism for gene regulation between adjacent chromosomes, while chromatin loops within TADs provide one way in which enhancers and promoters regulate gene expression, both in cis and in trans. The cytoskeleton spans the nucleus via SUN and CASH proteins and may exert drug-induced mechanical control of gene expression. Large transcriptional hubs contain multiple chromatin interaction loops and TADs.
Figure 5. . Overview of different spatial regions that determine transcriptional state in the 3D nucleome.
These include the fundamental unit of transcription, the TADs and larger transcriptional hubs, embedded in CTs. Active TADs tend to be located at the periphery of CTs. Enhancer–promoter looping in chromatin is an example of how distant regions in linear DNA sequence come together for regulation of gene expression. Heterochromatin located in perinucleolar and in LADs, associated with histone marks such as H3K9me2/3, correlates with repression of gene expression. Interchromosomal interactions define one type of spatial trans-interaction.
CT: Chromosome territory; LAD: Lamina-associated domain; TAD: Topologically associated domain.
Adapted with permission from [72] © Elsevier (2017).
The pharmacoepigenome can be defined as noncoding, regulatory regions of the genome that play an active role in the determination of medication efficacy, dose requirement and AE profile [18]. The pharmacoepigenome contains gene regulatory elements such as enhancers and promoters. Since variation in noncoding regions in the human genome accounts for over 80% of the genetic contribution to disease risk, apart from the few known common single variants that cause Mendelian disorders [17], it is likely that variations in complex traits such as drug response and susceptibility to adverse drug events are also controlled by the noncoding genome [66]. Subsumed in this set is treatment-resistance, for example, regulatory noncoding elements power chemotherapeutic habituation to vemurafenib in 90% of melanoma patients [73], and resistance to two or more antiepileptic drugs in approximately 30% of patients with epilepsy [74].
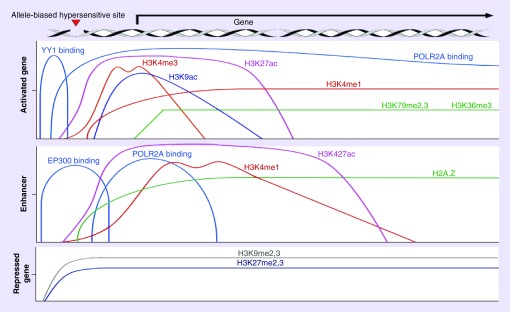
To infer attributes of noncoding regulatory SNPs and predict their impact on a phenotype, machine learning as well as probabilistic methods has been proposed. For example, to determine putative chromatin state annotation, applications based on Hidden Markov Models are still prevalent and are used to predict regulatory elements including promoters, enhancers, transcription start sites and gene bodies – among others. from SNPs found in genetic association studies [6]. These applications incorporate features such as histone marks that are characteristic of specific regulatory elements (see Table 1 & Figure 6), localization of regulatory elements in open chromatin as indicated by DNase I hypersensitivity, disruption of transcription factor binding sites and quantitative trait loci.
Table 1. . Table of associations between epigenomic elements and corresponding histone modifications.
| Element | Histones modifications |
|---|---|
| Transcriptional start site | H3K27ac, H3K4me3, open euchromatin |
| Active promoter | H3K27ac, H3K4me3, open euchromatin |
| Enhancer | H3K27ac, H3K4me1, H2A.Z, euchromatin |
| Gene body, toward 3′ end | H3K79me2/3, H3K36me3 |
| Large introns | H3K9ac, H3K18ac, H3K36me1 |
| Translation | H3K4me1, H3K79me1 |
| Strong polycomb repression | H3K9me2,3, H3k27me2/me3 |
Figure 6. . Selected histone modifications used in current applications for prediction and classification of noncoding variants that detect gene regulatory elements.
Distribution of histone modifications and other characteristics of epigenomic regulatory elements;.
Several machine learning applications have been developed for predicting the impact of noncoding SNPs in GWAS on phenotypes; however fewer than 40% of GWAS publications from 2015 utilized these tools [75]. Table 2 lists some examples of deep learning software that scores features, such as DNase I hypersensitivity for prioritization of regulatory function and protein annotation of chromatin loops, to predict functional enhancer–promoter interactions and drug–target inference.
Deep learning applications for detection of regulatory elements within the noncoding genome are beginning to emerge [7,8,76]. Most existing applications are based on CNN architecture, trained either from k-mers [77,78] or directly from genomic sequence data. For example, DeepSEA [47] is one of the first deep-learning-based algorithmic frameworks for predicting the chromatin effects of sequence alterations with single nucleotide sensitivity. In addition, it is trained on diverse sets of chromatin profiles from ENCODE and Roadmap Epigenomics Consortium projects [17,79]. DeepSEA can accurately predict the epigenetic state of a sequence, including transcription factor binding, DNase I sensitivities and histone marks in multiple cell types. In addition, it can further utilize its capabilities to predict the chromatin effects of sequence variants and prioritize regulatory variants. In another example, the DeepBind algorithm was implemented based on a deep CNN to calculate the ability of nucleotide sequences to bind transcription factors and RNA-binding proteins in order to characterize the effects of single point mutations on binding properties in various diseases [48]. More recently, the Basset CNN model was used to predict DNA accessibility within noncoding regions [41]; and is intended to predict allele-bias in DNA accessibility, which is indicative of causal variants. DNase-Seq data from 164 cell types that had been mapped by ENCODE and the Roadmap Epigenomics Consortium was used to create Basset. The Basset CNN learned both protein–DNA binding motif information, as well as the underlying regulatory knowledge that determines cell-specific DNA accessibility. In the analysis of GWAS SNPs that were determined to be casual autoimmune variants, Basset demonstrated that it could discriminate causal from noncasual SNPs in high linkage disequilibrium. In contrast to inference of regulatory elements using annotation based on predefined feature sets, models such as DeepSEA and Basset do not take handcrafted, preprocessed features. Instead, they adaptively learn them from raw sequence data during the training phase. This, combined with high expressive power, allows deep learning to outperform traditional machine learning models. More accurate prediction of noncoding variants and their functional annotations with deep learning methods promises to enable better understanding of pharmacoepigenomic variation and more accurate prediction of drug response and AEs.
Other recent applications of deep learning models to prediction of regulatory elements and their interactions with the state-of-the-art performance include: enhancer prediction [80–82]; classification of gene expression using histone modification data as input [55]; prediction of DNA methylation states from DNA sequence and incomplete methylation profiles in single cells [59]; prediction of enhancer–promoter interactions from genomic sequence [83]; prediction of DNA-binding residues in proteins [84]; global transcription start prediction [85]; and improved prediction of the impact of noncoding variants on DNA methylation [58,85]. In 2016, Google and Verily Life Sciences published a preprint describing ‘DeepVariant’ – a deep learning-based universal SNP and small indel variant caller. DeepVariant won the ‘highest performance’ award for the SNPs in the FDA-sponsored variant calling ‘Truth Challenge’ in May 2016 [86]. Recently, an updated, open-source version of DeepVariant has been further evaluated on a diverse set of additional tests by DNAnexus [87]. These tests showed that application of a general deep learning framework exceeded the accuracy of traditional methods for SNP and indel calling that has been developed over the last decade. Deep neural networks also demonstrated the ability to outperform conventional machine learning techniques in SNP–SNP interaction prediction [88,89].
