Summary
As most of the mitochondrial proteome is encoded in the nucleus, mitochondrial functions critically depend on nuclear gene expression and bidirectional mito-nuclear communication. However, mitochondria-to-nucleus communication pathways in mammals are incompletely understood. Here, we identify G-Protein Pathway Suppressor 2 (GPS2) as a mediator of mitochondrial retrograde signaling and a transcriptional activator of nuclear-encoded mitochondrial genes. GPS2 regulated translocation from mitochondria to nucleus is essential for the transcriptional activation of a nuclear stress response to mitochondrial depolarization and for supporting basal mitochondrial biogenesis in differentiating adipocytes and brown adipose tissue (BAT) from mice. In the nucleus, GPS2 recruitment to target gene promoters regulates histone H3K9 demethylation and RNA POL2 activation through inhibition of Ubc13-mediated ubiquitination. These findings, together, reveal an additional layer of regulation of mitochondrial gene transcription, uncover a direct mitochondria-nuclear communication pathway and indicate that GPS2 retrograde signaling is a key component of the mitochondrial stress response in mammals.
Introduction
Normal mitochondrial function is vital to the well-being of eukaryotic cells. Beyond ATP production, mitochondria generate key metabolites and biochemical signals central to apoptotic and metabolic pathways. Conversely, defects in mitochondrial functions are associated with a multitude of diseases, including primary inherited mitochondrial disorders, insulin resistance and Type 2 diabetes (T2D) (Altshuler-Keylin and Kajimura, 2017; Gorman et al., 2016; Kusminski and Scherer, 2012; Patti and Corvera, 2010; Vafai and Mootha, 2012; Wallace, 2013). Even though mitochondria contain multiple copies of their own genome, most of the mitochondrial proteome is encoded by nuclear genes and, thus, regulated by nuclear transcription factors (TFs) and associated cofactors (Fan and Evans, 2014; Hock and Kralli, 2009; Scarpulla et al., 2012). Thus, the biogenesis of mitochondria and the maintenance of mitochondrial homeostasis critically depend on nuclear transcription and signaling pathways that inform the nucleus of mitochondrial dysfunctions and changes in cellular metabolism (Finley and Haigis, 2009; Guha and Avadhani, 2013). This is partly achieved through regulation of nuclear TFs and cofactors via cytosolic mediators (i.e. Ca2+, ROS, NAD/NADH ratio)(Bohovych and Khalimonchuk, 2016; Chandel, 2015). However, much is still unknown about mitochondria-to-nuclear signaling pathways and the physiological effects of impaired communication networks in the development of diseases.
A direct mitochondrial retrograde response pathway was first described in response to mtDNA depletion in S. Cerevisiae (Jazwinski and Kriete, 2012). Partially overlapping functions of yeast retrograde factors Rtg1/Rtg3 with mammalian transcription factors (such as FOXOs, NFkB, ERα and Myc) speak to the functional conservation of this pathway, even in absence of clear mammalian orthologues of the Rtg genes (Germain, 2016; Jazwinski, 2013; Quiros et al., 2016). Characterization of mitochondria-to-nuclear translocation of transcription factor ATFS-1 in response to proteotoxic stresses (UPRmt) in worms (Lin et al., 2016; Nargund et al., 2015; Nargund et al., 2012) further supports that direct communication strategies might be employed across species. However, it is currently unknown whether direct mediators of retrograde signaling, yet to be identified, exist in mammals.
GPS2 is as an important regulator of inflammation and lipid metabolism, processes tightly linked to mitochondrial functions (Cardamone et al., 2012; Cardamone et al., 2014; Jakobsson et al., 2009; Toubal et al., 2013; Venteclef et al., 2010). GPS2 protein content is tightly controlled through protein stabilization/degradation (Huang et al., 2015) and GPS2 deficiency in mice is embryonic lethal (Guo et al., 2014), indicating the physiological importance of GPS2. Previous work indicates that nuclear GPS2 is a corepressor and coactivator for a number of TFs (Cheng and Kao, 2009; Natarajan et al., 2013; Peng et al., 2001; Zhang et al., 2008). Outside of the nucleus, GPS2 regulates insulin signaling and pro-inflammatory pathways (Cardamone et al., 2012; Cederquist et al., 2017). Both nuclear and cytosolic functions are linked to GPS2-mediated inhibition of Lysine 63 (K63) ubiquitination events (Cardamone et al., 2012; Cardamone et al., 2014; Cederquist et al., 2017; Lentucci et al., 2016). This raises a number of important questions regarding GPS2 function and regulation in the cell. For example, does GPS2 shuttles between different intracellular locations? Is its extra-nuclear residence important for informing and modulating GPS2-mediated transcriptional regulation? Do the opposing functions of GPS2 and ubiquitin-conjugating enzyme Ubc13 contribute to the coordinated regulation of metabolic and inflammatory pathways?
Results
Dual mitochondrial/nuclear localization of GPS2
To address these questions, we first sought out a detailed description of its subcellular localization. In agreement with its proposed functions, GPS2 was previously observed inside and outside of the nucleus (Cardamone et al., 2012). Closer inspection revealed that the extra-nuclear staining was both diffused in the cytosol as well as concentrated in the mitochondria (Fig. 1A, Supplemental Fig. S1A/S1B). Extensive HA mitochondrial staining was observed upon expression of HA-GPS2 (Supplemental Fig. S1C), confirming mitochondrial targeting of the fusion protein, and mitochondrial localization of endogenous GPS2 was confirmed by electron microscopy (Supplemental Fig. S1D). These results together identify GPS2 as a previously unrecognized mitochondrial protein.
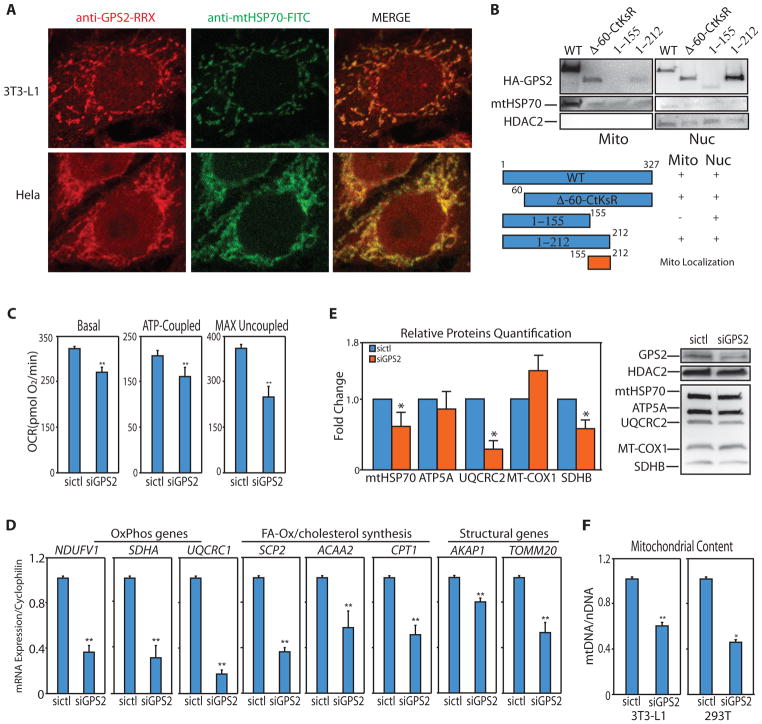
Figure 1. GPS2 localizes to mitochondria and regulates mitochondrial function through neMITO gene expression.
(A) GPS2 and mtHSP70 immunostaining in 3T3-L1 and Hela cells.
(B) WB of 293T cells are transfected with full length GPS2 (wt), GPS2 with deletion of first 60 aa and mutation of K254,300,327 (Δ60-CtKsR), GPS2 1–155 and 1–212.
(C) Decreased respiration and oxygen consumption in 3T3-L1 GPS2-KD cells.
(D) Relative expression of representative neMITO genes by RT-qPCR in 3T3-L1 cells.
(E) Mitochondrial proteins expression by WB with relative quantification, n=3 independent blots.
(F) Mitochondrial DNA content in WT and GPS2-KD 3T3-L1 and 293T cells.
* indicate p-value<0.05; ** indicate p-value<0.01.
To dissect the domain responsible for mitochondrial localization, we investigated the subcellular localization of different deletion constructs. Because N-terminal deletions lead to protein degradation, deletion of the first 60 aa was tested in association with mutations preventing protein ubiquitination (Huang et al., 2015). In this stabilized form, deletion of the N-terminus did not affect mitochondrial localization (Fig. 1B), confirming the absence of a classic N-terminal mitochondrial localization signal (MLS) as indicated by in silico prediction analyses (TargetP1.1 and Mitoprot). Likewise, the C-terminus domain was not required for mitochondrial localization. Deletion of aa.155–327, instead, severely impaired the expression of HA-GPS2 in the mitochondria (Fig. 1B), indicating that mitochondrial localization is driven by an internal import signal.
GPS2 regulate mitochondrial function through transcriptional regulation of nuclear-encoded mitochondrial genes
To investigate the physiological relevance of GPS2 in the regulation of mitochondrial functions, we measured mitochondrial respiratory capacity in intact cells. In both HeLa and 3T3-L1 cells, GPS2 downregulation (GPS2-KD) caused a significant decrease in maximal respiratory capacity and ATP-linked respiration (Fig. 1C, Supplemental Fig. S1E). Because of GPS2 well-known role in transcriptional regulation, we asked whether these phenotypes were caused by defective expression of mitochondrial genes. In agreement with this hypothesis, GPS2 was required for the expression of a variety of nuclear-encoded mitochondrial (neMITO) genes selected across different functional classes (Fig. 1D, distinct siRNAs validation and HA-GPS2 rescue in Supplemental Fig. S1F/S1G). In contrast, the expression of mtDNA-encoded genes was not affected by GPS2-KD (Supplemental Fig. S1H). These results were confirmed by corresponding changes in the levels of representative nuclear-encoded but not mitochondrial-encoded proteins (Fig. 1E). GPS2 downregulation also caused a significant reduction in mtDNA copy number (Fig. 1F). This suggests that the reduced respiratory capacity of GPS2-KD cells might result from a decline in the number of mitochondria, possibly caused by impaired expression of a broad mitochondrial gene program, rather than a specific defect in respiratory activity.
To further investigate GPS2 role in sustaining mitochondrial gene expression, we performed RNA-seq profiling of 3T3-L1 WT and GPS2-KD cells. Considering the full transcriptome, we identified 1086 Differentially Expressed (DE) genes, including 439 downregulated and 647 upregulated genes (Fig. 2A, left and Supplemental Table 1). Mitochondrial GO terms were one of the most significantly enriched categories among downregulated genes, whereas upregulated genes associated with non-mitochondrial terms (Fig. 2A, middle). Accordingly, mitochondrial genes represent approximately 20% of the downregulated genes, as compared to 10% of the upregulated genes (Fig. 2A, left). Also, only downregulated mitochondrial genes are enriched for canonical mitochondrial pathways like “TCA cycle” and “Electron transport chain” (Fig. 2A, right). Upregulated mitochondrial genes associate with “Interleukin signaling pathways”, “SREBP signaling” and “NFkB signaling” (Fig. 2A). This suggests their upregulation could result from a secondary response to mitochondrial dysfunction or from the upregulation of other cytosolic pathways, as expected based on GPS2 regulating insulin signaling and pro-inflammatory pathways (Supplemental Fig. S2A)(Cederquist et al., 2017; Lentucci et al., 2017). Notably, a significant depletion in genes associated with mitochondrial functions was similarly observed in the transcriptional signatures of GPS2-KD 293T cells (Supplemental Table 2)(Cardamone et al., 2012) and GPS2-deleted B cells (Lentucci et al., 2016). Thus, profiling of different cell types indicates that GPS2 is broadly required for the activation of mitochondrial gene expression, with its downregulation being associated with transcriptional reprogramming linked to cell growth and metabolism.
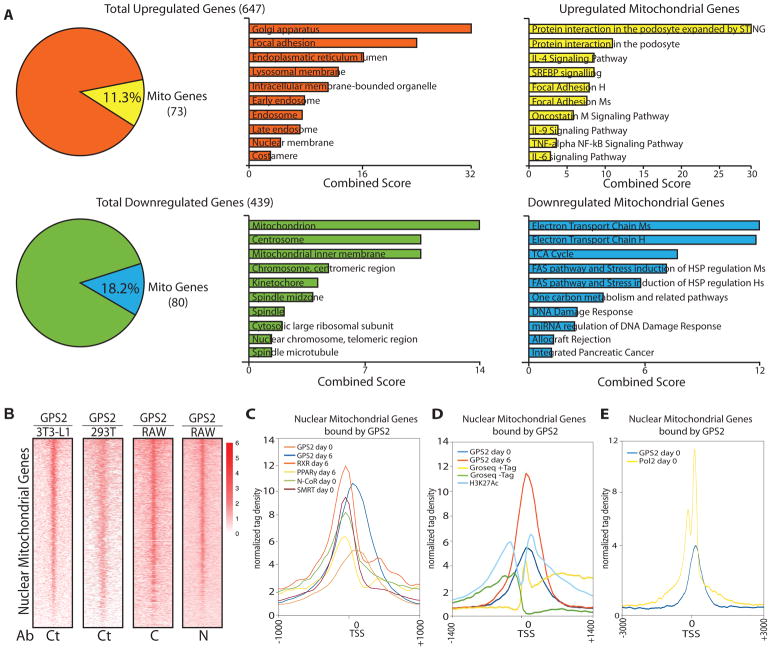
Figure 2. GPS2 binds to the core proximal promoter of neMITO genes to regulate their transcriptional activation.
