Abstract
Motivation
The emergence of high-throughput sequencing technologies revolutionized genomics in early 2000s. The next revolution came with the era of long-read sequencing. These technological advances along with novel computational approaches became the next step towards the automatic pipelines capable to assemble nearly complete mammalian-size genomes.
Results
In this manuscript, we demonstrate performance of the state-of-the-art genome assembly software on six eukaryotic datasets sequenced using different technologies. To evaluate the results, we developed QUAST-LG—a tool that compares large genomic de novo assemblies against reference sequences and computes relevant quality metrics. Since genomes generally cannot be reconstructed completely due to complex repeat patterns and low coverage regions, we introduce a concept of upper bound assembly for a given genome and set of reads, and compute theoretical limits on assembly correctness and completeness. Using QUAST-LG, we show how close the assemblies are to the theoretical optimum, and how far this optimum is from the finished reference.
Availability and implementation
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Modern DNA sequencing technologies cannot read the entire sequence of a chromosome. Instead, they generate large numbers of reads sampled from different parts of the genome. The emergence of low-cost and high quality second-generation sequencing (also known as next-generation sequencing or NGS) allowed scientists to decode numerous previously unsequenced organisms. However, the market of genome sequencing is rapidly developing and the recent domination of NGS technologies is now in question due to constantly improving third-generation (long-read) sequencing (LRS). The length of reads produced by LRS technologies, Pacific Biosciences and Oxford Nanopore Technologies, now can exceed hundred thousand bases, which is several orders of magnitude higher than the longest reads generated by NGS. At the same time, these technologies produce reads with the error rate significantly higher compared to the NGS competitors, and remain rather expensive. Thus, both kinds of DNA sequencing technologies suffer from major drawbacks, which can however be overcome with computational methods.
Genome assembly software combines the reads into larger sequences called contigs. Long repeats in the genomes impede reconstruction of full chromosomes, which is impossible in most cases when using only short reads produced by NGS technologies. However, these short read assemblies can be significantly improved with long-range mate-pair libraries (Chaisson et al., 2009; Vasilinetc et al., 2015). In contrast to paired-end libraries with typical insert size below 1 kilobase (kb), current mate-pairs protocols generate much longer inserts (up to 20 kb). Assembly software utilize that information to span over long repeats and coverage gaps, and extend contigs into longer sequences, usually referred to as scaffolds. At the same time, assembly algorithms for long reads typically do not require mate-pairs, since the read length is sufficient enough for resolving complex repeats. The most important drawback of the LRS technologies is the high per base error rate, that can be addressed either by investing into a higher sequencing depth (Koren et al., 2017), which may be expensive, or by correcting errors using high quality short reads (Walker et al., 2014).
Various assembly tools use different heuristic approaches to address the challenges of genome assembly, resulting in significant differences in the contigs and scaffolds they generate. It brings up the problem of comparing assemblies against each other. Assemblathon 1 (Earl et al., 2011) was one of the first attempts to assess the leading genome assembly software. The organizers provided a simulated short read dataset of an unknown simulated genome. The dataset was assembled by 17 groups of both assembler developers and users from around the globe and the resulting 41 assemblies were carefully evaluated. The subsequent study, Assemblathon 2 (Bradnam et al., 2013), used a similar competition model but provided the real sequencing data from three vertebrate species. Unfortunately, the evaluation scripts used in both assemblathon projects were mostly focused on those particular genomes and could not be easily applied for everyday quality assessment. The GAGE study (Salzberg et al., 2012) established a golden standard in the genome assembly evaluation and created a program that implements it. The GAGE tool was further extended and improved in the QUAST (Gurevich et al., 2013) software, which became a main evaluation tool in the subsequent GAGE-B competition for bacterial genome assemblers (Magoc et al., 2013). Recent critical assessment of metagenome interpretation study (Sczyrba et al., 2017) used MetaQUAST (Mikheenko et al., 2016b), an extension of QUAST for metagenomic assembly evaluation.
Despite the wide usage of the QUAST package, it can hardly be treated as a universal genome assembly evaluation tool. The reason for that is the original design aimed at bacterial and small eukaryotic assemblies which limits the tool application to genomes shorter than hundred megabases. In this work, we present QUAST-LG, a significantly upgraded version of QUAST capable of analyzing large genomic sequences. Our tool can evaluate mammalian-size assemblies within a few hours on a regular server. QUAST-LG includes both speed up improvements and new quality metrics that take into account specific features of the eukaryotic genomes, such as the abundance of transposons. In contrast to purely reference-free approaches like REAPR (Hunt et al., 2013), ALE (Clark et al., 2013) and LAP (Ghodsi et al., 2013), the original QUAST was limited in the absence of a known reference genome. QUAST-LG enables assessment for novel species by incorporating reference-free tools as a part of its pipeline; however, reference-free analysis is not the primary use case for QUAST-LG.
Another improvement in QUAST-LG is a concept of upper bound assembly which is based on the fact that the reference genome cannot be completely reconstructed from raw reads due to long genomic repeats and low covered regions. The previous studies on the near-optimal assembly problem (Bresler et al., 2013; Lam et al., 2014) estimate the read length and the coverage depth required for a successful genome reconstruction under some theoretical assumptions on shotgun sequencing data. We approach the opposite problem: given a dataset, QUAST-LG estimates the upper bound of completeness and contiguity that theoretically can be reached by assembly software from this particular set of reads. Moreover, our method takes into account that a dataset may consist of multiple sequencing libraries generated by both NGS and LRS technologies.
To demonstrate QUAST-LG functionality, we evaluate the ability of the state-of-the-art genome assembly tools to assemble medium-sized and large genomes. We use whole genome sequencing (WGS) datasets that include two libraries—Illumina paired-end reads, and either high-quality Illumina mate-pairs, or a long-read library. Both leading LRS technologies, Pacific Biosciences single molecule real time (PacBio SMRT) sequencing and Oxford Nanopore sequencing, are present among these test datasets. To enable convenient reproduction of our results, all datasets and software tools used in this study are freely available.
2 Materials and methods
2.1 Upper bound assembly construction
We construct upper bound assembly based on a reference genome and a given set of reads. At first, the construction algorithm maps all reads to the reference genome and detects zero-coverage regions (Fig. 1a). We use Minimap2 (Li, 2017) for aligning long error-prone reads (PacBio and Nanopore) and BWA-MEM (Li, 2013) for short Illumina reads (paired-ends and mate-pairs).