The application of AI for patient stratification
The FDA provided guidance in 2013 [90] that pharmacogenomic testing should be used in early-phase clinical trials for the identification of populations, cohorts and individuals “that should receive lower or higher doses of a drug, or longer titration intervals, based on genetic effects on drug exposure, dose-response, early effectiveness and/or common adverse reactions”. Although this approach has not been widely adopted by pharmaceutical companies, in part, for fear of reducing its potential market size and a lack of available large genomic data resources, applications of AI methods for patient stratification using clinical data are beginning to see usage and adoption [91]. The industry seems to be moving towards the direction of developing proprietary machine learning algorithms to stratify patients using both unstructured and structured data obtained from the client's electronic health records (EHRs), which can be applied to both clinical research in academia as well as clinical trials in pharmaceutical research. By leveraging genomic data, and information from HapMap and 1000 Genomes Project [92], ethnicity stratification can be performed on population, cohort and other levels. This can be further extended by using EHRs for improved patient stratification, potentially leading to more precise risk models designed to advance clinical and translational research. Indeed, AI continues to rapidly advance in the biomedical research domain, with examples of deep-learning-based methods recently gaining FDA clearance for the clinical usage with cardiac MRIs [16].
Patient stratification involves the complex integration of heterogeneous biomedical, demographic and sociometric data to categorize patients into subpopulations for design of clinical trials and clinical practice. In this context, data mining of EHRs has been proposed as an efficient relevance-based method to potentially identify eligible patients for clinical trials [93]. Despite not being designed for usage in research, substantial amounts of data within EHRs, such as surrogate disease phenotypes imputed from International Classification of Diseases (ICD) codes, have effectively been proven for use through several notable studies in GWAS and phenome-wide association studies analysis [94]. Furthermore, studies on EHR-linked DNA biorepositories have successfully shown that integration of such pharmacogenomic and sociometric data can be useful in predictive modeling for optimizing dosage and reducing dosing error [94]. By using clinically available information, such as age, gender and education, healthcare providers and clinical researchers can identify better treatment options and patient responses to maximize efficacy and cost–effectiveness [94,95], as shown in Figure 1.
However, there are several challenges associated with the effective integration of EHR data for pharmacogenomics applications. For example, due to high dimensionality of the EHR data structure, noise, heterogeneity, sparseness, incompleteness, random error and systematic biases [96], extraction of relevant clinical phenotypes may require extensive feature engineering and advanced computational models beyond traditional machine learning methodologies. Ongoing research in this domain and recent advances in deep learning demonstrate the potential of deep learning to overcome these challenges and learn patient data representations that are useful for treatment response and outcome prediction [91]. Recent applications in this area include extraction of general-purpose patient representations from EHRs, often performed with generative models trained either on static or temporal data [91]. These models are capable of uncovering patterns in sparse, complex, heterogeneous datasets and producing surrogate imputed patient phenotypes. For example, there are both unsupervised, for example, Deep Patient [96], and semisupervised, for example, Denoising Autoencoder for Phenotype Stratification [40], models that rely on a stacked autoencoder network architecture to model EHR data to derive patient representations that are predictive of final diagnosis, patient risk level and outcome (e.g., mortality, readmission) (Table 3). As generative deep model development progresses quickly, applications of novel architectures, such as Generative Adversarial Networks, to EHR data are starting to emerge, demonstrating improved performance for the disease prediction [97] and risk prediction given treatment [98].
Table 3. . Examples of recently published research and open-source deep learning software for applications of artificial intelligence in patient stratification and patient care coordination.
| Software | Source code | Description | Ref. |
|---|---|---|---|
| Deep Patient |
https://github.com/staplet14/DeepPatient https://github.com/natoromano/deep-patient |
Learns a general-purpose patient representation from EHR data in unsupervised manner that is broadly predictive of health status as assessed by predicting the probability of patients to develop various diseases. Results significantly outperforming those achieved using representations based on raw EHR data and alternative feature learning strategies | [96] |
| DeepCare | https://github.com/trangptm/DeepCare | Predicts unplanned readmission and high-risk patients for the diabetes and mental health patient cohorts using EHR data including diagnosis, procedure and medication codes. Outperforms SVM, random forests, ‘plain’ RNN and LSTM with logistic regression | [99] |
| Doctor AI | https://github.com/mp2893/doctorai | Implements a generic predictive model that covers observed medical conditions and medication uses from longitudinal time-stamped EHR data. Performance was judged on classification of the final diagnosis (aggregated to 1183 unique ICD-9 codes) and prediction of medical order (grouped into 595 unique GPI codes) | [100] |
| MIMIC trajectories | https://github.com/EpistasisLab/MIMIC_trajectories | Learns meaningful representations from a longitudinal sequence of a patient's interactions with the healthcare system (care events) in both unsupervised and supervised settings that are shown to be useful for patient survival prediction | [101] |
EHR: Electronic health record; GPI: Generic product identifier; LSTM: Long short-term memory network; RNN: Recurrent neural network; SVM: Support vector machine.
Overall, the promise of integrating pharmacogenomics with data-driven EHR analysis of population, cohort and individual patient data already shows usefulness for patient stratification and prediction of treatment response. A quickly growing body of work in the field demonstrates a great interest in applying deep networks to these problems [91], which allows one to learn from heterogeneous EHR data, extract temporal patterns, impute missing data and predict clinical outcomes and optimal treatment strategies while outperforming conventional machine leaning methods.
Deep learning for temporal patient data
Due to the longitudinal nature of EHR data, many applications employ deep network architectures that are capable of extraction of temporal patterns from it, such as RNNs, long short-term memory networks – among others. [91]. These networks are used for mapping patient trajectories with temporal predictions of clinical outcomes, outperforming conventional machine learning methods that typically require a single ‘snapshot’ in time, and are not as robust for longitudinal modeling [99–101]. Several methods have been proposed to deal with the complex nature of longitudinal EHR data, specifically because of temporality from clinical records. To account for possible interventions and predict optimal treatment strategy, deep learning approaches were shown to be efficient when combined with reinforcement learning. For example, Kale et al. demonstrated how this type of deep model can be used for discovery and analysis of causal phenotypes from clinical time series data [102]. Deep neural networks trained on EHR data with temporally dependent constraints and outputs have also been proposed to predict 3–12-month mortality of patients receiving improved palliative care [46]. Additional deep reinforcement learning models have been used to learn an optimal heparin dosing policy from sample dosing trials, their associated outcomes having been predicted from the publicly available MIMIC II intensive care unit database [103].
Looking forward, we also envision the incorporation of data from mobile devices and wearable sensors for measuring phenotypic markers and stratification of patients by these phenotypes. This type of continuously collected data allows researchers access to large-scale deep phenotyping of the human population, and to better assess patients’ prognosis by analyzing their real-time data. Rajpurkar et al. developed a 34-layer CNN which exceeded the performance of board certified cardiologists in detecting a wide range of heart arrhythmias from electrocardiograms recorded with a single-lead wearable monitor [104]. Apple's ResearchKit open-source framework enables access to enrolled patients’ heart rate, accelerometer and other mobile sensor data [105]. For example, the approach utilizing deep CNNs for feature extraction from accelerometry and gyroscope iPhone data has recently won the Parkinson's Disease Digital Biomarker DREAM challenge, an open crowd-sourced research project designed to benchmark the use of remote sensors to diagnose and track Parkinson's disease [106]. Similar studies with RNNs have also shown to be successful in classification of patients with bipolar disorder using NLP and accelerometer data collected from a patient's mobile device [107]. Although data from wearable sensors is not yet considered to be a part of a patient's EHR, this data have shown to be robust and usable with deep learning methods and will certainly contribute to the modernization of patient stratification.
