Abstract
The intracellular pH (pHi) of most cancers is constitutively higher than that of normal cells and enhances proliferation and cell survival. We found that increased pHi enabled the tumorigenic behaviors caused by somatic arginine-to-histidine mutations, which are frequent in cancer and confer pH sensing not seen with wild-type proteins. Experimentally raising the pHi increased the activity of R776H mutant epidermal growth factor receptor (EGFR-R776H), thereby increasing proliferation and causing transformation in fibroblasts. An Arg-to-Gly mutation did not confer these effects. Molecular dynamics simulations of EGFR suggested that decreased protonation of His776 at high pH causes conformational changes in the αC helix that may stabilize the active form of the kinase. An Arg-to-His, but not Arg-to-Lys, mutation in the transcription factor p53 (p53-R273H) decreased its transcriptional activity and attenuated the DNA damage response in fibroblasts and breast cancer cells with high pHi. Lowering pHi attenuated the tumorigenic effects of both EGFR-R776H and p53-R273H. Our data suggest that some somatic mutations may confer a fitness advantage to the higher pHi of cancer cells.
INTRODUCTION
Increased intracellular pH (pHi) is an established feature of most cancers regardless of tissue of origin or genetic background (1). This increased pHi can enable tumorigenic properties, such as increased proliferation, cell survival, and metastasis (1–5). Studies suggest increased pHi may be both a cause and a consequence of tumor cell evolution (6). Whereas the evolutionary theory of cancer has largely been shaped by genomic analysis of tumor samples (7, 8), cancer cell adaptation is mediated not by nucleotide changes but by proteomic changes that alter cell biology and enable cancer cell behaviors. Determining how distinct amino acid mutational signatures contribute to the physiological changes seen in cancer evolution is an understudied but important area of research. Recent work has analyzed cancers by amino acid substitution signatures (9–11) and found that arginine-to-histidine (Arg>His) mutations are dominant in a subset of cancers. Anoosha and colleagues (10) also showed that Arg>His mutations are enriched in driver mutations compared with passenger mutations. However, the physiological implications of this Arg>His amino acid mutation signature has not been determined or proposed. Arg>His mutations are of particular interest given recent work on the molecular mechanisms of His switches in pH sensors, or proteins with pH-sensitive functions or activities (12). Arginine with a pKa (where Ka is the acid dissociation constant) of ~12 should always be protonated, whereas histidine with a pKa ~ 6.5 can titrate within the narrow cellular pH range and exhibit a shift in population from the protonated to the neutral species at the higher pHi of cancer cells. With a gain in pH sensing, Arg>His substitutions could provide an adaptive advantage to cancer cells by altering protein binding or activity specifically at increased pHi.
To test this prediction, we examined the effects of two recurring Arg>His mutations in the epidermal growth factor receptor (EGFR-R776H) and the transcription factor p53 (p53-R273H). We selected these mutations because they are recurrent in human cancers (13), occur in functionally important regions of the proteins, and are candidates with predicted opposite effects induced by increased pHi—an increase in EGFR enzyme activity and a loss of p53 transcriptional activity. We found that these Arg>His mutations confer pH-sensitive functions that are not seen with the wild-type proteins. We also uncovered molecular mechanisms for how protonation changes at a single amino acid substitution can alter protein structure and activity of EGFR. Adaptive pH sensing conferred by these Arg>His mutations enabled cancer phenotypes specifically at increased pHi. Moreover, lowering pHi effectively limited the tumorigenic effects of the Arg>His mutants. Our findings suggest that recent broad analyses of somatic amino acid substitutions in cancer can inform and direct studies of functional effects of somatic mutations at the protein level and may open new avenues for better classification and treatment of cancers.
RESULTS
EGFR-R776H has pH-sensitive kinase activity in vitro
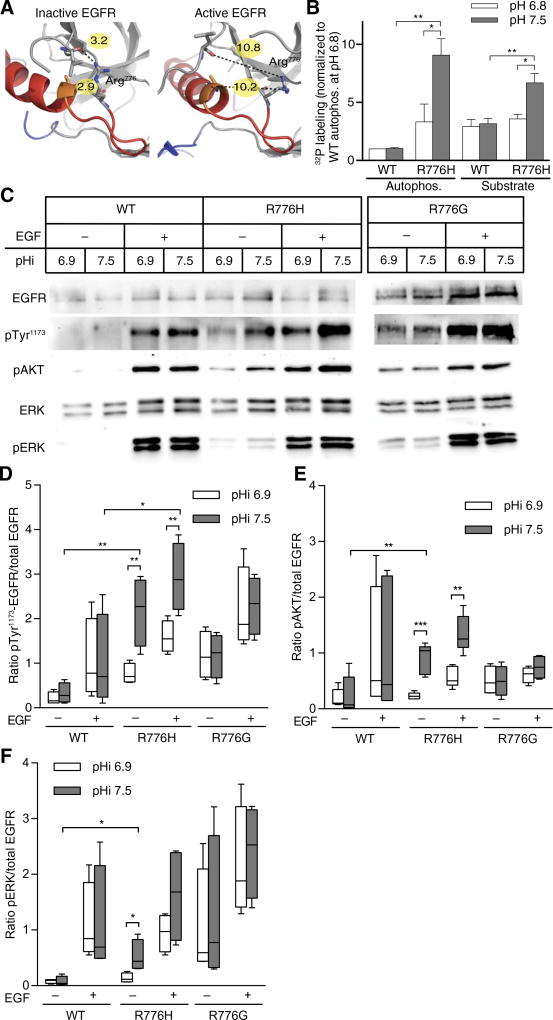
We first tested our hypothesis that some Arg>His mutations can confer a gain in pH sensing by investigating an EGFR-R776H mutant. Point mutations of Arg776 are recurrent in lung cancers, and 60% of mutations at this site are Arg>His (13). In the inactive crystal structure of wild-type EGFR (14), Arg776 is involved in hydrogen-bonding interactions with backbone carbonyl groups in the αC helix, which are not present in the active crystal structure (Fig. 1A) (15). These structural data suggest that Arg776 may help stabilize the inactive form of the kinase through a hydrogen-bonding network. Conformational reorganization of the αC helix is critical for kinase activity (16), and other activating EGFR mutations destabilize the inactive state (17, 18). We predicted that protonated His776 at lower pH might preserve the hydrogen-bonding network, whereas neutral His776 at higher pH would disrupt this network, causing a shift in the conformational equilibrium to active EGFR even in the absence of epidermal growth factor (EGF).
Fig. 1. EGFR-R776H has pH-sensitive activity and downstream signaling, with increased activity at higher pHi.
(A) Structures of inactive (left, 3GT8) and active (right, 2GS6) EGFR-WT. Arg776 is shown in stick. Red, αC helix; yellow, heteroatom distances (in Ångstrom). (B) In vitro kinase activity of WT and EGFR-R776H (R776H) was determined by [γ-32P]–adenosine 5′-triphosphate (ATP) incorporation into enzyme (autophosphorylation) or substrate peptide (substrate) at two pH values. Data were normalized to autophosphorylation of WT at pH 6.8. Data are from four independent experiments (means ± SEM). Representative gel and autoradiograph are shown in fig. S1A. (C) Representative immunoblots of lysates from quiescent MDA-MB-453 cells transfected with EGFR-WT, EGFR-R776H, or EGFR-R776G with (+) or without (−) EGF (50 ng/ml; 5 min) at indicated pHi values. Blots of total EGFR (EGFR), EGFR autophosphorylation (pTyr1173), AKT-pSer473 (pAKT), ERK1/2 (ERK), and ERK1/2-pThr202/Tyr204 (pERK). (D to F) Quantification of pTyr1173 (D), pAKT (E), and pERK (F), with data normalized to amount of total EGFR present. Data are from four independent experiments (Tukey box plots). For (B) and (D) to (F), Student’s t tests (unpaired, two-tailed) with Holm-Sidak multiple comparisons correction were used. *P < 0.05, **P < 0.01, ***P < 0.001.
