A missense mutation of KARRIKIN-INSENSITIVE2, KAI2ply2, compromises its ligand-binding activity, which subsequently impairs KAI2-signaling and multiple aspects of light-dependent responses.
Keywords: Arabidopsis, germination, KAI2, karrikin, photomorphogenesis, phytochrome
Abstract
A smoke-derived compound, karrikin (KAR), and an endogenous but as yet unidentified KARRIKIN INSENSITIVE2 (KAI2) ligand (KL) have been identified as chemical cues in higher plants that impact on multiple aspects of growth and development. Genetic screening of light-signaling mutants in Arabidopsis thaliana has identified a mutant designated as ply2 (pleiotropic long hypocotyl2) that has pleiotropic light-response defects. In this study, we used positional cloning to identify the molecular lesion of ply2 as a missense mutation of KAI2/HYPOSENSITIVE TO LIGHT, which causes a single amino acid substitution, Ala219Val. Physiological analysis and genetic epistasis analysis with the KL-signaling components MORE AXILLARY GROWTH2 (MAX2) and SUPPRESSOR OF MAX2 1 suggested that the pleiotropic phenotypes of the ply2 mutant can be ascribed to a defect in KL-signaling. Molecular and biochemical analyses revealed that the mutant KAI2ply2 protein is impaired in its ligand-binding activity. In support of this conclusion, X-ray crystallography studies suggested that the KAI2ply2 mutation not only results in a narrowed entrance gate for the ligand but also alters the structural flexibility of the helical lid domains. We discuss the structural implications of the Ala219 residue with regard to ligand-specific binding and signaling of KAI2, together with potential functions of KL-signaling in the context of the light-regulatory network in Arabidopsis thaliana.
Introduction
Smoke has long been recognized to contain chemical cues that impact on plant growth and development, and several phytoactive smoke compounds have been identified (Flematti et al., 2004, 2013; van Staden et al., 2004). Among them, the best characterized are karrikins (KARs), a group of butenolides. In plants, responses to KARs include enhancement of seed germination, potentiation of light-responsive seedling establishment, and de-etiolation (Chiwocha et al., 2009; Nelson et al., 2009, 2010). Molecular analysis has shown that KAR-dependent responses of plants entail transcriptional regulation of over one hundred of genes, which include primary KAR-target genes such as D14-LIKE2 (DLK2), KAR-UP F-BOX1 (KUF1), and SALT TOLERANCE HOMOLOG7 (STH7) (Nelson et al., 2009, 2010).
Recent studies have shown that plants are equipped with a smoke sensory system for karrikin (Morffy et al., 2016). A few components involved in KAR-signaling have been identified in Arabidopsis thaliana, including an α/β hydrolase fold protein, KARRIKIN INSENSITIVE2/HYPOSENSITIVE TO LIGHT (hereafter referred to as KAI2), an F-box protein, MORE AXILLARY GROWTH2 (MAX2, also referred to as ORESARA9/PLEIOTROPIC PHOTO SIGNALING/KARRIKIN INSENSITIVE1), and SUPPRESSOR OF MAX2 1 (SMAX1)/SMAX1-LIKE2 (SMXL2) (Nelson et al., 2011; Sun and Ni, 2011; Waters et al., 2012; Stanga et al., 2013, 2016). Crystallographic studies and biochemical analyses have demonstrated the role of KAI2 as a KAR receptor (Bythell-Douglas et al., 2013; Guo et al., 2013; Kagiyama et al., 2013; Zhao et al., 2013; Toh et al., 2014). By analogy to the well-defined mode of action of strigolactone (SL)-signaling, in which MAX2 forms a co-receptor with AtDWARF14 (AtD14), a paralog of KAI2, to target SMXLs/DWARF53 for ubiquitin-mediated degradation (Jiang et al., 2013; Zhou et al., 2013; Soundappan et al., 2015; Yao et al., 2016), it has been postulated that the KAI2–MAX2 complex would lead to ligand-dependent degradation of SMAX1/SMXL2, thus relieving a repressive gene regulation (Morffy et al., 2016; De Cuyper et al., 2017). However, KAI2 signaling may involve distinct biochemical mechanisms. For example, SL perception targets its receptor, AtD14, for MAX2-dependent ubiquitin-mediated degradation (Chevalier et al., 2014; Zhao et al., 2015), whereas KAR induces degradation of KAI2, independently of MAX2, via a non-proteosomal pathway as yet unknown (Waters et al., 2015a). In addition, it remains unclear how SMAX1/SMXL2 specifically repress the KAR-dependent responses whilst not affecting SL-dependent responses. Thus, the detailed mode of action of KAR-signaling remains to be elucidated.
Phylogenomic analyses combined with cross-species complementation assays using KAR- or SL-receptor mutants have not only dissected the biological impact of KAR but also raised an intriguing hypothesis, namely that plants have evolved to sense and respond to an endogenous, as yet hypothetical, KAI2-ligand (KL) (Waters et al., 2014; Conn and Nelson, 2016). Although the chemical nature of endogenous KL remains elusive, its biological roles appear to be broader than previously appreciated, including the control of germination, de-etiolation, light-dependent vegetative growth, formation of symbiotic associations with arbuscular mycorrhizal fungi (AMF), and, identified very recently, drought adaptation (Nelson et al., 2009, 2010; Sun and Ni, 2011; Gutjahr et al., 2015; Li et al., 2017). Despite our knowledge of the structure of KAI2 bound with KAR (Guo et al., 2013), we are still far from understanding how KAI2 senses its ligands, i.e. KAR and endogenous KL. For example, it has not been resolved how the catalytic triads (Ser-His-Asp) of KAI2 participate in its ligand-binding or signaling (Waters et al., 2015b). In addition, it remains to be determined how KL–KAI2 signaling interacts with other regulatory networks for proper developmental adjustment in higher plants.
Light is one of the most influential environment factors for plants, impacting on diverse aspects of growth and development throughout their life cycles (Smith, 2000; Chen et al., 2004; Franklin and Quail, 2010; Casal, 2013). Several classes of photoreceptors have been characterized that mediate both distinct and overlapping light-dependent responses, such as photoreceptors that absorb red/far-red (phytochromes), blue-light receptors (cryptochromes, phototropins, and ZTL/FKFs), and UV-B light receptors (UVR8) (Rockwell et al., 2006; Jenkins, 2014; Christie et al., 2015; Galvão and Fankhauser, 2015). The downstream signaling of activated photoreceptors has been extensively studied over recent decades, providing insights into the regulatory network of light-signaling. For example, light-activated phytochromes translocalize from the cytosol into the nucleus, and turn on early transcriptional circuitry (Nagatani and Matsushita, 2002; Leivar et al., 2009). Phytochrome-interacting proteins constitute diverse classes of transcriptional regulators (Bae and Choi, 2008), including a family of basic helix-loop-helix proteins, PHYTOCHROME INTERACTING FACTORs (PIF1-7) (Leivar et al., 2008; Shin et al., 2009), which regulate genome-wide gene expression cascades. For example, during germination of imbibed seeds, PIF1 activates direct target genes, such as SOMNUS (SOM) and the GA-signaling repressors, RGA and GAI, in the absence of light (Oh et al., 2009). The elevated level of SOM mediates the induction of ABA biosynthetic-/signaling-related genes, such as ABA1, NCED6/9, ABI3, and ABI5, but down-regulates GA-biosynthetic genes, such as GA3ox1 and GA3ox2. As such, light-responsive germination involves PIF1-regulated transcriptional cascades, which lead to increased GA-sensitivity/levels, but reduced ABA-sensitivity/levels. Recent genomics-based studies have also pointed to the extensive cross-talk between light-signaling and other endogenous or external stimuli (Seo et al., 2009; Lau and Deng, 2010; Wang et al., 2012). Indeed, several regulators of light signaling function as integrators of multiple signals, so that the output of light signaling can be co-ordinated with the ambient environment or the developmental/physiological context (de Lucas and Prat, 2014; Leivar and Monte, 2014). However, there are still many gaps in our understanding of the light-regulatory network (Galvão and Fankhauser, 2015; Wang and Wang, 2015).
As the kai2 mutant was originally identified by its reduced light-dependent responses (Sun and Ni, 2011), it has been suggested that endogenous KL–KAI2 signaling is integrated into the light-regulatory network. Here, we present the genetic identification and characterization of a mutant in Arabidopsis thaliana, designated as pleiotropic long hypocotyl2 (ply2), which affects multiple aspects of light responses, including photomorphogenic seedling development and germination. Identification of the molecular lesion of the ply2 mutation revealed that it is a missense allele of KAI2 that causes an amino acid substitution, Ala219Val. Physiological analyses together with a double-mutant analysis support the hypothesis that the pleiotropic effects of ply2 can be ascribed to severely reduced KL–KAI2 signaling, which could be suppressed by the smax1 mutation. Based on the results of biochemical and X-crystallographic analyses, we propose that the mutant KAI2ply2 protein has reduced ligand-binding activity as well as reduced structural flexibility, which thus compromise its sensitivity to KAR and endogenous KL. We discuss the structural features of KAI2ply2 with regard to ligand-binding and downstream signaling, together with specified roles of KL-signaling in the context of the light-regulatory network.
Materials and methods
Plant material
All of the Arabidopsis mutant lines used were in Col-0 background, unless otherwise stated. The ply2 mutant was initially identified as a long-hypocotyl mutant under short-day conditions from EMS-mutagenized pools in the gsd1-1D mutant (Ayele et al., 2014). After backcrossing four times to the Col wild-type, the ply2 single-mutant was isolated and used for further physiological and genetic studies. The phyA-211, phyB-9, CAB2::LUC, and ore9-1 lines have been described previously (Soh et al., 2000; Woo et al., 2001; Lee et al., 2006; Kim et al., 2008). The pif1-ko (SALK_072677), smax1-3 (SALK_097346C), and kai2-2 (NASC ID: N100282) seeds were obtained from the Arabidopsis Biological Resource Center (https://abrc.osu.edu/) or the Nottingham Arabidopsis Stock Centre (http://arabidopsis.info/).
Light treatments and hypocotyl elongation assays
Hypocotyl elongation assays were performed similar to as described by Yang et al. (2003). Seeds were surface-sterilized and cold-treated for 2–3 d. The seeds were then sown on half-strength MS (Murashige and Skoog) medium containing 0.8% agar. The plates were placed under white light for 12 h to improve germination and then transferred to the appropriate experimental light conditions. Monochromatic light was administered using light chambers equipped with blue, red, or far-red light-emitting diodes (Good Feeling, Korea), of which the spectral peaks are 470, 660, and 730 nm, respectively. After incubation at 23 °C for an additional 4 d, the seedlings were photographed. Hypocotyl lengths were measured using the ImageJ software (http://imagej.nih.gov/ij/). Fluence rates were measured using a LI-1400A detector (International Light, USA). For hormone-sensitivity tests, an appropriate concentration of 3-methyl-2H-furo[2,3-c]pyran-2-one (karrikin1, KAR1) or 2H-furo[2,3-c]pyran-2-one (karrikin2, KAR2) (Toronto Research Chemicals, Canada), or rac-GR24 (Chiralix, Netherlands) was added before pouring the MS or aqueous medium.