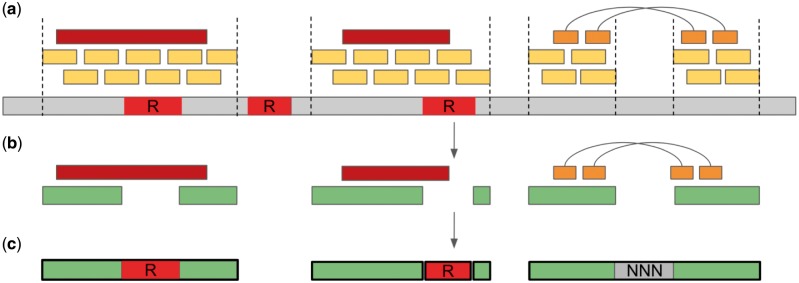
Fig. 1.
Upper bound assembly construction. (a) All available reads (brown for long reads, orange for mate-pairs, and yellow for paired-ends) are mapped to the reference (gray) to compute zero-coverage genomic regions. Repeat sequences (red) are detected using repeat finder software. Non-repetitive covered fragments are reported as upper bound contigs. (b) The overlaps between the contigs (green), and either long or mate-pair reads are detected, and contigs are further joined to form upper bound scaffolds. (c) The gaps between adjacent contigs within a scaffold are filled either with reference sequence (for covered regions) or with stretches of N nucleotides (for coverage gaps). Unresolved repeats are added as separate sequences
Further on, the lightweight Red (Girgis, 2015) de novo repeat finder is used to mark long genomic repeats in the reference (Fig. 1a). We call a repeat long if its length exceeds the median insert size of a paired-end library (when several paired-end libraries are available, the maximum median value is used). Among the detected repeated sequences, we select only those that occur at least twice in remote parts of the genome. Such long repeats cause ambiguities during the assembly, which may be resolved only by long reads or mate-pairs. Other long regions marked by Red appear to be short tandem repeats having multiple copies at the same genomic loci. To the best of our knowledge, such tandem repeats do not cause ambiguities and can be approximately resolved by the assemblers without using long-range information [e.g. using de Bruijn graph topology (Miller et al., 2010)].
Splitting the reference sequence by coverage gaps and long repeats results in a set of unique genomic fragments referred to as upper bound contigs, that however do not reflect the best possible assembly of the entire dataset. To achieve a more realistic upper bound, we detect the contigs that are connected by long reads or mate-pairs and further join them into upper bound scaffolds if the number of connections exceeds a small threshold n (Fig. 1b). In this study we used n = 1 for long reads and n = 2 for mate-pairs. We say that a long read connects two contigs if it simply overlaps with both contigs. A pair of reads connects contigs if the left read overlaps with the first contig and the right read overlaps with the second contig. During this analysis we ignore read pairs that map inconsistently or with abnormal insert size (in the first or the last decile). To enable efficient overlap detection between reads and upper bound contigs, we sort all reads according to their mapping positions. Thus, the scaffold construction algorithm requires time for read sorting and O(N) time for finding overlaps, where N is the total number of long and mate-pair reads used for scaffolding.
Once upper bound contigs are joined into scaffolds, the gaps between adjacent contigs are filled with the corresponding genomic sequences from the reference genome, or—in case of coverage gaps—with stretches of N’s (Fig. 1c). Remaining unresolved repeats are added to the final upper bound assembly as separate sequences.
2.2 Adaption of conventional metrics for large genomes
The key characteristics of the assembly quality are the assembly completeness (what fraction of the genome is assembled), correctness (how many errors the assembly contains) and contiguity (how many fragments the assembly consists of and how long they are). Both completeness and correctness can be accurately measured by QUAST-LG only when a high-quality reference genome is available. Some contiguity statistics, such as the well-known N50 metric, do not require a reference. However, when an estimate of the genome size is known, their more suitable analogues can be computed, namely NG50. If a reference sequence is available, we provide even more relevant insight by computing NGA50-like statistics (Gurevich et al., 2013), the contiguity measures based on error-free aligned assembly fragments rather than the initial contigs/scaffolds.
The alignment against the reference genome appears to be the most time consuming step in the assembly evaluation, especially for large genomes. To address this bottleneck, we replaced an accurate and slow NUCmer aligner [from MUMmer v3.23 package (Kurtz et al., 2004)] used in original QUAST with a faster Minimap2 aligner (Li, 2017). The recently released MUMmer 4 package (Marcais et al., 2018) was also outperformed by Minimap2 in our benchmark experiments, albeit the speed increase in this case was not as substantial as the Minimap2’s improvement over the previous MUMmer version. We have thoroughly chosen Minimap2 options in order to maintain the alignment speed-accuracy ratio for different scenarios. In standard mode QUAST-LG runs alignment with the parameters enabling accuracy comparable with NUCmer which is suitable for small genomes. In ‘‐‐large’ mode QUAST-LG configures Minimap2 to achieve adequate running times for large and complex inputs.
The assembly correctness is usually characterized by the number of large assembly errors, so-called misassemblies. Gurevich et al. (2013) define a misassembly breakpoint as a position in an assembled contig where the flanking sequences align to opposite strands (inversion), or to different chromosomes (translocation), or the inconsistency size δ (a gap or an overlap) between the alignments of the left and right flanking sequences on the reference is more than a predefined breakpoint threshold X (relocation). The alignments on the same strand of the same chromosome and having are considered as small errors and classified as local misassembly.
Eukaryotic genomes usually contain a lot of transposable elements (TEs) which may cause discrepancies between the reference genome and the genome actually being assembled. These short variations result in a huge number of false positive misassemblies if computed according to the definition given above. To distinguish between true misassemblies and the ones caused by TEs, QUAST-LG performs an additional check of each relocation and inversion to identify possible TEs (Fig. 2). The identification procedure depends on the size of the breakpoint threshold X which optimal value should slightly exceed the length of the largest TE in the genome (the processing of tandem TE insertions and deletions is out of scope of this paper). The optimal value thus depends on the subject organism and we allow users to set it manually. For the sake of consistency, we used the same X = 7 kb in all benchmark experiments in this study (see Supplementary Methods for details on the value choice). This is also the default value in QUAST-LG in contrast to regular QUAST which uses X = 1 kb.
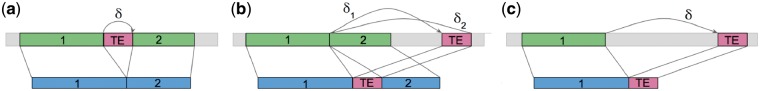
Fig. 2.