(A) Left: DE genes between WT and GPS2-KD 3T3-L1 cells by RNAseq. Center: most significant GO terms associated with total upregulated (orange) and downregulated (green) genes. Right: most significant pathways associated with upregulated (yellow) and downregulated (blue) mitochondrial genes. GO terms and WikiPathways enrichment is based on combined score between p-value and Z-score (EnrichR).
(B) Heatmaps of GPS2 binding on mitochondrial gene promoters by ChIPseq (2086 murine genes and 2296 human genes). GPS2 binding profiles in 3T3-L1 and 293T cells were generated using Ct antibody (GSE57779 and GSE35197). Profiles in RAW cells were generated using distinct antibodies against the N and C terminus (GSE66774).
(C) Tag density profile of GPS2 binding on mitochondrial genes compared to the binding for RXR, PPARγ, NCoR and SMRT (GSE13511 and Array Express E-MTAB-1031) in 3T3-L1 at day0 or day6 of differentiation.
(D) Tag density profile of GPS2 binding on mitochondrial genes compared to GROseq tags (GSE56745) and ChIPseq tags of H3K27Ac (GSE36965).
(E) Tag density profile of GPS2 binding on mitochondrial genes compared to POL2 binding (GSE13511) in undifferentiated 3T3-L1.
To investigate whether GPS2 directly regulates the expression of neMITO genes as a transcriptional cofactor, we re-analyzed previously published GPS2 ChIP-seq datasets by focusing on mitochondrial genes. Strikingly, GPS2 binding was observed on most mitochondrial genes in each cell model, including macrophages, adipocytes and embryonic kidney cells (Fig. 2B). In each model, GPS2 binding to neMITO genes was concentrated to promoter areas (Supplemental Fig. S2B), in contrast to the broader distribution to both proximal and distal regulatory regions previously reported genome-wide (Cardamone et al., 2012; Cardamone et al., 2014; Fan et al., 2016). Accordingly, GPS2 peaks on mitochondrial genes showed a significant enrichment in promoter-associated motifs (Supplemental Fig. S2C). Also, tag density plots showed that GPS2 binding is shifted towards the transcription start site (TSS) when compared to nuclear receptors and associated corepressors (Fig. 2C)(Lefterova et al., 2010; Raghav et al., 2012). This suggests that GPS2 recruitment to neMITO genes is distinct from what previously reported for other genomic targets. In keeping with this conclusion, comparative analysis with existing GRO-seq and H3K27Ac profiles in adipocytes (Harms et al., 2015; Step et al., 2014) confirmed that GPS2 binding concentrated around the TSS of mitochondrial genes (Fig. 2D) where it overlapped with the binding profile of RNA Polymerase II (POL2)(Siersbaek et al., 2011)(Fig. 2E). Thus, collectively, integration of RNA-seq and ChIP-seq data reveals that GPS2 regulates mitochondrial functions through proximal-promoter binding and transcriptional regulation of a vast mitochondrial program.
GPS2-mediated inhibition of Ubc13 activity is required for H3K9 demethylation and expression of neMITO genes
As GPS2 is a potent inhibitor of the ubiquitin-conjugating enzyme Ubc13 and regulates the expression of selected PPARγ target promoters through stabilization of an H3K9 demethylase (Cardamone et al., 2014), we then asked whether H3K9 demethylation and local ubiquitination are involved in GPS2-mediated regulation of neMITO genes. In 3T3-L1 GPS2-KD cells we detected a significant increase in the level of both K63 ubiquitin chains (Fig. 3A) and H3K9me3 occupancy (Fig. 3B) on the promoters of representative neMITO genes. In agreement with the regulation of PPARγ-bound promoters, the accumulation of H3K9me3 on Ndufv1 and Tomm20 promoters correlated with a significant decrease in binding of H3K9 demethylase JMJD2A/KDM4A (Supplemental Fig. S3A). KDM4A binding though is limited to a small subset of mitochondrial genes (Cardamone et al., 2014), whereas ChIP-seq profiling of H3K9 methylation upon GPS2-KD showed a specific and significant increase in H3K9me3 on the entire mitochondrial genes set, as compared to the whole genome or randomly selected groups of genes of comparable size (Fig. 3C, Supplemental Fig. S3B). Notably, the increase in H3K9me3 is not restricted to mitochondrial genes as significant accumulation is observed also on non-mitochondrial GPS2 target promoters. (Supplemental Fig. S3C/S3D). For both sets, the basal level of H3K9me3 in the surrounding area is significantly higher than for other genomic loci (Fig. 3C, Supplemental Fig. S3B–D). This indicates that, in normal cells, GPS2 target promoters are specifically marked by the exclusion of H3K9me3, which is lost, or at least significantly reduced, upon GPS2 downregulation.
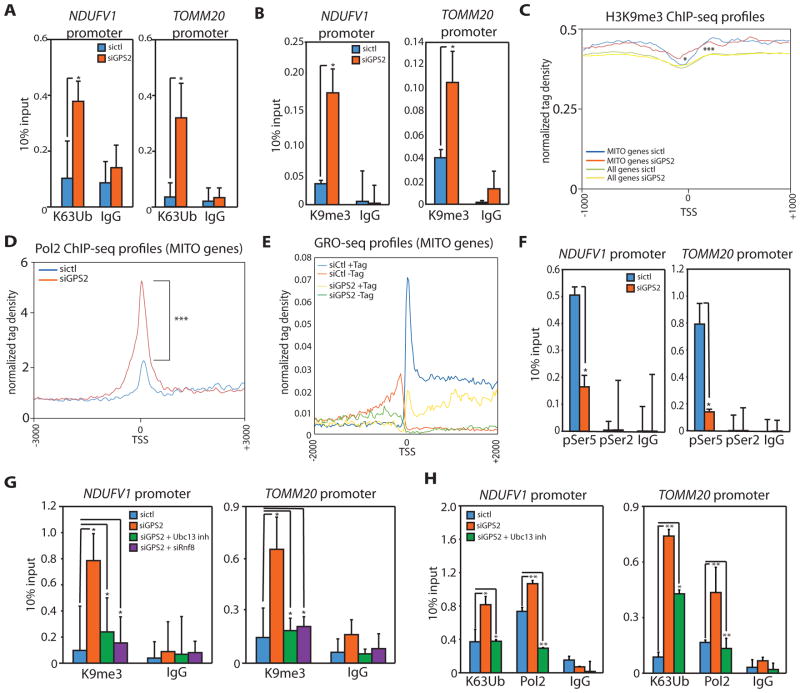
Figure 3. GPS2-mediated inhibition of K63 ubiquitination is required for H3K9 de-methylation, POL2 processivity and transcription of mitochondrial genes.
(A)(B) Increased K63 ubiquitination (A) and H3K9me3 (B) upon GPS2-KD by ChIP on Ndufv1 and Tomm20.
(C) Tag density profile of H3K9me3 showing the increase in H3K9me3 is specific to mitochondrial gene TSSes (2086 genes) compared to the TSSes of all genes (20950 genes in mm8 with length>1kb). * refers to the comparison between mitochondrial genes in sictl vs sigps2. *** refers to the comparison between mitochondrial genes and all mm8 genome in sictl.
(D) Tag density profile of POL2 ChIPseq peaks showing a significant increase in POL2 binding on mitochondrial TSSes in GPS2-KD cells.
(E) Tag density profile of GROseq data showing nascent sense (+Tag) and anti-sense (−Tag) transcription being impaired in GPS2-KD cells.
(F) ChIP analysis of POL2 phosphorylation (Ser5 and Ser2).
(G) Accumulation of H3K9me3 in GPS2-KD cells is rescued either by treatment with the Ubc13 inhibitor NSC697923 or by transient downregulation of ubiquitin ligase RNF8.
(H) Inhibition of Ubc13 activity by NSC697923 rescues the increase in K63 ubiquitin chains and POL2 binding upon GPS2-KD.
* indicate p-value<0.05; ** indicate p-value<0.01, *** indicate p-value<0.001. P-values were computed using the Wilcoxon test based on the difference in normalized counts +/−200bp around the TSS. All experiments are in 3T3-L1 cells.
Surprisingly, the accumulation of H3K9me3 was associated with increased POL2 occupancy (Fig. 3D, Supplemental Fig. S3E). A quantitatively significant increase in POL2 binding was observed for both the mitochondrial gene set and other GPS2-bound promoters, as observed for H3K9me3 (Supplemental Fig. S3F). This suggests that POL2 activity, rather that its recruitment to GPS2-target genes, is affected by the methylation status of the promoter. To interrogate POL2 status on GPS2-regulated genes, we measured nascent RNA by GRO-seq. In accord with the transcriptomic analysis, transcription of neMITO genes was found dramatically reduced upon GPS2 downregulation (Fig. 3E, Supplemental Table 3). Surprisingly, the lack of significant accumulation of nascent RNA detected over the first 50–100bp indicated that GPS2-KD does not promote an increase in POL2 pausing on neMITO genes, but rather inhibits POL2 activation (Fig. 3E). The read density pattern of GRO-seq tags in GPS2-KD cells in fact corresponds to that observed in cells treated with the initiation inhibitor Triptolide, rather than the pausing escape inhibitor Flavopiridol (Chen et al., 2015; Jonkers et al., 2014). In keeping with this conclusion, profiling the phosphorylation status of POL2 CTD in GPS2-KD cells confirmed that neither Ser5-P nor Ser2-P were detectable on representative promoters, despite the increase in POL2 accumulation (Fig. 3F). Notably, the transcription of mitochondrial genes upregulated in GPS2-KD cells by RNA-seq was also found impaired by GRO-seq (Supplemental Fig. S3G). This result confirms our initial hypothesis that the increased mRNA levels observed for a subset of genes by RNA-seq profiling (see Fig. 2A) results from a secondary response, possibly involving post-transcriptional mechanisms of RNA stabilization. In conclusion, integration of our ChIP-seq and GRO-seq experiments reveals that GPS2 depletion causes a robust accumulation on the promoters of neMITO genes of hypo-phosphorylated POL2, which is unable to escape from the core promoter and promote productive transcription, likely because of the altered chromatin environment aberrantly enriched in the repressive histone mark H3K9me3.
Finally, we asked to which extent GPS2 regulates chromatin remodeling and POL2 activation through modulation of Ubc13-mediated ubiquitination. Inhibition of Ubc13 enzymatic activity (Pulvino et al., 2012), or downregulation of the ubiquitin ligase RNF8 via siRNA (Cardamone et al., 2014), were indeed sufficient for restoring the normal level of ubiquitination and for rescuing the aberrant accumulation of H3K9me3 and the stalling of POL2 in GPS2-KD cells (Fig. 3G, 3H). Thus, our results together support the existence of a regulatory strategy, specific for neMITO genes and other GPS2 target promoters, which requires GPS2 presence to promote gene expression by preventing the establishment of a repressive chromatin environment and the stalling of POL2 via inhibition of Ubc13-mediated ubiquitination.
Retrograde translocation of GPS2 upon mitochondria depolarization
While these results establish that GPS2 is required to sustain the expression of nuclear-encoded mitochondrial components, our data indicates that GPS2 is not required for the expression of genes encoded in the mitochondrial genome, thus raising the question of what is the function of GPS2 physical presence in the mitochondria. As the expression of neMITO genes has to be regulated in response to changes in mitochondrial and cellular demands, we became intrigued by the possibility that mitochondrial GPS2 (mtGPS2) might play a role in integrating signals from the mitochondria with the regulation of nuclear transcription, a function known as mitochondrial retrograde signaling. To explore this hypothesis, we first used the uncoupling agent FCCP to depolarize mitochondria. Treatment with FCCP induces the activation of an integrated stress response program that requires mitochondria-to-nucleus signaling for promoting the transcriptional activation of neMITO and stress response genes (Catic et al., 2013; Quiros et al., 2017). Upon FCCP treatment, we rapidly observed a significant decrease in GPS2 localization in the mitochondria, as shown by loss of co-staining with mtHSP70 (Fig. 4A). As total GPS2 protein levels did not change (Supplemental Fig. S4A), this indicates that mtGPS2 redistributes within the cell. However, only a weak increase in nuclear staining was seen by IF, likely because of the significant amount of nuclear GPS2 in basal conditions (Fig. 4A).
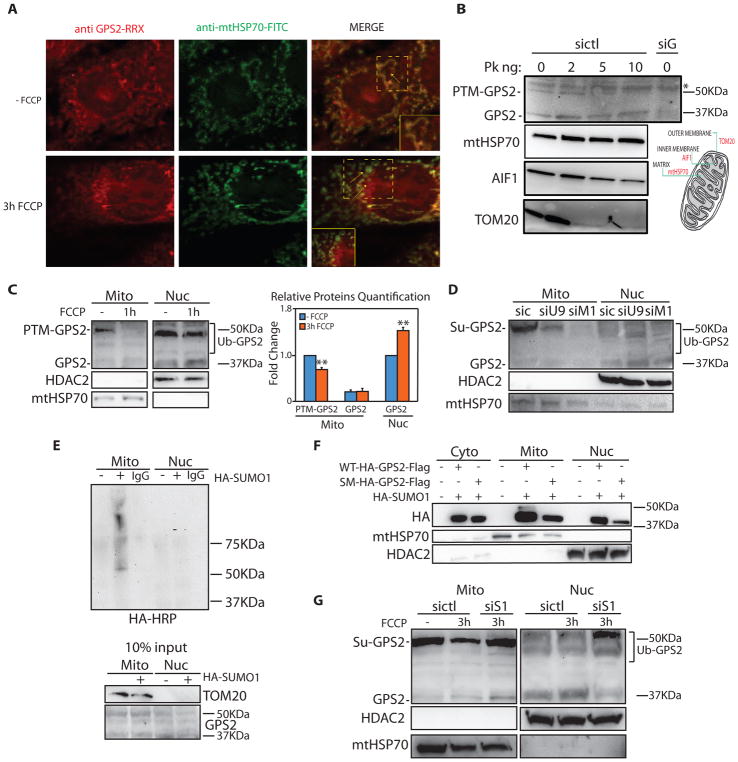
Figure 4. Mitochondria-to-nucleus regulated translocation of GPS2 in response to mitochondria depolarization.