Pharmacological applications for drug & target discovery, repurposing & interaction
Although the noncoding human genome represents the new source for drug targets and genetic variation discovery, so far, most approaches to ‘epigenetic’ drug discovery have focused on post-translational modification of histone proteins and DNA through enzymes (‘writers’ and ‘erasers’), and the recognition of these changes by adaptor proteins (‘readers’). There are now hundreds of identified chromatin remodeling proteins that aggregate into larger protein complexes, and exert complex functions such as chromatin-mediated neuroplasticity and neurogenesis in the human CNS. These chromatin remodeling proteins were first examined in the context of developmental decisions about cell fate, and in the adult, potential druggable targets were thought to consist of histone demethylases (e.g., KDM1A), histone methyltransferases such as EZH2 and bromodomain-containing proteins, that were thought to be the only ‘readers’ of the histone code. On closer inspection, these turned out to be super-families of related proteins, and there exist many other proteins that act as chromatin remodelers (Figure 5).
The realization that the human epigenome operates the fundamental regulatory machinery of transcription, many new druggable targets can be discovered that are not ‘writers’, ‘erasers’ or ‘readers’. Fundamental to this realization was the recognition that the linear genetic sequence was only the beginning, as the important mechanisms of gene regulation operate in the spatial and temporal dynamics between regulatory elements such as super-enhancers, enhancers and promoters along with the target genes they regulate. Additionally, several important characteristics of causal variants in GWAS have emerged, including properties such as allele-specific bias, location of gene variants in euchromatin and histone marks that are associated with genome elements that help define their function. Critical to transcriptional regulatory circuits was the realization that transcription factors are key drivers of phenotype, and they can be classified in a hierarchal manner. In addition, master transcription factors are controlled by super-enhancers for determination of cell-specific gene regulation and identity [108].
Ivanov et al. [109] first recognized the complexity of the molecular physiology responsible for regulation of ADME genes, including DNA methylation and hydroxymethylation, various histone modifications, miRNAs and lncRNAs. Since a xenobiotic substance that alters any of the myriad of enzymes and small RNAs involved in ADME gene regulation represents a novel therapeutic candidate, they emphasized the importance of ‘pharmacoepigenomics’ in drug discovery. Since 2012, our understanding of the druggable epigenome has increased exponentially, providing thousands of new druggable targets (Figures 5 & 6).
Although the human epigenome has yielded insight into pharmacogenomic regulatory mechanisms, translation of this wealth of data into drug discovery will not be trivial. Currently, although the single most valuable approach for detection of prospective druggable targets in the human epigenome is the application of deep learning methods for candidate identification (Table 2), the ability to test compound/drug–molecule pairs is hobbled by protracted preclinical screening in animal models [29,32]. The cost and time incurred by the brute force screening of thousands of small compounds for novel epigenome drug targets in animal models is a daunting challenge. Simulation of the mechanism of candidate drugs’ action in populations of ‘virtual humans’, which accurately represent the molecular physiology of actual humans is within reach [110], but is an underfunded application domain in biomedical informatics and computer science. Translation will require innovation in domains that have traditionally resisted change, including adoption of adaptive clinical trial design, transformation of federal regulatory governance and broad adoption in preclinical pharmaceutical research of genomics-based science and in silico strategies that have been shown to be effective in predicting the clinical success of drug targets [111].
Deep learning applications in drug discovery and repurposing are starting to emerge, and already show high potential in many tasks including virtual screening, prediction of ADME/Tox properties and prediction of novel drug–target interactions [8,28–32]. Generative deep models are also used for de novo design molecules with desired chemical properties [112–114]. In Table 4, we show those applications with publicly available open-source implementations.
Table 4. . Examples of open-source deep learning software applications for pharmacological applications.
| Software | Source code | Description | Ref. |
|---|---|---|---|
| DeepChem | https://deepchem.io/ | Implements low data learning method based on a novel iterative refinement long short-term memory architecture combined with graph convolutional neural networks to learn of meaningful distance metrics over small molecules. On the Tox21 and SIDER collections, one-shot learning methods strongly dominate simpler machine learning baselines, indicating the potential for strong performance on small biological datasets | [115] |
| DeepDTIs | https://github.com/Bjoux2/DeepDTIs_DBN | Training drug–target space extracted from DrugBank consisted of 1412 drugs and 1520 targets. Experimental drug–target pairs for testing were derived from DrugBank as well and consisted of 2528 targets and 4383 experimental drugs. DeepDTI gained the best performance in multiple performance metrics as compared with the Bernoulli Naive Bayesian, Decision Tree and Random Forest classifiers. In addition, DeepDTI showed potential to predict whether a new drug targets some existing targets, or whether a new target is interacting with some existing drugs | [116] |
| DeepSynergy | https://github.com/KristinaPreuer/DeepSynergy | Predicts synergistic drug combinations for cancer therapy by learning from chemical properties of the drugs and gene expression profiles of specific cancer cell lines. DeepSynergy significantly outperformed the other methods with an improvement of 7.2% over the second-best method at the prediction of novel drug combinations within the space of explored drugs and cell lines. Applying DeepSynergy for classification of these novel drug combinations resulted in a high predictive performance of an AUC of 0.90. | [117] |
AUC: Area under the receiver operating characteristic curve.
Deep learning & toxicology
The Tox21 Data Challenge has been the largest effort of the scientific community to compare computational methods for toxicity prediction [118]. This challenge comprised 12,000 environmental chemicals and drugs that were measured for 12 different toxic effects by specifically designed assays. In this challenge, deep learning-based DeepTox model had the highest performance of all computational methods winning the grand challenge, the nuclear receptor panel, the stress response panel and six single assays [118]. DeepTox also demonstrated the benefit of using a multitask network, which outperformed a single-task counterpart in ten out of 12 assays. Further studies also suggest that multitask deep networks show superior performance on a broad range of drug discovery datasets [31,119].
DeepAOT family of deep architectures for the compound acute oral toxicity prediction is based on molecular graph encoding CNNs [120]. These models implement regression, multiclassification and multitask networks that outperformed previously reported models for this task. Interpretation of these models was performed by exploration of networks’ internal features (referred to as deep fingerprints) that were highly correlated with topological structure-based fingerprints. Furthermore, one toxicity-related feature of each deep fingerprint was tracked back to the atomic level and the highlighted toxicity fragments were then compared with structural alerts for toxic chemicals and compounds with potential adverse reactions from ToxAlerts database [121]. Consistent results suggested that DeepAOT models could infer acute oral toxicity related toxic fragments from just the information on molecular shape and atomic bonds. Moreover, this model architecture is not limited to acute oral toxicity, and it could be applied for studying other end points induced by compounds in complex systems [120].
Other pharmacogenomic applications
Applications of deep learning recently demonstrated state-of-the-art performance for predicting cell phenotypes from transcriptomics data [122], drug response in cancer [123], seizure-inducing side effects of preclinical drugs [124], patient survival from multiomics data [38], drug-induced liver injury prediction [53] and classifying genomic variants into adverse drug reactions [125].
Future perspective
Impact on basic research in biology & pharmacology
We anticipate that as larger and more heterogeneous pharmacogenomic datasets become available in coming years, the predictive power of AI, and specifically deep learning models, will increase. Abundance of various types of data not only will enable more effective data-driven mining and discovery of important variants and markers, but allow as well for deeper investigation of corresponding interaction mechanisms by systematically considering underlying biological processes at different scales and biological data modalities. As already noted, discovery and characterization of noncoding regulatory elements in 4D nucleome already are becoming a major topic of pharmacogenomic studies. Further investigations will continue this trend, with a focus on causal relationship between elements of interest and increasing model specificity and predictive power originating from multiomics and pathway studies.
