Abstract
Motivation
Antimicrobial resistance (AMR) is becoming a huge problem in both developed and developing countries, and identifying strains resistant or susceptible to certain antibiotics is essential in fighting against antibiotic-resistant pathogens. Whole-genome sequences have been collected for different microbial strains in order to identify crucial characteristics that allow certain strains to become resistant to antibiotics; however, a global inspection of the gene content responsible for AMR activities remains to be done.
Results
We propose a pan-genome-based approach to characterize antibiotic-resistant microbial strains and test this approach on the bacterial model organism Escherichia coli. By identifying core and accessory gene clusters and predicting AMR genes for the E. coli pan-genome, we not only showed that certain classes of genes are unevenly distributed between the core and accessory parts of the pan-genome but also demonstrated that only a portion of the identified AMR genes belong to the accessory genome. Application of machine learning algorithms to predict whether specific strains were resistant to antibiotic drugs yielded the best prediction accuracy for the set of AMR genes within the accessory part of the pan-genome, suggesting that these gene clusters were most crucial to AMR activities in E. coli. Selecting subsets of AMR genes for different antibiotic drugs based on a genetic algorithm (GA) achieved better prediction performances than the gene sets established in the literature, hinting that the gene sets selected by the GA may warrant further analysis in investigating more details about how E. coli fight against antibiotics.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Antimicrobial-resistant (AMR) pathogens greatly undermine people’s ability to control pathogens and cure diseases. The ultra-fast mutation rates of these microbes render our existing drugs useless against superbugs, and ‘existing classes of antibiotics are probably the best we will ever have (Cormican and Vellinga, 2012).’ A study published in 2013 also identified that, due to AMR, additional economic costs may be as high as 55 billion USD and that trivial bacterial infections such as hip replacements may increase the death rate from approximately 0% to 30% (Smith and Coast, 2013). The rapid decrease in the number of new drugs further diminishes our chances of competing against these pathogens, and cutting-edge research in all dimensions direly needs to find a way to control these microbes.
Genomic information has been incorporated in order to understand why certain strains of pathogens are resistant to antibiotics, including Staphylococcus aureus, Mycobacterium tuberculosis, Klebsiella pneumoniae, Salmonella spp. and Pseudomonas aeruginosa (Bradley et al., 2015; Gordon et al., 2014; McDermott et al., 2016; Stoesser et al., 2013; Tyson et al., 2015). The PATRIC database is one of the most comprehensive antibiotic resistance databases that collects genes, proteins and genomic information related to the resistance or susceptibility of pathogens to various antibiotic drugs (Wattam et al., 2017). The collection of over 80 000 bacterial genomes available in the PATRIC database allows scientists to understand the mechanisms of AMR in terms of genes, proteins and genomes.
‘Pan-genome’, a term used to describe shared features of all strains of certain bacteria, has been applied to understand the strain-level diversity of these species (Medini et al., 2005). It was also used in analyzing the diversity, virulence and AMR phenotypes of Klebsiella pneumoniae, in which a genomic study found that K. pneumoniae can be split into three distinct groups, and that certain branches in the three groups may be either hyper-virulent or multi-drug-resistant (Holt et al., 2015). A computational approach, Scoary (Brynildsrud et al., 2016), was developed to associate the genetic components of the pan-genome with observed phenotypic traits and identify gene clusters that were associated with high-level AMR activities such as linezolid resistance in Staphylococcus epidermidis. These examples suggested that the pan-genome idea can be very useful in defining gene components that may contribute to phenotypes of the living organisms.
In this paper, we explored whether machine learning approaches can be applied on pan-genome to better define and predict AMR. We selected the model organism Escherichia coli as the exploration target due to its well-established gene profiles. Even though E. coli pan-genome has already been studied to compare commensal and pathogenic isolates (Rasko et al., 2008), no associations were established between its pan-genome and strain-level AMR activities. We therefore chose E. coli species as our primary target for pan-genome identification and incorporated AMR resolution into the analytical procedure. By analyzing the gene content within the pan-genome and building predictive models for the AMR activities we extracted the most likely gene sets that define whether E. coli strains are resistant or susceptible to antibiotic drugs. A genetic algorithm (GA) was also incorporated to select subsets of the genes that yielded outstanding performances and outperformed established genes described in the literature, highlighting the possibility of accurately predicting AMR strains from pan-genome content and opening up the potential of mining gene repertoire using machine learning algorithms to understand more about bacterial AMR mechanisms.
