Abstract
Defining units that can be afforded legal protection is a crucial, albeit challenging, step in conservation planning. As we illustrate with a case study of the red wolf (Canis rufus) from the southeastern United States, this step is especially complex when the evolutionary history of the focal taxon is uncertain. The US Endangered Species Act (ESA) allows listing of species, subspecies, or Distinct Population Segments (DPSs) of vertebrates. Red wolves were listed as an endangered species in 1973, and their status remains precarious. However, some recent genetic studies suggest that red wolves are part of a small wolf species (C. lycaon) specialized for heavily forested habitats of eastern North America, whereas other authors suggest that red wolves arose, perhaps within the last ~400 years, through hybridization between gray wolves (C. lupus) and coyotes (C. latrans). Using published genetic, morphological, behavioral, and ecological data, we evaluated whether each evolutionary hypothesis would lead to a listable unit for red wolves. Although the potential hybrid origin of red wolves, combined with abundant evidence for recent hybridization with coyotes, raises questions about status as a separate species or subspecies, we conclude that under any proposed evolutionary scenario red wolves meet both criteria to be considered a DPS: they are Discrete compared with other conspecific populations, and they are Significant to the taxon to which they belong. As population-level units can qualify for legal protection under endangered-species legislation in many countries throughout the world, this general approach could potentially be applied more broadly.
Keywords: de-listing, distinct population segment, hybrid policy, hybridization, listing criteria, taxonomy
What biological units merit special protection? This question is increasingly relevant as earth’s ecosystems are ever more strongly influenced by humans. It is a difficult question even for relatively straightforward scenarios, where at a minimum one must consider 1) uncertainties associated with estimates of extinction risk; 2) difficulty in predicting consequences of alternative intervention strategies; and 3) perceived values to humans and natural ecosystems. Prioritizing scarce conservation resources is even more challenging when the evolutionary history of the focal taxon is uncertain.
The red wolf (Canis rufus, C. lycaon rufus, or C. lupus rufus, depending on the authority), a small, wolf-like canid that historically occupied most of the United States east of the prairies and south of the Great Lakes, is a prime example of a taxon with an uncertain evolutionary history. Wolves, coyotes, jackals, dogs, and the dingo comprise the genus Canis, which first appeared in the fossil record in southwestern United States and Mexico in the Miocene (~6 MYA) (Wang and Tedford 2008). The gray wolf (C. lupus), which is the only wild Canis species that currently occurs in both the Old and New World, originated in Eurasia and appears in the North American fossil record by the late Rancholabrean period (~130000 ybp) or perhaps as early as the Illinoian period (~300000 ybp; Nowak 1979), but there is little evidence of its existence south of the glaciers until the late Rancholabrean (Nowak 2002). Historic range of the gray wolf included most of North America except the eastern United States (Chambers et al. 2012). In 1973, the gray wolf in the lower 48 states was listed as Endangered under the US Endangered Species Act (ESA), based on the last few remaining wolves in northern Minnesota and Michigan.
Several other wolf-like canids are restricted to North America. The small forms from areas of southern Ontario and Quebec centered on Algonquin Provincial Park are commonly referred to as “eastern wolves” (or more recently “Algonquin wolves” by the Committee on the Status of Species at Risk in Ontario) and are considered by some authors to be a separate species (C. lycaon) (Wilson et al. 2000; Baker et al. 2003) or subspecies (C. lupus lycaon) (Nowak 1995, 2002, 2003; Chambers et al. 2012). Wolves from the western Great Lakes area of the United States and Canada, sometimes referred to as “Great Lakes” wolves and included by some authorities as part of the subspecies C. l. nubilus, are generally intermediate in size to eastern wolves and gray wolves from western North America (Nowak 1995, 2002, 2003; Mech and Paul 2008; Mech et al. 2011). Red wolves historically occupied most of the eastern United States, outside of the range of the Canis lineage referred to as lycaon. Another small canid historically occurred in the American southwest and Mexico, and the last remnant populations were listed under the ESA in 1976 as the Mexican wolf, a subspecies of gray wolf (Canis lupus baileyi; aka “lobo”). Finally, the coyote (C. latrans), the smallest wild North American wolf-like canid, was historically restricted to the western half of the continent but rapidly expanded eastward following the functional extinction of eastern wolves and red wolves (Parker 1995; Fener et al. 2005; Kays et al. 2010; Levy 2012).
The precarious status of the red wolf is not in question: nearly driven to extinction by the middle of the 20th century, it exists today as a captive population and a small experimental wild population (Gese et al. 2015). Red wolves were listed as Endangered in 1967 under the US Endangered Species Preservation Act and remain listed (as C. rufus) under the ESA. However, this listing and associated recovery actions are controversial because of uncertainties and scientific disagreements about both the evolutionary history and contemporary history of red wolves. Recent hybridization between coyotes and both red wolves and eastern wolves, together with a paucity of historical genetic samples, has clouded interpretation of their evolutionary history.
As elaborated in the next section, several hypotheses have been proposed regarding the evolutionary history of these charismatic canids. One hypothesis suggests that the red wolf is a unique New World lineage that split off from the smaller coyote in the Pleistocene; a variation posits that red wolves and eastern wolves together form a species separate from coyotes and gray wolves. Another hypothesis is that red wolves were a subspecies, or ecotype, of C. lupus specialized on the eastern forests. Various alternative scenarios for the origin of the eastern wolf-like canids involve ancient and/or recent hybridization with coyotes, gray wolves, and potentially domestic dogs (C. familiaris or C. lupus familiaris) brought to the continent by humans (Anderson et al. 2009). All experts do agree that red wolves and coyotes hybridized in the southeastern United States as coyotes spread eastward in the 20th century and that hybridization remains a constant threat. However, there are diverse and strongly held views regarding what hybridization, old and new, means for conservation and management of red wolves.
Here, we tackle a question at the heart of the controversy: are red wolves a listable entity under the ESA? This is timely, as the red wolf’s status is under review as part of a periodic process mandated under Section 4(c)(2), and subsequent to the ESA listing of the red wolf, 2 events have changed the criteria for determining listability. First, the ESA has been amended several times, including the sections dealing with listing criteria. The 1978 ESA amendments [Public Law 95–632 (1978), 92 Stat. 3751] clarified what units can be considered “species” under the ESA and hence legally protected if they are determined to be Threatened or Endangered: an ESA “species” can be either 1) a recognized biological species, 2) a recognized subspecies, or 3) a “distinct population segment” (DPS). The provision to recognize DPSs applies only to vertebrate species. Although this language opened up new options for listing populations of vertebrates, the ESA provides no specific guidance on how to determine what constitutes a DPS.
The second major event was that, following almost 2 decades of applying the DPS provision on an ad hoc basis, the agencies that implement the ESA (USFWS and National Marine Fisheries Service, NMFS) developed a joint policy to guide DPS determinations (USFWS and NMFS 1996a). Under the joint species policy, to be a DPS a population or group of populations must meet 2 criteria: discreteness and significance. A population unit can be considered discrete if it satisfies either of the following conditions:
1. It is markedly separated from other populations of the same taxon as a consequence of physical, physiological, ecological, or behavioral factors. Quantitative measures of genetic or morphological discontinuity may provide evidence of this separation.
2. It is delimited by international governmental boundaries within which differences in the control of exploitation, management of habitat, conservation status, or regulatory mechanisms exist that are significant in light of Section 4(a)(1)(D) of the ESA.
According to the policy, information relevant to the “discrete” criterion includes (but is not necessarily limited to) physical, ecological, behavioral, and genetic data.
Once a population segment is deemed to be discrete, the next step in DPS evaluation is to determine whether it is also “significant” to the taxon to which it belongs. Factors that can be used to determine whether a discrete population segment is significant include:
1) Persistence of the discrete segment in an ecological setting unusual or unique for the taxon;
2) Evidence that loss of the discrete segment would result in a significant gap in the range of the taxon;
3) Evidence that the discrete segment represents the only surviving natural occurrence of a taxon that may be more abundant elsewhere as an introduced population outside its historical range; and
4) Evidence that the discrete segment differs markedly from other populations of the taxon in its genetic characteristics.
Determining whether the red wolf is a listable entity under the ESA, therefore, requires a determination whether they are a species, a subspecies, or a DPS. Our approach to this problem is as follows. First, we review the various published hypotheses regarding historical evolutionary relationships of red wolves and other North American wolf-like canids. Our objective is not to establish which hypothesis is most likely, but rather to enumerate the plausible hypotheses so that each can be considered from the ESA perspective. Next, we review recent information about hybridization among North American Canis. Finally, for each of the published hypotheses, we draw on the best available scientific information to evaluate whether the red wolf could be considered a listable unit under the ESA by virtue of its status as a species, subspecies, or DPS.
Evolutionary History of Red Wolves in Relation to Other NA Canids
Current Context: Captive Breeding and Recovery
By the early 1900s, a combination of direct persecution, forest clearing, road building, and perhaps the decline of deer herds had eliminated red wolves from most of their historic range (USFWS 1990), and hybridization between red wolves and coyotes had begun in central Texas (Nowak 2002; Phillips et al. 2003). By the time the USFWS initiated a captive breeding program in 1973, red wolves were confined to a single small population in Louisiana and Texas, surrounded by coyotes that had expanded their range eastward (Riley and McBride 1975; USFWS 1990). Over the next 7 years, more than 400 wild canids were captured from the area of the remaining red wolf population, and wild red wolves were extirpated from their historic range. A total of 42 captured animals were sent to the breeding facility as putatively pure red wolves; of them, only 14 became the founders of the captive population (USFWS 1990). Details of the captive breeding program are described elsewhere (Waddell and Long 2015).