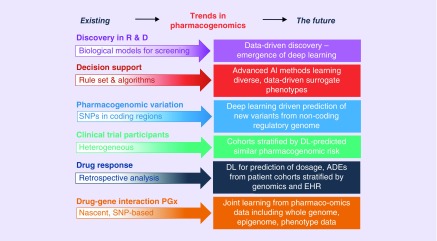
Increasing amounts of patient-specific data such as EHRs, environmental data and demographics, combined with pharmacogenomic targets and pharmacological knowledge bases will allow patient stratification into treatment groups with specificity at the population, cohort and individual levels. Advanced machine learning models such as deep learning will allow the researcher to jointly learn multiple objectives from heterogeneous, multimodal data and predict, for example, novel variants, their effects and functions, drug AE risk estimation, treatment and dosage recommendation and other pharmacological outputs (Figure 7). Given the growing amount of these data, AI methods, including deep learning, often demonstrate the best performance in addressing relevant methodological challenges [8]. However, as discussed above, applications of any machine learning algorithm, including deep learning, require careful selection of controls, training sets and appropriate validation schemes and metrics, and all of these should be combined with domain expertise to fully realize the potential of AI and deep learning.
Figure 7. . Future trends in pharmacogenomics.
We anticipate that as larger and more heterogeneous pharmacogenomic datasets become available in coming years, the predictive power of deep learning models will increase.
AI: Artificial inteligence; EHR: Electronic health records; PGx: Pharmacogenetics; R & D: Research and development.
Industrial perspective
Pharmaceutical companies were quick to recognize the potential application of AI methods such as deep learning for drug discovery and development. Market forecasts emphasized in 2017 that the “full potential healthcare service cost savings of AI-enabled initiatives would be $300 billion a year in the USA, or about 0.7 percent of GDP” [126], and “big pharma, biotech, contract research organizations and research institutes will spend $390M US on deep learning for drug discovery, including products, services and internal projects worldwide, and this market will grow to $1.25B by 2024” [127]. This led to a rush of investment into start-ups intending to offer AI consulting services and products offered to pharmaceutical companies, with a similarity to the rise of new companies offering companion diagnostics in the early part of the 21st century, which did not reach profitability in an entrepreneurial timeframe. There has been a reluctance of the large pharmaceutical companies, whose culture is often monolithic and conservative, to embrace innovation in the absence of pragmatic demonstration that AI methods would lead to success in clinical trials. More importantly, big pharma quickly realized that access to large quantities of genotyped patient data was a major priority for patient stratification and other applications, but in the USA, security concerns has limited partnerships among healthcare data owners – hospitals, insurers and drug makers. This has recently led big pharma to take the position of moving quickly to organize and curate their own datasets for internal use, and form partnerships with national biobanks and other entities for external forces of data, while engaging in watchful waiting for more realistic demonstrations of the practicality of deep learning for drug discovery and development before large investments are made. During this time, the industry as a whole has remained in vigorous discussions with various stakeholders about the potential and limitations of such methods. Now that there is some amount of critical evaluation of AI applications in drug discovery [128], the pharmaceutical industry is poised to embrace omics-driven drug discovery, phenotype-driven drug discovery and stratification in clinical trials on a massive scale as soon as convincing validation emerges from current efforts.
Open science considerations
A culture of data, code and model sharing promises to speed advances of deep learning applications in pharmacogenomics. The sharing of high quality, labeled datasets will be especially valuable; however, a clear asymmetry exists with government-sponsored academic researchers directed to share, while researchers in industry are often prohibited from sharing code, data and results due to proprietary and intellectual property protections. Availability of open-source solutions for the discovery of epigenomic regulatory interactions and variant annotation (Table 2) compared with those in patient stratification (Table 3) and pharmacological applications (Table 4) shows that the potentially translational character and patient privacy aspects of the study often prevent its details, data and implementation from being open to public. However, this situation is already changing in the machine learning and deep learning communities, that has witnessed acceleration of progress via public-posting of various datasets for benchmarking and software tools, including those developed and used in the industrial setting. Moreover, researchers who invest time to preprocess datasets to be suitable for deep learning can make the preprocessing code (e.g., Basset [38]) and cleaned data publicly available to catalyze further research. Code-sharing and open-source licensing are essential for continued progress in this domain. Because many deep learning models are often built using one of several popular software frameworks, it is also possible to directly share trained predictive models. A pretrained neural network can be quickly fine-tuned on new data and used in transfer learning, as discussed above. It is possible for models to be trained on competitive proprietary data without the release of such data, and a consortium model of joint training on proprietary data from multiple sources has been contemplated within the industry.
Conclusion
Deep learning models will be further improved to address current limitations such as training time, interpretability of results and requirements to training set size. Transfer learning that involves training deep learning models on one type of data and adaptation of a learnt representation to another type will be commonly used in experiments where data collection remains expensive. Models such as deep neural networks will be adapted to learn joint data representation from various omics data types, which will allow combining information from different experiments to provide better-informed predictions. As dataset sizes increase, we will see more of semi- and unsupervised machine learning applications, including generative models that will be able to produce or suggest new and testable biological hypotheses, such as potential novel pharmacogenomic markers (PK, PD and ADME), and also drug targets, based on information extracted from multimodal omics data.
Several decades from now, one can imagine the situation when machine learning and AI-based systems will shift their focus from ‘prediction’ to ‘prescription’, in other words, will not only provide insights, but will also provide recommendations for further action. Such changes not only have the potential to revolutionize pharmacogenomics and pharmaceutical research more broadly, but also will likely provide a wide impact on biological sciences, and on health, in general.
Executive summary.
Pharmacogenomics has promising applications in drug discovery & development, & medication optimization
Pharmacogenomic studies have established the importance of drug–genome interactions.
Pharmacogenomics offers promise for applications such as medication optimization for patients based on genotype in diagnostic testing, value as a companion diagnostic and drug discovery and development.
The noncoding regulatory genome is the current domain for the discovery of new genomic variants.
Brief overview of machine learning methodology
Machine learning methods have demonstrated the ability to identify novel regulatory variants located in noncoding domains that can inform pharmacogenomic response, prediction of drug–genome interactions and extraction of pharmacogenomic phenotype from clinical data, and drug discovery.
The predictive power of machine learning is realized mostly when it is combined with prior domain knowledge, such as gene networks and pathways.
Usage of traditional machine learning models is challenged by rapid growth of data volume, a need for combination of heterogeneous datasets from different experiments and is highly resource intensive in its preprocessing and feature handcrafting application(s).
Deep learning takes advantage of big data via representation learning
Deep learning is a subset of machine learning models composed of multiple processing layers to learn representations of data with multiple levels of abstraction, which eliminates the feature extraction step. These models improved the state-of-the-art in many machine learning tasks, including several examples in genomics and drug discovery.
Applications of deep learning in pharmacogenomics have started to emerge, but they are still in its infancy.
Deep learning models often require relatively large training sets, architecture design and careful choice of validation techniques to prevent overfitting.
Identification of regulatory pharmacogenomic variants & drug target discovery using deep learning
Significant molecular variation which accounts for human differences in medication response and adverse events may be based in the intricate organization of the 4D spatial genome (or 4D nucleome), which drives a need for deeper investigation of nuclear zones of transcriptional regulation.
Pharmacoepigenomic datasets contain information about gene regulatory elements such as promoters and enhancers, histone marks, disruption of transcription factor binding sites and quantitative trait loci. They are used in training of various machine learning models to infer regulatory attributes of noncoding SNPs, including chromatin state annotation, promoters, enhancers, transcription start sites, gene bodies – among others.
Deep learning models already have demonstrated state-of-the-art performance in several tasks such as predicting DNA accessibility within noncoding regions, potential transcription factor and RNA binding sites, and gene expression from histone modifications.