Detection of discrepancies caused by TEs. On each subfigure, we plot the reference genome R (top), the contig C (bottom), their matching fragments (blue and green bars for the positions in C and R, respectively) and locations of TEs (violet bars) causing discrepancies in the mapping. The inconsistencies in the alignments are shown by arrows and δ characters. (a) TE is present in R and missing in C. Since δ here is equal to the TE’s length, a specifically chosen breakpoint threshold X transforms classification of this discrepancy from a relocation to a local misassembly (). (b) TE is located inside C but its position in R is significantly away from the rest of C mappings and could also be located on the opposite strand. Original QUAST would treat this situation as two misassembly breakpoints (relocations or inversions) because δ1 and δ2 are usually much higher than X. In contrast, QUAST-LG classifies such pattern as possible TE since it computes , that is again equal to the TE’s length and could be prevailed by appropriate X. (c) TE is the first or the last alignment fragment in C, while its location on R is large distance δ away from the neighboring C fragment. QUAST-LG cannot reliably distinguish this situation from a real relocation/inversion: it would need to be able to recognize TE based on its genomic sequence, which is out of scope of this paper
2.3 Best set of alignments selection
Long contigs are rarely mapped to the reference perfectly as a single unambiguous alignment. An alignment software typically reports multiple alignment fragments from different locations of the genome. This may happen due to the presence of genomic repeats and TEs in the reference genome and in some cases because of algorithmic issues in the assembly or/and alignment software. QUAST-LG attempts to accurately assess each contig and select a set of non-overlapping alignments that maximizes the total alignment score, which is defined as a function of the alignment lengths, mapping qualities and side by side inconsistencies (misassemblies).
This problem is known as the collinear chaining problem (Myers and Miller, 1995) and it is usually solved by sequence aligners for a low-level chaining, that is joining short matching seeds into larger alignment fragments. For example, MUMmer (Kurtz et al., 2004) combines maximal unique matches and Minimap2 (Li, 2017) chains minimizers (Roberts et al., 2004). Here we implement a dynamic programming algorithm called BestSetSelection for a high-level chaining, that is combining alignment fragments (see Supplementary Methods).
Our algorithm is conceptually similar to delta-filter utility from MUMmer package (Kurtz et al., 2004) but our approach includes a comprehensive set of penalties for various misassembly events. This feature allows BestSetSelection to correctly resolve many complex sets of alignments, which are typical for eukaryotic assemblies, and produce a more accurate chaining than delta-filter in our benchmark experiments.
BestSetSelection is a quadratic algorithm with respect to the number of fragment alignments per contig which is usually fine since this number is generally small (up to 100). However, this may cause a significant slowdown in case of large genomes evaluation when the number of alignments may reach dozens of thousands in some contigs. Although there are chaining algorithms with sub-quadratic time complexity (Abouelhoda and Ohlebusch, 2005), they are not applicable to our gap cost function and associated with a large constant. Instead, we have implemented a simple heuristic that always finds the best alignment set or one of the several sets that maximize the score (Supplementary Methods). And even though the heuristic idea does not guarantee the speed up in theory, it significantly dropped the running time of all six benchmark dataset evaluations.
2.4 K-mer-based quality metrics
As shown above, the presence of many TEs and other specific features of eukaryotic genomes significantly complicates assembly evaluation. Although QUAST-LG adjustment of the conventional completeness and correctness measures improve the assessment, it may still not be good enough to form the complete picture of eukaryote assembly quality. Here we propose to assess assemblies using a completely different strategy inspired by the evaluation procedures in Putnam et al. (2016) and Chapman et al. (2016) and generalized for an arbitrary genome analysis in QUAST-LG. This strategy is based on the analysis of unique k-mers (non-repeated sequences of length k) both in the reference genome and in the assembly. If k value is sufficiently large (QUAST-LG uses 101-mers by default), unique k-mers appear to be widespread across the genome. For instance, the fruit fly genome contains 122 millions unique 101-mers out of 137 millions total 101-mers. The existence and the positions of such k-mers in the assembly describe its completeness and correctness.
We use KMC 3 (Kokot et al., 2017) to detect all unique k-mers in the reference genome. The percentage of these k-mers detected in the assembly is reported as its k-mer completeness. Compared to the genome fraction completeness measure, the k-mer-based value accounts for per-base quality of an assembly which is usually highly important for the downstream analysis such as genome annotation. The benchmarking below shows that assemblies with a very similar genome fraction may have completely different k-mer completeness due to a high mismatch and indel error rates.
The k-mer-based correctness is calculated based on a small uniformly distributed subset of all unique k-mers in order to speed up the computation. We select the subset in a way that any two k-mers from the subset are at least 1 kb apart from each other in the reference genome R. The subset is provided to KMC and it identifies contigs having at least two k-mers. The contig position of each detected k-mer is examined and we refer to a consecutive list of k-mers (where ) in a contig C as a marker if for any the distances between ki and in C and R are equivalent within a small error (5% of the distance in R by default). We further process contigs having at least two markers to check whether the relative positions of adjacent markers mj and correlate with their locations in R. QUAST-LG reports a k-mer-based translocation breakpoint if mj and are originated from different chromosomes and a k-mer-based relocation if the markers are from the same chromosome but the inconsistency between their positions in C and R is larger than a predefined threshold (we use 100 kb threshold by default). We further refer to k-mer-based translocations and relocations as k-mer-based misjoins to exclude confusion with regular QUAST misassemblies. K-mer-based misjoins are essentially similar to the regular misassembly metrics, except that they are focused on the most critical assembly errors. The key benefit of these measures is in their tolerance to inconsistencies caused by TEs, since TEs mostly correspond to genomic repeats and thus lack unique k-mers. For example, k-mer-based relocations can successfully resolve the situations when a contig starts or ends with a TE which cause an ambiguity in the regular misassembly detection algorithm (Fig. 2c).
2.5 Evaluation without a reference genome
In most real assembly projects a reference genome sequence is not available and the assembly quality assessment must rely on other sources of information. The primary purpose of QUAST-LG is the reference-based analysis but we also include a few de novo eukaryote-targeted completeness measures to make our tool useful in a wider set of applications. QUAST-LG is supplied with GeneMark-ES (Lomsadze et al., 2005) software for de novo gene prediction in eukaryotic genomes. However, despite the relevance of the gene finding in assessing downstream analysis perspectives its heuristic nature may result in a misleading output in some experiments. For instance, an assembly may contain multiple copies of one gene which will be reported several times. To counter this, we additionally use BUSCO (Simao et al., 2015) to find the number of assembled conserved genes that are present nearly universally in eukaryotes in a single copy. To demonstrate how BUSCO completeness correlates with more accurate reference-based quality metrics, we added its computation in all our benchmark experiments. Note that reference-free correctness metrics are out of scope of QUAST-LG and we recommend using specialized de novo evaluation tools for this scenario. For instance, REAPR (Hunt et al., 2013) identifies likely assembly and scaffolding errors based on paired reads mapping. Another example is KAT (Mapleson et al., 2017) that compares the k-mer spectrum of the assembly to the k-mer spectrum of the reads, which is quite useful in identifying missing sequence, collapsed repeats and expanded sequences in the assembly.
