ABSTRACT
Background: Brown rats (Rattus norvegicus) may carry pathogens that can be a risk for public health. Brown rats in the Netherlands were tested for the zoonotic pathogens Leptospira spp. and Seoul hantavirus (SEOV), in order to obtain insight in their prevalence. Methods and results: Cross-sectional studies were performed at four locations from 2011 to 2015. The rats were tested for Leptospira spp. using real-time PCR and/or culture resulting in a prevalence ranging between 33–57%. Testing for SEOV was done through an adapted human Seoul hantavirus ELISA and real-time RT-PCR. Although at several locations the ELISA indicated presence of SEOV antibodies, none could be confirmed by focus reduction neutralization testing. Conclusion: The results indicate a widespread presence of Leptospira spp. in brown rats in the Netherlands, including areas with a low leptospirosis incidence in humans. No evidence for circulation of SEOV was found in this study.
KEYWORDS: Leptospirosis, Seoul virus, hantavirus, Rattus norvegicus, epidemiology, prevalence
Introduction
Rodents are known to carry pathogens that may be a risk for public health [1–4]. Despite this, little is known about the prevalence and geographical distribution of zoonotic rodent-borne pathogens in the Dutch rodent population. This therefore limits opportunities for preventive measures and complicates the risk- assessment of transmission to humans.
Leptospirosis is (re-)emerging globally and numerous outbreaks have occurred worldwide during the past decade [5], resulting in a significant health burden, especially in resource-poor tropical countries [6]. In the Netherlands, human leptospirosis has been notifiable since 1928 [7]. In 2014 and 2015, 60 and 44 autochthonous human cases were notified respectively, representing a four-fold increase compared with 2010–2013 (mean: 13, range: 9–17 cases) [8–10]. Since rodent and Leptospira spp. survival are facilitated by warm weather, the increase in human leptospirosis cases could possibly be explained by 2014 and 2015 being the warmest years on record [11,12]. Areas with a high incidence of human leptospirosis can be found in the central and northern parts of the Netherlands. Most of the autochthonous Leptospira spp. cases are related to recreational, e.g. swimming, or occupational activities, mostly among farmers [8,9]. About half of the autochthonous cases from 2013–2015 for whom the probable infecting serogroup could be deduced were infected with serovars belonging to the serogroup Icterohaemorrhagiae. For this serogroup, rats are considered the main reservoir. Rats are considered a major source of Leptospira infection to humans through direct and indirect transmission, e.g. through contact of mucous membranes or broken skin with water or moist soil which is contaminated with the urine of Leptospira infected animals. However, reports on the presence of pathogenic Leptospira spp. and the various serovars in rats in the Netherlands are scarce. One report from 1934 quotes prevalences in rats ranging from 11–56%, emphasizing local differences [13]. In muskrats (Ondatra zibethicus), leptospires were isolated from 24 of 327 (7%) muskrats caught in the Netherlands [14].
In addition to pathogenic Leptospira spp., Rattus spp. may also carry Seoul virus (SEOV). SEOV is a hantavirus, a genus of mainly zoonotic RNA viruses that are carried, amongst others, by rodents [15]. Human SEOV infections often lead to mild or no disease [16] but some people may develop a form of haemorrhagic fever with renal syndrome, which has a case-fatality rate of 1–2% [17] Compared to Puumala virus, another hantavirus which is presumably responsible for a higher number of clinical cases in the Netherlands, the clinical manifestations of SEOV are more severe and give a higher mortality [17]. In Europe, a few severe clinical human cases of SEOV infections have been confirmed in the UK and France, where the source of infection was established as pet rats [18,19]. Furthermore, a serosurvey among farmers exposed to wild rats showed a seroprevalence of 8% in the UK [20]. Since 2000, the presence of SEOV in wild brown rats was demonstrated in Belgium, France and the UK [18,21–23]. In the Netherlands, SEOV infection was detected in wild brown rats captured in the area around Doetinchem in 2013 [24]. There has been no evidence for autochthonous human cases from wild rats yet [25], while some cases of SEOV in humans have occurred since 2016, with pet rats or feeder rats as the source of infection. Human hantavirus infections have been notifiable in the Netherlands since 2008.