Currently, a single wild population of red wolves exists in the red wolf recovery area (RWRA) on the Albemarle Peninsula in northeastern North Carolina. This was established as a nonessential, “experimental” population to allow additional management flexibility to reduce human conflicts. From 1987 to 1994, a total of 63 wolves were introduced to the RWRA from the captive facility. The RWRA encompasses about 4600 km2, roughly half in public and half in private ownership, with red wolves making use of about 47% of that area (Phillips et al. 2003; Gese et al. 2015). Coyotes were not present when introductions began but arrived soon after, and hybridization with red wolves was confirmed by the early 1990s (Adams et al. 2003). Recognizing that coyote hybridization was the greatest risk to recovery, USFWS implemented specific, ongoing actions to reduce coyote introgression into the red wolf population, including removing hybrid litters and euthanizing or sterilizing coyotes and hybrids (Kelly 2000; Gese et al. 2015). Although the wild population increased to more than 100 wolves, it has since declined to fewer than 50 individuals (Madison J, USFWS, personal communication)
Morphology
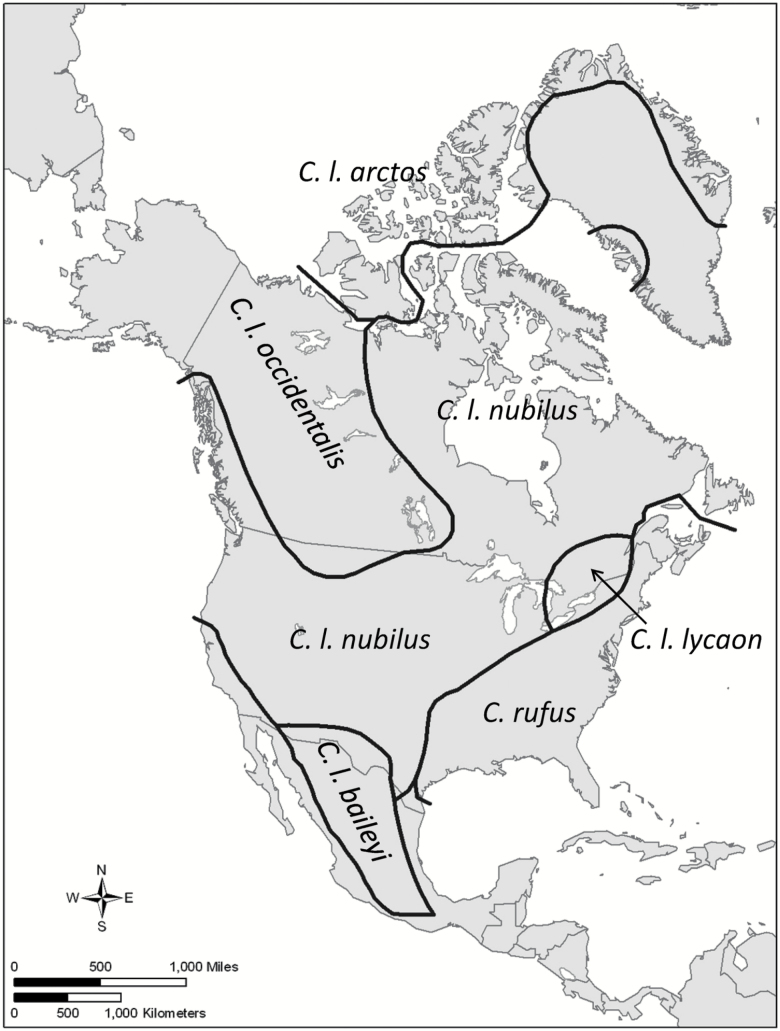
Canid taxonomy, like that for many mammalian groups, historically has focused on morphological analysis of teeth and bones of the skull. Early efforts to make sense of North American canid diversity (e.g., Goldman 1944) found morphological evidence for a large number of subspecies of the gray wolf. More recently, Nowak (1995, 2002) trimmed that to the red wolf (C. rufus) and five subspecies of gray wolf, including the Mexican wolf (C. l. baileyi), eastern wolves in southern Ontario and Quebec (C. l. lycaon), and a form that historically occupied much of the western United States and ranged northward to encompass all of Hudson’s Bay (C. l. nubilus) (Figure 1). Nowak’s (2002) paper focused on eastern canids and used only skulls collected prior to 1918. He found that a small wolf appeared in areas east of the Great Plains and south of the Great Lakes and St. Lawrence River near the end of the Pleistocene, at about the same time coyotes disappeared from eastern North America. He found this small wolf, which he considered to be C. rufus, to be distinct from both the coyote and gray wolf, with no evidence of hybridization. Prior to European settlement of North America, the geographic range of these “red wolves” had little overlap with that of coyotes, whose eastern limits largely coincided with the westerly plains (Nowak 2002). Historically, eastern North America was heavily forested, and these small wolves were presumably specialized to hunt the primary ungulate of the region, white-tailed deer (Odocoileus virginianus).
Figure 1.
Historical distribution of North American wolves from Chambers et al. (2012). In this version of the taxonomy, the red wolf (Canis rufus) is considered a separate species from gray wolves (Canis lupus), which are divided into 5 subspecies. The boundary between the red wolf and the eastern wolf, Canis l. lycaon, is uncertain, especially in the upper third of the C. rufus range shown here (from about Pennsylvania north). The other native wolf-like canid in North America, the coyote (C. latrans), is not shown here.
Nowak’s taxonomic conclusions are based on multivariate, discriminant function analysis (DFA) of morphological characters. DFA requires the user to pre-assign individuals into groups and then derives linear combinations of the original variables (the discriminant functions) that maximize differences among groups. DFA is most robust when group membership in the pre-defined groups is determined using independent information. When the same characters are used both to group the samples and to derive the discriminant functions (as Nowak did), the result can be an exaggeration of intergroup differences and overly-optimistic assessments of power to assign individuals to groups (Waples 2010).
More recently, Hinton and Chamberlain (2014) found that sympatric red wolves, coyotes, and their hybrids could be distinguished based on morphological characteristics: red wolves are the largest canid in the NC recovery region, coyotes the smallest, and hybrids are intermediate.
Ecology
Gray wolves historically inhabited most of North America except for the deciduous forests of eastern North America, an area occupied by red and eastern wolves, which are morphologically intermediate between gray wolves and coyotes (Nowak 1995; Kyle et al. 2006). The morphological and ecological differentiation of Canis taxonomic groups has been attributed to habitat and prey selection, as well as to interference competition (Mech 1970). Population structure is often associated with ecological differences in vegetation and prey type (Geffen et al. 2004; Carmichael et al. 2007). A striking example of ecological differentiation occurs for adjacent populations of gray wolves from coastal and inland British Columbia. The strong genetic structure between these groups does not correspond to geographic distance or physical dispersal barriers but rather to habitat differences, as coastal wolves obtain more than half of their protein from marine sources (Muñoz-Fuentes et al. 2009).
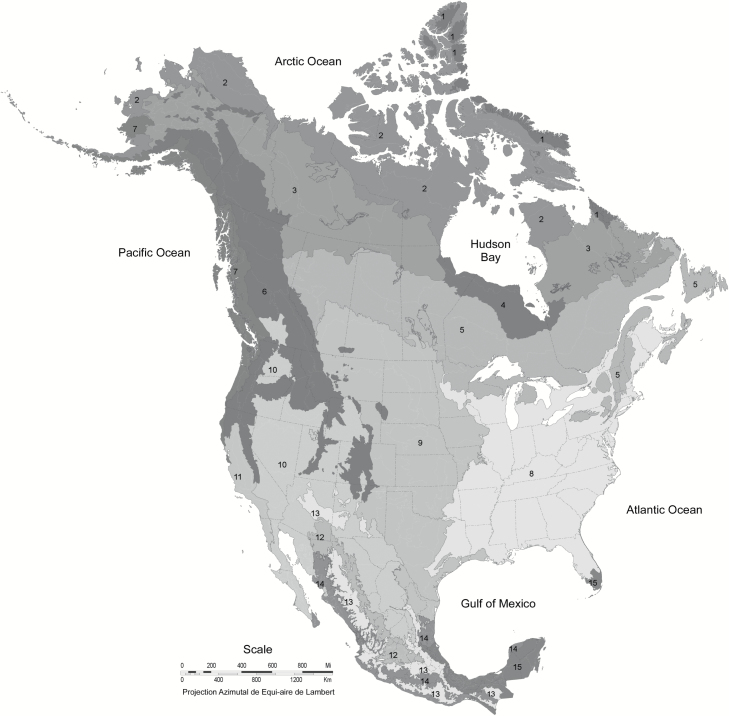
The historic distribution of red wolves (Figure 1) is largely congruent with North America’s Ecoregion 8, Eastern Temperate Forests (Figure 2). This ecological region is characterized by a relatively dense and diverse forest cover, an abundance of perennial streams and rivers, and a high diversity of many species, including birds, fish, reptiles, and amphibians; it is recognized as “a significant evolutionary area for the continent’s fauna” (Commission for Environmental Cooperation 1997). Summers are hot and humid, whereas winters exhibit a latitudinal gradient from subtropical temperatures in the south to cool, continental temperatures in the north. The north-central part of this ecoregion includes part of the historic range of C. lycaon (Figure 1).
Figure 2.
North American level-1 ecoregions (modified from Commission for Environmental Cooperation 1997). Ecoregion names: 1 = Arctic Cordillera; 2 = Tundra; 3 = Taiga; 4 = Hudson Plains; 5 = Northern Forests; 6 = Northwestern Forested Mountains; 7 = Marine West Coast Forests; 8 = Eastern Temperate Forests; 9 = Great Plains; 10 = North American Deserts; 11 = Mediterranean California; 12 = Southern Semi-Arid Highlands; 13 = Temperate Sierras; 14 = Tropical Dry Forests; 15 = Tropical Humid Forests.