Germination assays
Germination tests were performed with seeds that had been after-ripened for at least for 2 months, essentially as described Shinomura et al. (1994). For phyB-dependent germination assays, seeds were surface-sterilized and sown on aqueous medium containing 0.8% agar. Seeds were irradiated with FR light (1.325 μW cm–2) for 15 min and then kept in darkness with or without a single pulse of R light (5.72 μW cm–2) for 10 min. After 5 d, the germination rate was determined based on radicle emergence. Tests were performed with three experimental replicates with at least 200 seeds for each genotype. Similar results were obtained at least twice with independent seed batches.
Anthocyanin measurements
Arabidopsis seeds were plated on MS medium and grown under blue light (17.4 μW cm–2) for 3 d after irradiation with white light for 12 h. Fifty seedlings were used for each anthocyanin measurement. Total anthocyanins were determined by measuring absorption at 530 and 657 nm (A530 and A657) of the aqueous phase using a spectrophotometer, as described by Soh et al. (2000).
Construction of double-mutants
Double-mutants were generated by genetic crossing. The resulting F2 population derived from selfed F1 plants was screened to select the long-hypocotyl phenotype under short-day conditions (the ply2 phenotype) and grown to set F3 seeds. The F3 seeds were tested for the genotypes of the other mutant lines based on the mutant phenotypes or molecular markers if available, as listed in Supplementary Table S3 at JXB online. The selected F3 seedlings were further grown to select for homozygous lines at the next generation for double-mutant analysis. In the case of ply2ore9-1, increased axillary branching phenotype was used to identify the double-mutant in the F3 generation from F2 plants with a long-hypocotyl and normal branching phenotype. To confirm the genotype of ply2, we used a dCAPS marker, designed using the dCAPS finder (Neff et al., 1998).
RT-PCR and quantitative RT-PCR analyses
Total RNA was extracted from seeds or seedlings using a Spectrum Plant Total RNA kit (Sigma) following manufacturer’s instructions. After treatment with DNase I, 1 µg of total RNA was used for cDNA synthesis with Superscript II reverse transcriptase and an oligo(dT)18 primer (Invitrogen). cDNA diluted 10-fold was used for semi-quantitative RT-PCR analysis or real-time qRT-PCR analysis. Real-time qRT-PCR was conducted in an Eco™ Real-Time PCR System (Illumina), using QuantiMix SYBR Kits (PhileKorea) in a 10 µl volume. The reactions were performed in triplicate for each run. The comparative 2–ΔΔCt method was used to evaluate the relative quantities of each amplified product in the samples (Livak and Schmittgen, 2001). Relative expression levels were normalized according to Ct values for PP2A, as described by Shin et al. (2015). Primer sequences are listed in Supplementary Table S3. Three biological replicates were performed for each expression analysis.
Positional cloning of PLY2
To map the PLY2 locus, we crossed ply2 with Ler wild-type. From the segregating F2 population thus derived, seedlings with long hypocotyls were selected and subject to further phenotypic confirmation at the following F3 generation. Extraction of genomic DNA was performed using the 125 confirmed F3 lines with the long hypocotyl phenotype under short-day conditions (10/14 h light/dark). Using PCR-based molecular markers (Shin et al., 2015), initial mapping located PLY2 at the lower arm of chromosome 4. Fine-mapping was performed using newly designed SSLP/CAPS markers, listed in Supplementary Table S3, based on Cereon Polymorphism (http://www.arabidopsis.org). The molecular lesion of ply2 was identified by PCR amplification and sequencing analysis of genes annotated in the mapped region. For transgenic complementation of ply2, a full-length cDNA of wild-type KAI2 was amplified by RT-PCR and cloned into the pENTR1A vector (Invitrogen). After verifying no sequence alteration, it was recombined into a plant overexpression vector, pH2GW7, using gateway cloning technology (Invitrogen) (Karimi et al., 2002). The resulting binary vector was introduced into Agrobacterium tumefaciens GV3101. Transformation was performed with the ply2 mutant according to a modified floral dip method (Clough and Bent, 1998). More than ten T3 transgenic lines with homozygous single T-DNA insertions were isolated and used for physiological analysis. Phylogenetic analysis was performed using the MEGA6 program (Tamura et al., 2013), following the instructions, after retrieving sequences of KAI2 homologous proteins using the blastp program at NCBI (http://www.ncbi.nlm.nih.gov/).
Subcellular localization assays
The full-length KAI2 or KAI2ply2 ORF was amplified by PCR with primers containing appropriate restriction sites and then cloned into the pENTR1A vector (Invitrogen). After verification of no sequence errors, the entry clones were recombined into the Gateway version of pCsVMV-eGFP-N-999. For transient expression in Arabidopsis, mesophyll cell protoplasts were prepared from 3-week-old Arabidopsis Col-0 leaves and transfected with the construct using the PEG method as described previously (Kim et al., 2008). Fusion protein expression was observed using Zeiss LSM 510 Meta confocal microscopy (Carl Zeiss, Germany).
Assays of circadian rhythmic activity of CAB2::LUC
Plants carrying the CAB2::LUC reporter were grown on 1/2 B5 medium containing 1% sucrose with 12/12 h light/dark cycles under white light and transferred at ZT 0 to constant white light (20 μmol m–2 s–1). Luminescence activity was measured as described previously (Kim et al., 2008). Briefly, luminescence images were recorded using a Peltier-cooled CCD camera (Versarray; Roper Scientific). Image processing and quantification were performed using the MetaVue software program (Universal Imaging). Data were imported into the Biological Rhythms Analysis Software System (BRASS v2.14) and analysed with the FFT-NLLS suite of programs. Period lengths are shown as partially variance-weighted periods (±SE), which were estimated using bioluminescence data with a time window from 24 to 96 h under free-running conditions.
Preparation of recombinant proteins
The full-length cDNA of KAI2 or KAI2ply2 was subcloned into the pET28a vector (Novagen), and the protein was expressed in Escherichia coli strain BL21 (DE3) (Invitrogen). Cells at mid-log phase were further grown at 20 °C for 18 h following IPTG induction and then collected by centrifugation. Cell pellets were resuspended in lysis buffer (20 mM Tris-cl pH 7.5, 150 mM NaCl, 20 mM Imidazole, 2 mM β-mercaptoethanol) and lysed by sonication. The soluble lysates were passed through a Ni-NTA sepharose column and eluted with lysis buffer supplemented with 300 mM Imidazole. The N-terminal histidine tag was removed by treatment overnight with TEV protease. The protein was further purified by size-exclusion chromatography on a Superdex 75 column, and concentrated to final concentration of 20 mg ml–1.
Generation of antibody and immunoblot analysis
A polyclonal antibody was raised in rabbit against the recombinant KAI2 protein, purified from E. coli. For immunoblot analysis, the total proteins were extracted from approximately thirty 7-d-old seedlings grown under continuous light after treatment either with a mock solution (0.1% DMSO) or with 10 μM KAR2 for 2 h, essentially as described by Choi et al. (2014). After separation on 10% SDS-polyacrylamide gels, the proteins were transferred onto nitrocellulose membranes, and incubated with the anti-KAI2 antibody (1:1000). Antibody-bound proteins were detected by incubation with secondary antibody conjugated to horseradish peroxidase using the ECL system (Amersham Biosciences). Densitometric analysis was performed using ImageJ.
Crystallization and determination of structure
Crystals of KAI2 and KAI2ply2 were grown under the same conditions by vapor diffusion at 17 °C from 1:1 mixtures of protein solution and a reservoir solution (1 M Na-citrate, 100 mM HEPES pH 7.5, 5% glycerol). Crystals were flash-frozen by immersion in liquid nitrogen, then incubated in a cryoprotectant solution (reservoir solution supplemented with 20% ethylene glycol). The observed reflections were indexed, integrated, and scaled using HKL2000 (Otwinowski and Minor, 1997). Initial crystallographic phases for KAI2 structure were determined using MOLREP (Vagin and Teplyakov, 1997) in the CCP4 suite (http://www.ccp4.ac.uk/html/molrep.html). The starting search model used a structure of RsbQ (PDB code 1WOM) (Kaneko et al., 2005). Coot was used for visualization of electron density maps and for manual rebuilding of atomic models (Emsley and Cowtan, 2004). Refinement was performed with REFMAC in CCP4 (Vagin et al., 2004).
CPM assays
CPM assays were performed following the procedures described by Alexandrov et al. (2008). A stock solution of CPM dye (N-[4-(7-diethylamino-4-methyl-3-coumarinyl)phenyl]maleimide; Invitrogen) was prepared in DMSO (Sigma) at 4 mg ml–1 and diluted to 0.1 mg ml–1 in dilution buffer (20 mM HEPES pH 7.5, 150 mM NaCl) prior to use. The thermal denaturation assay was performed in a total volume of 130 μl. The KAI2 and KAI2ply2 proteins were diluted in the appropriate buffer to a final volume of 120 μl, adjusted to a final concentration of 51 μM. Then 10 μl of the diluted dye was added and thoroughly mixed with the protein. For the KAR-binding assay, 1000 μM (final concentration) of KAR1 was added. The reaction mixture was transferred within a 5 min period to a sub-micro quartz fluorometer cuvette (Starna Cells, Inc., Atascadero, CA) and heated in a controlled way with a ramp rate of 2 °C min–1 in a Cary Eclipse Spectrofluorometer. The excitation wavelength was set at 387 nm, while the emission wavelength was set to 463 nm. Assays were performed over a temperature range starting from 20 °C and ending at 90 °C.
Circular dichroism spectrometry
Circular dichroism (CD) spectra for KAI2 and KAI2ply2 were recorded on a Jasco J-815 spectropolarimeter. Molar ellipticity was calibrated with deionized water. Temperature was controlled with a NESLAB operation temperature system connected to a NESLAB water bath RTE-100. Using 1 mg of KAI2 or KAI2ply2 protein in 10 mM HEPES buffer (pH 7.5), thermal denaturation profiles were obtained at 220 nm, varying the temperatures from 10 °C up to 75 °C in 2 °C increments. The data were smoothed with SigmaPlot (Jandel Scientific), then the fraction of denatured protein as a function of temperature was calculated.
Isothermal titration calorimetry
Isothermal titration calorimetry (ITC) experiments were carried out at 25 °C in a MicroCal iTC200 calorimeter (GE Healthcare). The KAR1 compound dissolved in ITC buffer (20 mM HEPES pH 8.0, 50 mM sodium chloride) was injected in 2.0-μl increments into calorimetric cells containing 200 μl of KAI2 or KAI2ply2. The interval between injections was 150 s. Titration data were analysed using the Origin 7.0 data analysis software. Injections were integrated following manual adjustment of the baselines. Heats of dilution were determined from control experiments with the ITC buffer and subtracted prior to curve-fitting using a single set of binding site models.
Accession numbers
The co-ordinates of KAI2 and KAI2ply2 and the structure factors have been deposited in the Protein Data Bank with codes 5Z9G and 5Z9H, respectively. The accession numbers of genes used in this study are listed in Supplementary Table S3.