2 Materials and methods
2.1 Genome collection and annotation
Genome fasta files of 59 E. coli strains with resistance metadata to 38 antibiotic drugs were downloaded from the PATRIC ftp site (ftp://ftp.patricbrc.org/; Wattam et al., 2017) in July 2017. The genome IDs, genome sizes, genome status, number of contigs and N50 values as well as isolation sources, isolation countries and host information were listed in Supplementary Table S1. The antibiotic drug information, the classes of the drugs, along with the number of entries measured for different E. coli strains can be found in Supplementary Table S3. These genomes were re-annotated for their AMR activities by comparing their minimum inhibitory concentration (MIC; the lowest concentration of a chemical that prevents the bacterial growth) to the 2017 Clinical and Laboratory Standard Institute (CLSI)’s guidelines on AMR (M100 Performance Standards for Antimicrobial Susceptibility Testing), which offers the definition of breakpoints for antibiotic resistance. The designated annotations included ‘resistant’, ‘susceptible’, ‘intermediate’, ‘non-resistant’ and ‘non-susceptible’. The ‘intermediate’ label indicated that the dosage of an antibiotic drug required to kill the pathogenic strain was higher than those susceptible to the drug but not as high as resistant ones. ‘Non-resistant’ and ‘non-susceptible’, respectively, indicated that the microbial strain was not resistant and not susceptible to the antibiotic drug; however whether these microbes were indeed susceptible or resistant to the drugs cannot be determined (the reason we added these two categories is due to differences in maximum MIC experimented during serial dilution documented in different versions of CLSI, which resulted in insufficient information for adapting the AMR annotations to 2017 CLSI’s guideline). ezTree software, which was capable of identifying single-copy genes from a group of genomes, separately aligning the amino acid sequences of the single copy genes and concatenating the alignments to build a phylogenetic tree, was used to build the evolutionary tree for the involved E. coli strains (Wu, 2018). Default parameters (-evalue 1e–10 -model JTT) were employed to build the tree using ezTree. Visualization of the tree and AMR phenotypes was conducted using Evolview (He et al., 2016).
2.2 Pan-genome construction
To build the pan-genome, protein-coding genes were predicted from the genomes using Prodigal (Hyatt et al., 2010) with the ‘-p meta’ parameter (since more than 90% of the analyzed E. coli genomes were fragmented draft genomes), and CD-HIT (Fu et al., 2012) was utilized to group the predicted genes into gene clusters with 95% amino acid identity. [Even though PATRIC already consisted of predicted genes for the individual genomes using RASTtk (Wattam et al., 2014, 2017), which employed Glimmer3 (Brettin et al., 2015), the program Prodigal was evaluated to outperform other tools (Angelova et al., 2010; Hyatt et al., 2010) and was therefore adopted to predict genes for the downloaded genomes]. The 95% amino acid identity cutoff was determined by cross-comparing protein sequences from five E. coli strains randomly sampled from the 59 strains (including strains BIDMC 71, 17A, UCI 58, AR_0118 and MRSN388634) using BLAST (-p 1e–10 -max_target_seqs 1), in which we found that 95% identity served as a good cutoff value for grouping orthologous genes (Supplementary Fig. S5).
The extracted gene clusters were then classified into ‘core’ and ‘accessory’ genomes based on whether the clusters consisted of genes from all strains; only clusters with genes from all genomes were included in the ‘core’ set; otherwise they were classified into the ‘accessory’ set. Clusters of Orthologous groups (COGs) were predicted by searching the amino acid sequences of the genes against COG hidden Markov models downloaded from the eggNOG 4.5 database (Huerta-Cepas et al., 2016) using HMMER3 (Eddy, 2011) with an e-value cutoff of 1e–5. AMR genes were annotated by Resistance Gene Identifier (RGI) software provided by Comprehensive Antibiotic Resistance Database (CARD; Jia et al., 2017).
2.3 AMR phenotype prediction and performance evaluation
Predictive machine learning models, including Support Vector Machine (SVM; radial basis function kernel), Naïve Bayes (NB) (multi-variate Bernoulli models), Adaboost (based on an ensemble of 200 decision trees with a maximum depth of 2 and the SAMME algorithm) and Random Forest (RF; based on 200 decision trees with no limit on their maximum depths; tree-splitting criteria were Gini impurity), were built using Python scikit-learn (sklearn) machine learning API (http://scikit-learn.org/). Only two AMR phenotypes, ‘resistant’ and ‘susceptible’, were considered in the evaluation—‘intermediate’, ‘non-susceptible’ and ‘non-resistant’ phenotypes were not included due to our inability to determine whether those strains were indeed resistant or susceptible to an antibiotic. The evaluation was conducted using the leave-one-out cross validation method, in which one of the strains was used for validation while the remaining strains were recruited for training. For example, assuming that there are n bacterial strains numbered [1, 2, 3,…, n–1, n]; the leave-one-out process first uses [2, 3,., n–1, n] to build a prediction model and apply it on strain [1]; it then exhaustively builds models using data from n–1 strains and estimates the prediction accuracy on the left-out strain until all strains are evaluated. The performances of the predictions were estimated after all samples were iteratively predicted in the leave-one-out process using area under the receiver operating characteristics (ROC) curve (AUROC; also called AUC) metric. This process was repeated 10 times for each drug to get an averaged accuracy measurement.