(A) Reduced GPS2 localization to the mitochondria and corresponding increased nuclear staining (see inset and arrows) upon 3h of FCCP in 3T3-L1 cells.
(B) Proteinase K protection assay. Non-modified GPS2 and post-translationally modified GPS2 (PTM-GPS2) localize respectively to the matrix and OMM. Degradation patterns of TOM20, AIF1 and mtHSP70 are representative of proteins in the OMM, IMM and matrix.
(C) WB of fractionated extracts showing mitochondria-to-nucleus translocation of GPS2 in 3T3-L1 cells upon 1h FCCP (relative quantification on the right).
(D) GPS2-PTM is reduced by downregulation of either Ubc9 (siU9) or Mul1 (siM1).
(E) Sumoylation assay with HA-SUMO1 in 293T cells.
(F) Mitochondria localization of HA-GPS2-Flag WT or Sumo-mutant (SM: K45, K71R) in 293T cells.
(G) Impaired translocation of GPS2 in FCCP-treated cells transfected with siSENP1 (siS1).
To best appreciate the mitochondria-to-nucleus translocation, we assessed the changes in GPS2 localization upon FCCP through subcellular fractionation. This approach revealed that GPS2 is differentially modified in the various cellular compartments (see Supplemental Fig. S4B for bands specificity). In particular, within the mitochondrial compartment, we observed two distinct pools of GPS2. The most abundant pool ran significantly higher than the expected MW of 36kD, likely because of post-translational modifications (PTM-GPS2) (Fig. 4B, Supplemental Fig. S4B). Proteinase K protection assays indicated that PTM-GPS2 localizes to the OMM whereas a distinct pool of non-modified GPS2 localizes to the matrix (Fig. 4B). Differential sensitivity of the two pools to trypsin digestion confirmed their distinct localization (Supplemental Fig. S4C). Upon FCCP treatment, GPS2 was found to redistribute from the mitochondria to the nucleus, with the increase in nuclear non-modified GPS2 being matched by a striking decrease in PTM-GPS2 in the mitochondria (Fig. 4C).
Regulation of GPS2 intracellular localization through sumoylation/desumoylation
This result suggested that mitochondria-to-nucleus translocation of GPS2 could be modulated by post-translational modifications. Based on the size of the shift, on previous reports of GPS2 sumoylation (Bi et al., 2014; Huang et al., 2015), and consistent with the important role played by SUMO enzymes in the regulation of mitochondrial function and dynamics (Braschi et al., 2009; Fu et al., 2014; Prudent et al., 2015; Zunino et al., 2007), we asked whether GPS2 sumoylation accounts for the PTM observed in the mitochondria. This hypothesis was confirmed by the following experiments: i) downregulation of either the SUMO-conjugating enzyme UBC9 or the mitochondrial specific SUMO ligase MUL1 (Fig. 4D, Supplemental Fig. S4D–E); ii) in vivo sumoylation of endogenous GPS2 with HA-SUMO1 within the mitochondrial fraction (Fig. 4E). Also, in vivo sumoylation of overexpressed GPS2 was abrogated by specific mutations of the sumoylation sites (Fig. 4F). Together, these results confirm that sumoylation on K45/K71 accounts for the shift in GPS2 size in the mitochondria. Notably, sumoylation appears restricted to the mitochondria based on the specificity of the SUMO ligase and the fact that GPS2 higher molecular weight bands in the nucleus were not affected by UBC downregulation, despite a small but consistent increase in nuclear non-modified GPS2 (Fig. 4D). This observation, together with the decrease in SUMO-GPS2 observed upon FCCP-induced translocation of GPS2 (see Fig. 4C), suggested that GPS2 sumoylation status could be important for determining its intracellular localization and its nuclear translocation upon FCCP. Indeed, downregulation of the SUMO protease SENP1, which is induced upon mitochondria depolarization (Supplemental Fig. S4G) and was previously reported to desumoylate GPS2 (Bi et al., 2014), was sufficient to promote not only the stabilization of SUMO-GPS2, but also its retention in the mitochondria (Fig. 4G, Supplemental Fig. S4F). These results confirm that SENP1-mediated desumoylation of GPS2 regulates its retrograde translocation.
GPS2 is required for the transcriptional activation of the nuclear stress response program upon mitochondria depolarization
To dissect the transcriptional programs regulated by GPS2 upon nuclear translocation, we performed ChIP-seq for GPS2 upon FCCP treatment. Using GPS2 peaks in non-stimulated cells as background, we identified 11,308 genomic locations where GPS2 binding increases or occurs only upon FCCP treatment (Supplemental Table 4). These included a significant number of neMITO genes (42% of all neMITO genes) as well as non-mitochondrial genes. Pathway enrichment analysis of the non-mitochondrial targets revealed a striking enrichment in FOXO signaling, a known mediator of the mitochondrial stress response (Mouchiroud et al., 2013; Webb and Brunet, 2014), suggesting that GPS2 translocation might be important for regulating a broader transcriptional response to mitochondrial stress.
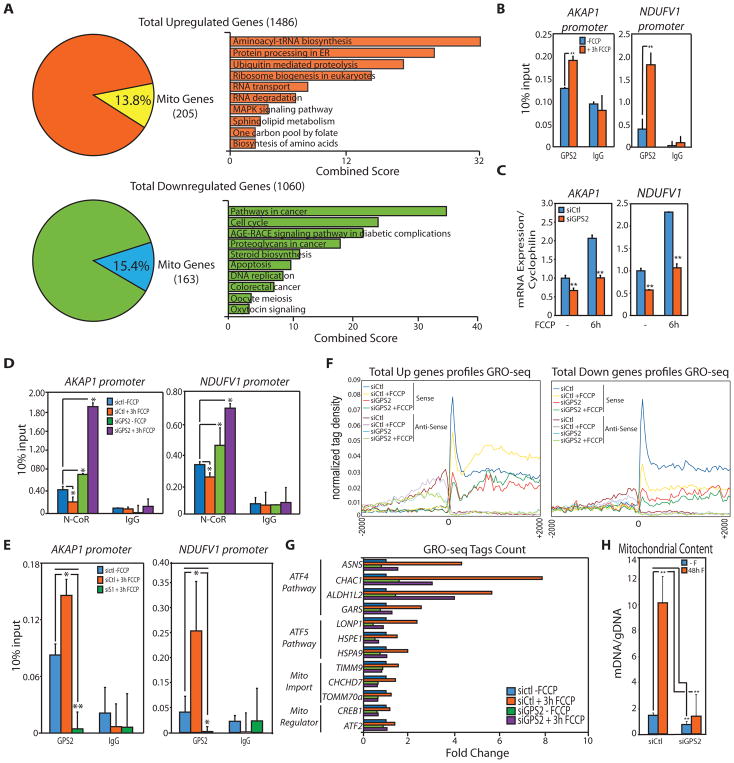
Recently, integrative analysis of HeLa cell proteomic, transcriptomic and metabolic profiling has defined a comprehensive stress response activated upon long term exposure to various mitochondrial stressors, including FCCP (Quiros et al., 2017). To discriminate between primary and secondary events within this response and assess to which extent GPS2 retrograde translocation is required for the transcriptional activation of a stress response program, we analyzed the early transcriptional events induced upon FCCP treatment by GRO-seq in WT and GPS2-KD 3T3-L1 cells. In WT cells, we identified 1486 upregulated and 1060 downregulated genes upon FCCP (Fig. 5A and Supplemental Table 3). In agreement with published work, FCCP stimulation induced the activation of genes associated with “Folate metabolism”, “tRNA aminoacylation”, “Aminoacid metabolism”, “Carbon metabolism” and “Ubiquitin-mediated proteolysis” (Fig. 5A, Supplemental Fig. S5A and Supplemental Table 3)(Quiros et al., 2017). Downregulated genes associated with “Cell cycle”, “Cholesterol and Lipid Metabolism”, “Glucose metabolism” and “Apoptosis” (Fig. 5A, Supplemental Fig. S5A, Supplemental Table 3). Mitochondrial genes were similarly represented among both groups (205 upregulated and 163 downregulated mitochondrial genes) and, despite the enrichment not reaching statistical significance, included in the downregulated set were numerous genes associated with “Oxidative phosphorylation” and “Mitochondrial Ribosomal Proteins” - categories reported decreased by proteomic analysis at later times (Quiros et al., 2017). Thus, our results indicate that transcriptional regulation represents an important part of the stress response to mitochondrial depolarization.
Figure 5. Impaired nuclear response to mitochondrial depolarization in GPS2-KD 3T3-L1 cells.
(A) DE genes after 3h FCCP as defined by GROseq with most enriched KEGG pathways.
(B) ChIP analysis of GPS2 recruitment to AKAP1 and NDUFV1 upon 3h of FCCP.
(C) Relative expression of AKAP1 and NDUFV1 upon 6h of FCCP.
(D) Dismissal of NCoR from AKAP1 and NDUFV1 promoters upon 3h FCCP is impaired in GPS2-KD cells.
(E) Increased GPS2 binding upon FCCP is impaired in cells transfected with siSENP1 (siS1).
(F) Tag density profiles of GRO-seq data showing that transcriptional activation of FCCP-upregulated genes is abrogated in GPS2-KD cells.
(G) GROseq tag count showing that GPS2 is required for the activation of representative neMITO and stress response genes.
(H) Increase in mtDNA content upon long term FCCP treatment (48h) is impaired in GPS2-KD 3T3-L1 cells.
* indicate p-value<0.05; ** indicate p-value<0.01.
To confirm that GPS2 translocation to the nucleus is a required component of this transcriptional response to mitochondrial stress, we first focused on neMITO genes. To our initial surprise, none of the neMITO genes activated in response to FCCP through regulation of NCoR stability (Catic et al., 2013) was included among the DE genes identified by GRO-seq. However, further characterization confirmed a significant increase in GPS2 binding to AKAP1 and Ndufv1 promoters upon 3h of FCCP (Fig. 5B), followed by GPS2-dependent increase in gene expression at later time points (Fig. 5C). Also, ChIP analysis confirmed that FCCP-induced activation of both genes associated with the dismissal of NCoR, whereas in GPS2-KD cells they were marked by increased NCoR binding (Fig. 5D). Most importantly, downregulation of SENP1 confirmed that their activation upon depolarization depended on GPS2 retrograde translocation (Fig. 5E).
Next, we investigated to which extent GPS2-mediated regulation extended to non-mitochondrial genes by comparing the GRO-seq signature of genes upregulated and downregulated upon FCCP in WT and GPS2-KD cells. Basal transcription of both upregulated and downregulated genes, as well as activation of the genes upregulated upon FCCP, were greatly impaired upon GPS2-KD (Fig. 5F). Notably, GPS2 target genes induced upon FCCP treatment include known targets of both the ATF4-dependent integrated stress response and ATF5-dependent mtUPR response (Fiorese et al., 2016; Quiros et al., 2016), in addition to proteins important for mitochondrial protein import and TFs previously associated with retrograde signaling and mitochondria biogenesis (Arnould et al., 2015; Biswas et al., 1999; Than et al., 2011; Vankoningsloo et al., 2006)(Fig. 5G, Supplemental Fig. S5B, Supplemental Table 4). This indicates that GPS2 plays a critical role in the transcriptional response to mitochondrial stress induced by depolarization. Accordingly, the increase in mitochondrial content induced during the recovery response to FCCP depolarization was dramatically impaired in GPS2-KD cells (Fig. 5H). These results confirm that GPS2 regulated translocation from mitochondria to the nucleus is required for the transcriptional activation of an extensive transcriptional program that includes both stress response genes as well as neMITO genes, as required for responding to mitochondrial depolarization through cellular reprogramming and regulation of mitochondrial functions.
Retrograde translocation of GPS2 during adipogenesis
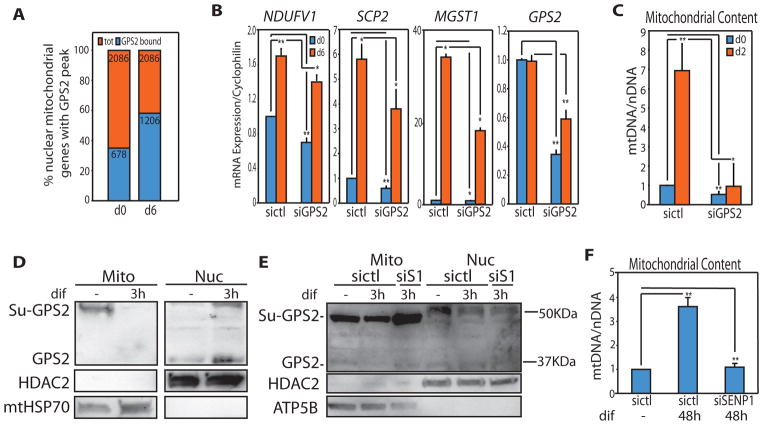
Because of GPS2 requirement in regulating basal mitochondrial gene expression, we reasoned that mitochondria-to-nucleus translocation of GPS2 might not be limited to stress-response events. To explore the role of GPS2 in regulating mitochondrial gene expression under physiological conditions, we focused on the adipogenic differentiation of 3T3-L1 cells, a process known to elicit significant mitochondrial biogenesis (Wilson-Fritch et al., 2003). In support of GPS2 relevance for the regulation of mitochondrial gene expression during adipogenesis we observed i) a 50% increase in GPS2 binding to the promoters of mitochondrial genes when comparing ChIP-seq data from 3T3-L1 preadipocytes or mature adipocytes (Cardamone et al., 2014)(Fig. 6A), and ii) a significant reduction in the activation of adipo-specific neMITO genes in GPS2-depleted, PPARγ-expressing preadipocytes (Fig. 6B). Accordingly, differentiation-induced increase in mitochondrial content was impaired in GPS2-KD cells (Fig. 6C). Most importantly, GPS2 engagement for the expression of neMITO genes correlated the translocation of SUMO-GPS2 from mitochondria to the nucleus (Fig. 6D) and downregulation of SENP1 inhibited both mitochondria-to-nucleus translocation of GPS2 and mitochondrial biogenesis during differentiation (Fig. 6E, 6F). Thus, we conclude that GPS2 retrograde signaling is important for regulating the expression of mitochondrial genes not only in response to stress, but also during the physiological reprogramming associated with cell differentiation, in this case of adipocytes.