Realization that many of genetic differences in drug response can be found via analysis of noncoding regulatory genome together with modern gene editing techniques opens great opportunities for applications of machine and deep learning to predict the key regulatory variants that impact drug response and induce adverse drug events.
Applications of deep learning for phenotype extraction from medical records and other patient data, including temporal, show the potential usefulness for pharmacogenomic patient stratification, individualized treatment outcome prediction and medication optimization.
The fact that human epigenome operates the fundamental regulatory machinery of transcription in the spatial and temporal dynamics suggests that discoveries of many new druggable targets that were not in the focus of traditional ‘epigenetic’ drug discovery are potentially realizable.
Deep learning can be a lead force in pharmacoepigenomics-based drug discovery, combining candidate prediction with virtual screening, in silico drug repurposing and evaluation.
Footnotes
Financial & competing interests disclosure
This work was partially supported by the NIH grants: P20 NR015331, U54 EB020406, P50 NS091856, P30 DK089503, P30 AG053760, and T32 GM070449; National Science Foundation grants: 1734853, 1636840, 1416953, 0716055 and 1023115; and the Elsie Andresen Fiske Research Fund. All authors are either faculty or trainees at the University of Michigan Medical School and are up to date in their conflict of interest (COI) management disclosures. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
- 1.Obermeyer Z, Emanuel EJ. Predicting the future – big data, machine learning, and clinical medicine. N. Engl. J. Med. 2016;375(13):1216–1219. doi: 10.1056/NEJMp1606181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dinov ID. Methodological challenges and analytic opportunities for modeling and interpreting big healthcare data. Gigascience. 2016;5:12. doi: 10.1186/s13742-016-0117-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belle A, Thiagarajan R, Soroushmehr SM, Navidi F, Beard DA, Najarian K. Big data analytics in healthcare. Biomed Res. Int. 2015:370194. doi: 10.1155/2015/370194. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Halevy A, Norvig P, Pereira F. The unreasonable effectiveness of data. IEEE Intell. Syst. 2009;24(2):8–12. [Google Scholar]
- 5.Lecun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 6.Leung MKK, Delong A, Alipanahi B, Frey BJ. Machine learning in genomic medicine: a review of computational problems and datasets. Proc. IEEE. 2016;104(1):176–197. [Google Scholar]
- 7.Angermueller C, Parnamaa T, Parts L, Stegle O. Deep learning for computational biology. Mol. Syst. Biol. 2016;12(7):878. doi: 10.15252/msb.20156651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ching T, Himmelstein DS, Beaulieu-Jones BK, et al. Opportunities and obstacles for deep learning in biology and medicine. J. R. Soc. Interface. 2018;15(141) doi: 10.1098/rsif.2017.0387. pii:20170387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patel JN. Cancer pharmacogenomics, challenges in implementation, and patient-focused perspectives. Pharmgenomics Pers. Med. 2016;9:65–77. doi: 10.2147/PGPM.S62918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith RM. Advancing psychiatric pharmacogenomics using drug development paradigms. Pharmacogenomics. 2017;18(15):1459–1467. doi: 10.2217/pgs-2017-0104. [DOI] [PubMed] [Google Scholar]
- 11.Adams SM, Conley YP, Wagner AK, et al. The pharmacogenomics of severe traumatic brain injury. Pharmacogenomics. 2017;18(15):1413–1425. doi: 10.2217/pgs-2017-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cavallari LH, Weitzel K. Pharmacogenomics in cardiology – genetics and drug response: 10 years of progress. Future Cardiol. 2015;11(3):281–286. doi: 10.2217/fca.15.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Filipski KK, Pacanowski MA, Ramamoorthy A, Feero WG, Freedman AN. Dosing recommendations for pharmacogenetic interactions related to drug metabolism. Pharmacogenet. Genomics. 2016;26(7):334–339. doi: 10.1097/FPC.0000000000000220. [DOI] [PubMed] [Google Scholar]
- 14.Dopazo J. Genomics and transcriptomics in drug discovery. Drug Discov. Today. 2014;19(2):126–132. doi: 10.1016/j.drudis.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 15.Biankin AV, Piantadosi S, Hollingsworth SJ. Patient-centric trials for therapeutic development in precision oncology. Nature. 2015;526(7573):361–370. doi: 10.1038/nature15819. [DOI] [PubMed] [Google Scholar]
- 16.Rosenblat JD, Lee Y, McIntyre RS. Does pharmacogenomic testing improve clinical outcomes for major depressive disorder? A systematic review of clinical trials and cost–effectiveness studies. J. Clin. Psychiatry. 2017;78(6):720–729. doi: 10.4088/JCP.15r10583. [DOI] [PubMed] [Google Scholar]
- 17.Roadmap Epigenomics Consortium. Kundaje A, Meuleman W, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518(7539):317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higgins GA, Allyn-Feuer A, Handelman S, Sadee W, Athey BD. The epigenome, 4D nucleome and next-generation neuropsychiatric pharmacogenomics. Pharmacogenomics. 2015;16(14):1649–1669. doi: 10.2217/pgs.15.111. [DOI] [PubMed] [Google Scholar]
- 19.Harper AR, Topol EJ. Pharmacogenomics in clinical practice and drug development. Nat. Biotechnol. 2012;30(11):1117–1124. doi: 10.1038/nbt.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nemutlu E, Zhang S, Juranic NO, Terzic A, Macura S, Dzeja P. 18O-assisted dynamic metabolomics for individualized diagnostics and treatment of human diseases. Croat. Med. J. 2012;53(6):529–534. doi: 10.3325/cmj.2012.53.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Husain SS, Kalinin A, Truong A, Dinov ID. SOCR data dashboard: an integrated big data archive mashing medicare, labor, census and econometric information. J. Big Data. 2015;2 doi: 10.1186/s40537-015-0018-z. pii:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalinin AA, Palanimalai S, Dinov ID. Proceedings of the 2nd Workshop on Human-In-the-Loop Data Analytics. ACM New York; NY, USA: 2017. SOCRAT platform design: a web architecture for interactive visual analytics applications; pp. 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan J, Liu H. Statistical analysis of big data on pharmacogenomics. Adv. Drug Deliv. Rev. 2013;65(7):987–1000. doi: 10.1016/j.addr.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li R, Kim D, Ritchie MD. Methods to analyze big data in pharmacogenomics research. Pharmacogenomics. 2017;18(8):807–820. doi: 10.2217/pgs-2016-0152. [DOI] [PubMed] [Google Scholar]
- 25.Vidyasagar M. Identifying predictive features in drug response using machine learning: opportunities and challenges. Annu. Rev. Pharmacol. 2015;55:15–34. doi: 10.1146/annurev-pharmtox-010814-124502. [DOI] [PubMed] [Google Scholar]
- 26.Lever J, Krzywinski M, Altman N. Points of significance: classification evaluation. Nat. Methods. 2016;13(8):603–604. [Google Scholar]
- 27.Mamoshina P, Vieira A, Putin E, Zhavoronkov A. Applications of deep learning in biomedicine. Mol. Pharm. 2016;13(5):1445–1454. doi: 10.1021/acs.molpharmaceut.5b00982. [DOI] [PubMed] [Google Scholar]
- 28.Baskin Ii, Winkler D, Tetko IV. A renaissance of neural networks in drug discovery. Expert Opin. Drug Discov. 2016;11(8):785–795. doi: 10.1080/17460441.2016.1201262. [DOI] [PubMed] [Google Scholar]
