Genome-Wide Association Studies of imaging-derived biomarkers in Alzheimer’s disease have focused on phenotypes derived from single imaging modalities. Scelsi et al. report the first use of a multimodal phenotype, derived through disease progression modelling, and identify a novel variant that is protective against conversion from healthy ageing to Alzheimer’s disease.
Keywords: dementia, amyloid imaging, structural MRI, GWAS, disease progression
Abstract
Identifying genetic risk factors underpinning different aspects of Alzheimer’s disease has the potential to provide important insights into pathogenesis. Moving away from simple case-control definitions, there is considerable interest in using quantitative endophenotypes, such as those derived from imaging as outcome measures. Previous genome-wide association studies of imaging-derived biomarkers in sporadic late-onset Alzheimer’s disease focused only on phenotypes derived from single imaging modalities. In contrast, we computed a novel multi-modal neuroimaging phenotype comprising cortical amyloid burden and bilateral hippocampal volume. Both imaging biomarkers were used as input to a disease progression modelling algorithm, which estimates the biomarkers’ long-term evolution curves from population-based longitudinal data. Among other parameters, the algorithm computes the shift in time required to optimally align a subjects’ biomarker trajectories with these population curves. This time shift serves as a disease progression score and it was used as a quantitative trait in a discovery genome-wide association study with n = 944 subjects from the Alzheimer’s Disease Neuroimaging Initiative database diagnosed as Alzheimer’s disease, mild cognitive impairment or healthy at the time of imaging. We identified a genome-wide significant locus implicating LCORL (rs6850306, chromosome 4; P = 1.03 × 10−8). The top variant rs6850306 was found to act as an expression quantitative trait locus for LCORL in brain tissue. The clinical role of rs6850306 in conversion from healthy ageing to mild cognitive impairment or Alzheimer’s disease was further validated in an independent cohort comprising healthy, older subjects from the National Alzheimer’s Coordinating Center database. Specifically, possession of a minor allele at rs6850306 was protective against conversion from mild cognitive impairment to Alzheimer’s disease in the National Alzheimer’s Coordinating Center cohort (hazard ratio = 0.593, 95% confidence interval = 0.387–0.907, n = 911, PBonf = 0.032), in keeping with the negative direction of effect reported in the genome-wide association study (βdisease progression score = −0.07 ± 0.01). The implicated locus is linked to genes with known connections to Alzheimer’s disease pathophysiology and other neurodegenerative diseases. Using multimodal imaging phenotypes in association studies may assist in unveiling the genetic drivers of the onset and progression of complex diseases.
Introduction
Alzheimer’s disease is the most common cause of dementia, affecting 46.8 million people worldwide (Prince et al., 2015). The pathophysiology of Alzheimer’s disease and its genetic drivers have been widely studied in recent years. The ɛ4 allele of the apolipoprotein E gene (APOE4) is the strongest known common genetic risk factor for sporadic late-onset Alzheimer’s disease (Saunders et al., 1993). Further genetic risk factors have been identified through genome-wide association studies (GWAS) in case-control datasets, with recent studies comprising 74 000 subjects (Lambert et al., 2013). However, case-control studies in a disorder where the prodromal stage spans decades neglect the fact that many participants labelled as controls may be future cases and therefore reduces statistical power of the study.
An increasingly popular alternative to genetic studies in case-control settings is the genome-wide screen for effects on quantitative disease biomarkers. These biomarkers, often referred to as endophenotypes, are believed to better reflect the underlying disease processes and also enable studies investigating the genetic influences on disease development in the prodromal phase. In the context of Alzheimer’s disease, various quantitative traits have been investigated. A considerable proportion of these studies focused on biomarkers derived from brain image analysis, such as grey matter density in several structures from MRI (Potkin et al., 2009; Shen et al., 2010), voxel-wise measures of brain atrophy (Stein et al., 2010), and cross-sectional and longitudinal amyloid burden from PET (Ramanan et al., 2014, 2015). Other studies have investigated plasma- or CSF-based biomarkers, which include levels of amyloid-β1-42, total and phosphorylated tau, and apolipoprotein J (Cruchaga et al., 2013; Deming et al., 2016; Piccio et al., 2016). These efforts have led to the identification of additional genetic risk factors, such as IL1RAP (Ramanan et al., 2015), which remained concealed even in large genome-wide case-controls studies.