Figure 6. GPS2 retrograde translocation supports mitochondrial biogenesis during 3T3-L1 adipogenesis.
(A) GPS2 binding to mitochondrial genes increases upon differentiation (GSE57779).
(B) Relative expression of adipo-specific neMITO genes by RT-qPCR.
(C) Mitochondrial biogenesis, as measured by increase in mtDNA content, is impaired in GPS2-KD cells.
(D) WB of fractionated extracts showing mitochondria-to-nucleus translocation of GPS2 after 3h of differentiation.
(E)(F) GPS2 desumoylation and translocation (E) and mitochondrial biogenesis (F) during adipogenesis are impaired in cells transfected with siSENP1 (siS1).
* indicate p-value<0.05; ** indicate p-value<0.01.
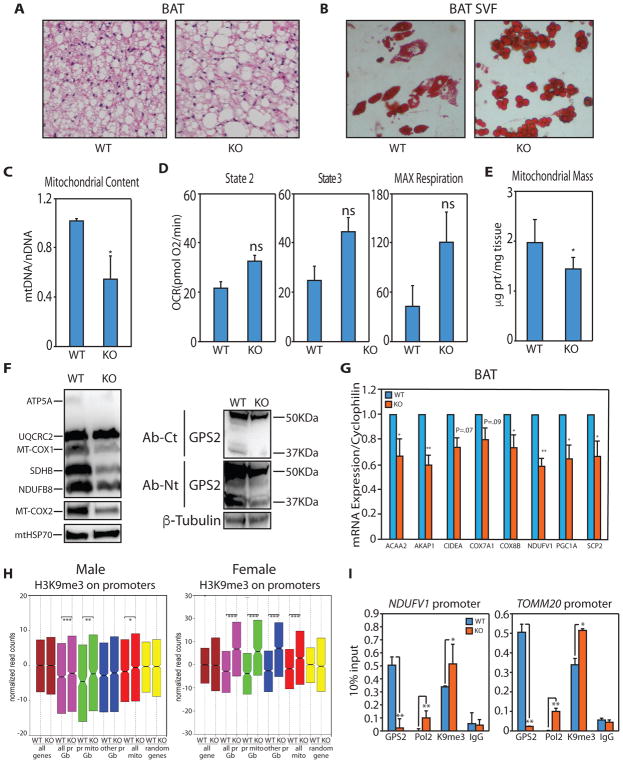
Reduced mitochondrial content in the BAT of GPS2-AKO mice
Adipose tissue-specific deletion of GPS2 promotes obesity associated with constitutive insulin signaling, increased lipid deposition in the white adipose tissue (WAT) and improved systemic insulin sensitivity (Cederquist et al., 2017). In agreement with the altered ratio of lipogenesis/lipolysis observed in the WAT, inspection of the BAT revealed that GPS2 deletion promoted whitening of the brown depot (Fig. 7A). A significant increase in lipid accumulation was also observed in adipocytes differentiated from the BAT stroma vascular fraction (SVF) of GPS2-AKO mice (Fig. 7B). To investigate whether this phenotype reflected a defect in mitochondrial activity, we first measured mtDNA copy number, and found that GPS2-deficient mice had 50% less mtDNA in the BAT than their WT littermates (Fig. 7C). On the contrary, oxygen consumption in isolated mitochondria was not affected by GPS2 absence (Fig. 7D). These results indicate while mitochondrial respiration is not impaired, GPS2-deficient BAT has a significant decrease in mitochondrial mass, which was confirmed by measuring the amount of mitochondrial proteins in the tissue (Fig. 7E, 7F). In accord with this conclusion and with our in vitro studies, we observed significant downregulation of representative neMITO genes (Fig. 7G). To confirm GPS2 role in the regulation of neMITO genes in vivo, we performed ChIP-seq for GPS2 in the BAT of WT and GPS2-AKO mice. Stringent definition of GPS2 peaks in brown adipocytes (40,040 peaks in male mice, 19,150 peaks in female mice) was achieved using the signal from GPS2-AKO BAT as background (Supplemental Table 5, Supplemental Fig. S6A/S6B). As previously seen in cell lines, GPS2 peaks distributed between proximal and distal regions (Supplemental Table 5). Notably, GPS2 binding to promoter locations associated with active transcription as indicated by significant overlap with the H3K27ac mark (77% of GPS2-bound promoters in male, 83% of GPS2-bound promoters in female mice are marked by H3K27ac)(Harms et al., 2015). On the contrary, no significant overlapping was observed on enhancers and other locations, confirming a promoter-specific role for GPS2 as a transcriptional activator. In addition, GPS2-bound promoters included about 30–40% of active mitochondrial genes (581 neMITO genes in male and 415 neMITO genes in female mice are marked by GPS2 peaks). Among them, we observed key regulators of BAT thermogenic capacity, such as PGC1α and UCP1, and genes representative of a variety of mitochondrial functions including import proteins, mitochondrial ribosomal proteins, components of all ETC complexes, fatty acid oxidation enzymes and two (Supplemental Table 5). Thus, our findings together support a key role for GPS2 in regulating mitochondrial functions and biogenesis in the adipose tissue through retrograde signaling and transcriptional regulation. Lastly, ChIP-seq analysis of H3K9me3 was performed in the BAT from WT and GPS2-AKO littermates to confirm the mechanistic details of GPS2-mediated transcriptional regulation. A significant increase in H3K9me3 accumulation on the promoters of neMITO genes and other GPS2-bound promoters was observed in GPS2-AKO mice of both sexes (Fig. 7H). Furthermore, the increase in H3K9 methylation correlated with aberrant accumulation of POL2 on the promoters of Ndufv1 and Tomm20 (Fig. 7I), thus fully confirming in vivo the molecular mechanism described in cells.
Figure 7. GPS2 regulates mitochondrial biogenesis in vivo in Brown Adipose Tissue.
(A) Increased lipid deposition in the BAT of GPS2-AKO by H&E staining.
(B) Oil Red O staining of in vitro differentiated adipocytes from BAT SVF.
(C) Decreased mtDNA content in the BAT of GPS2-AKO mice compared to WT littermates.
(D) Quantification of oxygen consumption rates (OCR) in BAT isolated mitochondria from chow-fed WT and GPS2-AKO littermates under the different respiratory states. Results represent average ± SEM for complex I-driven respiration (pyruvate-malate, n=6 mice per group), complex II-driven respiration (succinate-rotenone, n=6 mice per group).
(E) Reduced mitochondrial protein content in BAT isolated from GPS2-AKO compared to WT littermates.
(F) WB of mitochondrial proteins in BAT from GPS2-AKO and WT littermates. GPS2 deletion in GPS2-AKO mice is specific to mature adipocytes.
(G) RT-qPCR analysis of representative neMITO genes in the BAT of WT and KO mice.
(H) Box plot of H3K9me3 ChIPseq performed on BAT from WT and KO littermates showing a significant increase in H3K9me3 on neMITO and other GPS2-bound promoters in GPS2-AKO mice. In pink are represented all GPS2-bound promoters (all pr Gb), in green GPS2-bound mitochondrial promoters (pr mito Gb), in blue GPS2-bound non-mitochondrial promoters (other pr Gb), in red all mitochondrial promoters (all mito). Yellow represents a random set of genes.
(I) Increased promoter occupancy of POL2 and H3K9me3 by ChIP in GPS2-AKO BAT.
* indicate p-value<0.05; ** indicate p-value<0.01.
Discussion
Mitochondrial mass and functions differ considerably among tissues and are dynamically regulated in response to different physiological cues, including nutrient availability, cold exposure and endurance exercise. Mitochondrial biogenesis and remodeling are also critical for embryonic and adult stem cells differentiation (Hock and Kralli, 2009; Wanet et al., 2015). Because the majority of mitochondrial proteins are encoded in the nuclear genome, these functions critically depend on transcriptional regulation of mitochondrial gene expression in the nucleus (anterograde signaling) and feedback signaling from the mitochondria to the nucleus (retrograde signaling)(Finley and Haigis, 2009; Jazwinski, 2013; Quiros et al., 2016). Here, we have expanded the existing network of nuclear factors regulating the expression of mitochondrial gene expression to include GPS2. Notably, our data suggest that the complexity and specificity of mitochondrial gene expression is achieved through multiple levels of regulation mediated by distal (i.e. nuclear receptors and associated cofactors like PGC1α/β) and proximal regulators of transcription (i.e. GPS2). Future studies will be required to address how the different layers of regulation are coordinated to maintain cellular homeostasis.
Our work also indicates that GPS2 fulfills the role of a direct mediator of mitochondrial retrograde signaling in mammalian cells. Its transcriptional activity is in fact directly regulated and informed by the mitochondria functional status through regulated mitochondria-to-nucleus translocation. Pioneer work on mitochondria retrograde signaling in S. Cerevisiae identified a coordinated adaptation response to mitochondrial dysfunctions that includes the activation of metabolic and stress response genes. Key players in this pathway are DNA-binding TFs Rtg1/Rtg3 and a regulatory subunit, Rtg2, which senses changes in mitochondrial membrane potential, regulates the nuclear import of the TFs and directly contributes to the transcriptional regulation of the retrograde gene program in the nucleus as a component of the SLIK chromatin remodeling complex (Jazwinski, 2005; Liao and Butow, 1993; Miceli et al., 2011; Sekito et al., 2000). Based on the results reported here, we propose GPS2 as a functional homolog of Rtg2 in mammalian cells. As with Rtg2, GPS2 represents a direct link between the nuclear transcription factors regulating mitochondrial gene expression and the mitochondria. Similar to Rtg2 regulation, GPS2 nuclear translocation is triggered by mitochondria depolarization. Also, in accordance with Rtg2 contribution to chromatin remodeling through the SLIK (SAGA-like) complex, GPS2 directly binds to and modulates the chromatin environment of target gene promoters. Intriguingly, it does so by preventing the accumulation of H3K9me3, which is inhibitory against H3K9ac, a mark of active transcription deposited by Gcn5/PCAF, the catalytic subunit of the SAGA complex. Furthermore, Rtg2-mediated regulation of Rtg1–3 nuclear import via modulation of an inhibitory phosphorylation cascade closely resembles GPS2-mediated regulation of FOXO1 stabilization and nuclear functions via inhibition of AKT ubiquitination and activation (Cederquist et al., 2017; Lentucci et al., 2016). Lastly, our in vivo data indicate that GPS2 plays a critical role in the regulation of lipid metabolism and energy expenditure in the adipose tissue, just as RTG2 has proven to be an essential factor for the regulation of nutrient metabolism in yeast cells. In both cases, the phenotypic output is achieved through the coordinated regulation of an extensive transcriptional program, in the case of GPS2 including genes involved in mitochondrial biogenesis and functionality, adipocyte differentiation, lipid metabolism and stress response.
While these parallelisms are striking, they also raise numerous additional questions, including what is the nature of the relationship between GPS2 and the SAGA complex on nuclear target genes? What is the identity of GPS2 interacting partners in the mitochondria? And, how are GPS2 tethering to the mitochondria and the shuttling between nucleus and mitochondria regulated? Our results highlight a key role for the SUMO protease SENP1 in regulating GPS2 localization and activation in response to both developmental and mitochondrial stress. As SENP1 also regulates PGC1α via desumoylation (Cai et al., 2012), our results suggest that SENP1 induction upon mitochondrial stress might be an important step towards the activation of synergistic pathways. In addition, as both SENP1 and GPS2 are critically positioned at the intersection of metabolic and inflammatory pathways (Li et al., 2008; Shao et al., 2015; Xu et al., 2015), we speculate that SENP1-mediated regulation of GPS2 localization might important for crosstalk among these signaling cascades.
Interestingly, this study suggests that the crosstalk may extend to stress response pathways, as GPS2 emerges as a key regulator of the integrated mitochondrial stress response. Integration of our ChIP-seq and GRO-seq data in fact indicates that the early response to mitochondrial depolarization largely depends on GPS2 promoting the transcriptional activation of a nuclear program supporting mitochondrial recovery and reprogramming of cellular metabolism. This includes GPS2 recruitment to a number of key stress response gene activated through the ATF4/ATF5 mtUPR and stress response pathways (Fiorese et al., 2016; Quiros et al., 2017). Our results indicate that the promoters of neMITO genes and other GPS2 targets are specifically sensitive to the methylation status of H3K9. In mammalian cells, demethylation is partially achieved, but not limited to the action of the histone demethylase JMJD2/KDM4A, suggesting thatother demethylases might be important for providing specificity to the regulation of different gene subsets and contribute to fine-tuning the stress response (Duteil et al., 2016; Hino et al., 2012; Tian et al., 2016). Intriguingly, histone demethylases JMJD-1.2 and JMJD-3.1 are key mediators of lifespan extension and mtUPR response in worms (Merkwirth et al., 2016). Together these findings suggest that the expression of mitochondrial and stress response genes is controlled by a conserved regulatory strategy based on the modulation of H3K9 histone methylation/demethylation. They also reveal a role for H3K9me3 in the transcriptional regulation of active genes in addition to its well-known contribution to gene silencing and heterochromatin organization. Notably, as in the case of other stress response pathways, GPS2 availability in the nuclear compartment is tightly regulated by proteasomal degradation to avoid prolonged and possibly deleterious activation of the recovery program once homeostasis is re-established (Huang et al., 2015; Lamech and Haynes, 2015).