We tested pH-dependent EGFR kinase activity in vitro using recombinant EGFR containing the intracellular kinase domain and juxtamembrane segments (residues 645 to 998) (14). Activity of wild-type EGFR (EGFR-WT) was pH-insensitive, with similar amounts of autophosphorylation and substrate phosphorylation at pH 7.5 compared to pH 6.8 (Fig. 1B and fig. S1A). In contrast, EGFR-R776H activity was pH-sensitive, with greater autophosphorylation and substrate phosphorylation at pH 7.5 than at pH 6.8 (Fig. 1B and fig. S1A). To confirm that the pH-dependent activity observed is the result of titration at His776 and not due to the loss of Arg776, we tested a glycine substitution at position 776 (EGFR-R776G), which also occurs in human cancers (13). When Arg776 was mutated to a nontitratable glycine residue, autophosphorylation and substrate phosphorylation were pH-insensitive (fig. S1, B and C), suggesting that His776 specifically confers the pH-dependent activity observed for EGFR-R776H.
A pH titration revealed that EGFR-R776H was pH-sensitive within a narrow range of pH 7.3 to 7.6 (fig. S1D). These data suggest that the activity of EGFR-R776H is greater at the pHi of cancer cells (7.5 to 7.6) compared with the pHi of normal cells (7.2). Additionally, we observed a marked increase in EGFR-R776H activity between buffer pH of 7.3 and 7.6, which suggests that the histidine is titrating within that pH range. This result suggests that the pKa of His776 is upshifted compared to the pKa of histidine in solution (pKa = 6.5).
EGFR-R776H confers increased pathway activation at higher pHi
We confirmed pH-sensitive activity of EGFR-R776H when transiently expressed in MDA-MB-453 clonal breast cancer cells. Parental MDA-MB-453 cells are EGFR-null and have no response to EGF; but with transiently expressed EGFR-WT, we observed an EGF-dependent increase in phosphorylated extracellular signal–regulated kinases 1 and 2 (ERK1/2) (fig. S2A). We used incubation with ammonium chloride (NH4Cl) to increase pHi and incubation with 5-(N-ethyl-N-isopropyl) amiloride (EIPA), a selective inhibitor of the sodium proton exchanger NHE1, to decrease pHi (fig. S2B). Whereas EGFR-WT activity was pH-independent at the pH values we tested, EGFR-R776H was pH-sensitive with increased activity at higher pHi (Fig. 1C). In both the absence and presence of EGF, EGFR-R776H autophosphorylation (pTyr1173; Fig. 1, C and D) and downstream phosphorylation of AKT (pAKT) (Fig. 1, C and E) were greater at higher pHi. Moreover, in the absence of EGF, phosphorylation of both AKT and ERK in cells expressing EGFR-R776H was greater at pHi 7.5 compared to 6.9 and greater than observed with EGFR-WT (Fig. 1, E and F). This indicates that EGFR-R776H produces increased pathway activation at higher pHi in the absence of ligand.
In contrast to EGFR-R776H, EGFR-R776G did not show pH-dependent activity under any condition tested (Fig. 1, C to F). These data suggest that His776 specifically confers the pH-dependent autophosphorylation and downstream pathway activation observed with EGFR-R776H in mammalian cells. In the absence of extracellular EGF, EGFR-R776G did induce increased downstream pathway activation compared to EGFR-WT (Fig. 1, E and F). Additionally, we confirmed that EGFR-R776H had pH-dependent activity when expressed in Chinese hamster ovary (CHO) cells, which are also EGFR-null (fig. S2, C to F).
We also measured phosphorylation at three other EGFR autophosphorylation sites to determine whether the pH-sensitive activity of EGFR-R776H discriminated between the phosphoinositide 3-kinase (PI3K)–AKT and growth factor receptor-bound protein (Grb)–mitogen-activated protein kinase (MAPK) signaling pathways. In the absence of EGF, we found that EGFR-R776H had pH-dependent autophosphorylation of Tyr992, which couples to PI3K-AKT signaling, as well as Tyr1068 and Tyr1086, which activate the Grb-MAPK pathway (fig. S3, A to D). In the presence of EGF, EGFR-R776H autophosphorylation of Tyr992 and Tyr1068 was pH-dependent, with increased phosphorylation at increased pHi (fig. S3, B and D). Both EGFR-WT and EGFR-R776G had pH-independent autophosphorylation at all sites and under all conditions tested. These data suggest that the pH-dependent kinase activity observed with EGFR-R776H does not discriminate at the level of EGFR autophosphorylation for one downstream signaling pathway over another.
Molecular dynamics simulations reveal a mechanism for pH sensing by EGFR-R776H
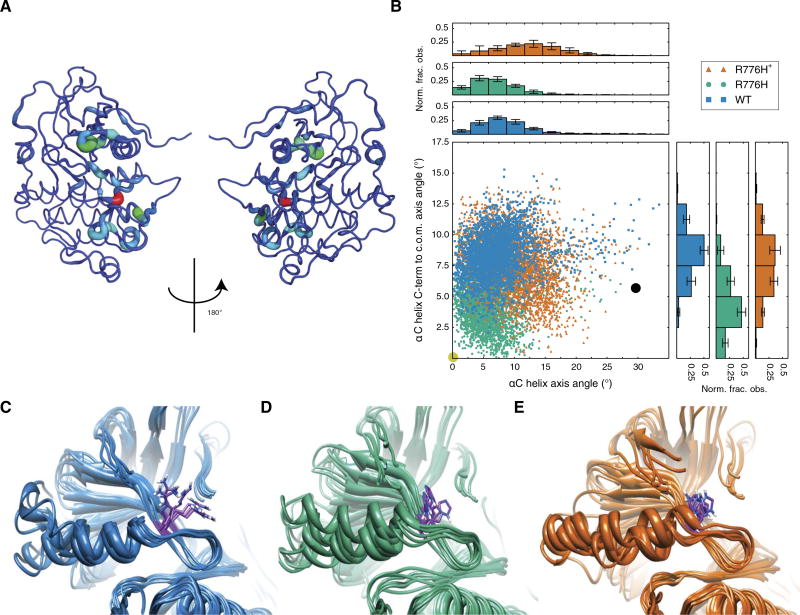
To identify a potential molecular mechanism for gain of pH sensing by EGFR-R776H, we used molecular dynamics (MD) simulations, an approach previously used to reveal EGFR transitions between active and inactive states (19, 20), regulatory mechanisms (21), and the impact of cancer mutations (22). A previous investigation of the activation mechanism of R776H in the EGFR kinase domain also focuses on the interactions around the αC-β4 loop but does not take into consideration the effect of different protonation states for the mutant (23). Here, we explore the pH-dependent activation mechanism by explicitly comparing WT and both protonated and neutral R776H mutants.