2.4 Genetic algorithm
To improve the prediction accuracy using the presence/absence patterns of the gene clusters identified from the 59 strains downloaded from the PATRIC database, a genetic algorithm (GA) was implemented to find the best subset of accessory gene clusters with CARD annotations (acc/card) for predicting resistant or susceptible strains. The ‘genomes’ of the GA (termed GA-genome hereafter) were defined as either including [1] or not including [0] certain gene clusters in predicting AMR activities and the GA-genome size was set to the length of the number of acc/card gene clusters. For example, for five gene clusters [A, B, C, D, E], the GA-genome [1, 0, 1, 0, 1] indicates that only gene clusters A, C and E are used in the prediction of AMR activities. The fitness function of the GA was selected to be the AUC estimated for the SVM training models using the partial gene cluster set selected by the GA-genomes, in which only gene clusters marked as ‘1’ in the GA-genome were included in the leave-one-out evaluation process as described before. The GA population size was 100. In the beginning of the GA the GA-genomes were randomly filled in either 0 or 1; each GA-genome was then evaluated for its AUC under SVM training. After the performance evaluation the best two GA-genomes were copied to the next iteration; all GA-genomes (including the two already-copied ones) were then sampled with probability proportional to their AUC score with base mutation probability 0.05 and crossover probability 0.1 until 100 GA-genomes were created. Only one crossover was allowed for any two sampled GA-genomes. This GA process was repeated 30 000 times to get the subset of gene clusters for each antibiotic drug for best predicting AMR activities (the performance improvements was shown in Supplementary Fig. S7, which indicated that AUC was maximized at around 15 000 GA runs).
To compare our prediction performances against previously-identified AMR genes and gene clusters identified by other pan-genome-based methods, AMR genes established in (Tyson et al., 2015) were checked for their presence in the 59 E. coli genomes using BLASTP (-evalue 1e–10). We chose genes related to four antibiotic drugs (ampicillin, ciprofloxacin, gentamicin and trimethoprim/sulfamethoxazole) that were also among the list of our 12 drug. The genes being detected were: blaTEM-1, blaOXA-1, blaCMY-2 and ampC for ampicillin; aac(3’)-Ia and aac(3’)-VI for gentamicin; dfrA1, dfrA5, dfrA12 and dfrA15 for trimethoprim/sulfamethoxazole, and qnrB2, qnrB6, qnrS2, gyrA, parC and parE for ciprofloxacin. We noted that gyrA, parC and parE were checked for non-synonymous mutations instead of their presence/absence patterns, as suggested by (Tyson et al., 2015), by comparing against E. coli K-12 MG1655 genes. Scoary (Brynildsrud et al., 2016) was used to establish gene clusters associated with AMR phenotypes of the 59 genomes by the following steps: a pan-genome was built using Roary (Page et al., 2015) with default settings (-i 95 -cd 99 -iv 1.5); the Roary pan-genome gene clusters associated with the AMR phenotypes were then identified using Scoary with default settings (-p 0.05 –c I). The Roary gene clusters were also annotated for their AMR activities using RGI/CARD as described before.
3 Results
We first checked whether patterns existed for the AMR phenotypes based on the phylogeny of the E. coli strains by building a phylogenetic tree and plotting the AMR phenotypes (resistant, susceptible, intermediate, non-resistant, non-susceptible, or no data; see Supplementary Table S2 for detailed information of the strains and the re-annotated antimicrobial activities). The results, as shown in Supplementary Figure S1, does not show clear clusters of the AMR phenotypes on the heatmap; however, we observed that drugs of the same class (for example, the four cephalosporins or the two aminoglycosides) were grouped together, suggesting that the strains reacted more similarly to drugs of the same classes. Note that even though there were totally 38 antibiotics annotated for E. coli strains, numbers of AMR annotations differed greatly among the drugs: only 12 drugs were annotated for more than 30 strains in the PATRIC database; and 13 drugs had fewer than 10 annotations (Supplementary Table S3). We therefore chose the 12 most-annotated antibiotic drugs or drug composites (meropenem, gentamicin, ciprofloxacin, trimethoprim/sulfamethoxazole, ampicillin, cefazolin, ampicillin/sulbactam, ceftazidime, cefepime, piperacillin/tazobactam, tobramycin and ceftriaxone) for further examination. The classes that these 12 drugs belonged to include: one carbapenem, one quinolone, two aminoglycosides, one penicillin, four cephalosporins, one folate pathway and two beta-lactam inhibitors.
We then applied the pan-genome approach to the 59 E. coli strains with various AMR activities. The number of unique gene clusters in the pan-genome was 15 950, in which 2874 belonged to the core part while the rest 13 076 belonged to the accessory part of the pan-genome. The growth of the gene cluster numbers that belong to the pan-genome, core genome and accessory genome were shown in Figure 1. The curve-fitting of the pan-genome growth was performed using a power law regression, which was based on Heaps’ law described in (Tettelin et al., 2005, 2008). The fitting was conducted using panGP (Zhao et al., 2014) to fit the equation (), in which y and x were pan-genome size and the number of genomes, respectively. was equivalent to the parameter for estimating whether a pan-genome is open or close in (Tettelin et al., 2008). Our estimated pan-genome profile curve was (), in which was estimated to be 0.38. R2 was 0.999554. Since a pan-genome was consider open when (and close otherwise), this result suggested that our constructed pan-genome was an open pan-genome. The core genome profile curve was also fitted to the equation () using panGP; the estimated parameters were (, and ). R2 was 0.887721. Previous studies such as (Medini et al., 2005) also suggested that species that colonized multiple environments and had numerous mechanisms for exchanging genetic elements (such as Streptococci, Meningococci, H. pylori, Salmonella, and E. coli) were more likely to have an open pan-genomes.
