Abstract
Background
The bacterial community present in the female lower genital tract plays an important role in maternal and neonatal health. Imbalances in this microbiota have been associated with negative reproductive outcomes, such as spontaneous preterm birth (sPTB), but the mechanisms underlying the association between a disturbed microbiota and sPTB remain poorly understood. An intrauterine infection ascending from the vagina is thought to be an important contributor to the onset of preterm labour. Our objective was to characterize the vaginal microbiota of pregnant women who had sPTB (n = 46) and compare to those of pregnant women who delivered at term (n = 170). Vaginal swabs were collected from women at 11–16 weeks of gestational age. Microbiota profiles were created by PCR amplification and pyrosequencing of the cpn60 universal target region.
Results
Profiles clustered into seven community state types: I (Lactobacillus crispatus dominated), II (Lactobacillus gasseri dominated), III (Lactobacillus iners dominated), IVA (Gardnerella vaginalis subgroup B or mix of species), IVC (G. vaginalis subgroup A dominated), IVD (G. vaginalis subgroup C dominated) and V (Lactobacillus jensenii dominated). The microbiota of women who experienced preterm birth (< 37 weeks gestation) had higher richness and diversity and higher Mollicutes prevalence when compared to those of women who delivered at term. The two groups did not cluster according to CST, likely because CST assignment is driven in most cases by the dominance of one particular species, overwhelming the contributions of more rare taxa. In conclusion, we did not identify a specific microbial community structure that predicts sPTB, but differences in microbiota richness, diversity and Mollicutes prevalence were observed between groups.
Conclusions
Although a causal relationship remains to be determined, our results confirm previous reports of an association between Mollicutes and sPTB and further suggest that a more diverse microbiome may be important in the pathogenesis of some cases.
Electronic supplementary material
The online version of this article (10.1186/s40168-018-0502-8) contains supplementary material, which is available to authorized users.
Keywords: Microbiome, Vagina, Lactobacillus, CST, Diversity, Richness, Mollicutes, Preterm birth, Pregnancy, Infection
Background
Preterm birth is defined as delivery before 37 completed weeks of gestational age [1] and can be further sub-categorized in extremely preterm (≤ 27+6 weeks+days), very preterm (28 to 31+6) and late preterm (32 to 36+6) [2]. Preterm birth comprises 11% of all livebirths worldwide, and its complications are estimated to cause 35% of world’s neonatal deaths, which represents 3.1 million deaths annually [3]. Children who are born prematurely also have higher rates of cardiovascular disorders, respiratory distress syndrome, neurodevelopmental disabilities and learning difficulties compared with children born at term [4].
Preterm birth is a complex multi-factorial condition with several known risk factors, such as low and high maternal ages [5–7], low BMI [8], black ethnicity [9], tobacco use, heavy alcohol intake, illicit drug use [4], close temporal proximity to a previous delivery [10], and multiple gestation [11]. Although studied extensively, some preterm cases remain unexplained for women with no known risk factors. Intrauterine infection with organisms ascending from the vagina has been hypothesized as an important contributor to preterm birth since many organisms isolated from the amniotic fluid/membranes of women who experienced preterm birth are also found in the lower genital tract of pregnant women [12–15]. A large number of studies support this hypothesis based on the strong association between intra-amniotic bacterial infection and preterm birth [12, 14–20].
The microbiological diagnosis of a ‘normal’ or disturbed vaginal microbiota has historically been based on the Nugent score, the current gold standard diagnostic method that relies on Gram stain of vaginal smears [21]. The ‘normal’ vaginal microbiota in non-pregnant reproductive aged women is understood to be dominated by Lactobacillus species, while an abnormal microbiota (defined as bacterial vaginosis) is characterized by low abundance of lactobacilli and an overgrowth of anaerobic bacteria, such as Gardnerella vaginalis, Prevotella spp., Bacteroides spp., Mobiluncus spp. and Mycoplasma hominis [22]. In low-risk pregnant women, it has been shown that the vaginal microbiota has reduced richness and diversity and increased abundance of lactobacilli compared to those of non-pregnant women [23–27]. An abnormal microbiota has been previously associated with preterm birth [28], but only a few in depth culture-independent studies of the vaginal microbiota of women who had preterm birth have been published, with inconsistent conclusions [29–32].
The objective of this study was to assess whether there are differences in the vaginal microbiota composition, early in gestation, of women who had spontaneous preterm birth (sPTB) and term delivery that could be further investigated as diagnostic indicators of preterm birth risk. Microbiome profiling was based on sequencing of the cpn60 universal target, which provides higher resolution than 16S rRNA variable regions [33] and allows the resolution of Gardnerella vaginalis subgroups, a hallmark bacteria in the disturbed microbiota [34].
Methods
Study population and sampling
This retrospective cohort study analysed the vaginal microbiota of women who experienced spontaneous preterm birth (sPTB) and compared the resulting microbial profiles to those of pregnant women who delivered at term. The bacterial profiles of pregnant Canadian women at low risk of sPTB who had term deliveries (n = 170) were previously generated by our research group [24]. The vaginal microbial profiles of Canadian women who had preterm birth originated from samples of this previous study (n = 7) [24] and from the Ontario Birth Study (n = 39), resulting in 46 samples. The Ontario Birth Study (ontariobirthstudy.com) is an open longitudinal pregnancy cohort at Mount Sinai Hospital, Toronto, Canada. It is a platform for studies of both pregnancy complications as well as Developmental Origins of Health and Disease related research. The PTB rates for the low-risk cohort and OBS cohorts were 4 and 6.2%, respectively. All biospecimens, including maternal vaginal swabs and maternal and infant blood, are collected concurrently with routine clinical specimens to reduce the burden on study participants. Detailed demographic and lifestyle characteristics are obtained from women during pregnancy and postpartum, and clinical information is extracted from the health records. For the purposes of this report, self-administered vaginal swabs were taken at 16 weeks gestation and placed in dry tubes prior to being placed in − 80 °C for storage in the Lunenfeld Tanenbaum Research Institute Biospecimen Storage and Processing Laboratory. Specimens from all cohorts were processed similarly in terms of sample collection, storage, DNA extraction, library preparation and sequencing.
Clinical and behavioural questionnaire data (pregnancy history, family and personal medical history, psychosocial health, demographic factors and other lifestyle and environmental exposures) were transferred to the Research Electronic Data Capture (REDCap) database protected by a secure server [35]. For the PTB group, eligible participants for this study were women who had undergone preterm delivery at greater than 20 weeks but less than 37 weeks gestational age, where onset of labour occurred spontaneously or in association with cervical incompetence or preterm premature rupture of membranes (PPROM). Vaginal swabs collected from pregnant women (both PTB and term groups) at 11–16 weeks of gestational age were used for bacterial genomic analysis.
Total nucleic acid was extracted from swabs using the MagMAX™ Total Nucleic Acid Isolation Kit (Life Technologies, Burlington, ON, Canada) as per manufacturer’s instructions. Kit reagents are aliquoted to eliminate repeated accessing of open reagents, and samples are processed in small batches using filter tips to prevent cross-contamination. Pipettes and other lab surfaces are regularly treated with DNA surface decontaminant (DNA Away, Thermo Fisher Scientific, Waltham, MA). Samples from both cohorts were processed in exactly the same way in terms of swab type, storage temperature (no stabilizer was used), DNA extraction, library preparation and sequencing.
Total bacterial DNA (qPCR) and detection of Mollicutes (PCR)
Quantitative PCR (qPCR)
Total bacterial DNA quantity in each sample was estimated using a SYBR Green assay based on amplification of the V3 region of the 16S rRNA gene. Primer sequences were as follows: SRV3-1 (5′-CGGYCCAGACTCCTAC-3′), SRV3-2 (5′-TTACCGCGGCTGCTGGCAC-3′) [36]. Reactions were run on a MyiQ thermocycler using the following cycling parameters: 95 °C for 3 min, followed by 30 cycles of 95 °C for 15 s, 62 °C for 15 s and 72 °C for 15 s, with a final extension at 72 °C for 5 min [37].
Conventional PCR
Some Mollicutes (Mycoplasma and Ureaplasma) species lack a cpn60 gene [38]. Thus, we performed a family-specific semi-nested PCR targeting the 16S rRNA gene to detect Mollicutes [39], and a PCR targeting the multiple-banded antigen gene to detect Ureaplasma spp. PCR products from U. parvum and U. urealyticum can be differentiated by size [40].
cpn60 universal target (UT) PCR and pyrosequencing
Universal primer PCR targeting the 549–567 bp cpn60 UT region was performed using a mixture of cpn60 primers consisting of a 1:3 M ratio of primers H279/H280:H1612/H1613, as described previously [41–43]. To allow multiplexing of samples in a single sequencing run, primers were modified at the 5′ end with one of 24 unique decamer multiplexing identification (MID) sequences, as per the manufacturer’s recommendations (Roche, Brandford, CT, USA). Amplicons were pooled in equimolar amounts for sequencing on the Roche GS Junior sequencing platform. The sequencing libraries were prepared using the GS DNA library preparation kit, and emulsion PCR (emPCR) was performed with a GS emPCR kit (Roche Diagnostics, Laval, Canada).