- 29.Gawehn E, Hiss JA, Schneider G. Deep learning in drug discovery. Mol. Inform. 2016;35(1):3–14. doi: 10.1002/minf.201501008. [DOI] [PubMed] [Google Scholar]
- 30.Goh GB, Hodas NO, Vishnu A. Deep learning for computational chemistry. J. Comput. Chem. 2017;38(16):1291–1307. doi: 10.1002/jcc.24764. [DOI] [PubMed] [Google Scholar]
- 31.Ramsundar B, Liu B, Wu Z, et al. Is multitask deep learning practical for pharma? J. Chem. Inf. Model. 2017;57(8):2068–2076. doi: 10.1021/acs.jcim.7b00146. [DOI] [PubMed] [Google Scholar]
- 32.Pérez-Sianes J, Pérez-Sánchez H, Díaz F. Virtual screening: a challenge for deep learning. In: Mohamad MS, Rocha MP, Fdez-Riverola F, Domínguez-Mayo FJ, De Paz JF, editors. 10th International Conference on Practical Applications of Computational Biology & Bioinformatics. Springer International Publishing; Cham, Switzerland: 2016. pp. 13–22. [Google Scholar]
- 33.Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. Med. Image Anal. 2017 doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 34.Iglovikov V, Rakhlin A, Kalinin A, Shvets A. Pediatric bone age assessment using deep convolutional neural networks. arXiv. 2017 1712.05053. [Google Scholar]
- 35.Rakhlin A, Shvets A, Iglovikov V, Kalinin AA. Deep convolutional neural networks for breast cancer histology image analysis. arXiv. 2018 1802.00752. [Google Scholar]
- 36.Shvets A, Rakhlin A, Kalinin A, Iglovikov V. Automatic instrument segmentation in robot-assisted surgery using deep learning. bioRxiv. 2018 [Google Scholar]
- 37.Liang M, Li Z, Chen T, Zeng J. Integrative data analysis of multi-platform cancer data with a multimodal deep learning approach. IEEE/ACM Trans. Comput. Biol. Bioinform. 2015;12(4):928–937. doi: 10.1109/TCBB.2014.2377729. [DOI] [PubMed] [Google Scholar]
- 38.Chaudhary K, Poirion OB, Lu L, Garmire LX. Deep learning based multi-omics integration robustly predicts survival in liver cancer. Clin. Cancer Res. 2017 doi: 10.1158/1078-0432.CCR-17-0853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aliper A, Plis S, Artemov A, Ulloa A, Mamoshina P, Zhavoronkov A. Deep learning applications for predicting pharmacological properties of drugs and drug repurposing using transcriptomic data. Mol. Pharm. 2016;13(7):2524–2530. doi: 10.1021/acs.molpharmaceut.6b00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beaulieu-Jones BK, Greene CS Pooled Resource Open-Access ALSCTC. Semi-supervised learning of the electronic health record for phenotype stratification. J. Biomed. Inform. 2016;64:168–178. doi: 10.1016/j.jbi.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 41.Kelley DR, Snoek J, Rinn JL. Basset: learning the regulatory code of the accessible genome with deep convolutional neural networks. Genome Res. 2016;26(7):990–999. doi: 10.1101/gr.200535.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu Z, Ramsundar B, Feinberg Evan N, et al. MoleculeNet: a benchmark for molecular machine learning. Chem. Sci. 2018;9(2):513–530. doi: 10.1039/c7sc02664a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lever J, Krzywinski M, Altman N. Points of significance: model selection and overfitting. Nat. Methods. 2016;13(9):703–704. [Google Scholar]
- 44.Saito T, Rehmsmeier M. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS ONE. 2015;10(3):e0118432. doi: 10.1371/journal.pone.0118432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang TH, Fan B, Rothschild MF, Hu ZL, Li K, Zhao SH. miRFinder: an improved approach and software implementation for genome-wide fast microRNA precursor scans. BMC Bioinformatics. 2007;8:341. doi: 10.1186/1471-2105-8-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Avati A, Jung K, Harman S, Downing L, Ng A, Shah NH. Improving palliative care with deep learning. arXiv. 2017 doi: 10.1186/s12911-018-0677-8. 1711.06402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou J, Troyanskaya OG. Predicting effects of noncoding variants with deep learning-based sequence model. Nat. Methods. 2015;12(10):931–934. doi: 10.1038/nmeth.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alipanahi B, Delong A, Weirauch MT, Frey BJ. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat. Biotechnol. 2015;33(8):831–838. doi: 10.1038/nbt.3300. [DOI] [PubMed] [Google Scholar]
- 49.Umarov RK, Solovyev VV. Recognition of prokaryotic and eukaryotic promoters using convolutional deep learning neural networks. PLoS ONE. 2017;12(2):e0171410. doi: 10.1371/journal.pone.0171410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shrikumar A, Greenside P, Kundaje A. Proceedings of the 34th International Conference on Machine Learning. Sydney, Australia: 6–11 August 2017. Learning important features through propagating activation differences. [Google Scholar]
- 51.Lanchantin J, Singh R, Wang B, Qi Y. Deep motif dashboard: visualizing and understanding genomic sequences using deep neural networks. Pac. Symp. Biocomput. 2016;22:254–265. doi: 10.1142/9789813207813_0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Deming L, Targ S, Sauder N, Almeida D, Ye CJ. Genetic architect: discovering genomic structure with learned neural architectures. arXiv. 2016 1605.07156. [Google Scholar]
- 53.Xu Y, Dai Z, Chen F, Gao S, Pei J, Lai L. Deep learning for drug-induced liver injury. J. Chem. Inf. Model. 2015;55(10):2085–2093. doi: 10.1021/acs.jcim.5b00238. [DOI] [PubMed] [Google Scholar]
- 54.Quang D, Xie X. DanQ: a hybrid convolutional and recurrent deep neural network for quantifying the function of DNA sequences. Nucleic Acids Res. 2016;44(11):e107. doi: 10.1093/nar/gkw226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Singh R, Lanchantin J, Robins G, Qi Y. DeepChrome: deep-learning for predicting gene expression from histone modifications. Bioinformatics. 2016;32(17):i639–i648. doi: 10.1093/bioinformatics/btw427. [DOI] [PubMed] [Google Scholar]
- 56.Qin Q, Feng JX. Imputation for transcription factor binding predictions based on deep learning. PLoS Comput. Biol. 2017;13(2):e1005403. doi: 10.1371/journal.pcbi.1005403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schreiber J, Libbrecht M, Bilmes J, Noble W. Nucleotide sequence and DNaseI sensitivity are predictive of 3D chromatin architecture. bioRxiv. 2017 [Google Scholar]
- 58.Zeng H, Gifford DK. Predicting the impact of non-coding variants on DNA methylation. Nucleic Acids Res. 2017;45(11):e99. doi: 10.1093/nar/gkx177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Angermueller C, Lee HJ, Reik W, Stegle O. DeepCpG: accurate prediction of single-cell DNA methylation states using deep learning. Genome Biol. 2017;18(1):67. doi: 10.1186/s13059-017-1189-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pan X, Shen HB. RNA-protein binding motifs mining with a new hybrid deep learning based cross-domain knowledge integration approach. BMC Bioinformatics. 2017;18(1):136. doi: 10.1186/s12859-017-1561-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pan X, Rijnbeek P, Yan J, Shen H-B. Prediction of RNA-protein sequence and structure binding preferences using deep convolutional and recurrent neural networks. bioRxiv. 2017 doi: 10.1186/s12864-018-4889-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Quang D, Xie X. FactorNet: a deep learning framework for predicting cell type specific transcription factor binding from nucleotide-resolution sequential data. bioRxiv. 2017 doi: 10.1016/j.ymeth.2019.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kelley DR, Reshef YA. Sequential regulatory activity prediction across chromosomes with convolutional neural networks. bioRxiv. 2017 doi: 10.1101/gr.227819.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Avsec Z, Barekatain M, Cheng J, Gagneur J. Modeling positional effects of regulatory sequences with spline transformations increases prediction accuracy of deep neural networks. bioRxiv. 2017 doi: 10.1093/bioinformatics/btx727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hiranuma N, Lundberg S, Lee S-I. DeepATAC: a deep-learning method to predict regulatory factor binding activity from ATAC-seq signals. bioRxiv. 2017 [Google Scholar]