Complex diseases, such as Alzheimer’s disease, are associated with sequences of changes in multiple disease-specific biomarkers. However, disease progression from the preclinical to advanced stage can take decades, and different biomarkers, reflecting different pathologies may show dynamic changes at specific disease stages (Jack et al., 2010, 2013). Thus, testing each biomarker independently in genetic studies provides insights into one specific disease-related process (i.e. atrophy, amyloid deposition, neurofibrillary tangles formation) in a limited time window. Recent methodological developments enable the estimation of data-driven models of disease progression involving multiple biomarkers (Donohue et al., 2014) and therefore provide more robust disease staging of patients. The approach by Donohue et al. (2014) models continuous long-term biomarker evolution from short-term longitudinal data. In contrast, the discrete event-based model by Fonteijn et al. (2012) focuses on the order in which biomarkers become abnormal. Both approaches provide an integrated view on the wide range of biomarker changes and aim to accurately stage patients and to predict future disease progression. For this reason, we introduce the disease progression score (DPS) as a quantitative phenotype for genetic association studies. DPS provides an integrated view on pathophysiological changes during disease development and potentially provides novel genetic insights, which are different from the ones gained by studying single biomarkers. DPS effectively collapses multiple sources of information and multiple time points into a single metric, and is therefore multi-modal by design. The rationale behind DPS as phenotypes is 2-fold: DPS enables multi-phenotype analyses, and also incorporates multiple time points for each biomarker, so as to have an holistic perspective of the patient’s disease history. This is something that existing multivariate analysis tools, such as MV-Plink (Ferreira and Purcell, 2009), SNPTEST (Marchini et al., 2007), BIMBAM (Servin and Stephens, 2007) or MultiPhen (O’Reilly et al., 2012), are unable to achieve, because they only allow for the combination of P-values from the testing of multiple biomarkers at the same time point in a cross-sectional setting.
In this work we present the first GWAS of an Alzheimer’s disease DPS derived from two imaging modalities [T1-weighted MRI and amyloid PET (18F-florbetapir)] on a large cohort from the ADNI database (Jack et al., 2008). We investigated the DPS properties as an endophenotype and compared GWAS results to single-modality cross-sectional biomarkers. We also compared association strengths to genome-wide polygenic risk scores across phenotypes. Lastly, we investigated our GWAS results in the National Alzheimer’s Coordinating Center (NACC) database, to assess their effect on the risk of conversion from healthy ageing to mild cognitive impairment (MCI) or Alzheimer’s disease.
Materials and methods
Data used in the preparation of this article were obtained from the ADNI database (http://adni.loni.usc.edu). The ADNI was launched in 2003 as a public-private partnership, led by Principal Investigator Michael W. Weiner, MD. The primary goal of the ADNI has been to test whether serial MRI, PET, other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of MCI and early Alzheimer’s disease. For up-to-date information, see www.adni-info.org.
Diagnosis
Diagnosis at the time of the first florbetapir PET scan was used in subsequent analyses. Diagnostic categories were healthy control, subjective memory complaints, early MCI/late MCI and Alzheimer’s disease. For the association analysis with polygenic risk scores (see ‘SNP effect on risk of conversion’ section), the latest diagnosis available in ADNI was used (healthy control/MCI/Alzheimer’s disease, date accessed 05/11/2015). Furthermore, diagnosis at baseline florbetapir PET scan and latest diagnosis were compared to define clinical progression (i.e. stable, conversion or reversion) for each subject.
Genotyping and imputation
At the time of this study single nucleotide polymorphism (SNP) genotyping data were available for 1674 subjects across all ADNI phases. Genotyping was conducted using three different platforms: Human610-Quad, HumanOmniExpress and Omni 2.5M (Illumina) (Saykin et al., 2010). The proportions of study participants genotyped with each platform and their relationship with diagnosis at baseline can be found in Supplementary Table 1.
Autosome imputation and quality control
Following recommendations by Roshyara et al. (2014), pre-imputation autosomal SNP filtering based on call rate was omitted. On the subject level we performed the following quality control steps: (i) sex checks were conducted on the original genotype files separately by platform and reported no mismatches; (ii) we computed subject-level call rate on the original genotype files separately by platform and reported no subject missing more than 10% genotypes; (iii) relatedness analysis was performed on genotyped SNPs before imputation: the Genetic Relationship Matrix (GRM) was computed in PLINK v1.9 (Chang et al., 2015) and pruned at 0.1, a threshold lower than the coefficient of relatedness for first cousins (via the –rel-cutoff 0.1 command). On the SNP level we conducted basic checks to ensure compatibility with the reference panel used for imputation. Specifically, we used a tool by W. Rayner (http://www.well.ox.ac.uk/∼wrayner/tools/), to check SNPs for strand consistency, allele names, position, Ref/Alt assignments and minor allele frequency (MAF) differences with the reference panel. The Sanger Imputation Server (https://imputation.sanger.ac.uk/) was used with SHAPEIT for phasing (Delaneau et al., 2011), Positional Burrows-Wheeler Transform (Durbin, 2014) for imputation and the Haplotype Reference Consortium version 1.1 (McCarthy et al., 2016) as reference panel. Data from the three different genotyping platforms were imputed separately. Quality control was performed on genotyped and imputed SNP calls with PLINK. Multi-allelic variants and SNPs with imputation INFO score < 0.3 were removed. Following the initial quality control, genotypes from the three platforms were merged. Genotype calls with posterior probability < 0.9 were set to missing. Next, SNPs with MAF < 5%, genotyping rate < 90%, or deviation from Hardy-Weinberg equilibrium (P < 5.7 × 10−7) were excluded. Finally, subjects missing 10% or more of the genotypes were removed.