Samples were handled in small batches to avoid cross-contamination, and experimental controls were included at several steps in the study. Regular monitoring of DNA extraction controls in our lab by universal PCR confirms that these procedures are sufficient to eliminate detectable template contamination of study samples. A no template control was also included in each set of PCR reaction as negative controls. Experimental controls were not sequenced as they did not yield any amplification.
Analysis of operational taxonomic units (OTU)
Raw sequence data was processed by using the default on-rig procedures from 454/Roche. Filter-passing reads were used in the subsequent analyses for each of the pyrosequencing libraries. MID-partitioned sequences were mapped using Bowtie 2 (http://bowtie-bio.sourceforge.net/bowtie2/) on to a manually curated reference set of 1561 OTU sequences representing the human vaginal microbiota. Bowtie 2 was run using the default end-to-end alignment mode.
The OTU reference set was generated originally by de novo assembly of cpn60 sequence reads from 546 vaginal microbiomes using the microbial Profiling Using Metagenomic Assembly pipeline (mPUMA, http://mpuma.sourceforge.net) [44] with Trinity as the assembly tool [45] (Additional file 1). OTU were labeled according to their nearest reference sequence determined by watered-Blast comparison [46] to the cpn60 reference database, cpnDB_nr (downloaded from http://www.cpndb.ca [38]). This reference assembly approach allows us to compare the microbial profiles from various cohorts under investigation, including the 46 pregnant women who had sPTB described in this study.
The result of mapping is an OTU frequency table (Additional file 2) that was used for microbiome data analysis. Some analyses were also performed at species level, i.e. combined OTU that have the same nearest neighbour.
Statistical analysis
Comparisons of socio-demographic characteristics of cohorts and participants were based on analysis of variance (ANOVA), t test and chi-square, performed in IBM SPSS (Statistical Package for the Social Sciences, version 21) at 5% level of significance. For analysis of associations between socio-demographic characteristics and microbiota profiles (CST), a false discovery rate (FDR) correction for multiple comparisons was applied [47].
Alpha (Shannon diversity and Chao1 estimated species richness) and beta diversity (jackknifed Bray–Curtis dissimilarity matrices) were calculated as the mean of 100 subsamplings of 1000 reads (or all reads available when less than 1000) in QIIME (Quantitative Insights Into Microbial Ecology) [48]. Plots of alpha diversity measures against bootstrap sample number were generated in R and visually inspected to ensure that an adequate sampling depth for each sample was achieved.
For community state type (CST) analysis, a Jensen–Shannon distance matrix was calculated using the ‘vegdist’ function in the vegan package [49] with a custom distance function that calculates the square root of the Jensen–Shannon divergence [50]. This distance matrix was used for hierarchical clustering using the ‘hclust’ function in R, with Ward linkage.
The function aldex.clr from the ALDEx2 package in R was used to compare the differential relative abundance of individual taxa in term and preterm groups [51]. Significant differences were determined based on the false discovery rate (FDR), which is the result of a Benjamini–Hochberg corrected p value from a Welch’s t test calculated within ALDEx2.
Results
Description of the study population and pregnancy outcomes
Socio-demographic characteristics of women who had spontaneous preterm birth (n = 46) and women who had term deliveries (n = 170) are summarized in Table 1. There were no significant differences in maternal age, BMI, ethnicity, smoking status, consumption of alcohol or use of probiotics between term and preterm groups (all p > 0.05). Average maternal age was 33 for participants in both cohorts. Average body mass index (BMI) was 22.9 and 24.2 for women in the term and preterm groups, respectively. Most women in both cohorts identified themselves as white ethnicity, followed by East Asian and South/Southeast Asian (Table 1). Consumption of tobacco (term 2.3%; preterm 0%), alcohol (term 5.9%; preterm 4.3%) or probiotic supplements (term 4.1%; preterm 6.5%) was low among women in both groups (chi-square, all p > 0.05).
Table 1.
Socio-demographic and microbiological characteristics of subjects
| Characteristics | Term pregnancies (n = 170) | Preterm pregnancies (n = 46) | p value |
|---|---|---|---|
| Age (mean ± SD, range)1 | 33.6 ± 4.2 (21–45) | 33.65 ± 4.1 (25–45) | 0.948 |
| 21–25 | 5 (2.9%) | 1 (2.1%) | |
| 26–35 | 114 (67.1%) | 32 (69.5%) | |
| 36–45 | 51 (30.0%) | 13 (28.2%) | |
| BMI (mean ± SD, range)1 | 22.9 ± 3.8 (17–40) | 24.2 ± 5.6 (19–43) | 0.125 |
| Underweight (< 18.50) | 7 (4.1%) | 0 (0%) | |
| Normal weight (18.51–24.9) | 131 (77.0%) | 33 (73.3%) | |
| Overweight (25.0–29.9) | 25 (14.7%) | 8 (17.7%) | |
| Obese (> 30) | 7 (4.1%) | 4 (9.0%) | |
| MD3 | 0 (0%) | 1 (2.2%) | |
| Ethnicity2 | 0.261 | ||
| White | 108 (63.5%) | 22 (47.8%) | |
| East Asian | 26 (15.3%) | 6 (13.0%) | |
| South/Southeast Asian | 15 (8.8%) | 4 (8.7%) | |
| Latin America/Hispanic | 8 (4.7%) | 3 (6.5%) | |
| Black | 3 (1.8%) | 2 (4.4%) | |
| Other/mixed ethnicity | 10 (5.9%) | 6 (13.0%) | |
| MD | 0 (0%) | 3 (6.5%) | |
| Community state type (CST)2 | 0.361 | ||
| I | 56 (32.9%) | 17 (37%) | |
| II | 9 (5.3%) | 5 (10.9%) | |
| III | 28 (16.5%) | 8 (17.4%) | |
| IVA | 31 (18.2%) | 6 (13%) | |
| IVC | 19 (11.2%) | 2 (4.3%) | |
| IVD | 11 (6.5%) | 1 (2.2%) | |
| V | 16 (9.4%) | 7 (15.2%) | |
| Estimated bacterial load (log copies of 16S rRNA gene)/swab (mean ± SD, range)1 | 7.78 ± 0.93 (4.89–10.67) | 8.07 ± 0.71 (6.32–10.33) | 0.049 |
| Presence of Mollicutes2 | 68 (40%) | 28 (60.8%) | 0.012 |
| Presence of Ureaplasma2 | 40 (23.4%) | 14 (30.4%) | 0.337 |
| U. parvum | 37 (21.7%) | 14 (30.4%) | |
| U. urealyticum | 3 (1.7%) | 0 (0%) | |
| Shannon diversity (mean ± SD, range)1 | 1.28 ± 0.86 (0.13–4.52) | 1.81 ± 1.13 (0.34–5.16) | 0.004 |
| Chao1 richness (mean ± SD, range)1 | 36.22 ± 14.80 (14.39–115.74) | 46.38 ± 24.19 (20.20–126.01) | 0.009 |
1t test; 2chi-square; 3MD = missing data
Most women in the preterm group had a Bachelor/graduate degree (29/46) and an average house income higher than CAD 100,000 per year (25/46). A minority of women who had preterm birth (5/46) reported consumption of substances without prescription prior pregnancy, of which 3/46 women consumed marijuana/hashish, 1/46 woman consumed tranquilizers/nerve pills and 1/46 woman consumed cocaine/crack. Approximately 74% of the participants in the preterm group reported a pre-existing condition. A total of 12/46 women had some condition related to mental health, such depression or anxiety. Seventeen percent (8/46) had a neurological condition, including migraine headaches, and 24% (11/46) had a genitourinary condition (corpus luteal cyst, bicornuate uterus, cervical polyp, cervical dysplasia (2), uterine polyp, ovarian cyst, polycystic ovarian syndrome, urinary tract infections with and without kidney stones (3).
Characteristics regarding pregnancy and neonatal outcomes are described in Table 2. Pregnancy outcome information was not available for one woman in the preterm group as she was lost to follow-up. There were no significant differences in gestational age at enrolment, mode of conception or fetal sex between groups (all p > 0.05). Average gestational age at delivery was 39+3 weeks for the women who delivered at term and 34+2 weeks for women who had preterm birth, most of which were considered late preterm, i.e. delivery between 32 and 36+6 weeks of gestational age. Women in the preterm group were more likely to have experienced preterm birth or miscarriage in their previous pregnancy (chi-square, p < 0.001). They also had higher percentage of caesarean sections than women who delivered at term. Number of previous gestations also differed between groups; women who had preterm birth were more likely to be primigravida (22/46) in comparison with women who had term deliveries (45/170). There was a significant difference between term and preterm groups regarding birth weight and number of infants admitted to level 3 neonatal intensive care unit (NICU) (Table 2). Apgar score at 1 (term 8.75 ± 0.6; preterm 8.38 ± 1.1) and 5 min (term 8.97 ± 0.17; preterm 8.76 ± 0.7) between groups also differed (t test, all p < 0.001). One preterm infant (1/46) died shortly after birth (20 weeks of gestational age).
Table 2.
