Abstract
Motivation
De novo genome assembly is a challenging computational problem due to the high repetitive content of eukaryotic genomes and the imperfections of sequencing technologies (i.e. sequencing errors, uneven sequencing coverage and chimeric reads). Several assembly tools are currently available, each of which has strengths and weaknesses in dealing with the trade-off between maximizing contiguity and minimizing assembly errors (e.g. mis-joins). To obtain the best possible assembly, it is common practice to generate multiple assemblies from several assemblers and/or parameter settings and try to identify the highest quality assembly. Unfortunately, often there is no assembly that both maximizes contiguity and minimizes assembly errors, so one has to compromise one for the other.
Results
The concept of assembly reconciliation has been proposed as a way to obtain a higher quality assembly by merging or reconciling all the available assemblies. While several reconciliation methods have been introduced in the literature, we have shown in one of our recent papers that none of them can consistently produce assemblies that are better than the assemblies provided in input. Here we introduce Novo&Stitch, a novel method that takes advantage of optical maps to accurately carry out assembly reconciliation (assuming that the assembled contigs are sufficiently long to be reliably aligned to the optical maps, e.g. 50 Kbp or longer). Experimental results demonstrate that Novo&Stitch can double the contiguity (N50) of the input assemblies without introducing mis-joins or reducing genome completeness.
Availability and implementation
Novo&Stitch can be obtained from https://github.com/ucrbioinfo/Novo_Stitch.
1 Introduction
De novo genome assembly is a fundamental problem in genomics and computational biology. Despite significant algorithmic progress, this problem remains challenging due to the high repetitive content of eukaryotic genomes, short read length, uneven sequencing coverage, non-uniform sequencing errors and chimeric reads. Compared to the second generation of sequencing technology, the third generation currently on the market, i.e. Pacific Biosciences (Eid et al., 2009) and Oxford Nanopore (Clarke et al., 2009), provides longer reads and more uniform sequencing coverage, but the sequencing error rate is much higher (although errors are more uniformly distributed than the second generation).
Several de novo genome assembly tools are available, for both second and third generation sequencing data. Most assemblers for second generation sequencing data rely on the de Bruijn graph (e.g. Idury and Waterman, 1995; Peng et al., 2010; Pevzner et al., 2001; Simpson et al., 2009; Zerbino and Birney, 2008) which allows one to avoid the pairwise overlap step on the massive number of short reads in input. Assemblers for third generation sequencing data mainly use the overlap graph to store prefix-suffix overlaps between the long (noisy) reads in input (Hernandez et al., 2008; Myers, 2005). Not only are these assembly tools fundamentally different at the algorithmic level, but their designers have made different choices in the tradeoff between maximizing assembly contiguity (e.g. N50) and minimizing the probability of misassemblies (e.g. mis-joins). Often these assembly tools have dozens of parameters that allow one to adjust these trade-offs, but these parameters can be difficult to optimize for a specific input dataset and target genome. As a result, it is common practice to generate as many assemblies as possible within the time frame of the sequencing project using different assemblers and/or parameter settings, and try to identify the highest quality assembly based on assembly statistics. However, it is difficult to identify the ‘best’ assembly just from the assembly statistics. For instance, the assembly with the highest N50 is likely to be the one with most chimeric contigs (i.e. contigs with mis-joins).
The concept of assembly reconciliation has been proposed recently as a more appealing alternative. Instead of selecting the ‘best’ assembly, assembly reconciliation tools take advantage of all the individual assemblies. They claim to produce a higher quality (consensus) assembly by merging all of the available assemblies, so that the contiguity of the assembly increases without introducing misassembles. It turns out that the problem of assembly reconciliation is also quite challenging. While several assembly reconciliation tools are available [see, e.g. Reconciliator (Zimin et al., 2008), CISA (Lin and Liao, 2013), GAA (Yao et al., 2012), GAM_NGS (Vicedomini et al., 2013), GARM (Mayela Soto-Jimenez et al., 2014), MIX (Soueidan et al., 2013), ZORRO (Argueso et al., 2009), Metassembler (Wences and Schatz, 2015)], we have recently shown in (Alhakami et al., 2017) that none of these tools can generate a reconciled assembly which has consistently better contiguity and correctness than the assemblies given in input.
Optical mapping technology allows life scientists to produce genome-wide maps by fingerprinting long DNA molecules, typically via nicking restriction enzymes. Linear DNA fragments are stretched on a glass surface or in a nano-channel array, then the locations of restriction sites are identified with the help of dyes or fluorescent labels. While the cost of an optical map depends on the genome size, it is not prohibitive in the overall budget of a sequencing project. For a eukaryotic genome of about 1 Gb, the cost ranges from $5000 to $10 000. Based on our survey of the literature in the last three years, about half of the major eukaryotic sequencing projects used an optical map in the assembly pipeline, e.g. goat (Bickhart et al., 2017), human (Pendleton et al., 2015), sea bass (Vij et al., 2016), Ae.tauschii (Zimin et al., 2017), apple (Daccord et al., 2017), barley (Mascher et al., 2017), maize (Jiao et al., 2017b), quinoa (Jarvis et al., 2017).
In this paper, we introduce an assembly reconciliation algorithm called Novo&Stitch that takes advantage of optical maps to accurately carry out assembly reconciliation. One or more optical maps are used to obtain coordinates for the assembled contigs, which are then stitched based on their alignments. The availability of the optical map dramatically reduces the complexity of the problem and the possibility of introducing mis-joins. To take advantage of the optical map, however, contigs have to be sufficiently long so that they can be reliably aligned (e.g. 50 Kbp or longer). The source code of Novo&Stitch can be obtained from https://github.com/ucrbioinfo/Novo_Stitch.