To obtain insight in Leptospira spp., brown rats from four geographic areas in the Netherlands were tested for its presence. This allows comparison between prevalence in rats within areas of high and low incidence of human leptospirosis (Figure 1), as well as the potential differences between urban and rural locations. To study the anticipated presence of SEOV in the Netherlands, these rats were also tested for SEOV. This paper describes the results of these studies.
Figure 1.
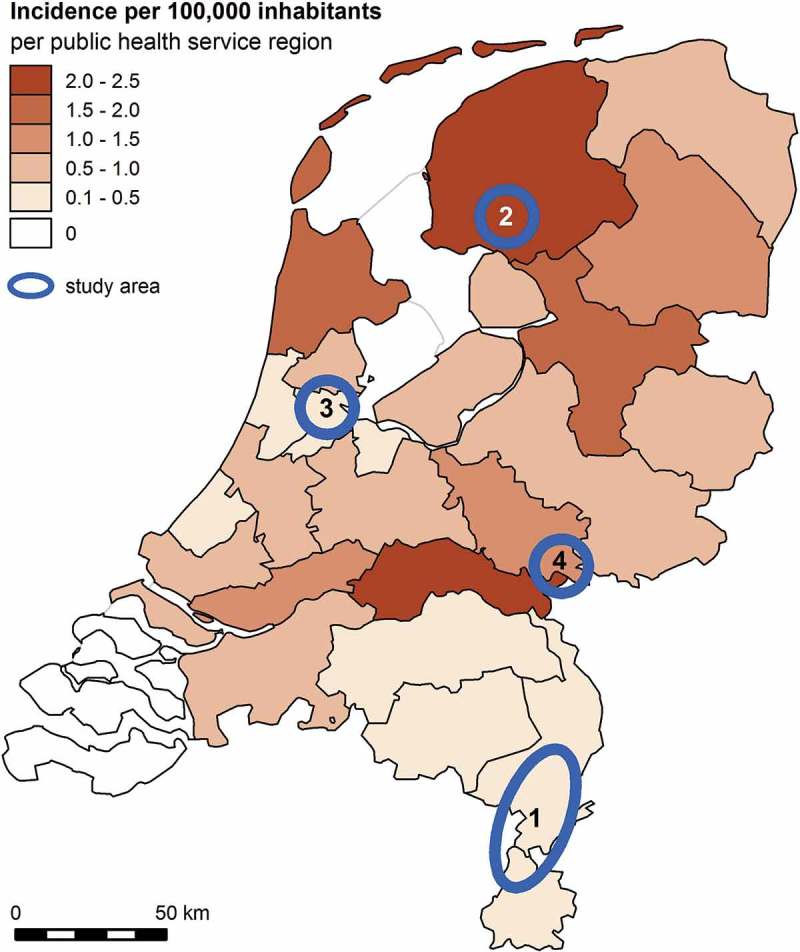
Incidence of autochthonous human leptospirosis cases in the Netherlands 2011–2015 by public health service region. Study areas are indicated with blue circles: 1: Limburg; 2 Friesland; 3 Amsterdam; 4 Nijmegen/Doetinchem.
Methods
Monitoring studies
From 2011–2015, four cross-sectional studies on Leptospira spp. and SEOV in brown rats were performed. From December 2011 to May 2012 the presence of Leptospira spp. and SEOV was studied in Limburg, from May 2012 to July 2013 in Friesland, from January 2014 to August 2015 in Amsterdam city center and from June to November 2015 in a rural area between Nijmegen and Doetinchem. Based on the geographic distribution of autochthonous human leptospirosis cases, Friesland is a region with a high risk for Leptospira spp. exposure and Limburg a region with a low risk (Figure 1). Amsterdam was selected, because it is a potential high exposure urban region with an increasing use of recreational water-rich and green spaces. Finally, Doetinchem was selected in the last phase of the project because SEOV positive rats were reported in this area [24].
