Abstract
Background
Beak deformity, typically expressed as the crossing of upper and lower mandibles, is found in several indigenous chicken breeds, including the Beijing-You chickens studied here. Beak deformity severely impairs the birds’ growth and welfare. Although previous studies shed some light on the genetic regulation of this complex trait, the genetic basis of this malformation remains incompletely understood.
Results
In this study, single SNP- and pathway-based genome-wide association studies (GWASs) were performed using ROADTRIPS and SNP ratio test (SRT), respectively. A total of 48 birds with deformed beaks (case) and 48 normal birds (control) were genotyped using Affymetrix 600 K HD genotyping arrays. As a result, 95 individuals and 429,539 SNPs were obtained after quality control. The P-value was corrected by a Bonferroni adjustment based on linkage disequilibrium pruning. The single SNP-based association study identified one associated SNP with 5% genome-wide significance and seven suggestively associated SNPs. Four high-confidence genes, LOC421892, TDRD3, RET, and STMN1, were identified as the most promising candidate genes underlying this complex trait in view of their positions, functions, and overlaps with previous studies. The pathway-based association study highlighted the association of six pathways with beak deformity, including the calcium signaling pathway.
Conclusions
Potentially useful candidate genes and pathways for beak deformity were identified, which should be the subject of further functional characterization.
Electronic supplementary material
The online version of this article (10.1186/s12864-018-4882-8) contains supplementary material, which is available to authorized users.
Keywords: Beijing-you chickens, Beak deformity, SNP, Pathway, GWAS
Background
The beak is an external anatomical structure of birds, which consists of an upper and a lower mandible [1]. In a similar way to lips and teeth in mammals, the beak functions in birds primarily for feeding and drinking. It also has differentuses because of its diversity in shape between bird species [2–4]. For example, crossbill species like Loxia are characterized by the mandibles with crossed beaks [5], which is an adaptation enabling them to extract seeds from the cones they live on. However, for most bird species, especially for poultry, crossed beaks represent a malformation or an abnormality [6–9]. According to our previous investigations, a frequency of beak deformity (normally the lower mandible crossed left or right randomly) of 1 to 3% was found in several indigenous chickens, including Silkies, Huxu, and Beijing-You (BJY) chickens. Figures 1 and 2 show examples of the deformed and normal beaks of BJY chickens. Birds with crossed beaks can be clearly determined within 4 weeks after birth and cannot be rehabilitated later. Birds with deformed beaks have reduced feed intake, inhibited growth, poor production performance, and shorter survival. In the absence of known environmental factors contributing to the malformation, birds with a deformed beak are present consistently in each generation and cannot be eliminated simply on the basis of the observed phenotype. Furthermore, pedigree information indicated that the birds with a deformed beak can be traced back to limited number of ancestors. This suggested that genetic effects underlie this complex trait [10]. In addition, our previous mating experiment suggested that beak deformity could be a complex trait regulated by multiple genes. Several recognized genetic factors associated with beak shapes include genes such as ALX1 [11], HMGA2 [12], FGF8 [13], Shh [14], BMP4 [15, 16], and CaM [17]. The over-expression of HOXA1 and HOXD3 genes may result in beak deformity in chicks [18]. Previous digital gene expression (DGE) analysis in our laboratory identified several new genes (e.g. LOC426217) and pathways (e.g. unsaturated fatty acid biosynthesis and glycerolipid metabolism) [10, 19] that are involved in beak deformity.
Fig. 1.

Examples of beak deformity birds of Beijing-You chickens used in the study
Fig. 2.

Skull anatomy of Beijing-You chickens with deformed and normal beaks. Compared with the normal beak, the lower mandible crossed left or right with an elongation of one of the rami
With the advance of high-throughput genotyping platforms and computing technology, molecular markers and genes related to complex traits or diseases have been identified using genome-wide association studies (GWASs) [20–23]. This approach is powerful to identify a single SNP with a notable effect, especially for phenotypes that are determined by a unique gene or mutation. However, more complex traits are determined by genetic variants that may have significant combined effects instead of a single-SNP effect [24]. Pathway-based GWAS [25, 26] is one of the strategies and statistical approaches that considers multiple genetic variants as SNPs or genes in a biological pathway to understand the genetic contributors of complex traits [27, 28].
In the present study, with the aim of identifying the genetic backgrounds of beak deformity in chickens, 48 BJY birds with deformed beaks and 48 normal ones were genotyped using the 600 K high-density (HD) genotyping array [29]. ROADTRIPS [30] and SNP ratio test (SRT) [31] software were used to identify the associated SNPs and pathways, respectively. The combined analyses extend our understanding of the genetic basis of this trait.
Results
Single SNP-based association study
In this study, a total of 48 birds with deformed beaks (case) and 48 normal birds (control) were genotyped using the Affymetrix Chicken 600 K HD genotyping arrays. According to the quality control (QC) criteria, one individual that had > 10% missing genotypes was removed. Additionally, 4505 SNPs with call rate < 90% and 120,261 SNPs with minor allele frequency (MAF) < 5% were excluded. As a result, 95 individuals and 429,539 SNPs were carried forward for subsequent analyses. The distribution of SNPs that passed QC in the chicken genome is presented in Table 1. A total of 21,984 independent SNP markers were obtained using multidimensional scaling (MDS) analysis using the first two principal components (Fig. 3), indicating no obvious population substructure in the birds.
Table 1.
