Abstract
Intestinal epithelial cells (IECs) absorb nutrients, respond to microbes, provide barrier function and help coordinate immune responses. We profiled 53,193 individual epithelial cells from mouse small intestine and organoids, and characterized novel subtypes and their gene signatures. We showed unexpected diversity of hormone-secreting enteroendocrine cells and constructed their novel taxonomy. We distinguished between two tuft cell subtypes, one of which expresses the epithelial cytokine TSLP and CD45 (Ptprc), the pan-immune marker not previously associated with non-hematopoietic cells. We also characterized how cell-intrinsic states and cell proportions respond to bacterial and helminth infections. Salmonella infection caused an increase in Paneth cells and enterocytes abundance, and broad activation of an antimicrobial program. In contrast, Heligmosomoides polygyrus caused an expansion of goblet and tuft cell populations. Our survey highlights new markers and programs, associates sensory molecules to cell types, and uncovers principles of gut homeostasis and response to pathogens.
Introduction
The intestinal mucosa dynamically interacts with the external milieu. Intestinal epithelial cells sense luminal contents and pathogens and secrete regulatory products that orchestrate appropriate responses. However, we do not yet know all the discrete epithelial cell types and sub-types in the gut; their molecular characteristics; how they change during differentiation; or respond to pathogenic insults.
A survey of RNA profiles of individual intestinal epithelial can help address these questions. Previous surveys that relied on known markers to purify cell populations1,2 cannot always fully distinguish between cell types, may identify only subsets of types in mixed populations or fail to detect rare cellular populations or intermediate states. Recent studies3–7 attempted to overcome these limitations using single-cell RNAseq (scRNA-seq), but have not yet extensively characterized intestinal epithelial cellular diversity.
Here, we perform a scRNA-seq survey of 53,193 epithelial cells of the small intestine (SI) in homeostasis and during infection. We identify gene signatures, key transcription factors (TFs) and specific G protein-coupled receptors (GPCRs) for each major small intestinal differentiated cell type. We distinguish proximal and distal enterocytes and their stem cells, establish a novel classification of different enteroendocrine subtypes, and identify previously unrecognized heterogeneity within both Paneth and tuft cells. Finally, we demonstrate how these cell types and states adaptively change is response to different infections.
Results
A single-cell census of SI epithelial cells
We profiled 53,193 individual cells (Supplementary Table 1) across the study. First, we used droplet-based massively-parallel single-cell RNA-Seq8 (Methods) to profile EpCAM+ epithelial cells from the small intestine of C57BL/6 wild-type and Lgr5-GFP knock-in mice1 (Fig. 1a). We estimated the required number based on a negative binomial model for random sampling (Methods). If we conservatively assume that 50 sampled cells are required to detect a subset, profiling 6,873 cells would allow us to detect all known IEC types and a hypothetical additional type present at 1% with 95% probability (Methods). We collected 8,882 profiles, removed 1,402 low quality cells (Methods) and 264 contaminating immune cells (Methods), retaining 7,216 cells for subsequent analyses (Extended Data Fig. 1a), with excellent reproducibility (n=6 mice, mean r=0.95, Extended Data Fig. 1c–f).
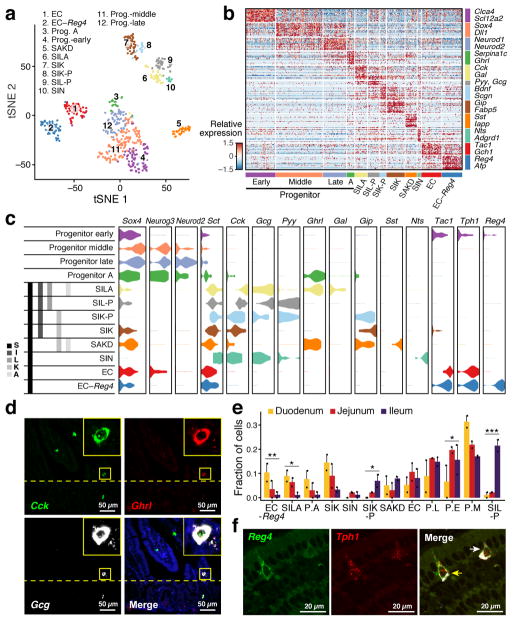
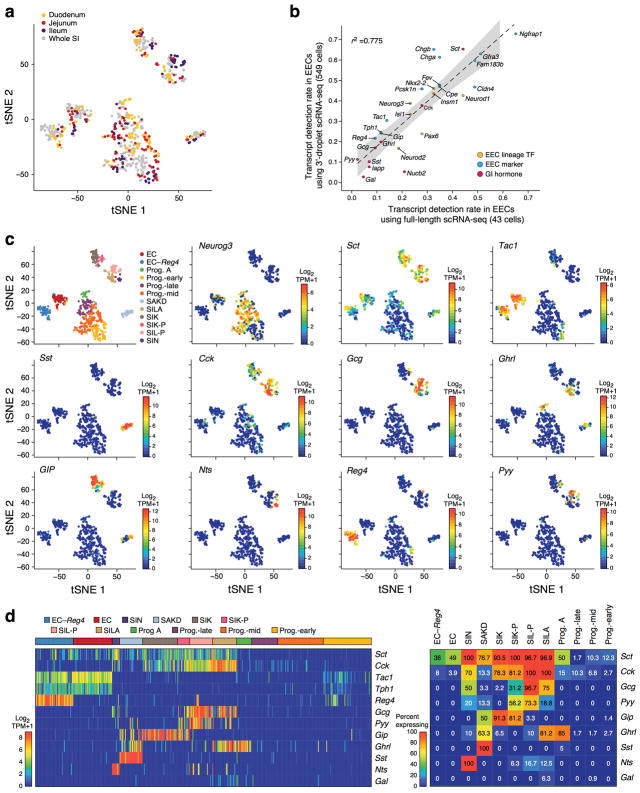
Figure 1. A single-cell expression survey of IECs.
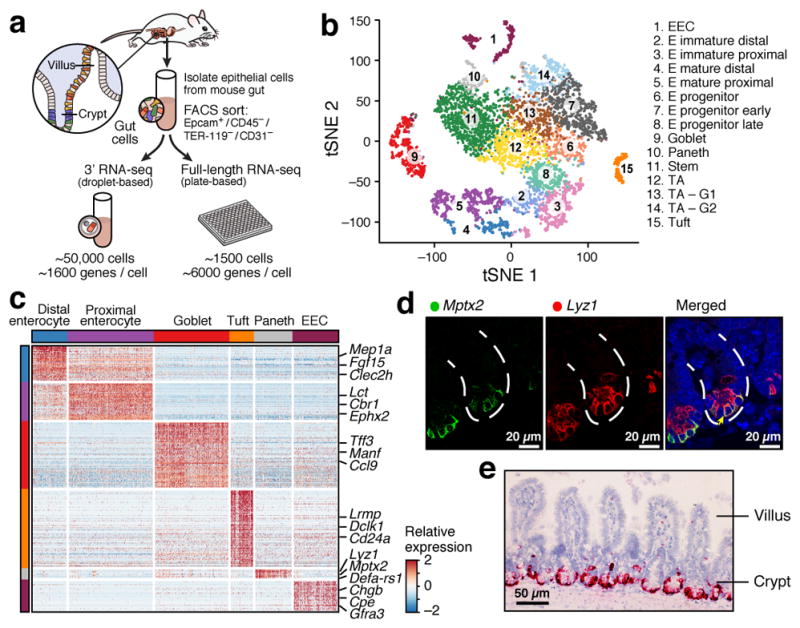
a. Overview. b. Cell type clusters. tSNE of 7,216 single cells (points), colored by cluster assignment (n=6 mice). E: Enterocyte. c. Cell type signatures. Relative expression level (row-wise Z-score of log2(TPM+1), color bar) of genes (rows) across cells (columns), sorted by types (color code). d,e. Mptx2 is a novel Paneth cell marker. (d) Combined smFISH of Mptx2 (green) and immunofluorescence assay (IFA) of the Paneth cell marker Lyz1 (red). Dashed line: Crypt, arrow: Paneth cell. Scale bar: 20μm. (e) In situ hybridization (ISH) of Mptx2 (red). Scale bar: 50μm.
Unsupervised graph clustering9,10 (Methods) partitioned the cells into 15 groups, which we visualized using t-stochastic neighborhood embedding10,11 (tSNE) (Fig. 1b), and labeled post hoc by the expression of known marker genes (Extended Data Fig. 1g). Each cluster was associated with a distinct cell type or state, including enterocyte (E), goblet, Paneth, enteroendocrine (EECs) and tuft cells (Fig. 1b). We identified proliferating cells using a cell-cycle signature12. The enteroendocrine, Paneth, goblet, stem and tuft cells were each represented by a single distinct cluster (Fig. 1b and Extended Data Fig. 1g). Absorptive enterocytes were partitioned across seven clusters representing distinct stages of maturation (Fig. 1b, Extended Data Fig. 1g). The proportions of most differentiated IEC types were consistent with expected abundances given our crypt-enriched isolation (Methods, Extended Data Fig. 1d), though Paneth cells were under-represented13 (3.6%), and enteroendocrine and tuft cells were higher than expected14,15 (4.3% and 2.3% respectively). To improve Paneth cell capture, we devised a sorting strategy to better capture large cells. Profiling an additional 10,396 epithelial cells identified 1,449 Paneth cells (13.9%) in two distinct clusters (Extended Data Fig. 3a), but no additional novel cell-types. We thus expect that all cell-types with >0.75% prevalence were detected in our survey at 99% confidence.
We validated our droplet-based data by independently analyzing 1,522 epithelial cells using full-length scRNA-seq16, with much higher coverage per cell (Fig. 1a, Extended Data Fig. 1b and 2a). Clustering (Methods) identified 8 clusters, which were generally congruent with the droplet-based clusters (Extended Data Fig. 2a) but without finer distinctions among the enterocytes - as expected given the smaller number of cells10.
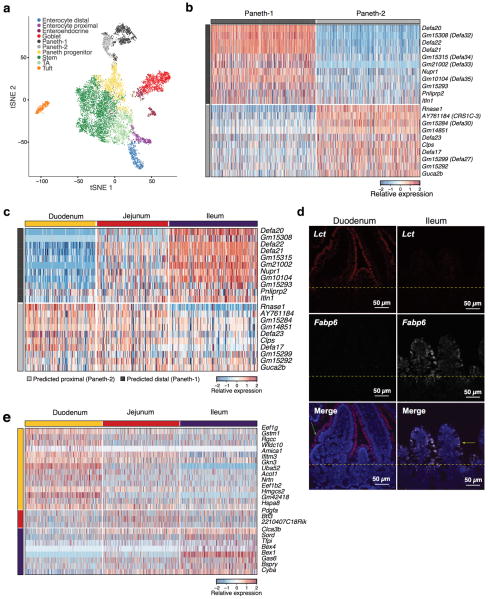
We then defined consensus expression signatures for each cell-type using both scRNA-seq datasets (Methods), highlighting known and novel markers (Fig. 1c, Extended Data Fig. 2b and Supplementary Tables 2–4). For example, the Paneth cell signature included Mptx2, a mucosal pentraxin with unknown function17 (Fig. 1c, Extended Data Fig. 2b,c, Supplementary Table 4), which we validated by single-molecule fluorescence in situ hybridization (smFISH, Methods, Fig. 1d,e). In the full-length scRNA-seq dataset, we also identified Paneth-specific expression of Mptx1 (FDR<0.001, Mann-Whitney U-test, Supplementary Table 3). Other Pentraxins, such as C-reactive protein (CRP) and serum amyloid P component protein (SAP), help defend against pathogenic bacteria18. In addition, the two Paneth cell subsets expressed distinct panels of anti-microbial alpha-defensins (Extended Data Fig. 3b).
Next, from the full-length scRNA-seq data, we identified enriched TFs, GPCRs and leucine-rich repeat (LRR) proteins (Methods) for each of the major cell-types (Extended Data Fig. 2d–f and Supplementary Table 5). Among TFs, these included Klf4, a known regulator of goblet cell development19, and novel Krüppel-like factors, including Klf15 in Paneth cells and Klf3 and Klf6 in tuft cells (Extended Data Fig. 2f). Among cell-type enriched GPCRs (Extended Data Fig. 2d,f and Supplementary Table 5), each of the sensory cell types (tuft and EECs) had more than 10 enriched receptors. These included many nutrient-sensing receptors (e.g., Gpbar1-a, a bile acid receptor20, and Gpr119, a sensor for food intake and glucose homeostasis21) in enteroendocrine cells, and Drd3, a dopamine receptor in tuft cells (Extended Data Fig. 2d). Pattern recognition receptors containing LRR domains were also variably expressed across subsets (Extended Data Fig. 2e).