Historically, sympatry between coyotes and wolves was restricted to western North America, where competition is reduced because large herbivores (moose, bison, and elk) provide abundant prey for wolves, whereas coyotes forage on deer and smaller species or scavenge wolf kills. As eastern forests were altered to more open, human-dominated habitats, wolf-control programs decimated historic populations of eastern wolves and red wolves, leaving a void that coyotes, being renowned generalists, readily exploited. By the mid 1900s, coyotes had expanded into most of North America (Parker 1995; Sears et al. 2003; Levy 2012). Subsequent hybridization between coyotes and remnant populations of wolves in eastern North America created an increasing coyote size gradient from west to east (Kays et al. 2010; Way et al. 2010). Hybridization between coyotes and eastern wolves has also created intermediate habitat preferences. Based on species distribution models, Otis et al. (2017) found that hybrids between the eastern wolves and eastern coyotes exhibited intermediate environmental niche characteristics compared with their progenitors.
Red wolf habitat use and prey types in the reintroduced North Carolina population overlap with those of the invasive coyote. However, the proportion of prey types differs, consistent with the larger body sizes of red wolves; in particular, red wolves consume more white-tailed deer and fewer small mammals and rabbits than coyotes do in the RWRA (Hinton et al. 2017). Likewise, current red wolf habitat use differs from other wolves, with preference for agricultural habitats over forest, perhaps tracking white-tailed deer densities (Dellinger et al. 2013; Hinton et al. 2016).
Behavior and Contemporary Hybridization
Recent studies in and near the RWRA provide insights into contemporary interactions between red wolves and coyotes. The red wolf social system is similar to that of gray wolves and differs from that of coyotes in the area. From 1999 to 2013, red wolves in the reintroduced population regularly displaced other red wolves and killed or displaced coyotes and hybrids (Gese and Terletzky 2015). This behavior is important because it may form the basis of a reproductive barrier between red wolves and coyotes (Fredrickson and Hedrick 2006). In addition, red wolves often formed packs by delayed dispersal of offspring or by inclusion of unrelated helpers into packs (Sparkman et al. 2011, 2012). Both of these are hallmarks of the gray wolf social system but rare among coyote populations.
In a 13-year study, Bohling and Waits (2015) found four times as many red-wolf litters as hybrid litters within the RWRA. About half of the hybridization events followed the death (typically caused by humans) of one member of a stable red-wolf breeding pair. Hybrid litters tended to be produced by first-time red-wolf breeders, away from the core RWRA. Red wolves that did not pair with other red wolves preferentially paired with admixed individuals rather than coyotes, even though coyotes vastly outnumbered hybrids within the study area. The authors concluded that social stability of red wolf family groups was an important factor in determining the probability of hybridization (see also Hinton et al. 2015).
Bohling et al. (2016) studied the spatial extent of hybridization within the RWRA and adjacent areas. They found that red wolf ancestry declined sharply across a transect leading outside the RWRA and that no red wolves were found outside the recovery area, whereas half of the canids within the RWRA were coyotes. In spite of the pervasive presence of coyotes, only 4% of the individuals surveyed by Bohling et al. (2016) were hybrids. This result, however, reflects at least in part success of the adaptive management plan to limit the consequences of hybridization.
In southern Quebec and Ontario, a study focused on eastern wolves from Algonquin Provincial Park (Rutledge et al. 2010a) found evidence that coyote mtDNA was widespread but coyote Y-chromosome haplotypes were absent, indicating that male coyotes were not involved in hybridization with eastern wolves. In contrast, eastern and gray-wolf Y-chromosome haplotypes were present, indicating some male-mediated introgression of gray-wolf genes via eastern wolves and possible sex-biased introgression mediated by males of the larger species breeding with females of the smaller species. These studies also found divergent Y-chromosome haplotypes unique to eastern Canada and the Great Lakes, supporting the hypothesis of a unique Canis taxon in the region (Wheeldon et al. 2010; Wilson et al. 2012).
Current high hybridization rates between coyotes and both eastern wolves and red wolves are associated with high kill rates of wolves by humans (Rutledge et al. 2012a; Bohling and Waits 2015). High human-caused death rates, particularly due to gunshot during the deer-hunting season, facilitate coyote introgression by removing resident red wolves just prior to the breeding season, in which case the remaining wolf of a pair is more likely to settle for a coyote or a hybrid as a mate. With social structure intact, red and eastern wolves exhibit positive assortative mating (Rutledge et al. 2010a; Bohling et al. 2016), and red wolves will exclude or displace coyotes from areas they occupy (Gese and Terletzky 2015). Death of transient red wolves removes individuals that might pair with other red wolves and displace coyotes as breeders (Hinton et al. 2015, 2016).
Although recent hybridization between coyotes and eastern wolves and red wolves is well documented, the extent of gray wolf × coyote hybridization is less clear. In western North America, gray wolves often kill coyotes, and no matings of the 2 species have been reported in the wild (Wheeldon et al. 2010; Rutledge et al. 2012a). In the western Great Lakes region, where the 2 species have co-existed since prior to European contact, studies have found evidence for little (Wheeldon et al. 2010) or no (Mech 2011) recent hybridization between gray wolves and coyotes. Wheeldon et al. (2010) concluded that wolves from the western Great Lakes are derived from hybridization between gray wolves and eastern wolves. Because eastern wolves have also hybridized with coyotes recently (and perhaps historically), they have potentially served as a conduit for indirect mixing of gray wolf and coyote genes (Rutledge et al. 2012a).
An experimental attempt at artificial insemination showed that gray wolves and coyotes are not completely incompatible reproductively, as one of 9 coyote females inseminated with gray wolf semen produced offspring (Mech et al. 2014). Two of these gray-wolf–coyote hybrids subsequently mated and produced F2 offspring (vonHoldt et al. 2017a), but whether the F2 generation is fertile is not known.
Genetics
The large (and growing) number of genetic studies of wolves and their relatives are challenging to summarize (see Chambers et al. 2012 for a recent attempt); these studies have only partially overlapping sets of samples, DNA markers, and analytical methods, which not surprisingly supports a variety of perspectives. Here we focus on studies most directly relevant to red wolves.
Wayne and Jenks (1991) evaluated genetic variation in captive red wolves in the broader context of patterns of genetic variation in North American coyotes and gray wolves. In maternally inherited mitochondrial DNA (mtDNA), the authors found that red wolves from the captive population carried a single haplotype that was phylogenetically similar to that of coyotes. Other potential founders that displayed at least a partial red-wolf phenotype but were not incorporated into the red-wolf captive population had either coyote, gray wolf, or Mexican wolf mtDNA, and a mismatch often was observed between mtDNA haplotype and the morphological classification. Haplotypes obtained from six “red wolf” pelts collected between 1905 and 1930 were similar or identical to known coyote or gray-wolf haplotypes. Wayne and Jenks (1991) hypothesized that the red wolf is either 1) wholly of hybrid origin or 2) has recently hybridized with other North American canids. This paper represents the first proposed 2-species hypothesis for North American canids (gray wolf and coyote), with eastern and red wolves being of mixed origin.
Using data for 10 nuclear gene loci previously reported by Roy et al. (1994), Reich et al. (1999) estimated that the coyote–gray-wolf hybridization event occurred no more than 12800 ybp, and likely less than 2500 ybp. Bertorelle and Excoffier (1998) found that the same data were consistent with red wolves and coyotes being sister species that diverged 10% as long ago as coyotes and gray wolves.
Wilson et al. (2000) proposed what has come to be known as the “three species” hypothesis based on the analysis of both nuclear and mtDNA data. At nuclear loci, Wilson et al. found that red wolves and eastern wolves were more similar to each other than either was to gray wolves. They identified coyote mtDNA in both eastern and red wolves but also found unique sequences in both of the latter forms that diverged from any coyote mtDNA haplotypes. The authors concluded that coyotes, red wolves, and eastern wolves all evolved in North America, with the eastern+red wolf lineage diverging from the coyote lineage 150000–300000 years ago. This three-species hypothesis considers eastern wolves and red wolves to be part of the same species, C. lycaon.
The first paper to take advantage of the revolution in genomics technology to study worldwide evolution of Canis was vonHoldt et al. (2011), who used over 48000 single-nucleotide-polymorphism (SNP) markers. These authors concluded that unique genetic features of the red-wolf population were less distinctive than for other recognized wolf species or subspecies. Bayesian clustering analysis suggested that this was primarily due to its mixed origin. Subsequent analyses based on haplotype block size suggested that the primary hybridization event between coyotes and gray wolves occurred 287–430 years ago; a similar analysis for Great Lakes wolves suggested initial admixture 546–963 years ago. vonHoldt et al. (2011) concluded that the close affinity of red wolves and eastern wolves proposed by Wilson et al. (2000) owed more to similar patterns of lupus × latrans hybridization than to a shared evolutionary history.
The global scope of the vonHoldt et al. (2011) analyses had both advantages and disadvantages. Including diverse canids from Eurasia and Africa provided a broad context for interpreting evolutionary distinctiveness of New World forms. On the other hand, this made it more difficult to discern fine-scale structure within North America of some forms (esp. eastern wolves) represented by relatively few samples. In particular, 75% of the individuals were domestic dogs, and it is well known that some methods (such as Bayesian clustering) can be very sensitive to unequal sample sizes (Kalinowski 2011) and most readily detect the strongest levels of genetic structuring (Evanno et al. 2005).