Distribution of SNPs that passed quality control on the chicken genome
| Chromosome | No. SNPs | Physical length (Mb)c | Marker density (kb/SNP) | 5% chromosome-wide significance thresholdd |
|---|---|---|---|---|
| 0a | 5979 | – | – | 8.36E-06 |
| 1 | 81,335 | 195.3 | 2.40 | 6.15E-07 |
| 2 | 50,755 | 148.8 | 2.93 | 9.85E-07 |
| 3 | 45,028 | 110.4 | 2.45 | 1.11E-06 |
| 4 | 33,760 | 90.2 | 2.67 | 1.48E-06 |
| 5 | 24,678 | 59.6 | 2.41 | 2.03E-06 |
| 6 | 16,436 | 35.0 | 2.13 | 3.04E-06 |
| 7 | 17,826 | 36.2 | 2.03 | 2.80E-06 |
| 8 | 13,667 | 28.8 | 2.10 | 3.66E-06 |
| 9 | 14,291 | 23.4 | 1.64 | 3.50E-06 |
| 10 | 14,631 | 19.9 | 1.36 | 3.42E-06 |
| 11 | 10,749 | 19.4 | 1.80 | 4.65E-06 |
| 12 | 11,590 | 19.9 | 1.72 | 4.31E-06 |
| 13 | 8589 | 17.8 | 2.07 | 5.82E-06 |
| 14 | 10,062 | 15.2 | 1.51 | 4.97E-06 |
| 15 | 7697 | 12.7 | 1.64 | 6.50E-06 |
| 16 | 273 | 0.5 | 1.96 | 1.83E-04 |
| 17 | 7113 | 10.5 | 1.47 | 7.03E-06 |
| 18 | 7292 | 11.2 | 1.54 | 6.86E-06 |
| 19 | 6517 | 10.0 | 1.53 | 7.67E-06 |
| 20 | 7321 | 14.3 | 1.95 | 6.83E-06 |
| 21 | 6786 | 6.8 | 1.00 | 7.37E-06 |
| 22 | 2877 | 4.1 | 1.42 | 1.74E-05 |
| 23 | 4584 | 5.7 | 1.25 | 1.09E-05 |
| 24 | 5541 | 6.3 | 1.14 | 9.02E-06 |
| 25 | 1860 | 2.2 | 1.18 | 2.69E-05 |
| 26 | 4485 | 5.3 | 1.19 | 1.11E-05 |
| 27 | 3784 | 5.2 | 1.38 | 1.32E-05 |
| 28 | 3862 | 4.7 | 1.23 | 1.29E-05 |
| LGE22C19W28_E50C23b | 124 | 1.0 | 7.78 | 4.03E-04 |
| LGE64b | 47 | 0.8 | 17.02 | 1.06E-03 |
| Total | 429,539 | 921.2 | 2.46 | 1.16E-07 |
aThese SNPs are not mapped to any chromosome
bTwo linkage groups
cThe physical length of the chromosome was based on the genome build Gallus_gallus-4.0/galGal4 (Nov. 2011)
dBonferroni-corrected 5% chromosome-wise significance threshold = 0.05/number of SNPs after quality control
Fig. 3.

Population structure evaluated by the first two principal components. Six colors indicate the progeny of six males
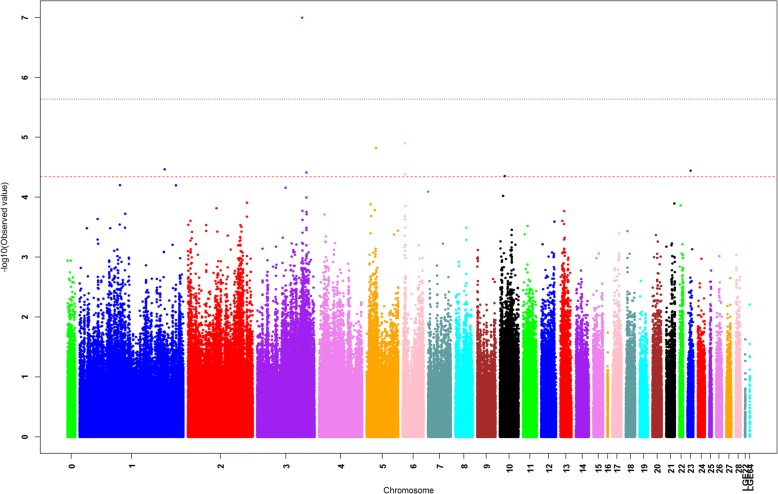
ROADTRIPS was used for the traditional SNP-based association analysis. In total, one associated SNP (5% genome-wide significance (2.27E-6, 0.05/21,984)) on GGA 3 (chicken chromosome 3), and seven suggestively associated SNPs (4.55E-5, 1/21,984) on GGAs 1, 3, 5, 6, 10, and 23 were detected. The detailed information of the SNP reaching 5% genome-wide significance and the seven suggestively associated SNPs is shown in Tables 2 and 3, respectively. For a SNP-based GWAS, the genomic inflation can be determined and calculated by a lambda (λ) value. If the analysis results of the data follow the normal chi-squared distribution (no inflation), the expected λ value is 1. In this study, for the Quantile-quantile (Q-Q) plot presented as Fig. 4, the λ value was 1.045, which means a minimal and acceptable inflation at the upper tail of the distribution. Therefore, we believe the result was reliable. The global view of P-values (in terms of -log10 (P-value)) for all SNPs was represented by a Manhattan plot, as shown in Fig. 5. The raw results of all the SNPs are described in Additional file 1: Table S1.
Table 2.
SNPs associated with beak deformity with 5% genome-wide significance
| SNP ID | Chromosome | Physical position (bp) | Nearbya genes | P-value |
|---|---|---|---|---|
| rs313625170 | 3 | 87,528,719 | LOC421892, TINAG, LOVL5, LRRC1, GCLC, KLHL31, LOC107053134 | 9.97E-08 |
aGenes located within 5 Mb upstream or downstream of the significant SNPs
Table 3.
SNPs suggested to be associated with beak deformity
| SNP ID | Chromosome | Physical position (bp) | Nearbya genes | P-value |
|---|---|---|---|---|
| rs316010119 | 6 | 4,029,716 | RET, MBL2, RASGEF1A | 1.25E-05 |
| rs317906090 | 5 | 18,672,451 | LDLRAD3, COMMD9, SLC1A2, RAG1 | 1.49E-05 |
| rs313002111 | 1 | 161,794,494 | OLFM4, DIAPH3, TDRD3 | 3.42E-05 |
| rs313486014 | 23 | 2,844,607 | PTPRU, STMN1 | 3.61E-05 |
| rs16327528 | 3 | 95,937,095 | YWHAQ, TAF1B | 3.87E-05 |
| rs317725514 | 6 | 4,455,292 | BMS1, RET | 4.16E-05 |
| rs14944750 | 10 | 5,358,795 | MIR204–2, APBA2, ADAL | 4.42E-05 |
aGenes located within 5 Mb upstream or downstream of the suggestively associated SNPs
Fig. 4.

Quantile-quantile (Q-Q) plot of the genome-wide association analysis using ROADTRIPS. The x-axis shows the expected P-values under the null hypothesis and the y-axis shows the observed P-values. The value of inflation factor λ was 1.045
Fig. 5.