Regional cell type diversity
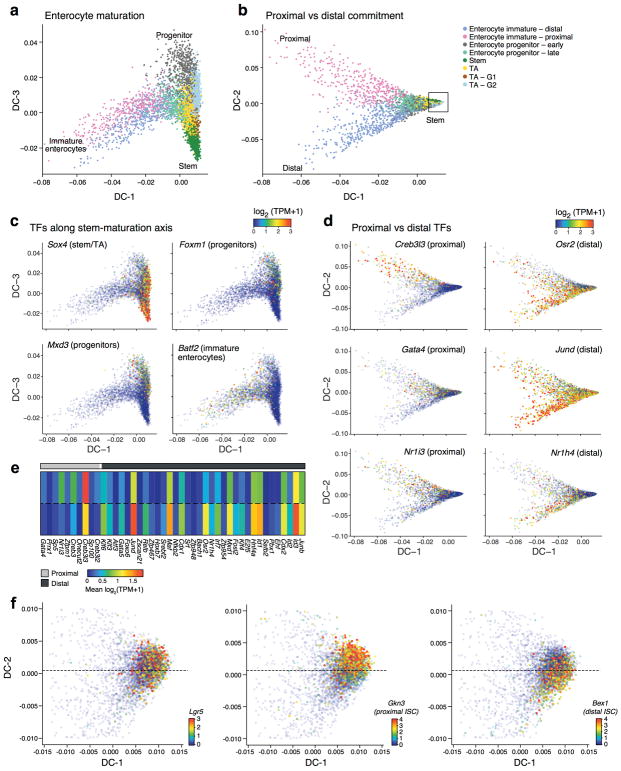
We next used diffusion maps22 to place the abundant population of enterocytes in pseudo-temporal order (Extended Data Fig. 4a–d), observing a trajectory from stem-like to progenitor to immature enterocytes (Extended Data Fig. 4a,c), and capturing (DC-2) distinct paths towards enterocytes of the proximal (duodenum and jejunum) and distal (ileum) small intestine (Extended Data Fig. 4b,d). By identifying TFs expressed in different regions of the diffusion map (Methods), we associated regulators with absorptive lineage commitment (known: Sox423, and novel: Batf2, Mxd3 and Foxm1) (Extended Data Fig. 4c,e), or with proximal vs. distal intestinal identity (known: Gata4, Nr1h424 and novel: Creb3l3, Jund, Osr2, Nr1i3; Extended Data Fig. 4d).
To test these predictions, in an independent experiment, we profiled 11,665 single cells from epithelial tissue extracted separately from the duodenum, jejunum and ileum (n=2 mice, Fig. 2a). Cells span a continuum that reflects both regional and differentiation ordering (Fig. 2a). Two separable subsets of differentiated enterocytes were populated by cells from either the duodenum or ileum (jejunum cells contributed to both). The signature genes for mature proximal and distal enterocytes that we identified computationally (Methods, Fig. 1c and Supplementary Table 2), were also differentially expressed between cells isolated separately from these regions (FDR < 0.05 Mann-Whitney U-test; Fig. 2b), and confirmed by smFISH (Extended Data Fig. 3d). Most marker genes of the two Paneth cell subsets (Extended Data Fig. 3b) were enriched (FDR<0.05) in proximal or distal gut respectively, confirming that they reflect regional distinctions (Extended Data Fig. 3c); however, the novel marker Mptx2 showed no regional specificity (Supplementary Table 10). Finally, the stem cells in each region also express region-specific markers (Extended Data Fig. 3e), which when examined in either the non-regional (Extended Data Fig. 4f) or the regional (Fig. 2c) diffusion maps mark distinct ISC subsets, each likely foreshadowing the eventual distinct enterocytes from the corresponding region (Fig. 2c).
Figure 2. Regional variation in cell types and differentiation.
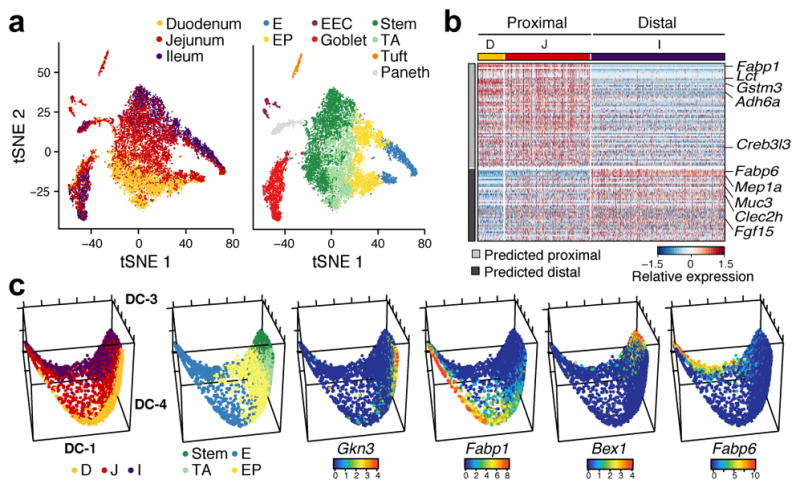
a. Regional surveys. tSNE of 11,665 cells from the duodenum, jejunum and ileum, colored by region (left) or post-hoc annotation (right). n=2 mice. b. Regional enterocyte signatures. Relative expression of genes (rows) across cells (columns), sorted by region. c. Regional differences in ISC differentiation. Diffusion-map embedding of 8,988 cells colored by region (left), cluster (center left), or expression of novel regional markers of ISCs (Gkn3, Bex1) or enterocytes (Fabp1, Fabp6). E: Enterocyte, EP: Enterocyte progenitor.
EEC subsets taxonomy and characterization
Enteroendocrine cells (EECs) are key sensors of nutrients and microbial metabolites14,25 that secrete diverse hormones and function as metabolic signal transduction units26. EECs have been reported to comprise 8 distinct sub-classes, such that cells expressing Sct, Cck, Gcg or GIP are traditionally termed S, I, L and K cells14. However, significant crossover between traditional subtypes has been observed14,27.
To define putative EEC subtypes, we partitioned the 553 EECs (Fig. 1b, 310 cells; Fig. 2a, 239 cells) into 12 clusters (Fig. 3a,b, Extended Data Fig. 5a, Supplementary Table 6, Methods). Four subsets expressed markers of EEC precursors (Neurog3, Neurod1, Sox4); the other eight represented mature EEC subsets. A recent study of scRNA-seq of organoid derived EECs showed EEC heterogeneity but with fewer EEC subsets4.
Figure 3. EEC taxonomy.
a. unsupervised clustering. tSNE of 533 EECs colored by sub-cluster. (n=8 mice) b. EEC subtype signatures. Relative expression of subtype-enriched genes (FDR < 0.01, rows) across cells (columns). c. Hormone-based EEC classification. Distribution of expression (x axis) of EEC TFs and hormones (columns) in cells from each subset (rows). Grey bars: traditional nomenclature by hormone expression. d. smFISH of Cck (green), Ghrl (red) and Gcg (white). Scale bar, 50μm. Inset (x5): triple-positive SILA cell e. Regional distribution of EEC subsets. Proportion (y axis) of each subset in the three regions (n=2 mice). P.A: Prog. -A, P.L: Prog. –late, P.E: Prog. –early, P.M: Prog. –middle. Error bars: SEM. * FDR<0.25, ** FDR<0.1, *** FDR<0.01, χ2 test (Methods) f. Enterochromaffin heterogeneity. smFISH of Reg4 (green) and Tph1 (red) co-stained with IFA of ChgA (white). Yellow and white arrows: Tph1+/Reg4+ and Tph1+/Reg4− EECs respectively. Scale bar: 20μm.
Comparing our ab initio subsets to the canonical classification (Fig. 3c, left), we found that several key hormones were expressed across multiple clusters (Extended Data Fig. 5c). Secretin (Sct), reported to be produced solely by S-cells14, was expressed by cells in all mature EEC subsets (Fig. 3c); cholecystokinin (Cck), the canonical marker for I-cells, was expressed in five subsets. This pattern was concordant in full-length scRNA-seq (Extended Data Fig. 5b).
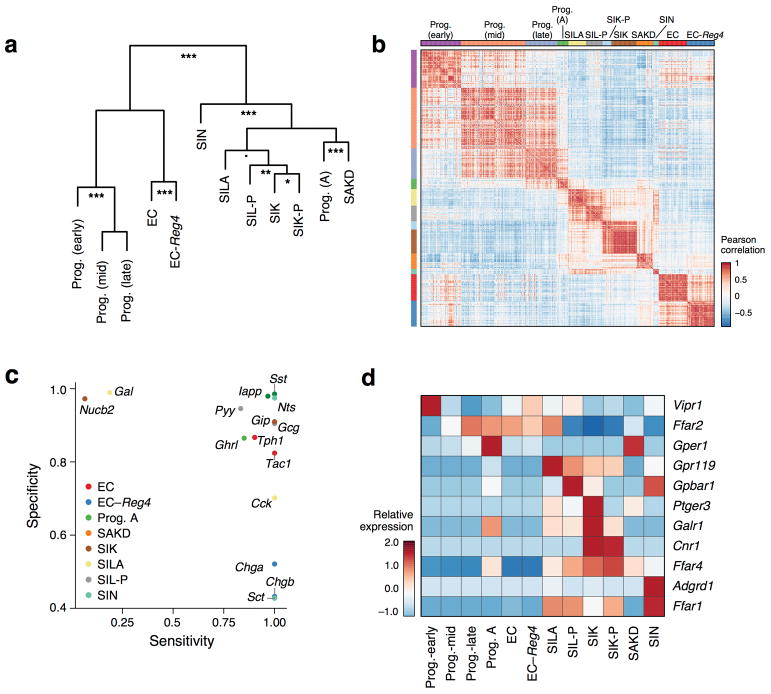
We placed each cluster in a new taxonomy (Fig. 3c and Extended Data Fig. 6a,b), and associated it with a canonical hormone if over 50% of cells expressed it (Extended Data Fig. 5d). Within each cluster, hormones were co-expressed in individual cells, without further partitioning (Extended Data Fig. 5c,d). Several hormones were subset-specific (Fig. 3c and Extended Data Fig. 6c): Galanin (Gal) to SILA, Neurotensin (Nts) to SIN, Nesfatin-1 (Nucb2) to SA, and Amylin (Iapp) and Somatostatin (Sst) to SAKD. Notably, we distinguished two subsets of enterochromaffin cells (ECs), which regulate gut motility and secretory reflexes28 (Fig. 3c and Extended Data Fig. 5c,d): one marked by Reg4 and Afp expression (“EC-Reg4”), whereas Reg4 is barely detectable in the other (“EC”) (Fig. 3b,c); we validated this in situ (Fig. 3f). The different subsets also vary in GPCR gene expression, which may reflect their role in luminal nutrient sensing (Extended Data Fig. 6d).
Some EEC subsets preferentially localized to specific regions (Fig. 3e). SILA, expressing ghrelin (Ghrl), the hunger hormone29, and proglucagon (Gcg, GLP-1), validated in situ (Fig. 3c,d) were enriched in the duodenum (FDR < 0.25, χ2 test, Methods), while SIL-P and SIK-P, both expressing the hormone peptide YY, which reduces appetite upon feeding30, were mainly found in the ileum (FDR < 0.1, χ2 test) (Fig. 3e and Extended Data Fig. 5a).
Two novel tuft cell subsets
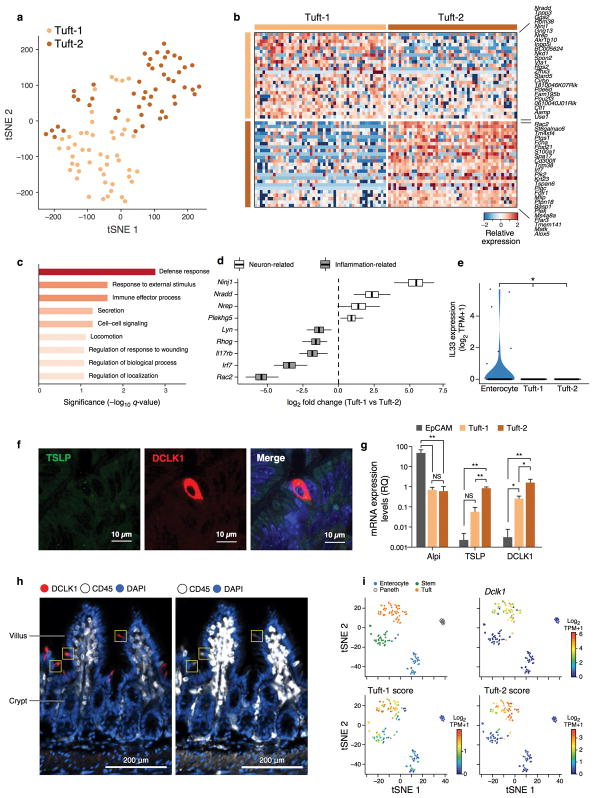
Tuft cells are the chemosensory cells of the gut and are enriched for taste-sensing molecules31. Recently, tuft cells were also shown to play a key role in the T helper 2 (Th2) response to Helminth infection, through Interleukin-25 (Il25)2,15,32. A previous tuft cell signature33 based on bulk profiles of Trpm5+ tuft cells contained both neuronal and inflammation gene programs; this could reflect either co-expression in the same cells or distinct subsets.
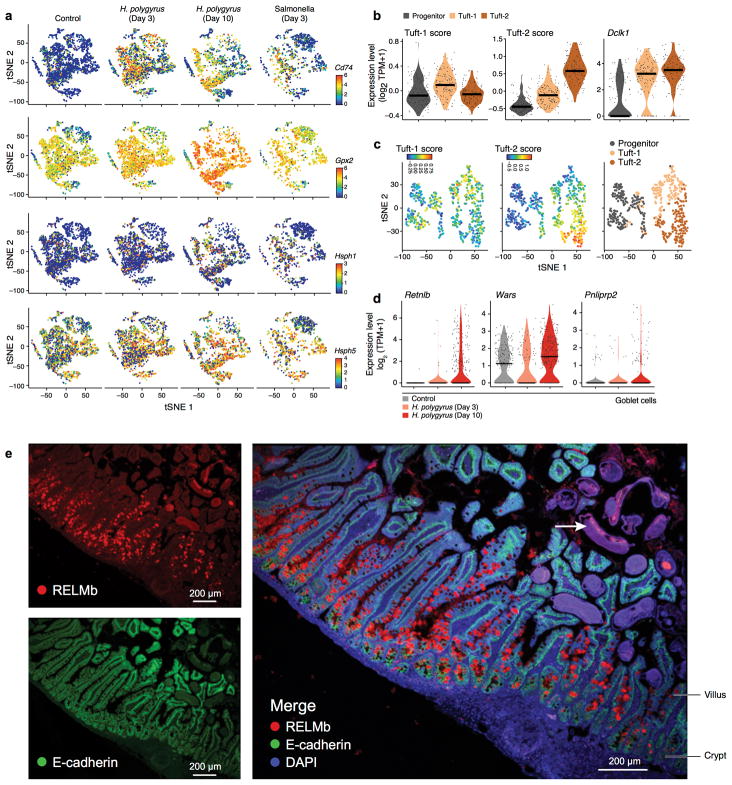
To distinguish these possibilities, we re-clustered the 166 cells in the 3′ droplet based tuft cell cluster (Fig. 1b, Extended Data Fig. 1g) into progenitors (early and late) and two mature tuft subsets (Methods), which we termed Tuft-1 and Tuft-2 (Fig. 4a). We confirmed the same sub-division in the tuft-cell enriched (CD24a+ sorted) full-length scRNA-seq dataset (Extended Data Fig. 7a). There was no significant distinction in Tuft-1 and Tuft-2 regional distribution (data not shown). We defined consensus signatures for the Tuft-1 and Tuft-2 clusters (FDR<0.01, Mann-Whitney U-test, Methods, Fig. 4b, Extended Data Fig. 7b and Supplementary Table 7).