We obtained 300 ns of MD trajectories for EGFR-WT and EGFR-R776H with protonated and neutral His776. We performed the information theory–based Jensen-Shannon (JS) divergence analysis (24) to identify statistically significant differences in protein structure and dynamics for EGFR-R776H with protonated and neutral His776. We identified changes in structure and dynamics in the vicinity of His776 as expected (fig. S4A). However, we also observed high JS divergence [0.5 to 0.61 nats (natural units of information)] induced by changing the protonation state of His776 localized around the αC helix N-terminal region (Fig. 2A and fig. S4A), suggesting that protonation changes the conformational dynamics of the entire αC helix.
Fig. 2. Protonation state of His776 alters EGFR aC helix conformations sampled in MD simulations.
(A) JS divergence of protonated versus neutral R776H highlights major nonlocal differences localized around the N-terminal end of the αC helix. (B) αC helix conformational distribution for 300-ns MD simulations defined by two angle measures (see also fig. S5 for comparisons with R776G) that describe tilting and displacement of the helix relative to the active crystal structure. Circles denote the tilt and displacement of active (yellow) and inactive (black) crystal structures. c.o.m., center of mass; Norm. frac. obs., normalized fraction observed. (C to E) Final snapshots of simulations for WT (C), neutral His776 (D), and protonated His776 (E).
We calculated average root mean square deviation (RMSD) of the αC helix in our simulations to the αC helix in the active crystal structure and obtained an RMSD of 2.07 ± 0.10 Å for neutral His776 compared with 2.27 ± 0.11 Å for WT Arg776 and 2.42 ± 0.16 Å for protonated His776 (fig. S4B). To characterize the αC helix conformation, we used two angle measures that quantify tilting and displacement of the helix relative to the conformation observed in the active crystal structure (fig. S5A). We observed again that the neutral His776 trajectory explored αC helix conformations that more closely resemble the active crystal structure than WT and protonated His776 (Fig. 2B). A superposition of final snapshots for each simulation shows the differences in conformations explored for WT and each protonation state of the R776H mutant (Fig. 2, C to E). αC helix conformations sampled by neutral His776 (Fig. 2D) were less constrained (fig. S5, B and C) than those sampled by protonated His776 (Fig. 2E). WT αC helix conformations were also constrained near Arg776 (Fig. 2C); however, the N-terminal region had more flexibility than protonated His776. In contrast, MD simulations with EGFR-R776G showed that this mutant explored a wider range of αC helix conformations sampling both active- and inactive-like conformations (fig. S5, B and C). These data suggest that the protonation state of His776 can induce localized and dynamic conformational changes in a functionally critical region of EGFR.
EGFR-R776H enhances cancer phenotypes at higher pHi
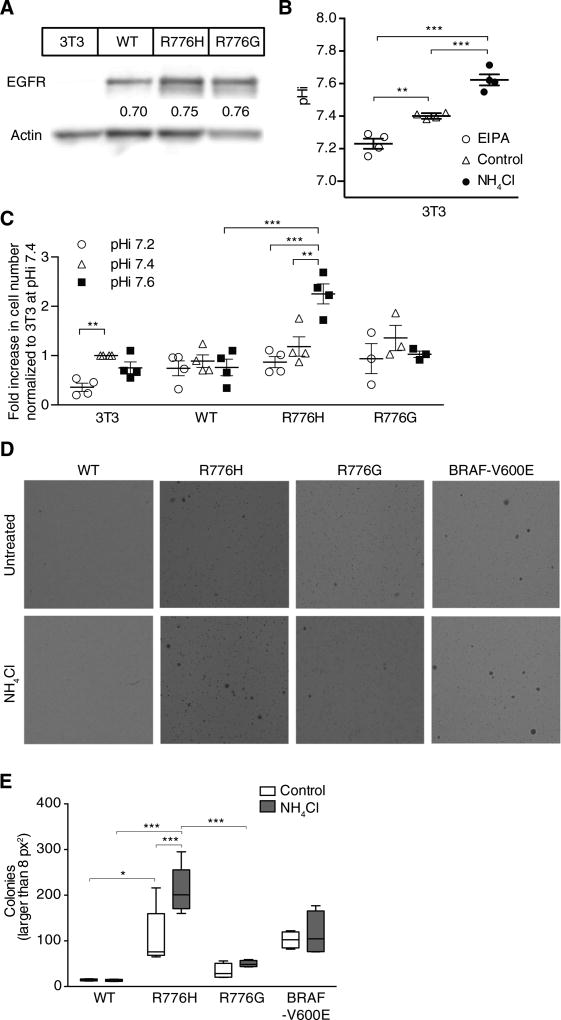
We also observed pH-dependent proliferation and transformation of cells expressing EGFR-R776H but not EGFR-WT or EGFR-R776G. For these studies, we stably expressed WT or mutant EGFR in NIH3T3 cells (Fig. 3A). We used NIH3T3 fibroblasts because they are a normal line, express extremely low amounts of EGFR (undetectable by immunoblot, although they are not true nulls), and undergo transformation with activating EGFR mutants but not EGFR-WT (25). We experimentally changed pHi in these NIH3T3 cells with NH4Cl and EIPA (Fig. 3B). We found that whereas proliferation of EGFR-WT and EGFR-R776G cells was comparable across tested pHi values, EGFR-R776H cells had greater proliferation at pHi 7.6 than at pHi 7.2 (Fig. 3C). Consistent with previous findings showing that decreased pHi slows proliferation (26), proliferation of parental cells was lower at pHi 7.2 than at 7.4, but there was no increase in proliferation between pHi 7.4 and 7.6. Additionally, transformation with EGFR-R776H, determined by colony formation in soft agar, was twofold greater with increased pHi induced by NH4Cl (Fig. 3, D and E). In contrast, there was no transformation with EGFR-WT, and transformation with EGFR-R776G and that with oncogenic BRAF-V600E were both pH-independent (Fig. 3E; representative full-well images in fig. S6). Together, these findings indicate that the dynamic pH-sensitive activity of EGFR-R776H results in increased pathway activation, proliferation, and transformation at higher pHi of cancer cells, which could confer an adaptive advantage to the cancer cells.
Fig. 3. EGFR-R776H has pH-sensitive proliferation and soft agar transformation.
(A) Immunoblot of lysates from parental NIH3T3 (3T3) cells and cells stably expressing vectors encoding WT or mutant EGFR. (B) pHi in 3T3 cells when cultured with ammonium chloride (NH4Cl) or EIPA. Data are means ± SEM from four independent experiments. (C) Fold increase in cell number at given pHi for stable WT or mutant EGFR cell lines. Data are means ± SEM from three or more independent experiments, each normalized to parental 3T3 at pHi 7.4. (D) Soft agar transformation assay in NIH3T3 cells stably expressing WT or mutant EGFR or BRAF-V600E. Cells were maintained for 15 days in complete medium or medium supplemented with 5 mM NH4Cl and then fixed and stained with crystal violet. (E) Colonies >8 pixel2 (px2) were counted using ImageJ particle analyzer. Data are from four independent experiments (Tukey box plots). Comparison of 3T3 at pHi 7.2 and 7.4 in (C) used a one-sample t test (two-tailed). All other comparisons in (B), (C), and (E) used Student’s t tests (unpaired, two-tailed) with Holm-Sidak multiple comparisons correction. *P < 0.05, **P < 0.01, ***P < 0.001.