Fig. 1.
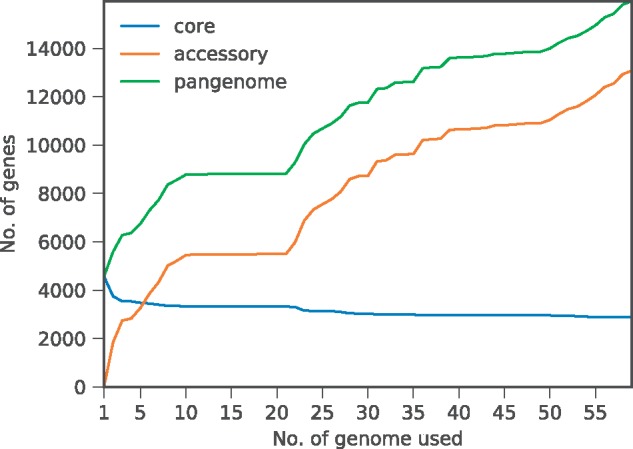
Growth rates of the pan-genome sizes, core gene cluster and accessory gene cluster numbers with the increasing number of E. coli genomes. The blue, orange and green lines, respectively, represent core-, accessory- and pan-genome sizes
In order to identify differences in the protein functional distributions of the core- and accessory-genomes, COG annotation was performed on the gene clusters. After quantifying the COG classes in the core and accessory genomes and selecting COGs with different distributions (at least 2-fold abundance differences) between the core and accessory parts of the pan-genome, we observed that protein classes essential to the core-genome were those responsible for translation and ribosomal structure (J), signal transduction (T), post-translational modification (O), energy production (C) and amino acid/nucleotide/coenzyme/lipid transportation and metabolism (E, F, H and I), as shown in Figure 2 (proportions of all COGs among the genes were reported in Supplementary Table S5). On the other hand, proteins for replication, recombination, and repair (L), intracellular trafficking, secretion and vesicular transport (U), defense mechanism (V) and cell motility (N) formed the majority of the accessory genome. These results suggested that genes responsible for metabolite transportation and metabolism, signal transduction, energy production and core nucleotide/amino acid processing were more critical to the survival of E. coli and were hence more conserved. Similar results were also reported by another pan-genome analysis on Bifidobacterium and Lactobacillus (Lukjancenko et al., 2012), in which classes J, E, G and O were enriched in the core genomes.
Fig. 2.
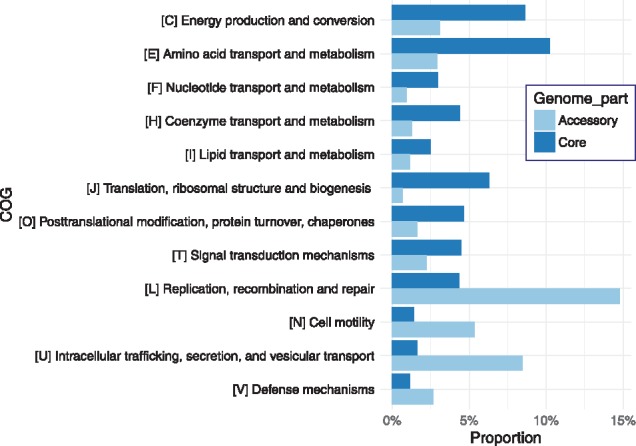
Differences in the COGs functional distributions between the core- and accessory-genomes. COG percentages were estimated by dividing COG numbers by the total gene cluster numbers in either the core- or accessory-genome. Only COGs differing by at least 2-fold between the core and accessory parts were included
The AMR potentials of the gene clusters were also annotated according to the CARD database. We discovered that totally 111 protein clusters (0.7% among all clusters) could be mapped to a CARD entry, suggesting that genes related to AMR may only account for a small proportion of the genome. After comparing the CARD annotation to the pan-genome, we found that only 61% (68 clusters) of CARD clusters belonged to the accessory genome, and that the most-annotated COG class (25%) of the 68 gene clusters was category V (defense mechanism). This suggested that there may be intrinsic resistance mechanisms shared by most E. coli strains, and that the 68 gene clusters may be more relevant to the distinctions of AMR phenotypes of the strains.