- 66.Higgins GA, Allyn-Feuer A, Athey BD. Epigenomic mapping and effect sizes of noncoding variants associated with psychotropic drug response. Pharmacogenomics. 2015;16(14):1565–1583. doi: 10.2217/pgs.15.105. [DOI] [PubMed] [Google Scholar]
- 67.Higgins GA, Allyn-Feuer A, Barbour E, Athey BD. A glutamatergic network mediates lithium response in bipolar disorder as defined by epigenome pathway analysis. Pharmacogenomics. 2015;16(14):1547–1563. doi: 10.2217/pgs.15.106. [DOI] [PubMed] [Google Scholar]
- 68.Higgins GA, Georgoff P, Nikolian V, et al. Network reconstruction reveals that valproic acid activates neurogenic transcriptional programs in adult brain following traumatic injury. Pharm. Res. 2017;34(8):1658–1672. doi: 10.1007/s11095-017-2130-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dekker J, Mirny L. The 3D genome as moderator of chromosomal communication. Cell. 2016;164(6):1110–1121. doi: 10.1016/j.cell.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kalinin AA, Allyn-Feuer A, Ade A, et al. 3D cell nuclear morphology: microscopy imaging dataset and voxel-based morphometry classification results. bioRxiv. 2017 [Google Scholar]
- 71.Zheng G, Kalinin AA, Dinov ID, Meixner W, Zhu S, Wiley JW. Rotational 3D mechanogenomic Turing patterns of human colon Caco-2 cells during differentiation. bioRxiv. 2018 doi: 10.1111/jcmm.13853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Higgins GA, Allyn-Feuer A, Georgoff P, Nikolian V, Alam HB, Athey BD. Mining the topography and dynamics of the 4D nucleome to identify novel CNS drug pathways. Methods. 2017;123:102–118. doi: 10.1016/j.ymeth.2017.03.012. [DOI] [PubMed] [Google Scholar]
- 73.Sanjana NE, Wright J, Zheng K, et al. High-resolution interrogation of functional elements in the noncoding genome. Science. 2016;353(6307):1545–1549. doi: 10.1126/science.aaf7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brodie MJ, Barry SJ, Bamagous GA, Norrie JD, Kwan P. Patterns of treatment response in newly diagnosed epilepsy. Neurology. 2012;78(20):1548–1554. doi: 10.1212/WNL.0b013e3182563b19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nishizaki SS, Boyle AP. Mining the unknown: assigning function to noncoding single nucleotide polymorphisms. Trends Genet. 2017;33(1):34–45. doi: 10.1016/j.tig.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Park Y, Kellis M. Deep learning for regulatory genomics. Nat. Biotechnol. 2015;33(8):825–826. doi: 10.1038/nbt.3313. [DOI] [PubMed] [Google Scholar]
- 77.Min X, Zeng WW, Chen N, Chen T, Jiang R. Chromatin accessibility prediction via convolutional long short-term memory networks with k-mer embedding. Bioinformatics. 2017;33(14):I92–I101. doi: 10.1093/bioinformatics/btx234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cao Z, Zhang S. gkm-DNN: efficient prediction using gapped k-mer features and deep neural networks. bioRxiv. 2017 [Google Scholar]
- 79.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kim SG, Harwani M, Grama A, Chaterji S. EP-DNN: a deep neural network-based global enhancer prediction algorithm. Sci. Rep. 2016;6:38433. doi: 10.1038/srep38433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xu M, Ning C, Ting C, Rui J. 2016 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) Shenzen, China: 15–18 December 2016. DeepEnhancer: predicting enhancers by convolutional neural networks. Presented at. [Google Scholar]
- 82.Liu F, Li H, Ren C, Bo X, Shu W. PEDLA: predicting enhancers with a deep learning-based algorithmic framework. Sci. Rep. 2016;6:28517. doi: 10.1038/srep28517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Singh S, Yang Y, Poczos B, Ma J. Predicting enhancer–promoter interaction from genomic sequence with deep neural networks. bioRxiv. 2016 doi: 10.1007/s40484-019-0154-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jiyun Z, Qin L, Ruifeng X, Lin G, Hongpeng W. 2016 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) Shenzen, China: 15–18 December 2016. CNNsite: prediction of DNA-binding residues in proteins using convolutional neural network with sequence features. Presented at. [Google Scholar]
- 85.Eser U, Churchman LS. FIDDLE: an integrative deep learning framework for functional genomic data inference. bioRxiv. 2016 [Google Scholar]
- 86.Poplin R, Newburger D, Dijamco J, et al. Creating a universal SNP and small indel variant caller with deep neural networks. bioRxiv. 2018 doi: 10.1038/nbt.4235. [DOI] [PubMed] [Google Scholar]
- 87.Carroll A, Thangaraj N. Evaluating DeepVariant: a new deep learning variant caller from the google brain team. 2017. https://blog.dnanexus.com/2017-12-05-evaluating-deepvariant-googles-machine-learning-variant-caller/
- 88.Uppu S, Krishna A. Improving Strategy for Discovering Interacting Genetic Variants in Association Studies. Springer; Cham, Switzerland: 2016. pp. 461–469. [Google Scholar]
- 89.Uppu S, Krishna A, Gopalan RP. A deep learning approach to detect SNP interactions. J. Software. 2016;11(10):965–975. [Google Scholar]
- 90.Food and Drug Administration. Silver Spring; MD, USA: 2013. Clinical pharmacogenomics: premarket evaluation in early-phase clinical studies and recommendations for labeling. US Department of Health and Human Services.www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/UCM337169.pdf [Google Scholar]
- 91.Shickel B, Tighe P, Bihorac A, Rashidi P. Deep EHR: a survey of recent advances on deep learning techniques for electronic health record (EHR) analysis. arXiv. 2017 doi: 10.1109/JBHI.2017.2767063. 1706.03446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Genomes Project C, Auton A, Brooks LD, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Miotto R, Weng C. Case-based reasoning using electronic health records efficiently identifies eligible patients for clinical trials. J. Am. Med. Inform. Assoc. 2015;22(e1):e141–e150. doi: 10.1093/jamia/ocu050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wei WQ, Denny JC. Extracting research-quality phenotypes from electronic health records to support precision medicine. Genome Med. 2015;7(1):41. doi: 10.1186/s13073-015-0166-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kannry JL, Williams MS. Integration of genomics into the electronic health record: mapping terra incognita. Genet. Med. 2013;15(10):757–760. doi: 10.1038/gim.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Miotto R, Li L, Kidd BA, Dudley JT. Deep patient: an unsupervised representation to predict the future of patients from the electronic health records. Sci. Rep. 2016;6:26094. doi: 10.1038/srep26094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hwang U, Choi S, Yoon S. Disease prediction from electronic health records using generative adversarial networks. arXiv. 2017 1711.04126. [Google Scholar]