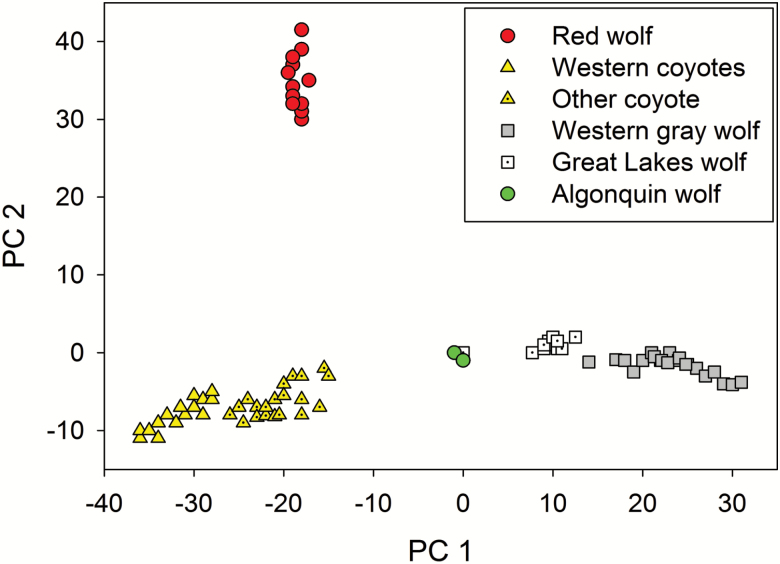
Rutledge et al. (2012b) re-analyzed vonHoldt et al’s (2011) SNP data in a principal components analysis (PCA), with a focus on 4 North American forms (gray wolves, coyotes, eastern wolves, and red wolves). They found red wolves to be the most distinctive of the 4 groups (Figure 3), which they pointed out could be due at least in part to founder effect and drift associated with the captive breeding program.
Figure 3.
Principal components analysis of 48000 SNPs for North American wolf-like canids (modified from Rutledge et al. 2012b, which is based on data from vonHoldt et al. 2011). PC 1 represents 15.7% of the variance and PC 2 represents 3.6%. For alternative ways of presenting these data, see Supplementary Figure S3 in vonHoldt et al. (2011).
Most recently, a whole-genome-sequencing study by vonHoldt et al. (2016) greatly expanded the number of loci (to 5.4 million SNPs), while focusing on a much smaller number of individuals (28). Red wolves were the most divergent group (FST = 0.177 with North American gray wolves and 0.107 with coyotes judged to be nonadmixed) but were genetically more similar to coyotes considered to be admixed. vonHoldt et al. (2016) noted that the amount of genetic divergence between North American gray wolves and coyotes (FST = 0.153) is not much larger than that between European and North American gray wolves (FST = 0.099), and they estimated the divergence time between gray wolves and coyotes at only about 50000 years ago. vonHoldt et al. (2016) further identified >16000 SNPs with fixed differences between coyotes and Eurasian gray wolves and used them to estimate coyote versus gray-wolf ancestry in putative admixed forms; results indicated that Great Lakes wolves derive slightly more of their genes from gray wolves and eastern wolves derive slightly more from coyotes, whereas at least 80% of red-wolf genes can be traced to coyotes. The authors argued that the percentage of novel alleles in eastern wolves and red wolves was lower than expected if they were distinct species. Using a genetic-demographic model that included divergence times and historical population sizes and an analysis that focused on 9 individuals, vonHoldt et al. (2016) estimated the divergence between red wolf and coyote as 55000–117000 ybp and divergence between Great Lakes wolf and gray wolf as 27000–32000 ybp.
Hohenlohe et al. (2017) criticized a number of aspects of the vonHoldt et al. (2016) study, including representativeness of the samples and their suitability for assessing admixture, pooling of eastern wolves from Algonquin Park with Great Lakes wolves in some analyses, interpretation of the PCA and rare-allele analyses, and conclusions about admixture. vonHoldt et al. (2017b) responded to these criticisms and reaffirmed conclusions of their 2016 study.
Genetic analysis of historical (pre-European contact) specimens could potentially resolve some of the uncertainties regarding canid evolution in North America, but to date these are limited to studies of mtDNA. Wilson et al. (2003) found nongrey-wolf mtDNA in 100-year-old samples from Maine and New York—long before the 20th Century range expansion of coyotes. Rutledge et al. (2010b) examined 4 Canis skull samples excavated from 16th Century middens in southern Ontario and found that none contained mtDNA of gray-wolf origin. They concluded that this area was historically occupied by the New World-evolved eastern wolf rather than the Old World-evolved gray wolf, but they could not rule out the possibility that the specimens analyzed were admixed forms of eastern and gray wolves. Brzeski et al. (2016) examined 3 wolf-like tooth samples dated to 350–1900 ybp collected within the historic range of the red wolf. Each specimen produced a previously undocumented mtDNA haplotype, all of which grouped within the coyote clade. This result is consistent with either an origin by ancient hybridization with coyotes or evolution of the red wolf from the coyote lineage, but does not support a recent hybrid origin following the invasion of the coyote into eastern North America.
Collectively, studies to date have produced a range of conflicting conclusions regarding the origin of red and eastern wolves, as well as the timing of important evolutionary events. Further complicating matters, Koblmüller et al. (2016), examined 105 complete gray-wolf mitogenomes, including 10 from Eurasian and North American wolves >14000 years old. The authors concluded that all extant North American gray wolves derive from a single gray-wolf colonization event from Eurasia about 23000 ybp. This is at odds with the gray wolf—coyote and gray wolf—Great Lakes wolf divergence-time estimates based on whole-genome sequences (vonHoldt et al. 2016), as well as the coyote—red/eastern-wolf divergence estimate based on control-region mtDNA sequence data (Wilson et al. 2000).
Summary
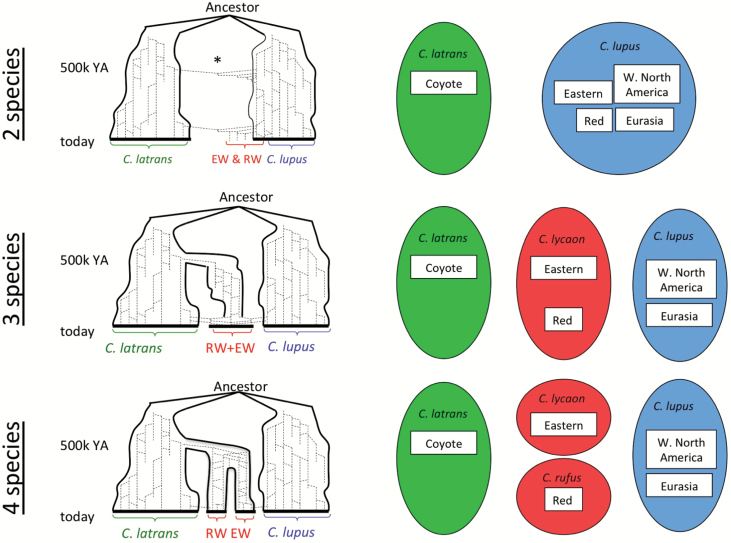
The diverse ideas summarized above about the evolutionary history of wolf-like canids in North America can be grouped into 3 general scenarios, referred to as the 4-, 3-, and 2-species hypotheses (Figure 4). The 4-species hypothesis generally follows existing taxonomy based on morphology and historical distributions (see Figure 1; the 4 species are gray wolf, eastern wolf, red wolf, and coyote, with both the red wolf and eastern wolf evolving from a coyote-like ancestor). The most comprehensive summary of this scenario can be found in Chambers et al. (2012). As noted above, all 4 of these groups show some level of genetic distinctiveness, in spite of acknowledged recent hybridization. However, the Chambers et al. (2012) study has been criticized because it adopted the 4-species taxonomic hypothesis as a framework for interpreting the genetic data, rather than allowing species delimitations to emerge directly from the analysis of the data (Dumbacher et al. 2014).
Figure 4.
Schematic evolutionary history (left) and resulting taxonomy (right) for 3 hypotheses for the origins of the red wolf (RW) and eastern wolf (EW). The evolutionary history diagrams show a timing of a coyote-wolf split around 1 MYA and subsequent speciation within North America around 500k YA, although as noted in the main text those dates are debatable. Under the 2-species hypothesis, the original RW and EW were forms of C. lupus that hybridized with coyotes when wolves were extirpated from most of their native range in eastern North America. This hypothesis also allows for the possibility of ancient hybridization between gray wolves and coyotes. The 3-species hypothesis recognizes RW and EW as a single species that diverged from the coyote lineage; C. lycaon is the older scientific name and so would have priority over C. rufus under this hypothesis. According to the 4-species hypothesis, EW and RW both evolved from the coyote lineage but diverged into separate species in the northeast and southeast.
The 3-species hypothesis originated with Wilson et al. (2000) and has been supported in various ways by several subsequent papers showing distinctiveness of eastern wolves and/or red wolves. Under this scenario, red wolves would be grouped along with eastern wolves within C. lycaon as a separate subspecies or some other subspecific population unit. In support of this hypothesis, eastern and red wolves are similar in size and (in theory) well suited to the heavily forested areas that historically dominated most of eastern North America. However, not all analyses have found a close genetic affinity between eastern wolves and red wolves.
The 2-species hypothesis suggests that all modern populations referred to as wolves are subspecies of C. lupus and/or recent hybrids. This is supported by the lack of distinctive genetic material in red wolves or eastern wolves. One version of this hypothesis proposes that eastern forests were populated by one or more smaller forms of C. lupus that specialized on deer, and these hybridized with coyotes as eastern wolf populations dwindled in the last 400 years. A variation proposes that red wolves and/or eastern wolves could have arisen from more ancient hybridization between gray wolves and coyotes but have not diverged enough to be considered full species.
Timing of the hypothesized historical hybridization events is highly uncertain, with estimates ranging from a few hundred years to over 100000 years. Under some scenarios of the 2-species hypothesis, red wolves and/or eastern wolves might be considered separate subspecies (within C. latrans and C. lupus, respectively). Some authors (Mech 2011; Rutledge et al. 2012a; Hohenlohe et al. 2017) argue that scant empirical evidence for recent hybridization of gray wolves and coyotes in the wild (in spite of abundant opportunities) poses a challenge for the 2-species hypothesis. However, it is well known that changing environments can promote hybridization between species that normally have effective isolating mechanisms, and there are ample examples of changing environments in North America during the Pleistocene and Holocene. Therefore, recent patterns of hybridization among North American canids are not necessarily a good indicator of historical patterns.