2 Problem definition
In the following, we will use to denote the set of contigs in the genome assembly, where each contig si is a string over the alphabet . Given our interest in assembly reconciliation, S is going to be the union of multiple assemblies, obtained from multiple assemblers and/or parameters settings. In other words, S is expected to be highly redundant, i.e. each genomic location is expected to be covered by multiple contigs (unless it is highly repetitive). Henceforth, we assume that the contigs in S are chimera-free (see ‘Materials and methods’ section below for details).
An optical map is composed by a set of optical molecules, each of which is represented by an ordered set of positions for the restriction enzyme sites. As described above, optical molecules are obtained by an assembly process similar to sequence assembly, but we will reserve the term ‘contig’ exclusively for sequenced contigs. We will use to denote the optical map, where each optical molecule oj is an ordered set of integers, corresponding to the distances in base pairs between two adjacent restriction enzyme sites on molecule oj. By digesting in silico the contig si using the same restriction enzyme used to produce the optical map and matching the ordered list of adjacent distances between sites, one can align the contigs in S to optical map M. High quality alignments allow some of the contigs to be anchored at specific coordinates on the optical map. In addition, contigs can be oriented with respect to each other. A reliable alignment requires contigs to be sufficiently long (e.g. 50 Kbp or longer). When multiple contigs align to the same optical molecule, an estimate of the distance between them can be obtained. If the distance is positive, a gap is introduced and a scaffold can be formed (Shelton et al., 2015). When the distance is negative (i.e. contigs are overlapping), it may be possible to stitch them.
Given our interest in merging multiple assemblies, here we focus on the case when contigs are overlapping. A series of practical factors make the problem of stitching overlapping contigs non-trivial. These factors include imprecisions in optical maps (e.g. mis-joins introduced during the assembly of the optical map), inaccurate alignment between contigs and optical molecules, and multiple anchoring positions for the same contigs that are not consistent with each other. As a consequence, it is appropriate to frame this problem as an optimization problem.
As said, we are given multiple chimera-free assemblies represented by a set of contigs S, an optical map M and a set of alignments of S to M, where is the alignment of contig si to optical molecule oj. The problem is to stitch overlapping contigs based on A and obtain a new set of longer contigs such that (i) T covers the same portion of the genome covered by S, (ii) k is as small as possible and (iii) the conflicts of T with respect to A are minimized. This optimization problem is not rigorously defined unless one defines precisely the concept of conflict, but this description captures the spirit of what we want to accomplish. Even if the notion of conflict could be made precise, this multi-objective optimization problem would be hard to solve. Instead of solving this problem, we propose an iterative method that accomplishes a similar objective.
3 Materials and methods
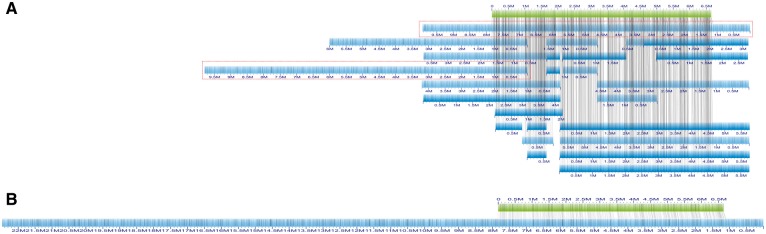
The proposed stitching method is an iterative algorithm. Each iteration is composed of three phases: data reduction, stitching and post-processing. The real example in Figure 1 will help understanding the phases. In (A) eight assemblies of cowpea were concatenated and aligned the contigs (blue) on the optical map (green) (see ‘Experimental results’ for details on these assemblies). As said, we assume that the input assemblies are free of chimeric contigs. Optical maps are routinely used to split chimeric contigs, which are easy to detect because they induce alignment conflicts. Unfortunately, since optical maps are obtained via an assembly process similar to sequence assembly, optical molecules can also be chimeric. According to Jiao et al. (2017a), ‘in around 7% of the (alignment) conflicts, the consensus map (optical map) was wrong’. Based on our experience, mis-joins in optical molecules typically occur in repetitive regions of the genome, which induce long stretches of regularly-spaced bands. For this reason, we have developed a new tool that can accurately split chimeric contigs (unpublished).
Fig. 1.
(A) Contigs (blue) of eight assemblies mapped to one optical molecule (green); minimum tiling path contigs are highlighted in red; (B) final stitched contig, at the end of the iterative stitching process (observe that the final contig spans a genomic region longer than the two MTP contigs)
Observe that among the eight assemblies, contigs produced by some assemblers can extend much further than others. In the first phase, the smallest subset of contigs that cover the same genomic region of the eight assemblies is selected (highlighted in red in the figure). In the second phase, the two MTP contigs are stitched to produce (B). Observe that in this case the resulting 22 Mb contig is much longer than the expected 12.5 Mb due to additional stitching that occurred in later iterations. In the third phase, stitched contigs are checked for consistency, then the entire process is iterated. The pipeline of the algorithm is illustrated in Figure 2.
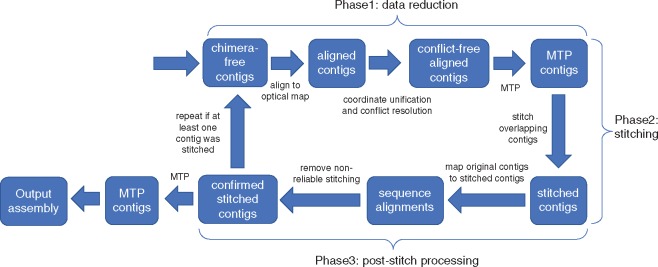
Fig. 2.