SNPweights (Chen et al., 2013) was used to infer genetic ancestry from genotyped SNPs. Ancestry adjustment in the ADNI cohort was implemented in two steps: (i) subjects were compared against a reference panel comprising Central European, Yoruba Africans and East Asian from HapMap 3 (Altshuler et al., 2010), and native Americans from Reich et al. (2012). Subjects with > 80% of Central European genetic ancestry were kept (157 subjects removed); (ii) SNPweights was used together with a reference panel from the Framingham Heart Study (Dawber et al., 1951) comprising north-western Europeans, south-eastern Europeans and Ashkenazi Jewish in order to compute two principal components of population structure to be used in the following association tests (Supplementary Fig. 1).
X chromosome variants
Variants on the X chromosome underwent recommended pre-imputation quality control provided with the X-wide analysis toolset (XWAS version 1.1) (Gao et al., 2015) using the following parameters: exclusion of genotype calls with MAF < 5%, missingness rate > 10%, deviation from Hardy-Weinberg equilibrium with P < 0.05 (Bonferroni corrected); exclusion of samples with >10% missing genotypes and high relatedness (GRM off-diagonal relatedness coefficient > 0.1). At the time of manuscript preparation, the Sanger server did not support imputation of X chromosome variants. Thus, we applied an in-house imputation pipeline for this task, using the 1000 Genomes Project data as a reference panel (Durbin et al., 2010), SHAPEIT (Delaneau et al., 2011) for phasing and IMPUTE2 for imputation (Marchini and Howie, 2010). Pseudo-autosomal regions were excluded, and the 1000 Genomes Project panel without pseudo-autosomal regions was used for imputation. Post-imputation quality control was performed with XWAS applying the following exclusion criteria: imputation INFO score < 0.3; MAF < 5%; genotyping (or imputation) rate < 90% per genotype; deviation from Hardy-Weinberg equilibrium with P < 0.05 (Bonferroni corrected).
Image processing
Cortical amyloid burden
At the time of this study, 2034 preprocessed longitudinal 18F-florbetapir PET scans for 1089 subjects were available in the ADNI database (date accessed 24/03/2016). Preprocessing of florbetapir PET scans in ADNI has been described in detail elsewhere (Jagust et al., 2010). Briefly, four 5-min frames were acquired 30–60 min post-injection; dynamic frames were co-registered to the base frame image, averaged, reoriented into a standard space (voxel grid size 160 × 160 × 96, voxel size 1.5 × 1.5 × 1.5 mm3), and smoothed down to the lowest resolution available in ADNI (8 mm full-width at half-maximum uniform isotropic).
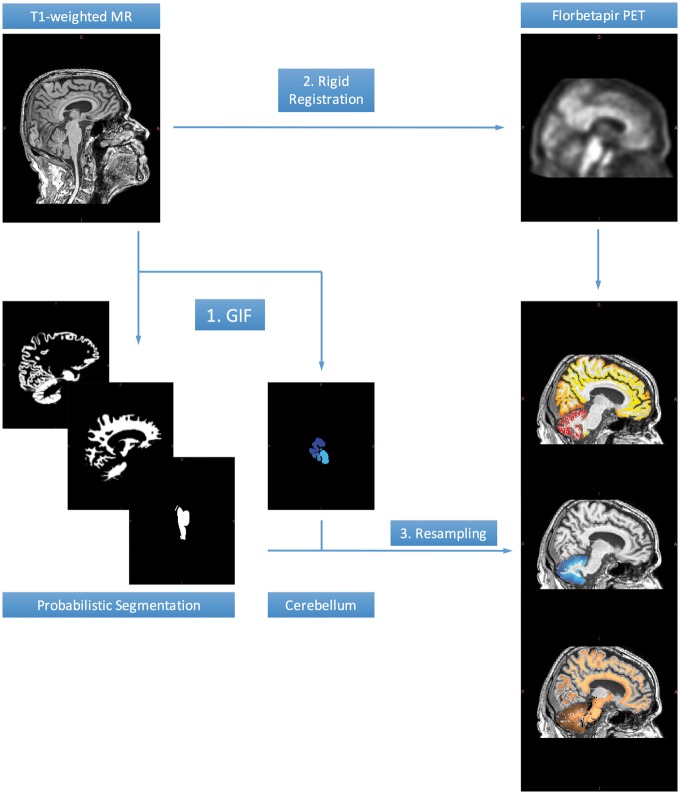
To compute the standardized uptake value ratio (SUVR), information from structural magnetic resonance scans was used. A total of 1931 preprocessed T1-weighted magnetic resonance scans (voxel size 1 × 1 × 1.2 mm3) were downloaded for the same subjects. Details about the magnetic resonance preprocessing can be found in Jack et al. (2008). Each florbetapir PET scan was time-matched to the closest-in-time magnetic resonance scan. However, because of missing concurrent MRI scans for ∼100 individuals, one or more florbetapir PET scans were time-matched to the same magnetic resonance scan. Figure 1 depicts a schematic of the implemented image processing pipeline. The GIF algorithm (Cardoso et al., 2015) and the Aladin algorithm implemented in NiftyReg (Ourselin et al., 2001) were used for segmentation and registration, respectively. PET-T1 registrations were visually assessed to ensure correct alignment.