Figure 4. CD45-positive Tuft-2 cells express TSLP.
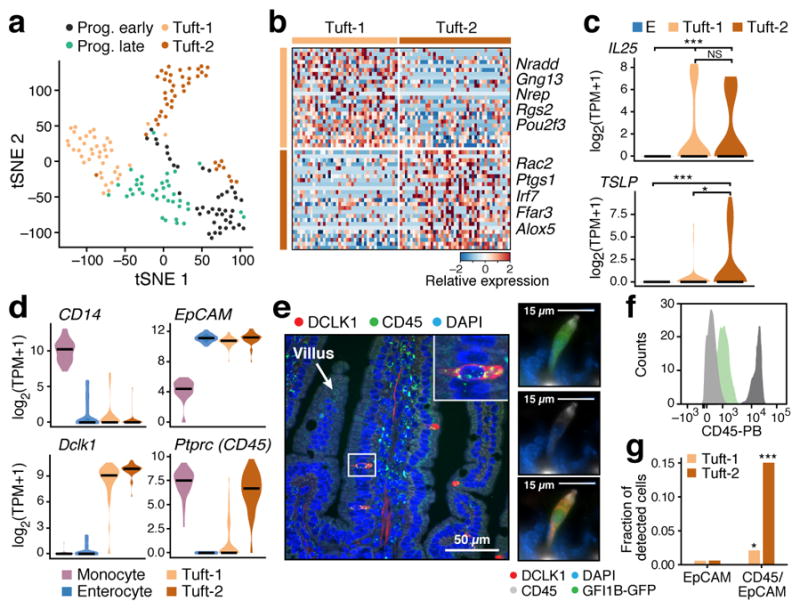
a. Tuft cell subsets. tSNE of 166 tuft cells colored by sub-cluster (n=6 mice). b. Tuft-1 and Tuft-2 gene signatures. Relative expression (in droplet-based data) of the top 25 genes (rows) for Tuft-1 and Tuft-2 cells (columns) (FDR < 0.01 in both datasets). c. Tuft-2 cells express TSLP. Distribution of expression of Il25 and TSLP in Enterocytes (E), Tuft-1 and Tuft-2 subsets (* FDR<0.1, *** FDR <0.0001, Mann-Whitney U-test). d–g. Tuft-2 cells express Ptprc (CD45). (d) Distribution of expression of Ptprc and known markers in indicated subsets (full-length scRNA-seq). (e) Left: smFISH of Ptprc (CD45, green) co-stained with DCLK1 antibody (red). Scale bar: 50μm. Right: IFA co-staining of DCLK1 (red), Gfi1b-GFP (green) and CD45 (white) in the same tuft cell. Merge in bottom panel. Scale bar: 15μm. (f) FACS histogram of CD45 protein levels in Gfi1b-GFP+ cells (green), background (light grey) and monocytes (dark grey). (g) Proportion (y axis) of tuft subsets in 3′ droplet scRNA-seq (n=3 pooled mice) of EpCAM+ (left) or EpCAM+/CD45+ cells (* p<0.05, *** p<0.0005, hypergeometric test).
The Tuft-2 cell signature was enriched for immune-related genes (FDR < 0.001, Extended Data Fig. 7c,d), whereas the Tuft-1 signature included genes related to neuronal development (Extended Data Fig. 7d). Thus, the inflammation and neuronal genes in the bulk signatures33 likely belonged to distinct cells.
Because tuft cells are important for communication with gut-resident immune cells2,15,32; we examined their expression of epithelial cytokine genes. Both subsets expressed Il25 (Fig. 4c), but neither expressed Il33 (Extended Data Fig. 7e). Importantly, Tuft-2 cells expressed significantly higher levels of the Th2 promoting cytokine, thymic stromal lymphopoietin (TSLP)34 (FDR<0.1, Mann-Whitney U-test, Fig. 4c), which we confirmed with smFISH and qPCR (Extended Data Fig. 7f,g). Tuft cells also specifically expressed receptors for the Th2-related cytokines Il4ra and Il13ra1 and for IL-25 (Il17rb), which could support autocrine signaling during Th2 responses (FDR < 0.05, Mann-Whitney U-test, Supplementary Table 2–4).
Surprisingly, Ptprc, encoding the pan-immune marker CD45, was expressed strongly and exclusively by Tuft-2 cells (Fig. 4d–f and Extended Data Fig. 7h). Consistently, Tuft-2 cells were strongly enriched in 3′ droplet-based scRNA-seq of EpCAM+/CD45+ cells (n=3 mice, Fig. 4g and Extended Data Fig. 7i, Methods). To our knowledge, this is the first finding of CD45+ cells from a non-hematopoietic lineage, and highlights the challenges related to even well-established markers.
Characterization of microfold (M) cells
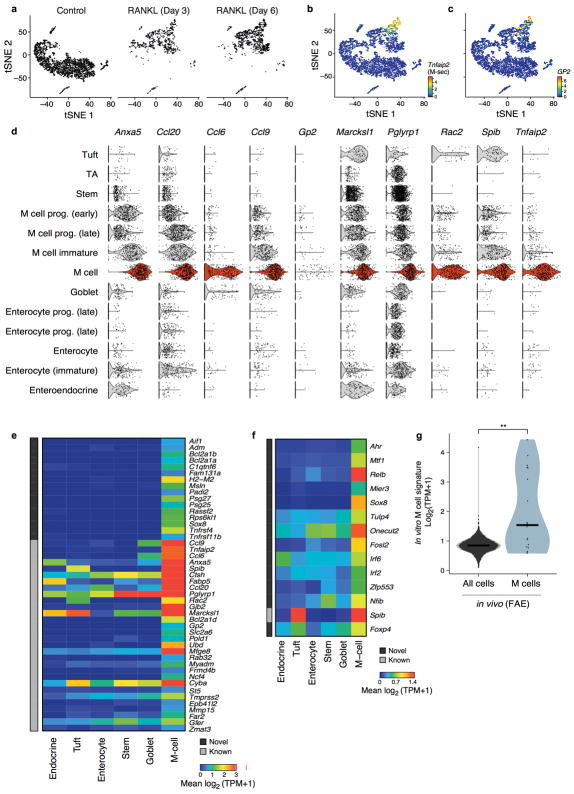
M cells are derived from Lgr5+ intestinal stem cells which reside in the rare follicle associated epithelia (FAE) of the small intestine35. Since M cells represent only about 10% of this rare structure36, they were not detected in our initial survey, as expected.
To identify and characterize M cells, we first used an ex vivo model of M cell differentiation, analyzing 5,434 cells from small intestinal organoids treated with RANKL35 for 0, 3, and 6 days (Fig. 5a,b, Extended Data Fig. 8a). We annotated a cluster of 378 cells (Fig. 5a, Methods) as differentiated M cells based on known marker gene expression37 (Extended Data Fig. 8b–d), and used it to construct in vitro M cell-specific signatures (Extended Data Fig. 8e,f, Supplementary Table 8, Methods).
Figure 5. Microfold (M) cell signatures.
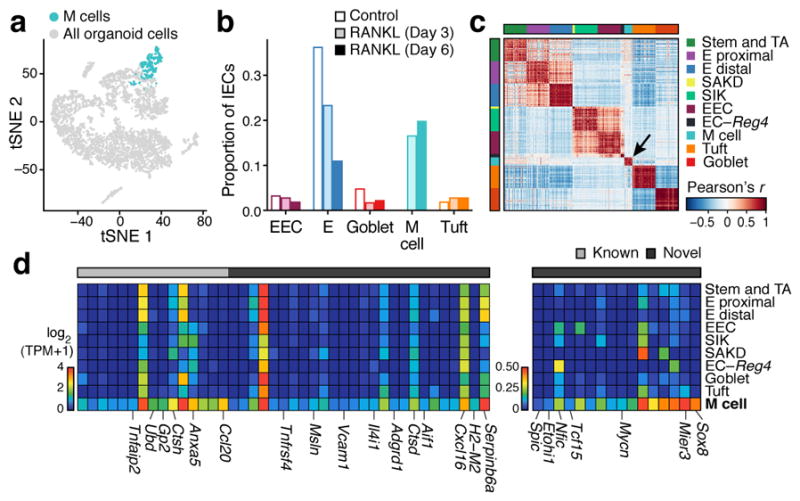
a,b. RANKL-induced organoid M cells. (a) 384 differentiated M cells (blue) in a tSNE of 5,434 epithelial cells from organoids (n=4 pooled wells per treatment). (b) Proportions (y axis) of IEC types (x axis) in control or RANKL-treated organoids. c–e. FAE M cells in vivo. (c) Pearson correlation coefficient (color bar) for each pair of 4,700 FAE cells (n=5 mice, large clusters down-sampled to 50 cells for visualization). Arrow: 18 M cells. (d). Mean expression (color bar) in each FAE cluster (rows) of genes (columns) for known (grey) or novel (black) markers (left) or TFs (right), enriched (FDR < 0.05, Mann-Whitney U-test) in M cells in vivo. E: enterocyte, EEC: enteroendocrine, EC: enterochromaffin.
We confirmed the in vivo relevance of these signatures by profiling 4,700 EpCAM+ cells from FAE of WT and Gfi1b-GFP labeled knock-in mice, a known marker for both tuft and M cells15,35 (n=5 mice). A cluster of 18 cells (Fig. 5c, Methods) was enriched for known M cell markers (FDR<0.05, Mann-Whitney U-test, Fig. 5d) and the in vitro M cell signature (p<10−4, Extended Data Fig. 8g). Next, we defined an in vivo signature of markers and TFs (Fig. 5d,e and Methods). Peyer’s patch M cells were indeed too rare to detect without specific FAE enrichment (only 1 of 7,216 cells in our initial sampling (Fig. 1b) was positive for the M cell signature). Thus, discovering any other, as yet unknown, subsets of cells of such exceptional rarity and unique location, would require additional stratification.
Epithelial response to pathogen infection
Immune and epithelial cell responses to pathogens play a key role in maintaining gut homeostasis38. We investigated the IEC responses to Salmonella enterica and to the parasitic helminth Heligmosomoides polygyrus. We profiled individual IECs using droplet-based 3′ scRNA-seq two days after Salmonella (n=2 mice, 1,770 cells) or 3 (n=2 mice, 2,121 cells) and 10 days (n=2 mice, 2,711 cells) after H. polygyrus infections and matched controls (n=4 mice, 3,240 cells). We also profiled 389 cells with full-length scRNA-seq. The response to each pathogen incorporated pathogen-specific and -shared changes in expression and shifts in cell proportions and cell-intrinsic programs.
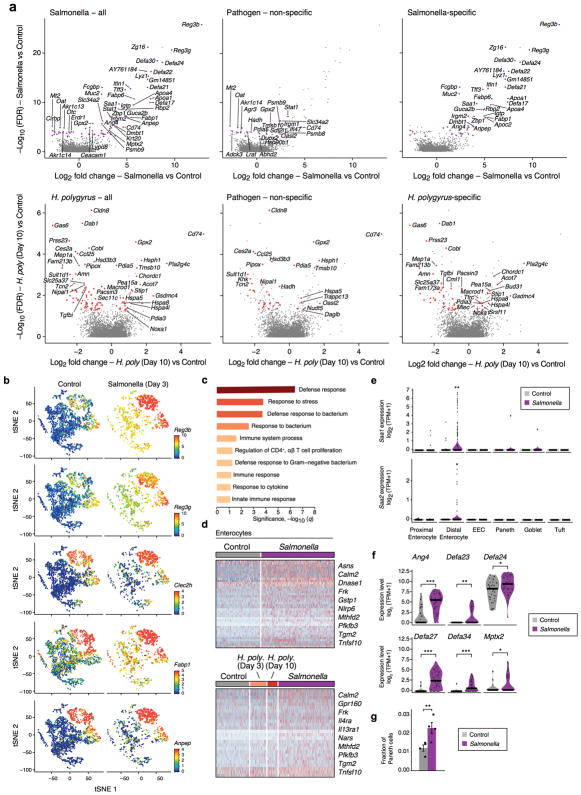
Salmonella-induced genes across all infected IECs (FDR<0.25, likelihood-ratio test, Extended Data Fig. 9a, top left and Supplementary Table 9) were enriched for pathways involved in defense response to bacterium (FDR<0.001, hypergeometric test Extended Data Fig. 9c), including Reg3b and Reg3g39, protective genes in Salmonella infection (Fig. 6c). Most H. polygyrus induced genes (62%) were specific to this pathogen and enriched for inflammatory response genes and tuft cell markers (FDR<0.25, likelihood-ratio test, Extended Data Fig. 9a, bottom and Supplementary Table 9). Other induced genes (112/571; 19%) comprised a non-specific, shared inflammatory response (FDR<0.25, likelihood-ratio test, Extended Data Fig. 9a, 10a middle panels and Supplementary Table 9). Stress gene modules were also up-regulated in stem cells following both Salmonella and day 10 helminth infection (FDR<0.05, data not shown).