p53-R273H has pH-sensitive transcriptional activity
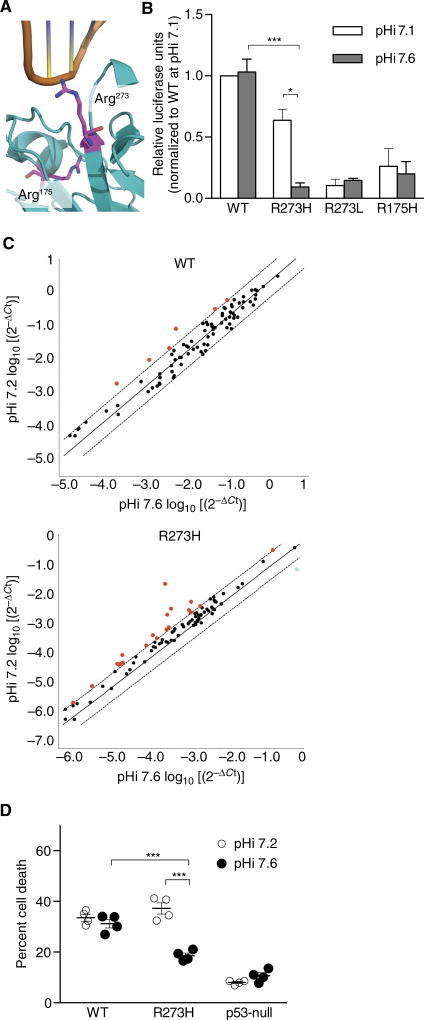
We showed with EGFR-R776H that increased pHi can enhance activity of an oncogenic mutation. To test the prediction that gain in pH sensing can decrease activity of a tumor suppressor at high pHi, we investigated p53-R273H, a recurrent somatic mutation in p53. Amino acid substitutions at Arg273 are the most frequent point mutations in p53, and 40% of these are Arg>His (13, 27). The cocrystal structure of p53 with DNA (28) suggests direct binding of positively charged Arg273 with the negatively charged phosphate backbone of DNA (Fig. 4A). We reasoned that protonated His273 could also form favorable electrostatic interactions with DNA but neutral His273 would not, thus conferring pH-sensitive DNA binding with decreased binding at higher pHi. Although several studies have measured decreased DNA binding by p53-R273H (29, 30), to our knowledge, pH-dependent binding and transcriptional activity have not been reported.
Fig. 4. p53-R273H has decreased DNA binding at higher pHi.
(A) Structure of p53 (4HJE) with Arg273 and Arg175 in magenta stick. (B) Luciferase assay in PS120 cells transfected with WT or mutant p53 at indicated pHi (see fig. S7A for pHi measurements). Luciferase was normalized to β-galactosidase control and then within each experiment to WT at pHi 7.1. Data are means ± SEM from four independent experiments for WT, R273H, and R175H and three independent experiments for R273L. (C) RT-PCR from etoposide-treated MDA-MB-157 cells stably expressing WT or p53-R273H at two pHi values (see fig. S7B for pHi measurements). Scatter plots of 2−ΔCt on log10 scale for WT and R273H show transcripts that are more (red) or less (blue) abundant at pHi 7.2 relative to 7.6 (see table S1 for the list of altered transcripts). Data from three independent experiments [mean, identity (solid) and threefold boundary (dotted) lines]. (D) Cells were treated as in (C), and percentage of dead cells was determined by trypan blue exclusion. Data are means ± SEM from four independent experiments. For (B) and (D), Student’s t tests (unpaired, two-tailed) with Holm-Sidak multiple comparisons correction were used. *P < 0.05, ***P < 0.001. For (C), a threefold boundary significance cutoff was used.
We used a luciferase reporter assay (31) to measure p53 transcriptional activity with experimentally changed pHi. We performed these assays first in PS120 fibroblasts deficient in the sodium-proton exchanger NHE1 but engineered to stably express an inactive NHE1 mutant (32, 33). These cells are a good model because they are untransformed and transfect very well, and we previously established protocols to manipulate pHi in these cells (fig. S7A) (34). We transiently expressed WT or mutant p53, altered pHi, and induced a DNA damage response with etoposide. Whereas transcriptional activity of p53-WT was pH-insensitive, activity of p53-R273H was pH-sensitive and decreased at higher pHi (Fig. 4B). In contrast, p53-R273L and p53-R175H both exhibited pH-independent and low transcriptional activity (Fig. 4B). These controls suggest that the pH-dependent transcriptional activity observed with p53-R273H is due to the titration of a histidine specifically located at the protein-DNA interface.
Cells expressing p53-R273H have pH-sensitive transcriptional profile and survival
Because PS120 fibroblasts have endogenous p53, we confirmed our findings on transcriptional activity using TP53-null MDA-MB-157 clonal breast cancer cells stably expressing p53-WT or p53-R273H. We changed pHi in these cell lines and induced a DNA damage response with etoposide. Etoposide treatment had no effect on pHi under the conditions used (fig. S7B). Using a reverse transcription polymerase chain reaction (RT-PCR) p53 profile array (Qiagen), we simultaneously quantified transcript abundance for 83 members of the p53 signaling pathway at pHi 7.2 and 7.6. We found a pH-dependent transcriptional profile in cells expressing p53-R273H, with 20 transcripts differentially expressed at lower pHi in p53-R273H–expressing cells (Fig. 4C). Nineteen transcripts including proapoptotic and arrest transcripts, such as ATM, BRCA1, CHEK1, and FADD, were more abundant at lower pHi in p53-R273H–expressing cells and 1 progrowth transcript, PTTG1, was less abundant at lower pHi (table S1). We also noted that a linear regression of 2−ΔCt values for pHi 7.2 versus pHi 7.6 gave a slope of 1.3 ± 0.09 (R2 = 0.83) for p53-R273H and 0.87 ± 0.02 (R2 = 0.99) for p53-WT. These data suggest that overall transcriptional activity of p53-R273H is decreased at higher pHi and that decreasing pHi can partially restore p53-R273H transcriptional activity of proapoptotic genes. At low pHi in cells expressing p53-R273H, we observed that transcript abundance of CHEK1 and BRCA1 was equivalent to that observed with p53-WT.
We also determined cell death in response to double-strand breaks under conditions identical to the RT-PCR assay. We found that the p53-WT cell line exhibits pH-insensitive induction of cell death but that p53-R273H has decreased cell death at increased pHi (Fig. 4D). Therefore, transcriptional activity and induction of cell death in response to DNA damage are decreased at higher pHi with p53-R273H, suggesting that transcriptional activity of this mutant is attenuated at the higher pHi of cancer cells. Furthermore, we were able to rescue functional p53-R273H response to double-strand breaks by lowering pHi.
DISCUSSION
Our study indicates that p53-R273H and EGFR-R776H substitutions confer a gain in pH sensing not seen with WT protein, enabling cancer cell behaviors at higher pHi. These findings add to an emerging list of tumorigenic behaviors enabled by the established higher pHi of cancer cells (5). We previously described how the higher pHi of cancers increases cell proliferation and promotes hyperplasia and metastatic progression (2, 4). Our new findings suggest that increased pHi can work in concert with mutant proteins to enhance oncogenic signaling and limit tumor suppression. Moreover, lowering pHi attenuates some of the oncogenic effects of EGFR-R776H and partially restores p53-R273H tumor suppressor functions.
Despite published work reporting a higher than expected frequency of Arg>His mutations in cancer (9–11), functional effects or physiological relevance of recurring Arg>His substitutions has not been reported. Our data show that Arg>His mutations can confer a gain in pH sensing to mutant proteins and suggest that Arg>His substitutions may provide a fitness advantage to the increased pHi of cancer cells. This work provides the first analysis of potential physiological relevance of the higher than expected frequency of Arg>His mutations observed in cancer (9–11). Furthermore, our functional assays in cells suggest that the tumorigenic effects of some somatic Arg>His cancer mutations become penetrant only at high pHi and suggest that lowering pHi in cancer cells may reduce the deleterious effects of some Arg>His mutations. These findings lay the groundwork for future studies on the functional effects of other amino acid substitutions that may allow adaptive and advantageous responses to either altered pHi dynamics or dynamic microenvironment pressures in cancers, such as oxidative stress, oxygen and nutrient availability, and metabolic reprogramming.