Manhattan plots showing association of all SNPs with the beak deformity trait using ROADTRIPS. SNPs are plotted on the x-axis according to their positions on each chromosome against their association with this trait on the y-axis (shown as -log10 (P-value)). The red dashed line indicates suggestive genome-wide significance (P-value = 4.55E-5), and the grey dashed line shows genome-wide 5% significance with a P-value threshold of 2.27E-6
Pathway-based association study
The same genotyping dataset was used in this analysis. Based on the QC and SNP selection criteria (see Methods), a total of 149 pathways were finally selected from chicken pathway dataset, covering 128,072 SNPs (with 115,222 SNPs and 12,850 SNPs involved in the unique and multiple pathways, respectively). After correction for genetic relationships, we conducted a standard association analysis in PLINK for both the original and 100 randomized phenotype datasets. The association tests for the original dataset resulted in about 5000 nominally significant SNPs (unadjusted P < 0.05, Additional file 2: Table S2). The Q-Q plot is shown in Fig. 6, and the λ value was 0.962.
Fig. 6.
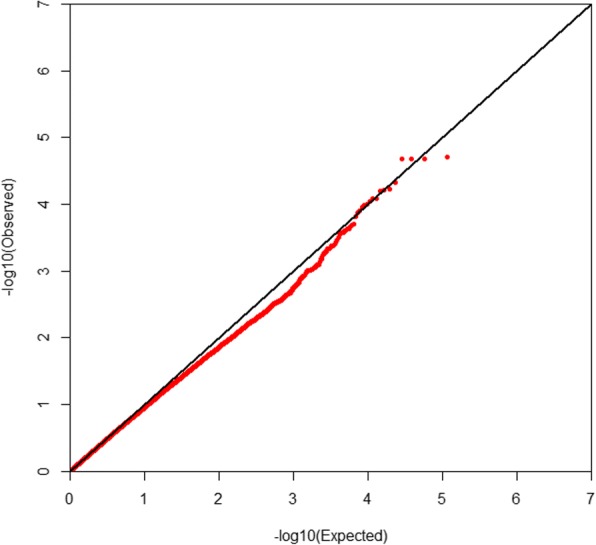
Quantile-quantile (Q-Q) plot for the SNPs used in the pathway-based GWAS. The x-axis shows the expected P-values under the null hypothesis and the y-axis shows the observed P-values. The value of inflation factor λ was 0.962
An SRT program was then applied to investigate the associations between beak deformity and the 149 pathways. As a result, we identified six associated pathways (PEMP < 0.05), including the ribosome pathway, the oocyte meiosis pathway, the pantothenate and CoA biosynthesis pathway, the pyruvate metabolism pathway, the glycine, serine, and threonine metabolism pathway, and the calcium signaling pathway (Table 4 and Fig. 7). The detailed information of each pathway is provided in Additional file 3: Table S3.
Table 4.
Six pathways significantly associated with beak deformity
| Pathway ID | Definition | PEMP-value | Ratio |
|---|---|---|---|
| gga03010 | Ribosome | 9.90E-03 | 194/1431 (13.6%) |
| gga04114 | Oocyte meiosis | 1.98E-02 | 98/433 (22.6%) |
| gga00770 | Pantothenate and CoA biosynthesis | 1.98E-02 | 32/95 (33.7%) |
| gga00620 | Pyruvate metabolism | 2.97E-02 | 63/231 (27.3%) |
| gga00260 | Glycine, serine and threonine metabolism | 3.96E-02 | 71/366 (19.4%) |
| gga04020 | Calcium signaling pathway | 4.95E-02 | 519/4106 (12.6%) |
Fig. 7.
The -log10 (PEMP-value) values of the 149 pathways for the beak deformity trait
Furthermore, we prepared plots of the number of significant SNPs, the number of total SNPs, the number of genes, the total length of genes, and the average length of genes for each pathway, against the -log10 PEMP-value of the selected pathways to investigate the effects of potential factors on SRT’s detection of the pathways. The associations between these factors and the significance of the pathways confirmed the utility of this strategy and permutations to reduce any bias caused by the SNPs, genes, gene sizes, and pathways (Additional file 4: Figure S1).
Discussion
Excluding other environmental contributing factors, beak deformity is observed in each generation of BJY chickens and cannot be eliminated based solely on the phenotype. The heritability of beak deformity was estimated at 0.12 (SE = 0.17) by ASReml-R 3 (https://www.vsni.co.uk/software/asreml-r/) using the SNP data in this study. This indicated that this trait is genetically determined. However, previous studies did not validate the role of any candidate genes for beak shapes and beak length in beak malformation [32, 33]. Birds with deformed beaks have much higher mortality and lower fertility rates, making it difficult to obtain individuals for genetic analysis. Eventually, a small-sized population with only 48 birds with deformed beaks was collected and used in the present study. Baird et al. [34] used 96 samples to identify three main chromosomal regions related to cranial cruciate ligament rupture in Newfoundland dogs. Fels and Distl [35] identified one significant and four suggestively significant SNPs related to canine hip dysplasia in 96 German shepherd dogs. These previous studies indicated that GWAS is also effective to study complex traits and diseases with a small sample size. In the present study, based on the case-control design, the genotype data of 48 birds with deformed beaks and 48 normal chickens were used for the single SNP and pathway-based GWAS analyses.