In conclusion, our work raises the intriguing possibility that GPS2 plays an important role in integrating multiple pathways regulating cell growth, inflammation, metabolism, and stress resistance through inhibition of ubiquitin signaling. Future studies addressing this hypothesis will be particularly relevant in the context of the adipose tissue where GPS2 downregulation is associated with human obesity and the establishment of inflammation within obese adipose tissue (Toubal et al., 2013). Our results, together with the recent characterization of GPS2 role in regulating insulin signaling (Cederquist et al., 2017), adds to the significance of GPS2 role in the adipose tissue and suggests that modulation of non-proteolytic ubiquitination might represent a hub for the integration of metabolic pathways modulating the balance between lipid storage and energy expenditure and promoting mitochondrial biogenesis to support tissue-specific functions, such as metabolites production and thermogenic activity. As therapeutic and lifestyle interventions designed for improving insulin sensitivity often rely on increased mitochondria biogenesis and oxidative metabolism (Kajimura and Saito, 2014; Zamora et al., 2015), unraveling the molecular mechanisms regulating mitochondrial functions is not only important for our basic understanding of cell biological processes, but have also the potential of leading to improved therapeutic treatments for obese and diabetic patients.
STAR*METHODS
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
|
| ||
| GPS2 rabbit polyclonal ct | custom | aa 307-327 |
| GPS2 rabbit polyclonal nt | custom | aa 1-11 |
| NCoR rabbit polyclonal | custom | (Jespen et al. 2000) |
| HDAC2 rabbit polyclonal | Santa Cruz | H-54 sc7899 |
| TOM20 mouse monoclonal | Santa Cruz | F-10 |
| Ubc9 goat polyclonal | Santa Cruz | N-19 |
| mtHSP70 mouse monoclonal | Thermo Scientific | JG1 |
| beta-Tubulin mouse monoclonal | Sigma | D66 |
| ATP5B mouse monoclonal | Molecular Probes | A21351 |
| Pol-II mouse monoclonal | Adgenode | ac-055-100 |
| K63-linkage-specific mouse monoclonal | Enzo Life Science | HWA4C4 |
| KDM4A rabbit polyclonal | Abcam | ab47984 |
| OXPHOs mouse monoclonal | Abcam | ab110413 |
| Pol-II phospho-ser5 mouse monoclonal | Abcam | ab5131 |
| Pol-II phospho-ser2 mouse monoclonal | Abcam | ab24758 |
| H3K9me3 rabbit polyclonal | Enzo Life Science | 6F12-H4 |
| SENP1 rabbit polyclonal | Cell Signaling | D16D7 |
| HA rabbit polyclonal | Santa Cruz | Y-11 |
| anti rabbit Rhodamine RedX | Jackson ImmunoReserch | 711-295-152 |
| anti mouse Fluorescein | Jackson ImmunoReserch | 715-095-151 |
|
| ||
| Bacterial and Virus Strain | ||
|
| ||
| Home-made E. Coli | This Paper | N/A |
|
| ||
| Chemicals, Peptides, and Recombinant Proteines | ||
|
| ||
| Lipofectamine 2000 | Invitrogen | 11668019 |
| Formaldehyde solution 37% | Sigma | 252549 |
| FCCP | TOCRIS | 04-531-1900 |
| Oligomycin A | Sigma | 75351 |
| Antimycin | Sigma | A8674 |
| IBMX | Acros | 228420010 |
| Dexamethasone | Sigma | D8893 |
| Triiodothyronine | Sigma | 709719 |
| Insulin | Sigma | I9278 |
| Protease Inhibitor Cocktail | Roche | 4693116001 |
| Collagenase I | Sigma | 1148089 |
| Dispase II | Sigma | D4693 |
| DMEM | Gibco | 11965 |
| FBS | Gibco | 10438 |
| Calf Serum | Gibco | 16010159 |
| Trizol | Invitrogen | 15596018 |
| T4 Plynucleotide Kinase | NEB | M0201S |
| Proteinase K | Sigma | P2308 |
| SUPERasein | Invitrogen | AM2694 |
| Trypsin-EDTA | Sigma | 59418C |
| Protein A-Sepharose | Invitrogen | 101041 |
| Poly(A) Polymerase | NEB | M0276 |
| Phusion® High-Fidelity DNA Polymerases | NEB | M0530 |
| BSA | Fisher | BP9706 |
| Ubc13 inhibitor | Sigma | 343351-67-8 |
|
| ||
| Critical commercial Assay | ||
|
| ||
| Quick Start™ Bradford Protein Assay | Biorad | 5000201 |
| ECL | Biorad | 1705060 |
| iScript™ Reverse Transcription Supermix | Biorad | 1708841 |
| Rneasy Plus | Qiagen | 74136 |
| Faster SYBER Green Master Mix | AB | 4385612 |
| QuickExtract DNA | Epicenter | QE09050 |
|
| ||
| Experimental Models: Cell Lines | ||
|
| ||
| Hela | ATCC | CCL2 |
| 3T3-L1 | ATCC | CL-173 |
| 293T | ATCC | CRL-3216 |
|
| ||
| Experimental Models: Organisms/Strains | ||
|
| ||
| GPS2-AKO | Cederquist et al. 2017 | N/A |
|
| ||
| Oligonucleotides | ||
|
| ||
| GPS2 Silencer Select siRNA | Ambion | s80309 |
| GPS2 Silencer Select siRNA | Ambion | s80311 |
| Ubc9 Silencer Select siRNA | Ambion | s75670 |
| Ubc13 Silencer Select siRNA | Ambion | s123784 |
| Senp1 Silencer Select siRNA | Ambion | s104660 |
| Mul1 Silencer Select siRNA | Ambion | s86602 |
| Rnf8 Silencer Select siRNA | Ambion | s81596 |
| NDUFV1 Fw | Sigma | GTGCGGGTATCTGTGCGTT |
| NDUFV1 Rv | Sigma | GGTTGGTAAAGATCCGGTCTTC |
| AKAP1 Fw | Sigma | ATGGCAATCCAGTTGCGTTC |
| AKAP1 Rv | Sigma | TCCACCTGCTTATCACTGCTG |
| Mgst1m Fw | Sigma | GAGCTCTGCGACCGCATTC |
| Mgst1m Rv | Sigma | ACGCGTTCCACCTTCTCGTC |
| SCP2 Fw | Sigma | AATGCTGCCCCACCTCAGAT |
| SCP2 Rv | Sigma | CAGGCCGTACTGCTGCACAA |
| Acaa2 Fw | Sigma | ACGCCTTGCAGTCTCAGCAG |
| Acaa2 Rv | Sigma | CCTCAATGGGTGCCATCTCC |
| SDHa Fw | Sigma | GGTGCAGAAGCTCGGAAGGA |
| SDHa Rv | Sigma | CCTCCAGTGTTCCCCAAACG |
| UQCRC1 Fw | Sigma | CTGCAGCTGGAGGTGTGGAA |
| UQCRC1 Rv | Sigma | CACTCGGGAGACACTGCTCAA |
| TOMM20 Fw | Sigma | CTGGGGTGGTCTTCTTGTGC |
| TOMM20 Rv | Sigma | CACGCCGCTGTCACTGTGTA |
| PGC-1alpha Fw | Sigma | GCACGAGAAGCGGGAGTCTG |
| PGC-1alpha Rv | Sigma | CTGTCCGCGTTGTGTCAGGT |
| CPT1a Fw | Sigma | CATGAAGCCCTCAAACAGAT |
| CPT1a Rv | Sigma | ATAAGCCAGCTGGAGGGACT |
| SENP1 Fw | Sigma | CACTCCAGCGTCAGGCTCAG |
| SENP1 Rv | Sigma | ACGCAGACATGTGGCAGTGG |
| TFAM Fw | Sigma | CTGCACTCTGCCCATCCAAA |
| TFAM Rv | Sigma | CTGAGCATTCGCAGGCCTTT |
| NADHI Fw | Sigma | GCCACCTTACAAATAAGCGCTCTC |
| NADHI Rv | Sigma | ACGCAATTTCCTGGCTCTGC |
| MUL1 Fw | Sigma | AGGTCGCGCAGGAACTCAAG |
| MUL1 Rv | Sigma | CGATGACAGCATAAGGCACACA |
| CIDEA fw | Sigma | TCCCAGTCTGCAAGCAACCA |
| CIDEA rv | Sigma | GTCCTTGGGGTTCAGCCTGT |
| COX7A1 fw | Sigma | GGGGAATGGACAACGTCCTG |
| COX7A1 rv | Sigma | AGGCCCAGCCCAAGCAGTAT |
| COX8B fw | Sigma | AGCCCATGTCTCTGCCAAGC |
| COX8B rv | Sigma | TGGAACCATGAAGCCAACGA |
| cyclophilA F | Sigma | AGCACTGGGGAGAAAGGATT |
| cyclophilA R | Sigma | CATGCCTTCTTTCACCTTCC |
| mt-Nd1FW | Sigma | TCCGAGCATCTTATCCACGC |
| mt-Nd1Rv | Sigma | GTATGGTGGTACTCCCGCTG |
| mt-Co1Fw | Sigma | TCGGAGCCCCAGATATAGCA |
| mt-Co1Rv | Sigma | TTTCCGGCTAGAGGTGGGTA |
| mt-Co2 Fw | Sigma | AACCATAGGGCACCAATGATAC |
| mt-Co2 Rv | Sigma | GGATGGCATCAGTTTTAAGTC |
| Chr NDUFV1 Fw | Sigma | CAGGACGATTCCGCTGTAGT |
| Chr NDUFV1 Rv | Sigma | GTAGTCCAGCTTGCCCAGAC |
| Chr AKAP1 Fw | Sigma | AGGTTACGGGAAAGCCAGAT |
| Chr AKAP1 Rv | Sigma | GGGTTCCTGGTGTCTGAGTG |
| Chr TOMM20 Fw | Sigma | ACGCGCAGATGGACCCAGT |
| Chr TOMM20 Rv | Sigma | AAACACGTGCTGCCCCTGAC |
| Chr ATF2 Fw | Sigma | GGCTTCCGTCCTCTCTCGCG |
| Chr ATF2 Rv | Sigma | GTGTGCGTGAAGGGGAGGGG |
| Chr HSPE1 Fw | Sigma | GGCTTCCGTCCTCTCTCGCG |
| Chr HSPE1 Rv | Sigma | CCCTCCCGGAAATGACGCCA |
| Chr ASNS Fw | Sigma | CAGCACATCCTCCGGCCTCC |
| Chr ASNS Rv | Sigma | CGGTCTTGTCACTGCGCTGC |
|
| ||
| Recombinant DNA | ||
|
| ||
| HA-SUMO1 | This paper | N/A |
| pCMX HA-GPS2 FL-FLAG | Huang et al 2015 | N/A |
| pCMX HA-GPS2 FL-FLAG K45R/K71R | Huang et al 2015 | N/A |
| pCMX HA-GPS2 Delta60-CtKsR-FLAG | Huang et al 2015 | N/A |
| pCMX HA-GPS2 1-155-FLAG | Huang et al 2015 | N/A |
| pCMX HA-GPS2 1-212-FLAG | Huang et al 2015 | N/A |
|
| ||
| Software and Algoritms | ||
|
| ||
| ChIP-seq and Gro-seq fastq files alignment | Langmead et al. 2009 | Bowtie2 |
| Normalization and differential gene expression | Robinson et al 2010 | edgeR |
| ChIPseq peaks calling, density profiles and reads count | Heinz et al. 2010 | HOMER |
| RNA-seq fastq files alignment | Kim et al. 2013 | Tophat |
| RNA-seq reads count | Trapnell et al. 2012 | Cufflinks |
| RNA-seq reads count | Liao et al. 2014 | FeatureCounts |
| RNA-seq reads count | Heinz et al. 2010 | HOMER |
| GO/pathway enrichment | Huang da et al. 2009 | DAVID |
| GO/pathway enrichment | Kuleshov et al. 2016 | enrichR |
| boxplots and scatterplots display | R | |
| boxplots and scatterplots display | ggplot2 | |
| boxplots and scatterplots display | limma | |
|
| ||
| Depositated Data | ||
|
| ||
| Raw data | This paper | GSE80994 |
| GPS2 ChIP-seq | Cardamone et al. 2014 | GSE35197 |
| GPS2 ChIP-seq | Cardamone et al. 2012 | GSE57777 |
| GPS2 ChIP-seq | Fan et al. 2016 | GSE66774 |
| PPARγ, RXR, and POL2 ChIP-seq | Nielsen et al. 2008 | GSE13511 |
| NCoR and SMRT ChIP-seq | Raghav et al. 2012 | ArrayExpress E-MTAB-103 |
| Gro-seq | Step et al. 2014 | GSE56747 |
| Mitochondrial genes set | Smith et al. 2016 | MITOMINER |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and request may be directed to and will be fulfilled by the lead contact, Valentina Perissi (vperissi@bu.edu)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
HeLa cell and 293T cell were grown in 10% FBS/DMEM supplemented with 0.1 mM MEM non-essential aminoacids, 2mM L-glutamine, 1mM sodium pyruvate. 3T3-L1 cells were grown in 10% BCS/DMEM supplemented with 0.1 mM MEM non-essential aminoacids, 2mM L-glutamine, 1mM sodium pyruvate. For cells transfection, Lipofectamine 2000 was used following the manufacturer’s protocol (Invitrogen). Induction of adipogenesis was performed in 10% FBS/DMEM high glucose supplemented with 0.1 mM MEM non-essential aminoacids, 2mM L-glutamine, 1mM sodium pyruvate, 0.1% 1mg/ml insulin, 0.01% 1mg/ml dexamethasone, 0.2% 55,6mg/ml isobutylmethylxanthine. 2μM Ubc13 inhibitor NSC697923 (Sigma) and 10μM or 25μM Carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP-TOCRIS) was used for cell treatments. Adipo-specific GPS2 knockout mice (GPS2-AKO) were generated using a cre/lox approach and backcrossed to a C57BL/6J background (Cederquist et al., 2017). Briefly, conditional Gps2 floxed mice were generated by inGenious Targeting Laboratory and adipose tissue specific deletion was achieved by crossing Gps2flox/flox mice with heterozygous Adipoq-Cre C57BL/6J transgenic mice expressing Cre recombinase under control of the adiponectin promoter (Eguchi et al., 2011). Male and female mice and littermate controls were used for all experiments. Mice were maintained on standard laboratory chow diet in temperature-controlled facility on a 12-hour light/dark cycle. All animal studies were approved by the Boston University Institutional Animal Care and Use Committee (IACUC) and performed in strict accordance of NIH guidelines for animal care.