3 Results
3.1 The datasets
We chose six WGS datasets representing four eukaryotic genomes: the yeast (YeastPB and YeastNP), the worm (WormPB), the fruit fly (FlyMP) and the human (HumanMP and HumanNP) (Table 1). Selected genomes represent a wide range of genome sizes, from 12.5 megabases (Mb) for yeast up to 3 gigabases (Gb) for human. The number of chromosomes ranges from just 4 (fruit fly) to 23 (human). Each dataset includes a single Illumina paired-end library and an additional long-read (PacBio SMRT or Oxford Nanopore) or Illumina mate-pair library which is associated with the subscripted abbreviations in the dataset names (PB, NP, MP, respectively). The use of two libraries in each dataset enables generation of both high quality and continuous assemblies. All the benchmarked species were previously sequenced and finished to a very high standard genome sequences, allowing us to deeply investigate the correctness of each assembly. All datasets and reference sequences were downloaded from the public sources, the links are available from our project web page.
Table 1.
Benchmark datasets details
| Dataset | YeastPB | YeastNP | WormPB | FlyMP | HumanMP | HumanNP | |
|---|---|---|---|---|---|---|---|
| Species | S.cerevisiae | S.cerevisiae | C.elegans | D.melanogaster | H.sapiens HG004 | H.sapiens HG001 | |
| Genome size | 12.1 Mb | 12.1 Mb | 100.3 Mb | 137.6 Mb | 3.1 Gb | 3.1 Gb | |
| Library 1 | RL, IS | 101 bp, 220 bp | 250 bp, 350 bp | 100 bp, 250 bp | 101 bp, 225 bp | 250 bp, 350 bp | 150 bp, 350 bp |
| Coverage | 1038× | 300× | 65× | 35× | 50× | 100× | |
| Library 2 | Type | PB | NP | PB | MP | MP | NP |
| RL, IS | 6 kb,— | 7.7 kb,— | 11 kb,— | 110 bp, 8 kb | 101 bp, 6 kb | 6.5 kb,— | |
| Coverage | 155× | 120× | 40× | 40× | 30× | 30× |
Note: Read lengths (RL) and insert sizes (IS) are represented by their median values, ‘—’ indicates no insert size for long-read libraries. Type stands for the sequencing technology used for generating Library 2, PB, NP, MP are for PacBio SMRT, Oxford Nanopore Technologies and Illumina mate-pairs data, respectively. Library 1 data were generated with Illumina sequencers for all datasets. HG001 and HG004 are human sample identifiers in the Genome in a Bottle Consortium (Zook et al., 2016).
In addition, we used previously annotated and curated structural variants (SVs) for both human individuals analyzed in this study. These SVs were provided to QUAST-LG using ‘‐‐sv-bedpe’ option (Mikheenko et al., 2016b) for muting misassemblies caused by true structural inconsistencies between the reference and the sequenced organism. SVs for the HumanMP (HG004) dataset were taken from Chaisson et al. (2014) (automated annotation of HG002, a close relative of HG004) and SVs for HumanNP (HG001) were downloaded from the curated database of genomic variants (MacDonald et al., 2014) and from the Wala et al. (2018) study [automated annotation of HG001 large deletions with LUMPY (Layer et al., 2014)].
3.2 The assemblers
Only a couple of the leading genome assembly tools can handle both long and short reads (paired-ends only or together with mate-pairs) data. Thus, we selected two separate groups of assemblers, one for each type of the input data (Table 2).
Table 2.
Assemblers used in the study
| Name | Reference | Version | Date |
|---|---|---|---|
| Short-read assemblers | |||
| ABYSS | Jackman et al. (2017) | 2.0.2 | Oct 2016 |
| DISCOVAR de novo | — | 52488 | Mar 2015 |
| MaSuRCA | Zimin et al. (2013) | 3.2.3 | Sep 2017 |
| Meraculous | Chapman et al. (2011) | 2.2.4 | Jun 2017 |
| Platanus | Kajitani et al. (2014) | 1.2.4 | Oct 2015 |
| SOAPdenovo | Luo et al. (2012) | 2.04 | Dec 2013 |
| SPAdes | Bankevich et al. (2012) | 3.11.1 | Oct 2017 |
| Long-read assemblers | |||
| Canu | Koren et al. (2017) | 1.6 | Jun 2017 |
| FALCON | Chin et al. (2016) | 0.7 | Jun 2016 |
| Flye | Kolmogorov et al. (2018) | 2.3 | Jan 2018 |
| MaSuRCA | Zimin et al. (2013) | 3.2.3 | Sep 2017 |
| Miniasm | Li (2016) | 0.2-r168 | Nov 2017 |
Note: The assemblers are divided into two groups based on the read types they can process. DISCOVAR de novo is the successor of popular ALLPATHS-LG (Gnerre et al., 2011) assembler but it is not published yet (indicated with ‘—’).
Most of the short-read assemblers (Table 2, top) utilize jumping libraries for repeat resolution and scaffolding. The only exception is DISCOVAR de novo (https://software.broadinstitute.org/software/discovar/blog/) that was scaffolded using external BESST (Sahlin et al., 2014) software.
The assemblers for long-read datasets are shown on the bottom of Table 2. We additionally used Pilon (Walker et al., 2014) polishing tool to improve Canu and FALCON assemblies based on the short reads data and Racon (Vaser et al., 2017) polishing tool to improve Miniasm assemblies. MaSuRCA utilizes both short and long reads simultaneously by design and Flye [the successor of ABruijn (Lin et al., 2016) assembler] has its own polishing tool based on long reads only. SPAdes package (Bankevich et al., 2012) includes a recently developed module for hybrid LRS and NGS data assembly (Antipov et al., 2016) which is however targeted at bacterial genomes in its current implementation and therefore was not included in this study. Thus, MaSuRCA is currently the only software capable to perform sufficiently well on both types of input datasets.
3.3 Data preprocessing
WGS data usually requires a preprocessing stage, such as adapter trimming and error correction. Some of the assemblers we benchmarked have its own error correction modules, while others rely on clean input. To fairly compare the assembly algorithms we performed an independent error correction, ran each assembler on both raw and corrected data and selected the best pipeline for each tool.
To correct sequencing errors in Illumina short reads, we performed quality trimming using Cutadapt v1.15 (Martin, 2011) with the option ‘-q 20’ and all other parameters set to the default values. All paired-ends libraries appeared to be free from the adapters. To clean adapters in mate-pairs, we ran NxTrim software (O’Connell et al., 2015). We did not perform additional correction of long-read libraries, since all four tools used for their assembly have intrinsic correction strategies.
3.4 The assemblies
The assemblies of the HumanMP dataset are taken from ABySS2 study (Jackman et al., 2017) (only the ones with scaffolds). The assemblies of the HumanNP dataset were generated by the corresponding assembler developers [Canu (https://genomeinformatics.github.io/NA12878-nanopore-assembly/), Flye (https://zenodo.org/record/1143753) and MaSuRCA (http://masurca.blogspot.ru/2017/05/human-na12878-hybrid-minionillumina.html)]. The assemblies of the rest four datasets were generated during this study. Most of the assemblers allow user to configure the assembly parameters in order to achieve better results. We ran all assemblers using different combinations of parameters. To select the best assembly in each case, we chose the one that produced the largest N50 for scaffolds, which is a common heuristic for selecting the best assembly when the true genome sequence is unknown. All our assemblies are available online from the project web page.