Results
Isolation of a pleiotropic long-hypocotyl mutant, ply2
To characterize genetic components that control light-dependent seedling development, we have screened long-hypocotyl mutants in Arabidopsis thaliana. In this study, we present genetic identification of a mutant designated as pleiotropic long hypocotyl2 (ply2). The ply2 mutant was originally identified from EMS-mutagenized pools under short-day conditions. When we grew the seedlings under different photoperiodic conditions, the elongated hypocotyl phenotype was more pronounced under short-day conditions compared to continuous light (see Supplementary Fig. S1A). Although the photoperiod-sensitive long-hypocotyl phenotype was reminiscent of mutants with an altered circadian rhythm, such as elf3-1 (Hazen et al., 2005), the ply2 mutation only marginally affected the circadian rhythm phenotype of CAB::LUC reporter activity (Supplementary Fig. S2). In line with these findings, unlike the mutants elf3-1 or phyB-9, the ply2 mutant exhibited normal flowering-time phenotypes under both long- and short-day conditions (Supplementary Fig. S1B). In addition, the ply2 mutant exhibited low germination rates under phytochrome B (phyB)- or phytochrome A (phyA)-dependent light conditions (Shinomura et al., 1994) (Supplementary Fig. S1C). We also found that the ply2 mutant was partially defective in light-responsive expression of some, but not all, light-regulated genes, including CAB, RBCS, STH7, and ELIP2 (Supplementary Fig. S1D). The pleiotropic effects suggested a critical role in light-dependent development, which led us to conduct further molecular analysis of the PLY2 gene.
ply2, a missense mutation of KAI2 that encodes a putative KAR receptor
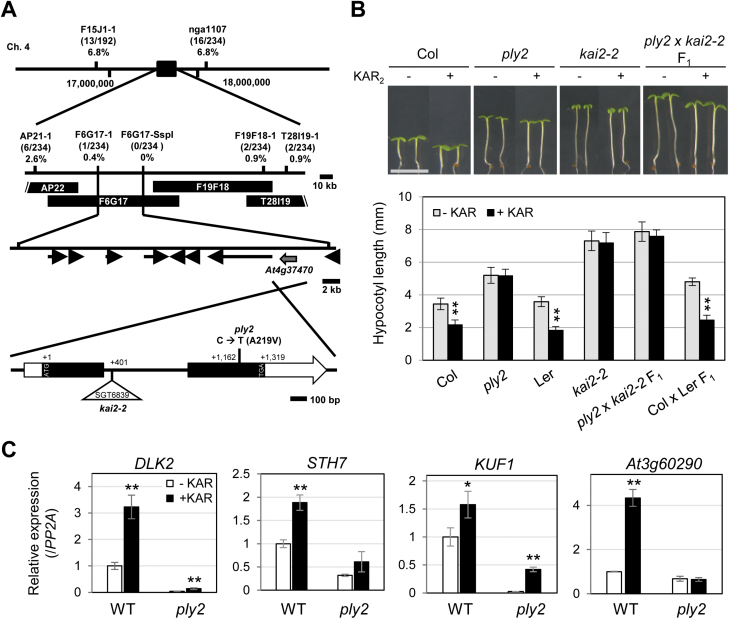
Genetic analysis indicated that the long-hypocotyl phenotype of ply2 under short-day conditions was inherited recessively. Subsequent F2 analysis showed a typical Mendelian segregation ratio of 3:1 (wild-type to mutant, 168:56), indicative of the monogenic nature of the ply2 mutation. To further understand the molecular nature of the PLY2 gene, we performed map-based cloning. Fine-mapping combined with candidate gene sequencing revealed that the ply2 mutant carried a C to T substitution mutation in the coding region of the At4g37470 gene, predicted to change Ala219 to Val in an α/β hydrolase fold protein (Fig. 1A). Next, we performed transgenic complementation by overexpressing the wild-type full-length cDNA of At4g37470 in the ply2 mutant. Multiple independent transgenic lines restored the long-hypocotyl phenotype as well as the defective light-induced germination phenotypes of the mutant (see Supplementary Fig. S3). These results confirmed that the missense mutation of At4g37470 was the responsible molecular lesion of the ply2 mutant. Two independent studies have identified the function of the same gene, At4g37470, designating it as HYPOSENSITIVE TO LIGHT (HTL) or KARRIKIN-INSENSITIVE2 (KAI2), which hereafter we refer to as KAI2 (Sun and Ni, 2011; Waters et al., 2012). To further confirm that the ply2 mutant phenotype was due to mutation of KAI2, we performed a complementation test between ply2 and kai2-2, a knock-out allele of KAI2. The F1 seedlings derived from crossing ply2 and kai2-2 exhibited a KAR-insensitive, long-hypocotyl phenotype, as did the parental single-mutants, indicating that ply2 is allelic to kai2-2 (Fig. 1B). The KAR-insensitivity of the ply2 mutant was also assessed with regards to the expression of the KAR-responsive genes STH7, DLK2, KUF1, and At3g60290 (Nelson et al., 2010; Stanga et al., 2013). As shown in Fig. 1C, the basal expression of these genes was down-regulated in the ply2 mutant compared with the wild-type. Furthermore, the KAR-inducibility of the genes was severely, if not completely, compromised in the ply2 mutant, suggesting a critical defect of KAI2ply2 in the KAR-responsive gene expression. These results suggested that the missense mutation Ala219Val of the KAI2 gene caused defects in KAR-sensitivity as well as in multiple aspects of light-responses in Arabidopsis.
Fig. 1.
Map-based cloning of PLY2. (A) Schematic diagram of map-based cloning. The PLY2 locus was initially mapped between the AP22 and T28I19-1 markers on chromosome 4. Fine-mapping narrowed the location to between the F6G17-1 and F6G17-SspI markers. The numbers in parenthesis indicate the numbers of recombinant chromatids. Candidate gene sequencing revealed that a substitution of 656th C to T occurred in the At4g37470 gene of the ply2 mutant, predicted to change the 219th alanine to valine (Ala219Val). The triangle indicates the transposon-insertion site in the kai2-2 mutant. (B) Complementation test between ply2 and kai2-2. The F1 plants of ply2 and kai2-2 were analysed for hypocotyl growth. The seedlings were sown on MS media and grown under short-day conditions (10/14 h light/dark) for 5 d with or without 10 μM KAR2 (KAR). The images show representative seedlings; scale bar, 5 mm. The graph shows the mean hypocotyl length (±SD) of at least 12 seedlings (n=12–18). Significant differences compared to the mock-treated control were determined using Student’s t-test; **P<0.01. (C) KAR-responsive gene expression in the ply2 mutant. Seedlings were grown under continuous light for 5 d. After treatment with either 10 μM KAR2 (+KAR) or 0.01% DMSO as control (–KAR) for 3 h, total RNAs were extracted and subjected to real-time RT-PCR analysis. The data are mean (±SD) values of relative expression level from experimental replicates (n=3), normalized to the level of PP2A expression in mock-treated wild-type (Col). Significant differences from the mock-treated control were determined using Student’s t-test; **P<0.01, *P<0.05.
Expression profiling of germination-related genes
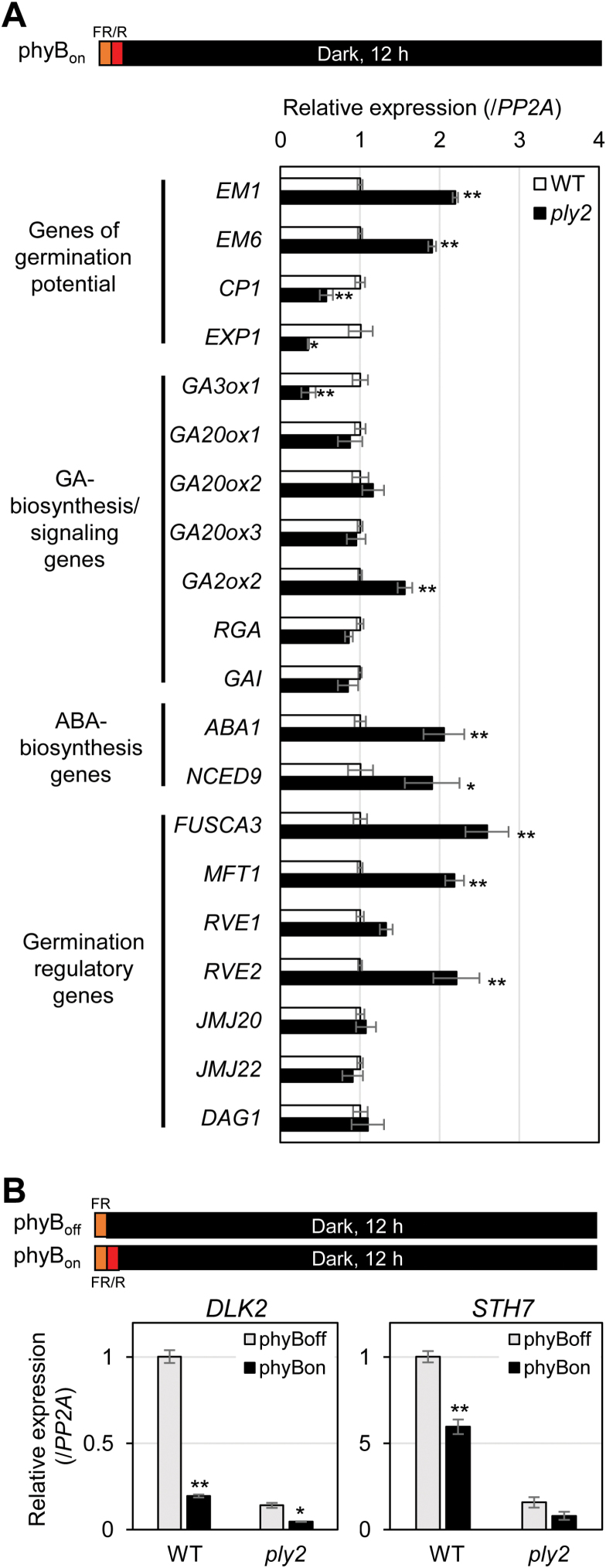
Several independent studies have reported the involvement of KL–KAI2 signaling in seed germination and light-dependent seedling development (Shen et al., 2007; Nelson et al., 2011; Sun and Ni, 2011; Waters et al., 2012). Shen et al. (2012) also showed that MAX2 controls phyB-dependent gene expression during germination in a PIF1-independent way, presumably through KL-signaling. To examine the effects of ply2 on light-responsive germination at the molecular level, we performed expression profiling under phyBon light conditions (treatment with FR light pulse/R light pulse followed by darkness) for genes with light-regulated expression (Fig. 2A and Supplementary Fig. S4). In accordance with the reduced germination rate, genes correlating with dormancy level or germination potential such as EM1, EM6, CP1, and EXP1 were significantly up- or down-regulated in the ply2 mutant. As shown in Fig. 2A, the expression of several genes including GA3ox1, GA2ox2, ABA1, NCED9, FUSCA3, MFT1, and RVE2 were significantly affected in the ply2 mutant compared to wild-type, being down- or up-regulated by at least 1.5-fold. In contrast, the ply2 mutation did not significantly affect the transcript level of other germination regulatory genes, including RGA, GAI, GA20ox1, GA20ox2, GA20ox3, JMJ20, JMJ22, DAG1, and RVE1. Together, these results showed that the reduced germination rate of ply2 under phyBon light conditions was accompanied by transcriptional changes of a subset of light-regulated genes, which included several ABA/GA-biosynthetic genes as well as germination regulatory genes. Notably, the altered expression profiles of the ply2 mutant largely overlapped with those of max2, as reported by Shen et al. (2012). In line with the hypothesis that KAI2–MAX2 mediated KL-signaling acts in a PIF1-independent pathway, we also found that ply2 could suppress the independent germination phenotype of the pif1 mutant (see Supplementary Fig. S5). Interestingly, when we investigated the effects of light on the expression of the primary KAR-target genes DLK2 and STH7, we found that they were down-regulated by red-light treatment (phyBon conditions). In the ply2 mutant, the reduced expression of DLK2 was further down-regulated by red-light treatment (Fig. 2B). The results implied that light and KAI2-signaling act largely independently with differential modes of interaction, functioning additively or oppositely, depending on the target genes.