We checked the best hit CARD annotations of the 68 gene clusters (Supplementary Table S6) and identified that most of the gene clusters were related to AMR activities of E. coli. For example, one of the gene clusters (cluster 12 174) was annotated as emrE, which belonged to the drug/metabolite transporter superfamily. Overexpression of emrE may provide resistance to various antibiotic drugs as well as a wide variety of toxic cationic hydrophobic compounds (Ma and Chang, 2004; Yerushalmi et al., 1995). Another example were two gene clusters (clusters 2965 and 5292), which were, respectively, annotated as mrx and mph(A) and were also reported to confer high-level drug resistance to erythromycin (Noguchi et al., 2000). Other examples included genes that provided resistance to sulfamethoxazole (sul1, sul2 and sul3; clusters 5871, 5926 and 6303), ampicillin (TEM1; cluster 5687), tetracycline (tetA and tetD; clusters 3154 and 10952), trimethoprim (dfrA12 and dfrA17; clusters 9729 and 8425), streptomycin (aadA; cluster 6352) and gentamicin [aac(3)-IV; cluster 6471], suggesting that most of the 68 gene clusters were relevant to AMR.
To check whether the 68 accessory gene clusters with CARD annotations (termed acc/card hereafter) may be used as viable predictors of AMR activities of the E. coli strains, we built predictive models from the presence/absence patterns of four gene sets (all core and accessory gene clusters, all accessory gene clusters, all acc/card gene clusters and all CARD gene clusters) and evaluated the performances of the models using the leave-one-out validation method. Four machine learning methods (including SVM, NB, RF and Adaboost) were incorporated into the evaluation process (see Section 2 for details). The results (in terms of the AUC), as shown in Figure 3, suggested that the 68 acc/card gene cluster were more suitable for predicting AMR phenotypes of E. coli strains (detailed prediction results were listed in Supplementary Tables S7–S10). We also noted that the Adaboost, RF and SVM algorithms performed slightly better than NB. Measurements of F1 scores were also similar to the AUC (Supplementary Fig. S2; precision and recall were shown in Supplementary Figs S3 and S4).
Fig. 3.
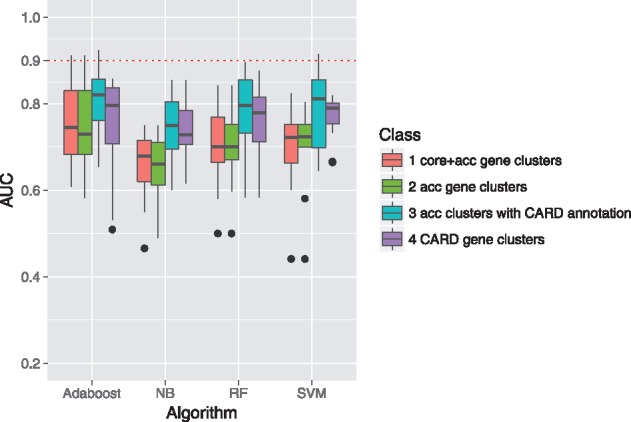
Prediction accuracies of the AMR activities [in terms of the area under the ROCs curve (AUC)] based on the presence/absence patterns of (i) all core and accessory gene clusters (core + acc); (ii) all accessory gene clusters (acc); (iii) accessory gene clusters with CARD annotations (acc/card) and (iv) all CARD gene clusters. The boxplots indicate the distribution of the predictive accuracy of 12 selected drugs (Section 2 and Section 3). The four blocks of boxplots represent four different machine learning algorithms, including Adaboost, NB, RF and SVM, used in the prediction process. Dashed red line indicates 0.9 AUC
Even though the 68 acc/card genes served as better predictors for AMR activities compared to other gene sets (including all core and accessory gene clusters, all accessory gene clusters and all CARD gene clusters), the prediction performances of the 68 acc/card genes on four selected drugs (ampicillin, gentamicin, trimethoprim/sulfamethoxazole and ciprofloxacin) were still not as good as genes proposed in the literature (Tyson et al., 2015) and gene clusters identified by another pan-genome-wide association tool, Scoary (Table 1; the number of gene clusters and AMR annotations identified by Scoary for each drug was shown in Supplementary Table S13; the SVM prediction performances evaluated for Scoary were listed in Supplementary Table S14). We therefore designed a genetic algorithm to select the subsets of the 68 acc/card genes for predicting AMR activities. As shown in Table 1 and Figure 4, the GA-selected gene cluster subsets for each drug clearly outperformed the 68 acc/card genes, suggesting that the gene clusters identified by GA may be associated with AMR phenotypes (detailed AUC measurements were listed in Supplementary Table S11; detailed information of the gene clusters selected by the GA was listed in Supplementary Table S12). Moreover, the GA-selected gene clusters outperformed genes established in the literature and Scoary, hinting that these genes may warrant more analysis in future AMR research for E. coli.
Table 1.