Gestation characteristics, pregnancy and neonatal outcomes
| Characteristics | Term pregnancies (n = 170) | Preterm pregnancies (n = 46) | p value |
|---|---|---|---|
| Gestational age in weeks+day | |||
| At enrolment (mean ± SD, range)1 | 13+ 2 ± 1+1 (11+1–16+6) | 13+3 ± 1+0 (11+6–16+0) | 0.641 |
| At delivery (mean ± SD, range) 1 | 39+3 ± 0+6 (39+3–41+2) | 34+2 ± 2+6 (20+0–36+6) | < 0.0001 |
| Late preterm (32 to 36+6) | NA | 39 (84.8%) | |
| Very preterm (28 to 31+6) | NA | 5 (10.8%) | |
| Extremely early (≤ 27+6) | NA | 1 (2.1%) | |
| Previous pregnancy history (excludes women in first pregnancy)2 | (n = 125) | (n = 24) | < 0.0001 |
| Livebirth, term | 89 (71.2%) | 9 (37.5%) | |
| Livebirth, preterm | 0 (0%) | 3 (12.5%) | < 0.0001 |
| Spontaneous abortion | 20 (16%) | 8 (33.3%) | 0.046 |
| Pregnancy termination | 16 (12.8%) | 3 (12.5%) | 0.968 |
| Ectopic pregnancy | 0 (0%) | 1 (4.1%) | |
| Mode of delivery2 | 0.042 | ||
| Vaginal delivery | 128 (75.3%) | 27 (58.7%) | |
| C-section | 42 (24.7%) | 18 (39.1%) | |
| Parity2 | 0.005 | ||
| 0 | 55 (32.3%) | 28 (60.8%) | |
| 1 | 94 (55.3%) | 12 (26%) | |
| 2–4 | 21 (12.3%) | 6 (13%) | |
| Assisted conception2 | 17 (10%) | 6 (13%) | 0.591 |
| Fetal sex (% male/% female) 2 | (48.8%)/(51.1%) | (41.3%)/(56.5%) | 0.430 |
| Birth weight (g) (mean ± SD, range)1 | 3398 ± 459 (1970–5200) | 2550 ± 559 (1300–3595) | < 0.0001 |
| Infant in NICU (n, %)2 | 1 (0.6%) | 24 (52.2%) | < 0.0001 |
1t test; 2chi-square
Among women who delivered preterm, 63% (29/46) had premature rupture of membranes (PPROM), 10.8% (5/46) had gestational diabetes and 4.3% (2/46) had anemia unresponsive to therapy. Twenty-four percent of women (11/46) presented one of the following conditions: maternal elevated liver enzymes, short cervix and incompetent cervix; fetal ascites, fetal distress and large foetus for gestational age; and placental findings of marginal cord insertion, two-vessel umbilical cord, placenta previa and low-lying placenta.
Sequencing results and OTU analysis
Raw sequence data files for the samples described in this study were deposited to the NCBI Sequence Read Archive (Accession SRP073152, BioProject PRJNA317763; BioProject PRJNA403856). Total dataset contained 1,635,072 cpn60 reads; median and average read count per sample were 4936 and 7569 (range 402–37,378), respectively. Average read length was 424 bp. Results of Bowtie2 mapping showed that these reads corresponded to 728 OTUs from the reference assembly (Additional file 1).
Microbiota profiles
Microbiota profiles were created by PCR amplification and pyrosequencing of the cpn60 universal target region. Hierarchical clustering of vaginal microbiota profiles resulted in seven community state types (CST): I (Lactobacillus crispatus dominated), II (Lactobacillus gasseri dominated), III (Lactobacillus iners dominated), IVA (Gardnerella vaginalis subgroup B or mix of different species), IVC (G. vaginalis subgroup A dominated), IVD (G. vaginalis subgroup C dominated) and V (Lactobacillus jensenii dominated) (Fig. 1). Each CST is defined by the dominance of one species of Lactobacillus (I, II, III, V), Gardnerella vaginalis (IVC, IVD) or a mixture of bacteria species (IVA), as previously described [52, 53].
Fig. 1.
Vaginal microbiota profiles of women who had sPTB and term deliveries. a Hierarchical clustering of Jensen–Shannon distance matrices with Ward linkage on the relative proportions of reads for each OTU within individual vaginal samples. b Community state type (CST). c Gestational age at delivery. d Heatmap of relative abundances of bacterial species within each vaginal microbiota. Each column represents a woman’s vaginal microbiota profile, and each row represents a bacteria species. Only species that are at least 1% abundant in at least one sample are shown. e Shannon diversity indices calculated for each sample
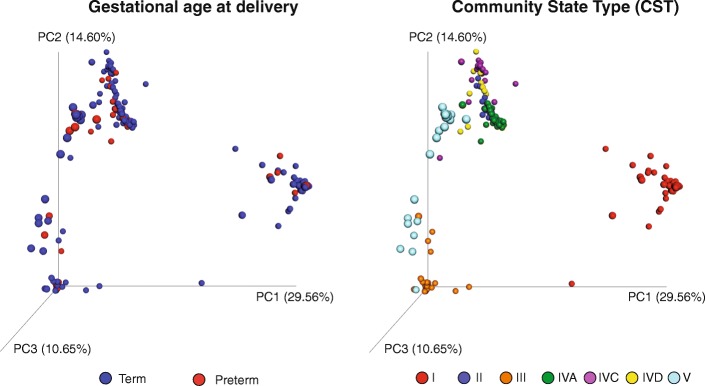
Overall microbiota profiles did not cluster together based on gestational age at delivery (Figs. 1 and 2). Most microbial profiles from the preterm group (80.5%) were assigned to Latobacillus-dominated CST: CST I (37% of profiles), CST III (17.4%), CST V (15.2%) and CST II (10.9%). The remaining profiles (19.5%) were assigned to CST IVA, IVC or IVD (Table 1). The CST IVA was the most heterogeneous group, represented by the dominance of Lactobacillus delbrueckii, Bifidobacterium dentium, Bifidobacterium infantis, Atopobium vaginae, Bifidobacterium breve or a mixture of different bacteria species. The CST IVC was dominated by G. vaginalis subgroup A and Megasphaera spp., and CST IVD was dominated by G. vaginalis subgroup C (Fig. 1).
Fig. 2.
Vaginal microbiota profiles coloured by gestational age at delivery or CST. Jackknifed principal coordinates analysis (PCoA) of Bray–Curtis distance matrices of microbial profiles from all participants in the study
Ecological analysis and total bacterial load
Assessment of alpha diversity revealed that microbiomes of women who delivered preterm were richer (Chao1 richness 46.3 ± 24.1) and more diverse (Shannon diversity index 1.8 ± 1.1) when compared to those of women in the term group (36.2 ± 14.8; 1.2 ± 0.8) (t test, p < 0.01) (Table 1). Total bacterial load was estimated based on qPCR targeting the 16S rRNA gene, and it was expressed as log 16S rRNA gene copy number per swab. Higher bacterial loads were detected in samples from the preterm group (7.7 ± 0.9) compared to term group (8.0 ± 0.7) (t test, p = 0.049) (Table 1).
Bacteria species relative abundance and prevalence
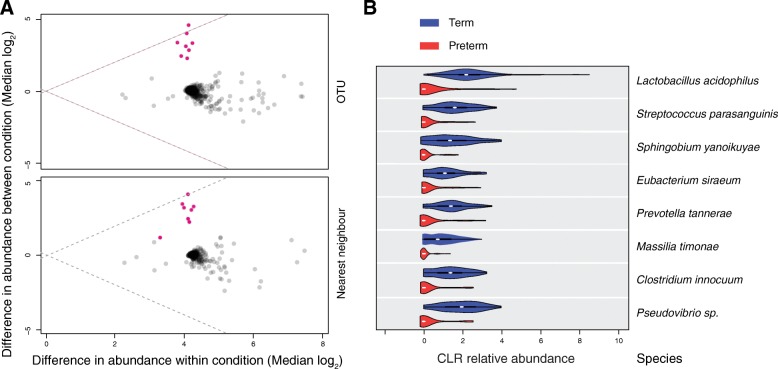
To investigate whether there was an association between individual taxa and sPTB, the abundance and prevalence of each species was evaluated. The ALDEx2 analysis assessed the relative abundance of each taxa (at the OTU and species level) in term and preterm groups. Eight OTU/species were more abundant in the term group in comparison with preterm, all of which were considered rare members of the bacterial community (Fig. 3). L. acidophilus represented 1% of the total reads in the dataset and had a low relative abundance average of 1.98% (range 0–69%) and 0.18% (range 0–0.87%) in samples from term and preterm groups respectively. All the other seven bacteria together represented only 0.4% of the total reads in the dataset.
Fig. 3.
Bacteria relative abundance differences between term and preterm groups represented by ALDEx2. a ALDEx2 between- and within-difference values for individual organisms across gestational age category. Organisms (at OTU and nearest neighbour species level) with significant p values are shown as pink circles (Welch’s t statistical test). b Violin plots showing the bacteria relative abundance (centre log transformed, CLR) in term and preterm groups. Only the eight bacteria with significant relative abundance differences between term and preterm groups are shown. In the violin plots, the white dot represents the median value, the black bar is the interquartile range, and the vertical width of the plot shows the density of the data along the X-axis
Bacteria prevalence (presence/absence) was also assessed (only species with at least 10 total reads were included). A total of 60 taxa had significant differences in prevalence between term and preterm groups; 11 species had greater prevalence in the term cohort and 49 species were more prevalent in the preterm cohort (Table 3). Bifidobacterium infantis, for example, was two times more prevalent in the term group in comparison with preterm, and Prevotella timonensis was 1.58 times more prevalent in the preterm group (Table 3). Several Prevotella spp. were associated with both term and preterm. Prevotella amnii and P. tannerae had greater prevalence in the term cohort, whereas P. timonensis, P. bivia, P. corporis and P. bucalis were more prevalent in the preterm group (Table 3). It is important to note that read depth distribution did not differ between term and preterm cohorts (t test, p > 0.05); therefore, the differences observed here in bacteria prevalence were unlikely to be driven by cohort sequencing bias.