Fig. 2.
Gene expression profiling during phytochrome B (phyB)-dependent germination. The experimental scheme is depicted at the top. For analysis of phyB-dependent gene expression, seeds were irradiated with far-red light for 15 min without (phyBoff) or with subsequent red light for 10 min (phyBon). After incubation in darkness for 12 h, total RNA was extracted and subject to real-time RT-PCR analysis. PP2A was used as the control. (A) Expression of germination-related genes under phyBon conditions. The relative expression of each gene in the ply2 mutant was normalized to that in the wild-type (WT), which was set to 1. Data are means (±SD) of experimental replicates (n=3). Significant differences from the expression level of the wild-type were determined using Student’s t-test; **P<0.01, *P<0.05. Similar trends were found with two independent seed batches and are shown in Supplementary Fig. S4. (B) Expression of the KAR-target genes DLK2 and STH7 under both phyBoff and phyBon conditions. The relative expression level of each gene was normalized to that of the wild-type under phyBoff conditions, which was set to 1. Data are means (±s.e.m.) from experimental replicates (n=6) with two independent seed batches. Significant differences from the expression level under phyBoff conditions were determined using Student’s t-test; **P<0.01, *P<0.05. (This figure is available in colour at JXB online.)
Double-mutant analysis of ply2ore9-1 and ply2smax1-3
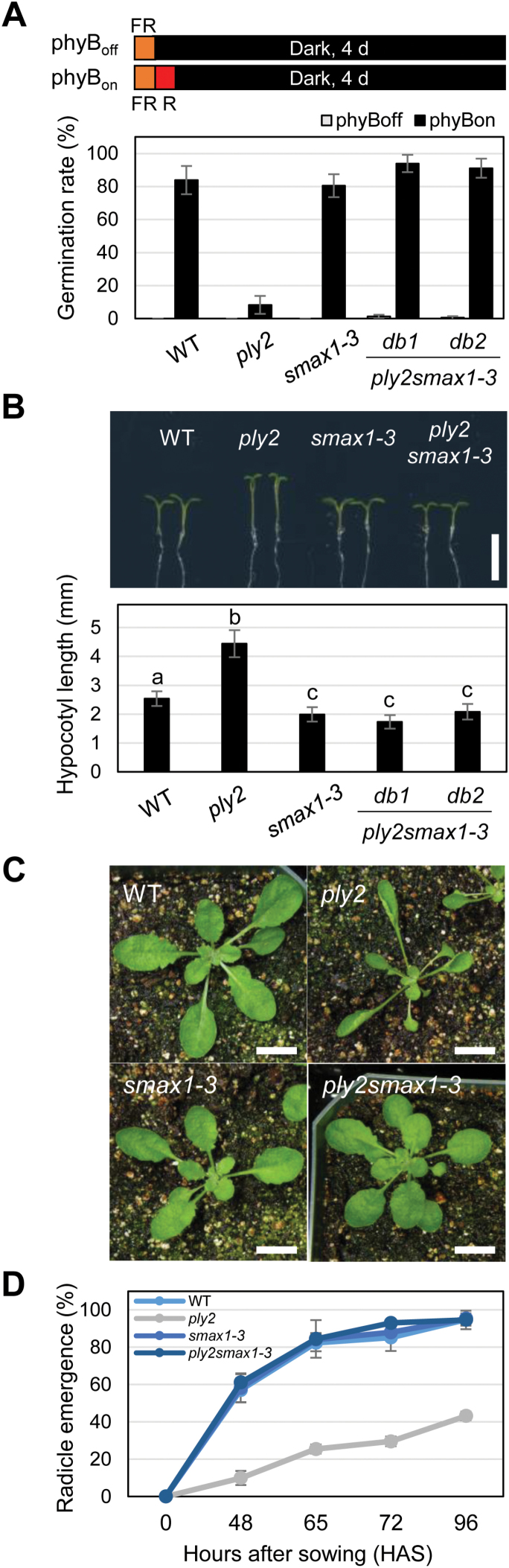
The SUPPRESSOR OF MAX2 1 (SMAX1) was originally identified as a genetic suppressor of the max2-1 mutant (Stanga et al., 2013) that rescues the KAR-dependent phenotypes of the mutant, while not affecting SL-dependent defects such as increased branching. Based on the working hypothesis that KAI2 acts together with MAX2-SMAX1 in the KL-signaling pathway (Morffy et al., 2016), we hypothesized that the smax1-3 mutant would restore all of the developmental defects of the ply2 mutant. To assess this hypothesis, we generated the double-mutant ply2smax1-3 by genetic crossing. The reduced germination phenotype of ply2 under phyBon light conditions was restored in the double-mutant (Fig. 3A). In addition, other phenotypic defects of ply2, including the long hypocotyl, elongated petiole, and deep seed dormancy, were also suppressed by the smax1-3 mutation (Fig. 3B–D). These results were in good agreement with the hypothesis that SMAX1 acts as a negative regulator downstream of KL–KAI2-signaling.
Fig. 3.
Double-mutant analysis of ply2 and smax1. (A) Phytochrome B (phyB)-dependent germination of the wild-type (WT, Col), ply2, smax1-3, and ply2smax1-3 double-mutant. After treatment with far-red light for 10 min, the seeds were then either irradiated (phyBon) or not (phyBoff) with red light for 10 min, and then kept in darkness for 4 d. Mean values (±SD) of germination rates were determined from three experimental replicates with at least 50 seeds per plate. (B) Hypocotyl elongation assay. The images show representative 6-d-old seedlings of each genotype grown on half-strength MS-SUC (0.5%) media under short-day conditions (10/14 h light/dark); scale bar, 5 mm. The graph shows the mean (±SD) hypocotyl length of the seedlings (n=15–18). Different letters indicate significant differences at P<0.01, analysed by one-way ANOVA with a post hoc Tukey HSD test. (C) Leaf morphology of 30-d-old plants. The plants were grown under short-day conditions (10/14 h light/dark); scale bars, 10 mm. (D) Primary dormancy assay. Freshly harvested seeds of each genotype were sown on aqueous media and incubated under continuous light conditions. Mean values (±SD) of germination rate were determined at different times after sowing for three experimental replicates with at least 50 seeds per plate.
To substantiate the hypothesis that the pleiotropic defects of ply2 are due to impaired KL–KAI2-signaling, we performed a double-mutant analysis of ply2 and the ore9-1 mutant, a null allele of MAX2 (Woo et al., 2001). The results showed that the ply2ore9-1 double-mutant exhibited essentially the same responses compared to the parental single-mutants, ply2 and ore9-1, without any noticeable additivity or synergism with regards to KAR-responsive hypocotyl elongation and light-responsive anthocyanin accumulation (Supplementary Fig. S6).
Functional aspects of the KAI2ply2 mutation
Given the phenotypic severity of the ply2 mutant, which was as severe as a null-mutant of KAI2 or MAX2 (Fig. 1, Supplementary Fig. S6), it was intriguing how a single amino acid substitution, Ala219Val, could compromise the function of KAI2. It is notable that the mutated residue in ply2, Ala219, is one of the highly conserved amino acids specifically among KAI2-clade homologous proteins (see Supplementary Fig. S7). Given that it has been proposed that the AtD14 and KAI2 clade proteins have evolved to sense and mediate distinct downstream signaling of different ligands, SL and KL, respectively (Conn et al., 2015; Toh et al., 2015; Waters et al., 2015b; Xu et al., 2016; Yao et al., 2017), it seemed plausible that the mutated residue might be functionally critical for ligand-specific binding/signaling. Therefore, we investigated the molecular and biochemical features of KAI2ply2 to determine how KAI2A219V affects its receptor function.
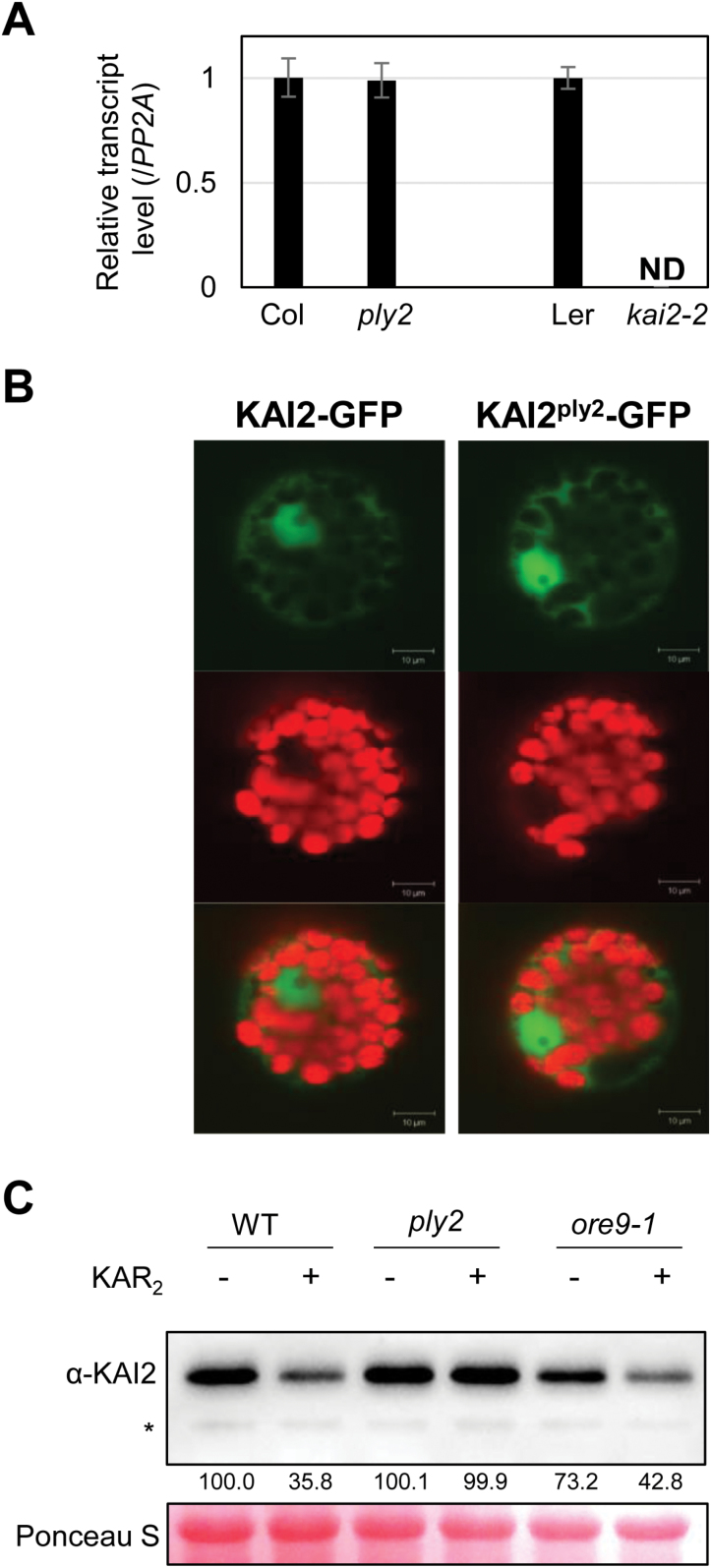
Expression analysis showed no significant differences in the KAI2 transcript level between the wild-type and ply2 mutant, precluding any effect of the mutation on the transcriptional expression of KAI2 (Fig. 4A). Next, we examined whether the ply2 mutation altered the subcellular localization of KAI2. After transient expression of KAI2-GFP or KAI2ply2-GFP constructs in protoplasts, we performed confocal microscopy analysis. As previously reported by Sun and Ni (2011), both KAI2 and KAI2ply2 were shown to be localized in the cytosol as well as in the nucleus (Fig. 4B), indicating that the ply2 mutation did not alter the subcellular localization of KAI2. When we performed immunoblot analysis, contrary to expectation, the amount of KAI2ply2 protein was not altered in the ply2 mutant compared to the wild-type (Fig. 4C). Taking into consideration recent results indicating that KAR triggers degradation of KAI2 (Waters et al., 2015a), we also checked whether the ply2 mutant was defective in this regard, and found it to be the case. The results suggested that the increased level of KAI2ply2 in the ply2 mutant might have resulted from defects in signaling for KAI2 turnover, which would constitute a negative feedback regulation of KAI2-signaling.