Figure 1.
Image processing pipeline for the amyloid load computation. GIF was used to obtain a probabilistic segmentation of the T1-weighted scans into background/skull, grey matter, white matter, CSF, subcortical structures, brainstem/pons and cerebellar nuclei. Each T1-weighted scan was rigidly registered to the closest-in-time florbetapir PET scan using the Aladin algorithm; a cubic spline interpolation scheme in two steps was used to resample the warped T1 image to the space of the closest-in-time lower-resolution PET. The GIF segmentations were resampled to the space of the PET scan, to define two key regions: (i) a cortical target region excluding the cerebellar grey matter; and (ii) a composite reference region comprising white matter, whole cerebellum, brainstem and pons, as proposed by Landau et al. (2015).
SUVRs were computed as a measurement of cortical amyloid deposition for each subject and time point. The SUVs were computed as the weighted sum of the florbetapir PET signal intensities with weights corresponding to voxel-wise cerebral grey matter probability. The SUVR is then defined as the SUV in the cortical region divided by the SUV in the reference region. The probability-weighted sum to compute SUVs controls for partial volume effects. A composite reference region comprising white matter, whole cerebellum, brainstem and pons was used (Landau et al., 2015). The SUVR cut-off used to determine amyloid status was obtained by transforming the value of 0.79, as recommended by Landau et al. (2015) by linear regression, yielding a value of 0.7585.
Hippocampal volume
Hippocampal volume longitudinal measurements for ADNI was performed by N. Schuff and colleagues at UCSF using FreeSurfer version 4.3 (Fischl et al., 2002) and accessed through the ADNI website (date accessed 05/11/2015). Hippocampal volumes were normalized by dividing by total intracranial volume.
Disease progression modelling
The GRACE algorithm (Donohue et al., 2014) was used for modelling disease progression curves from short-term longitudinal individual trajectories of imaging biomarkers. Briefly, GRACE is based on self-modelling regression, which assumes that the curves to be fitted share a common shape, e.g. a sigmoid in Alzheimer’s disease (Jack et al., 2013). GRACE iteratively applies regression splines to fit the following model:
| (1) |
where Yij(t) is the value of biomarker j for subject i at time t, gj is assumed to be a monotone function, α0ij and α1ij are a random intercept and slope, respectively, allowed for each subject, ɛij(t) is the fitting residual, and γi is an unknown subject-specific parameter. For each subject γi is estimated as the shift in time required to optimally align the subjects’ biomarker trajectories with the estimated population long-term progression curves. Once all subjects have been aligned, the population curves are re-estimated and a new set of γi is computed with respect to the updated population curves. This process is iterated until convergence. We utilise the final γi parameters as a continuous DPS. The DPS provides an estimate, based on the observed biomarker values, of how advanced in the disease process a subject is compared to the average of the cohort. The curves are all centred at t = 0, thus, individuals with negative time shift, i.e. residing on the left plateau of the sigmoid, are likely to be labelled as controls. Conversely, highly abnormal values for the biomarkers place a patient further in time on the standardized population trajectory, approaching the right plateau of the sigmoid (Supplementary Fig. 2). The DPS is computed by jointly considering the progression curves for all biomarkers. Thus, it natively incorporates information derived from different imaging modalities and renders it effectively as a multi-modal imaging-derived phenotype.
Florbetapir SUVR and hippocampal volumes were used as input for GRACE, after being Z-normalized with respect to healthy controls in the cohort:
| (2) |
where Yij is the value of the j-th biomarker for the i-th subject, and μj and σj are the mean and standard deviation of the j-th biomarker among the healthy controls (see above), respectively. This Z-score-based measure ranks the different biomarkers according to disease severity with respect to healthy controls. The independent variable for the fitting procedure was time from study entry (in years) because, from a computational perspective, GRACE implementation requires the input independent variable to start from t = 0.
Statistical analysis
Phenotypic effect sizes for clinical diagnosis
Effect sizes for differentiating diagnostic categories of the single-modality biomarkers (both cross-sectional baseline values and longitudinal rates of change) and the DPS were computed as Cohen’s d and 95% confidence intervals (CI), for three comparisons (healthy control versus Alzheimer’s disease, healthy control versus MCI, MCI versus Alzheimer’s disease).
Disease progression score distributions
A one-way ANOVA and a Tukey’s range test were performed to quantitatively assess pairwise differences in the DPS distributions among the diagnostic groups at first PET scan and the clinical progression of healthy control and MCI subjects.
Relation between disease progression score and Alzheimer’s disease genetic risk score
We hypothesized that the DPS would be able to give a more integrated view of the disease status than the single-modality phenotypes separately. To test this hypothesis, we looked at the association between the three phenotypes used in our GWAS and a set of Alzheimer’s disease genetic risk score (GRS). A GRS represents a ‘genetic summary’ of an individual’s disease risk, combining the weighted contribution of risk alleles across the whole genome into a single metric.