METHOD DETAILS
H&E Staining
Upon harvesting, brown adipose tissue was incubated at 4°C in Z-fix solution (Anatech LTD) overnight. Tissues were then transferred to 70% ethanol, paraffin embedded, sectioned, and stained with hematoxylin & eosin following standard protocols (Tuft University Pathology Core).
Immunogold-Electron Microscopy
For preparation of cryosections cells were rinsed in PBS, harvested and fixed in 4% paraformaldehyde/PBS for two hours then cells were infiltrated with 2.3M sucrose/PBS/glycin 0.2M) and frozen in liquid nitrogen. Frozen samples were sectioned at −120°C and transferred to formvar-carbon coated copper grids and gold-labeling was carried out following standard protocols (Harvard University EM Core).
Cell Stainings
Immunofluorescence was performed on cells fixed in 4% paraformaldehyde/PBS for 15 min at RT follow by membrane permeabilization in 0.3% Triton X/PBS 5 min at RT. Cells were then washed in Blocking Solution (0.5% BSA + 1:500 Donky Serum/PBS) for 30 min at RT. Staining was performed with specific primary antibody diluted 1:300 in Working Solution (1:5 Blocking Solution) for 1 hour at RT. After three washes of 5 min each with Working Solution secondary antibodies, Rhodamine RedX (RRX)-conjugated anti-rabbit and Fluorescein (FITC)-conjugated anti-mouse (Jackson ImmunoResearch), were then added for 1h at RT. Oil Red O staining was performed on primary adipocytes fixed in 10% formalin at RT for 30 min. In brief, fixed cells were incubated in Working Solution (1:3 oil Red O Stock Solution made with 0.3mg of oil Red power dissolved in 100 ml of isopropanol) for 10 min at RT then washed with 60% isopropanol followed by washed in water.
In vitro Adipocyte Differentiation
For the differentiation of primary mouse adipocytes, the stromal vascular fraction (SVF) was isolated from BAT after collagenase type I/dispase II digestion in 4% BSA KRH buffer for 45 minutes then washed, filtered and spun down at 900 rpm for 10 minutes. Cells were cultured in high glucose DMEM with 10% fetal bovine serum (Hyclone) and 1X pen/strep until confluence. Two days later, in vitro adipogenic differentiation was induced using the adipogenic cocktail described above (insulin, IBMX, DEX and T3) for 2 days, followed by maintenance media containing high glucose DMEM with 10% fetal bovine serum, pen/strep, and insulin for 12 additional days. 3T3-L1 cells were grown to confluence and then differentiated for 6 days using the same protocol.
Protein Extraction, Subcellular Fractionation, Submitochondrial Localization
For whole cell extracts preparation, cells were rinsed in PBS, harvested and incubated for 20′ on ice in IPH buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 5 mM EDTA, 0.5% NP-40, 50 mM NaF, 2 mM Na2VO3, 1mM PMSF and protease inhibitor mix). For cytoplasmatic, mitochondrial and nuclear extracts fractionation cells were rinsed in PBS, harvested and re-suspended in gradient buffer (10 mM HEPES pH 7.9, 1mM EDTA, 210 mM Mannitol, 70mM Sucrose, 10mM NEM, 50 mM NaF, 2 mM Na2VO3, 1mM PMSF and protease inhibitors cocktail) then homogenized via 10 passages through 25G syringe followed by low-speed centrifugation for 10 min. The nuclear pellet was incubated for 20 min in high-salt buffer (10 mM Hepes pH 7.9, 20% glycerol, 420 mM NaCl, 1.5 mM MgCl2, 0.2mM EDTA, 0.5mM DTT, 10mM NEM, 50 mM NaF, 2 mM Na2VO3, 1mM PMSF and protease inhibitor mix) while the supernatant was recovered and subjected to high-speed centrifugation to separate the mitochondrial pellet from the cytoplasmic fraction. The mitochondrial pellet was incubated for 15 min in lysis buffer (50 mM Tris/HCl pH 8, 300 mM NaCl, 1mM EDTA, 1% Triton X-100, 10mM NEM, 50 mM NaF, 2 mM Na2VO3, 1mM PMSF and protease inhibitor mix). Blotting with markers of the different fractions (HDAC2 for nuclear extract, mtHSP70 for mitochondrial extracts) was used to assess purity. To examine submitochondrial localization, the isolated mitochondria fraction was treated with either Proteinase K (2, 5 or 10 ng) in ice for 30 min or 50 μg/ml trypsin in ice for 30 min under either isotonic or hypotonic condition, reaction was terminated adding respectively 1mM PMSF or 10% TCA. Concentration of protein extracts was measured using the colorimetric BIORAD assay. Extracts were boiled in SDS sample buffer and loaded 10% Mini-PROTEAN TGX gels (Biorad), prior to transfer onto PVDF membranes (Millipore) and western blotting following standard protocols.
In Vivo Sumoylation Assay
For in vivo Sumoylation assay cells were transfected with HA-SUMO1 then mitochondrial and nuclear extracts were subjected to immunoprecipitation with antibody anti-GPS2 over night at 4C. Sumoylated GPS2 proteins were then isolated using Sepharose A beads (Life Technologies) and washed in 300nM NaCl/PBS three times for 15 min at 4C. Proteins were then eluted by boiling the samples for 10 min.
RNA Isolation, RT-PCR Analysis and RNA-seq
RNA was isolated from cell or mice tissue following the manufacturer protocol for the RNeasy Kit (QIAGEN). First strand cDNA synthesis from total RNA template was performed using the Biorad IScript cDNA Synthesis System, followed by SYBR-green qPCR amplification. Normalization was performed using specific amplification of CyclophilinA and qPCRs were performed in triplicate for each biological experiment. Data are shown as sample mean between triplicate experiments +/− standard deviation. Significance was calculated by paired student’s T-test. For the RNA-seq, cells were subjected to standard RNA isolation prior to quality control on Agilent Bioanalyzer and RNA library preparation following Illumina’s RNA-Seq Sample Preparation Protocol. Resulting cDNA libraries were sequenced on the Illumina’s HiSeq 2000.
ChIP Assay and ChIP-seq
For chromatin immunoprecipitation (ChIP) experiments in 3T3-L1 cells, approximately 107 cells were cross-linked with 1% formaldehyde at room temperature (~25 °C) for 10 min and neutralized with 0.125 M glycine. For ChIP experiments from mouse brown adipose tissue, the tissue was minced with curved scissors (slurry like) and put in 10ml tubes containing PBS 1% formaldehyde for crosslink for 15 min at RT and neutralized with 0.125 M glycine. Tissue was then shredded in lysis buffer using a Bullet Blender homogenizer at max power for 5 min 4°C. Sonication was performed using the Bioruptor UCD-200 for 2 cycles of 10 min (max power 30 sec on 30 sec off). After sonication, chromatin was incubated with 2μg of antibody at 4 °C overnight. Immunoprecipitated complexes were collected using Sepharose A beads (Life Technologies) and washed with 500 uL of respectively, Low Salt Wash Buffer (150 mM NaCl, Tris-HCl pH 8 20 mM, EDTA 2mM, Triton-X 1% and SDS 0.1%) High Salt Wash Buffer (500 mM NaCl, Tris-HCl pH 8 20 mM, EDTA 2mM, Triton-X 1% and SDS 0.1%), LiCl Wash (0.25 M LiCl, Tris-HCl pH 8 10 mM, EDTA 1mM, Igepal 1% and Deoxicholete 1%), 1X TE (Tris-HCl pH 8 10 mM, EDTA). DNA was then extracted and purified by phenol/chloroform extraction. Hand ChIP experiments were repeated at least three times and representative results are shown as samples mean between technical replicates +/− standard deviation. Significance was calculated by paired student’s T-test. For ChIP-Seq, the extracted DNA was ligated to specific adaptors followed by deep sequencing on the Illumina’s HiSeq 2000 according to the manufacturer’s instructions. ChIP-seq for GPS2 upon FCCP treatment was performed on two separate replicate experiments. For ChIP-seqs from BAT tissue, four biological replicate ChIPs from individual littermate mice were pooled for sequencing. Samples from male and female mice were grouped together and analysis performed separately.
Gro-seq Assay
For GRO-seq experiments, 3T3-L1 cells were swelled in swelling buffer (10mM Tris-Cl pH 7.5, 2mM MgCl2, 3mM CaCl2) for 5min on ice and then lysed in lysis buffer (swelling buffer with 0.5% IGEPAL and 10% glycerol, 4U/ml of superase in) in ice for 5 min then nuclei were pellet and re-suspended in 100μl of freezing buffer (50mM Tris-Cl pH 8.3, 40% glycerol, 5mM MgCl2, 0.1mM EDTA). For the run-on assay, re-suspended nuclei were mixed with an equal volume of reaction buffer (10mM Tris-Cl pH 8.0, 5mM MgCl2, 1mM dithiothreitol (DTT), 300mM KCl, 20 units of Superase-In, 1% sarkosyl, 500μM ATP, GTP, Br-UTP and 2μM CTP) and incubated for 5min at 30°C. The nuclear-run-on RNA (NRO-RNA) was then extracted with TRIzol LS reagent (Invitogen) following the manufacturer’s instructions. After base hydrolysis on ice for 40min and followed by treatment with DNase I and antarctic phosphatase, the Br-UTP-labelled NRO-RNA was purified by anti-BrdU agarose beads (Santa Cruz Biotech) in binding buffer (0.5× SSPE, 1mM EDTA, 0.05% Tween) for 3h at 4°C while rotating. Then T4 PNK (NEB) was used to repair the end of NRO-RNA. The RNA fragments were subjected to the poly-A-tailing reaction by poly-A polymerase (NEB) for 30min at 37°C. Reverse transcription was then performed using superscript III (Invitrogen) with oNTI223 primer (available on request). The cDNA products were separated on a 10% polyacrylamide TBE-urea gel with the right product (~100–500bp) being excised and recovered by gel extraction. After that, the first-strand cDNA was circularized by CircLigase (Epicentre) and re-linearized by Ape1 (NEB). Re-linearized single-strand cDNA was separated by TBE gel and the products of the desired size were excised (~120–320bp) for gel extraction. Finally, the cDNA template was amplified by PCR using the Phusion High-Fidelity enzyme (NEB) with primers oNTI200 and oNTI201 for deep sequencing on Illumina’s HiSeq 2000.
Bioinformatics Analysis of ChIP-seq, RNA-seq and GRO-seq Datasets
For the bioinformatics analyses, we have built comprehensive lists of mouse and human mitochondrial genes by combining the sets of mitochondrial genes from MITOMINER database (Smith and Robinson, 2016) with the sets of genes that are annotated as mitochondrial from NCBI, AMIGO, and ENSEMBL databases. Previously published GPS2 ChIP-seq datasets were downloaded from NCBI GEO database (GSE35197 (Cardamone et al., 2014), GSE57777 (Cardamone et al., 2012) and GSE66774 (Fan et al., 2016)), PPARγ, RXR, and POL2 ChIP-seq datasets were downloaded from NCBI GEO series GSE13511(Nielsen et al., 2008), NCoR and SMRT ChIP-seq datasets were downloaded from ArrayExpress E-MTAB-103 (Raghav et al., 2012).
For ChIP-seq analysis, Bowtie2 (Langmead et al, Nature Methods 2012) was used to align the fastq file to mm8 mouse genome, and the ChIP-seq reads on specific genomics regions were counted by using Bedtools (Quinlan et al, Bioinformatics 2010) or HOMER. Normalization on specific genomics regions was done using the TMM normalization procedure in edgeR (Robinson et al., 2010), and the normalized counts were obtained by using cpm() function. HOMER software suite(Heinz et al., 2010) was used to call the ChIP-seq peaks and to compute the heatmaps, the read density profiles, the motif density profiles on promoters, and to find the enriched motifs. The alignment of POL2 and H3K9me3 ChIP-seq samples was performed by using Bowtie2 (Langmead et al., 2009) to mm8 assembly of the mouse genome, and sets of equal number of reads were randomly extracted from siCTL and siGPS2 aligned samples. The heatmaps of ChIP-seq datasets were displayed by using R packages pheatmap and gplots, and the boxplots were displayed in ggplot2. The GPS2 peaks in the WT mice samples were called against the GPS2-AKO mice samples as background using a modified set of parameters in HOMER (-F 2 -L 2 -fdr < 0.05). H3K9me3 levels in both WT and KO samples were normalized by using edgeR on the counts in the promoter areas, and the input DNA was subtracted.