Rats
Within the study area, locations were selected based on known presence of rats. Because the total rat populations were unknown, the capturing of rats was continued for each study area until about 50 rats (for Amsterdam 30 rats) were captured or when capturing efforts were not effective anymore. Rats were captured outside buildings by employees of a professional rat control agency using live traps (Killgerm Inc., Turnhout, Belgium) baited with food, though rat infestations were not necessarily present. Rats were transported alive to either the National Institute of Public Health and the Environment (RIVM) (rats from Limburg, Amsterdam, Doetinchem) or Wageningen Bioveterinary Research (WBVR) (rats from Friesland). Procedures at WBVR were similar to those at RIVM. Rats were anesthetized using isoflurane (RIVM) or CO2 (WBVR), weighed, and blood was collected, after which they were euthanized by cervical dislocation or isoflurane overdosing. Rats were classified in age groups (adult, subadult, juvenile) according to their appearance and weight. During post mortem inspection, kidney and urine samples were collected to test for Leptospira spp. and lungs were collected to test for SEOV. All procedures were approved by the Dutch Animal Ethics Committee (DEC project numbers 200900164, 201200208, and 201500089).
Leptospira spp. detection
Rats were tested by real-time PCR and/or culture, and rats were considered positive if one of the tests was positive. The diagnostic methods differed slightly between the four field studies: culture was introduced after the first study, because it enabled additional serotyping. In Limburg (2011–2012), kidneys were collected and were stored in 90% ethanol and sent to the National Collaborating Centre for Reference and Research on Leptospirosis (NRL) in Amsterdam for real-time PCR testing for leptospires. In Friesland (2012–2013), after euthanization of the rat, one of its kidneys, and if possible also urine, was immediately added to liquid EMJH culture medium [26]. The next day, the culture tubes were sent to the NRL for further incubation. Additionally, of 18 rats, a kidney sample in 90% ethanol was sent to the NRL for real-time PCR testing for leptospires. In Amsterdam (2014–2015) and Nijmegen-Doetinchem (2015), kidneys from all captured rats were both tested by culture as well as by real-time PCR at the NRL. At the NRL, culturing, real-time PCR analyses and Leptospira species determination were performed by previously described methods [27]. Prevalence rates and confidence intervals were determined using R version 3.2.0 [28].
Seoul hantavirus diagnostics
For the detection of SEOV IgG in rat sera, a human SEOV ELISA (Hantavirus Dobrava/Hantaan IgG Elisa, Progen Biotechnik GmbH, Heidelberg, Germany) was adapted to enable detection of rat IgG. Rabbit-α-rat IgG horseradish peroxidase labeled (Sigma–Aldrich Chemie B.V. Zwijndrecht, The Netherlands) was used as conjugate at a 1:5,000 dilution. Samples were tested in duplicate and the average optical density (OD) value of the samples was used for analyses. The frequency distribution of the ODc-values of field samples was analyzed and the optimal cut-off value was determined by use of a binary mixture model [29]. This model used ELISA results of 161 brown and 61 black rats captured from 2008–2010, of which a subset was confirmed by an in-house ELISA performed at the Belgian Reference Laboratory for Vector-borne diseases, by focus reduction neutralization test (FRNT) [30] performed at the Swedish Institute for Communicable Disease Control, by immuno-fluorescence assay (IFA) based on SEOV infected cells and/or by real-time RT-PCR for SEOV on total RNA isolated from lung tissue, as previously described [31,32].
In the current study, if a positive ELISA result was obtained, lung tissue was tested by a SEOV–specific real-time RT-PCR [33]. For the rats from Nijmegen-Doetinchem, this real-time RT-PCR was performed on all samples, to be confident that no early-stage infections were missed. This was only performed for the rats of this location, because of the findings of SEOV in rats in this area by Verner-Carlsson et al. [24]. Three samples from Limburg with positive ELISA results were tested by FRNT.
Results
Rats
The numbers of live rats captured were as follows: 42 in Limburg (2011–2012), 24 in Friesland (2012–2013), 31 in Amsterdam (2014–2015) and 53 in Nijmegen-Doetinchem (2015).