Pipeline of Novo&Stitch; the input to the pipeline is a set of chimera-free contigs and an optical map
3.1 Phase 1: Coordinate unification, conflict resolution and MTP
At high level, phase one has three major steps. In step 1, we align in silico-digested chimera-free contigs to the optical map (e.g. for a Bionano optical map, we use RefAligner), but not all alignments are used. We only consider alignments that (i) meet a minimum confidence level (typically confidence 25 in the case of RefAligner) and (ii) do not create conflict with each other (see below for details). Since some contigs can have high-quality conflict-free alignments to more than one optical molecule (which could indicate alternative overlaps), a ‘unification’ step is necessary (step 2, see below for details). Finally in step 3, we compute the minimum tiling path (MTP) of the contigs. Formally, let S be the initial set of contigs, and M be the optical map. Let be the set of high-quality conflict-free alignments of S to M, where is the alignment of contig si to optical molecule oj. Let be the set of intervals of M covered by the contigs S through the set of alignments A. A minimum tiling path of S is the smallest set such that P covers every interval in R.
3.1.1 Selecting conflict-free alignments
Our algorithm for reducing false alignments relies on a conflict graph. The conflict graph is an undirected hypergraph in which each vertex represents an alignment, and each hyperedge connects four vertices when the corresponding four alignments conflict with each other. Nodes of the hypergraph are weighted by the confidence of the alignments. Let us call the alignments of contig p and q on optical molecule i and j, respectively. We say that is a conflict if any of them have an orientation conflict or a coordinate conflict. An orientation conflict occurs when the orientations of and , and the orientations of and are neither both 5′–3′ nor both 3′–5′ (depending on whether i and j are from the same strand of the genome or not). A coordinate conflict occurs when the distance between and is significantly different from the distance between and . When four alignments have a conflict, at least one must be a false alignment. We model the problem of removing false alignments as a weighted vertex cover problem on the conflict hypergraph.
Since the weighted vertex cover problem on hypergraph is NP-hard, we use an approximation algorithm. We formulate weighted vertex cover as an integer program, as follows
where V and E are the vertex set and hyperedge set of the conflict hypergraph, respectively.
In order to solve the integer program we relax it to a linear program, and solve the linear program by standard software packages (e.g. GLPK or CPLEX). The solution of the linear program is transformed into an integer solution as follows. We sort each variable in decreasing order and we add the corresponding vertex to the solution if the new vertex covers at least one hyperedge that was not covered previously. This greedy algorithm is a 4-approximation algorithm (see below), which is the best known approximation achievable in polynomial time for hypergraph with hyperedges of constant size (Cardinal et al., 2012).
Theorem 1. The LP-based greedy algorithm for the weighted vertex cover on hypergraph gives an approximation ratio of 4.
Proof. Let C be a vertex cover. Consider a hyperedge . Since , either or or or . Therefore, is covered. If is an optimum vertex cover, then because
First inequality: LP is a relaxation of ILP. Second inequality: . Third inequality: . Fourth inequality: . □
Details of the approximation algorithm are shown in Algorithm 1. In Novo&Stitch, the conflict graph is first divided into connected components. The approximation algorithm is run on connected components with more than twenty vertices. For components with at most twenty nodes, we run the exhaustive (optimal) algorithm.
Algorithm 1 .
Greedy algorithm for weighted vertex cover problem on hypergraph
1: procedure LP_Round_Greedy(V, E)
2: Compute the optimum solution to LP relaxation (2).
3:
4:
5:
6: fordo
7: ifnew = False
8: fordo
9: ifthen
10: ifnew = True
11: remove e from
12: if ifnew = True then
13: add i to C ▹ pick i, if it appears in at least one new superedges
14: returnC
3.1.2 Unifying coordinates and computing the MTP
Our algorithms for computing the MTP and coordinate unification use the association graph between optical molecules and contigs. The association graph is an undirected graph in which each vertex represents an optical molecule and an edge indicates that the two molecules share at least one contig aligned to both of them. The weight of edge (oi, oj) between molecule oi and oj is obtained from the confidences of the alignments of all common contigs, that is where Si and Sj are the sets of contigs aligned to oi and oj, respectively, and is the confidence score provided by RefAligner between contig s and molecule o. The confidence score represents the quality of the alignment (higher is better). For the MTP and unification step, we do not use association graph directly, rather the minimum spanning forest (MSF) of the association graph. By construction, MSF identifies the most reliable alignments (i.e. highest total confidence) between contigs and optical molecules.
We first unify the coordinates of all contigs with respect to the optical molecules they are aligned to using the MSF of A, as follows. We traverse each MST, starting from the vertex that represents the molecule that has the highest total alignment confidence score (for the contigs aligned to it). That node becomes the root of the MST and it defines the origin of the coordinate system. As we traverse the MST, we assign the coordinates of each contig on a molecule based on average position of all the common contigs. Specifically, the position of molecule x with respect to molecule r is where C is the set of contigs aligned to both r and x, and gives the middle points of contig c’s alignment on molecule m.
Once the unification process is complete, we rebuild the association graph using the updated coordinates. At this stage we also remove contigs which are completely contained in other contigs, since they will not be used in the stitching. In order to compute the MTP we build another graph, called the overlap graph. The overlap graph O is an unweighted directed acyclic graph (DAG) in which vertices represent contigs and directed edges indicate overlaps between the corresponding contigs (oriented left to right along the coordinates induced by the alignment). Each optical molecule induces an overlap (sub)graph for the contigs aligned to it, but since a contig can align to multiple optical molecules, some of the overlap subgraphs can be connected. In order to efficiently connect the subgraphs in O, we use the MSF of A. Recall that by construction the nodes (which are optical molecules) in the same MST of A share common contigs. We process each minimum spanning tree in A, as we did above. As we traverse the MST we connect the corresponding subgraphs in O. Edges are added to O only if no cycles are introduced.