The GRS computation requires two data sources: (i) individual level SNP data; and (ii) the SNPs effect sizes, which were obtained from the largest Alzheimer’s disease GWAS study conducted so far by the International Genomics of Alzheimer's Project (IGAP; Lambert et al., 2013). Specifically, we used the published results on 7 055 881 SNPs of the discovery phase comprising 54 162 subjects (17 008 Alzheimer’s disease cases and 37 154 control subjects). To avoid over-fitting we only analysed ADNI participants who did not contribute to the IGAP study. Further, we restricted the sample to subjects with >80% probability of being of Caucasian ancestry according to SNPweights (as detailed in the ‘Autosome imputation and quality control’ section), resulting in n = 990 study subjects. Briefly, we used 15 P-value thresholds in the range 0.95–10−5. SNPs in the extended APOE locus (44 400–46 500 kb on chromosome 19; human genome release hg19) were excluded from the GRS construction to enable investigations of genome-wide risk independent from APOE. For each of the P-value thresholds we selected the final set of SNPs for the score by using linkage disequilibrium (LD) clumping implemented in PLINK resulting in only the most significant SNP above the P-value threshold within an LD block to be selected for the score [PLINK parameters: –clump-r2 0.2 –clump-kb 1000 as used in Escott-Price et al. (2015)]. Next, we computed the GRS for all 15 thresholds and all 990 independent ADNI subjects. To compute the GRS, for each SNP the number of effect alleles (i.e. 0, 1 or 2) is multiplied by the effect size obtained from the IGAP results. The GRS is then simply the sum of all those products and was previously used to investigate longitudinal changes in PET biomarkers (Altmann et al., 2016). Here, we tested the GRS for association with the latest diagnosis available in ADNI (healthy control/MCI/Alzheimer’s disease), and with the three quantitative phenotypes described above, controlling for age, sex, number of APOE4 alleles, years of education, and two principal components of population structure (Supplementary Fig. 1).
Genome-wide association analysis
Genome-wide association tests were conducted with PLINK on the imputed SNP data (5 137 219 markers). We tested three quantitative phenotypes (via the –linear command): bilateral hippocampal volume at first PET scan (926 subjects), florbetapir SUVR at first PET scan (936 subjects) and DPS (944 subjects). Sample sizes differences resulted from exclusion of poorly registered scans. The larger sample size for DPS was due to the model’s ability to deal with missing data: if a baseline value of hippocampus or amyloid was excluded from the cross-sectional GWAS, GRACE was still able to work with the remaining time points and return a DPS (see ‘Results’ section). Table 1 reports demographics and clinical outcomes for the DPS GWAS sample.
Table 1.
Demographics and clinical measures for the DPS GWAS sample
| HC | SMC | EMCI | LMCI | AD | Total | |
|---|---|---|---|---|---|---|
| Sample size, n (%) | 226 (23.9) | 92 (9.7) | 267 (28.2) | 234 (24.7) | 125 (13.2) | 944 |
| Age at baseline PET, mean (SD) | 76.5 (6.5) | 72.6 (5.7) | 71.5 (7.2) | 74.7 (8.1) | 75.1 (7.9) | 74.1 (7.5) |
| Females, n (%) | 107 (47.3) | 55 (59.7) | 116 (43.4) | 95 (40.5) | 51 (40.8) | 424 (44.9) |
| Education, years, mean (SD) | 16.6 (2.7) | 16.8 (2.6) | 16.1 (2.7) | 16.3 (2.8) | 15.6 (2.7) | 16.3 (2.7) |
| APOE ɛ4+, n (%) | 60 (26.5) | 29 (31.5) | 118 (44.1) | 123 (52.5) | 83 (66.4) | 413 (43.7) |
| Amyloid+ at baseline PET | 85 (37.6) | 29 (31.5) | 133 (49.8) | 162 (69.2) | 110 (88.0) | 519 (54.9) |
| MMSE at baseline PET | 28.3 (2.5) | 28.8 (1.6) | 27.6 (2.7) | 23.4 (6.2) | 20.1 (4.8) | 25.9 (5.0) |
| Florbetapir SUVR at baseline PET | 0.74 (0.07) | 0.74 (0.07) | 0.77 (0.08) | 0.82 (0.09) | 0.87 (0.08) | 0.78 (0.09) |
| Hippocampal volume at baseline PET (normalized by intracranial volume) | 4.8 × 10−3 (6.9 × 10−4) | 5 × 10−3 (6 × 10−4) | 4.8 × 10−3 (7.3 × 10−4) | 4.1 × 10−3 (7.9 × 10−4) | 3.8 × 10−3 (6.2 × 10−4) | 4.5 × 10−3 (8.2 × 10−4) |
Values are mean ± standard deviation (SD), or n (% of the diagnostic group). Diagnosis at time of the first PET scan. The SUVR cut-off to determine amyloid status was obtained transforming the value of 0.79 recommended by Landau et al. (2015) by linear regression, yielding a value of 0.7585. AD = Alzheimer’s disease; EMCI = early MCI; HC = healthy control; LMCI = late MCI; SMC = subjective memory complaints.