Regardless of what conclusions are reached regarding species- and subspecies-level taxonomy of these North American canids, it is noteworthy that, in spite of recent introgression, and in spite of being surrounded and vastly outnumbered by coyotes or gray wolves, red wolves and eastern wolves both exhibit positive assortative mating—at least when harvest pressure and other anthropogenic mortality factors are low enough that social groups remain intact.
Analysis of Evolutionary Hypotheses in an ESA Context
In this section, we review the 3 major evolutionary hypotheses and discuss the implications of each for the status of red wolves as a potentially listable entity (species, subspecies, or DPS) under the ESA.
Four-species Hypothesis
In this scenario, the red wolf is considered a full species (C. rufus; see Goldman 1944; Nowak 2002; Baker et al. 2003; Chambers et al. 2012), so it could continue to be listed on that basis (Figure 4).
Three-species Hypotheses
In the various 3-species scenarios, the red wolf is not a full species; instead, it and the eastern wolf are considered to be synonymous with or subspecific units of C. lycaon. Opinions about whether to define subspecies—and if so, how—differ widely (Mayr 1982; Burbrink et al. 2000; Haig et al. 2006; Taylor et al. 2017). We are not aware of any published paper formally proposing that the red wolf be considered a subspecies of C. lycaon, although Chambers et al. (2012) discussed this idea hypothetically and some of the nomenclatural issues it would entail. As described above, however, the red wolf and eastern gray wolf meet several of the criteria that are most commonly used to delimit subspecies: they are geographically allopatric (currently and perhaps historically) and are genetically and morphologically different from each other, as well as from coyotes and gray wolves.
If not considered a separate subspecies, the red wolf would be evaluated as a potential DPS of C. lycaon. Because the only other extant population of C. lycaon is in Canada, the red wolf is “delimited by international governmental boundaries” and therefore meets Element 2 of the Discreteness criterion. PCA results (Figure 3) and other results presented by Rutledge et al. (2012b) show that the 2 populations are genetically distinct, so the red wolf likely also meets Discreteness Element 1 (marked separation from other populations of the taxon). After meeting the Discreteness test, Significance of the red wolf would be evaluated “with respect to the taxon to which it belongs”—that is, C. lycaon. The red wolf would presumably meet Significance Element 2 (only 2 conspecific populations are extant, so loss of the red wolf would create a major gap in the range of C. lycaon) and arguably would meet Significance Element 1 (occurrence in an unusual ecological setting). Although eastern wolves are found in part of the Eastern Temperate Forests Ecoregion historically occupied by red wolves, habitats for red wolves are more temperate/subtropical, and the northern range of eastern wolves also extended into the colder Northern Forests Ecoregion (Figure 2). Whether the red wolf would also meet Significance Element 4 (marked genetic differences from other conspecifics) is more subjective. The joint DPS policy does not clarify whether the same genetic data can be used for both Discreteness Element 1 and Significance Element 4. In applying a similar 2-part test (reproductive isolation and evolutionary significance) to Pacific salmon populations, as well as in applying the joint DPS policy to a variety of marine species, NOAA Fisheries has typically used presumably neutral molecular genetic data primarily to address the discreteness/reproductive isolation criterion and has largely relied on proxies for adaptive genetic differences (e.g., behavior/life history/ecology) to meet the significance criterion (Waples 2006).
Although meeting multiple Significance elements might make the case stronger, if Discreteness has been established it is not necessary to meet more than a single Significance element to be considered a DPS. Under a scenario in which both red wolves and eastern wolves are considered subspecific units of C. lycaon, therefore, we conclude that red wolves would qualify as at least a DPS because they meet both Discreteness elements and at least 1 or 2 Significance elements of the joint DPS policy.
Two-species Hypotheses
The various 2-species hypotheses agree that the gray wolf and coyote are the 2 species but differ in other details. In the following, we consider 3 variations of the 2-species hypothesis.
Red Wolves are Derived From Gray Wolves
Under this scenario, red wolves are not a full species but might be a subspecies of C. lupus, as proposed by Lawrence and Bossert (1975) and Wozencraft (2005).
If red wolves are not a separate subspecies, they could be a DPS of C. lupus. For that evaluation, it would be necessary to consider red wolves in the context of all other subspecific units of C. lupus. As noted above, red wolves can be considered Discrete compared with eastern (Algonquin) wolves according to both Discreteness elements. Some Great Lakes wolves occur in the United States, so the international border element does not apply to the Great Lakes wolves × red wolves comparison. In the PCA analysis shown in Figure 3, red wolves are the most genetically distinctive of all the North American wolf-like canids (the Mexican wolf, C. l. baileyi, was not represented in these samples, but its genetic distinctiveness has been established by many studies—see review by Chambers et al. 2012). This is also consistent with results presented by vonHoldt et al. (2016), who found that red wolves had the highest FST values (all FST > 0.1) for pairwise comparisons with NA gray wolves, Great Lakes wolves, and coyotes. Collectively, these results support the conclusion that red wolves are Discrete compared with all other North American wolves.
With respect to Significance, there is little or no overlap in the historical distribution of red wolves with gray wolves, eastern wolves, Great Lakes wolves, and Mexican wolves, so loss of the red wolf would likely represent a significant gap in the range of C. lupus (Significance Element 2). As the red wolf is the last remaining small-wolf population in the large area of Ecoregion 8 in the United States east of the prairies and south of the Great Lakes (Figure 2), it could also be argued that under this hypothesis, the red wolf occupies an ecological setting that is unusual or unique for the species (Significance Element 1). As noted above, it is possible that the red wolf might also meet Significance Element 4, but that is more speculative.
Red Wolves are Derived From Coyotes
Like the Chambers et al. (2012) version of the 4-species hypothesis, this scenario has red wolves being derived from the coyote lineage, but more recently. It does not appear that anyone has formally proposed that the red wolf be considered a subspecies of C. latrans, although that is one possibility that might be evaluated. If not, and if red wolves were to remain a listable entity under this version of the 2-species hypothesis, it would have to be as a DPS of the coyote.
In vonHoldt et al. (2016), the red wolf was genetically more similar to coyotes than to gray wolves or Great Lakes wolves, but the level of divergence (FST = 0.108) was still substantial and larger than the values found for many other vertebrate populations that have been considered to be Discrete under the joint DPS policy (e.g., Gustafson et al. 2006; Seminoff et al. 2015). The PCA analysis (Figure 3) also provides strong evidence that red wolves are genetically distinctive compared with coyotes. Furthermore, although by all accounts substantial hybridization between red wolves and coyotes has occurred for at least a century, recent studies within and around the RWRA demonstrate that red wolves can be resistant to hybridization if anthropogenic pressures do not compromise their social structure. Current data therefore indicate that, under this scenario, red wolves could be considered discrete from other populations of C. latrans.
Until about 1900, the distributions of coyotes and red wolves were largely nonoverlapping, with coyotes being restricted to the west and red wolves filling a niche for a small, wolf-like canid in the deciduous forests of the east and southeast. Under those historical conditions, therefore, it is likely that (compared with other C. latrans) red wolves would have satisfied Significance Elements 1 and 2. Following near-extirpation of the red wolf, coyotes have greatly expanded their range eastward in the past century, so the contemporary situation is quite different.
Red Wolves are the Product of Recent Hybridization Between Gray Wolves and Coyotes
Hybridization is a well-known mechanism for creating new species; although more common in plants (Rieseberg 1997), it also occurs in animals, including mammals (Larsen et al. 2010) and birds (Lamichhaney et al. 2018). Evolutionary hypotheses II, IIIA, and IIIB do not specify an ancient hybrid origin for the lineage leading to contemporary red wolves, but they do not exclude this possibility. Hypothesis IIIC differs from the others in postulating that red wolves are not an ancient lineage, but rather arose recently from hybridization between gray wolves and coyotes. Timing of the putative hybridization event has been variously estimated as likely less than 2500 ybp (Reich et al. 1999) and 287–430 ybp (vonHoldt et al. 2011). Under this scenario, if the hybrid entity is not recognized as a formal species or subspecies to be listable under the ESA, it would have to be considered a DPS. In this case, DPS evaluations would be somewhat problematic because the taxon to which the putative DPS belongs is an important reference point. However, based on currently available information, we have concluded above that red wolves could be considered both Discrete and Significant with respect to either coyotes or gray wolves. This could be used to argue that even if red wolves are a hybrid-origin taxon, they nevertheless meet the criteria to be considered a DPS, regardless which formal taxon they are considered to be associated with. This scenario would also raise some legal/policy issues that are considered in the next section.
Discussion
The red wolf is currently listed under the ESA as a full species (C. rufus), which is consistent with traditional taxonomic treatments and the most recent review of the taxonomy of Canis in North America (Chambers et al. 2012). However, a number of more recent genetic studies have called into question the existing taxonomy of wolf-like canids, and the evolutionary history of the red wolf remains controversial (Dumbacher et al. 2014). Under some scenarios, the red wolf would not be a valid species and perhaps not a valid subspecies, in which case any ESA listing would have to be as a DPS of a valid taxon. In the section Evolutionary History of Red Wolves in Relation to Other NA Canids of this paper, we have summarized the relevant genetic and nongenetic data, but we have not attempted to resolve the uncertainties or disagreements. Instead, in the section Analysis of Evolutionary Hypotheses in an ESA Context, we have considered whether the red wolf would be a listable unit under the ESA under each of the major evolutionary scenarios that have been proposed, which can be characterized as 4-species (C. lupus, C lycaon, C. rufus, C. latrans), 3-species (C. lupus, C lycaon, C. latrans), and 2-species (C. lupus, C. latrans) hypotheses.