Once the overlap graph is finalized, we compute the MTP on each connected component of the graph. In the ideal case, each connected component Oi of O (which is a DAG by construction) is expected to have exactly one source and one sink because the genome is one-dimensional and the chain of overlaps is expected to have exactly one leftmost contig (source of the DAG) and exactly one rightmost contig (sink). When a connected component Oi has one source s and one sink t, the MTP problem reduces to finding the path from s to t with the smallest number of vertices. In practice however, Oi may have a set OS of sources and a set OT of sinks. A simple example explains why this could happen. Assume three staggered contigs A, B, C of the same length which overlap two disjoint optical molecules: the first molecule overlaps A, B, C in their prefixes, while the second molecule overlap only B, C on their suffixes. One should expect the overlap DAG to be . But now assume that the quality of the alignment of B with the first molecule is poor, so in the first molecule we get , in the second molecule we get . When we merge them, we end up with a DAG with two sources. When either or , the MTP problem requires finding the smallest subgraph Pi of Oi such that for any source-sink pair in which t is reachable from s in Oi, t is also reachable from s in Pi. We call this problem the smallest sub-DAG problem, defined as follow.
Definition 2 (Smallest SubDAG). Input: A connected directed acyclic graph , with source set S, and sink set T. Output: A subgraph of G such that (i) is a connected directed acyclic graph, (ii) (iii) , (iv) is the smallest among all the subgraphs satisfying (i–iii).
Theorem 3. Smallest SubDAG is NP-hard.
Proof. We show that Set Cover Smallest SubDAG. Let be an instance of Set Cover, where U is the universe of sets and C represents the collection of sets. Given we build an instance of Smallest SubDAG as follows. For each element in U, build a vertex in V and in T. For each set in C, build a vertex in V. Let and add s to V. For each set c in C and each element e in U, if e belongs to c, build an edge in E from the vertex corresponding to c to the vertex corresponding to e. For each set c in C, build an edge in E from s to the vertex corresponding to c. The equivalence between these two problems is obvious. □
Given the hardness of the Smallest SubDAG problem, we propose a greedy heuristic. First, we find the shortest path from each source to each sink. The shortest path among all these paths is chosen as the initial path. Then, the source and sink vertices not covered by the chosen path are added to the solution iteratively by calculating the shortest path between them to the current sub-DAG. Details of this greedy algorithm are shown in Algorithm 2.
Algorithm 2 .
Greedy algorithm for Smallest SubDAG problem
1: procedure GA()
2: current_set
3: subgraph
4: fors in S and t in Tdo
5: path BFS(G, s, t) ▹ compute the shortest path in G from s to t
6: if no_vertices(path) < no_vertices(subgraph) then
7: subgraph, path, s, t
8: current_set current_set
9: while current_set do
10: path* ← G
11: forx in current_set do
12: fory in subgraph do
13: ifthen ▹ x is a source
14: path BFS(G, x, y)
15: else ▹ x is a sink
16: path BFS(G, y, x)
17: if no_vertices(path) < no_vertices(path*) then
18: path*, x* ← path, x
19: current_set, subgraph current_set , subgraph path*
20: return subgraph
3.2 Phase 2 and 3: Contig stitching and post-processing
In phase 2 we first compute the sequence alignments for MTP contigs that are overlapping according to coordinates obtained in phase 1. For each pair of overlapping contigs, we determine the best alignment between the corresponding sequences. If the best alignment is (i) above a certain length and (ii) of sufficient quality (e-value), and (iii) consistent with the optical map coordinates, the stitching is carried out. When stitching two aligned contigs c1 and c2, both c1 and c2 are composed of three parts, left overhang l1, l2, right overhang r1, r2 and common region (aligned region) m1, m2. The new stitched contig d is formed by the concatenation of (i) the longest between l1 and l2 (ii) either m1, m2 depending which one is closer to the 5′ of its respective contig and (iii) the longest between r1 and r2. If c2 is stitched with c1, neither c1 or c2 will be used for stitching with the other contig in this iteration. Contig d could be stitched to other contigs in later iterations.
In Phase 3, we check the correctness of the stitching by aligning the two original contigs to the new stitched contig. The difficulty of this step stems from the possible fragmentation of alignments. Sequence alignment tools (e.g. Blast) can generate a large set of alignments, many of which are not informative. To determine the best overall alignment, we find the subset of mutually compatible alignments (from the set of all alignments) which has the longest total length. We say that two alignments are compatible if their overlap is smaller than a given fraction of the shorter alignment.
Definition 4 (Optimal set of mutually compatible alignments). Input: A list A of n alignments and their lengths, and a compatibility matrix C in which True if alignment i is compatible with alignment j. Output: A subset of A in which (i) for any pair of alignments True and (ii) the total length of the alignments in is the largest among all the subsets of A satisfying (i).
To solve this problem, we use a dynamic programming algorithm. All alignments are first sorted by their starting positions. Let be the total length of the alignments selected from that includes alignment . First we initialize to the length of first alignment. The rest of the dynamic programming vector can be filled using the following recurrence relation:
Details of this dynamic programming algorithm are described in Algorithm 3. The time complexity of this dynamic programming algorithm is . To speed it up, we remove alignments shorter than a given threshold to reduce the value of n.
After the optimal set of compatible alignments is computed, we determine the fraction of the two original overlapping contigs mapped to the stitched contig. If the fraction is below a predefined threshold, the stitching is cancelled.
Phase 1, 2 and 3 are repeated iteratively until no further stitching takes place. Recall that when we unify the coordinates, we compute the average position of all common contigs. When a contig appears in multiple fragments, it can affect the coordinates of other contigs, which in turn can change the detection of overlapping contigs. When we stitch contigs, the coordinates of several contigs can change, which can reveal overlaps that were not detected before. That is why we use an iterative strategy: later iterations can ‘make up’ for stitches that were missed in earlier iterations.
The final assembly produced in output is the MTP of the latest stitched assembly.