Table 3.
Bacteria prevalence in the vaginal microbiomes of women who delivered preterm and at term
| Species | Total reads | Preterm (n = 46) | Term (n = 170) | Prevalence ratio | FDR* | ||
|---|---|---|---|---|---|---|---|
| Reads | Prevalence (%) | Reads | Prevalence (%) | ||||
| A | term/preterm | ||||||
| Bifidobacterium infantis | 56,246 | 409 | 19.57 | 55,837 | 40.00 | 2.04 | 0.033 |
| Lactobacillus delbrueckii subsp. lactis | 48,063 | 9 | 4.35 | 48,054 | 28.82 | 6.63 | 0.005 |
| Lactobacillus acidophilus | 17,440 | 569 | 89.13 | 16,871 | 97.65 | 1.10 | 0.033 |
| Prevotella amnii | 13,339 | 68 | 4.35 | 13,271 | 21.76 | 5.01 | 0.023 |
| Pseudovibrio sp. | 1634 | 52 | 15.22 | 1582 | 91.76 | 6.03 | 0.000 |
| Streptococcus parasanguinis | 1199 | 63 | 19.57 | 1136 | 88.82 | 4.54 | 0.000 |
| Sphingobium yanoikuyae | 1174 | 16 | 13.04 | 1158 | 80.00 | 6.13 | 0.000 |
| Prevotella tannerae | 930 | 58 | 15.22 | 872 | 88.24 | 5.80 | 0.000 |
| Clostridium innocuum | 814 | 42 | 13.04 | 772 | 81.18 | 6.22 | 0.000 |
| Eubacterium siraeum | 641 | 43 | 17.39 | 598 | 78.24 | 4.50 | 0.000 |
| Massilia timonae | 426 | 6 | 8.70 | 420 | 70.00 | 8.05 | 0.000 |
| B | preterm/term | ||||||
| Prevotella timonensis | 11,450 | 4211 | 71.74 | 7239 | 45.29 | 1.58 | 0.005 |
| Dialister micraerophilus | 7381 | 2850 | 67.39 | 4531 | 44.12 | 1.53 | 0.021 |
| Prevotella sp. | 2216 | 626 | 47.83 | 1590 | 14.71 | 3.25 | 0.000 |
| Bacteroides coagulans | 1283 | 1212 | 41.30 | 71 | 7.06 | 5.85 | 0.000 |
| Corynebacterium accolens | 1083 | 999 | 52.17 | 84 | 13.53 | 3.86 | 0.000 |
| Porphyromonas uenonis | 1040 | 795 | 28.26 | 245 | 12.94 | 2.18 | 0.038 |
| Actinomyces neuii subsp. anitratus | 877 | 738 | 43.48 | 139 | 10.00 | 4.35 | 0.000 |
| Peptoniphilus harei | 875 | 811 | 45.65 | 64 | 7.65 | 5.97 | 0.000 |
| Lactobacillus fermentum | 729 | 664 | 17.39 | 65 | 5.29 | 3.29 | 0.026 |
| Facklamia hominis | 711 | 709 | 15.22 | 2 | 1.18 | 12.93 | 0.000 |
| Prevotella bivia | 638 | 228 | 26.09 | 410 | 8.24 | 3.17 | 0.005 |
| Prevotella corporis | 535 | 463 | 23.91 | 72 | 5.88 | 4.07 | 0.000 |
| Corynebacterium timonense | 503 | 355 | 34.78 | 148 | 10.00 | 3.48 | 0.000 |
| Corynebacterium genitalium | 372 | 323 | 34.78 | 49 | 7.06 | 4.93 | 0.000 |
| Tepidanaerobacter sp. | 329 | 303 | 23.91 | 26 | 4.12 | 5.81 | 0.000 |
| Corynebacterium amycolatum | 264 | 228 | 32.61 | 36 | 4.71 | 6.93 | 0.000 |
| Parvimonas micra | 193 | 183 | 10.87 | 10 | 1.18 | 9.24 | 0.005 |
| Mobiluncus curtsii subsp. curtsii | 183 | 171 | 21.74 | 12 | 2.35 | 9.24 | 0.000 |
| Finegoldia magna | 155 | 149 | 13.04 | 6 | 3.53 | 3.70 | 0.038 |
| Coprococcus eutactus | 154 | 68 | 47.83 | 86 | 27.06 | 1.77 | 0.026 |
| Brevibacterium linens | 147 | 142 | 10.87 | 5 | 0.59 | 18.48 | 0.000 |
| Rothia dentocariosa | 144 | 120 | 30.43 | 24 | 5.29 | 5.75 | 0.000 |
| Streptococcus thermophilus | 132 | 126 | 19.57 | 6 | 2.35 | 8.32 | 0.000 |
| Magnetospirillum magnetotacticum | 116 | 40 | 43.48 | 76 | 24.12 | 1.80 | 0.033 |
| Dethiobacter alkaliphilus | 101 | 97 | 10.87 | 4 | 1.76 | 6.16 | 0.017 |
| Brevibacterium massiliense | 99 | 86 | 15.22 | 13 | 1.18 | 12.93 | 0.000 |
| Eremococcus coleocola | 90 | 82 | 10.87 | 8 | 1.76 | 6.16 | 0.017 |
| Anaeromusa acidaminophila | 86 | 85 | 15.22 | 1 | 0.59 | 25.87 | 0.000 |
| Arthrobacter globiformis | 66 | 48 | 10.87 | 18 | 1.76 | 6.16 | 0.017 |
| Staphylococcus epidermidis | 54 | 47 | 13.04 | 7 | 2.35 | 5.54 | 0.009 |
| Corynebacterium simulans | 52 | 41 | 26.09 | 11 | 1.76 | 14.78 | 0.000 |
| Anaeroglobus geminatus | 52 | 49 | 15.22 | 3 | 1.18 | 12.93 | 0.000 |
| Cellvibrio gilvus | 42 | 26 | 15.22 | 16 | 1.76 | 8.62 | 0.000 |
| Prosthecochloris vibrioformis | 41 | 40 | 6.52 | 1 | 0.59 | 11.09 | 0.028 |
| Prevotella buccalis | 33 | 33 | 15.22 | 0 | 0.00 | – | 0.000 |
| Acidaminococcus fermentans | 31 | 29 | 13.04 | 2 | 1.18 | 11.09 | 0.000 |
| Brevibacterium casei | 25 | 24 | 8.70 | 1 | 0.59 | 14.78 | 0.005 |
| Streptococcus sanguinis | 23 | 18 | 13.04 | 5 | 2.35 | 5.54 | 0.009 |
| Atopobium parvulum | 23 | 20 | 8.70 | 3 | 1.18 | 7.39 | 0.023 |
| Peptoniphilus duerdenii | 22 | 20 | 10.87 | 2 | 0.59 | 18.48 | 0.000 |
| Pelobacter propionicus | 21 | 18 | 6.52 | 3 | 0.59 | 11.09 | 0.028 |
| Staphylococcus hominis | 21 | 17 | 15.22 | 4 | 1.76 | 8.62 | 0.000 |
| Atopostipes suicloacalis | 18 | 8 | 10.87 | 10 | 1.18 | 9.24 | 0.005 |
| Corynebacterium pyruviciproducens | 18 | 13 | 13.04 | 5 | 1.76 | 7.39 | 0.005 |
| Corynebacterium jeikeium | 14 | 11 | 13.04 | 3 | 0.59 | 22.17 | 0.000 |
| Sporichthya polymorpha | 13 | 11 | 8.70 | 2 | 1.18 | 7.39 | 0.023 |
| Rhodococcus jostii | 13 | 7 | 8.70 | 6 | 1.18 | 7.39 | 0.023 |
| Nitrospina gracilis | 10 | 5 | 8.70 | 5 | 0.59 | 14.78 | 0.005 |
| Megasphaera sp. BV3C16-1 | 10 | 9 | 6.52 | 1 | 0.59 | 11.09 | 0.028 |
Panel A: species with greater prevalence in the term group (ratio term/preterm)
Panel B: species with greater prevalence in the preterm group (ratio preterm/term)
*FDR (false discovery rate) represents the corrected p value for multiple comparisons
Mollicutes (Mycoplasma and/or Ureaplasma) were detected by family-specific conventional PCR in 28/46 (60%) of pregnant women who delivered preterm (Table 1). Ureaplasma species were detected by genus-specific PCR in samples of 14/46 (30%) women who had PTB, with all women testing positive for U. parvum and none for U. urealyticum. Women who delivered at term were less likely to be PCR positive for Mollicutes compared to women who had PTB (Table 1). No significant differences were observed in Ureaplasma prevalence between the two groups (Table 1). An association between Mollicutes/Ureaplasma detection and the composition of the vaginal microbiota, represented as CST, was also investigated. Detection of Mollicutes and Ureaplasma was not associated with any CST in particular when investigated in the term cohort, preterm cohort or both groups together (chi-square, p > 0.05).