SNP-trait associations were tested under an additive model. Sex, age at first PET scan, years of education, two principal components of population structure, and number of APOE ɛ4 alleles were included as covariates. Since we regard DPS as a longitudinal measure, associations were tested controlling also for baseline levels of hippocampal volume and cortical amyloid. Variants passing the threshold for genome-wide significance were tested for a bias introduced by the additional covariates using the method by Aschard et al. (2015).
X chromosome variants were tested for sex-stratified association using XWAS (Gao et al., 2015): for each SNP, linear models were built separately for males and females, and the resulting Z-statistics combined via Stouffer’s method.
Genome-wide significance was defined at a P-value of 5 × 10−8 or lower. Manhattan, quantile-quantile and beeswarm plots were generated in R (R Development Core Team, 2005), and regional association plots with LocusZoom (Pruim et al., 2011). Variants passing the genome-wide suggestive threshold (P = 10−5) were annotated using the Ensembl Variant Effect Predictor (McLaren et al., 2016). Genome-wide significant variants were tested for effects on gene expression levels in 10 brain tissues by accessing data from the UKBEC (www.braineac.org, Ramasamy et al., 2014) and followed up in the GTEx database (Aguet et al., 2017) (date accessed: 9 May 2017) with a targeted query for a specific triplet SNP-gene-tissue and correcting for the number of tests using the Bonferroni method.
SNPs effect on risk of conversion
Following up on our GWAS results, we hypothesized that variants related to disease progression through significant association with DPS might exert an influence on clinical conversion from healthy control to MCI or Alzheimer’s disease. To test this hypothesis, we ran a Competing Risks regression analysis using the model developed by Fine and Gray (1999) to evaluate the risk of conversion to MCI or Alzheimer’s disease of top SNPs, while accounting for death as a competing risk. This analysis compared carriers of the minor allele (heterozygotes and homozygotes) to non-carriers, and was carried out using the cmprsk package in R (Scrucca et al., 2010). Competing risks analysis is a type of time-to-event analysis that aims to accurately estimate the marginal probability of an event in the presence of competing events such as death. This approach is relevant in this study because participants are elderly and death may occur before the event of interest (i.e. conversion to MCI or Alzheimer’s disease) is observed, which can produce bias in risk estimates. This analysis used data from three NACC Alzheimer’s disease centres, collected between 2005 and 2015. We included all participants from the NACC dataset who were healthy at baseline, had genotyping data available, and had at least 1 year of follow-up, to assess the effect of top SNPs on risk of conversion. Age at entry, APOE ɛ4 status, sex, years of education, first two principal components (Supplementary Fig. 1), and Alzheimer’s disease centre were included as covariates in the analysis (n = 911, Table 2). P-values were corrected for multiple comparisons using the Bonferroni procedure.
Table 2.
Demographics for the NACC participants included in the competing risks regression analysis
| Total (n = 911) | |
|---|---|
| APOE ɛ4, carriers/non-carriers | 249/662 |
| Age, years (±SD) | 74.9 (8.75) |
| Education, years (±SD) | 15.9 (2.70) |
| Sex, female/male | 549/362 |
Results
SNP imputation and quality control
After imputation and quality control, the available genetic data comprised 5 082 878 autosomal and 54 340 X- chromosome SNP markers and 1674 subjects, with a total genotyping rate of 0.99. No subjects were missing more than 10% of the genotypes. After ancestry adjustment and removal of related individuals (144 individuals that appeared to be related), the final sample comprised 1499 unrelated Central European individuals. For the X chromosome, there were 1329 Central European unrelated subjects because of additional X-specific pre-imputation quality control (Gao et al., 2015).
Quantitative traits
Registration failure between PET scans and time-matched T1-weighted MRI scans was detected in 16 subjects. Hippocampal volumes concurrent with a correctly registered baseline PET scan were available for 1065 subjects. Of these, genetic data were available for 926 unrelated Central European subjects. SUVR values were available for 1089 subjects. For 11 of them, registration failure occurred on the baseline PET scan; consequently, these SUVR values were excluded. Of the remaining 1078 individuals, genetic data were available for 936 unrelated Central European subjects.