Figure 6. Epithelial response to pathogen infections.
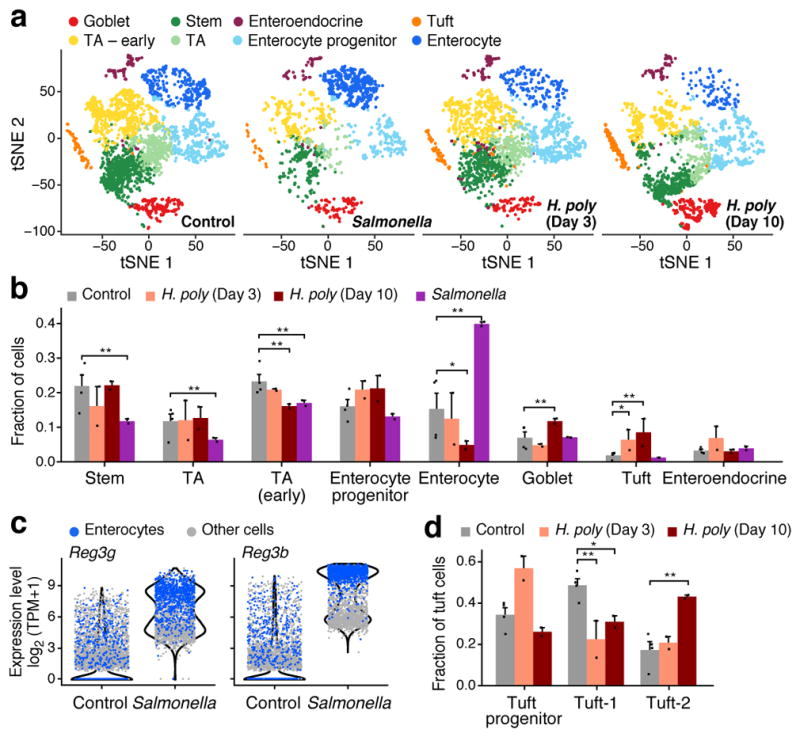
a,b. Changes in cell composition. (a) IEC subsets (colored by clusters) in control (n=4), Salmonella-infected (n=2), and helminth-infected mice (3 and 10 days; n=2 each). b. Frequencies (y axis) of each cell type in each mouse (dots) under each condition (* FDR < 10−5; ** FDR < 10−10, Wald test). Error bars: SEM. c. Anti-microbial lectin induction in Salmonella infection. Distribution of expression (y axis) in enterocytes (blue) and all other cells (grey). d. Shifts in tuft cell proportions in helminth infection. Frequencies (y axis) of each tuft subset in each mouse (dots, n=2 mice). Error bars: SEM. (* FDR < 0.25; ** FDR < 0.05, Wald test).
Additional responses to Salmonella were cell-type-specific: an increase in the expression of antimicrobial peptides (AMPs) and Mptx2 in Paneth cells (Extended Data Fig. 9f); 40 genes induced in enterocytes, mostly (65%) in a Salmonella-specific manner (Extended Data Fig. 9d, Methods) including the pattern-recognition receptor Nlrp6; and induction in distal enterocytes of the pro-inflammatory apolipoproteins Serum Amyloid A1 and 2 (Saa1 and Saa2)40 (Extended Data Fig. 9a,e). Some AMPs, such as Reg3a-g, that are normally enterocyte-specific were induced in all cell-types following Salmonella infection (Fig. 6c; Extended Data Fig. 9b and Supplementary Tables 2,3,9).
We distinguished the contribution of changes in cell-intrinsic expression programs vs. shifts in cell composition (determined by unsupervised clustering, Fig. 6a,b). Following Salmonella infection, the frequency of mature enterocytes increased substantially (from 13.1% on average in control to 21.7% in infection; Fig. 6b), whereas the proportion of TA (52.9% to 18.3%) and stem (20.7% to 6.4%) cells significantly decreased (FDR<10−10). In agreement with a previous study41, mature Paneth cell proportions also increased significantly (from 1.1% to 2.3%, FDR<0.01). (We used another 2,029 cells with sorting optimized to avoid loss of the large Paneth cells; Methods; n=4 infected mice, Extended Data Fig. 9f–g).
During infection with H. polygyrus there was a striking increase in the number of goblet cells, known to respond to the parasite42, and a reduction in enterocytes (Fig. 6b). Tuft cell proportions increased substantially at day three (1.9% to 6.3%, FDR<10−5, Wald test), and further by day 10 (to 8.5%, FDR<10−10, Wald test, Fig. 6b), with a significant increase of Tuft-2 cells within them by day 10 (17.2% to 43.0%, FDR<0.05, Wald test, Fig. 6d, Extended Data Fig. 10b,c). There were also cell-intrinsic changes: within goblet cells, induction of genes previously implicated in anti-parasitic immunity42 (FDR < 1×10−5, likelihood-ratio test; Extended Data Fig. 10d,e) some of which (e.g., Wars and Pnlipr2) were not previously known to be expressed by goblet cells.
Discussion
The intestinal epithelium is the most diverse epithelial tissue in the body. A high-resolution single-cell survey of the mouse intestinal epithelium revealed further diversity, as well as coherent cell-specific transcriptional programs, some revising canonical marker expression such as CD45, which we validated in situ and in prospectively isolated cells. For example, we discovered two subsets of tuft cells, expressing neuron-related and Th2-recruiting epithelial cytokines, respectively, which may provide insight into mechanisms underlying food allergies.
Our survey resolved the cellular populations that are implicated in key sensory pathways at high resolution. For example, we provide a detailed profile of the GPCRs expressed by IECs, including EEC subsets. Notably, the important cannabinoid receptor Gpr11921 was enriched in the novel SILA subset (FDR < 0.05, Extended Data Fig. 6d), which co-expresses Ghrl and Gcg, genes encoding gut hormones that regulate appetite and satiety. Tuft cells were also enriched for GPCR expression, supporting studies on their specialized chemosensory properties.
Although many studies have shown an expansion of goblet cells and, more recently, tuft cells in response to parasites2,15,34, our analysis revealed that this restructuring of the epithelial barrier is specific to the identity of the pathogen. Helminth infection led to a dramatic expansion of secretory cell-types, whereas Salmonella infection induced a strong expansion of absorptive enterocytes and Paneth cells. These compositional changes were accompanied and enhanced by cell-intrinsic changes to regulatory programs. Moreover, we uncovered a novel epithelial cell response to Salmonella, where the expression of genes that are cell-type-specific in homeostatic conditions was broadened across multiple cell-types during infection. Overall, our study provides a detailed reference dataset and specific hypotheses for follow-up studies, including cell-type-specific markers, TFs and GPCRs, which may lead to novel interventions in inflammatory, metabolic and proliferative gut pathologies.
Materials and Methods
Mice
All mouse work was performed in accordance with the Institutional Animal Care and Use Committees (IACUC) and relevant guidelines at the Broad Institute and MIT, with protocols 0055-05-15 and 0612-058-18, respectively. Seven to ten weeks old female or male wild-type C57BL/6J or Lgr5-EGFP-IRES-CreERT2 mice, obtained from the Jackson Laboratory (Bar Harbor, ME) or Gfi1beGFP/+ (Gfi1b-GFP)43 were housed under specific-pathogen-free (SPF) conditions at the Broad Institute, MIT or at the Harvard T.H. Chan School of Public Health animal facilities.
Salmonella enterica and H. polygyrus infection
C57BL/6J mice (Jackson Laboratory) were infected with 200 third-stage larvae of H. polygyrus or 108 Salmonella enterica at the laboratory of Dr. HN Shi, maintained under specific pathogen-free conditions at Massachusetts General Hospital (Charlestown, MA), with protocol 2003N000158. H. polygyrus was propagated as previously described44. Mice were sacrificed 3 and 10 days after H. polygyrus infection. For Salmonella enterica, mice were infected with a naturally streptomycin-resistant SL1344 strain of S. Typhimurium (108 cells) as described44 and were sacrificed 48 hours after infection.
Cell dissociation and crypt isolation
Crypt isolation
The small intestine of C57BL/6J wild-type, Lgr5-GFP or Gfi1b-GFP mice was isolated and rinsed in cold PBS. The tissue was opened longitudinally and sliced into small fragments roughly 2 mm long. The tissue was incubated in 20mM EDTA-PBS on ice for 90 min, while shaking every 30 min. The tissue was then shaken vigorously and the supernatant was collected as fraction 1 in a new conical tube. The tissue was incubated in fresh EDTA-PBS and a new fraction was collected every 30 min. Fractions were collected until the supernatant consistent almost entirely of crypts. The final fraction (enriched for crypts) was washed twice in PBS, centrifuged at 300g for 3 min, and dissociated with TrypLE express (Invitrogen) for 1 min at 37°C. The single cell suspension was then passed through a 40μm filter and stained for FACS sorting for either scRNA-seq method (below) or used for organoid culture. We confirmed the robustness of this method by testing additional single-cell isolation methods: either “whole” (scraping the epithelial lining) or “villus-enriched” (fraction 1, see above) and found that due to the high mortality rate (via anoikis) of post-mitotic differentiated cells – the primary component of which is mature enterocytes – crypt-enriched single-cell suspension represents faithfully the composition of the small intestine cell types (data not shown).
FAE isolation
Epithelial cells from the follicle-associated epithelia (FAE) were isolated by extracting small sections (0.2–0.5cm) containing Peyer’s patches from the small intestine of C57Bl/6J or Gfi1beGFP/+ mice.
Cell sorting
For plate-based scRNA-seq experiments, a fluorescence-activated cell sorting (FACS) machine (Astrios) was used to sort a single cell into each well of a 96-well PCR plate containing 5μl of TCL buffer with 1% 2-mercaptoethanol. For EpCAM+ isolation, cells were stained for 7AAD− (Life Technologies), CD45− (eBioscience), CD31− (eBioscience), Ter119− (eBioscience), EpCAM+ (eBioscience), and for specific epithelial cells we also stained for CD24+/− (eBioscience) and c-Kit+/− (eBioscience). To enrich for specific IEC populations, cells were isolated from Lgr5-GFP mice, stained with the antibodies mentioned above and gated on GFP-high (stem cells), GFP-low (TAs), GFP−/CD24+/c-Kit+/− (secretory lineages) or GFP−/CD24−/EpCAM+ (epithelial cells). For better Paneth cell recovery, we allowed higher side scatter and forward scatter parameters in combination with CD24+/c-Kit+ to verify Paneth cell recovery in EpCAM+ cells. For Tuft-2 isolation, epithelial cells from 3 different mice were stained as above only this time we used EpCAM+/CD45+ and sorted 2000 single cells. Note that we used a lenient sorting gate to ensure we obtained sufficient numbers of these rare Tuft-2 cells, which led to a higher contamination rate of T cells, which we removed later in our single cell analysis using unsupervised clustering.
For full length scRNA-seq sorting, the 96 well plate was sealed tightly with a Microseal F and centrifuged at 800g for 1 min. The plate was immediately frozen on dry ice and kept at −80°C until ready for the lysate cleanup. Bulk population cells were sorted into an Eppendorf tube containing 100μl solution of TCL with 1% 2-mercaptoethanol and stored at −80°C.
For droplet-based scRNA-seq, cells were sorted with the same parameters as described for plate-based scRNA-seq, but were sorted into an Eppendorf tube containing 50μl of 0.4% BSA-PBS and stored on ice until proceeding to the GemCode Single Cell Platform.
Plate-based scRNA-seq
Single cells
Libraries were prepared using a modified SMART-Seq2 protocol as previously reported16. Briefly, RNA lysate cleanup was preformed using RNAClean XP beads (Agencourt) followed by reverse transcription with Maxima Reverse Transcriptase (Life Technologies) and whole transcription amplification (WTA) with KAPA HotStart HIFI 2× ReadyMix (Kapa Biosystems) for 21 cycles. WTA products were purified with Ampure XP beads (Beckman Coulter), quantified with Qubit dsDNA HS Assay Kit (ThermoFisher), and assessed with a high sensitivity DNA chip (Agilent). RNA-seq libraries were constructed from purified WTA products using Nextera XT DNA Library Preperation Kit (Illumina). On each plate, the population and no-cell controls were processed using the same method as the single cells. The libraries were sequenced on an Illumina NextSeq 500.
Bulk samples
Bulk population samples were processed by extracting RNA with RNeasy Plus Micro Kit (Qiagen) per the manufacturer’s recommendations, and then proceeding with the modified SMART-Seq2 protocol following lysate cleanup, as described above.
Droplet-based scRNA-seq
Single cells were processed through the GemCode Single Cell Platform using the GemCode Gel Bead, Chip and Library Kits (10X Genomics, Pleasanton, CA), following the manufacturer’s protocol. Briefly, single cells were sorted into 0.4% BSA-PBS. An input of 6,000 cells was added to each channel of a chip with a recovery rate of 1,500 cells in average. The cells were then partitioned into Gel Beads in Emulsion (GEMs) in the GemCode instrument, where cell lysis and barcoded reverse transcription of RNA occurred, followed by amplification, shearing and 5′ adaptor and sample index attachment. Libraries were sequenced on an Illumina NextSeq 500.