Algorithm 3 .
Dynamic programming algorithm for optimal set of compatible alignments
1: procedure Compatible(A, L, C)
2:
3: for to ndo
4:
5: for to i – 1 do
6: ifthen
7: ifthen
8:
9:
10:
11: for to ndo
12: ifthen
13:
14:
15: whiledo
16:
17: return
4 Experimental results
We tested Novo&Stitch on multiple PacBio assemblies of (i) cowpea (Vigna unguiculata) and (ii) Phytophthora infestans. Both sequencing projects are currently underway at UC Riverside. Cowpea is a legume crop that is resilient to hot and drought-prone climates. This legume is the primary source of protein in sub-Saharan Africa and other parts of the developing world. P.infestans is responsible for the late blight diseases of tomato and potato. It was the major culprit for the European potato famines of the 19th century. Worldwide the disease causes around $6 billion of damage to crops each year.
We considered running Novo&Stitch on the assemblies provided for the Assemblathon 2 competition (Bradnam et al., 2013) in which optical maps were used to evaluate the quality of the assemblies. Unfortunately, the authors of (Bradnam et al., 2013) could not locate the optical maps. We also considered comparing the results of Novo&Stitch against published assembly reconciliation tools, e.g. CISA (Lin and Liao, 2013), GAA (Yao et al., 2012), GAM_NGS (Vicedomini et al., 2013), GARM (Mayela Soto-Jimenez et al., 2014), MIX (Soueidan et al., 2013), ZORRO (Argueso et al., 2009), Metassembler (Wences and Schatz, 2015). However, (1) we have shown in Alhakami et al. (2017) that none of these tools can generate assemblies that are consistently better than the assemblies provided in input, and (2) the comparison would not be entirely fair because these tools do not take advantage of the optical map.
4.1 Experimental results on cowpea assemblies
Cowpea (Vigna unguiculata) is a diploid with a chromosome number 2n = 22 and an estimated genome size of 620 Mb. The genome has very low heterozygosity; in practice it can be considered haploid. We sequenced an elite African variety (IT97K-499-35) using single-molecule real-time sequencing (Pacific Biosciences RSII). A total of 87 SMRT cells yielded about 6 M reads for a total of 56.84 Gbp (91.7× genome equivalent). To test Novo&Stitch we generated several assemblies with a mix of parameters, polishing qualities and assembly tools. We used Canu (Berlin et al., 2015; Koren et al., 2017), Falcon (Chin et al., 2016) and ABruijn (Lin et al., 2016) to generate eight assemblies. Canu was run with different parameters to generate six of the eight assemblies (parameters shown in Table 1). Canu1, Canu2 and Canu6 were polished with Quiver. Chimeric contigs were detected and split using a tool currently under development in our lab (unpublished). Our tool can also detect mis-joins in the optical maps. This step could have been carried out manually, by inspecting the alignments of contigs against the optical map via IrysView. The final eight assemblies were inspected visually to ensure that they were indeed chimera-free.
Table 1.
Parameter choices for Canu v1.3
| CANU assembly | cMS | cMEE | cOC | Quiver |
|---|---|---|---|---|
| 1 | Low | Default | Default | ✓ |
| 2 | Low | Default | 100 | ✓ |
| 3 | High | Default | Default | |
| 4 | High | 0.15 | 100 | |
| 5 | Normal | 0.15 | 100 | |
| 6 | High | Default | 100 | ✓ |
Note: Column cMS reports the value of corMhapSensitivity, cMEE reports corMaxEvidenceErate, cOC reports corOutCoverage. Three of these assemblies were polished with Quiver.
The basic statistics for the eight assemblies are provided in Table 2. In addition to standard contiguity statistics [N50 (length for which the set of contigs of that length or longer accounts for at least half of the assembly size), L50 (minimum number of contigs accounting for at least half of the assembly), NG50 (length for which the set of contigs of that length or longer accounts for at least half of the 620Mb genome) and LG50 (minimum number of contigs accounting for at least half of the 620Mb genome)], total assembled size and contig length distributions, we evaluated the assemblies using several other independent metrics. We mapped (i) about 129K cowpea WGS contigs assembled from short reads (Muñoz-Amatriaín et al., 2017; assembly v.0.03), (ii) about 200 M 2× 100 paired-end Illumina reads generated at UCR in 2014 and (iii) transcripts assembled from RNA-Seq short reads. In Table 2 we report the percentage of DNA sequenced mapped with BWA with a minimum MapQ of 30. Finally, we compared the assemblies against the high-density genetic map available from (Muñoz-Amatriaín et al., 2017). To evaluate possible chimeric contigs, we BLASTed 121 bp-long design sequence for the 51 128 genome-wide SNPs described in (Muñoz-Amatriaín et al., 2017) against each assembly, then we identified which contigs had SNPs mapped to them, and what linkage group (chromosome) of the genetic map those mapped SNPs belonged to. Chimeric contigs are revealed when their mapped SNPs belong to more than one linkage group. The last line of Table 2 reports the total size of contigs in each assembly for which (i) they have at least one SNPs mapped to it and (ii) all SNPs belong to the same linkage group (i.e. likely to be non-chimeric). Observe in Table 2 that there is no single assembly that is the ‘best’ in each row. Canu6 has the highest N50 and the lowest L50, but Canu2 has the longest contig. Canu1 has the highest NG50. ABruijn has the smallest number of contigs.
Table 2.