3.5 QUAST-LG performance
We compared the performance of QUAST-LG against the conventional QUAST v4.5 on all six benchmark datasets (Table 3). Note that original QUAST cannot generate an upper bound assembly, so input assemblies of each dataset were supplied with the corresponding upper bound assemblies generated with QUAST-LG for a fair comparison. Also, QUAST-LG functionality in these benchmarks was limited to the metrics computed by QUAST for the sake of consistency. At the same time, the conventional QUAST tool was configured accordingly to the QUAST-LG settings on the minimal contig and aligned fragment lengths which gave a considerable speed up comparing to the default parameters.
Table 3.
QUAST and QUAST-LG performance
| Dataset | Genome | # asm. | QUAST |
QUAST-LG |
||
|---|---|---|---|---|---|---|
| size (Mb) | Time | RAM | Time | RAM | ||
| YeastPB | 12.1 | 5 | 00:06 | 1.2 | 00:01 | 1.1 |
| YeastNP | 12.1 | 4 | 00:04 | 1.2 | 00:01 | 0.6 |
| WormPB | 100.3 | 5 | 02:51 | 8.4 | 00:08 | 6.3 |
| FlyMP | 137.6 | 6 | 04:55 | 13.8 | 00:21 | 9.8 |
| HumanMP | 3088.3 | 4 | — | — | 03:55 | 135.2 |
| HumanNP | 3088.3 | 4 | — | — | 04:05 | 135.4 |
Note: # asm. stands for the number of assemblies being processed. Running time is in hh:mm format; maximal RAM consumption is in GB; ‘—’ indicates the fact that conventional QUAST was not able to process the human datasets. All benchmarking was done on a server with Intel Xeon X7560 2.27 GHz CPUs using 8 threads.
Table 3 shows that QUAST-LG in all situations runs faster than conventional QUAST. The speed up becomes more significant on large genomes. The smallest datasets (YeastPB and YeastNP) give 4–6 fold speed up whereas FlyMP and WormPB datasets are processed 14× and 21× times faster respectively by QUAST-LG comparing to the original QUAST software. While QUAST was not able to process two human datasets at all, QUAST-LG did it in about four hours. The maximal RAM consumption in QUAST-LG runs is always smaller than the one in the corresponding QUAST runs. Note that Table 3 does not include time and RAM needed by QUAST-LG for computing novel metrics (k-mer-based statistics and BUSCO) and the upper bound assembly generation. The full performance benchmark is shown in Supplementary Table S1.
To illustrate that Minimap2 -based QUAST-LG demonstrates a reasonable accuracy comparing to NUCmer-engined QUAST, we additionally implemented an artificial version of QUAST-LG based on NUCmer aligner and compared it against QUAST-LG on three medium-size datasets (Supplementary Table S2). This comparison shows that the choice of aligner does not significantly affect the metrics computed by QUAST-LG.
3.6 The assemblies comparison
Assemblers performance on all six benchmark datasets is shown in Table 4. The visual form of this table is available in Supplementary Figure S1. The full QUAST-LG reports are in Supplementary Tables S3–S8 and also available online in the interactive form. Scaffold alignment viewers (Mikheenko et al., 2016a) are depicted in Supplementary Figures S2–S7 in form of Circos visualizations (Krzywinski et al., 2009). In all six test cases there is no clear winner by all metrics, since some assemblers tend to generate more accurate but less complete assemblies and vise versa. Note that FALCON assembler is originally designed for PacBio data and it failed to process the Nanopore datasets (YeastNP and HumanNP). Furthermore, DISCOVAR de novo was not able to assemble FlyMP dataset since it does not satisfy DISCOVAR’s requirements on the input data. Finally, the current version of SPAdes assembler is not designed for mammalian-size datasets, so it is missed in the HumanMP benchmark.
Table 4.
Comparison of assemblies of six benchmark datasets
| Assembler | LGA50 | Largest alignment (Mb) | Genome fraction (%) | # mis. | NGA50 (Mb) | Mismatches per 100 kb | Indels per 100 kb | K-mer-based |
BUSCO | |
|---|---|---|---|---|---|---|---|---|---|---|
| compl. (%) | # misjoins | compl. (%) | ||||||||
| YeastPB (genome size 12 157 105 bp) | ||||||||||
| Canu | 7 | 1.512 | 98.77 | 35 | 0.669 | 579.50 | 48.25 | 64.39 | 3 | 99.31 |
| FALCON | 7 | 1.502 | 96.07 | 19 | 0.694 | 184.09 | 92.09 | 86.31 | 3 | 95.18 |
| Flye | 7 | 1.083 | 98.04 | 24 | 0.677 | 118.27 | 30.23 | 91.72 | 7 | 99.31 |
| MaSuRCA | 14 | 0.686 | 97.41 | 60 | 0.346 | 680.43 | 50.04 | 62.36 | 10 | 99.31 |
| Miniasm | 7 | 1.512 | 97.31 | 35 | 0.663 | 155.74 | 104.48 | 85.46 | 3 | 97.93 |
| UpperBound | 6 | 1.524 | 99.92 | 0 | 0.777 | 0.00 | 0.00 | 99.90 | 0 | 99.31 |
| YeastNP (genome size 12 157 105 bp) | ||||||||||
| Canu | 7 | 1.090 | 98.84 | 12 | 0.658 | 565.83 | 101.41 | 62.04 | 1 | 98.96 |
| Flye | 8 | 1.081 | 97.71 | 5 | 0.663 | 56.14 | 649.09 | 52.09 | 3 | 67.93 |
| MaSuRCA | 6 | 1.522 | 99.52 | 14 | 0.782 | 12.39 | 2.87 | 99.12 | 5 | 99.31 |
| Miniasm | 8 | 1.057 | 98.26 | 7 | 0.639 | 52.60 | 754.08 | 48.08 | 1 | 66.89 |
| UpperBound | 6 | 1.524 | 99.87 | 0 | 0.777 | 0.00 | 0.00 | 99.94 | 0 | 99.31 |
| Reference | 6 | 1.532 | 100.00 | 0 | 0.924 | 0.00 | 0.00 | 100.00 | 0 | 99.31 |
| WormPB (genome size 100 286 401 bp) | ||||||||||
| Canu | 27 | 3.374 | 99.54 | 147 | 1.292 | 41.18 | 7.29 | 99.09 | 1 | 96.37 |
| FALCON | 29 | 3.052 | 98.67 | 94 | 1.176 | 65.11 | 126.13 | 88.94 | 8 | 94.39 |
| Flye | 26 | 3.354 | 99.31 | 122 | 1.306 | 19.77 | 43.50 | 95.23 | 6 | 96.37 |