In the single SNP-based study, ROADTRIPS was used to increase the power of the association study [30]. PLINK was also used in the same way, however, it only identified one suggestively associated SNP (Additional file 5: Figure S2), which was exactly the 5% genome-wide significant SNP (rs313625170) identified by ROADTRIPS. This SNP is located on GGA 3, 6.7 kb, 24 kb, 63 kb, and 0.13 Mb upstream of LOC421892 (Transcription elongation factor B polypeptide 3-like,TCEB3-like), KLHL31 (Kelch like family member 31), GCLC (Glutamate-cysteine ligase catalytic subunit), and ELOVL5 (ELOVL fatty acid elongase 5) genes, respectively, and 26 kb and 0.21 Mb downstream of LRRC1 (Leucine rich repeat containing 1) and TINAG (Tubulointerstitial nephritis antigen) genes, respectively. This SNP is also located in the intron of LOC107053134, which is a predicted gene. The seven suggestively associated SNPs, rs313002111, rs16327528, rs317906090, rs316010119, rs317725514, rs14944750, and rs313486014, are located on GGAs 1, 3, 5, 6, 6, 10, and 23, respectively. They are located within, downstream or upstream of genes including TDRD3 (Tudor domain containing 3), RET (Ret proto-oncogene), and STMN1 (Stathmin 1). Two genes identified here, LOC421892 and TDRD3, were also highlighted in our former transcriptome study with a false discover rate (FDR) < 0.01 and |log2-Ratio (deformed/normal)| ≥ 1.5 [10], where LOC421892 was down-regulated and TDRD3 was up-regulated in the deformed beaks. The LOC421892 gene is a homolog of TCEB3 (also known as ELOA) gene, which is related to von Hippel-Lindan (VHL) [36] and nonsyndromic cleft lip and palate (NSCLP) [37] in human. It is the gene nearest to the significant SNP (rs313625170) identified here and could be the first candidate gene contribute to the formation of a deformed beak. TDRD3, located 0.19 Mb downstream of a suggestively significant SNP (rs313002111), is related to chromatin binding and methylated histone binding [38]. Mutation of TDRD3 is associated with human Fragile-X syndrome [39] and primary ovarian insufficiency [40]. RET, located 0.26 Mb upstream and 92 kb downstream of two suggestively significant SNPs, rs316010119 and rs317725514, respectively, might also play an important role in the beak shaping. Its gene product is involved in neural crest development [41]. RET can undergo oncogenic activation in vivo and in vitro by cytogenetic rearrangement [42]. As reported by Schneider and Helms [43], the origin and evolution of the beak in birds is strongly associated with neural crest cells. Moreover, mutations in RET are also associated with the disorders such as multiple endocrine neoplasia, type IIA, multiple endocrine neoplasia, type IIB, Hirschsprung disease, and medullary thyroid carcinoma [44]. In addition, its interacting gene, LRIG2, was also identified as one of the most promising candidate genes underlying this trait in our previous CNV study within the same genotyped data used here [45]. Thus, the RET and LRIG2 genes might jointly contribute to the development of deformed beaks. Similarly, STMN1, also named Oncoprotein 18, located 0.35 Mb downstream of another suggestively significant SNP (rs313486014), is involved in myelodysplastic syndromes [46], and is related to chick retina [47] and nasopharyngeal carcinoma [48]. These four genes may be related to beak deformity in chickens according to their positions, known functions, and our former studies.
In contrast to the single-SNP GWAS, pathway-based GWAS can identify the biological pathways that are useful to interpret the genetic basis of complex traits. The limits of pathway annotation and the power of the GWAS dataset, mean that adequate SNP coverage is essential for a pathway to be effectively tested [49]. This method has two primary advantages [31]: (1) it avoids issues arising from linkage disequilibrium (LD) by using the same SNPs in all simulations. Only pathways with additional significant SNPs, not merely arising from LD, are deemed as significant; (2) it uses individual level data in its simulations, which maximizes the information available to test pathway hypotheses. Furthermore, we also conducted some plots to confirm that the SNP number, gene number, and gene length in each pathway did not influence the results of SRT.
Six associated pathways were obtained, of which, the calcium signaling pathway (Additional file 6: Figure S3) has the most potential to be involved in beak deformity. Calcium ions are important for cellular signaling, because once they enter the cytoplasm they exert allosteric regulatory effects on many enzymes and proteins [50]. Calcium can act in signal transduction resulting from activation of ion channels or as a second messenger caused by indirect signal transduction pathways, such as those of G protein-coupled receptors [51]. In neurons, concomitant increases in cytosolic and mitochondrial calcium are important for the synchronization of neuronal electrical activity with mitochondrial energy metabolism. Thus, calcium plays an important role in regulating various neuronal processes [52]. Several studies have shown that calcium was highly associated with beak shape in birds. A previous study demonstrated differential expression of calmodulin between finches with different beak types [17]. The structure of a toucan beak was found to be a sandwich composite with an exterior of keratin and a fibrous network of closed cells comprising calcium-rich proteins [53]. Lamichhaney [11] found that among the 15 most significant genomic regions related to beak shape, six harbored genes associated with craniofacial and/or beak development in mammals or birds were identified, including calmodulin. From the previous studies in our laboratory, Zhu et al. [54] revealed that the calcium content of the beak was 7.5–12%. Liu et al. [55] observed that the over-expression of parvalbumin, a calcium ion-binding protein, could result in beak deformity in chickens, as assessed using iTRAQ-based proteomic analysis. Taken together, these results suggest that the calcium signaling pathway could be a key factor related to the beak deformity trait in chickens.
Conclusions
In conclusion, one 5% genome-wide significant SNP and seven suggestively significant SNPs that may be involved in the beak deformity trait were identified, using the single SNP GWAS. Four candidate genes, LOC421892, TDRD3, RET, and STMN1, were identified as the most promising genes underlying this trait. Simultaneously, six pathways were found to be associated with this trait using the pathway-based GWAS, where the calcium signaling pathway may be the most important. Overall, our findings are worthy of further functional characterization to reveal the pinpoint causes of beak deformity and the underlying the mechanism of this disorder in chickens.
Methods
Animals and DNA samples collection
Six males and 12 females with deformed beaks from a BJY chicken population, a local breed conserved by Institute of Animal Science, Chinese Academy of Agricultural Sciences (IAS, CAAS, Beijing, China), were used for a random mating experiment to produce 921 offspring. The incidence of beak deformity birds was 7.82%, which was higher than that observed previously in the normal population (up to 3%). Forty-eight birds with deformed beaks and 48 normal birds were randomly selected from the offspring at 20 days of age, when the crossed beaks could be clearly identified, and used for genotyping. All of these birds were observed until 90 days of age before blood collection to make sure that the phenotype was stable. Blood samples were collected from the brachial vein by venipuncture. Genomic DNA (gDNA) was isolated from blood samples using the phenol-chloroform method. The purity and concentration of the gDNA samples were measured using a Nanodrop ND-2000 spectrophotometer (Thermo Scientific, MA, USA). The final concentration was adjusted to 50 ng/μL. gDNA samples with an A260/280 ratio of 1.8–2.0 were submitted for genotyping.
Genotyping and quality control
gDNA samples were genotyped using 600 K Affymetrix Axiom HD genotyping arrays containing 580,954 SNPs based on the Affymetrix GeneChip platform [29]. The genotypes were identified using Affymetrix Genotyping Console (Version 4.2, Affymetrix Inc., Santa Clara, CA, USA) and a custom cluster file developed from the 96 (48 deformed and 48 normal) samples. Following the case-control design, stringent QC procedures were performed for the genotype data using PLINK [56] (Version 1.07, http://zzz.bwh.harvard.edu/plink/). First, individuals with > 10% missing genotypes were excluded (n = 1). Second, out of the initial full set of 554,305 effective SNPs in this study, we discarded those with a call rate < 90% (n = 4505) and those having a minor allele frequency (MAF) < 0.05 in all birds (n = 120,261). The Hardy-Weinberg equilibrium (HWE) was not used to filter data in view of the small population.