MATERIALS AND METHODS
Cloning and expression
Complementary oligos for the EGFR substrate peptide TAENAEYLRVAPQ (15) flanked by Eco RI and Xho I sites were ordered, annealed, and ligated into pGEX6P digested with Eco RI and Xho I. Sequences were as follows: (forward primer) AATTCCCACCGCGGAAAACGCGGAATATCTGCGCGTGGCGCCGCAGTGAC and (reverse primer) TCGAGTCACTGCGGCGCCACGCGCAGATATTCCGCGTTTTCCGCGGTGGG.
Construct was transformed into and expressed in BL21-DE3 Escherichia coli. Cells were grown in Luria broth + ampicillin (100 µg/ml; 37°C, with shaking) until optical density at 600 nm (OD600) = 0.5 and induced (1 mM isopropyl-β-d-1-thiogalatapyranoside; for 12 hours at room temperature, with shaking). Cells were pelleted (6000g; 10 min at 4°C), resuspended in binding buffer [25 mM tris (pH 7.5), 150 mM NaCl, 1 mM EDTA, protease inhibitor cocktail (Roche)], and lysed by passing six times through Microfluidizer (Microfluidics, M110-S), cooling lysate between passages. Clarified supernatant (14,000g; 10 min at 4°C) was loaded onto to a pre-equilibrated glutathione agarose column (Thermo Fisher) and washed with 20 column volumes of binding buffer, and the protein was eluted in binding buffer with 15 mM reduced glutathione (pH 8.0). Glutathione S-transferase (GST) peptide was concentrated and dialyzed into storage buffer: 10 mM tris, 50 mM NaCl, 1 mM dithiothreitol (DTT), 5% glycerol (pH 7.5). Aliquots were flash-frozen in liquid nitrogen and stored at −80°C.
For EGFR, the pFASTBAC-EGFR (residues 645 to 998) plasmid (a gift from the N. Jura laboratory) was mutated using QuikChange (Stratagene) to create pFASTBAC-EGFR-R776H and EGFR-R776G. Recombinant baculovirus production, expression, and purification of both WT and mutant EGFR (residues 645 to 998) from Hi5 insect cells were performed as described (15). We note that expression of EGFR-R776Gat high abundance required increased multiplicity of infection (5 versus 2) and larger expression volumes. For EGFR-R776G, expression was performed in 6 × 500 ml flasks before pooling and purifying. Proteins were concentrated to 5 to 10 mg/ml and dialyzed into storage buffer: 20 mM tris, 50 mM NaCl, 2 mM DTT, 2 mM tris(2-carboxyethyl)phosphine, and 10% glycerol (pH 8.0). Aliquots were flash-frozen in liquid nitrogen and stored at −80°C.
EGFR in vitro kinase assays
Reactions were assembled with 1 µM purified EGFR and 1 mM ATP with 25,000 cpm mol−1 of [γ-32P]-ATP in kinase buffer [50 mM Hepes, 10 mM MgCl2, and 100 µM DTT (pH 6.8 or 7.5)] and preincubated at 25°C for 10 min. After preincubation, 3 µM GST substrate peptide [TAENAEYLRVAPQ (15)] fusion (Cf) was added, and reactions were incubated at 25°C for 10 min and then quenched (EDTA; 25 mM Cf). Reactions were mixed with Laemmli buffer, boiled, run on 10% SDS–polyacrylamide gel electrophoresis (SDS-PAGE) gels, and dried using a gel dryer (Bio-Rad, model 853). Autoradiograph film (Denville HyBlot CL, E3018) was exposed overnight at −20°C. Using ImageJ software, background-subtracted autoradiography intensities were determined and normalized within each experiment to autophosphorylation observed with EGFR-WT or EGFR-R776H at pH 6.8. For pH titration, reactions were performed as described above in kinase buffers prepared at a range of pH values. Quenched reactions were divided in half, captured on duplicate Whatman P81 disks, and washed as described previously (35). [32P] incorporation was measured by scintillation counting (Packard, Tri-Carb 2100TR) and averaged.
Cell culture
For MDA-MB-453 [American Type Culture Collection (ATCC)] and MDA-MB-157 (J.M. Bishop Lab), we used Leibovitz L15 medium with 15% fetal bovine serum (FBS), grown in atmospheric conditions at 37°C. For CHO (ATCC), we used minimum essential medium–α with 10% FBS, grown in 5% CO2 at 37°C. For NIH3T3 (J. Roose Lab), we used Dulbecco’s modified Eagle’s medium (DMEM; Gibco) with 10% FBS, grown in 5% CO2 at 37°C. For PS120-E266I, we used DMEM (Gibco) with 5%FBS, grown in 5%CO2 at 37°C.
EGFR activity
Cells were plated (1 × 105 per well; six-well plate) and transfected 24 hours later with 1 µg per well of pcDNA3–EGFR-WT (or EGFR-R776H or EGFR-R776G) using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. Twenty-four hours after transfection, medium was changed to complete medium for 24 hours, and then the cells were serum-starved for 24 hours. After serum starvation, to raise pHi, the cells were incubated with serum-free medium (SFM) containing 15 mM NH4Cl for 10 min. To lower pHi, the cells were incubated with SFM + 15 mM NH4Cl for 10 min and then incubated with SFM + 1 µM EIPA for 5 min. Where indicated, cells were treated with EGF (50 ng/ml; 5 min) added to pHi control media. Cells were washed twice with ice-cold phosphate-buffered saline (PBS) and incubated with 100 µl of ice-cold lysis buffer [50 mM tris, 150 mM NaCl, 1 mM NaF, 1% Triton X-100, and protease inhibitor cocktail (Roche) (pH 7.5)] for 15 min on ice. Cells were scraped, and clarified lysate (10,000 rpm; 10 min) was used immediately for immunoblotting or flash-frozen and stored at −80°C.
Immunoblotting and quantification
For each sample, 10 µg of total protein per lane (Bradford assay) was prepared (with addition of Laemmli loading buffer, boiled) and loaded on duplicate 10% SDS-PAGE gels. Proteins were transferred to 0.45-µm polyvinylidene difluoride membranes (Immobilon) (50 V, 2.5 hours, on ice). Membranes were blocked in 5%fat-free milk in TBST (0.1% Tween in tris-buffered saline) for 1 hour at room temperature and then divided for blotting on the basis of prestained protein ladder. We used 100+ kDa for total EGFR (Cell Signaling, 2232), EGFR-pTyr1173 (Cell Signaling, 4407), EGFR-pTyr992 (Cell Signaling, 2235), EGFR-pTyr1068 (Cell Signaling, 2234), and EGFR-pTyr1086 (Cell Signaling, 2220), all 1:1000 in 5% bovine serum albumin in TBST. We used 50 to 100 kDa for AKT-pSer473 (Cell Signaling, 9271; 1:1000 in 5% milk/TBST). We used 10 to 50 kDa for total ERK (Santa Cruz Biotechnology, sc93; 1:5000 in 5% milk/TBST) and ERK1/2–pThr202/Tyr204 (Cell Signaling, 4377; 1:5000 in 5% milk/TBST). Where indicated, actin was blotted (Millipore, MAB1501; 1:10,000 in 5% milk/TBST). Primary antibodies were incubated overnight (4°C, with shaking). Membranes were washed (3 × 5 min; TBST) with shaking. Secondary horseradish peroxidase–conjugated antibodies against mouse and rabbit (Bio-Rad, 1706515 and 1706516; 1:10,000 in 5% milk/TBST) were incubated for 1 hour (room temperature, with shaking). Membranes were washed with shaking (3 × 5 min; TBST), developed (SuperSignal West Femto, Pierce), and visualized (Alpha Innotech FluorChem Q). Using ImageJ software, background-subtracted densitometries for phosphorylated antibodies (EGFR-pTyr1173, EGFR-pTyr992, EGFR-pTyr1068, EGFR-pTyr1086, AKT-pSer473, or ERK1/2–pThr202/Tyr204) were normalized to background-subtracted densitometries for total EGFR for each condition.