| MaSuRCA | 32 | 2.542 | 99.18 | 138 | 1.016 | 33.28 | 7.79 | 97.45 | 25 | 96.37 |
| Miniasm | 29 | 2.839 | 99.41 | 262 | 1.215 | 54.47 | 143.88 | 87.41 | 5 | 96.04 |
| UpperBound | 8 | 12.667 | 99.95 | 0 | 3.507 | 0.00 | 0.00 | 99.96 | 0 | 96.37 |
| Reference | 3 | 20.924 | 100.00 | 0 | 17.494 | 0.00 | 0.00 | 100.00 | 0 | 96.37 |
| FlyMP (genome size 137 567 484 bp) | ||||||||||
| ABySS | 94 | 2.694 | 79.45 | 266 | 0.331 | 1166.59 | 92.10 | 60.50 | 3 | 99.01 |
| MaSuRCA | 186 | 1.807 | 84.61 | 922 | 0.157 | 1316.66 | 90.11 | 63.35 | 66 | 100.00* |
| Meraculous | 111 | 1.586 | 82.58 | 305 | 0.316 | 1241.33 | 91.06 | 63.51 | 6 | 99.01 |
| Platanus | 97 | 2.811 | 81.07 | 280 | 0.371 | 1288.45 | 91.18 | 62.36 | 24 | 99.01 |
| SOAPdenovo | 155 | 1.631 | 84.64 | 713 | 0.238 | 1308.22 | 91.12 | 63.50 | 12 | 99.67 |
| SPAdes | 123 | 1.656 | 80.41 | 388 | 0.287 | 1173.67 | 93.08 | 61.39 | 108 | 99.01 |
| UpperBound | 44 | 3.558 | 99.16 | 0 | 1.015 | 0.00 | 0.00 | 97.28 | 0 | 99.67 |
| Reference | 3 | 32.079 | 100.00 | 0 | 25.281 | 0.00 | 0.00 | 100.00 | 0 | 99.67* |
| HumanMP (genome size 3 088 286 401 401 bp) | ||||||||||
| ABySS | 263 | 20.392 | 93.56 | 820 | 3.326 | 100.49 | 27.44 | 86.92 | 572 | 93.73 |
| DISCOVAR | 138 | 31.629 | 94.81 | 508 | 6.094 | 106.24 | 25.87 | 88.15 | 535 | 93.40 |
| SOAPdenovo | 3725 | 2.193 | 85.10 | 670 | 0.210 | 129.15 | 50.41 | 77.73 | 93 | 90.43 |
| UpperBound | 112 | 35.878 | 99.06 | 0 | 8.309 | 0.00 | 0.00 | 99.24 | 0 | 92.74 |
| HumanNP (genome size 3 088 286 401 bp) | ||||||||||
| Canu | 296 | 25.751 | 92.25 | 853 | 2.745 | 258.95 | 68.03 | 83.93 | 523 | 92.08 |
| Flye | 266 | 21.735 | 91.91 | 673 | 3.172 | 580.26 | 1125.37 | 26.59 | 97 | 69.64 |
| MaSuRCA | 226 | 22.413 | 93.71 | 13227 | 3.932 | 184.06 | 31.94 | 85.72 | 892 | 87.79 |
| UpperBound | 105 | 75.724 | 99.07 | 0 | 7.862 | 0.00 | 0.00 | 99.51 | 0 | 92.74 |
| Reference | 9 | 248.956 | 100.00 | 0 | 144.769 | 0.00 | 0.00 | 100.00 | 0 | 93.75 |
Note: All statistics are given for scaffolds kb. The best value for each column is indicated in bold (upper bound assembly and reference genome statistics are excluded from the best value determination). LGA50 is the minimal number of aligned fragments that cover half of the reference genome. NGA50 corresponds to the shortest length among the LGA50 aligned fragments. # mis. stands for the total number of misassemblies. K-mer-based compl. is for the fraction of unique reference 101-mers present in the assemblies. K-mer-based # misjoins is the total number of k-mer-based misjoins. BUSCO compl. stands for the total number of conserved genes completely or partially identified in the assembly, divided by the total number of BUSCO genes. UpperBound and Reference stand for the upper bound assembly and the reference genome statistics, respectively. Note that Reference is given once per unique genome, that is only four times per six datasets. We manually checked the overestimated BUSCO compl. measure for MaSuRCA assembly of FlyMP which outperformed the reference value (100.00 versus 99.67% completeness; marked ‘*’ in the table). The D.melanogaster reference sequence misses a single short BUSCO gene which is different from the BUSCO core sequence in a few SNPs and is not identified by the tool. At the same time, MaSuRCA assembled this gene in a form more similar to the BUSCO version which enabled its identification. The similar situation happened on the HumanMP dataset, where ABySS and DISCOVAR partially assembled two BUSCO genes missed in the reference genome. These two assemblers thus were able to exceed the upper bound estimate of the BUSCO completeness.
3.6.1 The yeast datasets
The yeast genome was clearly the simplest one to assemble for all benchmarked assemblers. Flye and Canu have the largest number of the best quality metric values on the YeastPB dataset. The most accurate assembly was constructed by FALCON with 19 misassemblies, the largest NGA50 value (694 kb) and just 3 k-mer-based misjoins (together with Canu and Miniasm results). Canu has the highest percentage of the genome (98.77%) and assembled the largest aligned fragment (1.512 Mb, together with Miniasm) which length is very close to the upper bound value (1.524 Mb). However, Canu assembly is rather inaccurate both in terms of per-base quality (579.5 mismatches and 48 indels per each 100 kb on average) and the number of misassemblies (35, last but one result). MaSuRCA shows rather average quality on this dataset. Nevertheless, MaSuRCA performed clearly the best on YeastNP and generated the best values in all but two quality metrics. The only exceptions are accuracies measures won by Flye (5 misassemblies) and Canu/Miniasm (1 k-mer-based misjoin) on this dataset. Still, MaSuRCA's NGA50 is the best (782 kb) that even slightly exceeds our upper bound limit (777 kb, see the Discussion section for details).
3.6.2 The worm dataset
Canu demonstrated the best results in six out of ten considered metrics on the WormPB dataset. Its assembly has tremendous genome fraction (99.5%) and k-mer-based completeness (99.1%). FALCON has the smallest number of misassemblies (94) but its k-mer-based correctness is only the forth with Canu being the clear winner by this parameter (just 1 misjoin). Three out of five assemblies demonstrate the perfect BUSCO completeness equivalent to the reference genome value (96.37%).