Relationships between microbiological and socio-demographic characteristics within the preterm group
The association between CST (I, II, III, IVC, IVD, V) from profiles of women who delivered preterm and several microbiologic-socio-demographic characteristics was investigated. Only two associations were significant: CST and microbiota richness, and CST and microbiota diversity (ANOVA, p < 0.001). There was no significant association between CST and the remaining following metadata: microbiota richness and diversity (continuous variable), presence of Mollicutes and Ureaplasma (yes/no), log 16S rRNA gene copies (continuous variable), maternal age (continuous variable; 18–25, 26–35, 36–45), BMI category (underweight, normal, overweight, obese; < 25, ≥ 25), ethnicity (White, East Asian, South Asian, Black, Hispanic, Other), natural conception (yes/no), parity (0, > 1), gestational age (continuous variable), mode of delivery (vaginal, C-section), pre-existing condition (yes/no), folic acid intake before or during pregnancy (yes/no), drinking alcohol (yes/no), neonate in high level care (yes/no), birth weight (continuous variable) and Apgar score at 1 and 5 min (1–9).
Discussion
In this study, we determined the composition of the vaginal microbiota of women who had spontaneous preterm birth and compared these profiles to those of women who delivered at term, previously reported by our research group [24]. The availability of foundational data on women who delivered at term and the infeasibility of collecting large numbers of samples at 11–16 weeks gestation from women who would go on to deliver pre-term, our study design included comparison of samples collected in a previously published study [24] and from the OBS. To minimize any batch effects, we were rigorous in implementation of consistent sample processing and did extensive analysis of the clinical and demographic characteristics to ensure they were well matched (Table 1). The cohorts were comparable in terms of maternal age, BMI, ethnicity, consumption of tobacco, alcohol and probiotics, which is of interest given that several of these characteristics have been previously associated with preterm delivery. In particular, previous described factors included low and high maternal ages [5–7], low BMI [8], black ethnicity [9], high levels of tobacco, alcohol and illicit drugs consumption [4], close temporal proximity to a previous delivery [10] and multiple gestation [11]. This cohort is unique in that it did offer the opportunity to have gestational age at delivery as the main characteristic distinguishing these two groups recognizing that the majority of preterm births occurred beyond 32 weeks gestation.
A difference in number of previous gestations was observed between groups, with women who experienced preterm birth more likely to be primigravida in comparison with women who had term deliveries. It has been recently demonstrated that women with a prior conception, regardless of whether or not this proceeded to a birth, have a decrease in the relative abundance of L. crispatus and a concomitant increase in the abundance of other Lactobacillus species as well as Gardnerella [54].
Other known risk factors for sPTB include maternal medical disorders like hypertension, asthma, diabetes and thyroid disease [4]. Although some women in both cohorts reported these conditions, there were not enough participants to stratify the data based on the individual disorder and therefore was not possible to investigate the interaction between those medical conditions and gestation outcome. We were, however, able to confirm previous reports of history of prematurity as a risk factor for preterm birth [55].
Since many organisms isolated from the amniotic cavity of women who experienced preterm birth are also found in the genital tract [12–15], an intrauterine infection ascending from the vagina is one of the currently hypothesized triggers of PTB [56]. In this study, however, we did not identify a signature microbiota composition (CST) associated with preterm birth. This observation is consistent with the results presented by others [29, 30]. CST assignments are largely driven by the dominance of a single species, which may mask differences in rare taxa that would differentiate term and preterm groups, and indeed, further analysis revealed that the vaginal microbiota of women who experienced preterm birth was richer and more diverse than those of women who delivered at term. Also, most women (84.8%) in our study were considered late preterm and although we cannot address this question, it is possible that sPTB driven by an ascending infection would be more evident in a high-risk cohort or extreme preterm cases. A recent study of a high-risk pregnant cohort has reported that L. iners was strongly associated with short cervix and preterm birth, as L. crispatus was associated with term deliveries [57]. Those differences in study outcomes indicate that the pathogenesis of sPTB in low- and high-risk groups might be different. Identifying differences in the causes of early and late sPTB and the role of the vaginal microbiota in those processes will require further study.
One controversy that challenges the current hypothesis of preterm caused by an ascending infection is that antibiotic administration to pregnant women with a disturbed vaginal microbiota does not improve outcome in most cases, as demonstrated by study trials [58, 59] and systematic reviews [60–62]. One explanation for the inefficacy of antibiotic treatment in the prevention of preterm birth relies is the high rates of antibiotic resistance among bacterial-vaginosis-associated bacteria [63, 64]. In this case, antibiotics not only do not kill the targeted bacteria, but might also reduce the vaginal Lactobacillus population leading to an even more disturbed microbiota, as recently demonstrated [65].
In addition to differences in richness and diversity, differences in the microbiota between the two cohorts regarding bacterial abundance and prevalence were also identified. The ALDEx2 analysis indicated that eight rare taxa were more abundant in the term group, which does not necessarily mean they are associated with a ‘healthier’ state or implicated in preventing sPTB. Since these bacteria are detected at very low abundance within the microbiota profiles, their biological significance in the vaginal microbiome is questionable. Differences in the prevalence of several other taxa between groups were also observed. For example, more women in the term group had Prevotella amnii and P. tannerae detected in their vaginal samples, whereas P. timonensis, P. bivia, P. corporis and P. bucalis were more frequently detected in samples from women in the preterm group (Table 3). Prevotella spp. have been previously associated with bacterial vaginosis and preterm labour [22, 66, 67], and our results indicate that different Prevotella species might have different roles in sPTB. Several of the taxa that were significantly different in their prevalence among women in the two groups also had low sequence read counts (Table 3). Further investigation would be required to determine if these rare members of the microbial community play a yet unknown role in sPTB.
It is also important to note that the number of bacterial species with greater prevalence in the preterm (49/60) was higher than in the term (11/60) cohort (Table 3), which is consistent with our results of increased microbial richness and diversity in the samples from women who experienced preterm birth. This might indicate that increased richness, rather than the presence of specific taxa, might be associated with sPTB. Those differences might also be an indicator of physiological/biochemical dissimilarities in the vaginal microbiomes of women who deliver at term or preterm. In other words, the physiological state that leads to sPTB might also create an environment that supports a richer/more diverse microbiota.
Our results also confirmed previous reports of an association between Mycoplasma and preterm birth [68]. Mollicutes were detected significantly more often in women in the preterm group compared to women in the term group, but no differences were observed in Ureaplasma prevalence between groups indicating that the difference in Mollicutes prevalence is primarily driven by the presence of Mycoplasma spp. Although individual Mycoplasma species could not be discerned based on assay used in our study, both Mycoplasma genitalium [69–71] and Mycoplasma hominis [72–75] have been previously associated with negative reproductive outcomes including PTB.
Collectively, our overall findings were similar to other two studies, which provided us the opportunity to compare different study designs (based on different cohorts and barcode gene) that addressed the same research question. Hyman and colleagues [30] described the vaginal microbiota of 83 pregnant women (term n = 66, preterm n = 17) based on Sanger sequencing of cloned 16S rRNA genes. Samples were collected at each trimester and preterm was defined as delivery before 37 weeks of gestation. There was no correlation between preterm and absence/low abundance of Lactobacillus in the microbiota; in other words, preterm outcome could not be predicted based on CST. Similar to our results, they found an association between increased microbiota diversity and preterm delivery among women of white ethnicity (n = 40) (data from women of others ethnicities was not included in the analysis because of small sample sizes). Although there was no association between CST and ethnicity, it is important to note that most women enrolled in this study described themselves as being white, and it is possible that an increased sample size of participants of other ethnicities could result in a different conclusion.
Romero and colleagues [29] also investigated the vaginal microbiota of pregnant women who experienced preterm, defined as delivery before 34 weeks of gestation (term n = 72, preterm n = 18). The profiles were created by 16S rRNA amplicon sequencing, and samples were collected every 4 weeks until 24 weeks of gestation and then every 2 weeks. They found no differences in the frequency of different CST between women who had term and preterm deliveries. Likewise, no differences in bacteria relative abundance were observed between the two cohorts, although only bacteria that were present in at least 25% of samples were included in the analysis. These results are consistent with our findings of bacterial abundance based on the ALDEx analysis since we only found significant differences in relative abundance for eight rare bacteria. Unlike Hyman et al. [30] and our results, Romero et al. [29] did not find differences in microbiota diversity between women who delivered preterm and at term. One possible explanation for this contradictory result might be related to differences in participant ethnicity among these studies. While most women in our study and the Hyman et al. study described themselves as white, the majority of participants in the Romero et al. study described themselves as African American. It has been reported that the composition of the vaginal microbiota is strongly associated with a woman’s ethnicity [52, 76]. Other studies have also demonstrated that black ethnicity is associated with an increased microbiota diversity in comparison with white ethnicity [77], which could have masked differences in bacterial diversity between term and preterm cohorts in the Romero study.