Leptospira spp. detection
In Limburg, 14 of 42 (33%) rats tested positive for Leptospira spp. by real-time PCR. Sequence analysis of three positive PCR products showed Leptospira interrogans species.
Of the 24 rats captured in Friesland, 8 (33%) tested positive for Leptospira spp.: 7 (of 24) were positive by culture and 5 (of 18) were positive by real-time PCR, including four that were positive in both culture and real-time PCR. All the obtained Leptospira isolates were serotyped as serovar Icterohaemorrhagiae belonging to serogroup Icterohaemorrhagiae.
Twelve of 31 (39%) rats captured in Amsterdam were positive by culture, including 10 rats that were also positive by real-time PCR. The positive isolates were identified as serovar Icterohaemorrhagiae (6) or serovar Copenhageni (5) belonging to serogroup Icterohaemorrhagiae and one isolate was lost.
Of the 53 rats captured in Nijmegen-Doetinchem, 30 (57%) tested positive for Leptospira spp.: 27 were positive by culture and 26 were positive by real-time PCR, including 23 that were positive in both culture and real-time PCR. The isolates were typed as serovar Icterohaemorrhagiae (9/27) and serovar Copenhageni (18/27) belonging to serogroup Icterohaemorrhagiae. Results are summarized in Table 1.
Table 1.
Results of Leptospira spp. and SEOV diagnosed in cohorts of rats in four geographical regions in the Netherlands. Leptospira spp. results are based on combined results of culture and RT-PCR. The SEOV results are based on ELISA and RT-PCR of seropositive animals. Of the Nijmegen-Doetinchem study, all animals were tested by SEOV RT-PCR. Seropositive results for SEOV could not be confirmed by RT-PCR or a neutralization test, and were therefore considered negative.
|
Leptospira spp. |
SEOV |
|||||||
|---|---|---|---|---|---|---|---|---|
| Study area | Number of brown rats tested | Nr. of positives/number tested (prevalence; 95% confidence interval)a | Culture positive/number tested | PCR positive/number tested | Serovar species (number) | Nr. of positives/number tested (prevalence; 95% confidence interval) | ELISA positives/number tested (average OD values of positive samples) |
RT-PCR positives/number tested |
| 1 Limburg | 42 (34 in 2011, 8 in 2012) | 14/42 (33%; 21–49%) | Not done | 14/42 | L. interrogans (3)b | 0/42 (0%; 0–8%) | 6/42 (2.4; 1.3; 0.9; 0.8; 1.9; 2.0) | 0/6 |
| 2 Friesland | 24 (7 in 2012, 17 in 2013) | 8/24 (33%; 18–53%) | 7/24 | 5/18 | serovar Icterohaemorrhagiae (7) (serogroup Icterohaemorrhagiae) |
0/24 (0%; 0–14%) | 1/24 (0.8) |
0/1 |
| 3 Amsterdam | 31 (9 in 2014, 22 in 2015) | 12/31 (39%; 24–56%) | 12/31 | 10/31 | serovar Icterohaemorrhagiae (6) and serovar Copenhageni (5) (serogroup Icterohaemorrhagiae) | 0/31 (0%; 0–11%) | 0/31 | Not done |
| 4 Nijmegen-Doetinchem | 53 (2015) | 30/53 (57%; 43–69%) | 27/53 | 26/53 | serovar Icterohaemorrhagiae (9/27) and serovar Copenhageni (18/27) (serogroup Icterohaemorrhagiae) | 0/53 (0%; 0–7%) | 3/53 (0.8;1.0;1.1) |
0/53 |
a Combined prevalence, culture and PCR results if available.
b Three positive amplicons were further sequenced as being derived from L. interrogans.