3.6.3 The fruit fly dataset
ABySS assembly has the smallest LGA50 (94) and the smallest number of misassemblies (266) with Platanus being a close second (97 and 280, respectively). At the same time, Platanus has the largest aligned fragment (2.811 Mb) and the highest NGA50 (371 kb). Meraculous has the best k-mer-based completeness (63.51%) and the second-best correctness (6 misjoins), however, it demonstrates rather average results in all other quality metrics. Important to note that all assemblies have very high mismatch and indel rates (>1100 and >90 per 100 kb, respectively) and relatively low genome fraction (<85%) which may indicate a significant difference between the reference genome and the actually sequenced organism.
3.6.4 The human datasets
The upper bound assemblies of HumanMP and HumanNP indicate that it should be almost equally difficult to assemble both datasets. We may see that the majority of quality metrics for real assemblies of HumanMP are indeed very close to the results of HumanNP assemblies. However, the performance of the leading mate-pair assembly (DISCOVAR) is significantly better than the best values of Nanopore-based assemblies in several important statistics, namely LGA50 (138 versus MaSuRCA’s 226), largest alignment (31.6 Mb versus Canu’s 25.8 Mb) and NGA50 (6.1 Mb versus MaSuRCA’s 3.9 Mb).
While Flye generated the most accurate assembly of the HumanNP dataset in terms of the number of misassemblies (673) and k-mer-based misjoins (97), MaSuRCA appears to dominate by almost all other quality metrics. In particular, this assembler produced the highest genome fraction (93.71%) and k-mer-based completeness (85.72%) among other HumanNP assemblies. It may seem like the main drawback of these remarkable MaSuRCA results is the huge number of misassemblies (13 227), which is an order of magnitude higher that the values of all the competitors on the both human datasets. However, the detailed analysis of the corresponding scaffold alignment viewer (Supplementary Fig. S7) revealed that the vast majority of these misassemblies are located in the human centromeres, which were barely assembled by the other tools. Taking into account the fact that centromeres have an extremely complex repeat structures, the high number of misassemblies can be caused not only by the assembler’s errors, but also by mistakes in the reference sequence or contigs misalignments.
4 Discussion
Since our benchmarks demonstrate that the upper bound estimates are much more realistic than the finished reference genome statistics, we find them useful. However, it is important to note that the proposed approach for their computation is heuristic and has some important limitations.
The algorithm for upper bound assembly construction depends on third-party software, namely Minimap2, BWA-MEM and Red, that are the authors’ choice of tools based on their expertise. Using another set of tools may result in a different upper bound assembly: for instance, a user might want to predict genomic repeats using RepeatMasker (Smit et al., 2013).
To ensure that the upper bound assembly is indeed an upper bound, we use rather conservative settings for Red to exclude false positive repeats, and relaxed settings for the alignment software to retain all possible mappings. On top of that, we use a very naive scaffolding strategy that requires just a single long read or two mate-pairs to join upper bound contigs into a scaffold. The real-life scaffolding algorithms (Boetzer and Pirovano, 2014; Sahlin et al., 2014; Vasilinetc et al., 2015) do not usually exploit such weak evidence alone. Likewise, a very low read coverage may prevent the reconstruction of the source sequence in practice, while we penalize only completely zero covered fragments, treating 1-fold coverage as sufficient. Therefore, our correctness and completeness estimates are the optimistic upper bounds on the real assembly measures and could be potentially strengthen. For instance, we provide users an option for setting the minimal number of connections needed for joining upper bound contigs.
The upper bound estimates are relevant to the alignment-based metrics only, such as genome fraction, NGA50, etc. At the same time, it is not possible to compute theoretical limits on the reference-free analogues of these quality metrics, namely total assembly length, NG50, etc. For example, an assembler may randomly concatenate contigs into a huge scaffold which will presumably contain many misassemblies. Using this strategy, the assembler may always outperform the upper bound assembly in terms of the largest scaffold length. If we compute the corresponding alignment-based metric (the largest alignment length) by splitting assemblies at misassembly breakpoints, the upper bound estimate will most likely be equal to or better than the assembler’s value.
However, some genome assembly heuristics may result in a real assembly that overcomes the upper bound assembly even in alignment-based metrics. For instance, a random repeat resolution may possibly be correct in some cases and thus result in better alignment-based metric values than the upper bound estimates. Indeed, real-life assemblers usually do not concatenate sequences randomly without any experimental evidence making this scenario rather unrealistic. Another, a more plausible situation, when a real assembly may have a bigger NGA50 or largest alignment, may appear due to the fact that unresolved repeats are reported as separate scaffolds in the upper bound assembly. On the other hand, an assembler may join unresolved repeats to their adjacent unique fragments (randomly to the left or to the right one). Although such kind of junction will not result in a misassembly, it may lead to undesired results such as incorrect multiplicities of repeats in the assembly. An real example of such situation may be found in Table 4 where MaSuRCA assembly of YeastNP dataset has NGA50 0.6% (5 kb) higher than the corresponding upper bound value.
Another novelty of QUAST-LG, k-mer-based statistics, may report misleading results if its key parameter k is selected improperly. This group of metrics is heavily affected by the density of SNPs for the evaluated species. For example, a single SNP is expected roughly once per kb in the human genome. Using our default k = 101 bp, we have roughly 10% of the reference k-mers missing in the assemblies due to SNPs. Thus, the maximally possible k-mer completeness for human species is about 90% which correlates with assemblers performance in Table 4. This issue can be somewhat solved by using smaller values of k (can be set with ‘‐‐k-mer-size’ QUAST-LG option). However, very small k-mer sizes may give irrelevant results for repeat-rich genomes.
5 Conclusion
In this work, we compared the ability of the state-of-the-art genome assembly tools to assemble four eukaryotic genomes of various size range. This study analyzed datasets which were recently sequenced using conventional NGS technologies and both leading LRS market players, Pacific Biosciences and Oxford Nanopore Technologies. We, however, excluded several other important data types, such as Hi-C and Optical mapping, which are now often in use for chromosome-scale scaffolding. Thus, the shown assembler performance may be further improved using additional data not considered here.
The vast majority of the assemblers evaluated in this study are under constant development. Thus, the snapshot of their performance presented here will soon become obsolete. The major benefit of this work is the development of QUAST-LG, a universal tool for large scale genome assembly evaluation. QUAST-LG makes it easy to reproduce this or similar benchmarking in the future and compare any other genome assembly programs on any other LRS or NGS dataset. We believe that the presented tool will also be suitable for everyday quality control in ongoing research studies of eukaryotic genomes.
Supplementary Material
Acknowledgements
We would like to thank Anton Korobeynikov for fruitful discussions and Heng Li for his huge help in configuring Minimap2.
Funding
This work was supported by Russian Science Foundation (grant 14-50-00069).
Conflict of Interest: none declared.