Immunofluorescence and single-molecule fluorescence in situ hybridization
Immunofluorescence (IFA): staining of small intestinal tissues was conducted as described34. Briefly, tissues were fixed for 14 hours in formalin, embedded in paraffin and cut into 5 μm thick sections. Sections were deparaffinized with standard techniques, incubated with primary antibodies overnight at 4°C and then with secondary antibodies at RT for 30 min. Slides were mounted with Slowfade Mountant+DAPI (Life Technologies, S36964) and sealed.
Single-molecule fluorescence in situ hybridization (smFISH)
RNAScope Multiplex Flourescent Kit (Advanced Cell Diagnostics) was used per manufacturer’s recommendations with the following alterations. Target Retrieval boiling time was adjusted to 12 minutes and incubation with Protease IV at 40°C was adjusted to 8 minutes. Slides were mounted with Slowfade Mountant+DAPI (Life Technologies, S36964) and sealed.
Combined IFA and smFISH was implemented by first performing smFISH as described above, with the following changes. After Amp 4, tissue sections were washed in washing buffer, incubated with primary antibodies overnight at 4°C, washed in 1x TBST 3 times and then incubated with secondary antibodies for 30 min at room temperature. Slides were mounted with Slowfade Mountant+DAPI (Life Technologies, S36964) and sealed.
Image analysis
Images of tissue sections were taken with a confocal microscope Fluorview FV1200 using Kalman and sequential laser emission to reduce noise and signal overlap. Scale bars were added to each image using the confocal software FV10-ASW 3.1 Viewer. Images were overlaid and visualized using Image J software45.
Antibodies and probes
Antibodies used for IFA: rabbit anti-DCLK1 (1:200, Abcam ab31704), rat anti-CD45 (1:100, Biolegend 30-F11), goat anti-ChgA (1:100, Santa Cruz Sc-1488), mouse anti-E-cadherin (1:100, BD Biosciences 610181), rabbit anti-RELMβ (1:200, Peprotech 500-p215), rat anti-Lysozyme (1:200, Dako A0099), rat anti-CD45 (1:100, Biolegend 30-F11, cat: 103101), Alexa Fluor 488-, 594-, and 647-conjugated secondary antibodies were used and obtained from Life Technologies.
Probes used for single-molecule RNAscope (Advanced Cell Diagnostics)
Cck (C1), Ghrl (C2), GCG (C3), Tph1 (C1), Reg4 (C2), TSLP (C1), Ptprc (C1) and Mptx2 (C1).
Intestinal organoid cultures
Following crypt isolation, the single cell suspension was resuspended in Matrigel (BD Bioscience) with 1μM Jagged-1 peptide (Ana-Spec). Roughly 300 crypts embedded in 25μl of Matrigel were seeded onto each well of a 24-well plate. Once solidified, the Matrigel was incubated in 600μl culture medium (Advanced DMEM/F12, Invitrogen) with streptomycin/penicillin and glutamatax and supplemented with EGF (100 ng/mL, Peprotech), R-Spondin-1 (600ng/mL, R&D), Noggin (100ng/mL, Prepotech), Y-276432 dihydrochloride monohydrate (10μM, Tochris), N-acetyl-1-cysteine (1μM, Sigma-Aldrich), N2 (1X, Life Technologies), B27 (1X, Life Technologies) and Wnt3A (25ng/mL, R&D Systems). Fresh media was replaced on day 3, and organoids were passaged by dissociation with TrypLE and resuspended in new Matrigel on day 6 with a 1:3 split ratio. For selected experiments, organoids were additionally treated with RANKL (100 ng/mL, Biolegends). Treated organoids were dissociated and subjected to scRNA-seq using both methods.
Computational Analysis
Pre-processing of droplet (10X) scRNA-seq data
Demultiplexing, alignment to the mm10 transcriptome and UMI-collapsing were performed using the Cellranger toolkit (version 1.0.1) provided by 10X Genomics. For each cell, we quantified the number of genes for which at least one read was mapped, and then excluded all cells with either fewer than 800 detected genes. Expression values Ei,j for gene i in cell j were calculated by dividing UMI count values for gene i by the sum of the UMI counts in cell j, to normalize for differences in coverage, and then multiplying by 10,000 to create TPM-like values, and finally taking log transform to compute log2(TPM+1) values. Batch correction was performed using ComBat46 as implemented in the R package sva47, using the default parametric adjustment mode. The output was a corrected expression matrix, which was used as input to further analysis.
Selection of variable genes was performed by fitting a generalized linear model to the relationship between the squared co-efficient of variation (CV) and the mean expression level in log/log space, and selecting genes that significantly deviated (P<0.05) from the fitted curve, as previously described48.
Pre-processing of SMART-Seq2 scRNA-seq data
BAM files were converted to merged, de-multiplexed FASTQs using the Illumina provided Bcl2Fastq software package v2.17.1.14. Paired-end reads were mapped to the UCSC hg19 human transcriptome using Bowtie49 with parameters “-q --phred33-quals -n 1 -e 99999999 -l 25 -I 1 -X 2000 -a -m 15 -S -p 6”, which allows alignment of sequences with one mismatch. Expression levels of genes were quantified as using transcript-per-million (TPM) values calculated by RSEM50 v1.2.3 in paired-end mode. For each cell, we quantified the number of genes for which at least one read was mapped, and then excluded all cells with either fewer than 3,000 detected genes or a transcriptome-mapping of less than 40%. We then identified highly variable genes as described above.
Dimensionality reduction using PCA and tSNE
We restricted the expression matrix to the subsets of variable genes and high-quality cells noted above, and values were centred and scaled before input to PCA, which was implemented using the R function ‘prcomp’ from the ‘stats’ package for the SMART-seq2 dataset. For the droplet dataset, we used a randomized approximation to PCA, implemented using the ‘rpca’ function from the ‘rsvd’ R package, with the parameter k set to 100. This low-rank approximation was used as it is several orders of magnitude faster to compute for very wide matrices. Given that many principal components (PCs) explain very little of the variance, the signal to noise ratio can be substantially improved by selecting a subset of n ‘significant’ PCs. After PCA, significant PCs were identified using the permutation test described in 51, implemented using the ‘permutationPA’ function from the ‘jackstraw’ R package. This test identified 13 and 15 significant PCs in the 10X and SMART-Seq2 datasets of Fig. 1, respectively. Only scores from these significant PCs were used as the input to further analysis.
For visualization, the dimensionality of the datasets was further reduced using the ‘Barnes-hut’ approximate version of the t-distributed stochastic neighbor embedding (tSNE)52,53. This was implemented using the ‘Rtsne’ function from the ‘Rtsne’ R package using 20,000 iterations and a perplexity setting that ranged from 10 to 30 depending on the size of the dataset.
Identifying cell differentiation trajectories using diffusion maps
Prior to running diffusion-map dimensionality reduction we selected highly variable genes in the data as follows. We first fit a null model for baseline cell-cell gene expression variability in the data based on a power-law relationship between coefficient of variation (CV) and the mean of the UMI-counts of all the expressed genes, similar to 54. Next, we calculated for each gene the difference between the value of its observed CV and that expected by the null model (CVdiff). The histogram of CVdiff exhibited a “fat tail”. We calculated the mean μ and standard deviation σ of this distribution, and selected all genes with CVdiff > μ + 1.67σ, yielding 761 genes that were used for further analysis.
We performed dimensionality reduction using the diffusion map approach22. Briefly, a cell-cell transition matrix was computed using the Gaussian kernel where the kernel width was adjusted to the local neighborhood of each cell, following 55. This matrix was converted to a Markovian matrix after normalization. The right eigenvectors vi(i = 0,1,2,3,...) of this matrix were computed and sorted in the order of decreasing eigenvalues λi(i = 0,1,2,3,...) after excluding the top eigenvector v0, corresponding to λ0 = 1 (which reflects the normalization constraint of the Markovian matrix). The remaining eigenvectors vi(i = 0,1,2...) define the diffusion map embedding and are referred to as diffusion components (DCk(k = 1,2,...)). We noticed a spectral gap between the λ4 and the λ5, and hence retained DC1 − DC4, for both the initial dataset (Extended Data Fig. 4) and the data extracted from distinct intestinal regions (Fig. 2c).
Removing contaminating immune cells and doublets
Although cells were sorted prior to sequencing using EpCAM, a small number of contaminating immune cells were observed in the 10X dataset. These 264 cells were removed by an initial round of unsupervised clustering (density-based clustering of the tSNE map using ‘dbscan’ 56 from the R package ‘fpc’) as they formed an extremely distinct cluster. In the case of the SMART-Seq2 dataset, several cells were outliers in terms of library complexity, which could possibly correspond to more than one individual cell per sequencing library or ‘doublets’. These cells were then removed by calculating the top quantile 1% of the distribution of genes detected per cell and removing any cells in this quantile.
Cluster analysis
To cluster single cells by their expression, we used an unsupervised clustering approach, based on the Infomap graph-clustering algorithm9, following approaches recently described for single-cell CyTOF data57 and scRNA-seq10. Briefly, we constructed a k-nearest-neighbor (kNN) graph on the data using, for each pair of cells, the Euclidean distance between the scores of significant PCs to identify k nearest neighbors. The parameter k was chosen to be consistent with the size of the dataset. Specifically, k was set to 200 and 80 for the droplet dataset of 7,216 cells (Fig. 1a), the SMART-Seq2 dataset of 1,522 cells (Extended Data Fig. 2a). RANKL-treated organoids contained 5434 cells and k was set to 200, while the Salmonella and H. polygyrus dataset contained 9842 cells and k was set to 500. For cluster analyses within celltypes, specifically the EEC and tuft cell subsets, we used the Pearson correlation distance instead of Euclidean, and set k=15, k=30 and k=40 for the enteroendocrine subtypes (533 cells), and 166 and 102 tuft cells in the 10X and SMART-Seq2 datasets respectively. The nearest neighbor graph was computed using the function ‘nng’ from the R package ‘cccd’. The k-NN graph was then used as the input to Infomap9, implemented using the ‘infomap.community’ function from the ‘igraph’ R package.
Detected clusters were mapped to cell-types or intermediate states using known markers for intestinal epithelial cell subtypes. (Extended Data Fig. 1g and Extended Data Fig. 2a). In the case of the enteroendocrine cell (EEC) sub-analysis (Figure 3), any group of EEC progenitor clusters with average pairwise correlations between significant PC scores r>0.85 was merged, resulting in 4 clusters, which were annotated as Prog. (A) based on high levels of Ghrl and Prog. (early), (mid) and (late) – based on decreasing levels of stem (Slc12a2, Ascl2, Axin2) and cell-cycle genes and increasing levels of known EEC regulatory factors (Neurod1, Neurod2 and Neurog3) from early to late (Extended Data Fig. 5c). For the SMART-Seq2 dataset, two clusters expressing high levels of stem cell marker genes (Extended Data Fig. 2a) were merged to form a ‘Stem’ cluster and two other clusters were merged to form a ‘TA’ cluster.
For the cluster analysis of the follicle-associated epithelium (FAE) dataset of 4700 cells, the M cells were exceedingly rare (0.38%), and therefore the ‘ClusterDP’ method58 was used to identify them, as it empirically performed better than the kNN-graph algorithm on this dataset containing such a rare subgroup. As with the kNN methods, ClusterDP was run using significant (p<0.05) PC scores (19 in this case) as input, and was implemented using the ‘findClusters’ and ‘densityClust’ functions from the ‘densityClust’ R package using parameters rho=1.1 and delta=0.25.
Extracting rare cell-types for further analysis
The initial clustering of the whole-gut dataset (7,216 cells, Fig. 1b) showed a cluster of 310 EECs and 166 tuft cells. The tuft cells were taken ‘as is’ for the sub-analysis (Fig. 4a–b), while the EECs were combined with a second cluster of 239 EECs identified in the regional dataset (Fig. 2a, right) for a total of 533 EECs. A group of 16 cells co-expressed EEC markers Chga, Chgb with markers of Paneth cells including Lyz1, Defa5 and Defa22, and were therefore interpreted as doublets, and removed from the analysis, leaving 533 EECs, which were the basis for the analysis in Fig. 3. To compare expression profiles of enterocytes from proximal and distal small intestine (Fig. 2b), the 1,041 enterocytes identified from 11,665 cells in the regional dataset (Fig. 2a) were used.
Defining cell-type signatures
To identify maximally specific genes for cell-types, we ran differential expression tests between each pair of clusters for all possible pairwise comparisons. Then, for a given cluster, putative signature genes were filtered using the maximum FDR Q-value and ranked by the minimum log2 fold-change. The minimum fold-change and maximum Q-value represent the weakest effect-size across all pairwise comparisons, therefore this a stringent criterion. Cell-type signature genes shown in (Fig. 1c, Extended Data Fig. 8e, and Supplementary Tables 2–4 and 8) were obtained using a maximum FDR of 0.05 and a minimum log2 fold-change of 0.5.
In the case of signature genes for subtypes within cell-types (Fig 3b, Fig 4b and Extended Data Fig. 7b), a combined p-value (across the pairwise tests) for enrichment was computed using Fisher’s method - a more lenient criterion than simply taking the maximum p-value - and a maximum FDR Q-value of 0.01 was used, along with a cutoff of minimum log2 fold-change of 0.25 for tuft cell subsets (Fig. 4b, Extended Data Fig. 7b and Supplementary Table 7) and 0.1 for enteroendocrine subsets (Fig. 3b and Supplementary Table 6). Due to low cell numbers (n=18), Fisher’s combined p-value was also used for the in vivo M cell signature, with an FDR cutoff of 0.001 (Fig. 5d), Supplementary Table 8). Marker genes were ranked by minimum log2 fold-change. Differential expression tests were carried out using the Mann-Whitney U-test (also known as the Wilcoxon rank-sum test) implemented using the R function ‘wilcox.test’. For the infection experiments (Fig. 6), we used a two part ‘hurdle’ model to control for both technical quality and mouse-to-mouse variation. This was implemented using the R package MAST59, and p-values for differential expression were computed using the likelihood-ratio test. Multiple hypothesis testing correction was performed by controlling the false discovery rate60 using the R function p.adjust.