SVM prediction performances (based on the AUC) measured for ampicillin, gentamicin, trimethoprim/sulfamethoxazole and ciprofloxacin
| Drugs | 68 acc/carda | Tyson 2005b | Scoaryc | Scoary/cardd | GAe |
|---|---|---|---|---|---|
| Ampicillin | 0.64g | 0.86 | 0.75 | 0.79 | 0.97 |
| Gentamicin | 0.78 | 0.83 | 0.85 | 0.68 | 0.98 |
| Trim/sulfaf | 0.87 | 0.82 | 0.76 | 0.87 | 0.94 |
| Ciprofloxacin | 0.71 | 0.78 | 0.93 | 0.87 | 0.93 |
68 accessory gene clusters with CARD annotations.
Genes established in (Tyson et al., 2015).
Gene clusters that were associated with phenotypes extracted by Scoary.
Gene clusters that can be mapped to the CARD database extracted by Scoary.
Gene clusters selected by the GA.
Trimethoprim/sulfamethoxazole.
AUC measured from the leave-one-out evaluation process using SVM.
Fig. 4.
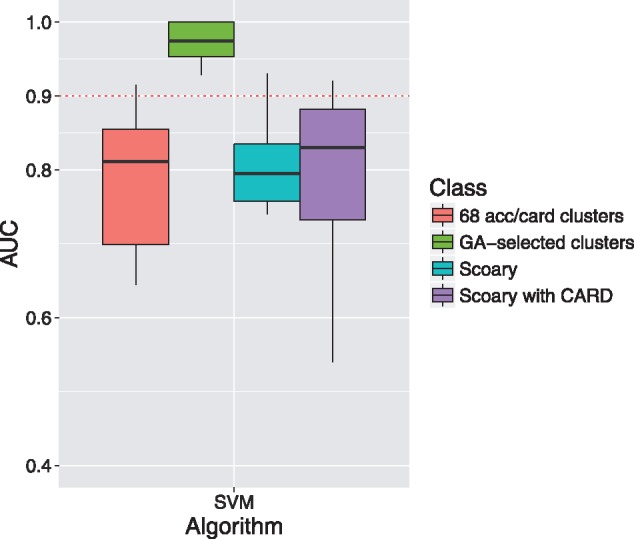
SVM prediction accuracies of the antimicrobial resistance (AMR) activities (in terms of the area under the receiver operating characteristics curve (AUC)) based on 1) 68 accessory genes with CARD annotations (68 acc/card genes); 2) gene clusters selected for each drug based on the genetic algorithm (GA-selected clusters); 3) gene clusters identified by Scoary; and 4) gene clusters with CARD annotations identified by Scoary (Scoary with CARD). The boxplot indicates the distribution of the prediction accuracies for the 12 selected drugs. Dashed red line indicates 0.9 AUC
4 Discussion
In this paper, we attempted to identify and classify AMR activities of E. coli through the use of the pan-genome. Although the pan-genome idea has previously been applied to identify commensal and pathogenic strains, no previous pan-genome-based works were related to E. coli AMR phenotypes. The construction of pan-genome allowed us to simultaneously inspect all strains and their AMR activities. The pan-genome idea also made it possible to unearth potential gene clusters that may confer resistance to antibiotic drugs or toxic materials and measure variations between resistant and susceptible groups. We noted that Brynildsrud et al. also employed Scoary, a tool for scoring genes in microbial pan-genome-wide association studies, to identify gene clusters that were associated with high-level AMR activities, including linezolid resistance in Staphylococcus epidermidis (Brynildsrud et al., 2016).
After downloaded the E. coli genomes and AMR metadata from PATRIC, we deliberately re-annotated the E. coli AMR activities using the 2017 CLSI’s guideline. This was because the PATRIC metadata that we downloaded (as of July 2017) did not specify which criteria it used to annotate the resistance profiles of the bacterial strains. For example, E. coli strain 5CRE51 was annotated as resistant to gentamicin (MIC > 8); however the CLSI 2017 specified that MIC needs to be >= 16 to be resistant to gentamicin. Therefore, we re-annotated the 5CRE51 strain, which was only tested for its MIC values up to 8, as ‘non-susceptible’ to gentamicin, indicating that we knew it was not susceptible to gentamicin but could not determine whether it was indeed resistant to gentamicin. Another example is E. coli strain AR_0104, which was annotated as resistant to norfloxacin (MIC > 8) in the PATRIC database; the CLSI 2017 however noted that MIC needed to be >= 32 to be resistant to norfloxacin. Therefore we can only say that the AR_0104 strain was ‘not susceptible’ to norfloxacin without knowing whether it was resistant to this drug.
To determine the best amino acid cutoff threshold for building the pan-genome, we cross-compared the amino acid sequences of the genes from five randomly-sampled E. coli strains and found that 95% amino acid identity served as a good cutoff value. This cutoff was consistent with another pan-genomic study on Bacillus, which also used 95% as the identity cutoff (Kim et al., 2017). We therefore used 95% cutoff identity to build the pan-genome, which yielded 15 950 gene clusters, among which 2876 gene clusters belonged to the core genome and 13 076 gene clusters belonged to the accessory genome. The proportion of core gene clusters was about 18%. These pan-genome statistics were similar to those in a previous report (Rasko et al., 2008). We also compared our pan-genome to a much larger E. coli pan-genome (which consisted of 307 strains) available on panX (Ding et al., 2018), in which the core gene cluster proportion was 13% (totally 23 128 gene clusters, among which 3199 gene clusters belonged to the core genome and 19 929 gene clusters belonged to the accessory genome) after adjusted the clustering criterion to 95%. We noted that the core gene clusters identified by panX (3199) was slightly larger than ours (2876), probably due to different clustering algorithms between panX and ours.