Under the 4-species hypothesis, the red wolf would remain a full species and could continue to be listed on that basis. We also conclude that the 3-species hypothesis, which would group eastern wolves and red wolves under C. lycaon, would be relatively straightforward to evaluate. That scenario considers these 2 populations to be the last remnants of a biological species. Given that they are geographically disjunct and demonstrably differ in genetic and other characteristics, we conclude that eastern wolves and red wolves might be considered separate subspecies, and if not the red wolf would at least qualify as a DPS of C. lycaon.
The various 2-species hypotheses are more challenging to evaluate, both because of their diversity and the fact that most would require evaluation of DPS status. Nevertheless, our overall conclusion remains the same: if the red wolf is not considered to be a valid subspecies of either gray wolf or coyote, it would at least qualify as a DPS of its respective taxon. This conclusion is based on hypothetical application of the 2 criteria in the joint 1996 interagency DPS policy: Discreteness and Significance. Available data indicate that the red wolf is genetically the most distinctive wolf-like canid in North America, which establishes Discreteness. Congruence of the historical distribution of red wolf with the Eastern Temperate Forests Ecoregion (Figure 2) provides strong evidence for Significance Element 1 (persistence in an unusual ecological setting), and a case could be made for Elements 2 and 4 as well.
Some caveats are important to note regarding our evaluations. First, are red wolves genetically distinctive primarily because of their recent bottleneck and/or effects of the captive breeding program? This is a reasonable question, but we are not aware of any quantitative analyses that attempt to answer it. The finding by Brzeski et al. (2016) of unique haplotypes in pre-Columbian samples presumed to be red wolf suggests that the genetic distinctiveness of red wolves is not merely a recent phenomenon, but more studies of this type would be useful to better clarify historical patterns. The Mexican wolf does not have the same hybridization issues as the red wolf, but it underwent an even more extreme bottleneck that has also undoubtedly affected recent genetic samples (Chambers et al. 2012); this, however, has not prevented the Mexican wolf from being recognized as a valid subspecies and listed as such under the ESA.
The second caveat has to do with hybridization, which by all accounts has been extensive recently between red wolves and coyotes, and which by some accounts is responsible for producing the red-wolf phenotype in the first place (through hybridization of gray wolves and coyotes). These evaluations would be easier if the Services had a formal policy outlining how hybridization and hybrids should be considered in ESA listing and recovery. However, although the Services announced a proposed ESA intercross policy 2 decades ago (USFWS and NMFS 1996b), it was never implemented or finalized. In the absence of specific policy guidance regarding hybridization, we applied the criteria in the joint DPS policy to existing data and concluded that the red wolf could be considered a DPS regardless of whether the taxon to which it belongs is considered to be the gray wolf or the coyote.
All of the DPS evaluations discussed above focus on the most recent data for red wolves and their relatives. These data, therefore, reflect the consequences of any hybridization that has occurred recently or historically. In spite of evidence for introgression of genes from coyote and perhaps gray wolf into the red wolf, we conclude that the current population meets both criteria to be a DPS, if not a subspecies or full species.
We have not attempted to grapple with questions of the following type:
Can a biological entity that arises through hybridization be considered a “species” under the ESA (i.e., a named species or subspecies or a DPS)?
If so, how far in the past must the hybridization event have taken place?
If a biological entity historically would have qualified as an ESA “species,” could it lose that status through hybridization with another biological entity?
If so, how much hybridization is too much? What metrics should be monitored to determine whether a threshold of too much hybridization has been reached?
Could or should the captive breeding program be modified to select for particular traits, such as larger (more wolf-like) size, which might help minimize hybridization with smaller coyotes?
Given uncertainties about taxonomic status, if all plausible scenarios lead to either a species, subspecies, or DPS designation, could the red wolf continue to be listed without specifying which taxonomic category it fits into?
These are interesting questions but they are difficult or impossible to address in a strictly objective framework, because they involve societal values as well as legal and policy issues.
Broader Relevance
As a case study of listability under the US ESA, evaluations discussed here have necessarily focused on details specific to red wolves and to the legal/legislative context of one particular piece of legislation. However, many of the themes covered here resonate more broadly for biodiversity conservation and management. Taxonomic uncertainty is a pervasive issue, which is not surprising given that at least 2 dozen species concepts have been proposed in the literature (Mayden 1997; de Queiroz 2007). This uncertainty creates challenges for those trying to implement endangered species legislation (national laws exist for a number of countries besides the United States, including Australia, Canada, Costa Rica, and South Africa), which typically afford legal protection only to pre-defined entities like species or subspecies. One level of taxonomic uncertainty arises from the imperfect understanding of particular evolutionary histories; another reflects the fact that evolution is a dynamic process and its products occur along a continuum rather than fitting neatly into discrete categories (Hey et al. 2003). Only the first type of uncertainty can be resolved with more and better data. Both levels of taxonomic uncertainty are relevant for evaluations of red wolves: We can continue to refine our understanding of the evolutionary history, but even perfect understanding would not resolve all uncertainties about whether this enigmatic taxon should be considered a species, subspecies, or something else.
Jackson et al. (2017) recently faced a similar challenge regarding the Australian dingo—another problematical canid whose taxonomic status has been in dispute since the 18th Century. Jackson et al. disagree with some recent authors who have proposed that the dingo be considered a separate species (Canis dingo), pointing to strong evidence that it is a feral form of domestic dog. Nonetheless, and despite evidence for recent hybridization between dingos and domestic dogs, Jackson et al. (2017) conclude that Australian dingos are of great conservation significance because of the ecological roles they now play and the insights they can provide about early stages of the domestication of dogs.
In addition to considering taxonomic uncertainty, a second theme of our analyses focuses on conservation relevance of population-level units below the species level (Soule 1986). Populations are routinely the focus of conservation efforts, and numerous laws provide for their protection. Although “Distinct Population Segment” is not a generally recognized biological term and is not in general use outside the United States, almost identical criteria (Discreteness and Evolutionary Significance) have been adopted by Canada to help identify population units that qualify for protection as Designatable Units under the Species at Risk Act, SARA http://www.cosewic.gc.ca/default.asp?lang=en&n=DD31EAEE-1). This means that the criteria used here to evaluate DPSs of red wolves play a large role in determining formal conservation priorities across most of North America.
Finally, hybridization, both natural (Genovart 2009) and human-mediated (Vilà et al. 2000), is an issue of global conservation concern. Several decades of wrestling with complex problems associated with hybridization and conservation have produced a diversity of viewpoints (Hedrick 1995; Allendorf et al. 2001; Haig and Allendorf 2006; Stronen and Paquet 2013; Wayne and Shaffer 2016). However, no strong consensus on practical application has emerged, no doubt in part because the ramifications of hybridization for conservation are very context specific. If a consensus is to eventually emerge, it will have to be built by synthesizing a series of detailed case studies like the one here for red wolves.
Supplementary Material
Supplementary material can be found at https://academic.oup.com/jhered/.
Funding
Funding for the science workshop that initiated this paper, and for publication costs, was provided by the Science Support Program of the U.S. Geological Survey and the U.S. Fish and Wildlife Service (agreement number G16AC00049).
Acknowledgments
Motivation for this study came from a 2016 workshop with the following participants: Fred Allendorf, Luigi Boitani, David Cobb, Jaime Collazo, Holly Doremus, Rich Fredrickson, Dale Goble, Sue Haig, Roland Kays, Scott Mills, Krishna Pacifici, Mike Phillips, Linda Rutledge, Mike Schwartz, David Smith, Lisette Waits, Robin Waples, Robert Wayne, Michael Morse, Pete Benjamin, and Bridgett vonHoldt. We thank Holly Doremus, Paul Hohenlohe, Marty Kardos, Mike Phillips, Michael Runge, Linda Rutledge, Luigi Univ, Bridgett vonHoldt, Lisette Waits, and Bob Wayne for useful discussions and/or comments on earlier versions of this manuscript, and Kathleen Neely for help preparing figures. We also acknowledge logistical support of the North Carolina Cooperative Fish and Wildlife Research Unit, Department of Applied Ecology at North Carolina State University, Jaime Collazo, Ruby Valeton, and Kairsten Fay. The scientific results and conclusions, as well as any views or opinions expressed herein, are those of the authors and do not necessarily reflect the views of NOAA, USGS, or any agency involved in red wolf restoration efforts.