Scoring cells using signature gene sets
To obtain a score for a specific set of n genes in a given cell, a ‘background’ gene set was defined to control for differences in sequencing coverage and library complexity between cells in a manner similar to 12. The background gene set was selected to be similar to the genes of interest in terms of expression level. Specifically, the 10n nearest neighbors in the 2-D space defined by mean expression and detection frequency across all cells were selected. The signature score for that cell was then defined as the mean expression of the n signature genes in that cell, minus the mean expression of the 10n background genes in that cell.
Estimates of cell type sampling frequencies
For each cell-type the probability of observing at least n cells in a sample of size k is modeled using the cumulative distribution function of a negative binomial NBcdf(k, n, p), where p is the relative abundance of this cell type. For m cell types with the same parameter p the overall probability of seeing each type at least n times is NBcdf(k; n, p)^m. Such analysis can now be performed with user specified parameters at http://satijalab.org/howmanycells.
EEC dendrogram
Average expression vectors were calculated for all 12 EEC subset clusters, using log2(TPM+1) values, and restricted to the subset of 1,361 genes identified as significantly variable between EEC susbsets (p<0.05), as described above. The average expression vectors including these genes were hierarchically clustered using the R package pvclust (Spearman distance, ward.D2 clustering method), which provides bootstrap confidence estimates on every dendrogram node, as an empirical p-value over 100,000 trials (Extended Data Fig. 6a).
Cell-type specific TFs, GPCRs and LRRs
A list of all genes identified as acting as transcription factors in mice was obtained from AnimalTFDB 61, downloaded from: http://www.bioguo.org/AnimalTFDB/BrowseAllTF.php?spe=Mus_musculus. The set of G-protein coupled receptors (GPCRs) was obtained from the UniProt database, downloaded from: http://www.uniprot.org/uniprot/?query=family%3A%22g+protein+coupled+receptor%22+AND+organism%3A%22Mouse+%5B10090%5D%22+AND+reviewed%3Ayes&sort=score. Functional annotations for each protein (Extended Data Fig. 2d) were obtained from the The British Pharmacological Society (BPS) and the International Union of Basic and Clinical Pharmacology (IUPHAR) data, downloaded from: http://www.guidetopharmacology.org/GRAC/GPCRListForward?class=A. The list of leucine-rich repeat proteins (LRRs) was taken from 62. To map from human to mouse gene names, human and mouse orthologs were downloaded from Ensembl (latest release 86, http://www.ensembl.org/biomart/martview), and human and mouse gene synonyms from NCBI (ftp://ftp.ncbi.nlm.nih.gov/gene/DATA/GENE_INFO/Mammalia/). For each human LRR gene, all human synonyms were mapped to the orthologous gene in mouse using the ortholog list, and mouse gene names were mapped to those in the single-cell data using the synonym list.
Cell-type enriched TFs, GPCRs and LRRs were then identified by intersecting the list of genes enriched in to each cell type with the lists of TFs, GPCRs and LRRs defined above. Cell-type enriched genes were defined using the SMART-Seq2 dataset, as those with a minimum log2 fold-change of 0 and a maximum FDR of 0.5, retaining a maximum of 10 genes per cell type in Extended Data Fig. 2e,f, while complete lists are provided in Supplementary Table 5. In addition, a more extensive panel of cell-type specific GPCRs was identified (Extended Data Fig. 2d) by selecting a more lenient threshold. This was achieved by comparing each cell-type to all other cells, instead of the pairwise comparisons described in the previous section, and selecting all GPCR genes differentially expressed (FDR < 0.001).
Testing for changes in cell type proportions
We model the detected number of each cell-type in each analyzed mouse as a random count variable using a Poisson process. The rate of detection is then modeled by providing the total number of cells profiled in a given mouse as an offset variable, while the condition of each mouse (treatment or control) was provided as a covariate. The model was fit using the R command ‘glm’ from the ‘stats’ package. The p-value for the significance of the effect produced by the treatment was then assessed using a Wald test on the regression coefficient.
In the case of the assessment of the significance of spatial distributions of enteroendocrine (EEC) subsets (Fig. 3e), the comparison involved more than two groups. In particular, our null hypothesis was that the proportion of each EEC subset detected in the three intestinal regions (duodenum, jejunum, and ileum) was equal. To test this hypothesis, we used analysis of variance (ANOVA) with a χ2-test on the Poisson model fit described above, implemented using the ‘anova’ function from the ‘stats’ package.
Gene set enrichment and GO analysis
GO analysis was performed using the ‘goseq’ R package63, using significantly differentially expressed genes (FDR <0.05) as target genes, and all genes expressed with log2(TPM+1) > 3 in at least 10 cells as background.
Data Availability
All data is deposited in GEO (GSE92332) and in the Single Cell Portal for visualization and download (https://portals.broadinstitute.org/single_cell).
Code Availability
R markdown scripts enabling the main steps of the analysis to be performed will be made available on request.
Extended Data
Extended Data Figure 1. Identifying intestinal epithelial cell-types in scRNA-seq data by unsupervised clustering, related to Figure 1.
a,b. Quality metrics for scRNA-seq data. Shown are distributions of the number of reads per cell (left), the number of genes detected with non-zero transcript counts per cell (center) and the fraction of reads mapping to the mm10 mouse transcriptome per cell (right) in the droplet-based 3′ scRNA-seq data (a) and the plate-based full-length scRNA-Seq data (b). c–f. Agreement across batches. (c) Contribution of batches to each cluster. Each pie chart shows the batch composition (color coded legend) of each detected cluster (post-hoc annotation and number of cells are marked on top) in the droplet-based 3′ scRNA-seq dataset. All 10 replicates contribute to all clusters, and no major batch effect is observed. (n=6 mice). (d) Cell type proportions across batches. Shown is the proportion of detected cells (y axis) in each major cell type (x axis) in the droplet-based 3′ scRNA-seq dataset in each of 10 batches (dots, n=6 mice). Grey bar: mean; error bars: standard error of the mean (SEM). (e) Agreement in expression profiles across mice. Box and whisker plot shows the Pearson correlation coefficients (x axis) in average expression profiles (average log2(TPM+1)) for cells in each cluster (y axis), across all pairs of mice. Black bar indicates median value, box edges correspond to the 25th and 75th percentiles, while whiskers indicate a further 1.5*IQR where IQR is the interquartile range. Note that clusters with additional sub-types (e.g., Tuft, enteroendocrine cells) show more variation, as expected. (f) Scatter plots comparing the average log2(TPM+1) gene expression values between two scRNA-seq experiments from the droplet-based 3′ scRNA-seq dataset (top, x and y axis), two scRNA-seq experiments from the plate-based full length scRNA-seq dataset (center, x and y axis), or between the average of a plate-based full-length scRNA-seq (x axis) and a population control (y axis, bottom). Pearson correlation is marked top left. g. Additional QC metrics and post-hoc cluster annotation by the expression of known cell type markers. tSNE visualization of 7,216 single cells, where individual points correspond to single cells. Top left corner to bottom right corner, in order: Cells are colored by their assignment to clusters (top left, identical to Fig. 1b), mean expression (log2(TPM+1), color bar) of several known marker genes for a particular cell type or state (indicated on top), the mouse from which they originate (color legend), the number of reads per cell (color bar), the number of genes detected per cell (color bar) and the number of transcripts as measured by unique molecular identifiers (UMIs) per cell.
Extended Data Figure 2. Identification and characterization of intestinal epithelial cell types in plate-based full-length scRNA-seq data by unsupervised clustering, related to Figure 1.
a. QC metrics and post-hoc cluster annotation by the expression of known cell type markers. tSNE visualization of 1,522 single cells where individual points correspond to single cells. Top left corner to bottom right corner, in order: Cells are colored by their assignment to clusters, using a k-nearest neighbor (kNN) graph-based algorithm (Methods; Legend shows the cluster post-hoc annotation to cell types); mean expression (log2(TPM+1), color bar) of several known marker genes for a particular cell type or state (indicated on top; same as in Extended Data Fig. 1g); the mouse from which they originate (color legend) and its genotype, the FACS gate used to sort them (color legend), the number of reads per cell (color bar) and the number of genes detected per cell (color bar). n=8 mice. b. Cell-type-specific signatures. Heatmap shows the relative expression level (row-wise Z-scores, color bar) of genes (rows) in consensus cell-type-specific signatures (same genes as Figure 1c, with the exception of enterocytes), across the individual post-mitotic IECs (columns) in the full-length scRNA-seq data. Color code marks the cell types and their associated signatures. c. Mptx2, a novel Paneth cell marker. tSNE of the cells from the droplet-based 3′ scRNA-seq (left, as in Fig. 1b) and plate-based full-length scRNA-seq (right, as in a) datasets, colored by expression (log2(TPM+1), color bar) of the mucosal pentraxin Mptx2. d. Cell-type-enriched GPCRs. Heatmap shows the relative expression (row-wise Z-scores, color bar) of genes encoding GPCRs (rows) that are significantly (FDR < 0.001, Mann-Whitney U-test, Methods) up- or down-regulated in the cells (columns) in a given cell type (top, color coded as in a) compared to all other cells, in the plate-based full-length scRNA-seq data. e. Cell-type-specific Leucine-rich repeat (LRR) proteins. Heatmap depicts the mean relative expression (column-wise Z-score of mean log2(TPM+1) values, color bar) of genes (columns) encoding LRR proteins that are significantly (FDR < 0.001, Mann-Whitney U-test) up- or down-regulated in a given cell type (rows) compared to all other cells, in the plate-based full length scRNA-seq data. f. Cell type TFs and GPCRs. Average relative expression (Z-score of mean log2(TPM+1), color bar) of the top TFs (left) and GPCRs (right, columns) enriched in each cell type (rows).
Extended Data Figure 3. Regional variation in Paneth cell sub-types and stem cell markers.
a,b. Paneth cell subsets. (a) tSNE of 10,396 single cells (points) obtained using a large cell-enriched protocol (Methods), colored by clusters annotated post-hoc. n=2 mice. b,c. Paneth cell subset markers. (b) Expression (row-wise Z-score, color bar) of genes specific (FDR<0.05, Mann-Whitney U-test, log2 fold-change > 0.5) to each of the two Paneth cell subsets (average of 724.5 cells per subtype, down-sampled to 500 for visualization) shown in (a). c. Two Paneth subsets reflect regional diversity. Expression of the same genes (rows) as in (b) in Paneth cells from each of three small intestinal regions (176.3 cells obtained per each of the regions on average, columns; Fig. 2a); 11 of 11 Paneth-1 markers are enriched in the ileal Paneth cells, while 7/10 Paneth-2 markers are enriched in duodenal or jejunal Paneth cells (FDR <0.05, Mann-Whitney U-test). d. Validation of regional enterocyte markers. smFISH of Lct (red) and Fabp6 (white) in the duodenum (proximal, left) and ileum (distal, right). Dotted line: boundary between crypt and villi, green and yellow arrows: proximal and distal enterocytes, respectively. Scale bar, 50μm. e. Regional variation of intestinal stem cells. Expression (row-wise Z-score) of genes specific to stem cells from each intestinal region (FDR<0.05, Mann-Whitney U-test, log2 fold-change > 0.5). There are 1,226.3 obtained cells per each of the three regions on average, down-sampled to 500 for visualization (columns).
Extended Data Figure 4. Differentiation from stem cells to mature enterocytes.
a–d. Diffusion-map embedding of 5,282 cells (points) progressing through stages of enterocyte differentiation (Methods). a,b Cells are colored by their cluster assignment (Fig. 1b). Diffusion component 1 and 3 (DC-1 and DC-3) are associated with the transition from stem cells to progenitors (a), while DC-2 distinguishes between proximal and distal enterocyte fate commitment (b). c,d Cells are colored by the expression (log2(TPM+1), color bar) of known and novel TFs associated with stages of differentiation (c), or with proximal or distal enterocyte differentiation (d). e. TF genes differentially expressed between proximal and distal cell fate. Heatmap shows the mean expression level (color bar) of 44 TFs differentially expressed between the proximal and distal (color legend) enterocyte clusters of Fig. 1b (FDR < 0.05, Mann-Whitney U-test). f. Novel regional stem cell markers (Extended Data Fig. 3e) identify distinct populations in diffusion map space. Close-up of stem-cell region of diffusion space (b, inset square) colored by expression level (log2(TPM+1), color bars) pan-ISC marker Lgr5 (left), proximal ISC marker Gkn3 (center) and distal ISC marker (Bex1). Dashed line helps visualize separation of ISCs.
Extended Data Figure 5. Heterogeneity within EECs, related to Figure 3.
a. EEC subset discovery and regional location. tSNE of the 533 enteroendocrine cells (EECs) identified from the droplet-based datasets for whole SI and regional samples (color legend, n=8 mice, Methods). b. Agreement in hormone detection rates between 3′ droplet and full-length scRNA-seq. Scatter plot shows the detection rate (fraction of cells with non-zero expression of a given transcript) for a set of known EEC hormones, TFs and marker genes (color legend) in EECs from the full-length dataset (x axis), and from the 3′ droplet-based dataset (y axis). Linear fit (dashed line) and 95% confidence interval (shaded) are shown. c. Expression of key genes across subset clusters. tSNE plot shows cells colored by their assignment to the 12 clusters (top left plot; identical to Fig. 3a) or by the expression (log2(TPM+1), color bar) of markers of immature EECs (Neurog3), genes encoding gut hormones (Sct, Sst, Cck, Gcg, Ghrl, GIP, Nts, PYY) or markers of enterochromaffin cells (Tac1, Reg4). d. Co-expression of GI hormones by individual cells. Left: Heatmap shows the expression (color bar) of canonical gut hormone genes (rows) in each of 533 individual EECs (columns), colored based on their assignment to the clusters in Fig 3a (color bar, top). Right: Heatmap shows for each cluster (columns) the percentage of cells (color bar, inset text) in which the transcript for each hormone (rows) is detected.