Seoul hantavirus
Based on the cut-off (0.74) determined with the binomial mixture model, six rats from Limburg, one rat from Friesland and three rats from Nijmegen-Doetinchem were serologically positive for SEOV by the adapted human SEOV IgG ELISA. No rats from Amsterdam were seropositive for SEOV. Because extensive cross-reactivity exists within the genus hantavirus and due to limited validation of the in-house adapted SEOV ELISA on wild rats, serum samples from three seropositive rats from Limburg were tested further by FRNT in Uppsala, Sweden, for the presence of SEOV neutralizing antibodies. The presence of SEOV specific IgG could not be confirmed by FRNT and none of the lung tissues from these rats tested positive in the real-time RT-PCR for SEOV RNA either. Results are summarized in Table 1.
Discussion
Brown rats were captured at four locations in the Netherlands to obtain information about the prevalence and geographical distribution of Leptospira spp. and SEOV. The prevalence of Leptospira spp. ranged between 33–57%. Serovars Copenhageni and Icterohaemorrhagiae were isolated, which are known to belong to species Leptospira interrogans in the Netherlands.
It was unexpected to find the same prevalence of leptospires (33%) in rats from Limburg and Friesland, since Friesland has a much higher leptospirosis incidence in humans than Limburg. Possible explanations might be (1) more awareness amongst Frisian physicians for human leptospirosis than in Limburg, (2) a higher exposure risk because of water recreational differences between Friesland and Limburg or (3) differences in environmental conditions, such as type of ground soil, which could influence Leptospira spp. survival. Also, it needs to be taken into account that sample sizes differed between locations and that there may have been seasonal or yearly variation in Leptospira spp. circulation. Furthermore, in Limburg rats were only tested by real-time PCR of the kidney, whereas in the other studies culture was also performed, which could potentially influence the prevalence found.
The relatively high prevalence of Leptospira spp. found in different areas in the Netherlands could be relevant for public health. General practitioners and other medical professionals need to be aware of possible human cases of leptospirosis. This knowledge also emphasizes the importance of personal protection for people that occupationally come into contact with rats or frequent wet outdoor environments. Since the increase in human leptospirosis cases in 2014, various actions have been undertaken to enhance awareness and ensure early detection and reporting of new cases. For example, alerts at the start of leptospirosis season (July-September) were sent in a weekly report by the Netherlands Early Warning Committee to professionals working in the field of infectious diseases in the Netherlands. In addition, veterinarians were informed through a Dutch veterinary journal and microbiologists through Labinf@ct, a message service for professionals working in medical microbiological laboratories.
Although currently most human leptospirosis cases contract the disease outside cities, the Leptospira spp. prevalence of 39% in rats in Amsterdam suggests there is also a risk of infection within cities. This may be particularly relevant for (large) swimming events in the city, which are becoming increasingly popular, and for smaller swim play areas that are present in the city.
Although results of the in-house adapted SEOV ELISA suggest the presence of SEOV antibodies in sera of rats captured in Limburg, Friesland and Nijmegen-Doetinchem, the presence of SEOV could not be confirmed by genome detection or virus neutralization tests. The latter two tests are considered as confirmation of an infection, although they depend on samples taken within a limited time-frame from the start of infection. Because of this, there is no confirmation of SEOV infection in the brown rats in this study.
Recently, the first detection of SEOV in wild brown rats in the Netherlands was reported, with 3 of 16 (19%) rats testing positive for SEOV specific antibodies and viral RNA [24]. The rats in our study in 2015 were captured in a larger area, encompassing the area where these three rats were captured. However, we were unable to detect SEOV. One potential explanation is that SEOV is only present in very focal areas, as patchy distributions are also known for other hantaviruses depending on habitat connectivity of their hosts [34].
All four studies were cross-sectional studies, which limits further analyses on for example the influence of seasonal and yearly changes in Leptospira spp. prevalence in rats. However, the results do indicate a high prevalence of Leptospira spp. in brown rats throughout the Netherlands. SEOV was found to be either absent, present at low prevalence, or present in specific hot-spots only.
Funding Statement
The studies were financed by the Netherlands Food and Consumer Product Safety Authority and the Ministry of Health, Sports and Welfare.