Contrary to our overall findings, DiGiulio and colleagues [31] found a strong association between the non-Lactobacillus-dominated CST IV and preterm birth in a case–control study based on the 16S rRNA amplicon sequencing. Pregnant women (preterm n = 34, term n = 15), mostly of white ethnicity, were sampled weekly throughout gestation. Interestingly, the authors pointed out that if samples had been collected less frequently, short-term ‘excursions’ to CST IV would have been missed and probably the association between CST IV and preterm birth would have been less obvious. The detection of a temporary microbiota disturbance represented by a change from a Lactobacillus-dominated CST to CST IV may have been missed in our study since samples were not collected longitudinally. Moreover, a recent study has demonstrated that PTB–microbiota associations are population-dependent [32]; lower Lactobacillus and higher Gardnerella abundances were associated with PTB in a low-risk predominantly Caucasian cohort, but not in a high-risk predominantly African American cohort. These population-dependent associations might contribute to explain contradictory conclusions among different studies and emphasize the importance of investigating the vaginal microbiota of different populations with varying ethnic backgrounds and from different geographical locations.
Most samples in the preterm group were dominated by Lactobacillus, yet, they collectively had higher richness and diversity compared to samples from the term group. The increased microbiota richness/diversity might indicate a transient state between Lactobacillus-dominated CST and non-Lactobacillus-dominated, i.e., CST IV (A, C or D). In other words, the increased richness and diversity we observed might be a remnant characteristic of the previous disturbed microbiota. In summary, although we did not “detect” a specific microbial community structure that is associated with preterm birth, the increased microbiota richness/diversity was associated with preterm birth. In addition, the association with differences in Prevotella species and Mycoplasma presence may point to signature species associated with preterm birth.
Conclusions
Taken together, our results suggest that the differences in the microbiota of women who had preterm deliveries, such as increased microbiota richness and diversity and greater prevalence of Mollicutes and other bacteria, may have a role in sPTB. Other differences between cohorts might have been masked by the presence of highly dominant bacteria like Lactobacillus. At the overall level, we did not identify a specific vaginal microbial community structure at 11–16 weeks gestation age that predicts sPTB. Also, differences in relative abundance of bacterial species between term and preterm groups were only significant for a few low abundance species. Although a causal relationship remains to be determined, our results confirm previous reports of an association between Mollicutes and preterm birth, and further suggest that a diverse bacterial community may contribute to the microbiome’s role in sPTB. Alternatively, the more rich and diverse microbiotas of the preterm group may reflect physiological differences between the groups that affect selection of bacteria. This study provides valuable evidence of subtle alterations in the microbiome associated with preterm birth that requires further study utilizing sequencing methodology. In addition, future study should include evaluation of the microbial metabolite production and host response to further elucidate factors leading to sPTB and identify women at risk early in pregnancy.
Additional files
cpn60 OTU sequences. Multiple fasta file containing 728 OTU sequences. (TXT 336 kb)
Summary of OTU analysed in this study. OTU ID, percentage of identity, length, cpnDB name, species, and abundance in each library are shown. (XLSX 500 kb)
Acknowledgements
The authors are grateful to the women who participated in the study and acknowledge the contribution and support of Ontario Birth Study Team members and the participants. The VOGUE Research Group is Deborah Money, Alan Bocking, Sean Hemmingsen, Janet Hill, Gregor Reid, Tim Dumonceaux, Gregory Gloor, Matthew Links, Kieran O’Doherty, Patrick Tang, Julianne van Schalkwyk and Mark Yudin.
Funding
Financial support was provided by a joint Canadian Institutes of Health Research (CIHR) Emerging Team Grant and a Genome British Columbia (GBC) grant awarded to DMM, AB and JEH (grant reference #108030) as well as a CIHR grant MOP-82799 to AB. ACF was supported by a University of Saskatchewan graduate scholarship. Funding for the Ontario Birth Study was provided by Mount Sinai Hospital Foundation, Lunenfeld-Tanenbaum Research Institute and the Department of Obstetrics and Gynecology at Mount Sinai Hospital.
Availability of data and materials
The dataset supporting the results of this article is available in the NCBI SRA repository (Accession SRP073152, BioProject PRJNA317763; BioProject PRJNA403856).
Authors’ contributions
DM, AB, JEH and the other members of the VOGUE Research Group conceived the study. DM, AB and JEH oversaw and contributed to data collection and participated in writing and manuscript review. ACF generated the Mollicutes PCR, 16S rRNA qPCR and the cpn60 microbiome profiles data, performed data analysis and wrote the paper. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study received ethical approval from the University of British Columbia Children’s and Women’s Research Ethics Board (Approval Number H14-01954), and Mount Sinai Hospital Research Ethics Board (Approval Number 15-0184-E). All participants provided written consent at enrolment.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s40168-018-0502-8) contains supplementary material, which is available to authorized users.
Contributor Information
Aline C. Freitas, Email: aline.freitas@usask.ca
Alan Bocking, Email: abocking@mtsinai.on.ca.
Janet E. Hill, Email: janet.hill@usask.ca
Deborah M. Money, Email: deborah.money@ubc.ca
the VOGUE Research Group:
Deborah Money, Alan Bocking, Sean Hemmingsen, Janet Hill, Gregor Reid, Tim Dumonceaux, Gregory Gloor, Matthew Links, Kieran O’Doherty, Patrick Tang, Julianne van Schalkwyk, and Mark Yudin
References
- 1.WHO. March of Dimes. PMNCH. Save the Children . In: Born too soon: the global action report on preterm birth. Howson CP, Kinney MV, Lawn JE, editors. Geneva: World Health Organization; 2012. [Google Scholar]
- 2.Quinn J, Munoz FM, Gonik B, Frau L, Cutland C, Mallett-Moore T, et al. Preterm birth: case definition & guidelines for data collection, analysis, and presentation of immunisation safety data. Vaccine. 2016;34:6047–6056. doi: 10.1016/j.vaccine.2016.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blencowe H, Cousens S, Oestergaard MZ, Chou D, Moller AB, Narwal R, et al. National, regional, and worldwide estimates of preterm birth rates in the year 2010 with time trends since 1990 for selected countries: a systematic analysis and implications. Lancet. Elsevier Ltd. 2012;379:2162–2172. doi: 10.1016/S0140-6736(12)60820-4. [DOI] [PubMed] [Google Scholar]
- 4.Behrman RE, Butler AS. In: Preterm birth: causes, consequences, and prevention. Behrman RE, Butler AS, editors. Washington, D.C.: National Academies Press; 2007. [PubMed] [Google Scholar]
- 5.Fraser AM, Brockert JE, Ward RH. Association of young maternal age with adverse reproductive outcomes. N Engl J Med. 1995;332:1113–1118. doi: 10.1056/NEJM199504273321701. [DOI] [PubMed] [Google Scholar]
- 6.Jacobsson B, Ladfors L, Milsom I. Advanced maternal age and adverse perinatal outcome. Obstet Gynecol. 2004;104:727–733. doi: 10.1097/01.AOG.0000140682.63746.be. [DOI] [PubMed] [Google Scholar]
- 7.Schempf AH, Branum AM, Lukacs SL, Schoendorf KC. Maternal age and parity-associated risks of preterm birth: differences by race/ethnicity. Paediatr Perinat Epidemiol. 2007;21:34–43. doi: 10.1111/j.1365-3016.2007.00785.x. [DOI] [PubMed] [Google Scholar]
- 8.Han Z, Mulla S, Beyene J, Liao G, McDonald SD. Maternal underweight and the risk of preterm birth and low birth weight: a systematic review and meta-analyses. Int J Epidemiol. 2011;40:65–101. doi: 10.1093/ije/dyq195. [DOI] [PubMed] [Google Scholar]
- 9.Kistka ZAF, Palomar L, Lee KA, Boslaugh SE, Wangler MF, Cole FS, et al. Racial disparity in the frequency of recurrence of preterm birth. Am J Obstet Gynecol. 2007;196:1–6. doi: 10.1016/j.ajog.2006.06.093. [DOI] [PubMed] [Google Scholar]
- 10.Conde-Agudelo A, Rosas-Bermúdez A, Kafury-Goeta AC. Birth spacing and risk of adverse perinatal outcomes. JAMA. 2006;295:1809–1823. doi: 10.1001/jama.295.15.1809. [DOI] [PubMed] [Google Scholar]
- 11.Stock S, Norman J. Preterm and term labour in multiple pregnancies. Semin Fetal Neonatal Med Elsevier Ltd. 2010;15:336–341. doi: 10.1016/j.siny.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 12.Gardella C, Riley DE, Hitti J, Agnew K, Krieger JN, Eschenbach DA. Identification and sequencing of bacterial rDNAs in culture-negative amniotic fluid from women in premature labor. Am J Perinatol. 2004;21:319–323. doi: 10.1055/s-2004-831884. [DOI] [PubMed] [Google Scholar]