MD structure preparation
A complete structure for initiating MD simulations was created using 2J6M (18) and by adding missing residues (Arg984 to Asp1003) from another crystallographic structure (3POZ) (36). The two structures were aligned, missing residues were built in, and energy minimization was performed on the composite model in PLOP (Protein Local Optimization Program) (37). For histidine mutants with fixed protonation states, the Arg776 in the WT model structure file was mutated.
MD simulations
Amber’s LEaP program was used with the Amber ff99SB force field and GAFF (general Amber force field). The TIP3P water model was used to solvate the system in a cubic periodic box, such that the closest distance between any atom in the system and the periodic boundary is 10Å. Net negative charge on EGFR was neutralized by adding counterions (Na+). Energy minimization was performed in two steps: using harmonic restraints on the protein (500.0 kcal mol−1 Å−2) and using unrestrained minimization. For each step, we ran 500 steps of steepest descent and 500 steps of conjugate gradient minimization at a constant volume with a nonbonded cutoff of 9 Å. The equilibration was done in three steps. First, the system was heated from 0 to 300 K with restrained equilibration (10.0 kcal mol−1 Å−2) for 20 ps at constant volume with a nonbonded cutoff of 9 Å, using the SHAKE algorithm to constrain bonds involving hydrogens, and the Andersen thermostat. The second round of equilibration was performed lowering the harmonic restraints (1.0 kcal mol−1 Å−2) on the system for 20 ps (other parameters were identical). The third round was performed for 10 ns at a constant pressure of 1.0 bar with a nonbonded cutoff of 9 Å at 300 K with the Andersen thermostat. Simulations were performed without restraints using new velocities with random seeds at a constant pressure of 1 bar with a nonbonded cutoff distance of 9 Å. Six independent 50-ns simulations were run with 2-fs time step, for a total of 300 ns of simulation time per EGFR construct. Coordinates and energy were saved every picosecond (500 steps).
MD analysis
Convergence of the simulations was confirmed by RMSD plots. For each trajectory, the structures were aligned to a reference structure (active, 2GS6). To characterize the conformation of the αC helix, we calculated the angle of the axis of the αC helix (amide nitrogens’ center of mass, residues 757 to 761, to amide nitrogens’ center of mass, residues 764 to 768) and the angle of the axis defined by the vector from the αC helix center of mass to the center of mass of the full tyrosine kinase domain to the corresponding axes in the active crystal structure. Additionally, we calculated the distance between the center of mass of the αC helix (residues 756 to 767) to the center of mass of the tyrosine kinase. Conformational ensembles from MD simulations were compared using the JS divergence, which is a measure of dissimilarity between probability distributions. A discrete distribution of sampled torsion angles for each dihedral angle in the system is created for trajectories at each condition (WT and protonated and neutral R776H). These distributions are then compared using the JS divergence, based on Kullback-Leibler (KL) divergence:
where .
With the JS divergence, we can quantify the degree of dissimilarity between two dihedral angle distributions simulated under different conditions. Our calculations take into consideration sample variability using a statistical bootstrap approach with the full six independent simulations as reference (24).
Stable expression of EGFR in cells
NIH3T3 cells were transfected with pcDNA3–EGFR-WT, EGFR-R776G, or EGFR-R776H (3.5-mm dish; Lipofectamine 2000, Invitrogen). Twenty-four hours after transfection, cells were trypsinized and replated for selection [10-cm dish; zeocin (800 µg/ml), Invitrogen]. When visible, individual colonies were isolated (cloning cylinders, Corning), EGFR expression was analyzed, and clones with similar expression were propagated.
Cell proliferation
NIH3T3 parental or stable NIH3T3 cell lines expressing EGFR-WT, EGFR-R776G, or EGFR-R776H were plated (1 × 103 per well; 24-well plate) in pHi control medium (6 wells per condition). To assess pHi, control medium was composed of DMEM + 10% FBS, to which 5 mM NH4Cl was added to increase the pHi, or 10 µM EIPA was added to decrease the pHi. Twenty-four and 72 hours after plating, three wells of each condition were trypsinized, and living cells were counted using the trypan blue exclusion method.
Cell transformation
Cells were plated on a solidified bed of 0.5% agarose in growth medium in 3.5-cm plates. For plating, 1 × 104 3T3–EGFR-WT, 3T3–EGFR-R776G, 3T3–EGFR-R776H, or 3T3–BRAF-V600E stable cell lines were resuspended in 0.35% agarose in growth medium. Agar was topped with 2 ml of growth medium and incubated overnight. Twenty-four hours after plating, top medium was removed and replaced with pHi control medium (“Control,” DMEM + 10% FBS; “NH4Cl, ” DMEM + 10% FBS + 5 mM NH4Cl). For BRAF plates, BRAF-V600E expression was induced with 1 µM tamoxifen. Medium was replaced every 3 days until day 15, when cells were washed twice in PBS, fixed with 4% paraformaldehyde in PBS for 1 hour, washed twice in PBS, and stained with 0.0005% crystal violet for 2 hours. After PBS washes to remove excess crystal violet, images were acquired (Alpha Innotech FluorChem Q). Colonies were counted using Analyze Particles function on ImageJ software. Particles larger than 8 pixel2 were quantified and averaged from two technical replicates per condition.
Luciferase assay
PS120 cells stably expressing NHE1-E266I (32) were plated (4 × 104 per well; 24-well plate). Twenty-four hours later, cells were transfected (Lipofectamine 2000, Invitrogen) with 100 ng each of PG13-luciferase (31) and pRK–β-galactosidase (control reporter) as well as 500 ng of p53 plasmid (pCB6-p53, p53-R175H, p53-R273L, or p53-R273H; WT plasmid was provided by K. Vousden, Beatson Institute). Conditions with a nonspecific reporter plasmid (MG13-luciferase) (31) were used for background subtraction. Twenty-four hours after transfection, pHi was altered. To raise or lower pHi, 5 mM NH4Cl or 0.1 µM vacuolar-type H+-ATPase inhibitor concanamycin A (Sigma, C9705), respectively, was added for 48 hours. To induce a p53 response, 10 µM etoposide (Millipore, 341205) was added for the final 12 hours. Luciferase and β-galactosidase activities were measured using Dual-Light Luciferase Assay (Roche) according to the manufacturer’s instructions. Briefly, 10µl of lysate was measured in duplicate for each condition, and background-subtracted ratios of luciferase/β-galactosidase relative light units were determined.