Acknowledgments
The study of rats in Nijmegen-Doetinchem was performed in collaboration with the Union of Water Boards (Unie van Waterschappen). We thank the collaboration of van Eck bedrijfshygiëne, in particular Johan van Rooij, for their excellent work trapping live rats. We also like to thank Yvonne Dijkstra from WBVR for successfully performing the first crucial steps in the microbiological analyses for Leptospira. We are grateful to Ben Bom, RIVM, for preparing the map.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1]. Backhans A, Fellström C.. Rodents on pig and chicken farms - a potential threat to human and animal health. Infect Ecol Epidemiol. 2012;2(1):17093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2]. Meerburg BG, Singleton GR, Kijlstra A. Rodent-borne diseases and their risks for public health. Crit Rev Microbiol. 2009;35(3):221–7. [DOI] [PubMed] [Google Scholar]
- [3]. van de Giessen AW, van Santen-Verheuvel MG, Hengeveld PD, et al. Occurrence of methicillin-resistant Staphylococcus aureus in rats living on pig farms. Prev Vet Med. 2009;91(2–4):270–273. [DOI] [PubMed] [Google Scholar]
- [4]. Easterbrook JD, Kaplan JB, Vanasco NB, et al. A survey of zoonotic pathogens carried by Norway rats in Baltimore, Maryland, USA. Epidemiol Infect. 2007;135(07):1192–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5]. World Health Organization Report of the second meeting of the leptospirosis burden epidemiology reference group. Geneva: World Health Organization; 2011. [Google Scholar]
- [6]. Torgerson PR, Hagan JE, Costa F, et al. Global burden of leptospirosis: estimated in terms of disability adjusted life years. PLoS Negl Trop Dis. 2015;9(10):e0004122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7]. van Vliet J. History of notification [in dutch]). Tijdschrift Voor Infectieziekten. 2009;4(2):51–60. [PubMed] [Google Scholar]
- [8]. Pijnacker R, Goris M, Te Wierik M, et al. Marked increase in leptospirosis infections in humans and dogs in the Netherlands, 2014. Euro Surveillance: Bulletin Européen Sur Les Maladies Transmissibles= European Communicable Disease Bulletin. 2016;21:17. [DOI] [PubMed] [Google Scholar]
- [9]. Goris M, Boer KR, Duarte T, et al. Human leptospirosis trends, the Netherlands, 1925-2008. Emerg Infect Dis. 2013;19(3):371–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10]. Uiterwijk M, de Rosa M, Friesema I, et al. Staat van zoonosen 2015. RIVM report. 2016. (2016–2139).
- [11]. Photiadou C, van der Schrier G, van Oldenborgh GJ, et al. Warmest years on record in Europe. In: EURO4M, editor. 2014. KNMI, The Netherlands and Deutscher Wetterdienst, Germany. 2015. [Google Scholar]
- [12]. van den Besselaar E, Cornes R, Photiadou C, et al. Joint warmest year on record in Europe. In: EURO4M, editor. 2015. KNMI, The Netherlands.2016. [Google Scholar]
- [13]. Schüffner W. Recent work on leptospirosis. Trans R Soc Trop Med Hyg. 1934;28(1):7–31. [Google Scholar]
- [14]. Steinen ACM, Schuurman JL, Gravekamp C, et al. Muskrats as carriers of pathogenic leptospires in The Netherlands. Antonie Van Leeuwenhoek. 1992;61(1):43–50. [DOI] [PubMed] [Google Scholar]
- [15]. Reusken C, Heyman P. Factors driving hantavirus emergence in Europe. Curr Opin Virol. 2013;3(1):92–99. [DOI] [PubMed] [Google Scholar]
- [16]. Hart CA, Bennett M. Hantavirus infections: epidemiology and pathogenesis. Microbes Infect. 1999;1(14):1229–1237. [DOI] [PubMed] [Google Scholar]
- [17]. Goeijenbier M, Verner-Carlsson J, Van Gorp E, et al. Seoul hantavirus in brown rats in the Netherlands: implications for physicians-epidemiology, clinical aspects, treatment and diagnostics. Neth J Med. 2015;73(4):155–160. [PubMed] [Google Scholar]