References
- Abouelhoda M.I., Ohlebusch E. (2005) Chaining algorithms for multiple genome comparison. J. Discret. Algorithms, 3, 321–341. [Google Scholar]
- Antipov D. et al. (2016) hybridSPAdes: an algorithm for hybrid assembly of short and long reads. Bioinformatics, 32, 1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A. et al. (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol., 19, 455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boetzer M., Pirovano W. (2014) SSPACE-LongRead: scaffolding bacterial draft genomes using long read sequence information. BMC Bioinformatics, 15, 211.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradnam K.R. et al. (2013) Assemblathon 2: evaluating de novo methods of genome assembly in three vertebrate species. Gigascience, 2, 10.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresler G. et al. (2013) Optimal assembly for high throughput shotgun sequencing. BMC Bioinformatics, 14, S18.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisson M.J. et al. (2014) Resolving the complexity of the human genome using single-molecule sequencing. Nature, 517, 608–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisson M.J. et al. (2009) De novo fragment assembly with short mate-paired reads: does the read length matter? Genome Res., 19, 336–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman J.A. et al. (2011) Meraculous: de novo genome assembly with short paired-end reads. PLoS ONE, 6, e23501.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman J.A. et al. (2016). Meraculous2: fast accurate short-read assembly of large polymorphic genomes. ArXiv e-prints.
- Chin C. et al. (2016) Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods, 13, 1050–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S. et al. (2013) ALE: a generic assembly likelihood evaluation framework for assessing the accuracy of genome and metagenome assemblies. Bioinformatics, 29, 435–443. [DOI] [PubMed] [Google Scholar]
- Earl D. et al. (2011) Assemblathon 1: a competitive assessment of de novo short read assembly methods. Genome Res., 21, 2224–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghodsi M. et al. (2013) De novo likelihood-based measures for comparing genome assemblies. BMC Res. Notes, 6, 334.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girgis H.Z. (2015) Red: an intelligent, rapid, accurate tool for detecting repeats de-novo on the genomic scale. BMC Bioinformatics, 16, 227.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnerre S. et al. (2011) High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc. Natl. Acad. Sci. USA, 108, 1513–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich A. et al. (2013) QUAST: quality assessment tool for genome assemblies. Bioinformatics, 29, 1072–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt M. et al. (2013) REAPR: a universal tool for genome assembly evaluation. Genome Biol., 14, R47.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman S.D. et al. (2017) ABySS 2.0: resource-efficient assembly of large genomes using a Bloom filter. Genome Res., 27, 768–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajitani R. et al. (2014) Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res., 24, 1384–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokot M. et al. (2017) KMC 3: counting and manipulating k-mer statistics. Bioinformatics, 33, 2759–2761. [DOI] [PubMed] [Google Scholar]
- Kolmogorov M. et al. (2018) Assembly of long error-prone reads using repeat graphs. doi.org/10.1101/247148. [DOI] [PubMed]
- Koren S. et al. (2017) Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res., 27, 722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M. et al. (2009) Circos: an information aesthetic for comparative genomics. Genome Res., 19, 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S. et al. (2004) Versatile and open software for comparing large genomes. Genome Biol., 5, R12.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam K.K. et al. (2014) Near-optimal assembly for shotgun sequencing with noisy reads. BMC Bioinformatics, 15, S4.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layer R. et al. (2014) LUMPY: a probabilistic framework for structural variant discovery. Genome Biol., 15, R84.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv: 1708.01492.
- Li H. (2016) Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics, 32, 2103–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. (2017) Minimap2: fast pairwise alignment for long nucleotide sequences. arXiv: 1708.01492.
- Lin Y. et al. (2016) Assembly of long error-prone reads using de Bruijn graphs. Proc. Natl. Acad. Sci. USA, 113, E8396–E8405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomsadze A. et al. (2005) Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res., 33, 6494–6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R. et al. (2012) SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience, 1, 18.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald J.R. et al. (2014) The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res., 42, D986–D992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoc T. et al. (2013) GAGE-B: an evaluation of genome assemblers for bacterial organisms. Bioinformatics, 29, 1718–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapleson D. et al. (2017) KAT: a K-mer analysis toolkit to quality control NGS datasets and genome assemblies. Bioinformatics, 33, 574–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcais G. et al. (2018) MUMmer4: a fast and versatile genome alignment system. PLoS Comput. Biol., 14, e1005944.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal, 17, 10. [Google Scholar]
- Mikheenko A. et al. (2016a) Icarus: visualizer for de novo assembly evaluation. Bioinformatics, 32, 3321–3323. [DOI] [PubMed] [Google Scholar]
- Mikheenko A. et al. (2016b) MetaQUAST: evaluation of metagenome assemblies. Bioinformatics, 32, 1088–1090. [DOI] [PubMed] [Google Scholar]
- Miller J.R. et al. (2010) Assembly algorithms for next-generation sequencing data. Genomics, 95, 315–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers G., Miller W. (1995). Chaining multiple-alignment fragments in sub-quadratic time. In: Proceedings of the Sixth Annual ACM-SIAM Symposium on Discrete Algorithms, SODA’95. Society for Industrial and Applied Mathematics, Philadelphia, PA, USA, pp. 38–47.
- O’connell J. et al. (2015) NxTrim: optimized trimming of Illumina mate pair reads. Bioinformatics, 31, 2035–2037. [DOI] [PubMed] [Google Scholar]
- Putnam N.H. et al. (2016) Chromosome-scale shotgun assembly using an in vitro method for long-range linkage. Genome Res., 26, 342–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts M. et al. (2004) Reducing storage requirements for biological sequence comparison. Bioinformatics, 20, 3363–3369. [DOI] [PubMed] [Google Scholar]
- Sahlin K. et al. (2014) BESST–efficient scaffolding of large fragmented assemblies. BMC Bioinformatics, 15, 281.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzberg S.L. et al. (2012) GAGE: a critical evaluation of genome assemblies and assembly algorithms. Genome Res., 22, 557–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sczyrba A. et al. (2017) Critical Assessment of Metagenome Interpretation-a benchmark of metagenomics software. Nat. Methods, 14, 1063–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simao F. et al. (2015) BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics, 31, 3210–3212. [DOI] [PubMed] [Google Scholar]
- Smit A. et al. (2013) RepeatMasker Open-4.0. http://www.repeatmasker.org.
- Vaser R. et al. (2017) Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res., 27, 737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilinetc I. et al. (2015) Assembling short reads from jumping libraries with large insert sizes. Bioinformatics, 31, 3262–3268. [DOI] [PubMed] [Google Scholar]
- Wala J.A. et al. (2018) SvABA: genome-wide detection of structural variants and indels by local assembly. Genome Res, 28, 581–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker B.J. et al. (2014) Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE, 9, e112963.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimin A.V. et al. (2013) The masurca genome assembler. Bioinformatics, 29, 2669–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zook J.M. et al. (2016) Extensive sequencing of seven human genomes to characterize benchmark reference materials. Sci. Data, 3, 160025.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.