Assembly statistics of eight assemblies for cowpea; all reads/transcripts/BAC assemblies were mapped with BWA, MapQ30; number in boldface are the best statistics (min or max) across assemblies; for # contigs 100kbp and 1Mbp it is not obvious whether to report min or max
| Canu1 | Canu2 | ABruijn | Falcon | Canu3 | Canu4 | Canu5 | Canu6 | |
|---|---|---|---|---|---|---|---|---|
| Contig N50 (bp) | 4 859 617 | 4 498 063 | 1 896 002 | 2 869 362 | 3 280 469 | 2 797 949 | 2 666 731 | 5 340 274 |
| Contig L50 | 30 | 32 | 74 | 49 | 42 | 51 | 55 | 29 |
| Contig NG50 (bp) | 3 767 556 | 3 417 577 | 1 330 435 | 1 737 012 | 2 431 239 | 1 949 515 | 2 068 575 | 3 451 071 |
| Contig LG50 | 43 | 45 | 119 | 73 | 63 | 73 | 77 | 42 |
| Total assembled (bp) | 506 154 442 | 516 817 613 | 478 230 679 | 511 933 729 | 503 187 311 | 516 537 734 | 515 949 175 | 507 773 747 |
| # contigs | 894 | 928 | 538 | 1820 | 1038 | 1110 | 1140 | 897 |
| # contigs 100kbp | 220 | 288 | 437 | 404 | 299 | 354 | 334 | 278 |
| # contigs 1Mbp | 104 | 107 | 151 | 118 | 128 | 142 | 145 | 103 |
| # contigs 10Mbp | 7 | 8 | 0 | 1 | 2 | 2 | 0 | 9 |
| Longest contig (bp) | 18 473 372 | 18 498 533 | 8 846 014 | 10 554 495 | 14 090 735 | 14 331 160 | 9 775 097 | 17 211 165 |
| WGS contigs 500bp, % mapped (129K) | 98.27412% | 98.77014% | 88.30652% | 97.84959% | 98.30618% | 98.25853% | 98.23673% | 98.73930% |
| UCR2014 reads, % properly paired (202M) | 92.59433% | 92.64181% | 92.30106% | 91.95107% | 92.52969% | 92.63057% | 92.62330% | 92.59763% |
| UCR2014 reads, % mapped (202M) | 64.35764% | 63.50279% | 64.21367% | 59.49035% | 64.38425% | 63.00587% | 63.22414% | 62.84466% |
| Assembled transcripts, % mapped (157K) | 92.60644% | 94.83972% | 94.95582% | 94.16235% | 92.65416% | 92.52276% | 92.46959% | 94.85657% |
| Total length with 100% consistent LG (bp) | 331 956 528 | 338 556 993 | 379 029 914 | 312 593 019 | 356 505 616 | 349 534 672 | 347 586 448 | 425 812 490 |
Novo&Stitch was run on the eight assemblies in Table 2 using two Bionano Genomics optical maps, the first obtained using the BspQI nicking enzyme (which recognizes ‘GCTCTTC’), and the second obtained with the BssSI nicking enzyme (‘CACGAG’). The BspQI optical map had 508 assembled optical molecules with a molecule N50 of 1.62 Mb and a total length of 622.21 Mb. The BssSI optical map had 743 assembled optical molecules with a molecule N50 of 1.02 Mb and a total length of 577.76 Mb. Both optical maps were assembled at UC Davis using the Bionano assembler. For each optical map we used two sets of parameters, called ‘strict’ (-a 3000 -b 0.1 -c 10000 -d 0.5 -e 0.9 -h 25 -r 0.2) and ‘loose’ (-a 0 -b 0.2 -c 5000 -d 0.5 -e 0.8 -h 25 -r 0.2). Please refer to the README at https://github.com/ucrbioinfo/Novo_Stitch for details about these parameters. For convenience, in first column of Table 3, we copied the best statistics across the eight assemblies in Table 2. Note that no individual assembly, however, has these statistics. Observe in Table 3 that Novo&Stitch almost doubled the N50, reduced the L50 from 29 to 19, increased the number of contigs 10 Mb from 9 to 17–18. Mapping statistics remained unaltered, as well as the agreement with the genetic map. Taken all together, these statistics indicate that Novo&Stitch produced a much more contiguous assembly, with no more chimeric contigs than the best of the eight input assemblies. On this dataset Novo&Stitch converges in two-three iterations and takes only a few hours on our 32-core server.
Table 3.