Fig. 4.
Functional assay of the KAI2ply2 mutation. (A) Expression analysis of the KAI2 transcript. Total RNA from 5-d-old light-grown seedlings was used for qRT-PCR analysis. PP2A was used as a loading control. The relative expression of the KAI2 transcript in the ply2 or kai2-2 mutant was compared to that in the wild-type, which was set to 1. Data are means (±SD) of experimental replicates (n=3). ND, not detected. (B) Subcellular localization of KAI2. The wild-type KAI2-GFP or KAI2ply2-GFP constructs were transfected to Arabidopsis mesophyll protoplasts. Representative images taken by confocal microscopy are shown: GFP signal (upper), chloroplast auto-fluorescence (middle), merged image (lower). (C) Immunoblot analysis of the KAI2 protein. Total proteins were extracted from seedlings grown under continuous light for 7 d after treatment with a mock solution (–) or 10 μM KAR2 (+) for 2 h. The KAI2 protein (29.8 kDa) was detected using an α-KAI2 antibody. The transferred proteins were stained with Ponceau S solution as a loading control. Numbers below the bands indicate the relative densitometry values of the KAI2 protein band, normalized to those of the loading control. The asterisk indicates a non-specific protein.
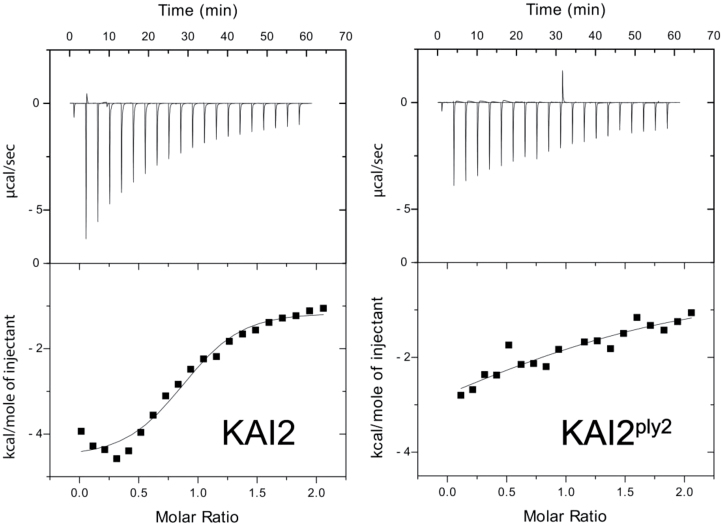
Being a KAR-receptor, the KAI2 protein has been shown to bind with KAR by various biochemical assays (Guo et al., 2013; Kagiyama et al., 2013; Toh et al., 2014). To assess the possibility that KAI2ply2 was defective in ligand-binding activity, an isothermal titration calorimetry (ITC) assay was used to determine the binding properties of KAI2 and KAI2ply2 towards KAR1 (3-methyl-2H-furo[2,3-c]pyran-2-one). As shown in Fig. 5, KAI2 and KAI2ply2 exhibited different thermodynamic properties upon application of KAR1. The results showed that the dissociation constant (Kd) of KAI2ply2 towards KAR1 was dramatically increased to 2857 μM, about 20-fold higher than that of KAI2 (147 μM) (see Supplementary Table S1), indicating that the KAI2ply2 mutation impaired its binding activity for KAR. Under the same experimental conditions, the KAI2S95A mutant protein displayed very low (indeed hardly detected) binding activity toward KAR1 (results not shown), supporting the idea that the Ser95 residue, one of the catalytic triad of KAI2, is critical for the biological function and ligand-binding activity (Waters et al., 2015b). The results suggested that the mutant protein KAI2ply2 had greatly reduced ligand-binding activity. We also assessed the ligand-binding activity by a N-[4-(7-diethylamino-4-methyl-3-coumarinyl)phenyl]maleimide (CPM) assay. Upon addition of KAR1, the wild-type KAI2 protein exhibited a shifted thermal function of fluorescence, indicating that KAI2 undergoes changes in conformational flexibility caused by KAR1 (Supplementary Fig. S8). In contrast, KAI2ply2 showed only a marginal change upon application of KAR1, implying that KAI2ply2 was defective in ligand-induced conformational changes. It was also notable that the KAI2 and KAI2ply2 proteins exhibited different values for melting temperature (Tm) in the absence of KAR1; the Tm values of KAI2 and KAI2ply2 were calculated to be 36.39 °C and 45.60 °C, respectively, indicating that thermal stability of KAI2ply2 was increased (see below).
Fig. 5.
Reduced ligand binding activity of KAI2ply2. Isothermal titration calorimetry (ITC) experiments were performed by titrating KAR1 into recombinant KAI2 or KAI2ply2 protein. The upper panels show raw ITC data. The lower panels present data for the enthalpy change derived from the upper panels. Corresponding thermodynamic properties of KAI2 and KAI2ply2 are shown in Supplementary Table S1.
Structural comparison of KAI2 and KAI2ply2
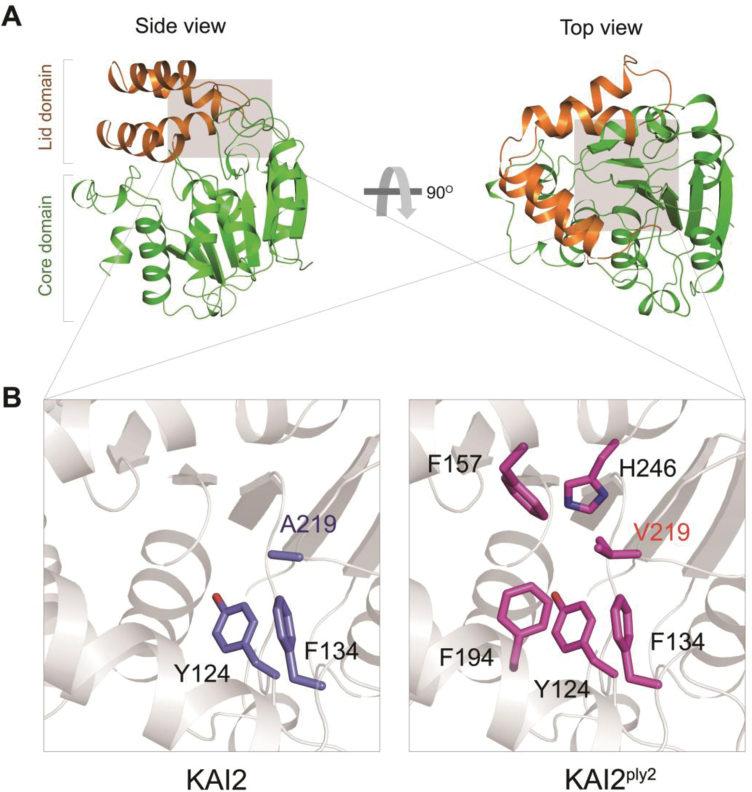
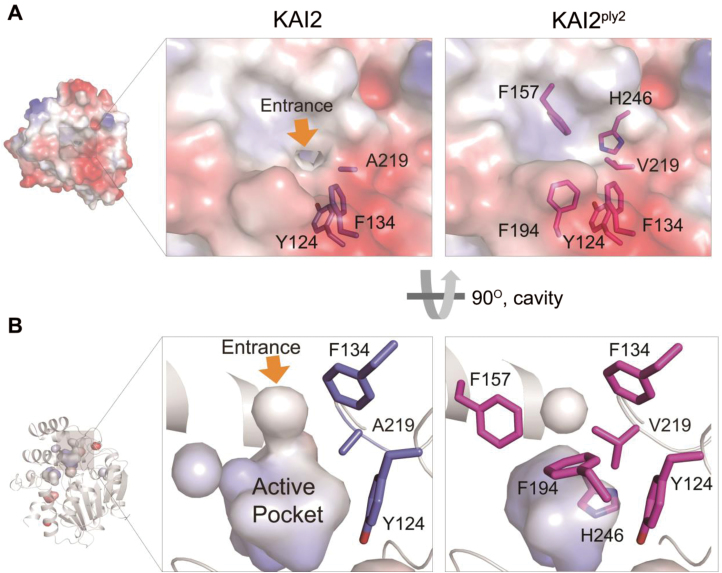
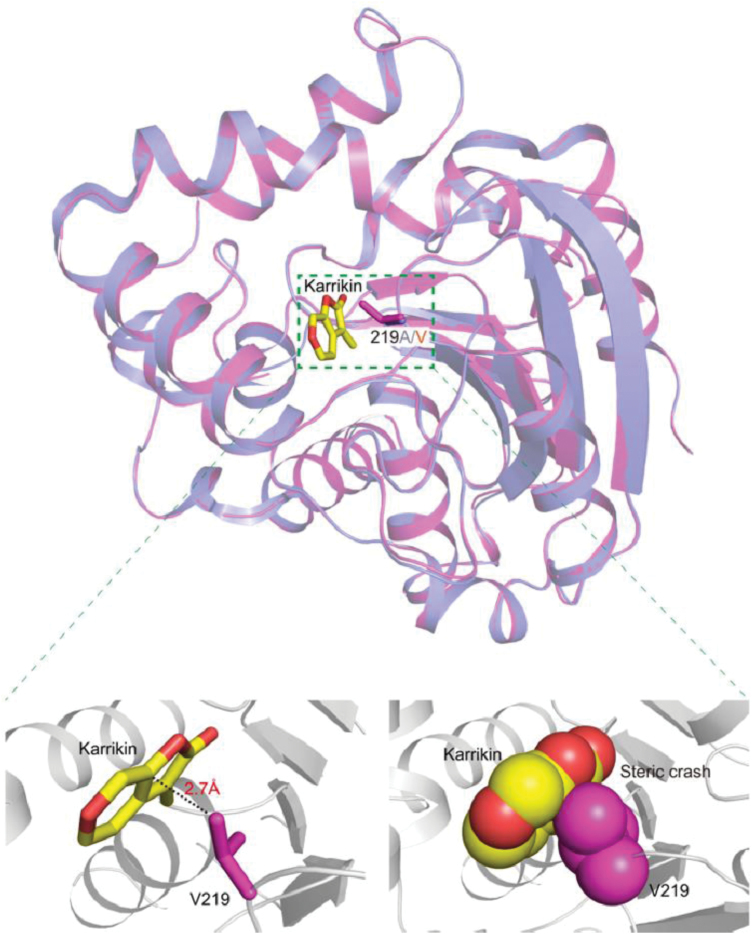
To gain further insight into the molecular lesion of the KAI2ply2 mutant protein, we performed X-ray crystallography to assess any structural alterations. Recombinant proteins of KAI2 and KAI2ply2, purified from E. coli, were crystallized. The crystal structures were solved and refined at 1.9 Å by molecular replacement (MR) using a search model based on the bacterial signaling protein RsbQ (PDB ID code 1WOM) (Kaneko et al., 2005) (Supplementary Table S2). As reported previously (Bythell-Douglas et al., 2013; Guo et al., 2013; Kagiyama et al., 2013; Zhao et al., 2013; Xu et al., 2016), the structure of KAI2 displayed a canonical compact α/β-hydrolase domain that contained an α/β ‘core’ domain and a ‘lid’ domain (Fig. 6A) (Nardini and Dijkstra, 1999). Comparison of KAI2 and KAI2ply2 showed that there was little structural difference (r.m.s.d., 0.2 Å) in overall structure. However, in detail, we found that the residue A219 of KAI2 was bound to aromatic residues as Y124 and F134 of the lid domain by hydrophobic interactions, while the residue V219 of KAI2ply2 was bound to extended aromatic residues as Y124, F134, F157, F194, and H246 of the lid domain (Fig. 6B). Thus, the mutation Ala219Val caused stronger hydrophobic interactions with more residues. Moreover, the electrostatic potential surface model and cavity view showed another structural alteration of KAI2ply2. The entrance of KAI2 presents an opened pocket and hydrophobic charge for the ligand, whereas the entrance of KAI2ply2 forms a closed pocket due to C-gamma 1 and C-gamma 2 of V219, which is a bulkier residue than alanine (Fig. 7). Thus, the narrowed entrance gate might contribute to the reduced ligand-binding ability of KAI2ply2. Furthermore, when we performed superimposition with the KAR1-bound KAI2 model previously reported by Guo et al. (2013), the V219 was predicted to cause a steric clash with the bound KAR (Fig. 8). Thus, collectively, these structural alterations may underlie the reduced binding affinity of KAI2ply2 to KAR (Fig. 5). While it is still open to question how KL binds to KAI2, the A219 residue may take part in ligand-specific binding of KAI2.