- 13.Krohn MA, Hillier SL, Nugent RP, Cotch MF, Carey JC, Gibbs RS, et al. The genital flora of women with intraamniotic infection. J Infect Dis. 1995;171:1475–1480. doi: 10.1093/infdis/171.6.1475. [DOI] [PubMed] [Google Scholar]
- 14.Hillier SL, Martius J, Krohn M, Kiviat N, Holmes KK, Eschenbach DA. A case–control study of chorioamnionic infection and histologic chorioamnionitis in prematurity. N Engl J Med. 1988;319:972–978. doi: 10.1056/NEJM198810133191503. [DOI] [PubMed] [Google Scholar]
- 15.Romero R, Sirtori M, Oyarzun E, Avila C, Mazor M, Callahan R, et al. Infection and labor V. Prevalence, microbiology, and clinical significance of intraamniotic infection in women with preterm labor and intact membranes. Am J Obstet Gynecol. 1989;161:817–824. doi: 10.1016/0002-9378(89)90409-2. [DOI] [PubMed] [Google Scholar]
- 16.Watts HD, Krohn MA, Hillier SL, Eschenbach DA. The association of occult amniotic fluid infection with gestational age and neonatal outcome among women in preterm labor. Obstet Gynecol. 1992;79:351–357. doi: 10.1097/00006250-199203000-00005. [DOI] [PubMed] [Google Scholar]
- 17.Yoon BH, Romero R, Bin MJ, Shim S-S, Kim M, Kim G, et al. Clinical significance of intra-amniotic inflammation in patients with preterm labor and intact membranes. Am J Obstet Gynecol. 2001;185:1130–1136. doi: 10.1067/mob.2001.117680. [DOI] [PubMed] [Google Scholar]
- 18.DiGiulio DB, Romero R, Amogan HP, Kusanovic JP, Bik EM, Gotsch F, et al. Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: a molecular and culture-based investigation. PLoS One. 2008;3:e3056. doi: 10.1371/journal.pone.0003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hillier SL, Witkin SS, Krohn MA, Watts DH, Kiviat NB, Eschenbach DA. The relationship of amniotic fluid cytokines and preterm delivery, amniotic fluid infection, histologic chorioamnionitis, and chorioamnion infection. Obstet Gynecol. 1993;81:941–948. [PubMed] [Google Scholar]
- 20.Jacobsson B, Mattsby-Baltzer I, Hagberg H. Interleukin-6 and interleukin-8 in cervical and amniotic fluid: relationship to microbial invasion of the chorioamniotic membranes. BJOG An Int. J. Obstet. Gynaecol. 2005;112:719–724. doi: 10.1111/j.1471-0528.2005.00536.x. [DOI] [PubMed] [Google Scholar]
- 21.Nugent RP, Krohn MA, Hillier SL. Reability of diagnosing bacterial vaginosis is improved by standardized method of gram stain interpretation. J Clin Microbiol. 1991;29:297–301. doi: 10.1128/jcm.29.2.297-301.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hill GB. The microbiology of bacterial vaginosis. Am J Obstet Gynecol. 1993;169:450–454. doi: 10.1016/0002-9378(93)90339-k. [DOI] [PubMed] [Google Scholar]
- 23.Romero R, Hassan SS, Gajer P, Tarca AL, Fadrosh DW, Nikita L, et al. The composition and stability of the vaginal microbiota of normal pregnant women is different from that of non-pregnant women. Microbiome. 2014;2:1–19. doi: 10.1186/2049-2618-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freitas AC, Chaban B, Bocking A, Rocco M, Yang S, Hill JE, et al. The vaginal microbiome of pregnant women is less rich and diverse, with lower prevalence of Mollicutes, compared to non-pregnant women. Sci Rep. 2017;7:1–16. doi: 10.1038/s41598-017-07790-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aagaard KM, Riehle K, Ma J, Segata N, Mistretta TA, Coarfa C, et al. A metagenomic approach to characterization of the vaginal microbiome signature in pregnancy. PLoS One. 2012;7:e36466. doi: 10.1371/journal.pone.0036466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walther-António MRS, Jeraldo P, Berg Miller ME, Yeoman CJ, Nelson KE, Wilson BA, et al. Pregnancy’s stronghold on the vaginal microbiome. PLoS One. 2014;9:e98514. doi: 10.1371/journal.pone.0098514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacIntyre DA, Chandiramani M, Lee YS, Kindinger L, Smith A, Angelopoulos N, et al. The vaginal microbiome during pregnancy and the postpartum period in a European population. Sci Rep. 2015;5:1–9. doi: 10.1038/srep08988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leitich H, Bodner-Adler B, Brunbauer M, Kaider A, Egarter C, Husslein P. Bacterial vaginosis as a risk factor for preterm delivery: a meta-analysis. Am J Obstet Gynecol. 2003;189:139–147. doi: 10.1067/mob.2003.339. [DOI] [PubMed] [Google Scholar]
- 29.Romero R, Hassan SS, Gajer P, Tarca AL, Fadrosh DW, Bieda J, et al. The vaginal microbiota of pregnant women who subsequently have spontaneous preterm labor and delivery and those with a normal delivery at term. Microbiome. 2014;2:1–15. doi: 10.1186/2049-2618-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hyman RW, Fukushima M, Jiang H, Fung E, Rand L, Johnson B, et al. Diversity of the vaginal microbiome correlates with preterm birth. Reprod Sci. 2014;21:32–40. doi: 10.1177/1933719113488838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DiGiulio DB, Callahan BJ, Mcmurdie PJ, Costello EK, Lyell DJ, Robaczewska A, et al. Temporal and spatial variation of the human microbiota during pregnancy. Proc Natl Acad Sci U S A. 2015;112:11060–11065. doi: 10.1073/pnas.1502875112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Callahan BJ, DiGiulio DB, Goltsman DSA, Sun CL, Costello EK, Jeganathan P, et al. Replication and refinement of a vaginal microbial signature of preterm birth in two racially distinct cohorts of US women. Proc Natl Acad Sci U S A. 2017;114:9966–9971. doi: 10.1073/pnas.1705899114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Links MG, Dumonceaux TJ, Hemmingsen SM, Hill JE. The chaperonin-60 universal target is a barcode for bacteria that enables de novo assembly of metagenomic sequence data. PLoS One. 2012;7:e49755. doi: 10.1371/journal.pone.0049755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paramel Jayaprakash T, Schellenberg JJ, Hill JE. Resolution and characterization of distinct cpn60-based subgroups of Gardnerella vaginalis in the vaginal microbiota. PLoS One. 2012;7:e43009. doi: 10.1371/journal.pone.0043009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform Elsevier Inc. 2009;42:377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee DH, Zo YG, Kim SJ. Nonradioactive method to study genetic profiles of natural bacterial communities by PCR—single-strand-conformation polymorphism. Appl Environ Microbiol. 1996;62:3112–3120. doi: 10.1128/aem.62.9.3112-3120.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chaban B, Albert AYK, Links MG, Gardy J, Tang P, Hill JE. Characterization of the upper respiratory tract microbiomes of patients with pandemic H1N1 influenza. PLoS One. 2013;8:e69559. doi: 10.1371/journal.pone.0069559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hill JE, Penny SL, Crowell KG, Goh SH, Hemmingsen SM. cpnDB: a chaperonin sequence database. Genome Res. 2004;14:1669–1675. doi: 10.1101/gr.2649204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Kuppeveld FJM, van der Logt JTM, Angulo AF, van Zoest MJ, Quint WGV, Niesters HGM, et al. Genus- and species-specific identification of mycoplasmas by 16S rRNA amplification. Appl Environ Microbiol. 1992;58:2606–2615. doi: 10.1128/aem.58.8.2606-2615.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watson HL, Blalock DK, Cassell GH. Variable antigens of Ureaplasma urealyticum containing both serovar-specific and serovar-cross-reactive epitopes. Infect Immun. 1990;58:3679–3688. doi: 10.1128/iai.58.11.3679-3688.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chaban B, Links MG, Jayaprakash TP, Wagner EC, Bourque DK, Lohn Z, et al. Characterization of the vaginal microbiota of healthy Canadian women through the menstrual cycle. Microbiome. 2014;2:1–12. doi: 10.1186/2049-2618-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hill JE, Town JR, Hemmingsen SM. Improved template representation in cpn60 polymerase chain reaction (PCR) product libraries generated from complex templates by application of a specific mixture of PCR primers. Environ Microbiol. 2006;8:741–746. doi: 10.1111/j.1462-2920.2005.00944.x. [DOI] [PubMed] [Google Scholar]
- 43.Schellenberg JJ, Links MG, Hill JE, Dumonceaux TJ, Kimani J, Jaoko W, et al. Molecular definition of vaginal microbiota in East African commercial sex workers. Appl Environ Microbiol. 2011;77:4066–4074. doi: 10.1128/AEM.02943-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Links MG, Chaban B, Hemmingsen SM, Muirhead K, Hill JE. mPUMA: a computational approach to microbiota analysis by de novo assembly of operational taxonomic units based on protein-coding barcode sequences. Microbiome. 2013;1:1–7. doi: 10.1186/2049-2618-1-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schellenberg JJ, Links MG, Hill JE, Dumonceaux TJ, Peters GA, Tyler SD, et al. Pyrosequencing of the chaperonin-60 universal target as a tool for determining microbial community composition. Appl Environ Microbiol. 2009;75:2889–2898. doi: 10.1128/AEM.01640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc. 1995;57:289–300. [Google Scholar]
- 48.Caporaso JG, Kuczynski J, Stombaugh JI, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oksanen AJ, Blanchet FG, Kindt R, Legendre P, Minchin PR, Hara RBO, et al. Community Ecology Package “vegan”. R version 2.3-1. Available: https://cran.r-project.org/web/packages/vegan/. 2015.