Population stratification assessment
To evaluate the existence of a population substructure among the individuals, the classical MDS was performed in PLINK using the following procedures: (a) autosomal SNPs were subjected to LD-based pruning to ensure that uncorrected LD did not distort the analysis. Thus, the remaining SNPs within a window size of 50 SNPs and a step of 10 SNPs had pairwise r2 < 0.2; (b) the pairwise identical-by-state (IBS) distance among the 95 individuals was calculated using the remaining 21,984 SNPs; (c) the first two MDS dimensions were extracted via the “MDS-plot” command and visualized in R (version 3.2.0, www.r-project.org).
Single SNP-based association analysis
The RM test was performed for the association analysis in ROADTRIPS (Version 1.2) [30]. An important advantage of ROADTRIPS is that it can deal with data with a known pedigree structure as well as population admixture in an association test by constructing an empirical covariance matrix from genome-wide SNP data to adjust for potential population admixture and for genetic connectedness among individuals in both the control and case groups. The P-value was corrected using a strict Bonferroni adjustment based on LD pruning [57]. The sum of the independent LD blocks plus singleton markers were used to define the number of independent statistical comparisons [58]. Finally, 21,984 independent tests were used to determine the P-value thresholds, including 5% genome-wide significance (2.27E-6, 0.05/21,984) and suggestive association (4.55E-5, 1/21,984). The Q-Q plot and the Manhattan plot of GWAS for this trait were produced using the “gap” package [59] in R.
Pathway-based association analysis
We retrieved all the pathways in chicken (Additional file 7: Table S4) from the KEGG [60] pathway database (http://www.genome.jp/kegg/) to identify pathways that potentially contribute to the beak deformity trait in the chicken. A total of 162 annotated pathways were collected for analysis. Only SNPs located within or 50 kb upstream or downstream of a gene were selected to create a file linking pathways and SNP information (Additional file 8: Table S5). In addition, if a SNP was involved in multiple pathways, the SNP and the pathways were both included in the analysis. A SRT program (Version 3, https://sourceforge.net/projects/snpratiotest/) was employed for the analysis. The SRT compared the proportion of the significant SNPs (unadjusted P < 0.05 in the single SNP analysis) to all the SNPs that are part of a pathway and computed an empirical P-value (PEMP) based on comparisons to ratios in simulated datasets where the assignment of case/control status had been randomized. The simulated datasets were constructed from the original dataset, preserving the original case/control ratio, but randomizing the assignment of case/control status among individuals. The SRT accepts files in the PLINK binary format and allows the user to prepare randomized phenotype datasets. The PEMP for a particular pathway, = (s + 1)/(N + 1), where s is the number of simulated datasets that produce a ratio greater than or equal to the original ratio and N is the number of permutations [61]. The pipeline of SRT is shown in Additional file 9: Figure S4.
Additional files
Figure S1. Significance of the pathway (−log10 (PEMP-value)) versus: (a) the number of significant SNPs in the pathways, (b) the number of SNPs in the pathways, (c) the number of genes in the pathways, (d) total length (kb) of genes in the pathways, and (e) average length (kb) of the genes in the pathways. The P = 0.05 cut-off is highlighted by a vertical red line. (JPG 105 kb)
Figure S2. Manhattan plots showing the association of all SNPs with beak deformity trait using PLINK. SNPs are plotted on the x-axis according to their positions on each chromosome against their association with this trait on the y-axis (shown as -log10 (P-value)). The red dashed line indicates suggestive genome-wide significance (P-value = 4.55E-5). (TIF 2715 kb)
Figure S3. The calcium signaling pathway. (TIF 27 kb)
Figure S4. The pipeline of SRT (Referred to the SRT manual). (TIF 100 kb)
Table S1. Raw results of the single SNP-based association study. (XLSX 21782 kb)
Table S2. Single SNP-based association analysis of the selected SNPs used in the pathway-based association study. (XLSX 5391 kb)
Table S3. The detailed information of each pathway based on the pathway-based association analysis. (XLSX 15 kb)
Table S4. The detailed lists of all the chicken pathways used in the pathway-based association analysis. (XLSX 14 kb)
Table S5. The detailed lists of pathways and SNPs information used in the pathway-based association analysis. (XLSX 3841 kb)
Acknowledgements
We would like to thank the native English speaking scientists of Elixigen Company (Huntington Beach, California) for editing our manuscript.
Funding
This work was supported by the China Agriculture Research Systems [grant number CARS-40]; the National Natural Science Foundation of China [grant number 31501949]; the Beijing Municipal Science and Technology Project [grant number D171100007817005]; and the Agricultural Science and Technology Innovation Program [grant numbers ASTIPIAS04, CAAS-XTCX2016010–03]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Availability of data and materials
All the raw data of SNP genotypes have been deposited in the NCBI Gene Expression Omnibus (GEO, accession number: GSE101703).
Abbreviations
- ALX1
ALX homeobox 1
- BJY
Beijing-You
- BMP4
Bone morphogenetic protein 4
- CaM
Calmodulin
- CNV
Copy number variation
- DGE
Digital gene expression
- ELOA
Elongin A
- ELOVL5
ELOVL fatty acid elongase 5
- FGF8
Fibroblast growth factor 8
- GCLC
Glutamate-cysteine ligase catalytic subunit
- gDNA
Genomic DNA
- GWAS
Genome-wide association study
- HD
High-density
- HMGA2
High mobility AT-hook 2
- HOXA1
Homeobox A1
- HOXD3
Homeobox D3
- HWE
Hardy-Weinberg equilibrium
- IBS
Identical-by-state
- iTRAQ
Isobaric tags for relative and absolute quantification
- KLHL31
Kelch like family member 31
- LD
Linkage disequilibrium
- LRRC1
Leucine rich repeat containing 1
- MAF
Minor allele frequency
- MDS
Multidimensional scaling
- PEMP
Empirical P-value
- QC
Quality control
- Q-Q
Quantile-quantile
- RET
Ret proto-oncogene
- Shh
Sonic hedgehog
- SNPs
Single nucleotide polymorphisms
- SRT
SNP ratio test
- STMN1
Stathmin 1
- TDRD3
Tudor domain containing 3
- TINAG
Tubulointerstitial nephritis antigen
Authors’ contributions
JC, HB, and YS conceived and designed the experiments. HB, NL, FX, YL, SX, JY, and YC performed the experiments and participated in the data collection. HB and LZ analyzed the data. HB and YS wrote the paper. JC, HB, YS, and JY revised the manuscript. All authors read and approved the final manuscript.
Ethics approval
All of the animal procedures were conducted in strict accordance with the guidelines proposed by the China Council on Animal Care and the Ministry of Agriculture of the People’s Republic of China. Animal experiments were approved by the Science Research Department of IAS, CAAS (Beijing, China).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Hao Bai and Yanyan Sun contributed equally to this work.