To explore the effect of clustering identity cutoffs, we tried several identity cutoffs (95%, 90%, 80% and 70%) to build different pan-genomes. As expected, the decrease of the identity cutoff increased the core gene clusters and reduced the accessory gene clusters and total pan-genome sizes, as shown in Supplementary Table S4. The growth rate of the gene clusters also slowed down. Similar observation were also made on panX, in which adjusting the identity cutoff from 95% to 90% resulted in the number of core gene clusters increased from 3199 to 3345 while the amount of accessory gene clusters reduced from 19 929 to 19 783. We noted that the reduction of accessory gene cluster numbers from 95% identity cutoff to 90% on panX was not as much as our analysis, probably because (i) different clustering algorithms were adopted for producing gene clusters; or (ii) the sources of the panX E. coli strains were very diverse (including dogs, cats, mice, cattle and human) while the PATRIC E. coli strain sources were strictly human.
From the pan-genome we identified core and accessory gene clusterss and annotated AMR genes. Cross comparisons between AMR genes and core/accessory genomes revealed that only 61% of the genes belonged to the accessory genome. One of the possible reasons for this phenomenon is that some resistance factors may be intrinsic to E. coli, resulting in shared AMR genes among most strains. It was also possible that the number of E. coli genomes (which is 59) was not large enough, or that the majority of the E. coli strains in the PATRIC database carried resistance to multiple drugs and hence shared some AMR genes.
By building machine learning models to check the predictive abilities of four different cluster sets, we found that the set of acc/card genes yielded the best prediction results compared to other gene sets. This result suggested that AMR genes that did not appear universally in every genome may be a good predictor for forecasting AMR activities of the E. coli strains. The finding that SVM, RF and Adaboost outperformed NB also suggested that the problem of predicting AMR activities from genomic information may be a non-linear problem.
By designing a genetic algorithm to pick subsets of acc/card genes with the most AMR predictive power for the antibiotic drugs, we extracted gene clusters that were able to better predict the resistance profiles of the E. coli strains for each of the 12 drugs. To check whether patterns exist for the GA-selected gene clusters, we made a heatmap from the GA-selected patterns, as shown in Supplementary Figure S6. No clear patterns were observed from this heatmap, and some drugs of the same class were not grouped together (e.g. the four cephalosporins). One of the possible reasons was that there may be some weakly-associated genes that the GA also recruited in order to maximize the prediction accuracy and therefore disturbed the patterns of the heatmap. We also observed some gene clusters picked by the GA cannot be fully associated with known genetic functions despite the outstanding prediction performances. For example, gene clusters 1202, 1513, 3412, 3408 and 3397 were annotated as either pmrC or pmrE, which were related to polymyxin resistance (Olaitan et al., 2014); the GA however selected either pmrC or pmrE or both in the prediction of strains resistant to drugs with different antibiotic mechanisms such as ampicillin, cefazolin, or trimethoprim/sulfamethoxazole, to name just a few. There were two possible explanations: either the GA ‘found’ that the inclusion of these gene clusters could improve the prediction accuracy, or that there were two or more equally good combinations of selected gene clusters that may lead to similar prediction performances. We also cannot rule out the possibility that unknown genetic mechanisms may be associated with the seemingly-unrelated AMR genes. Further investigation is still needed to fully interpret the results obtained by the GA. We noted that Scoary also associated pmrE with cefazolin and trimethoprim/sulfamethoxazole (Supplementary Table S13), lending support to the hypothesis that there may be some associations between seemingly-unrelated genes and the antibiotic drugs.
One of the limitations of our study is that there were only 59 E. coli strains with AMR annotations in the PATRIC database—far less than the number of E. coli strains in the NCBI database. We however argue that the successful determination of the pan-genome and crucial AMR gene sets from the 59 E. coli strains showcase the potential of the proposed pan-genome approach for predicting AMR activities from genomic content. We look forward to testing our approach on larger E. coli genome set with AMR profiles and checking whether the pan-genome-based machine learning method is robust. We also plan to extend our approach into other pathogenic species such as Klebsiella pneumoniae or Samonella enterica. We hope that by establishing crucial AMR gene cluster sets for the species we can better understand how these microorganisms fight against antibiotics.
5 Conclusion
In this study, we constructed the pan-genome of E. coli strains with AMR annotations and identified key factors for predicting whether or not a strain was resistant to certain antibiotics. Specifically, we found that a very small set of accessory genes with antimicrobial activity annotations achieved the best predictive accuracy. To the best of our knowledge, this is the first study to employ a pan-genome as an essential guide in predicting E. coli AMR activities and we hope that this study can serve as a stepping stone in dealing with AMR pathogens using genomic information.