Assembly statistics of Novo&Stitch on the eight cowpea assemblies using either the BspQI or the BssSI optical map, ‘best of 8’ is a copy the best statistics (boldface) among the eight assemblies in Table 2—no individual assembly, however, has these statistics; see text about strict and loose parameters; all DNA sequences were mapped with BWA, MapQ30
| Best of 8 | BspQI (loose) | BspQI (strict) | BssSI (loose) | BssSI (strict) | |
|---|---|---|---|---|---|
| Contig N50 (bp) | 4 859 617 | 9 944 851 | 9 944 851 | 9 584 779 | 9 584 779 |
| Contig L50 | 29 | 19 | 19 | 19 | 19 |
| Contig NG50 (bp) | 3 767 556 | 9 944 851 | 8 187 172 | 7 956 155 | 7 826 863 |
| Contig LG50 | 42 | 19 | 24 | 24 | 24 |
| Total assembled (bp) | 516 817 613 | 522 393 141 | 523 526 657 | 520 162 831 | 523 249 509 |
| # contigs | 538 | 791 | 798 | 791 | 798 |
| # contigs 100kbp | N/A | 211 | 218 | 211 | 218 |
| # contigs 1Mbp | N/A | 72 | 72 | 66 | 69 |
| # contigs 10Mbp | 9 | 18 | 18 | 17 | 17 |
| Longest contig (bp) | 18 498 533 | 21 980 320 | 21 980 320 | 22 385 362 | 22 385 362 |
| WGS contigs 500bp, % mapped (129K) | 98.77014% | 97.77496% | 97.79009% | 97.40359% | 97.02018% |
| UCR2014 reads, % properly paired (202M) | 92.64181% | 92.57437% | 92.58778% | 92.47305% | 92.50176% |
| UCR2014 reads, % mapped (202M) | 64.38425% | 62.20807% | 62.11027% | 61.82553% | 61.63417% |
| Assembled transcripts, % mapped (157K) | 94.95582% | 93.93669% | 93.90570% | 94.01125% | 93.46803% |
| % contigs with 100% consistent LG | 425 812 490 | 429 367 225 | 430 234 966 | 423 454 837 | 434 621 644 |
4.2 Experimental results on P.infestans assemblies
We sequenced a strain of P.infestans from California called ‘1306’. Strain 1306 is a diploid (other P.infestans strains are triploid or aneuploid), has 11–14 chromosomes and an estimated genome size of 220 Mb. P.infestans has a much higher rates of heterozygosity compared to V.unguiculata, thus the former has a more challenging genome to assemble. P.infestans 1306 was sequenced using single-molecule real-time sequencing (Pacific Biosciences RSII). A total of 17 SMRT cells yielded about 3.1 M reads for a total of 24.87 Gbp (113× genome equivalent). We tested Novo&Stitch on six assemblies of P.infestans. We generated two assemblies with Canu v1.5, one on the entire dataset (Canufull, 113× coverage) and one on PacBio reads longer than 10Kb (Canu10K, 80.9× coverage). We generated three assemblies with ABruijn v0.4 on three datasets, namely (i) PacBio reads longer than 10Kb (ABruijn10K, k = 17, 80.9× coverage), (ii) all PacBio reads corrected by Canu (ABruijncorr, k = 17, 75.4× coverage) and (iii) all PacBio reads corrected and trimmed by Canu (ABruijntrim, k = 17, 73.6× coverage). One assembly was produced with Falcon on the whole dataset by the UC Davis core facility. We detected and split chimeric contigs using Chimericognizer, a tool currently under development (unpublished). We ensured that the six assemblies were free of chimeric contigs by visually inspecting the alignments.
Novo&Stitch was run on the six assemblies in Table 4 using a Bionano Genomics optical map. To evaluate the quality of these assemblies, we mapped about 47 M miSeq reads and 202 M Dovetail read using BWA. We also mapped a fraction of those reads using Blast, which does not penalize the mapping quality in case of alignment of a read to multiple locations. The last two columns report the statistics of Novo&Stitch using strict and loose parameters. Observe again how Novo&Stitch significantly improved the contiguity of the assembly (N50, L50, longest contig, etc.) while maintaining mapping statistics similar to the six input assemblies.
Table 4.
Statistics of six input assemblies for P.infestans and two stitched assemblies (N&S = Novo&Stitch) with strict and loose parameters; all reads were mapped with Bwa, except for 1% of miSeq and 0.1% of Dovetail which were mapped using Blast (e-value < 1e-30)
| Falcon | Canu10K | Canufull | ABruijn10K | ABruijncorr | ABruijntrim | N&Sloose | N&Sstrict | |
|---|---|---|---|---|---|---|---|---|
| Contig N50 (bp) | 481 068 | 131 313 | 135 263 | 356 459 | 293 280 | 302 893 | 769 322 | 730 890 |
| Contig L50 | 107 | 462 | 473 | 142 | 171 | 169 | 74 | 82 |
| Total assembled (bp) | 215 910 203 | 305 686 040 | 292 352 599 | 195 768 168 | 177 232 870 | 175 149 119 | 240 150 657 | 250 416 680 |
| # contigs | 1364 | 3496 | 2863 | 835 | 888 | 867 | 1304 | 1329 |
| # contigs 100kbp | 445 | 667 | 725 | 561 | 539 | 522 | 398 | 423 |
| # contigs 1Mbp | 36 | 12 | 7 | 19 | 9 | 10 | 54 | 55 |
| longest contig (bp) | 4 206 720 | 1 810 393 | 1 813 497 | 2 437 907 | 2 004 950 | 1 638 783 | 4 930 683 | 4 797 067 |
| miSeq reads, % mapped (47M) | 98.4995% | 98.6503% | 98.2370% | 98.0305% | 98.3051% | 98.2923% | 98.0928% | 98.0958% |
| miSeq reads, % properly paired (47M) | 96.3825% | 97.6855% | 95.5383% | 93.5911% | 94.8410% | 94.8218% | 95.6384% | 95.7194% |
| 1% miSeq reads, % mapped (0.47M) | 97.6510% | 97.9313% | 96.7831% | 96.4854% | 97.3612% | 97.3092% | 97.1461% | 97.1521% |
| Dovetail reads, % mapped (202M) | 97.7712% | 97.8934% | 97.6519% | 97.5093% | 97.6161% | 97.5835% | 97.4953% | 97.4989% |
| Dovetail reads, % properly paired (202M) | 38.7274% | 37.5416% | 37.4140% | 38.6057% | 38.5723% | 38.4578% | 37.9324% | 37.7392% |
| 0.1% Dovetail reads, % mapped (0.2M) | 91.6264% | 92.0876% | 91.3643% | 90.8826% | 91.3535% | 91.2612% | 91.1237% | 91.1447% |
5 Conclusions
We presented a new assembly reconciliation tool called Novo&Stitch for improving the contiguity of de novo genome assemblies using optical maps. Novo&Stitch leverages the alignments of contigs from multiple input assemblies to an optical map to detect overlaps between contigs and drive the stitching process. A requirement of Novo&Stitch is that assembled contigs have to be sufficiently long (e.g. 50 Kbp or longer) in order to be reliably aligned to the optical maps. This requirement is not very limiting for assemblies based on 3rd generation sequencing data, but it can be for Illumina-based assemblies. Experimental results on V.unguiculata and P.infestans clearly demonstrates that the addition of the optical map can significantly improve the contiguity of genome assemblies. The optical map can be used again on the improved stitched assembly to create scaffolds.