Disease progression scores
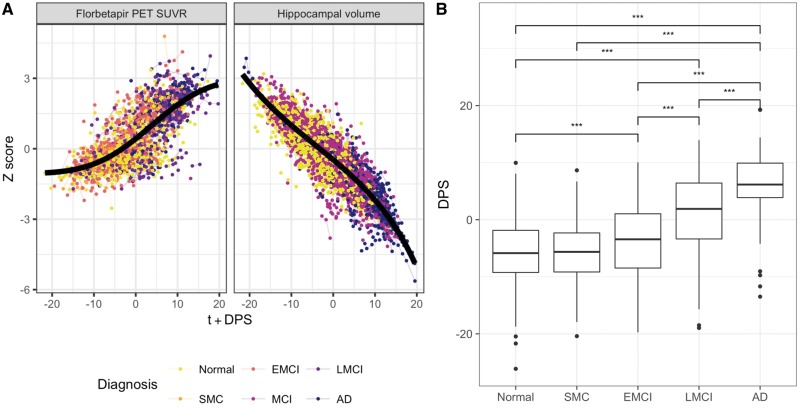
Figure 2A shows the long-term dynamic trajectories fitted by GRACE to the florbetapir SUVR and hippocampal volume data. DPS values were available for 1085 subjects; genetic data were available for 944 unrelated Central European subjects. Mean DPS was significantly higher in early MCI (P = 9 × 10−4) and late MCI (P = 4.36 × 10−13) than in healthy control, higher in late MCI than in early MCI (P = 4.81 × 10−13), and higher in Alzheimer’s disease than in healthy control (P = 4.36 × 10−13), subjective memory complaints (P = 4.36 × 10−13), early MCI (P = 4.36 × 10−13) and late MCI (P = 5.23 × 10−13, Fig. 2B). Supplementary Fig. 3 shows the DPS distributions for the different MCI progression status. DPS showed better effect sizes (Cohen’s d) in separating healthy control and Alzheimer’s disease subjects as well as MCI and Alzheimer’s disease subjects compared to both cross-sectional hippocampal volume and cortical amyloid alone, and their longitudinal rate of change (Supplementary Fig. 4 andSupplementary Table 2). There was no difference across cross-sectional biomarkers in separating healthy controls and MCI, whereas DPS outperforms rates of change in hippocampal volume and amyloid for this task.
Figure 2.
Disease progression modelling results. (A) Long-term progression curves for two Alzheimer’s disease biomarkers. Every point in the plot represents a biomarker measurement; longitudinal data from the same subject are connected by lines. The subjects’ clinical diagnosis at the initial PET scan is colour-coded. The x-axis shows the time from study entry plus the estimated DPS, values on the y-axis are the Z-score normalized individual biomarker measurements: florbetapir PET SUVR and intracranial-volume-normalized bilateral hippocampal volume. (B) Disease progression scores stratified by diagnosis at baseline PET scan. The y-axis shows the DPS and the x-axis corresponds to different diagnostic groups of increasing severity from left (Normal) to right (Alzheimer’s disease). Each box shows the DPS distribution for the corresponding diagnostic group. Annotations represent the level of statistical significance for pairwise tests, after correction for multiple comparisons (*** P < 0.001). EMCI = early MCI; LMCI = late MCI; SMC = subjective memory complaints.
Effect of APOE4 status on quantitative traits
APOE4 allele count was strongly correlated with all three phenotypes. Without any covariate adjustments, hippocampal volume exhibited a negative correlation (Pearson’s r = −0.12, P = 1.8 × 10−4), while cortical amyloid (r = 0.44, P < 2.2 × 10−16) and DPS (r = 0.35, P < 2.2 × 10−16) were positively correlated. When adjusting for age and sex, the correlations with APOE4 allele count were strengthened for hippocampal volume (Pearson’s r = −0.21, P < 2.2 × 10−16), cortical amyloid (r = 0.47, P < 2.2 × 10−16) and DPS (r = 0.41, P < 2.2 × 10−16).
Relation between phenotypes and Alzheimer’s disease polygenic risk score
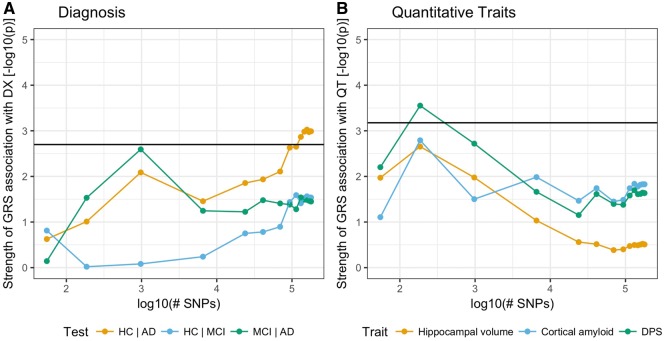
As a preliminary point, we verified that the GRS that incorporated many weakly associated variants were significantly associated with Alzheimer’s disease diagnosis, when discriminating healthy controls from MCI (minimal P = 1.44 × 10−3) and healthy controls from Alzheimer’s disease (minimal P = 2 × 10−4; Fig. 4A). This behaviour is consistent with results reported in the literature (Escott-Price et al., 2015). The quantitative traits showed the strongest association with GRS at low P-value cut-offs (P = 10−5, 10−4, 10−3), i.e. scores that include established Alzheimer’s disease risk variants (Fig. 4B). Only the association between DPS and GRS at P-value cut-off 10−4 was significant after Bonferroni correction. Furthermore, at the same P-value cut-off, the GRS exhibits a stronger association with DPS than the GRS at high P-value cut-offs (e.g. 0.7) in the healthy control versus Alzheimer’s disease setting.