Fig. 6.
Three-dimensional X-ray crystal structure of KAI2 and KAI2ply2. (A) Left, a side view of KAI2. The ribbon representation of the overall structure of KAI2 is shown. Brown and green represent the lid and core domains, respectively. Right, a top view of KAI2: a 90° rotation of KAI2 relative to the side view is shown. (B) Magnified views of the areas in (A) shaded grey, showing residues forming hydrophobic interactions with A219 of KAI2 (left, indicated in blue) and V219 of KAI2ply2 (right, indicated in magenta).
Fig. 7.
Structural comparison of KAI2 and KAI2ply2. (A) Surface views of KAI2 and KAI2ply2. KAI2 exhibits a small opening at the top, forming an entrance hole into the ligand-binding pocket (yellow arrow). Note that the entrance is closed in the KAI2ply2 protein. (B) Close-up view of the cavity on the active, putative ligand-binding pocket. Note that KAI2 shows an open-gate conformation with continuous space, whereas KAI2ply2 shows a closed gate with discontinuous space.
Fig. 8.
Structural comparison between KAI2ply2 and the KAI2–KAR1 complex (PDB:4JYM). Superimposition of KAI2ply2 (magenta) and KAI2–KAR1 (light blue; karrikin, yellow). The r.m.s.d. score between KAI2ply2 and KAI2-KAR1 is 0.2. The close-up views (below) shows a stick (left) and a sphere (right) model of KAR1 and the Val219 residue.
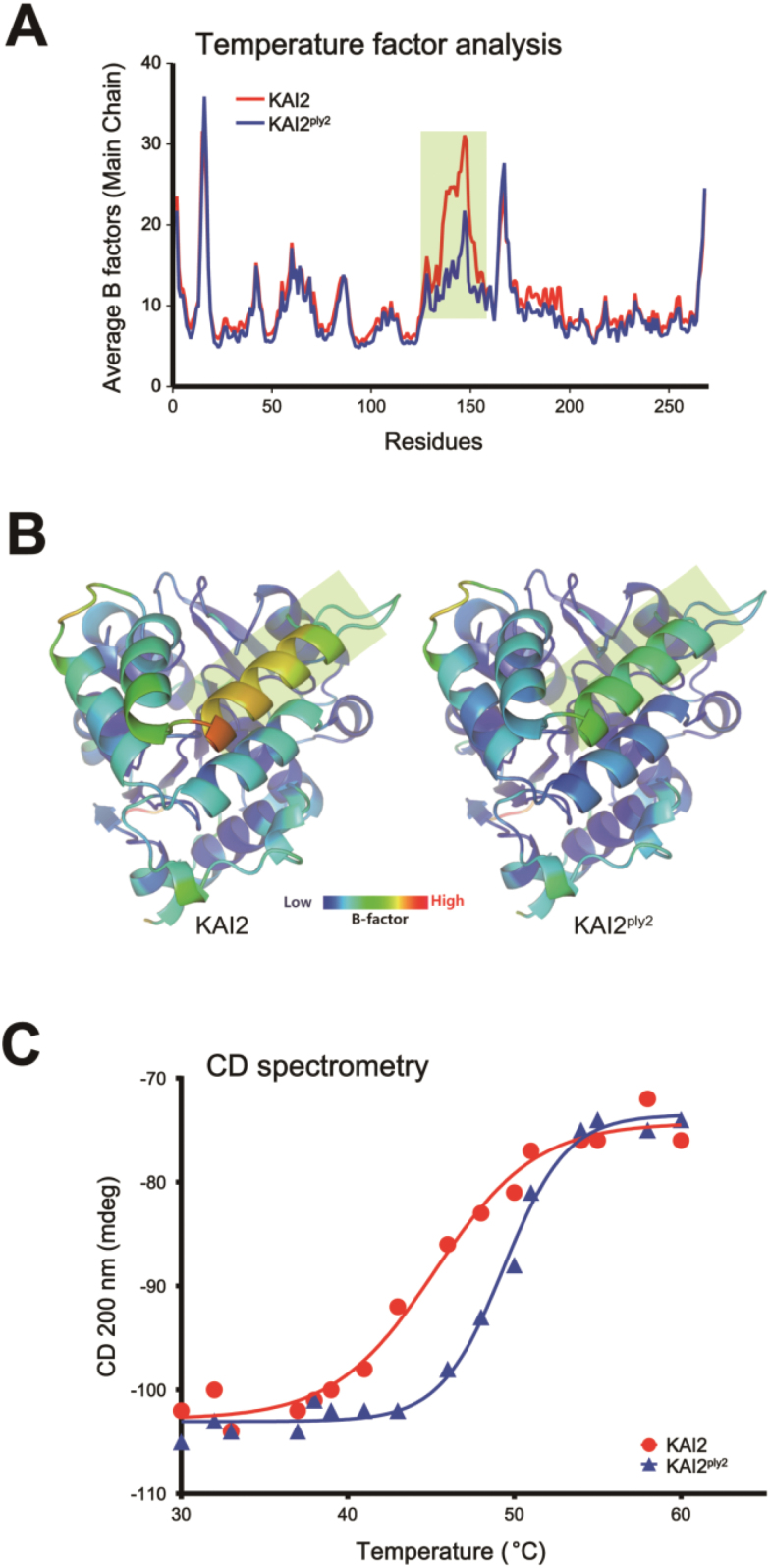
To investigate any structural impacts of the predicted increased hydrophobic interaction in KAI2ply2 (Fig. 6B), we performed B-factor analysis with electron density maps from protein crystals. The results showed that the amino acid residues of KAI2 ranging from 125–150 exhibited a higher B-factor than those of KAI2ply2, implying that this region, corresponding to the helical ‘lid’ domain, might be more rigid with lower motional amplitude in the KAI2ply2 protein (Fig. 9A, B). To assess the predicted biochemical features of KAI2ply2, we examined the thermal stability of KAI2 and KAI2ply2 by circular dichroism (CD) spectroscopy. We found that the Tm of the KAI2ply2 protein was increased compared to that of the wild-type KAI2 under CD assay conditions; the Tm values of KAI2 and KAI2ply2 were calculated to be 45.23 °C and 49.36 °C, respectively (Fig. 9C). The results suggested that KAI2ply2 was more thermally stable than the wild-type KAI2 protein, presumably due to increased hydrophobic interactions and reduced flexibility. Taken together, these results suggested that the Ala219Val substitution mutation of KAI2 not only impaired ligand-binding but also reduced the conformational flexibility of the lid domain, both of which would be critical for ligand perception and downstream signaling.
Fig. 9.
Comparison of thermal stability between KAI2 and KAI2ply2. (A) B-factor analysis of protein crystals for KAI2 and KAI2ply2. The residues ranging from 120–150 (highlighted) of KAI2 showed higher B factor values compared to those of KAI2ply2. (B) Ribbon diagram of B-factors of KAI2 and KAI2ply2. Note that the residues showing clear differences between KAI2 and KAI2 ply2 in (A) correspond to the helix of the cap domains (shaded areas). (C) Circular dichroism (CD) spectrometry assay. The melting temperature (Tm) of KAI2 is 45 °C and for KAI2ply2 it is 50 °C.
Discussion
A hypothetical molecule, termed KL, for KAI2-ligand, has recently been recognized as a potentially new plant hormone that is sensed by a receptor, KAI2 (Flematti et al., 2013; Conn and Nelson, 2016). Despite its unknown chemical nature and its biosynthetic pathway, a series of molecular genetic analyses of the kai2 mutant have uncovered the biological functions of KL, including promotion of seed germination, reduced seedling de-etiolation under light conditions, drought adaptations, and formation of AMF associations (Waters et al., 2014; De Cuyper et al., 2017). However, the detailed mode of action of KAI2 in the ligand-binding and signaling remains to be identified.