Acknowledgements
The authors thank Tao Jiang for helping with the NP-hardness proof for the smallest sub-DAG problem, and Timothy Close and María Muñoz-Amatriaín for helpful discussions as this work developed and for critically reading the manuscript.
Funding
This work was supported in part by the US National Science Foundation [IOS-1543963, IIS-1526742].
Conflict of Interest: none declared.
References
- Alhakami H. et al. (2017) A comparative evaluation of genome assembly reconciliation tools. Genome Biol., 18, 93.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argueso J.L. et al. (2009) Genome structure of a Saccharomyces cerevisiae strain widely used in bioethanol production. Genome Res., 19, 2258–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin K. et al. (2015) Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat. Biotechnol., 33, 623–630. [DOI] [PubMed] [Google Scholar]
- Bickhart D.M. et al. (2017) Single-molecule sequencing and chromatin conformation capture enable de novo reference assembly of the domestic goat genome. Nat. Genet., 49, 643–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradnam K.R. et al. (2013) Assemblathon 2: evaluating de novo methods of genome assembly in three vertebrate species. Gigascience, 2, 10.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardinal J. et al. (2012) Approximating vertex cover in dense hypergraphs. J. Discret. Algorithms, 13, 67–77. [Google Scholar]
- Chin C.-S. et al. (2016) Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods, 13, 1050–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke J. et al. (2009) Continuous base identification for single-molecule nanopore DNA sequencing. Nat. Nanotechnol., 4, 265–270. [DOI] [PubMed] [Google Scholar]
- Daccord N. et al. (2017) High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat. Genet., 49, 1099–1106. [DOI] [PubMed] [Google Scholar]
- Eid J. et al. (2009) Real-time DNA sequencing from single polymerase molecules. Science, 323, 133–138. [DOI] [PubMed] [Google Scholar]
- Hernandez D. et al. (2008) De novo bacterial genome sequencing: millions of very short reads assembled on a desktop computer. Genome Res., 18, 802–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idury R.M., Waterman M.S. (1995) A new algorithm for DNA sequence assembly. J. Comput. Biol., 2, 291–306. [DOI] [PubMed] [Google Scholar]
- Jarvis D.E. et al. (2017) The genome of chenopodium quinoa. Nature, 542, 307–312. [DOI] [PubMed] [Google Scholar]
- Jiao W.-B. et al. (2017b) Improving and correcting the contiguity of long-read genome assemblies of three plant species using optical mapping and chromosome conformation capture data. Genome Res., 27, 778–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y. et al. (2017b) Improved maize reference genome with single-molecule technologies. Nature, 546, 524–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S. et al. (2017) Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res., 27, 722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.-H., Liao Y.-C. (2013) CISA: contig integrator for sequence assembly of bacterial genomes. PloS ONE, 8, e60843.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. et al. (2016) Assembly of long error-prone reads using de Bruijn graphs. Proc. Natl. Acad. Sci. USA, 113, E8396–E8405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher M. et al. (2017) A chromosome conformation capture ordered sequence of the barley genome. Nature, 544, 427–433. [DOI] [PubMed] [Google Scholar]
- Muñoz-Amatriaín M. et al. (2017) Genome resources for climate-resilient cowpea, an essential crop for food security. Plant J., 89, 1042–1054. [DOI] [PubMed] [Google Scholar]
- Myers E.W. (2005) The fragment assembly string graph. Bioinformatics, 21, ii79–ii85. [DOI] [PubMed] [Google Scholar]
- Pendleton M. et al. (2015) Assembly and diploid architecture of an individual human genome via single-molecule technologies. Nat. Methods, 12, 780–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y. et al. (2010). IDBA–a practical iterative de bruijn graph de novo assembler. In: Proceeedings of Annual International Conference on Research in Computational Molecular Biology (RECOMB). Springer, pp. 426–440.
- Pevzner P.A. et al. (2001) An eulerian path approach to DNA fragment assembly. Proc. Natl. Acad. Sci. USA, 98, 9748–9753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelton J.M. et al. (2015) Tools and pipelines for BioNano data: molecule assembly pipeline and FASTA super scaffolding tool. BMC Genomics, 16, 734.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson J.T. et al. (2009) ABySS: a parallel assembler for short read sequence data. Genome Res., 19, 1117–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto-Jimenez L. et al. (2014) GARM: genome assembly, reconciliation and merging pipeline. Curr. Top. Med. Chem., 14, 418–424. [DOI] [PubMed] [Google Scholar]
- Soueidan H. et al. (2013) Finishing bacterial genome assemblies with mix. BMC Bioinformatics, 14, S16.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicedomini R. et al. (2013) GAM-NGS: genomic assemblies merger for next generation sequencing. BMC Bioinformatics, 14, S6.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vij S. et al. (2016) Chromosomal-Level assembly of the asian seabass genome using long sequence reads and multi-layered scaffolding. PLoS Genet., 12, e1005954.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wences A.H., Schatz M.C. (2015) Metassembler: merging and optimizing de novo genome assemblies. Genome Biol., 16, 207.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao G. et al. (2012) Graph accordance of next-generation sequence assemblies. Bioinformatics, 28, 13–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino D.R., Birney E. (2008) Velvet: algorithms for de novo short read assembly using de bruijn graphs. Genome Res., 18, 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimin A.V. et al. (2008) Assembly reconciliation. Bioinformatics, 24, 42–45. [DOI] [PubMed] [Google Scholar]
- Zimin A.V. et al. (2017) Hybrid assembly of the large and highly repetitive genome of aegilops tauschii, a progenitor of bread wheat, with the MaSuRCA mega-reads algorithm. Genome Res., 27, 787–792. [DOI] [PMC free article] [PubMed] [Google Scholar]