Electronic supplementary material
The online version of this article (10.1186/s12864-018-4882-8) contains supplementary material, which is available to authorized users.
Contributor Information
Hao Bai, Email: bhowen1027@163.com.
Yanyan Sun, Email: yanyansun2014@163.com.
Nian Liu, Email: liunian.14@163.com.
Fuguang Xue, Email: xueguang123@126.com.
Yunlei Li, Email: mailyunlei@163.com.
Songshan Xu, Email: 1023302752@qq.com.
Jianhua Ye, Email: yjh6scxh@163.com.
Lei Zhang, Email: lz@capitalbio.com.
Yu Chen, Email: chenyu.cncn@163.com.
Jilan Chen, Email: chen.jilan@163.com.
References
- 1.Seki Y, Bodde SG, Meyers MA. Toucan and hornbill beaks: a comparative study. Acta Biomater. 2010;6(2):331–343. doi: 10.1016/j.actbio.2009.08.026. [DOI] [PubMed] [Google Scholar]
- 2.Badyaev AV, Young RL, Oh KP, Addison C. Evolution on a local scale: developmental, functional, and genetic bases of divergence in bill form and associated changes in song structure between adjacent habitats. Evolution. 2008;62(8):1951–1964. doi: 10.1111/j.1558-5646.2008.00428.x. [DOI] [PubMed] [Google Scholar]
- 3.Herrel A, Podos J, Huber S, Hendry A. Bite performance and morphology in a population of Darwin's finches: implications for the evolution of beak shape. Funct Ecol. 2005;19(1):43–48. doi: 10.1111/j.0269-8463.2005.00923.x. [DOI] [Google Scholar]
- 4.Smith TB. Resource use by bill morphs of an African finch: evidence for intraspecific competition. Ecology. 1990;71(4):1246–1257. doi: 10.2307/1938261. [DOI] [Google Scholar]
- 5.Jobling JA. Helm dictionary of scientific bird names. London: A&C Black: Bloomsbury Publishing; 2010.
- 6.Hemert CV, Handel CM. Beak deformities in northwestern crows: evidence of a multispecies epizootic. Auk. 2010;127(4):746–751. doi: 10.1525/auk.2010.10132. [DOI] [Google Scholar]
- 7.Pomeroy D. Birds with abnormal bills. British Birds. 1962;55(2):49–72. [Google Scholar]
- 8.Williamson AP, Blattner RJ, Lutz HR. Abnormalities in Chick embryos following thalidomide and other insoluble compounds in the amniotic cavity. P Soc Exp Biol Med. 1963;112(4):1022–1025. doi: 10.3181/00379727-112-28241. [DOI] [PubMed] [Google Scholar]
- 9.Cheung MO, Gilbert EF, Peterson RE. Cardiovascular teratogenicity of 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin in the chick embryo. Toxicol Appl Pharm. 1981;61(2):197–204. doi: 10.1016/0041-008X(81)90409-9. [DOI] [PubMed] [Google Scholar]
- 10.Bai H, Zhu J, Sun Y, Liu RT, Liu N, Li DL, Wen J, Chen JL. Identification of genes related to beak deformity of chickens using digital gene expression profiling. PLoS One. 2014;9(9):e107050. doi: 10.1371/journal.pone.0107050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamichhaney S, Berglund J, Almén MS, Maqbool K, Grabherr M, Martinez-Barrio A, Promerová M, Rubin C-J, Wang C, Zamani N. Evolution of Darwin's finches and their beaks revealed by genome sequencing. Nature. 2015;518(7539):371–375. doi: 10.1038/nature14181. [DOI] [PubMed] [Google Scholar]
- 12.Lamichhaney S, Han F, Berglund J, Wang C, Almén MS, Webster MT, Grant BR, Grant PR, Andersson L. A beak size locus in Darwin’s finches facilitated character displacement during a drought. Science. 2016;352(6284):470–474. doi: 10.1126/science.aad8786. [DOI] [PubMed] [Google Scholar]
- 13.MacDonald ME, Abbott UK, Richman JM. Upper beak truncation in chicken embryos with the cleft primary palate mutation is due to an epithelial defect in the frontonasal mass. Dev Dynam. 2004;230(2):335–349. doi: 10.1002/dvdy.20041. [DOI] [PubMed] [Google Scholar]
- 14.Hu D, Marcucio RS, Helms JA. A zone of frontonasal ectoderm regulates patterning and growth in the face. Development. 2003;130(9):1749–1758. doi: 10.1242/dev.00397. [DOI] [PubMed] [Google Scholar]
- 15.Abzhanov A, Protas M, Grant BR, Grant PR, Tabin CJ. Bmp4 and morphological variation of beaks in Darwin's finches. Science. 2004;305(5689):1462–1465. doi: 10.1126/science.1098095. [DOI] [PubMed] [Google Scholar]
- 16.Wu P, Jiang TX, Shen JY, Widelitz RB, Chuong CM. Morphoregulation of avian beaks: comparative mapping of growth zone activities and morphological evolution. Dev Dynam. 2006;235(5):1400–1412. doi: 10.1002/dvdy.20825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abzhanov A, Kuo WP, Hartmann C, Grant BR, Grant PR, Tabin CJ. The calmodulin pathway and evolution of elongated beak morphology in Darwin's finches. Nature. 2006;442(7102):563–567. doi: 10.1038/nature04843. [DOI] [PubMed] [Google Scholar]
- 18.Jaszczak K, Malewski T, Parada R, Malec H. Expression of Hoxal and Hoxd3 genes in chicken embryos with exencephaly. J Anim Feed Sci. 2006;15(3):463–469. doi: 10.22358/jafs/66917/2006. [DOI] [Google Scholar]
- 19.Bai H, Sun Y, Zhu J, Liu N, Li DL, Xue FG, Li YL, Chen JL. Study on LOC426217 as a candidate gene for beak deformity in chicken. BMC Genet. 2016;17(1):44. doi: 10.1186/s12863-016-0353-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gudmundsson J, Sulem P, Gudbjartsson DF, Blondal T, Gylfason A, Agnarsson BA, Benediktsdottir KR, Magnusdottir DN, Orlygsdottir G, Jakobsdottir M. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat Genet. 2009;41(10):1122–1126. doi: 10.1038/ng.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Einarsdottir E, Hafrén L, Leinonen E, Bhutta MF, Kentala E, Kere J, Mattila PS. Genome-wide association analysis reveals variants on chromosome 19 that contribute to childhood risk of chronic otitis media with effusion. Sci Rep. 2016;6:33240. doi: 10.1038/srep33240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giri AK, Banerjee P, Chakraborty S, Kauser Y, Undru A, Roy S, Parekatt V, Ghosh S, Tandon N, Bharadwaj D. Genome wide association study of uric acid in Indian population and interaction of identified variants with type 2 diabetes. Sci Rep. 2016;6:21440. doi: 10.1038/srep21440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun Y, Huang Y, Yin A, Pan Y, Wang Y, Wang C, Du Y, Wang M, Lan F, Hu Z. Genome-wide association study identifies a new susceptibility locus for cleft lip with or without a cleft palate. Nat Commun. 2015;6:6414. doi: 10.1038/ncomms7414. [DOI] [PubMed] [Google Scholar]