Extended Data Figure 6. Classification and specificity of enteroendocrine subsets related to Figure 3.
a–b. Relationships between EEC subsets. (a) Dendrogram shows the relationship between EEC clusters as defined by hierarchical clustering of mean expression profiles of all the cells in a subset (Methods). Estimates for the significance of each split are derived from 100,000 bootstrap iterations using the R package pvclust (■ p<0.1, * p < 0.05; ** p < 0.01, p < 0.001, χ2 test). (b) Heatmap shows cell-cell similarities (Pearson’s r, color bar) between the 11 significant PCs scores (p<0.05, Methods) across the 533 EECs (rows, columns). Rows and columns are ordered using cluster labels obtained using unsupervised clustering (Methods). c. Subset specificity of gut hormones and related genes. Scatter plot shows each gene’s specificity to its marked cell subset (y axis; defined as the proportion of cells not in a given subset which do not express a given gene) and its sensitivity in that subset (defined as the fraction of cells of a given type which do express the gene, Methods). Subsets are color coded as in the legend. Genes are assigned to the subset where they are most highly expressed on average. Genes were chosen based on their known annotation as gut hormones (Cck, Gal, Gcg, Ghrl, GIP, Iapp, Nucb2, Nts, Pyy, Sct, Sst), enterochromaffin markers (Tph1, Tac1) and canonical EEC markers (Chga, Chgb). d. GPCRs enriched in different EEC subtypes. Heatmap shows the expression levels (row-wise Z-score, color bar) averaged across the cells in each of the EEC sub-types (columns) of 11 GPCR-encoding genes (rows) that are differentially expressed (FDR <0.25, Mann-Whitney U-test) in one of the EEC subtype clusters. The free fatty acid receptors (Ffar) 1 and 4 show specific expression patterns: Ffar1 highest in SIN cells, and also expressed by the Cck-expressing subsets previously termed I-cells (SIL-P, SILA and SIK-P), while Ffar4 is highest in the GIP-expressing subsets (SIK and SIK-P). These receptors are known to induce the expression of GIP and Gcg to maintain energy homeostasis1. Ffar2 was expressed by some progenitors and by EC cells, but notably absent from GIP-expressing cells, while the oleoylethanolamide receptor Gpr119, important for food intake and glucose homeostasis2, is expressed highest in SILA cells.
Extended Data Figure 7. Characterization of tuft cell heterogeneity and identification of TSLP and the hematopoietic lineage marker Ptprc (CD45) in a subset of tuft cells, related to Figure 4.
a. Tuft-1 and Tuft-2 cells. tSNE visualization of 102 tuft cells (dots) from the plate-based full-length scRNA-seq dataset (Extended Data Fig. 2a), labeled by their sub-clustering into Tuft-1 (orange) and Tuft-2 (brown) subtypes. n=8 mice. b. Gene signatures for Tuft-1 and Tuft-2 cells. Heatmap shows the relative expression (row-wise Z-scores, color bar) of the consensus Tuft-1 and Tuft-2 marker genes (rows; orange and brown, respectively), across single cells from the plate-based dataset (columns) assigned to Tuft-1 and Tuft-2 cell clusters (orange and brown, respectively). Top 25 genes shown for each subtype (all FDR < 0.01 and log2 fold change > 0.1 in both plate- and droplet-based datasets). c. Tuft-2 signature genes are enriched in immune functions. Shown are the significantly enriched (Methods, FDR <0.1, -log10(q-value), x axis) GO terms (y axis) in the gene signature for the Tuft-2 subset. d. Expression of neuron- and inflammation-related genes in Tuft-1 and Tuft-2 subsets, respectively. Plot shows for each gene (y axis) its differential expression (x axis) between Tuft-1 and Tuft-2 cells. Bar indicates Bayesian bootstrap3 estimates of log2 (fold change), and hinges and whiskers indicate 25% and 95% confidence intervals, respectively. e. IL-33 not detected in tuft cells. Distribution of expression of Il33 in cell subsets (x axis), in full-length scRNA-seq. (* FDR<0.1, *** FDR <0.0001, Mann-Whitney U-test). f–g. Tuft-2 cells enriched for TSLP. f. Combined smFISH and IFA of TSLP (green) with DCLK1 (red), scale bar 10μm. g. Relative quantification (RQ) of mRNA expression by qPCR of Alpi, TSLP and Dclk1 (tuft cell markers) from Tuft-1, Tuft-2 or randomly selected EpCAM+ single cells identified from full-length scRNA-seq 96-well plate (16 cells per group). (* p<0.05, ** p<0.005, t-test). h. Validation of CD45 expression in Tuft-2 cells. IFA showing co-expression of the tuft cell marker, DCLK1 and CD45 (left) and CD45 (right, with higher intensity), yellow boxes show three representative tuft cells. Scale bar, 200μm. i. Isolation of Tuft-2 cells based on CD45 expression using FACS. tSNE of 332 EpCAM+/CD45+ FACS-sorted single cells (points, n=3 pooled mice), colored by unsupervised clustering (top left), the expression of the Tuft cell marker Dclk1 (top right), or the signature scores for Tuft-1 and Tuft-2 cells (bottom left and right, respectively).
Extended Data Figure 8. Microfold (M) cells from RANKL-treated intestinal organoids and in vivo, related to Figure 5.
a–d. M cells in RANKL treated organoids. a–c tSNE of 5,434 single cells (dots) from control (left) or RANKL-treated (middle, right) intestinal organoids; or coloring each cell (b–c) by the expression (log2(TPM+1), color bar) of the canonical M cell markers TNF-alpha induced protein 2 (Tnfaip2, M-sec, b) and glycoprotein 2 (Gp2, c). n=4 pooled wells per treatment condition. d. Expression of M cell marker genes4–6 in each of the organoid cell clusters. Violin plots show the distribution of expression levels (log2(TPM+1)) for each of 10 previously reported M cell marker genes5 (columns), in the cells (dots) in each of 13 clusters, including mature M cells (red), identified by k-NN clustering of the 5,434 scRNA-seq profiles from organoids. e,f. M cell gene signature in vitro. Heatmaps show for each cell type cluster of organoid-derived intestinal epithelial cells (columns) the mean expression (color bar) of genes (rows) for known (grey bars) or novel (black bars) M cell markers (e) or transcription factors (f), identified as specific (FDR<0.05, Mann-Whitney U-test) to M cells both in vitro and in vivo (Methods). g. Congruence of in vitro and in vivo-derived M cell gene signatures. Violin plot shows the distribution of the mean expression of the in vitro-derived signature genes (y-axis) across the in vivo M cells (blue) and all other cells derived from the FAE (grey).
Extended Data Figure 9. Intestinal epithelial cell response to pathogenic stress, related to Figure 6.
a. Generalized and pathogen-specific response genes. Volcano plots show for each gene (dot) the differential expression (DE, x axis), and its associated significance (y axis; (-log10(Q value); Likelihood-ratio test) in response to either Salmonella (top) or H. polygyrus (bottom). Genes strongly up-regulated in Salmonella (FDR < 10−6) or H. polygyrus (FDR < 5×10−3) are highlighted in purple or red, respectively. All highlighted genes are significantly differentially expressed (FDR < 0.05) in both the 3′ scRNA-seq and the higher depth full-length scRNA-seq datasets. Left panels: all genes differentially expressed in the noted pathogen infection vs. uninfected controls; middle panels: the subset differentially expressed in both pathogens vs. control; right panels: the subset differentially expressed only in the noted pathogen but not the other (Methods). b. Global induction of enterocyte-specific genes across cells during Salmonella infection. tSNE of 9,842 single IECs from control wild-type mice (left) and mice infected with Salmonella (right). Cells are colored by the expression of the indicated genes, all specific to enterocytes in control mice (Supplementary Tables 2–4) and strongly up-regulated by infection (FDR < 10−10 in both the 3′ scRNA-seq datasets and in the higher depth full length scRNA-seq dataset). c. IEC programs in Salmonella infection. Enriched (–log10(q), x axis) GO terms in genes induced in Salmonella-treated IECs vs. control. d. Cell-intrinsic changes following Salmonella infection. Relative expression (row-wise Z-scores, color bar) of 104 genes (top) of which 58 (bottom) are specific to Salmonella infection, significantly up-regulated (FDR < 0.05, Mann-Whitney U-test, log2 fold-change > 0.1) in enterocytes (columns) from Salmonella infection. 10 representative genes are labeled. e. Up-regulation of pro-inflammatory apolipoproteins Serum Amyloid A 1 and 2 (Saa1 and Saa2) in distal enterocytes under Salmonella infection. Violin plot shows log2(TPM+1) expression level (y axis) of Saa1 (top) and Saa2 (bottom) across all post-mitotic cell-types from control and Salmonella-treated mice (n=4 mice, sample identity shown by color legend) (* FDR < 0.01; ** FDR < 0.0001, Mann-Whitney U-test). f. Up-regulation of anti-microbial peptides (AMPs) by Paneth cells following Salmonella infection. Violin plots show log2 (TPM+1) expression levels (y axis) of genes encoding AMPs (panels) and the mucosal pentraxin Mptx2 (bottom right) in the cells (dots) from control and Salmonella–infected mice (n=4 mice, sample identity shown by color legend) (* FDR < 0.1; ** FDR < 0.01, ** FDR < 0.0001, Mann-Whitney U-test). g. Paneth cell numbers detected (using graph-clustering, Methods) after Salmonella infection. Frequencies (y-axis) of Paneth cells in each mouse (dots) under each condition (color legend). Error bars: standard error of the mean (SEM). (** FDR < 0.01, Wald test).
Extended Data Figure 10. Goblet and tuft cell responses to H. polygyrus show a unique defense mechanism, related to Figure 6.
a. Genes significantly induced in response to infection in a non-cell-type specific manner. tSNE visualization of 9,842 single IECs (dots) from control wild-type mice (left), mice infected with H. polygyrus for three or 10 days (middle) and mice infected with Salmonella (right). Cells are colored by the expression (log2(TPM+1), color bar) of the indicated genes. Genes were selected as significantly differentially expressed in response to infection in a non-cell-type specific manner (FDR < 0.001 in both the 3′ scRNA-seq and full-length scRNA-seq datasets). b,c. Expression of the Tuft-1 and Tuft-2 signatures in the dataset of control, Salmonella and H. polygyrus infected cells. (b) Violin plots of the distribution of the respective signature scores (left and middle) and the expression of Dclk1 (right, log2 (TPM+1, y axis) in cells (dots) in each of the tuft subsets (x axis). (c) tSNE mapping of the 409 tuft progenitor, Tuft-1 and Tuft-2 cells, colored by the scores for each signature (color bar, left and middle) and their assignment to subtype clusters via kNN-graph clustering (right). d. Induction of anti-parasitic genes by goblet cells in helminth infection. Distribution of expression (log2 (TPM+1), y axis) of three anti-parasitic immunity genes7 up-regulated by goblet cells in response to H.polygyrus infection (FDR < 0.05, Mann-Whitney U-test), in control and infected mice. e. Anti-parasitic protein secretion by goblet cells during H. polygyrus infection. Immunofluorescence assay (IFA) of FFPE sections of RELMβ (top-left, red), E-cadherin (Bottom left, green) and their merged view (right) after 10 days of helminth infection. White arrow: sections of H. polygyrus. Scale bar, 200μm.
Supplementary Material
Acknowledgments
We thank Leslie Gaffney for help with figure preparation; the Broad Flow Cytometry Facility: Patricia Rogers, Stephanie Saldi and Chelsea Otis; Christoph Hafemeister and Rahul Satija for use of the ‘How Many Cells’ tool; and Tim Tickle for help with the Single Cell Portal. This study was supported by the Klarman Cell Observatory at the Broad Institute, NIH RC2DK114784 (AR and RJX), HHMI (AR), Food Allergy Science Initiative (FASI) at the Broad Institute (AR and RJX), and a Broadnext10 award (AR and RJX). MB is supported by a postdoctoral fellowship from the Human Frontiers Science Program (HFSP). RJX is supported by NIH DK43351, DK097485 and Helmsley Charitable Trust.
Footnotes
Author contributions
A.L.H., M.B. and N.R. contributed equally to this study; M.B., R.J.X and A.R. co-conceived the study; M.B., N.R., A.L.H., R.J.X and A.R. designed experiments and interpreted the results; N.R. and M.B. carried out all experiments; G.B., T.M.D., M.R.H., S.B., D.D., M.Z. and R.R. assisted with experiments; A.L.H. designed and performed computational analysis with assistance from R.H.H., K.S., C.S., Y.K., I.T., and A.R.; M.R.H. and W.S.G. assisted with tuft and FAE experiments; M.Z. and H.N.S. assisted with pathogen infections; S.B. and O.Y. assisted with epithelial cell sorting; D.D., and O.R.R. assisted with scRNA-seq; A.L.H., M.B., N.R., R.J.X and A.R. wrote the manuscript with input from all authors.
The authors declare competing financial interests: A.R. is a member of the scientific advisory board of ThermoFisher, Syros Pharmaceuticals, and Driver Group. R.J.X is a consultant at Novartis, Janssen and Celgene. A.H., M.B., N.R., R.H., K.S., C.S., O.R., R.X. and A.R. are co-inventors on provisional patent application filed by the Broad Institute relating to this manuscript.