The RNA-seq fastq files (siCTL, siGPS2) were aligned by using Tophat(Kim et al., 2013) and the sequencing reads were counted over transcripts by using Cufflinks(Trapnell et al., 2012), FeatureCounts(Liao et al., 2014) and HOMER(Heinz et al., 2010). Differential expressed genes across two independent RNAseq replicate experiments were identified by edgeR(Robinson et al., 2010) using FDR<0.05. GO/pathway enrichment was computed with DAVID(Huang da et al., 2009a, 2009b), ToppFun, and in enrichR(Chen et al., 2013; Kuleshov et al., 2016). The boxplots and scatterplots were displayed in R standard libraries, in ggplot2, and in limma.
The GRO-seq fastq files (siCTL, siGPS2) were aligned to mouse genome mm8 by using bowtie2; we have used the same number (4.2 millions) of randomly extracted reads for computing the gene expression levels. The aligned reads were counted over the RefSeq gene bodies (after excluding a TSS 400bp-proximal region, and TTS 400bp-distal region) by using bedtools (Quinlan and Hall, 2010). The statistically significant differentially expressed genes were defined by edgeR(Robinson et al., 2010) for a BCV of 0.01. We have also used a GRO-seq dataset from NCBI GEO (GSE56747 (Step et al., 2014)) for computing the read density profiles on TSSes.
Data and Software Availability
New RNA-seq, ChIP-seq and GRO-seq datasets have been deposited to NCBI GEO (GSE80994).
Mitochondrial Content
Total DNA was extracted from cells using QuickExtract DNA Extraction Solution 1.0 (Epicenter) following manufacturer’s instructions. DNA amplification of the mitochondrial-encoded NADH dehydrogenase 1 (mt-ND1) relative to nuclear TFAM was used to determine mitochondrial DNA copy number.
Mitochondrial Isolation and Mass
BAT was isolated from GPS2-WT and KO mouse and weighted. Isolated BAT was rinsed and minced in ice-cold PBS then homogenized in a glass-Teflon Dounce homogenizer containing SHE buffer pH=7.2 (250 mM Sucrose, 5mM HEPES, 2 mM EGTA, BSA 2%.) + BSA. After 9–10 strokes through the pestle the homogenate was centrifuged at 900xg for 10 min at 4°C. The resulting supernatant was then centrifuged at 9000xg for 10 min at 4°C and the pellet was washed once and then re-suspended in SHE buffer without BSA. Protein content was measured by BCA. Mitochondrial mass was calculated as ratio between protein concentrations expressed in μg and tissue weight expressed in mg.
Respirometry
Cells were plated in Seahorse V.7 multi-well culture plates. The next day, media was replaced by running media (XF Seahorse Assay Media supplemented with 5.5mM glucose, 0.5mM pyruvate and 1mM glutamine) and the plate was placed at 37°C for 1 h (no carbon dioxide). Oxygen consumption was measured at 37°C using a Seahorse XF24 Extracellular Flux Analyzer (Seahorse Bioscience, Billerica, MA). Mitochondrial stress test compounds (10 μM oligomycin, 2.5 μM FCCP and 10 μM antimycin A) were injected through ports A, B, and C, respectively, to measure mitochondrial respiration linked to ATP synthesis, leak, maximal respiratory capacity and non-mitochondrial oxygen consumption according to the manufacturer’s instructions. For mitochondrial activity, 4μg of mitochondrial protein fractions were loaded per well for complex I driven respiration (pyruvate+malate) and 2μg for complex II-driven respiration (succinate+rotenone) in 25 μl of Mitochondrial Assay Solution (MAS) pH 7.2 (MAS: 100mM KCl, 10mM KH2PO4, 2mM MgCl2, 5mM HEPES, 1mM EGTA, 0.1% BSA and 1mM GDP) per Seahorse XF96 well. The plate was centrifuged at 4°C, 5 min at 3400 rpm. Then, 110 μl of MAS with the respective fuels were carefully added per well. Plate was warmed at 37°C for 4 min then respirometry assay within the XF96 was performed as described above for the XF24. Pyruvate was used at 5mM, malate 5mM, succinate 5mM and rotenone 2μM. ADP was injected at port A (3.5mM), Oligomycin A at port B (3.5μM), FCCP at port C (4μM) and Antimycin A at port D (4μM).
Statistical Analysis
All of the data shown in the histograms are the results of at least three independent experiments and are presented as the mean +/− SEM. ChIP-seq data in 3T3-L1 is representative of two independent biological replicates. ChIP-seq data in BAT is representative of four independent biological replicates. Hand ChIPs data are representative of three independent experiments, bar graphs represent the sample mean of three technical replicates +/− SD. The differences between groups were compared using Student’s t test. Imaging results and western blot are representative of three independent experiments.
Supplementary Material
Acknowledgments
We are grateful to our colleagues at Boston University and past/present members of the Perissi lab for shared reagents and insightful discussions and to Dr. M. Picard for valuable comments on the manuscript. We thank K. Ohgi and the UCSD Sequencing Core, the BU Analytical Instrumentation Core, the BU Confocal Microscopy Core and the Electron Microscopy Core at Harvard University for their excellent work and Dr. G. Gaietta for initial guidance on imaging assays. Work was supported by BNORC and BU/Joslin P&F Awards to V.P. and M.D.C. (P30DK046200, P30DK036836), F31DK108571 NRSA Predoctoral Fellowship to C.T.C. and Research Grants R01DK100422 and DoD-BC160363 to V.P.
Footnotes
Authors Contribution
Conceptualization, V.P. and M.D.C.; Investigation, M.D.C, C.T.C, J.H., and K.M.; Formal Analysis, B.T.; Methodology, W.L.; Writing – Original Draft, V.P. and M.D.C.; Writing – Review & Editing, V.P., M.L. and M.G.R; Resources, M.G.R; Supervision, V.P. and M.L.
Declaration of Interests
The authors declare no competing interests.
References
- Altshuler-Keylin S, Kajimura S. Mitochondrial homeostasis in adipose tissue remodeling. Science signaling. 2017;10 doi: 10.1126/scisignal.aai9248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnould T, Michel S, Renard P. Mitochondria Retrograde Signaling and the UPR mt: Where Are We in Mammals? International journal of molecular sciences. 2015;16:18224–18251. doi: 10.3390/ijms160818224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi H, Li S, Wang M, Jia Z, Chang AK, Pang P, Wu H. SUMOylation of GPS2 protein regulates its transcription-suppressing function. Molecular biology of the cell. 2014;25:2499–2508. doi: 10.1091/mbc.E13-12-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas G, Adebanjo OA, Freedman BD, Anandatheerthavarada HK, Vijayasarathy C, Zaidi M, Kotlikoff M, Avadhani NG. Retrograde Ca2+ signaling in C2C12 skeletal myocytes in response to mitochondrial genetic and metabolic stress: a novel mode of inter-organelle crosstalk. The EMBO journal. 1999;18:522–533. doi: 10.1093/emboj/18.3.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohovych I, Khalimonchuk O. Sending Out an SOS: Mitochondria as a Signaling Hub. Front Cell Dev Biol. 2016;4:109. doi: 10.3389/fcell.2016.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braschi E, Zunino R, McBride HM. MAPL is a new mitochondrial SUMO E3 ligase that regulates mitochondrial fission. EMBO reports. 2009;10:748–754. doi: 10.1038/embor.2009.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai R, Yu T, Huang C, Xia X, Liu X, Gu J, Xue S, Yeh ET, Cheng J. SUMO-specific protease 1 regulates mitochondrial biogenesis through PGC-1alpha. The Journal of biological chemistry. 2012;287:44464–44470. doi: 10.1074/jbc.M112.422626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardamone MD, Krones A, Tanasa B, Taylor H, Ricci L, Ohgi KA, Glass CK, Rosenfeld MG, Perissi V. A protective strategy against hyperinflammatory responses requiring the nontranscriptional actions of GPS2. Molecular cell. 2012;46:91–104. doi: 10.1016/j.molcel.2012.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardamone MD, Tanasa B, Chan M, Cederquist CT, Andricovich J, Rosenfeld MG, Perissi V. GPS2/KDM4A pioneering activity regulates promoter-specific recruitment of PPARgamma. Cell reports. 2014;8:163–176. doi: 10.1016/j.celrep.2014.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catic A, Suh CY, Hill CT, Daheron L, Henkel T, Orford KW, Dombkowski DM, Liu T, Liu XS, Scadden DT. Genome-wide map of nuclear protein degradation shows NCoR1 turnover as a key to mitochondrial gene regulation. Cell. 2013;155:1380–1395. doi: 10.1016/j.cell.2013.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cederquist CT, Lentucci C, Martinez-Calejman C, Hayashi V, Orofino J, Guertin D, Fried SK, Lee MJ, Cardamone MD, Perissi V. Systemic insulin sensitivity is regulated by GPS2 inhibition of AKT ubiquitination and activation in adipose tissue. Mol Metab. 2017;6:125–137. doi: 10.1016/j.molmet.2016.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandel NS. Evolution of Mitochondria as Signaling Organelles. Cell metabolism. 2015;22:204–206. doi: 10.1016/j.cmet.2015.05.013. [DOI] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma’ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Gao X, Shilatifard A. Stably paused genes revealed through inhibition of transcription initiation by the TFIIH inhibitor triptolide. Genes & development. 2015;29:39–47. doi: 10.1101/gad.246173.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Kao HY. G protein pathway suppressor 2 (GPS2) is a transcriptional corepressor important for estrogen receptor alpha-mediated transcriptional regulation. The Journal of biological chemistry. 2009;284:36395–36404. doi: 10.1074/jbc.M109.062109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duteil D, Tosic M, Lausecker F, Nenseth HZ, Muller JM, Urban S, Willmann D, Petroll K, Messaddeq N, Arrigoni L, et al. Lsd1 Ablation Triggers Metabolic Reprogramming of Brown Adipose Tissue. Cell reports. 2016;17:1008–1021. doi: 10.1016/j.celrep.2016.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi J, Wang X, Yu S, Kershaw EE, Chiu PC, Dushay J, Estall JL, Klein U, Maratos-Flier E, Rosen ED. Transcriptional control of adipose lipid handling by IRF4. Cell metabolism. 2011;13:249–259. doi: 10.1016/j.cmet.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan R, Toubal A, Goni S, Drareni K, Huang Z, Alzaid F, Ballaire R, Ancel P, Liang N, Damdimopoulos A, et al. Loss of the co-repressor GPS2 sensitizes macrophage activation upon metabolic stress induced by obesity and type 2 diabetes. Nat Med. 2016 doi: 10.1038/nm.4114. [DOI] [PubMed] [Google Scholar]
- Fan W, Evans R. PPARs and ERRs: molecular mediators of mitochondrial metabolism. Current opinion in cell biology. 2014;33C:49–54. doi: 10.1016/j.ceb.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley LW, Haigis MC. The coordination of nuclear and mitochondrial communication during aging and calorie restriction. Ageing research reviews. 2009;8:173–188. doi: 10.1016/j.arr.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorese CJ, Schulz AM, Lin YF, Rosin N, Pellegrino MW, Haynes CM. The Transcription Factor ATF5 Mediates a Mammalian Mitochondrial UPR. Curr Biol. 2016;26:2037–2043. doi: 10.1016/j.cub.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Yu HM, Chiu SY, Mirando AJ, Maruyama EO, Cheng JG, Hsu W. Disruption of SUMO-specific protease 2 induces mitochondria mediated neurodegeneration. PLoS genetics. 2014;10:e1004579. doi: 10.1371/journal.pgen.1004579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain D. Sirtuins and the Estrogen Receptor as Regulators of the Mammalian Mitochondrial UPR in Cancer and Aging. Adv Cancer Res. 2016;130:211–256. doi: 10.1016/bs.acr.2016.01.004. [DOI] [PubMed] [Google Scholar]
- Gorman GS, Chinnery PF, DiMauro S, Hirano M, Koga Y, McFarland R, Suomalainen A, Thorburn DR, Zeviani M, Turnbull DM. Mitochondrial diseases. Nat Rev Dis Primers. 2016;2:16080. doi: 10.1038/nrdp.2016.80. [DOI] [PubMed] [Google Scholar]
- Guha M, Avadhani NG. Mitochondrial retrograde signaling at the crossroads of tumor bioenergetics, genetics and epigenetics. Mitochondrion. 2013;13:577–591. doi: 10.1016/j.mito.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Li Y, Gow CH, Wong M, Zha J, Yan C, Liu H, Wang Y, Burris TP, Zhang J. The optimal corepressor function of nuclear receptor corepressor (NCoR) for peroxisome proliferatoractivated receptor gamma requires G-protein pathway suppressor 2. The Journal of biological chemistry. 2014 doi: 10.1074/jbc.M114.598797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms MJ, Lim HW, Ho Y, Shapira SN, Ishibashi J, Rajakumari S, Steger DJ, Lazar MA, Won KJ, Seale P. PRDM16 binds MED1 and controls chromatin architecture to determine a brown fat transcriptional program. Genes & development. 2015;29:298–307. doi: 10.1101/gad.252734.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hino S, Sakamoto A, Nagaoka K, Anan K, Wang Y, Mimasu S, Umehara T, Yokoyama S, Kosai K, Nakao M. FAD-dependent lysine-specific demethylase-1 regulates cellular energy expenditure. Nature communications. 2012;3:758. doi: 10.1038/ncomms1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock MB, Kralli A. Transcriptional control of mitochondrial biogenesis and function. Annual review of physiology. 2009;71:177–203. doi: 10.1146/annurev.physiol.010908.163119. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009a;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009b;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang J, Cardamone MD, Johnson HE, Neault M, Chan M, Floyd ZE, Mallette FA, Perissi V. Exchange Factor TBL1 and Arginine Methyltransferase PRMT6 Cooperate in Protecting GPS2 from Proteasomal Degradation. The Journal of biological chemistry. 2015 doi: 10.1074/jbc.M115.637660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson T, Venteclef N, Toresson G, Damdimopoulos AE, Ehrlund A, Lou X, Sanyal S, Steffensen KR, Gustafsson JA, Treuter E. GPS2 is required for cholesterol efflux by triggering histone demethylation, LXR recruitment, and coregulator assembly at the ABCG1 locus. Molecular cell. 2009;34:510–518. doi: 10.1016/j.molcel.2009.05.006. [DOI] [PubMed] [Google Scholar]