- 24.Goldstein DB. Common genetic variation and human traits. New Engl J Med. 2009;360(17):1696. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- 25.Wang K, Li M, Bucan M. Pathway-based approaches for analysis of genomewide association studies. Am J Hum Genet. 2007;81(6):1278–1283. doi: 10.1086/522374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang K, Li M, Hakonarson H. Analysing biological pathways in genome-wide association studies. Nat Rev Genet. 2010;11(12):843–854. doi: 10.1038/nrg2884. [DOI] [PubMed] [Google Scholar]
- 27.Fan H, Wu Y, Zhou X, Xia J, Zhang W, Song Y, Liu F, Chen Y, Zhang L, Gao X. Pathway-based genome-wide association studies for two meat production traits in simmental cattle. Sci Rep. 2015;5:18389. doi: 10.1038/srep18389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiao H, Wang K, Yang F, Grant SF, Hakonarson H, Price RA, Li WD. Pathway-based genome-wide association studies for plasma triglycerides in obese females and normal-weight controls. PLoS One. 2015;10(8):e0134923. doi: 10.1371/journal.pone.0134923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kranis A, Gheyas AA, Boschiero C, Turner F, Yu L, Smith S, Talbot R, Pirani A, Brew F, Kaiser P. Development of a high density 600K SNP genotyping array for chicken. BMC Genomics. 2013;14(1):59. doi: 10.1186/1471-2164-14-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thornton T, McPeek MS. ROADTRIPS: case-control association testing with partially or completely unknown population and pedigree structure. Am J Hum Genet. 2010;86(2):172–184. doi: 10.1016/j.ajhg.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Dushlaine C, Kenny E, Heron EA, Segurado R, Gill M, Morris DW, Corvin A. The SNP ratio test: pathway analysis of genome-wide association datasets. Bioinformatics. 2009;25(20):2762–2763. doi: 10.1093/bioinformatics/btp448. [DOI] [PubMed] [Google Scholar]
- 32.Bai H. The mRNA Expression and SNPs Analysis of Candidate Genes associated with Beak Deformity Trait in Beijing-You chickens. Degree of Master. Yangzhou University, Yangzhou, Jiangsu, China. China Academic Journal (CD) Electronic Journals Publishing House Co., Ltd, 36 District of Tsinghua University, Haidian District, Beijing, China; 2013.
- 33.Bai H, Chen JL, Liu RR, Zhu J, Bi YL, Tang S, Qin N, Chang GB, Zheng MQ, Zhao GP, Wen J, Chen GH. Study on polymorphisms of GLA as a candidate gene for the beak deformity trait in Beijing-you chickens. Chinese. J Anim Sci. 2013;49(15):1–6. [Google Scholar]
- 34.Baird A, Carter S, Innes J, Ollier W, Short A. Genome-wide association study identifies genomic regions of association for cruciate ligament rupture in Newfoundland dogs. Anim Genet. 2014;45(4):542–549. doi: 10.1111/age.12162. [DOI] [PubMed] [Google Scholar]
- 35.Fels L, Distl O. Identification and validation of quantitative trait loci (QTL) for canine hip dysplasia (CHD) in German shepherd dogs. PLoS One. 2014;9(5):e96618. doi: 10.1371/journal.pone.0096618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohh M, Takagi Y, Aso T, Stebbins CE, Pavletich NP, Zbar B, Conaway RC, Conaway JW, Kaelin WG., Jr Synthetic peptides define critical contacts between elongin C, elongin B, and the von Hippel-Lindau protein. J Clin Invest. 1999;104(11):1583–1591. doi: 10.1172/JCI8161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Araujo TK, Secolin R, Félix TM, de Souza LT, Fontes MÍ, Monlleó IL, de Souza J, Fett-Conte AC, Ribeiro EM, Xavier AC, de Rezende AA, Simioni M, Ribeiro-dos-Santos ÂK, dos Santos SE, Gil-da-Silva-Lopes VL. A multicentric association study between 39 genes and nonsyndromic cleft lip and palate in a Brazilian population. J Cranio Maxill Surg. 2016;44(1):16–20. doi: 10.1016/j.jcms.2015.07.026. [DOI] [PubMed] [Google Scholar]
- 38.Yang Y, Lu Y, Espejo A, Wu J, Xu W, Liang S, Bedford MT. TDRD3 is an effector molecule for arginine-methylated histone marks. Mol Cell. 2010;40(6):1016–1023. doi: 10.1016/j.molcel.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Linder B, Plöttner O, Kroiss M, Hartmann E, Laggerbauer B, Meister G, Keidel E, Fischer U. TDRD3 is a novel stress granule-associated protein interacting with the fragile-X syndrome protein FMRP. Hum Mol Genet. 2008;17(20):3236–3246. doi: 10.1093/hmg/ddn219. [DOI] [PubMed] [Google Scholar]
- 40.Stolk L, Perry JR, Chasman DI, He C, Mangino M, Sulem P, Barbalic M, Broer L, Byrne EM, Ernst F. Meta-analyses identify 13 loci associated with age at menopause and highlight DNA repair and immune pathways. Nat Genet. 2012;44(3):260–268. doi: 10.1038/ng.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robertson K, Mason I. Expression of ret in the chicken embryo suggests roles in regionalisation of the vagal neural tube and somites and in development of multiple neural crest and placodal lineages. Mech Develop. 1995;53(3):329–344. doi: 10.1016/0925-4773(95)00449-1. [DOI] [PubMed] [Google Scholar]
- 42.Grieco M, Santoro M, Berlingieri MT, Melillo RM, Donghi R, Bongarzone I, Pierotti MA, Della Ports G, Fusco A, Vecchiot G. PTC is a novel rearranged form of the ret proto-oncogene and is frequently detected in vivo in human thyroid papillary carcinomas. Cell. 1990;60(4):557–563. doi: 10.1016/0092-8674(90)90659-3. [DOI] [PubMed] [Google Scholar]