Reverse transcription polymerase chain reaction
MDA-MB-157 cells stably expressing either p53-WT or p53-R273H were plated in 10-cm dishes and grown to ~50% confluency before beginning experimental modulation of pHi. To raise or lower pHi, medium was supplemented with 5 mM NH4Cl or 10 µM EIPA, respectively, for 48 hours. To induce a p53 response, 10 µM etoposide was added for the final 12 hours. Cells were harvested, and total RNA was extracted using the RNeasy Mini Kit (Qiagen). For each condition, 0.5 µg total RNA was used. Synthesis of complementary DNA was performed using the RT2 First Strand Synthesis Kit (Qiagen), and the 96-well RT2 Profiler PCR Array plates (Qiagen, PAHS-027ZA) were prepared according to the manufacturer’s instructions. The assay was performed on three independent preparations of cells for each condition. The RT2 PCR Array data sets were processed using the SAB PCR Array Data Analysis Web portal (http://pcrdataanalysis.sabiosciences.com/pcr/arrayanalysis.php), with a boundary cutoff of threefold increase in expression.
Cell death assays
Cells were prepared as described in the RT-PCR section and treated identically to alter pHi. Three wells of each condition were trypsinized and mixed with trypan blue for counting. Cells were counted using a hemocytometer, and percentage of dead cells was reported and normalized within each experiment to that observed with cells expressing p53-WT at low pHi.
pHi measurements
For each condition, triplicate wells were plated (2 × 104 per well; 24-well plates). Steady-state pHi was measured (38) under conditions outlined in pHi control–related sections.
Supplementary Material
Acknowledgments
We thank B. Vogelstein (Johns Hopkins University), K. Vousden (Beatson Institute), and N. Jura [University of California, San Francisco (UCSF)] for sharing plasmids; J. Roose (UCSF) for sharing the BRAF-V600E–inducible NIH3T3 cell line; and J. M. Bishop (UCSF) for sharing the MDA-MB-157 cell line. We also thank N. Jura (UCSF) for helpful discussions on EGFR. Funding: This work was supported by the NIH grants CA178706 (to D.L.B. and R.D.H.), CA197855 (to D.L.B.), and HG007644 (to R.D.H.) and by the NIH F32 grant CA177085 (to K.A.W.). The consulting statistician (S. Gansky) was funded through the National Center for Advancing Translational Sciences grant UL1 TR001872 (to UCSF–Clinical and Translational Science Institute).
Footnotes
www.sciencesignaling.org/cgi/content/full/10/495/eaam9931/DC1
Fig. S1. EGFR-R776H activity is pH-sensitive in vitro and titrates with increasing pH.
Fig. S2. pHi control and pH-dependent EGFR-R776H activity in EGFR-null cells.
Fig. S3. EGFR-R776H activity is pH-sensitive at distinct autophosphorylation sites.
Fig. S4. Quantification of C helix flexibility and trajectory RMSD.
Fig. S5. EGFR MD simulations metrics for analysis.
Fig. S6. Full-well images of transformation assays.
Fig. S7. pHi control for p53 assays.
Table S1. pH-dependent transcriptional profiles.
Author contributions: D.L.B. conceived the hypothesis, which was developed in collaboration with K.A.W., R.D.H., and M.P.J. K.A.W. directed collaborations and performed all biochemical and cell-based assays. D.L.B. performed pHi measurements. D.G.R. performed MD simulations and analyses. K.A.W. and D.G.R. performed data analysis. Z.A.S. and N.B.S. contributed discussions of data and new ideas. All authors contributed to the writing of the manuscript.
Competing interests: M.P.J. is a consultant to and stockholder of Schrödinger LLC, which distributes some of the software used in this work. All other authors declare that they have no competing interests.
References
- 1.Cardone RA, Casavola V, Reshkin SJ. The role of disturbed pH dynamics and the Na+/H+ exchanger in metastasis. Nat. Rev. Cancer. 2005;5:786–795. doi: 10.1038/nrc1713. [DOI] [PubMed] [Google Scholar]
- 2.Grillo-Hill BK, Choi C, Jimenez-Vidal M, Barber DL. Increased H+ efflux is sufficient to induce dysplasia and necessary for viability with oncogene expression. eLife. 2015;4:e03270. doi: 10.7554/eLife.03270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harguindey S, Orive G, Luis Pedraz J, Paradiso A, Reshkin SJ. The role of pH dynamics and the Na+/H+ antiporter in the etiopathogenesis and treatment of cancer. Two faces of the same coin—One single nature. Biochim. Biophys. Acta. 2005;1756:1–24. doi: 10.1016/j.bbcan.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Webb BA, Chimenti M, Jacobson MP, Barber DL. Dysregulated pH: A perfect storm for cancer progression. Nat. Rev. Cancer. 2011;11:671–677. doi: 10.1038/nrc3110. [DOI] [PubMed] [Google Scholar]
- 5.White KA, Grillo-Hill BK, Barber DL. Cancer cell behaviors mediated by dysregulated pH dynamics at a glance. J. Cell Sci. 2017;130:663–669. doi: 10.1242/jcs.195297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gillies RJ, Verduzco D, Gatenby RA. Evolutionary dynamics of carcinogenesis and why targeted therapy does not work. Nat. Rev. Cancer. 2012;12:487–493. doi: 10.1038/nrc3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bignell GR, Greenman CD, Davies H, Butler AP, Edkins S, Andrews JM, Buck G, Chen L, Beare D, Latimer C, Widaa S, Hinton J, Fahey C, Fu B, Swamy S, Dalgliesh GL, Teh BT, Deloukas P, Yang F, Campbell PJ, Futreal PA, Stratton MR. Signatures of mutation and selection in the cancer genome. Nature. 2010;463:893–898. doi: 10.1038/nature08768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SAJR, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Børresen-Dale A-L, Boyault S, Burkhardt B, Butler AP, Caldas C, Davies HR, Desmedt C, Eils R, Eyfjörd JE, Foekens JA, Greaves M, Hosoda F, Hutter B, Ilicic T, Imbeaud S, Imielinsk M, Jäger N, Jones DTW, Jones D, Knappskog S, Kool M, Lakhani SR, López-Otín C, Martin S, Munshi NC, Nakamura H, Northcott PA, Pajic M, Papaemmanuil E, Paradiso A, Pearson JV, Puente XS, Raine K, Ramakrishna M, Richardson AL, Richter J, Rosenstiel P, Schlesner M, Schumacher TN, Span PN, Teague JW, Totoki Y, Tutt ANJ, Valdés-Mas R, van Buuren MM, van ’t Veer L, Vincent-Salomon A, Waddell N, Yates LR, Australian Pancreatic Cancer Genome Initiative; ICGC Breast Cancer Consortium; ICGC MMML-Seq Consortium; ICGC PedBrain. Zucman-Rossi J, Futreal PA, McDermott U, Lichter P, Meyerson M, Grimmond SM, Siebert R, Campo E, Shibata T, Pfister SM, Campbell PJ, Stratton MR. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan H, Bao J, Zhou X. Genome-wide mutational spectra analysis reveals significant cancer-specific heterogeneity. Sci. Rep. 2015;5:12566. doi: 10.1038/srep12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anoosha P, Sakthivel R, Michael Gromiha M. Exploring preferred amino acid mutations in cancer genes: Applications to identify potential drug targets. Biochim. Biophys. Acta. 2016;1862:155–165. doi: 10.1016/j.bbadis.