- Jazwinski SM. Rtg2 protein: at the nexus of yeast longevity and aging. FEMS Yeast Res. 2005;5:1253–1259. doi: 10.1016/j.femsyr.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Jazwinski SM. The retrograde response: when mitochondrial quality control is not enough. Biochimica et biophysica acta. 2013;1833:400–409. doi: 10.1016/j.bbamcr.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazwinski SM, Kriete A. The yeast retrograde response as a model of intracellular signaling of mitochondrial dysfunction. Frontiers in physiology. 2012;3:139. doi: 10.3389/fphys.2012.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkers I, Kwak H, Lis JT. Genome-wide dynamics of Pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. Elife. 2014;3:e02407. doi: 10.7554/eLife.02407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajimura S, Saito M. A new era in brown adipose tissue biology: molecular control of brown fat development and energy homeostasis. Annu Rev Physiol. 2014;76:225–249. doi: 10.1146/annurev-physiol-021113-170252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusminski CM, Scherer PE. Mitochondrial dysfunction in white adipose tissue. Trends in endocrinology and metabolism: TEM. 2012;23:435–443. doi: 10.1016/j.tem.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamech LT, Haynes CM. The unpredictability of prolonged activation of stress response pathways. The Journal of cell biology. 2015;209:781–787. doi: 10.1083/jcb.201503107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefterova MI, Steger DJ, Zhuo D, Qatanani M, Mullican SE, Tuteja G, Manduchi E, Grant GR, Lazar MA. Cell-specific determinants of peroxisome proliferator-activated receptor gamma function in adipocytes and macrophages. Molecular and cellular biology. 2010;30:2078–2089. doi: 10.1128/MCB.01651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentucci C, Belkina A, Cederquist CT, Chan M, Johnson HE, Prasad S, Lopacinski A, Nikolajczyk BS, Monti S, Snyder-Cappione J, et al. Inhibition of Ubc13-mediated ubiquitination by GPS2 regulates multiple stages of B cell development. The Journal of biological chemistry. 2016 doi: 10.1074/jbc.M116.755132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentucci C, Belkina AC, Cederquist CT, Chan M, Johnson HE, Prasad S, Lopacinski A, Nikolajczyk BS, Monti S, Snyder-Cappione J, et al. Inhibition of Ubc13-mediated Ubiquitination by GPS2 Regulates Multiple Stages of B Cell Development. The Journal of biological chemistry. 2017;292:2754–2772. doi: 10.1074/jbc.M116.755132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Luo Y, Yu L, Lin Y, Luo D, Zhang H, He Y, Kim YO, Kim Y, Tang S, et al. SENP1 mediates TNF-induced desumoylation and cytoplasmic translocation of HIPK1 to enhance ASK1-dependent apoptosis. Cell death and differentiation. 2008;15:739–750. doi: 10.1038/sj.cdd.4402303. [DOI] [PubMed] [Google Scholar]
- Liao X, Butow RA. RTG1 and RTG2: two yeast genes required for a novel path of communication from mitochondria to the nucleus. Cell. 1993;72:61–71. doi: 10.1016/0092-8674(93)90050-z. [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Lin YF, Schulz AM, Pellegrino MW, Lu Y, Shaham S, Haynes CM. Maintenance and propagation of a deleterious mitochondrial genome by the mitochondrial unfolded protein response. Nature. 2016;533:416–419. doi: 10.1038/nature17989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkwirth C, Jovaisaite V, Durieux J, Matilainen O, Jordan SD, Quiros PM, Steffen KK, Williams EG, Mouchiroud L, Tronnes SU, et al. Two Conserved Histone Demethylases Regulate Mitochondrial Stress-Induced Longevity. Cell. 2016;165:1209–1223. doi: 10.1016/j.cell.2016.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miceli MV, Jiang JC, Tiwari A, Rodriguez-Quinones JF, Jazwinski SM. Loss of mitochondrial membrane potential triggers the retrograde response extending yeast replicative lifespan. Frontiers in genetics. 2011;2:102. doi: 10.3389/fgene.2011.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchiroud L, Houtkooper RH, Moullan N, Katsyuba E, Ryu D, Canto C, Mottis A, Jo YS, Viswanathan M, Schoonjans K, et al. The NAD(+)/Sirtuin Pathway Modulates Longevity through Activation of Mitochondrial UPR and FOXO Signaling. Cell. 2013;154:430–441. doi: 10.1016/j.cell.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargund AM, Fiorese CJ, Pellegrino MW, Deng P, Haynes CM. Mitochondrial and Nuclear Accumulation of the Transcription Factor ATFS-1 Promotes OXPHOS Recovery during the UPR(mt) Molecular cell. 2015;58:123–133. doi: 10.1016/j.molcel.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, Haynes CM. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science. 2012;337:587–590. doi: 10.1126/science.1223560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan M, Schiralli Lester GM, Lee C, Missra A, Wasserman GA, Steffen M, Gilmour DS, Henderson AJ. Negative elongation factor (NELF) coordinates RNA polymerase II pausing, premature termination, and chromatin remodeling to regulate HIV transcription. The Journal of biological chemistry. 2013;288:25995–26003. doi: 10.1074/jbc.M113.496489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, Pedersen TA, Hagenbeek D, Moulos P, Siersbaek R, Megens E, Denissov S, Borgesen M, Francoijs KJ, Mandrup S, et al. Genome-wide profiling of PPARgamma:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev. 2008;22:2953–2967. doi: 10.1101/gad.501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patti ME, Corvera S. The role of mitochondria in the pathogenesis of type 2 diabetes. Endocrine reviews. 2010;31:364–395. doi: 10.1210/er.2009-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng YC, Kuo F, Breiding DE, Wang YF, Mansur CP, Androphy EJ. AMF1 (GPS2) modulates p53 transactivation. Molecular and cellular biology. 2001;21:5913–5924. doi: 10.1128/MCB.21.17.5913-5924.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudent J, Zunino R, Sugiura A, Mattie S, Shore GC, McBride HM. MAPL SUMOylation of Drp1 Stabilizes an ER/Mitochondrial Platform Required for Cell Death. Molecular cell. 2015;59:941–955. doi: 10.1016/j.molcel.2015.08.001. [DOI] [PubMed] [Google Scholar]
- Pulvino M, Liang Y, Oleksyn D, DeRan M, Van Pelt E, Shapiro J, Sanz I, Chen L, Zhao J. Inhibition of proliferation and survival of diffuse large B-cell lymphoma cells by a small-molecule inhibitor of the ubiquitin-conjugating enzyme Ubc13-Uev1A. Blood. 2012;120:1668–1677. doi: 10.1182/blood-2012-02-406074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiros PM, Mottis A, Auwerx J. Mitonuclear communication in homeostasis and stress. Nat Rev Mol Cell Biol. 2016;17:213–226. doi: 10.1038/nrm.2016.23. [DOI] [PubMed] [Google Scholar]
- Quiros PM, Prado MA, Zamboni N, D’Amico D, Williams RW, Finley D, Gygi SP, Auwerx J. Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. The Journal of cell biology. 2017 doi: 10.1083/jcb.201702058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghav SK, Waszak SM, Krier I, Gubelmann C, Isakova A, Mikkelsen TS, Deplancke B. Integrative genomics identifies the corepressor SMRT as a gatekeeper of adipogenesis through the transcription factors C/EBPbeta and KAISO. Molecular cell. 2012;46:335–350. doi: 10.1016/j.molcel.2012.03.017. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpulla RC, Vega RB, Kelly DP. Transcriptional integration of mitochondrial biogenesis. Trends in endocrinology and metabolism: TEM. 2012;23:459–466. doi: 10.1016/j.tem.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekito T, Thornton J, Butow RA. Mitochondria-to-nuclear signaling is regulated by the subcellular localization of the transcription factors Rtg1p and Rtg3p. Molecular biology of the cell. 2000;11:2103–2115. doi: 10.1091/mbc.11.6.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao L, Zhou HJ, Zhang H, Qin L, Hwa J, Yun Z, Ji W, Min W. SENP1-mediated NEMO deSUMOylation in adipocytes limits inflammatory responses and type-1 diabetes progression. Nature communications. 2015;6:8917. doi: 10.1038/ncomms9917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siersbaek R, Nielsen R, John S, Sung MH, Baek S, Loft A, Hager GL, Mandrup S. Extensive chromatin remodelling and establishment of transcription factor ‘hotspots’ during early adipogenesis. The EMBO journal. 2011;30:1459–1472. doi: 10.1038/emboj.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AC, Robinson AJ. MitoMiner v3.1, an update on the mitochondrial proteomics database. Nucleic Acids Res. 2016;44:D1258–1261. doi: 10.1093/nar/gkv1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Step SE, Lim HW, Marinis JM, Prokesch A, Steger DJ, You SH, Won KJ, Lazar MA. Anti-diabetic rosiglitazone remodels the adipocyte transcriptome by redistributing transcription to PPARgamma-driven enhancers. Genes Dev. 2014;28:1018–1028. doi: 10.1101/gad.237628.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Than TA, Lou H, Ji C, Win S, Kaplowitz N. Role of cAMP-responsive element-binding protein (CREB)-regulated transcription coactivator 3 (CRTC3) in the initiation of mitochondrial biogenesis and stress response in liver cells. The Journal of biological chemistry. 2011;286:22047–22054. doi: 10.1074/jbc.M111.240481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Garcia G, Bian Q, Steffen KK, Joe L, Wolff S, Meyer BJ, Dillin A. Mitochondrial Stress Induces Chromatin Reorganization to Promote Longevity and UPR(mt) Cell. 2016;165:1197–1208. doi: 10.1016/j.cell.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toubal A, Clement K, Fan R, Ancel P, Pelloux V, Rouault C, Veyrie N, Hartemann A, Treuter E, Venteclef N. SMRT-GPS2 corepressor pathway dysregulation coincides with obesity-linked adipocyte inflammation. The Journal of clinical investigation. 2013;123:362–379. doi: 10.1172/JCI64052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vafai SB, Mootha VK. Mitochondrial disorders as windows into an ancient organelle. Nature. 2012;491:374–383. doi: 10.1038/nature11707. [DOI] [PubMed] [Google Scholar]
- Vankoningsloo S, De Pauw A, Houbion A, Tejerina S, Demazy C, de Longueville F, Bertholet V, Renard P, Remacle J, Holvoet P, et al. CREB activation induced by mitochondrial dysfunction triggers triglyceride accumulation in 3T3-L1 preadipocytes. Journal of cell science. 2006;119:1266–1282. doi: 10.1242/jcs.02848. [DOI] [PubMed] [Google Scholar]
- Venteclef N, Jakobsson T, Ehrlund A, Damdimopoulos A, Mikkonen L, Ellis E, Nilsson LM, Parini P, Janne OA, Gustafsson JA, et al. GPS2-dependent corepressor/SUMO pathways govern anti-inflammatory actions of LRH-1 and LXRbeta in the hepatic acute phase response. Genes & development. 2010;24:381–395. doi: 10.1101/gad.545110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial bioenergetic etiology of disease. The Journal of clinical investigation. 2013;123:1405–1412. doi: 10.1172/JCI61398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanet A, Arnould T, Najimi M, Renard P. Connecting Mitochondria, Metabolism, and Stem Cell Fate. Stem Cells Dev. 2015 doi: 10.1089/scd.2015.0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb AE, Brunet A. FOXO transcription factors: key regulators of cellular quality control. Trends Biochem Sci. 2014;39:159–169. doi: 10.1016/j.tibs.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson-Fritch L, Burkart A, Bell G, Mendelson K, Leszyk J, Nicoloro S, Czech M, Corvera S. Mitochondrial biogenesis and remodeling during adipogenesis and in response to the insulin sensitizer rosiglitazone. Molecular and cellular biology. 2003;23:1085–1094. doi: 10.1128/MCB.23.3.1085-1094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Sun HY, Xiao FJ, Wang H, Yang Y, Wang L, Gao CJ, Guo ZK, Wu CT, Wang LS. SENP1 inhibition induces apoptosis and growth arrest of multiple myeloma cells through modulation of NF-kappaB signaling. Biochemical and biophysical research communications. 2015;460:409–415. doi: 10.1016/j.bbrc.2015.03.047. [DOI] [PubMed] [Google Scholar]
- Zamora M, Pardo R, Villena JA. Pharmacological induction of mitochondrial biogenesis as a therapeutic strategy for the treatment of type 2 diabetes. Biochemical pharmacology. 2015 doi: 10.1016/j.bcp.2015.06.032. [DOI] [PubMed] [Google Scholar]
- Zhang D, Harry GJ, Blackshear PJ, Zeldin DC. G-protein pathway suppressor 2 (GPS2) interacts with the regulatory factor X4 variant 3 (RFX4_v3) and functions as a transcriptional co-activator. The Journal of biological chemistry. 2008;283:8580–8590. doi: 10.1074/jbc.M708209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zunino R, Schauss A, Rippstein P, Andrade-Navarro M, McBride HM. The SUMO protease SENP5 is required to maintain mitochondrial morphology and function. Journal of cell science. 2007;120:1178–1188. doi: 10.1242/jcs.03418. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.