In this study, we showed that the ply2 mutation, a missense allele with Ala219Val substitution, caused loss of function of KAI2 (Fig. 1, Supplementary Figs S1, S3). The pleiotropic developmental defects of ply2 were reminiscent of phenotypes of mutants defective in KL-signaling, kai2 and max2, as reported previously (Shen et al., 2007; Sun and Ni, 2011; Waters et al., 2012; Toh et al., 2014) (Figs 1, 2, Supplementary Figs S1, S5). In addition, the genetic epistasis between ply2 and smax1-3 suggested that the pleiotropic defect of ply2 is due to impaired KL-signaling (Fig. 3), providing genetic evidence that that SMAX1 acts in a pathway downstream of KAI2 as a negative regulator of KL-signaling (Stanga et al., 2013).
It was notable that the phenotypic severity of the ply2 mutation was comparable to kai2-2 and ore9-1, null-mutants of KAI2 and MAX2, respectively (Fig. 1, Supplementary Fig. S5), suggesting that KAI2ply2 is a strong, if not null, loss-of-function allele. Intriguingly, the Ala219 of KAI2 is highly conserved in the KAI2-clade proteins among KAI2 homologs (see Supplementary Fig. S7) and has been annotated as one of the putative substrate-determining residues among KAI2/D14 family proteins (Chevalier et al., 2014). Our results from biochemical and structural analyses provided several clues as to how the single amino acid substitution (Ala219Val) impairs the receptor function of KAI2. ITC experiments with recombinant KAI2 and KAI2ply2 proteins indicated that the KAR-binding activity of KAI2 was greatly (~20-fold) reduced by the ply2 mutation (Fig. 5, Supplementary Table S1). The reduced KAR-binding ability of KAI2ply2 was also verified by CPM assays (Supplementary Fig. S8). In addition, X-ray crystallography studies including B-factor analyses showed that KAI2ply2 forms a narrower gate for the entrance into the ligand-binding pocket, and forms a more rigid helical lid domain, presumably due to enhanced hydrophobic interactions, which do not affect the overall structural integrity (Figs 6, 7, 9). Thus, the Ala219 of KAI2 appears to be important in forming space for entry of the ligand as well as maintaining flexibility of the lid domain. By analogy with the current working model of strigolactone and its receptor, D14 (Yao et al., 2016), it is conceivable that the lid domain of KAI2 may take part in a ligand-dependent protein–protein interaction. In this scenario, the more rigid lid domain of KAI2ply2 would hamper the conformational changes upon ligand-binding, resulting in reduced KAI2 signaling. Furthermore, our homology modelling of KAI2ply2 with the KAR1-bound KAI2 model proposed by Guo et al. (2013) suggested that the substituted Val219 might impair ligand binding through steric hindrance (Fig. 8). However, this should be considered with caution, as it has not been unequivocally demonstrated that the KAR1-bound KAI2 model represents the active state of KAI2. Overall, these results indicate that the single missense mutation Ala219Val altered multiple biochemical features of KAI2ply2, accounting for its severe, if not complete, insensitivity to KAR or endogenous KL.
To date, only a few missense mutations of KAI2 have been characterized, including G133E and S95A (Waters et al., 2015b). While G133 was shown to be important for protein integrity, S95, together with amino acids forming the catalytic triad of the α/β hydrolase fold, has been suggested to be critical for ligand binding. However, it remains unknown whether S95 catalyses hydrolysis of the pro-ligand, as D14 does with SL (Yao et al., 2016) or if it participates in binding directly with the ligand via bridging molecule, as ShKAI2iB does (Xu et al., 2016). Our identification of A219 as a critical residue for ligand binding and possibly signaling should help to determine the binding mode of KL–KAR–KAI2. Since the endogenous KL has not yet been identified, it is still speculative how KAI2 senses its ligand in order to transmit downstream signaling. Characterization of additional informative missense mutations of KAI2 by structure-guided design, mutant screening, or targeting-induced local lesions in genomes (TILLING) would help to further identify the molecular consequences during ligand perception and signal transmission of KAI2, as elegantly demonstrated by recent analysis of the d14-5 allele, AtD14G158E, a missense mutant of a SL receptor (Yao et al., 2016).
Taking together with recent studies showing that KAI2-signaling is also required for AMF formation and drought tolerance (Gutjahr et al., 2015; Li et al., 2017), it is tempting to speculate that endogenous KL levels and subsequent KAI2-signaling may represent a hormonal/metabolic signal that sensitizes or desensitizes plants to various environmental fluctuations. For example, as shown in Fig. 2 and by Shen et al. (2012), the loss of KL–KAI2 signaling may set a higher threshold of light for phytochrome-dependent seed germination by up-regulation of germination-inhibitory genes such as FUSCA3, RVE2, GA2ox2, and ABA1 as well as by down-regulation of germination-promoting genes such as GA3ox1. As such, KAI2-signaling may intersect with other endogenous/environmental stimuli, controlling a subset of downstream target genes. More sophisticated experimental designs together with identification of genome-wide KL-target genes under specific physiological conditions will shed light on the additional regulatory functions of KL–KAI2-signaling in higher plants.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Identification of the ply2 mutant with a long-hypocotyl phenotype.
Fig. S2. The circadian rhythm phenotype of the ply2 mutant.
Fig. S3. Transgenic complementation of the ply2 mutant.
Fig. S4. Additional biological replications of gene expression profiling under phyB-dependent germination conditions.
Fig. S5. Double-mutant analysis using ply2 and pif1 mutants.
Fig. S6. Double-mutant analysis using ply2 and ore9-1 mutants.
Fig. S7. Multiple sequence alignments of the KAI2, AtD14, and DLK2 homologs.
Fig. S8. CPM assays of KAI2 and KAI2ply2.
Table S1. Thermodynamic properties determined by ITC.
Table S2. Statistics for X-ray data collection and refinement.
Table S3. Primers used in this study.
Acknowledgements
We thank ABRC and NASC for providing the Arabidopsis mutant lines. We are also grateful to the staff scientists at the BL-5C beamline of the Pohang Accelerator Laboratory for data collection. This work was supported by grants from the Basic Science Research Program through a National Research Foundation (NRF) grant funded by the Korean government (2015R1D1A1A01058039 to MSS, 2015R1C1A2A01052950 to SL), the Mid-career Researcher Program through a NRF grant funded by the Korean government (MEST) (nos. NRF-2016R1A2B2013305, -2016R1A5A1010764, to HSC), the Strategic Initiative for Microbiomes in Agriculture and Food funded by the Ministry of Agriculture, Food and Rural Affairs (916006-2, to HSC), the Rural Development Administration [Next-Generation Biogreen21 program (Agricultural Biotechnology Research Center no. PJ01369001) to SP] and by the Institute for Basic Science (IBS-R013-D1 to HGN).
Glossary
Abbreviations:
- CPM
N-[4-(7-diethylamino-4-methyl-3-coumarinyl)phenyl]maleimide
- FR
far-red
- ITC
isothermal titration calorimetry
- KAI2
KARRIKIN INSENSITIVE2
- KAR
karrikin
- KL
KAI2-ligand
- MAX2
MORE AXILLARY GROWTH2
- MS medium
Murashige–Skoog medium
- phyB
phytochrome B
- PIF1
PHYTOCHROME INTERACTING FACTOR1
- PLY2
PLEIOTRIPIC LONG HYPOCOTYL2
- R
red
- SL
strigolactone
- SMAX1
SUPPRESSOR OF MAX2 1
References
- Alexandrov AI, Mileni M, Chien EY, Hanson MA, Stevens RC. 2008. Microscale fluorescent thermal stability assay for membrane proteins. Structure 16, 351–359. [DOI] [PubMed] [Google Scholar]
- Ayele B, Magome H, Lee S, Shin K, Kamiya Y, Soh M-S, Yamaguchi S. 2014. GA-sensitive dwarf1-1D (gsd1-1D) defines a new mutation that controls endogenous GA levels in Arabidopsis. Journal of Plant Growth Regulation 33, 340–354. [Google Scholar]
- Bae G, Choi G. 2008. Decoding of light signals by plant phytochromes and their interacting proteins. Annual Review of Plant Biology 59, 281–311. [DOI] [PubMed] [Google Scholar]
- Bythell-Douglas R, Waters MT, Scaffidi A, Flematti GR, Smith SM, Bond CS. 2013. The structure of the karrikin-insensitive protein (KAI2) in Arabidopsis thaliana. PLoS ONE 8, e54758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ. 2013. Photoreceptor signaling networks in plant responses to shade. Annual Review of Plant Biology 64, 403–427. [DOI] [PubMed] [Google Scholar]
- Chen M, Chory J, Fankhauser C. 2004. Light signal transduction in higher plants. Annual Review of Genetics 38, 87–117. [DOI] [PubMed] [Google Scholar]
- Chevalier F, Nieminen K, Sánchez-Ferrero JC, Rodríguez ML, Chagoyen M, Hardtke CS, Cubas P. 2014. Strigolactone promotes degradation of DWARF14, an α/β hydrolase essential for strigolactone signaling in Arabidopsis. The Plant Cell 26, 1134–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiwocha SDS, Dixon KW, Flematti GR, Ghisalberti EL, Merritt DJ, Nelson DC, Riseborough J-AM, Smith SM, Stevens JC. 2009. Karrikins: a new family of plant growth regulators in smoke. Plant Science 177, 252–256. [Google Scholar]
- Choi H, Jeong S, Kim DS, Na HJ, Ryu JS, Lee SS, Nam HG, Lim PO, Woo HR. 2014. The homeodomain-leucine zipper ATHB23, a phytochrome B-interacting protein, is important for phytochrome B-mediated red light signaling. Physiologia Plantarum 150, 308–320. [DOI] [PubMed] [Google Scholar]
- Christie JM, Blackwood L, Petersen J, Sullivan S. 2015. Plant flavoprotein photoreceptors. Plant & Cell Physiology 56, 401–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Conn CE, Bythell-Douglas R, Neumann D, et al. . 2015. Convergent evolution of strigolactone perception enabled host detection in parasitic plants. Science 349, 540–543. [DOI] [PubMed] [Google Scholar]
- Conn CE, Nelson DC. 2016. Evidence that KARRIKIN-INSENSITIVE2 (KAI2) receptors may perceive an unknown signal that is not karrikin or strigolactone. Frontiers in Plant Science 6, 1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Cuyper C, Struk S, Braem L, Gevaert K, De Jaeger G, Goormachtig S. 2017. Strigolactones, karrikins and beyond. Plant, Cell & Environment 40, 1691–1703. [DOI] [PubMed] [Google Scholar]
- de Lucas M, Prat S. 2014. PIFs get BRright: PHYTOCHROME INTERACTING FACTORs as integrators of light and hormonal signals. New Phytologist 202, 1126–1141. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallographica. Section D, Biological Crystallography 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Flematti GR, Ghisalberti EL, Dixon KW, Trengove RD. 2004. A compound from smoke that promotes seed germination. Science 305, 977. [DOI] [PubMed] [Google Scholar]
- Flematti GR, Waters MT, Scaffidi A, Merritt DJ, Ghisalberti EL, Dixon KW, Smith SM. 2013. Karrikin and cyanohydrin smoke signals provide clues to new endogenous plant signaling compounds. Molecular Plant 6, 29–37. [DOI] [PubMed] [Google Scholar]
- Franklin KA, Quail PH. 2010. Phytochrome functions in Arabidopsis development. Journal of Experimental Botany 61, 11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvão VC, Fankhauser C. 2015. Sensing the light environment in plants: photoreceptors and early signaling steps. Current Opinion in Neurobiology 34, 46–53. [DOI] [PubMed] [Google Scholar]