References
- 1.Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 2.von Moltke J, Ji M, Liang HE, Locksley RM. Tuft-cell-derived IL-25 regulates an intestinal ILC2-epithelial response circuit. Nature. 2016;529:221–225. doi: 10.1038/nature16161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barriga FM, et al. Mex3a Marks a Slowly Dividing Subpopulation of Lgr5+ Intestinal Stem Cells. Cell Stem Cell. 2017;20:801–816 e807. doi: 10.1016/j.stem.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basak O, et al. Induced Quiescence of Lgr5+ Stem Cells in Intestinal Organoids Enables Differentiation of Hormone-Producing Enteroendocrine Cells. Cell Stem Cell. 2017;20:177–190 e174. doi: 10.1016/j.stem.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 5.Grun D, et al. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature. 2015;525:251–255. doi: 10.1038/nature14966. [DOI] [PubMed] [Google Scholar]
- 6.Yan KS, et al. Non-equivalence of Wnt and R-spondin ligands during Lgr5+ intestinal stem-cell self-renewal. Nature. 2017;545:238–242. doi: 10.1038/nature22313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan KS, et al. Intestinal Enteroendocrine Lineage Cells Possess Homeostatic and Injury-Inducible Stem Cell Activity. Cell Stem Cell. 2017;21:78–90 e76. doi: 10.1016/j.stem.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng GX, et al. Haplotyping germline and cancer genomes with high-throughput linked-read sequencing. Nature biotechnology. 2016;34:303–311. doi: 10.1038/nbt.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosvall M, Bergstrom CT. Maps of random walks on complex networks reveal community structure. Proceedings of the National Academy of Sciences. 2008;105:1118–1123. doi: 10.1073/pnas.0706851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shekhar K, et al. Comprehensive Classification of Retinal Bipolar Neurons by Single-Cell Transcriptomics. Cell. 2016;166:1308–1323.e1330. doi: 10.1016/j.cell.2016.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amirel AD, et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nature biotechnology. 2013;31:545–552. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kowalczyk MS, et al. Single-cell RNA-seq reveals changes in cell cycle and differentiation programs upon aging of hematopoietic stem cells. Genome Research. 2015;25:1860–1872. doi: 10.1101/gr.192237.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garabedian EM, Roberts LJ, McNevin MS, Gordon JI. Examining the role of Paneth cells in the small intestine by lineage ablation in transgenic mice. J Biol Chem. 1997;272:23729–23740. doi: 10.1074/jbc.272.38.23729. [DOI] [PubMed] [Google Scholar]
- 14.Gribble FM, Reimann F. Enteroendocrine Cells: Chemosensors in the Intestinal Epithelium. Annual review of physiology. 2016;78:277–299. doi: 10.1146/annurev-physiol-021115-105439. [DOI] [PubMed] [Google Scholar]
- 15.Howitt MR, et al. Tuft cells, taste-chemosensory cells, orchestrate parasite type 2 immunity in the gut. Science. 2016;351:1329–1333. doi: 10.1126/science.aaf1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Picelli S, et al. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 17.van der Meer-van Kraaij C, et al. Dietary modulation and structure prediction of rat mucosal pentraxin (Mptx) protein and loss of function in humans. Genes & nutrition. 2007;2:275–285. doi: 10.1007/s12263-007-0058-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du Clos TW. Pentraxins: structure, function, and role in inflammation. ISRN inflammation. 2013;2013:379040. doi: 10.1155/2013/379040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katz JP, et al. The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development. 2002;129:2619–2628. doi: 10.1242/dev.129.11.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duboc H, Tache Y, Hofmann AF. The bile acid TGR5 membrane receptor: from basic research to clinical application. Dig Liver Dis. 2014;46:302–312. doi: 10.1016/j.dld.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Overton HA, Fyfe MC, Reynet C. GPR119, a novel G protein-coupled receptor target for the treatment of type 2 diabetes and obesity. Br J Pharmacol. 2008;153(Suppl 1):S76–81. doi: 10.1038/sj.bjp.0707529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coifman RR, et al. Geometric diffusions as a tool for harmonic analysis and structure definition of data: diffusion maps. Proc Natl Acad Sci U S A. 2005;102:7426–7431. doi: 10.1073/pnas.0500334102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Basak O, et al. Mapping early fate determination in Lgr5+ crypt stem cells using a novel Ki67-RFP allele. EMBO J. 2014;33:2057–2068. doi: 10.15252/embj.201488017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beuling E, et al. GATA factors regulate proliferation, differentiation, and gene expression in small intestine of mature mice. Gastroenterology. 2011;140:1219–1229. e1211–1212. doi: 10.1053/j.gastro.2011.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Furness JB, Rivera LR, Cho HJ, Bravo DM, Callaghan B. The gut as a sensory organ. Nature reviews. Gastroenterology & hepatology. 2013;10:729–740. doi: 10.1038/nrgastro.2013.180. [DOI] [PubMed] [Google Scholar]
- 26.Worthington JJ, Reimann F, Gribble FM. Enteroendocrine cells-sensory sentinels of the intestinal environment and orchestrators of mucosal immunity. Mucosal Immunol. 2017 doi: 10.1038/mi.2017.73. [DOI] [PubMed] [Google Scholar]
- 27.Habib AM, Richards P, Rogers GJ, Reimann F, Gribble FM. Co-localisation and secretion of glucagon-like peptide 1 and peptide YY from primary cultured human L cells. Diabetologia. 2013;56:1413–1416. doi: 10.1007/s00125-013-2887-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gershon MD, Tack J. The serotonin signaling system: from basic understanding to drug development for functional GI disorders. Gastroenterology. 2007;132:397–414. doi: 10.1053/j.gastro.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 29.Klok MD, Jakobsdottir S, Drent ML. The role of leptin and ghrelin in the regulation of food intake and body weight in humans: a review. Obes Rev. 2007;8:21–34. doi: 10.1111/j.1467-789X.2006.00270.x. [DOI] [PubMed] [Google Scholar]
- 30.Karra E, Chandarana K, Batterham RL. The role of peptide YY in appetite regulation and obesity. J Physiol. 2009;587:19–25. doi: 10.1113/jphysiol.2008.164269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gerbe F, Jay P. Intestinal tuft cells: epithelial sentinels linking luminal cues to the immune system. Mucosal Immunol. 2016;9:1353–1359. doi: 10.1038/mi.2016.68. [DOI] [PubMed] [Google Scholar]
- 32.Gerbe F, et al. Intestinal epithelial tuft cells initiate type 2 mucosal immunity to helminth parasites. Nature. 2016;529:226–230. doi: 10.1038/nature16527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bezencon C, et al. Murine intestinal cells expressing Trpm5 are mostly brush cells and express markers of neuronal and inflammatory cells. The Journal of comparative neurology. 2008;509:514–525. doi: 10.1002/cne.21768. [DOI] [PubMed] [Google Scholar]
- 34.Biton M, et al. Epithelial microRNAs regulate gut mucosal immunity via epithelium-T cell crosstalk. Nat Immunol. 2011;12:239–246. doi: 10.1038/ni.1994. [DOI] [PubMed] [Google Scholar]
- 35.de Lau W, et al. Peyer’s patch M cells derived from Lgr5(+) stem cells require SpiB and are induced by RankL in cultured “miniguts”. Molecular and cellular biology. 2012;32:3639–3647. doi: 10.1128/MCB.00434-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mabbott NA, Donaldson DS, Ohno H, Williams IR, Mahajan A. Microfold (M) cells: important immunosurveillance posts in the intestinal epithelium. Mucosal Immunol. 2013;6:666–677. doi: 10.1038/mi.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Terahara K, et al. Comprehensive gene expression profiling of Peyer’s patch M cells, villous M-like cells, and intestinal epithelial cells. Journal of immunology. 2008;180:7840–7846. doi: 10.4049/jimmunol.180.12.7840. [DOI] [PubMed] [Google Scholar]
- 38.Peterson LW, Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nature reviews. Immunology. 2014;14:141–153. doi: 10.1038/nri3608. [DOI] [PubMed] [Google Scholar]
- 39.Loonen LM, et al. REG3gamma-deficient mice have altered mucus distribution and increased mucosal inflammatory responses to the microbiota and enteric pathogens in the ileum. Mucosal Immunol. 2014;7:939–947. doi: 10.1038/mi.2013.109. [DOI] [PubMed] [Google Scholar]
- 40.Eckhardt ER, et al. Intestinal epithelial serum amyloid A modulates bacterial growth in vitro and pro-inflammatory responses in mouse experimental colitis. BMC Gastroenterol. 2010;10:133. doi: 10.1186/1471-230X-10-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinez Rodriguez NR, et al. Expansion of Paneth cell population in response to enteric Salmonella enterica serovar Typhimurium infection. Infect Immun. 2012;80:266–275. doi: 10.1128/IAI.05638-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Artis D, et al. RELMbeta/FIZZ2 is a goblet cell-specific immune-effector molecule in the gastrointestinal tract. Proc Natl Acad Sci U S A. 2004;101:13596–13600. doi: 10.1073/pnas.0404034101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vassen L, Okayama T, Moroy T. Gfi1b:green fluorescent protein knock-in mice reveal a dynamic expression pattern of Gfi1b during hematopoiesis that is largely complementary to Gfi1. Blood. 2007;109:2356–2364. doi: 10.1182/blood-2006-06-030031. [DOI] [PubMed] [Google Scholar]
- 44.Su L, et al. Coinfection with an intestinal helminth impairs host innate immunity against Salmonella enterica serovar Typhimurium and exacerbates intestinal inflammation in mice. Infect Immun. 2014;82:3855–3866. doi: 10.1128/IAI.02023-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics (Oxford, England) 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 47.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brennecke P, et al. Accounting for technical noise in single-cell RNA-seq experiments. Nature Methods. 2013;10:1093–1095. doi: 10.1038/nmeth.2645. [DOI] [PubMed] [Google Scholar]
- 49.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology. 2009 doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buja A, Eyuboglu N. Remarks on Parallel Analysis. Multivariate Behavioral Research. 1992;27:509–540. doi: 10.1207/s15327906mbr2704_2. [DOI] [PubMed] [Google Scholar]
- 52.van der Maaten L. Accelerating t-SNE using Tree-Based Algorithms. The Journal of Machine Learning Research. 2014;15:3221–3245. [Google Scholar]
- 53.van der Maaten L, Hinton G. Visualizing Data using t-SNE. The Journal of Machine Learning Research. 2008;9:2579–2605. [Google Scholar]
- 54.Zeisel A, et al. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 55.Haghverdi L, Buettner F, Theis FJ. Diffusion maps for high-dimensional single-cell analysis of differentiation data. Bioinformatics. 2015;31:2989–2998. doi: 10.1093/bioinformatics/btv325/-/DC1. [DOI] [PubMed] [Google Scholar]
- 56.Ester M, Kriegel HP, Sander J, Xu X. A density-based algorithm for discovering clusters in large spatial databases with noise. Kdd. 1996 [Google Scholar]
- 57.Levine JH, et al. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell. 2015:1–15. doi: 10.1016/j.cell.2015.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rodriguez A, Laio A. Machine learning Clustering by fast search and find of density peaks. Science. 2014;344:1492–1496. doi: 10.1126/science.1242072. [DOI] [PubMed] [Google Scholar]
- 59.Finak G, et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015;16:278. doi: 10.1186/s13059-015-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B Methodological. 1995;57:289–300. [Google Scholar]
- 61.Zhang H-M, et al. AnimalTFDB: a comprehensive animal transcription factor database. Nucleic Acids Research. 2012;40:D144–149. doi: 10.1093/nar/gkr965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ng A, Eisenberg JM, Heath R. Proceedings of the …. 2011 [Google Scholar]
- 63.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biology. 2010;11 doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 1.Ichimura A, Hirasawa A, Hara T, Tsujimoto G. Free fatty acid receptors act as nutrient sensors to regulate energy homeostasis. Prostaglandins Other Lipid Mediat. 2009;89:82–88. doi: 10.1016/j.prostaglandins.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Overton HA, Fyfe MC, Reynet C. GPR119, a novel G protein-coupled receptor target for the treatment of type 2 diabetes and obesity. Br J Pharmacol. 2008;153(Suppl 1):S76–81. doi: 10.1038/sj.bjp.0707529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rubin DB. The Bayesian bootstrap. The Annals of Statistics. 1981;9:130–134. [Google Scholar]
- 4.de Lau W, et al. Peyer’s patch M cells derived from Lgr5(+) stem cells require SpiB and are induced by RankL in cultured “miniguts”. Molecular and cellular biology. 2012;32:3639–3647. doi: 10.1128/MCB.00434-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Terahara K, et al. Comprehensive gene expression profiling of Peyer’s patch M cells, villous M-like cells, and intestinal epithelial cells. Journal of immunology. 2008;180:7840–7846. doi: 10.4049/jimmunol.180.12.7840. [DOI] [PubMed] [Google Scholar]
- 6.Kobayashi A, et al. Identification of novel genes selectively expressed in the follicle-associated epithelium from the meta-analysis of transcriptomics data from multiple mouse cell and tissue populations. DNA research : an international journal for rapid publication of reports on genes and genomes. 2012;19:407–422. doi: 10.1093/dnares/dss022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Datta R, et al. Identification of novel genes in intestinal tissue that are regulated after infection with an intestinal nematode parasite. Infect Immun. 2005;73:4025–4033. doi: 10.1128/IAI.73.7.4025-4033.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data is deposited in GEO (GSE92332) and in the Single Cell Portal for visualization and download (https://portals.broadinstitute.org/single_cell).