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 11.Szpiech ZA, Strauli NB, White KA, Garrido Ruiz D, Jacobson MP, Barber DL, Hernandez RD. Prominent features of the amino acid mutation landscape in cancer. PLOS ONE. 2017;12:e0183273. doi: 10.1371/journal.pone.0183273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schönichen A, Webb BA, Jacobson MP, Barber DL. Considering protonation as a posttranslational modification regulating protein structure and function. Annu. Rev. Biophys. 2013;42:289–314. doi: 10.1146/annurev-biophys-050511-102349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, Tate J, Cole CG, Ward S, Dawson E, Ponting L, Stefancsik R, Harsha B, Kok CY, Jia M, Jubb H, Sondka Z, Thompson S, De T, Campbell PJ. COSMIC: Somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017;45:D777–D783. doi: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jura N, Shan Y, Cao X, Shaw DE, Kuriyan J. Structural analysis of the catalytically inactive kinase domain of the human EGF receptor 3. Proc. Natl. Acad. Sci. U.S.A. 2009;106:21608–21613. doi: 10.1073/pnas.0912101106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 16.Jura N, Zhang X, Endres NF, Seeliger MA, Schindler T, Kuriyan J. Catalytic control in the EGF receptor and its connection to general kinase regulatory mechanisms. Mol. Cell. 2011;42:9–22. doi: 10.1016/j.molcel.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non–small-cell lung cancer to gefitinib. N. Engl. J. Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 18.Yun C-H, Boggon TJ, Li Y, Woo MS, Greulich H, Meyerson M, Eck MJ. Structures of lung cancer-derived EGFR mutants and inhibitor complexes: Mechanism of activation and insights into differential inhibitor sensitivity. Cancer Cell. 2007;11:217–227. doi: 10.1016/j.ccr.2006.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, Li X, Ma W, Dong Z. Conformational transition pathways of epidermal growth factor receptor kinase domain from multiple molecular dynamics simulations and Bayesian clustering. J. Chem. Theory Comput. 2014;10:3503–3511. doi: 10.1021/ct500162b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Papakyriakou A, Vourloumis D, Tzortzatou-Stathopoulou F, Karpusas M. Conformational dynamics of the EGFR kinase domain reveals structural features involved in activation. Proteins. 2009;76:375–386. doi: 10.1002/prot.22353. [DOI] [PubMed] [Google Scholar]
- 21.Mustafa M, Mirza A, Kannan N. Conformational regulation of the EGFR kinase core by the juxtamembrane and C-terminal tail: A molecular dynamics study. Proteins. 2011;79:99–114. doi: 10.1002/prot.22862. [DOI] [PubMed] [Google Scholar]
- 22.Dixit A, Verkhivker GM. Hierarchical modeling of activation mechanisms in the ABL and EGFR kinase domains: Thermodynamic and mechanistic catalysts of kinase activation by cancer mutations. PLOS Comput. Biol. 2009;5:e1000487. doi: 10.1371/journal.pcbi.1000487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruan Z, Kannan N. Mechanistic insights into R776H mediated activation of epidermal growth factor receptor kinase. Biochemistry. 2015;54:4216–4225. doi: 10.1021/acs.biochem.5b00444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McClendon CL, Hua L, Barreiro A, Jacobson MP. Comparing conformational ensembles using the Kullback–Leibler divergence expansion. J. Chem. Theory Comput. 2012;8:2115–2126. doi: 10.1021/ct300008d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Velu TJ, Beguinot L, Vass WC, Willingham MC, Merlino GT, Pastan I, Lowy DR. Epidermal-growth-factor-dependent transformation by a human EGF receptor protooncogene. Science. 1987;238:1408–1410. doi: 10.1126/science.3500513. [DOI] [PubMed] [Google Scholar]
- 26.Putney LK, Barber DL. Na-H exchange-dependent increase in intracellular pH times G2/M entry and transition. J. Biol. Chem. 2003;278:44645–44649. doi: 10.1074/jbc.M308099200. [DOI] [PubMed] [Google Scholar]
- 27.Joerger AC, Fersht AR. The tumor suppressor p53: From structures to drug discovery. Cold Spring Harb. Perspect. Biol. 2010;2:a000919. doi: 10.1101/cshperspect.a000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Y, Zhang X, Dantas Machado AC, Ding Y, Chen Z, Qin PZ, Rohs R, Chen L. Structure of p53 binding to the BAX response element reveals DNA unwinding and compression to accommodate base-pair insertion. Nucleic Acids Res. 2013;41:8368–8376. doi: 10.1093/nar/gkt584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ang HC, Joerger AC, Mayer S, Fersht AR. Effects of common cancer mutations on stability and DNA binding of full-length p53 compared with isolated core domains. J. Biol. Chem. 2006;281:21934–21941. doi: 10.1074/jbc.M604209200. [DOI] [PubMed] [Google Scholar]
- 30.Brázdová M, Navrátilová L, Tichý V, Němcová K, Lexa M, Hrstka R, Pečinka P, Adámik M, Vojtesek B, Paleček E, Deppert W, Fojta M. Preferential binding of hot spot mutant p53 proteins to supercoiled DNA in vitro and in cells. PLOS ONE. 2013;8:e59567. doi: 10.1371/journal.pone.0059567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.El-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 32.Denker SP, Huang DC, Orlowski J, Furthmayr H, Barber DL. Direct binding of the Na–H exchanger NHE1 to ERM proteins regulates the cortical cytoskeleton and cell shape independently of H+ translocation. Mol. Cell. 2000;6:1425–1436. doi: 10.1016/s1097-2765(00)00139-8. [DOI] [PubMed] [Google Scholar]
- 33.Karydis A, Jimenez-Vidal M, Denker SP, Barber DL. Mislocalized scaffolding by the Na-H exchanger NHE1 dominantly inhibits fibronectin production and TGF-β activation. Mol. Biol. Cell. 2009;20:2327–2336. doi: 10.1091/mbc.E08-08-0842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Webb BA, White KA, Grillo-Hill BK, Schönichen A, Choi C, Barber DL. A Histidine cluster in the cytoplasmic domain of the Na-H exchanger NHE1 confers pH-sensitive phospholipid binding and regulates transporter activity. J. Biol. Chem. 2016;291:24096–24104. doi: 10.1074/jbc.M116.736215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barker SC, Kassel DB, Weigl D, Huang X, Luther MA, Knight WB. Characterization of pp60c-src tyrosine kinase activities using a continuous assay: Autoactivation of the enzyme is an intermolecular autophosphorylation process. Biochemistry. 1995;34:14843–14851. doi: 10.1021/bi00045a027. [DOI] [PubMed] [Google Scholar]
- 36.Aertgeerts K, Skene R, Yano J, Sang B-C, Zou H, Snell G, Jennings A, Iwamoto K, Habuka N, Hirokawa A, Ishikawa T, Tanaka T, Miki H, Ohta Y, Sogabe S. Structural analysis of the mechanism of inhibition and allosteric activation of the kinase domain of HER2 protein. J. Biol. Chem. 2011;286:18756–18765. doi: 10.1074/jbc.M110.206193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jacobson MP, Pincus DL, Rapp CS, Day TJF, Honig B, Shaw DE, Friesner RA. A hierarchical approach to all-atom protein loop prediction. Proteins. 2004;55:351–367. doi: 10.1002/prot.10613. [DOI] [PubMed] [Google Scholar]
- 38.Choi C-H, Webb BA, Chimenti MS, Jacobson MP, Barber DL. pH sensing by FAK-His58 regulates focal adhesion remodeling. J. Cell Biol. 2013;202:849–859. doi: 10.1083/jcb.201302131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.