- 50.Endres DM, Schindelin JE. A new metric for probability distributions. IEEE Trans Inf Theory. 2003;49:1858–1860. [Google Scholar]
- 51.Fernandes AD, Macklaim JM, Linn TG, Reid G, Gloor GB. ANOVA-like differential expression (ALDEx) analysis for mixed population RNA-Seq. PLoS One. 2013;8:e67019. doi: 10.1371/journal.pone.0067019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ravel J, Gajer P, Abdo Z, Schneider GM, Koenig SSK, McCulle SL, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A. 2011;108:4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Albert AYK, Chaban B, Wagner EC, Schellenberg JJ, Links MG, van Schalkwyk J, et al. A study of the vaginal microbiome in healthy Canadian women utilizing cpn60-based molecular profiling reveals distinct Gardnerella subgroup community state types. PLoS One. 2015;10:e0135620. doi: 10.1371/journal.pone.0135620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nasioudis D, Forney LJ, Schneider GM, Gliniewicz K, France M, Boester A, et al. Influence of pregnancy history on the vaginal microbiome of pregnant women in their first trimester. Sci. Rep. Springer US; 2017;7:1–6. [DOI] [PMC free article] [PubMed]
- 55.Mercer BM, Goldenberg RL, Moawad AH, Meis PJ, Iams JD, Das AF, et al. The preterm prediction study: effect of gestational age and cause of preterm birth on subsequent obstetric outcome. Am J Obstet Gynecol. 1999;181:1216–1221. doi: 10.1016/s0002-9378(99)70111-0. [DOI] [PubMed] [Google Scholar]
- 56.Goldenberg RL, Hauth JC, Andrews WW. Intrauterine infection and preterm delivery. N Engl J Med. 2000;342:1500–1507. doi: 10.1056/NEJM200005183422007. [DOI] [PubMed] [Google Scholar]
- 57.Kindinger LM, Bennett PR, Lee YS, Marchesi JR, Smith A, Cacciatore S, et al. The interaction between vaginal microbiota, cervical length, and vaginal progesterone treatment for preterm birth risk. Microbiome. 2017;5:1–14. doi: 10.1186/s40168-016-0223-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kekki M, Kurki T, Pelkonen J, Kukinen-Raty M, Cacciatore B, Paavonen J. Vaginal clindamycin in preventing preterm birth and peripartal infections in asymptomatic women with bacterial vaginosis: a randomized, controlled trial. Obstet Gynecol. 2001;97:643–648. doi: 10.1016/s0029-7844(01)01321-7. [DOI] [PubMed] [Google Scholar]
- 59.Andrews WW, Goldenberg RL, Hauth JC, Cliver SP, Copper R, Conner M. Interconceptional antibiotics to prevent spontaneous preterm birth: a randomized clinical trial. Am J Obstet Gynecol. 2006;194:617–623. doi: 10.1016/j.ajog.2005.11.049. [DOI] [PubMed] [Google Scholar]
- 60.Morency A, Bujold E. The effect of second-trimester antibiotic therapy on the rate of preterm birth. J Obstet Gynaecol Canada. 2007;29:35–44. doi: 10.1016/s1701-2163(16)32350-7. [DOI] [PubMed] [Google Scholar]
- 61.Okun N, Gronau KA, Hannah ME. Antibiotics for bacterial vaginosis or Trichomonas vaginalis in pregnancy: a systematic review. Obstet Gynecol. 2005;105:857–868. doi: 10.1097/01.AOG.0000157108.32059.8f. [DOI] [PubMed] [Google Scholar]
- 62.Thinkhamrop J, Hofmeyr GJ, Adetoro O, Lumbiganon P, Ota E. Antibiotic prophylaxis during the second and third trimester to reduce adverse pregnancy outcomes and morbidity. Cochrane Database Syst Rev. 2015;1:CD002250. doi: 10.1002/14651858.CD002250.pub2. [DOI] [PubMed] [Google Scholar]
- 63.Beigi RH, Austin MN, Meyn LA, Krohn MA, Hillier SL. Antimicrobial resistance associated with the treatment of bacterial vaginosis. Am J Obstet Gynecol. 2004;191:1124–1129. doi: 10.1016/j.ajog.2004.05.033. [DOI] [PubMed] [Google Scholar]
- 64.Budd W, Bostwick DG, Woody J, Hunt C. Antimicrobial resistance genes and modelling of treatment failure in bacterial vaginosis: clinical study of 289 symptomatic women. J Med Microbiol. 2016;65:377–386. doi: 10.1099/jmm.0.000236. [DOI] [PubMed] [Google Scholar]
- 65.Brown RG, Marchesi JR, Lee YS, Smith A, Lehne B, Kindinger LM, Terzidou V, Holmes E, Nicholson JK, Bennett PR, et al. Vaginal dysbiosis increases risk of preterm fetal membrane rupture, neonatal sepsis and is exacerbated by erythromycin. BMC Med. 2018;16(1):9. doi: 10.1186/s12916-017-0999-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Paramel Jayaprakash T, Wagner EC, van Schalkwyk J, Albert AYK, Hill JE, Money DM. High diversity and variability in the vaginal microbiome in women following preterm premature rupture of membranes (PPROM): a prospective cohort study. PLoS One. 2016;11:e0166794. doi: 10.1371/journal.pone.0166794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ling Z, Kong J, Liu F, Zhu H, Chen X, Wang Y, et al. Molecular analysis of the diversity of vaginal microbiota associated with bacterial vaginosis. BMC Genomics. 2010;11:1–16. doi: 10.1186/1471-2164-11-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Foxman B, Wen A, Srinivasan U, Goldberg D, Marrs CF, Owen J, et al. Mycoplasma, bacterial vaginosis-associated bacteria BVAB3, race, and risk of preterm birth in a high-risk cohort. Am J Obstet Gynecol. 2014;210:226.e1–226.e7. doi: 10.1016/j.ajog.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hitti J, Garcia P, Totten P, Paul K, Astete S, Holmes KK. Correlates of cervical Mycoplasma genitalium and risk of preterm birth among Peruvian women. Sex Transm Dis. 2010;37:81–85. doi: 10.1097/OLQ.0b013e3181bf5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Edwards RK, Ferguson RJ, Reyes L, Brown M, Theriaque DW, Duff P. Assessing the relationship between preterm delivery and various microorganisms recovered from the lower genital tract. J. Matern. Neonatal Med. 2006;19:357–363. doi: 10.1080/00207170600712071. [DOI] [PubMed] [Google Scholar]
- 71.Lis R, Rowhani-Rahbar A, Manhart LE. Mycoplasma genitalium infection and female reproductive tract disease: a meta-analysis. Clin Infect Dis. 2015;61:418–426. doi: 10.1093/cid/civ312. [DOI] [PubMed] [Google Scholar]
- 72.Donders GG, Van Calsteren K, Bellen G, Reybrouck R, Van Den Bosch T, Riphagen I, et al. Predictive value for preterm birth of abnormal vaginal flora, bacterial vaginosis and aerobic vaginitis during the first trimester of pregnancy. BJOG An Int J Obstet Gynaecol. 2009;116:1315–1324. doi: 10.1111/j.1471-0528.2009.02237.x. [DOI] [PubMed] [Google Scholar]
- 73.Kwak D-W, Hwang H-S, Kwon J-Y, Park Y-W, Kim Y-H. Co-infection with vaginal Ureaplasma urealyticum and Mycoplasma hominis increases adverse pregnancy outcomes in patients with preterm labor or preterm premature rupture of membranes. J Matern Neonatal Med. 2014;27:333–337. doi: 10.3109/14767058.2013.818124. [DOI] [PubMed] [Google Scholar]
- 74.Hillier SL, Nugent RP, Eschenbach DA, Krohn MA, Gibbs RS, Martin DH, et al. Association between bacterial vaginosis and preterm delivery of a low-birth-weight infant. N Engl J Med. 1995;333:1737–1742. doi: 10.1056/NEJM199512283332604. [DOI] [PubMed] [Google Scholar]
- 75.Lamont RF, Taylor-Robinson D, Wigglesworth JS, Furr PM, Evans RT, Elder MG. The role of mycoplasmas, ureaplasmas and chlamydiae in the genital tract of women presenting in spontaneous early preterm labour. J Med Microbiol. 1987;24:253–257. doi: 10.1099/00222615-24-3-253. [DOI] [PubMed] [Google Scholar]
- 76.Zhou X, Brown CJ, Abdo Z, Davis CC, Hansmann MA, Joyce P, et al. Differences in the composition of vaginal microbial communities found in healthy Caucasian and black women. ISME J. 2007;112:121–133. doi: 10.1038/ismej.2007.12. [DOI] [PubMed] [Google Scholar]
- 77.Fettweis JM, Brooks JP, Serrano MG, Sheth NU, Girerd PH, Edwards DJ, et al. Differences in vaginal microbiome in African American women versus women of European ancestry. Microbiology. 2014;160:2272–2282. doi: 10.1099/mic.0.081034-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
cpn60 OTU sequences. Multiple fasta file containing 728 OTU sequences. (TXT 336 kb)
Summary of OTU analysed in this study. OTU ID, percentage of identity, length, cpnDB name, species, and abundance in each library are shown. (XLSX 500 kb)
Data Availability Statement
The dataset supporting the results of this article is available in the NCBI SRA repository (Accession SRP073152, BioProject PRJNA317763; BioProject PRJNA403856).