References
- Adams JR, Kelly BT, Waits LP. 2003. Using faecal DNA sampling and GIS to monitor hybridization between red wolves (Canis rufus) and coyotes (Canis latrans). Mol Ecol. 12:2175–2186. [DOI] [PubMed] [Google Scholar]
- Allendorf RF, Leary P, Spruell JK, Wenburg JK. 2001. The problems with hybrids: setting conservation guidelines. Trends Ecol Evolut. 16:613–622. [Google Scholar]
- Anderson TM, vonHoldt BM, Candille SI, Musiani M, Greco C, Stahler DR, Smith DW, Padhukasahasram B, Randi E, Leonard JA, et al. . 2009. Molecular and evolutionary history of melanism in North American gray wolves. Science. 323:1339–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker RJ, Bradley LC, Bradley RD, Dragoo JW, Engstrom MD, Hoffman FS, Jones CA, Reid F, Rice DW, Jones C. 2003. Revised checklist of North American mammals north of Mexico. Lubbock (TX): Museum of Texas Tech University; Occasional Papers No. 229. [Google Scholar]
- Bertorelle G, Excoffier L. 1998. Inferring admixture proportions from molecular data. Mol Biol Evol. 15:1298–1311. [DOI] [PubMed] [Google Scholar]
- Bohling JH, Dellinger J, McVey JM, Cobb DT, Moorman CE, Waits LP. 2016. Describing a developing hybrid zone between red wolves and coyotes in eastern North Carolina, USA. Evol Appl. 9:791–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohling JH, Waits LP. 2015. Factors influencing red wolf-coyote hybridization in eastern North Carolina. Biol Conserv. 184:108–116. [Google Scholar]
- Brzeski KE, DeBiasse MB, Rabon DR Jr, Chamberlain MJ, Taylor SS. 2016. Mitochondrial DNA variation in Southeastern Pre-Columbian canids. J Hered. 107:287–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbrink FT, Lawson R, Slowinski JB. 2000. Mitochondrial DNA phylogeography of the polytypic North American rat snake (Elaphe obsoleta): a critique of the subspecies concept. Evolution. 54:2107–2118. [DOI] [PubMed] [Google Scholar]
- Carmichael LE, Krizan J, Nagy JA, Fuglei E, Dumond M, Johnson D, Veitch A, Berteaux D, Strobeck C. 2007. Historical and ecological determinants of genetic structure in arctic canids. Mol Ecol. 16:3466–3483. [DOI] [PubMed] [Google Scholar]
- Chambers SM, Fain SR, Fazio B, Amaral M. 2012. An account of the taxonomy of North American wolves from morphological and genetic analyses. North Am Fauna. 77:1–67. [Google Scholar]
- Commission for Environmental Cooperation 1997. Ecological regions of North America: toward a common perspective. Montreal (Quebec): CEC Secretariat, ISBN 2-922305-18-X Available from: http://www.cec.org. [Google Scholar]
- De Queiroz K. 2007. Species concepts and species delimitation. Syst Biol. 56:879–886. [DOI] [PubMed] [Google Scholar]
- Dellinger JA, Proctor C, Steury TD, Kelly MJ, Vaughan MR. 2013. Habitat selection of a large carnivore, the red wolf, in a human-altered landscape. Biol Conserv. 157:324–330. [Google Scholar]
- Dumbacher J, Fallon S, Murdoch W, Patton J, Wayne R, Wilson P, Courtney S. 2014. Review of proposed rule regarding status of the wolf under the endangered species act. Report to US Fish and Wildlife Service. Santa Barbara (CA): National Center for Ecological Analysis and Synthesis; Available from: https://www.fws.gov/science/pdf/Peer-Review-Report-of-Proposed-rule-regarding-wolves.pdf. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 14:2611–2620. [DOI] [PubMed] [Google Scholar]
- Fener HM, Ginsberg JR, Sanderson EW, Gompper ME. 2005. Chronology of range expansion of the coyote, Canis latrans, in New York. Can Field-Nat. 119:1–5. [Google Scholar]
- Fredrickson RJ, Hedrick PW. 2006. Dynamics of hybridization and introgression in red wolves and coyotes. Conserv Biol. 20:1272–1283. [DOI] [PubMed] [Google Scholar]
- Geffen E, Anderson MJ, Wayne RK. 2004. Climate and habitat barriers to dispersal in the highly mobile grey wolf. Mol Ecol. 13:2481–2490. [DOI] [PubMed] [Google Scholar]
- Genovart M. 2009. Natural hybridization and conservation. Biodivers Conserv. 18:1435. [Google Scholar]
- Gese EM, Knowlton FF, Adams JR, Beck K, Fuller TK, Murray DL, Steury TD, Stoskopf MK, Waddell WT, Waits LP. 2015. Managing hybridization of a recovering endangered species: the red wolf Canis rufus as a case study. Curr Zool. 61:191–205. [Google Scholar]
- Gese EM, Terletzky PA. 2015. Using the “placeholder” concept to reduce genetic introgression of an endangered carnivore. Biol Conserv. 192:11–19. [Google Scholar]
- Goldman EA. 1937. The wolves of North America. J Mammal. 18:37–45. [Google Scholar]
- Goldman EA. 1944. Classification of wolves: part II. In: Young SP, Goldman EA, editors. The wolves of North America. Washington, D.C: The American Wildlife Institute; p. 389–636. [Google Scholar]
- Gustafson RG, Drake J, Ford MJ, Myers JM, Holmes EE, Waples RS. 2006. Status review of Cherry point pacific herring (Clupea pallasii) and updated status review of the Georgia Basin Pacific herring distinct population segment under the Endangered Species Act. Seattle, WA: U.S. Dept. of Commerce, NOAA Tech; Memo., NMFS-NWFSC-76, 182 p. [Google Scholar]
- Haig SM, Allendorf FW. 2006. Hybrids and policy. In: Michael Scott J, Goble DD, Davis FW, editors. The endangered species act at thirty, volume 2: conserving biodiversity in human-dominated landscapes. Washington: Island Press; p. 150–163. [Google Scholar]
- Haig SM, Beever EA, Chambers SM, Draheim HM, Dugger BD, Dunham S, Elliott-Smith E, Fontaine JB, Kesler DC, Knaus BJ, et al. . 2006. Taxonomic considerations in listing subspecies under the U.S. Endangered Species Act. Conserv Biol. 20:1584–1594. [DOI] [PubMed] [Google Scholar]
- Hedrick PW. 1995. Gene flow and genetic restoration: the Florida panther as a case study. Conserv Biol. 9:996–1007. [DOI] [PubMed] [Google Scholar]
- Hey J, Waples RS, Arnold ML, Butlin RK, Harrison RG. 2003. Understanding and confronting species uncertainty in biology and conservation. Trends Ecol Evol. 18:597–603. [Google Scholar]
- Hinton JW, Ashley AK, Dellinger JA, Gittleman JL, vanManen F, Chamberlain MJ. 2017. Using diets of Canis breeding pairs to assess resource partitioning between sympatric red wolves and coyotes. J Mammal. 98:475–488. [Google Scholar]
- Hinton JW, Brzeski KE, Rabon DR, Chamberlain MJ. 2015. Effects of anthropogenic mortality on critically endangered red wolf Canis rufus breeding pairs: implications for red wolf recovery. Oryx. 51:174–181. [Google Scholar]
- Hinton JW, Chamberlain MJ. 2014. Morphometrics of Canis taxa in eastern North Carolina. J Mammal. 95:855–861. [Google Scholar]
- Hinton JW, Proctor C, Kelly MJ, van Manen FT, Vaughan MR, Chamberlain MJ. 2016. Space use and habitat selection by resident and transient red wolves (Canis rufus). PLoS One. 11:e0167603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohenlohe PA, Rutledge LY, Waits LP, Andrews KR, Adams JR, Hinton JW, Nowak RM, Patterson BR, Wydeven AP, Wilson PA, et al. . 2017. Comment on “Whole-genome sequence analysis shows two endemic species of North American wolf are admixtures of the coyote and gray wolf”. Sci Adv. 3:e1602250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SM, Groves CP, Fleming PJ, Aplin KP, Eldridge MD, Gonzalez A, Helgen KM. 2017. The Wayward dog: is the Australian native dog or Dingo a distinct species?Zootaxa. 4317:201–224. [Google Scholar]
- Kalinowski ST. 2011. The computer program STRUCTURE does not reliably identify the main genetic clusters within species: simulations and implications for human population structure. Heredity (Edinb). 106:625–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kays R, Curtis A, Kirchman JJ. 2010. Rapid adaptive evolution of northeastern coyotes via hybridization with wolves. Biol Lett. 6:89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly BT. 2000. Red wolf recovery program adaptive work plan FY00-FY02. Atlanta, GA: US Fish and Wildlife Service; 17 pp. [Google Scholar]
- Koblmüller S, Vilà C, Lorente-Galdos B, Dabad M, Ramirez O, Marques-Bonet T, Wayne RK, Leonard JA. 2016. Whole mitochondrial genomes illuminate ancient intercontinental dispersals of grey wolves (Canis lupus). J Biogeogr. 43:1728–1738. [Google Scholar]
- Kyle CJ, Johnson AR, Patterson BR, Wilson PJ, Shami K, Grewal SK, White BN. 2006. Genetic nature of eastern wolves: past, present, and future. Conserv Genet. 7:273–287. [Google Scholar]
- Lamichhaney S, Han F, Webster MT, Andersson L, Grant BR, Grant PR. 2018. Rapid hybrid speciation in Darwin’s finches. Science. 359:224–228. [DOI] [PubMed] [Google Scholar]
- Larsen PA, Marchán-Rivadeneira MR, Baker RJ. 2010. Natural hybridization generates mammalian lineage with species characteristics. Proc Natl Acad Sci USA. 107:11447–11452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence B, Bossert W. 1975. Relationships of North American Canis shown by a multiple character analysis of selected populations. In: Fox MW, editor. The wild canids: their systematic, behavioral ecology, and evolution. New York: Van Nostrand Reinhold; p. 73–86. [Google Scholar]
- Levy S. 2012. Rise of the coyote: the new top dog. Nature. 485:296–297. [DOI] [PubMed] [Google Scholar]
- Mayden RL. 1997. A hierarchy of species concepts: the denouement in the saga of the species problem. In: Claridge MF, Dawah HA, Wilson MR, editors. Species: the units of biodiversity. New York, NY: Chapman & Hall; p. 381–424. [Google Scholar]
- Mayr E. 1982. Of what use are subspecies?The Auk. 99:593–595. [Google Scholar]