Supplementary Material
Acknowledgement
The authors thank the three anonymous reviewers for their constructive comments for improving this manuscript.
Funding
This work was supported by Taipei Medical University grant TMU105-AE1-B19 to YWW, Taiwan Ministry of Science and Technology (MOST) grant MOST106-2218-E-038-002-MY2 to YWW and MOST grant 106-2813-C-038-010-E (undergraduate research grant) to HLH.
Conflict of Interest: none declared.
References
- Angelova M. et al. (2010) Computational methods for gene finding in prokaryotes In: Gusev M. (ed.) ICT Innovations 2010. Ohrid, Macedonia, Springer, pp. 11–20. [Google Scholar]
- Bradley P. et al. (2015) Rapid antibiotic-resistance predictions from genome sequence data for Staphylococcus aureus and Mycobacterium tuberculosis. Nat. Commun., 6, 10063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brettin T. et al. (2015) RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep., 5, 8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brynildsrud O. et al. (2016) Rapid scoring of genes in microbial pan-genome-wide association studies with Scoary. Genome Biol., 17, 238.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormican M., Vellinga A. (2012) Existing classes of antibiotics are probably the best we will ever have. Brit. Med. J., 344, e3369.. [DOI] [PubMed] [Google Scholar]
- Ding W. et al. (2018) panX: pan-genome analysis and exploration. Nucleic Acids Res., 46, e5.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S.R. (2011) Accelerated profile HMM searches. PLoS Comput. Biol., 7, e1002195.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L. et al. (2012) CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics, 28, 3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon N.C. et al. (2014) Prediction of Staphylococcus aureus antimicrobial resistance by whole-genome sequencing. J. Clin. Microbiol., 52, 1182–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z. et al. (2016) Evolview v2: an online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res., 44, W236–W241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt K.E. et al. (2015) Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc. Natl. Acad. Sci. USA, 112, E3574–E3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J. et al. (2016) eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res., 44, D286–D293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt D. et al. (2010) Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics, 11, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia B. et al. (2017) CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res., 45, D566–D573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. et al. (2017) Pan-genome analysis of Bacillus for microbiome profiling. Sci. Rep., 7, 10984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukjancenko O. et al. (2012) Comparative genomics of Bifidobacterium, Lactobacillus and related probiotic genera. Microb. Ecol., 63, 651–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott P.F. et al. (2016) Whole-genome sequencing for detecting antimicrobial resistance in nontyphoidal Salmonella. Antimicrob. Agents Chemother., 60, 5515–5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Chang G. (2004) Structure of the multidrug resistance efflux transporter EmrE from Escherichia coli. Proc. Natl. Acad. Sci. USA, 101, 2852–2857. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Medini D. et al. (2005) The microbial pan-genome. Curr. Opin. Genet. Dev., 15, 589–594. [DOI] [PubMed] [Google Scholar]
- Noguchi N. et al. (2000) Regulation of transcription of the mph(A) gene for macrolide 2 '-phosphotransferase I in Escherichia coli: characterization of the regulatory gene mphR(A). J. Bacteriol., 182, 5052–5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olaitan A.O. et al. (2014) Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front. Microbiol., 5, 643.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page A.J. et al. (2015) Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics, 31, 3691–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasko D.A. et al. (2008) The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J. Bacteriol., 190, 6881–6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R., Coast J. (2013) The true cost of antimicrobial resistance. BMJ-Brit. Med. J., 346, f1493.. [DOI] [PubMed] [Google Scholar]
- Stoesser N. et al. (2013) Predicting antimicrobial susceptibilities for Escherichia coli and Klebsiella pneumoniae isolates using whole genomic sequence data. J. Antimicrob. Chemother., 68, 2234–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H. et al. (2005) Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial ‘pan-genome’. Proc. Natl. Acad. Sci. USA, 102, 13950–13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H. et al. (2008) Comparative genomics: the bacterial pan-genome. Curr. Opin. Microbiol., 11, 472–477. [DOI] [PubMed] [Google Scholar]
- Tyson G.H. et al. (2015) WGS accurately predicts antimicrobial resistance in Escherichia coli. J. Antimicrob. Chemother., 70, 2763–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattam A.R. et al. (2014) PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res., 42, D581–D591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattam A.R. et al. (2017) Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res., 45, D535–D542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.W. (2018) ezTree: an automated pipeline for identifying phylogenetic marker genes and inferring evolutionary relationships among uncultivated prokaryotic draft genomes. BMC Genomics, 19, 921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yerushalmi H. et al. (1995) Emre, an Escherichia-Coli 12-Kda multidrug transporter, exchanges toxic cations and H+ and is soluble in organic-solvents. J. Biol. Chem., 270, 6856–6863. [DOI] [PubMed] [Google Scholar]
- Zhao Y. et al. (2014) PanGP: a tool for quickly analyzing bacterial pan-genome profile. Bioinformatics, 30, 1297–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