- Guo Y, Zheng Z, La Clair JJ, Chory J, Noel JP. 2013. Smoke-derived karrikin perception by the α/β-hydrolase KAI2 from Arabidopsis. Proceedings of the National Academy of Sciences, USA 110, 8284–8289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutjahr C, Gobbato E, Choi J, et al. . 2015. Rice perception of symbiotic arbuscular mycorrhizal fungi requires the karrikin receptor complex. Science 350, 1521–1524. [DOI] [PubMed] [Google Scholar]
- Hazen SP, Schultz TF, Pruneda-Paz JL, Borevitz JO, Ecker JR, Kay SA. 2005. LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proceedings of the National Academy of Sciences, USA 102, 10387–10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins GI. 2014. The UV-B photoreceptor UVR8: from structure to physiology. The Plant Cell 26, 21–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Liu X, Xiong G, et al. . 2013. DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504, 401–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagiyama M, Hirano Y, Mori T, Kim SY, Kyozuka J, Seto Y, Yamaguchi S, Hakoshima T. 2013. Structures of D14 and D14L in the strigolactone and karrikin signaling pathways. Genes to Cells 18, 147–160. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Tanaka N, Kumasaka T. 2005. Crystal structures of RsbQ, a stress-response regulator in Bacillus subtilis. Protein Science 14, 558–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. 2002. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science 7, 193–195. [DOI] [PubMed] [Google Scholar]
- Kim J, Kim Y, Yeom M, Kim JH, Nam HG. 2008. FIONA1 is essential for regulating period length in the Arabidopsis circadian clock. The Plant Cell 20, 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau OS, Deng XW. 2010. Plant hormone signaling lightens up: integrators of light and hormones. Current Opinion in Plant Biology 13, 571–577. [DOI] [PubMed] [Google Scholar]
- Lee S, Lee S, Yang KY, Kim YM, Park SY, Kim SY, Soh MS. 2006. Overexpression of PRE1 and its homologous genes activates gibberellin-dependent responses in Arabidopsis thaliana. Plant & Cell Physiology 47, 591–600. [DOI] [PubMed] [Google Scholar]
- Leivar P, Monte E. 2014. PIFs: systems integrators in plant development. The Plant Cell 26, 56–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E, Quail PH. 2008. Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Current Biology 18, 1815–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Tepperman JM, Monte E, Calderon RH, Liu TL, Quail PH. 2009. Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. The Plant Cell 21, 3535–3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Nguyen KH, Chu HD, et al. . 2017. The karrikin receptor KAI2 promotes drought resistance in Arabidopsis thaliana. PLoS Genetics 13, e1007076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCt method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Morffy N, Faure L, Nelson DC. 2016. Smoke and hormone mirrors: action and evolution of karrikin and strigolactone signaling. Trends in Genetics 32, 176–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatani A, Matsushita T. 2002. Light-induced nuclear targeting of phytochromeB-sgreen fluorescent protein in plants. In: Hicks BW, ed. Methods in molecular biology, vol. 183. Humana Press, 163–170. [DOI] [PubMed] [Google Scholar]
- Nardini M, Dijkstra BW. 1999. α/β Hydrolase fold enzymes: the family keeps growing. Current Opinion in Structural Biology 9, 732–737. [DOI] [PubMed] [Google Scholar]
- Neff MM, Neff JD, Chory J, Pepper AE. 1998. dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. The Plant Journal 14, 387–392. [DOI] [PubMed] [Google Scholar]
- Nelson DC, Flematti GR, Riseborough J-A, Ghisalberti EL, Dixon KW, Smith SM. 2010. Karrikins enhance light responses during germination and seedling development in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 107, 7095–7100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DC, Riseborough JA, Flematti GR, Stevens J, Ghisalberti EL, Dixon KW, Smith SM. 2009. Karrikins discovered in smoke trigger Arabidopsis seed germination by a mechanism requiring gibberellic acid synthesis and light. Plant Physiology 149, 863–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DC, Scaffidi A, Dun EA, Waters MT, Flematti GR, Dixon KW, Beveridge CA, Ghisalberti EL, Smith SM. 2011. F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 108, 8897–8902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Kang H, Yamaguchi S, Park J, Lee D, Kamiya Y, Choi G. 2009. Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. The Plant Cell 21, 403–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Rockwell NC, Su YS, Lagarias JC. 2006. Phytochrome structure and signaling mechanisms. Annual Review of Plant Biology 57, 837–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M, Nambara E, Choi G, Yamaguchi S. 2009. Interaction of light and hormone signals in germinating seeds. Plant Molecular Biology 69, 463–472. [DOI] [PubMed] [Google Scholar]
- Shen H, Luong P, Huq E. 2007. The F-box protein MAX2 functions as a positive regulator of photomorphogenesis in Arabidopsis. Plant Physiology 145, 1471–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Zhu L, Bu QY, Huq E. 2012. MAX2 affects multiple hormones to promote photomorphogenesis. Molecular Plant 5, 750–762. [DOI] [PubMed] [Google Scholar]
- Shin J, Kim K, Kang H, Zulfugarov IS, Bae G, Lee C-H, Lee D, Choi G. 2009. Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proceedings of the National Academy of Sciences, USA 106, 7660–7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin K, Lee S, Song WY, et al. . 2015. Genetic identification of ACC-RESISTANT2 reveals involvement of LYSINE HISTIDINE TRANSPORTER1 in the uptake of 1-aminocyclopropane-1-carboxylic acid in Arabidopsis thaliana. Plant & Cell Physiology 56, 572–582. [DOI] [PubMed] [Google Scholar]
- Shinomura T, Nagatani A, Chory J, Furuya M. 1994. The induction of seed germination in Arabidopsis thaliana is regulated principally by phytochrome B and secondarily by phytochrome A. Plant Physiology 104, 363–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. 2000. Phytochromes and light signal perception by plants—an emerging synthesis. Nature 407, 585–591. [DOI] [PubMed] [Google Scholar]
- Soh MS, Kim YM, Han SJ, Song PS. 2000. REP1, a basic helix-loop-helix protein, is required for a branch pathway of phytochrome A signaling in arabidopsis. The Plant Cell 12, 2061–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soundappan I, Bennett T, Morffy N, Liang Y, Stanga JP, Abbas A, Leyser O, Nelson DC. 2015. SMAX1-LIKE/D53 family members enable distinct MAX2-dependent responses to strigolactones and karrikins in Arabidopsis. The Plant Cell 27, 3143–3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanga JP, Morffy N, Nelson DC. 2016. Functional redundancy in the control of seedling growth by the karrikin signaling pathway. Planta 243, 1397–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanga JP, Smith SM, Briggs WR, Nelson DC. 2013. SUPPRESSOR OF MORE AXILLARY GROWTH2 1 controls seed germination and seedling development in Arabidopsis. Plant Physiology 163, 318–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XD, Ni M. 2011. HYPOSENSITIVE TO LIGHT, an alpha/beta fold protein, acts downstream of ELONGATED HYPOCOTYL 5 to regulate seedling de-etiolation. Molecular Plant 4, 116–126. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 30, 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh S, Holbrook-Smith D, Stogios PJ, Onopriyenko O, Lumba S, Tsuchiya Y, Savchenko A, McCourt P. 2015. Structure–function analysis identifies highly sensitive strigolactone receptors in Striga. Science 350, 203–207. [DOI] [PubMed] [Google Scholar]
- Toh S, Holbrook-Smith D, Stokes ME, Tsuchiya Y, McCourt P. 2014. Detection of parasitic plant suicide germination compounds using a high-throughput Arabidopsis HTL/KAI2 strigolactone perception system. Chemistry & Biology 21, 988–998. [DOI] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A. 1997. MOLREP: an automated program for molecular replacement. Journal of Applied Crystallography 30, 1022–1025. [Google Scholar]
- Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN. 2004. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallographica. Section D, Biological Crystallography 60, 2184–2195. [DOI] [PubMed] [Google Scholar]
- van Staden J, Jager AK, Light ME, Burger BV. 2004. Isolation of the major germination cue from plant-derived smoke. South African Journal of Botany 70, 654–659. [Google Scholar]
- Wang ZY, Bai MY, Oh E, Zhu JY. 2012. Brassinosteroid signaling network and regulation of photomorphogenesis. Annual Review of Genetics 46, 701–724. [DOI] [PubMed] [Google Scholar]
- Wang H, Wang H. 2015. Phytochrome signaling: time to tighten up the loose ends. Molecular Plant 8, 540–551. [DOI] [PubMed] [Google Scholar]
- Waters MT, Nelson DC, Scaffidi A, Flematti GR, Sun YK, Dixon KW, Smith SM. 2012. Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 139, 1285–1295. [DOI] [PubMed] [Google Scholar]
- Waters MT, Scaffidi A, Flematti G, Smith SM. 2015a. Substrate-induced degradation of the α/β-fold hydrolase KARRIKIN INSENSITIVE2 requires a functional catalytic triad but is independent of MAX2. Molecular Plant 8, 814–817. [DOI] [PubMed] [Google Scholar]
- Waters MT, Scaffidi A, Moulin SL, Sun YK, Flematti GR, Smith SM. 2015b. A Selaginella moellendorffii ortholog of KARRIKIN INSENSITIVE2 functions in arabidopsis development but cannot mediate responses to karrikins or strigolactones. The Plant Cell 27, 1925–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MT, Scaffidi A, Sun YK, Flematti GR, Smith SM. 2014. The karrikin response system of Arabidopsis. The Plant Journal 79, 623–631. [DOI] [PubMed] [Google Scholar]
- Woo HR, Chung KM, Park JH, Oh SA, Ahn T, Hong SH, Jang SK, Nam HG. 2001. ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. The Plant Cell 13, 1779–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Miyakawa T, Nakamura H, Nakamura A, Imamura Y, Asami T, Tanokura M. 2016. Structural basis of unique ligand specificity of KAI2-like protein from parasitic weed Striga hermonthica. Scientific Reports 6, 31386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang KY, Kim YM, Lee S, Song PS, Soh MS. 2003. Overexpression of a mutant basic helix-loop-helix protein HFR1, HFR1-ΔN105, activates a branch pathway of light signaling in Arabidopsis. Plant Physiology 133, 1630–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao R, Ming Z, Yan L, et al. . 2016. DWARF14 is a non-canonical hormone receptor for strigolactone. Nature 536, 469–473. [DOI] [PubMed] [Google Scholar]
- Yao R, Wang F, Ming Z, Du X, Chen L, Wang Y, Zhang W, Deng H, Xie D. 2017. ShHTL7 is a non-canonical receptor for strigolactones in root parasitic weeds. Cell Research 27, 838–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao LH, Zhou XE, Wu ZS, et al. . 2013. Crystal structures of two phytohormone signal-transducing α/β hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14. Cell Research 23, 436–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao LH, Zhou XE, Yi W, et al. . 2015. Destabilization of strigolactone receptor DWARF14 by binding of ligand and E3-ligase signaling effector DWARF3. Cell Research 25, 1219–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Lin Q, Zhu L, et al. . 2013. D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature 504, 406–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.