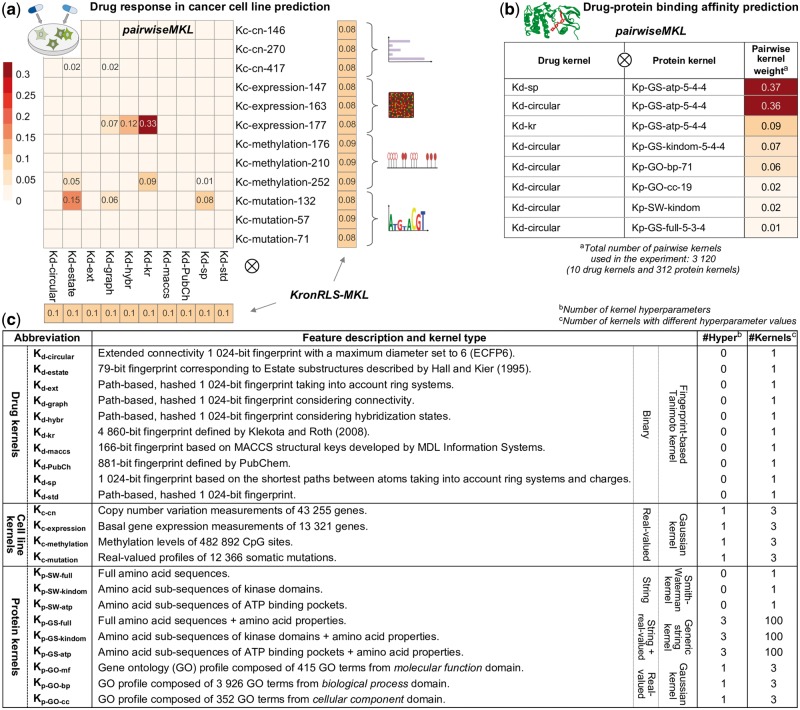
Fig. 2.
Pairwise kernel mixture weights obtained with pairwiseMKL and KronRLS-MKL (average across 10 outer CV folds) in the task of (a) drug response in cancer cell line prediction and (b) drug–protein binding affinity prediction (note: KronRLS-MKL did not execute with 1 TB memory); only the weights different from 0 are shown. KronRLS-MKL finds separate weights for drug kernels and cell line (protein) kernels instead of pairwise kernels. Numbers at the end of kernel names indicate the kernel hyperparameter values, in particular (i) kernel width hyperparameter in case of Gaussian kernels (e.g. Kc-cn-146 with ), and (ii) maximum sub-string length L, σ1 controlling for the shifting contribution term and σ2 controlling for the amino acid similarity term in case of GS kernels (e.g. Kp-GS-atp-5-4-4 with , see Section 2.3.2 for details). (c) Summary of drug, cell line and protein kernels used in this work for the two prediction problems.