- Mech LD. 1970. The wolf: the behavior and ecology of an endangered species. New York: Natural History Press and Doubleday Publishing Co; 389 pp. [Google Scholar]
- Mech LD. 2011. Non-genetic data supporting genetic evidence for the eastern wolf. Northeast Nat. 18:521–526. [Google Scholar]
- Mech LD, Christensen BW, Asa CS, Callahan M, Young JK. 2014. Production of hybrids between western gray wolves and western coyotes. PLoS One. 9:e88861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mech LD, Nowak RM, Weisberg S. 2011. Use of cranial characters in taxonomy of the Minnesota wolf (Canis sp.). Can J Zool. 89:1188–1194. [Google Scholar]
- Mech LD, Paul WJ. 2008. Wolf body mass cline across Minnesota related to taxonomy?Can J Zool. 86:933–936. [Google Scholar]
- Muñoz-Fuentes V, Darimont CT, Wayne RK, Paquet PC, Leonard JA. 2009. Ecological factors drive differentiation in wolves from British Columbia. J Biogeogr. 36:1516–1531. [Google Scholar]
- Nowak RM. 1979. North American quaternary canis, monograph of the museum of natural history. Lawrence: The University of Kansas. [Google Scholar]
- Nowak RM. 1995. Another look at wolf taxonomy. In: Carbyn LN, Fritts SH, Seip DR, editors. Proceedings of the second North American symposium on wolves; 1992 Edmonton, Alberta: Canadian Circumpolar Institute, University of Alberta; p. 375–397. [Google Scholar]
- Nowak RM. 2002. The original status of wolves in eastern North America. Southeast Nat. 1:95–130. [Google Scholar]
- Nowak RM. 2003. Wolf evolution and taxonomy. In Mech LD, Boitani L, editors. Wolves, behavior, ecology, and conservation. Chicago: University of Chicago Press; p. 239–258. [Google Scholar]
- Otis J, Thornton D, Rutledge L, Murray DL. 2017. Ecological niche differentiation across a wolf-coyote hybrid zone in eastern North America. Divers Distrib. 23:529–539. [Google Scholar]
- Parker GR. 1995. Eastern coyote, the story of its success. Halifax Canada: Nimbus Publishing. [Google Scholar]
- Phillips MK, Henry VG, Kelly BT. 2003. Restoration of the red wolf. In: Mech LD, Boitani L, editors. Wolves: behavior, ecology, and conservation. Chicago: University of Chicago Press; p. 272–288. [Google Scholar]
- Reich DE, Wayne RK, Goldstein DB. 1999. Genetic evidence for a recent origin by hybridization of red wolves. Mol Ecol. 8:139–144. [DOI] [PubMed] [Google Scholar]
- Rieseberg L. 1997. Hybrid origins of plant species. Ann Rev Ecol Syst. 28:359–389. [Google Scholar]
- Riley GA, McBride RT. 1975. A survey of the red wolf (Canis rufus). In: Fox MW, editor. The wild canids: their systematics, behavioral ecology and evolution. New York: Van Nostrand Reinhold; p. 263–277. [Google Scholar]
- Roy MS, Geffen E, Smith D, Ostrander EA, Wayne RK. 1994. Patterns of differentiation and hybridization in North American wolflike canids, revealed by analysis of microsatellite loci. Mol Biol Evol. 11:553–570. [DOI] [PubMed] [Google Scholar]
- Rutledge LY, Bos KI, Pearce RJ, White BN. 2010b. Genetic and morphometric analysis of sixteenth century Canis skull fragments: implications for historic eastern and gray wolf distribution in North America. Conserv Genet. 11:1273–1281. [Google Scholar]
- Rutledge LY, Garroway CJ, Loveless KM, Patterson BR. 2010a. Genetic differentiation of eastern wolves in Algonquin Park despite bridging gene flow between coyotes and grey wolves. Heredity (Edinb). 105:520–531. [DOI] [PubMed] [Google Scholar]
- Rutledge LY, White BN, Row JR, Patterson BR. 2012a. Intense harvesting of eastern wolves facilitated hybridization with coyotes. Ecol Evol. 2:19–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutledge LY, Wilson PJ, Klütsch CF, Patterson BR, White BN. 2012b. Conservation genomics in perspective: a holistic approach to understanding Canis evolution in North America. Biol Conserv. 155:186–192. [Google Scholar]
- Sears H, Theberge J, Therberge M, Thornton I, Campbell G. 2003. Landscape influence on Canis morphological and ecological variation in a coyote-wolf C. lupus x latrans hybrid zone, Southeastern Ontario. Can Field-Nat. 117:589–600. [Google Scholar]
- Seminoff JA, Allen CD, Balazs GH, Dutton PH, Eguchi T, Haas HL, Hargrove SA, Jensen MP, et al. . 2015. Status review of the green turtle (Chelonia mydas) under the U.S. Endangered Species Act. La Jolla, CA: NOAA Technical Memorandum, NOAA-NMFS-SWFSC-539. 571pp. [Google Scholar]
- Soule ME. 1986. Viable populations for conservation. Cambridge (UK): University Press. [Google Scholar]
- Sparkman AM, Adams J, Beyer A, Steury TD, Waits L, Murray DL. 2011. Helper effects on pup lifetime fitness in the cooperatively breeding red wolf (Canis rufus). Proc Biol Sci. 278:1381–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkman AM, Adams J, Steury TD, Waits L, Murray DL. 2012. Evidence for a genetic basis for delayed dispersal in a cooperatively breeding canid. Anim Behav. 83:1091–1098. [Google Scholar]
- Stronen AV, Paquet PC. 2013. Perspectives on the conservation of wild hybrids. Biol Conserv. 167:390–395. [Google Scholar]
- Taylor BL, Perrin WF, Reeves RR, Rosel PE, Wang JY, Cipriano F, Baker SC, Brownell RL. 2017. Why we should develop guidelines and quantitative standards for using genetic data to delimit subspecies for data‐poor organisms like cetaceans. Mar Mamm Sci. 33:12–26. [Google Scholar]
- United States Fish and Wildlife Service (USFWS) 1990. Red wolf recovery/ species survival plan. Atlanta (GA): USFWS. [Google Scholar]
- USFWS (U.S. Fish and Wildlife Service) and NMFS (National Marine Fisheries Service) 1996a. Policy regarding the recognition of distinct vertebrate population segments under the Endangered Species Act. Fed Reg. 61:4721–4725. [Google Scholar]
- USFWS (U.S. Fish and Wildlife Service) and NMFS (National Marine Fisheries Service) 1996b. Proposed policy on the treatment of intercrosses and intercross progeny (the issue of ‘‘hybridization’’). Fed Reg. 61:4709–4713. [Google Scholar]
- Vilà M, Weber E, Antonio CM. 2000. Conservation implications of invasion by plant hybridization. Biol Invasions. 2:207–217. [Google Scholar]
- vonHoldt BM, Cahill JA, Fan Z, Gronau I, Robinson J, Pollinger JP, Shapiro B, Wall J, Wayne RK. 2016. Whole-genome sequence analysis shows that two endemic species of North American wolf are admixtures of the coyote and gray wolf. Sci Adv. 2:e1501714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- vonHoldt BM, Cahill JA, Gronau I, Shapiro B, Wall J, Wayne RK. 2017b. Response to Hohenlohe et al. Sci Adv. 3:e1701233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- vonHoldt B, Heppenheimer E, Petrenko V, Croonquist P, Rutledge LY. 2017a. Ancestry-specific methylation patterns in admixed offspring from an experimental coyote and gray wolf cross. J Hered. 108:341–348. [DOI] [PubMed] [Google Scholar]
- vonHoldt BM, Pollinger JP, Earl DA, Knowles JC, Boyko AR, Parker H, Geffen E, Pilot M, Jedrzejewski W, Jedrzejewska B, et al. . 2011. A genome-wide perspective on the evolutionary history of enigmatic wolf-like canids. Genome Res. 21:1294–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddell W, Long S. 2015. Population analysis & breeding and transfer plan red wolf (Canis rufus gregoryi). Point Defiance Zoo and Aquarium. [Google Scholar]
- Wang X, Tedford RH. 2008. Dogs: their fossil relatives and evolutionary history. New York: Columbia University Press. [Google Scholar]
- Waples RS. 2006. Distinct population segments. In: Scott JM, Goble DD, Davis FW, editors. The endangered species act at thirty: conserving biodiversity in human-dominated landscapes. Washington, D.C: Island Press; p. 127–149. [Google Scholar]
- Waples RS. 2010. High-grading bias: subtle problems with assessing power of selected subsets of loci for population assignment. Mol Ecol. 19:2599–2601. [DOI] [PubMed] [Google Scholar]
- Way JG, Rutledge L, Wheeldon T, White BN. 2010. Genetic characterization of eastern coyotes in eastern Massachusetts. Northeast Nat. 17:189–204. [Google Scholar]
- Wayne RK, Jenks SM. 1991. Mitochondrial DNA analysis implying extensive hybridization of the endangered red wolf Canis rufus. Nature. 351:565–568. [Google Scholar]
- Wayne RK, Shaffer HB. 2016. Hybridization and endangered species protection in the molecular era. Mol Ecol. 25:2680–2689. [DOI] [PubMed] [Google Scholar]
- Wheeldon TJ, Patterson BR, White BN. 2010. Sympatric wolf and coyote populations of the western Great Lakes region are reproductively isolated. Mol Ecol. 19:4428–4440. [DOI] [PubMed] [Google Scholar]
- Wilson PJ, Grewal S, Lawford ID, Heal JNM, Granacki AG, Pennock D, Theberge JB, Theberge MT, Voigt DR, Waddell W, et al. . 2000. DNA profiles of the eastern Canadian wolf and the red wolf provide evidence for a common evolutionary history independent of the gray wolf. Can J Zool. 78:2156–2166. [Google Scholar]
- Wilson PJ, Grewal S, McFadden T, Chambers RC, White BN. 2003. Mitochondrial DNA extracted from eastern North American wolves killed in the 1800s is not of gray wolf origin. Can J Zool. 81:936–940. [Google Scholar]
- Wilson PJ, Rutledge LY, Wheeldon TJ, Patterson BR, White BN. 2012. Y-chromosome evidence supports widespread signatures of three-species Canis hybridization in eastern North America. Ecol Evol. 2:2325–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozencraft WC. 2005. Order Carnivora. In Wilson DE, Reeder DM, editors. Mammal species of the world. 3rd ed Johns Hopkins University Press. [Google Scholar]