- [18]. Jameson L, Logue C, Atkinson B, et al. The continued emergence of hantaviruses: isolation of a Seoul virus implicated in human disease, UK, October 2012. Euro Surveill. 2013;18(1):4–7. [PubMed] [Google Scholar]
- [19]. Macé G, Feyeux C, Mollard N, et al. Severe Seoul hantavirus infection in a pregnant woman, France, October 2012. Euro Surveill. 2013;18:20464. [PubMed] [Google Scholar]
- [20]. Jameson LJ, Newton A, Coole L, et al. Prevalence of antibodies against hantaviruses in serum and saliva of adults living or working on farms in Yorkshire, UK. Viruses. 2014;6(2):524–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21]. Heyman P, Plyusnina A, Berny P, et al. Seoul hantavirus in Europe: first demonstration of the virus genome in wild rattus norvegicus captured in France. Eur J Clin Microbiol Infect Dis. 2004;23(9):711–717. [DOI] [PubMed] [Google Scholar]
- [22]. Heyman P, Baert K, Plyusnina A, et al. Serological and genetic evidence for the presence of Seoul hantavirus in rattus norvegicus in Flanders, Belgium. Scand J Infect Dis. 2009;41(1):51–56. [DOI] [PubMed] [Google Scholar]
- [23]. Dupinay T, Pounder KC, Ayral F, et al. Detection and genetic characterization of Seoul virus from commensal brown rats in France. Virol J. 2014;11(1):32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24]. Verner-Carlsson J, Lõhmus M, Sundström K, et al. First evidence of Seoul hantavirus in the wild rat population in the Netherlands. Infect Ecol Epidemiol. 2015;5(1):27215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25]. Goeijenbier M, Hartskeerl RA, Reimerink J, et al. The hanta hunting study: underdiagnosis of Puumala hantavirus infections in symptomatic non-travelling leptospirosis-suspected patients in the Netherlands, in 2010 and April to November 2011. Euro Surveill. 2014;19(32):20878. [DOI] [PubMed] [Google Scholar]
- [26]. World Health Organization Human leptospirosis: guidance for diagnosis, surveillance and control. Geneva: World Health Organization and the International Leptospirosis Society; 2003. [Google Scholar]
- [27]. Ahmed A, Engelberts MF, Boer KR, et al. Development and validation of a real-time PCR for detection of pathogenic leptospira species in clinical materials. PLoS One. 2009;4(9):e7093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28]. R Core Team R: A language and environment for statistical computing. Version 3.2.0. Vienna, Austria: R Foundation for Statistical Computing; 2015. [Google Scholar]
- [29]. Opsteegh M, Teunis P, Mensink M, et al. Evaluation of ELISA test characteristics and estimation of toxoplasma gondii seroprevalence in dutch sheep using mixture models. Prev Vet Med. 2010;96(3–4):232–240. [DOI] [PubMed] [Google Scholar]
- [30]. Lundkvist A, Hukic M, Horling J, et al. Puumala and dobrava viruses cause hemorrhagic fever with renal syndrome in bosnia-herzegovina: evidence of highly cross-neutralizing antibody responses in early patient sera. J Med Virol. 1997;53(1):51–59. [PubMed] [Google Scholar]
- [31]. Reusken C. Towards a monitoring and surveillance system for rodent-borne diseases in the Netherlands. RIVM Letter report 145/10 LZO/CR. 2010.
- [32]. Reusken C, Maas M, De Vries A, et al. Rodents and rodentborne zoonoses: towards a generic rodent monitoring in the Netherlands. RIVM progress report 006/13 Z&O. 2012. (- available upon request).
- [33]. Kramski M, Meisel H, Klempa B, et al. Detection and typing of human pathogenic hantaviruses by real-time reverse transcription-PCR and pyrosequencing. Clin Chem. 2007;53(11):1899–1905. [DOI] [PubMed] [Google Scholar]
- [34]. Olsson GE, Leirs H, Henttonen H. Hantaviruses and their hosts in Europe: reservoirs here and there, but not everywhere? Vector Borne Zoonotic Dis. 2010;10(6):549–561. [DOI] [PubMed] [Google Scholar]