- 98.Che Z, Cheng Y, Zhai S, Sun Z, Liu Y. Boosting deep learning risk prediction with generative adversarial networks for electronic health records. arXiv. 2017 1709.01648. [Google Scholar]
- 99.Pham T, Tran T, Phung D, Venkatesh S. DeepCare: a deep dynamic memory model for predictive medicine. In: Bailey J, Khan L, Washio T, Dobbie G, Huang JZ, Wang R, editors. Advances in Knowledge Discovery and Data Mining: 20th Pacific-Asia Conference, PAKDD 2016, Auckland, New Zealand, April 19–22, 2016, Proceedings, Part II. Springer International Publishing; Cham, Switzerland: 2016. pp. 30–41. [Google Scholar]
- 100.Choi E, Bahadori MT, Schuetz A, Stewart WF, Sun J. Doctor AI: predicting clinical events via recurrent neural networks. In: Doshi-Velez F, Fackler J, Kale D, Wallace B, Wiens J, editors. Proceedings of the 1st Machine Learning for Healthcare Conference. PMLR Children's Hospital LA; CA, USA: 2016. pp. 301–318. [PMC free article] [PubMed] [Google Scholar]
- 101.Beaulieu-Jones BK, Orzechowski P, Moore JH. Mapping patient trajectories using longitudinal extraction and deep learning in the MIMIC-III critical care database. In: Altman RB, Dunker AK, Hunter L, Ritchie MD, Murray TA, Klein TE, editors. Biocomputing 2018. World Scientific; Singapore: 2018. pp. 123–132. [PubMed] [Google Scholar]
- 102.Kale DC, Che Z, Bahadori MT, Li W, Liu Y, Wetzel R. Causal phenotype discovery via deep networks. AMIA Annu. Symp. Proc. 2015:677–686. 2015. [PMC free article] [PubMed] [Google Scholar]
- 103.Nemati S, Ghassemi MM, Clifford GD. Optimal medication dosing from suboptimal clinical examples: a deep reinforcement learning approach. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2016:2978–2981. doi: 10.1109/EMBC.2016.7591355. 2016. [DOI] [PubMed] [Google Scholar]
- 104.Rajpurkar P, Hannun AY, Haghpanahi M, Bourn C, Ng AY. Cardiologist-level arrhythmia detection with convolutional neural networks. arXiv. 2017 1707.01836. [Google Scholar]
- 105.ResearchKit. http://researchkit.org/
- 106.Mangravite L. Sage bionetworks in collaboration with The Michael J. Fox Foundation announce winners in the DREAM Parkinson's Disease Digital Biomarker Challenge. 2018. www.businesswire.com/news/home/20180117006187/en/Sage-Bionetworks-Collaboration-Michael-J.-Fox-Foundation
- 107.The Mood Challenge. 2017. www.moodchallenge.com/
- 108.Saint-Andre V, Federation AJ, Lin CY, et al. Models of human core transcriptional regulatory circuitries. Genome Res. 2016;26(3):385–396. doi: 10.1101/gr.197590.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ivanov M, Kals M, Kacevska M, Metspalu A, Ingelman-Sundberg M, Milani L. In-solution hybrid capture of bisulfite-converted DNA for targeted bisulfite sequencing of 174 ADME genes. Nucleic Acids Res. 2013;41(6):e72. doi: 10.1093/nar/gks1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hunter P, Coveney PV, De Bono B, et al. A vision and strategy for the virtual physiological human in 2010 and beyond. Philos. Trans. A Math. Phys. Eng. Sci. 2010;368(1920):2595–2614. doi: 10.1098/rsta.2010.0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhu F, Li XX, Yang SY, Chen YZ. Clinical success of drug targets prospectively predicted by in silico study. Trends Pharmacol. Sci. 2018;339(3):229–231. doi: 10.1016/j.tips.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 112.Kadurin A, Aliper A, Kazennov A, et al. The cornucopia of meaningful leads: applying deep adversarial autoencoders for new molecule development in oncology. Oncotarget. 2017;8(7):10883–10890. doi: 10.18632/oncotarget.14073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kadurin A, Nikolenko S, Khrabrov K, Aliper A, Zhavoronkov A. druGAN: an advanced generative adversarial autoencoder model for de novo generation of new molecules with desired molecular properties in silico . Mol. Pharm. 2017;14(9):3098–3104. doi: 10.1021/acs.molpharmaceut.7b00346. [DOI] [PubMed] [Google Scholar]
- 114.Blaschke T, Olivecrona M, Engkvist O, Bajorath J, Chen H. Application of generative autoencoder in de novo molecular design. Mol. Inform. 2017;37(1–2):1700123. doi: 10.1002/minf.201700123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Altae-Tran H, Ramsundar B, Pappu AS, Pande V. Low data drug discovery with one-shot learning. ACS Cent. Sci. 2017;3(4):283–293. doi: 10.1021/acscentsci.6b00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wen M, Zhang Z, Niu S, et al. Deep-learning-based drug-target interaction prediction. J. Proteome Res. 2017;16(4):1401–1409. doi: 10.1021/acs.jproteome.6b00618. [DOI] [PubMed] [Google Scholar]
- 117.Preuer K, Lewis RPI, Hochreiter S, Bender A, Bulusu KC, Klambauer G. DeepSynergy: predicting anticancer drug synergy with deep learning. Bioinformatics. 2018;34(9):1538–1546. doi: 10.1093/bioinformatics/btx806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mayr A, Klambauer G, Unterthiner T, Hochreiter S. DeepTox: toxicity prediction using deep learning. Front. Environ. Sci. 2016 [Google Scholar]
- 119.Kearnes S, Goldman B, Pande V. Modeling industrial ADMET data with multitask networks. arXiv. 2016 1606.08793. [Google Scholar]
- 120.Xu Y, Pei J, Lai L. Deep learning based regression and multiclass models for acute oral toxicity prediction with automatic chemical feature extraction. J. Chem. Inf. Model. 2017;57(11):2672–2685. doi: 10.1021/acs.jcim.7b00244. [DOI] [PubMed] [Google Scholar]
- 121.Sushko I, Salmina E, Potemkin VA, Poda G, Tetko IV. ToxAlerts: a web server of structural alerts for toxic chemicals and compounds with potential adverse reactions. J. Chem. Inf. Model. 2012;52(8):2310–2316. doi: 10.1021/ci300245q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Guo W, Xu YE, Feng X. DeepMetabolism: a deep learning system to predict phenotype from genome sequencing. bioRxiv. 2017 [Google Scholar]
- 123.Vougas K, Krochmal M, Jackson T, et al. Deep learning and association rule mining for predicting drug response in cancer. A personalised medicine approach. bioRxiv. 2017 [Google Scholar]
- 124.Gao M, Igata H, Takeuchi A, Sato K, Ikegaya Y. Machine learning-based prediction of adverse drug effects: an example of seizure-inducing compounds. J. Pharmacol. Sci. 2017;133(2):70–78. doi: 10.1016/j.jphs.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 125.Liang ZH, Huang JX, Zeng X, Zhang G. DL-ADR: a novel deep learning model for classifying genomic variants into adverse drug reactions. BMC Med. Genomics. 2016;9(Suppl. 2):48. doi: 10.1186/s12920-016-0207-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bughin J, Hazan E, Ramaswamy S, et al. Artificial intelligence the next digital frontier? McKinsey and Company Global Institute. 2017. www.mckinsey.com/∼/media/McKinsey/Industries/Advanced Electronics/Our Insights/How artificial intelligence can deliver real value to companies/MGI-Artificial-Intelligence-Discussion-paper.ashx
- 127.Sullivan F. Bioinformatics and advanced analytics powering drug discovery. 2017. www.researchandmarkets.com/reports/4308287/bioinformatics-and-advanced-analytics-powering
- 128.Benhenda M. Outsourcing AI for drug discovery: independent expertise is key to avoid overhyped claims. 2017. www.biopharmatrend.com/post/49-research-in-ai-for-drug-discovery-is-overhyped-and-what-to-do-about-it/