- 43.Schneider RA, Helms JA. The cellular and molecular origins of beak morphology. Science. 2003;299(5606):565–568. doi: 10.1126/science.1077827. [DOI] [PubMed] [Google Scholar]
- 44.Eng C, Clayton D, Schuffenecker I, Lenoir G, Cote G, Gagel RF, van Amstel HKP, Lips CJ, Nishisho I, Takai S-I. The relationship between specific RET proto-oncogene mutations and disease phenotype in multiple endocrine neoplasia type 2: international RET mutation consortium analysis. Jama. 1996;276(19):1575–1579. doi: 10.1001/jama.1996.03540190047028. [DOI] [PubMed] [Google Scholar]
- 45.Bai H, Sun Y, Liu N, Liu YF, Xue FG, Li YL, Xu SS, Ni AX, Ye JH, Chen Y, Chen JL. Genome-wide detection of CNVs associated with beak deformity in chickens using high-density 600K SNP arrays. Anim Genet. 2018;49:226–236. doi: 10.1111/age.12652. [DOI] [PubMed] [Google Scholar]
- 46.Machado-Neto JA, de Melo Campos P, Favaro P, Lazarini M, Lorand-Metze I, Costa FF, Saad STO, Traina F. Stathmin 1 is involved in the highly proliferative phenotype of high-risk myelodysplastic syndromes and acute leukemia cells. Leukemia Res. 2014;38(2):251–257. doi: 10.1016/j.leukres.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 47.Godbout R. Identification and characterization of transcripts present at elevated levels in the undifferentiated chick retina. Exp Eye Res. 1993;56(1):95–106. doi: 10.1006/exer.1993.1014. [DOI] [PubMed] [Google Scholar]
- 48.Hsu HP, Li CF, Lee SW, Wu WR, Chen TJ, Chang KY, Liang SS, Tsai CJ, Shiue YL. Overexpression of stathmin 1 confers an independent prognostic indicator in nasopharyngeal carcinoma. Tumor Biol. 2014;35(3):2619–2629. doi: 10.1007/s13277-013-1345-3. [DOI] [PubMed] [Google Scholar]
- 49.Bolormaa S, Neto L, Zhang Y, Bunch R, Harrison B, Goddard M, Barendse W. A genome-wide association study of meat and carcass traits in Australian cattle. J Anim Sci. 2011;89(8):2297–2309. doi: 10.2527/jas.2010-3138. [DOI] [PubMed] [Google Scholar]
- 50.Clapham DE. Calcium signaling. Cell. 2007;131(6):1047–1058. doi: 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 51.Clapham DE. Calcium signaling. Cell. 1995;80(2):259–268. doi: 10.1016/0092-8674(95)90408-5. [DOI] [PubMed] [Google Scholar]
- 52.Berridge MJ. Neuronal calcium signaling. Neuron. 1998;21(1):13–26. doi: 10.1016/S0896-6273(00)80510-3. [DOI] [PubMed] [Google Scholar]
- 53.Seki Y, Schneider MS, Meyers MA. Structure and mechanical behavior of a toucan beak. Acta Mater. 2005;53(20):5281–5296. doi: 10.1016/j.actamat.2005.04.048. [DOI] [Google Scholar]
- 54.Zhu J, Chen JL, Liu RR, Zheng MQ, Hu J, Liu GF, Wang ZW, Bi YL, Tang S, Bai H, Zhao GP, Wen J. Chemical analysis of chicken beak tissue. China Poultry. 2012;34(10):59–60. [Google Scholar]
- 55.Liu N, Sun Y, Bai H, Hua DK, Xue FG, Liu RR, Li DL, Wen J, Chen JL. iTRAQ-based proteomic analysis for identification of candidate proteins underlying beak deformity in chickens. Sci Agric Sin. 2015;48(21):4390–4396. [Google Scholar]
- 56.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, De Bakker PI, Daly MJ. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Johnson RC, Nelson GW, Troyer JL, Lautenberger JA, Kessing BD, Winkler CA, O'Brien SJ. Accounting for multiple comparisons in a genome-wide association study (GWAS) BMC Genomics. 2010;11(1):724. doi: 10.1186/1471-2164-11-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nicodemus KK, Liu W, Chase GA, Tsai YY, Fallin MD. Comparison of type I error for multiple test corrections in large single-nucleotide polymorphism studies using principal components versus haplotype blocking algorithms. BMC Genet. 2005;6:S78. doi: 10.1186/1471-2156-6-S1-S78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao JH. Gap: Genetic analysis package. J Stat Softw. 2007;23(8):1–18. doi: 10.18637/jss.v023.i08. [DOI] [Google Scholar]
- 60.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.North BV, Curtis D, Sham PC. A note on the calculation of empirical P values from Monte Carlo procedures. Am J Hum Genet. 2002;71(2):439–441. doi: 10.1086/341527. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Significance of the pathway (−log10 (PEMP-value)) versus: (a) the number of significant SNPs in the pathways, (b) the number of SNPs in the pathways, (c) the number of genes in the pathways, (d) total length (kb) of genes in the pathways, and (e) average length (kb) of the genes in the pathways. The P = 0.05 cut-off is highlighted by a vertical red line. (JPG 105 kb)
Figure S2. Manhattan plots showing the association of all SNPs with beak deformity trait using PLINK. SNPs are plotted on the x-axis according to their positions on each chromosome against their association with this trait on the y-axis (shown as -log10 (P-value)). The red dashed line indicates suggestive genome-wide significance (P-value = 4.55E-5). (TIF 2715 kb)
Figure S3. The calcium signaling pathway. (TIF 27 kb)
Figure S4. The pipeline of SRT (Referred to the SRT manual). (TIF 100 kb)
Table S1. Raw results of the single SNP-based association study. (XLSX 21782 kb)
Table S2. Single SNP-based association analysis of the selected SNPs used in the pathway-based association study. (XLSX 5391 kb)
Table S3. The detailed information of each pathway based on the pathway-based association analysis. (XLSX 15 kb)
Table S4. The detailed lists of all the chicken pathways used in the pathway-based association analysis. (XLSX 14 kb)
Table S5. The detailed lists of pathways and SNPs information used in the pathway-based association analysis. (XLSX 3841 kb)
Data Availability Statement
All the raw data of SNP genotypes have been deposited in the NCBI Gene Expression Omnibus (GEO, accession number: GSE101703).