Figure 4.
Significance of association between Alzheimer’s disease polygenic risk score at different SNP inclusion thresholds and binary and continuous phenotypes. (A) Diagnosis coded as three different logistic regressions [healthy control (HC) versus Alzheimer’s disease (AD), healthy control versus MCI, MCI versus Alzheimer’s disease]. (B) The three quantitative traits used as outcomes in GWASs. The x-axis shows the number of SNPs included in the computation of the GRS (on logarithmic scale). We selected from the results of the IGAP GWAS SNPs that exceeded a P-value cut-off ranging from 10−5 (55 SNPs) to 0.95 (179 211 SNPs). The y-axis represents the strength of association (P-value for the regression coefficient in a general linear model, logarithmic scale) between the GRS and the outcome variables. The black line is the 0.01 significance threshold after Bonferroni correction for the effective number of independent tests performed. The effective number of independent GRS (Meff,GRS) and phenotypes (Meff,phen) tested was computed following the simpleM method in Gao et al. (2008). (A) Significance level adjusted for Meff,GRS only. (B) Significance level adjusted for both Meff,GRS and Meff,phen.
Genome-wide association tests
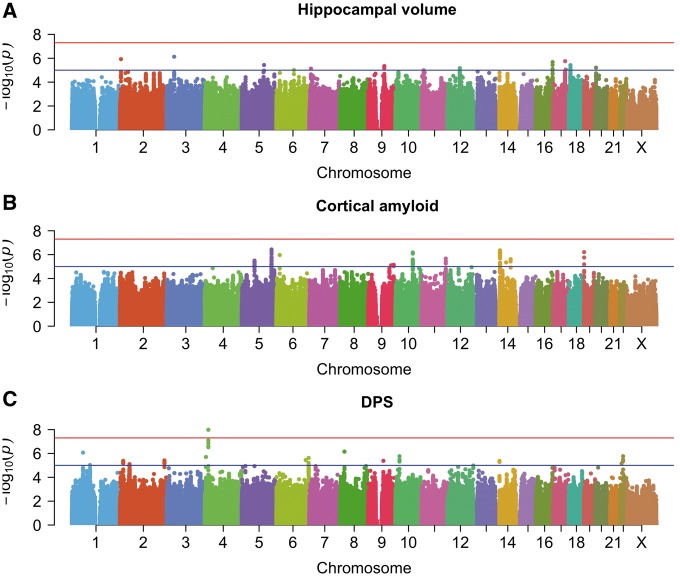
A genome-wide significant association was found for the DPS on chromosome 4 at rs6850306 (MAF = 0.15, P = 1.03 × 10−8; Fig. 3C and Supplementary Table 3). The second most strongly associated locus was rs114368656 on chromosome 22 (sample MAF = 0.08, P = 1.70 × 10−6, Fig. 3C). Supplementary Figs 6 and 7 show the regional association plots. Supplementary Fig. 8 shows the DPS distributions stratified by rs6850306 and rs114368656 genotype, after adjusting for the covariates used in the GWAS. No significant associations were found for either hippocampal volume or cortical amyloid (Fig. 3A and B). Supplementary Table 3 lists the 13, 14 and 12 suggestive independent loci for hippocampal volume, cortical amyloid and DPS, respectively. The DPS results include genetic loci that were neither suggestive by amyloid burden (rs6850306: P = 0.87; rs114368656: P = 0.96) nor by hippocampal volume (rs6850306: P = 0.21; rs114368656: P = 0.16) alone, and that have not been reported in the case-control meta-analysis by Lambert et al. (2013) (rs6850306: P = 0.17; rs114368656 was not tested). Association significance with DPS remained almost unchanged when testing imputed dosages in place of genotypes (i.e. for rs6850306 P = 1.22 × 10−8; for rs114368656 P = 1.92 × 10−6). There was no evidence for P-value inflation with λ < 1.062 (Supplementary Fig. 5). Summary statistics are publicly available at https://doi.org/10.6084/m9.figshare.5603203.v5 (autosomes and chromosome X separately; summary statistics for males and females separately as returned by XWAS can be found in the X chromosome files).
Figure 3.
Manhattan plots for the three GWASs. Cross-sectional hippocampal volume (A), cross-sectional amyloid burden (B), disease progression score (C), after correcting for age, sex, number of APOE ɛ4, years of education, baseline cortical amyloid and hippocampal volume, and two principal components of population structure. The red line is the genome-wide significance threshold at P = 5 × 10−8; the blue line is a threshold for suggestive associations at P = 10−5.
The SNP rs6850306 acts as expression quantitative trait locus (cis-eQTL) for the LCORL gene in the hippocampus (minimal P = 0.035, GTEx), and specifically for the exon probe sets 2720253 and 2720265 of the Affymetrix Human Exon 1.0 ST array in the same brain tissue (minimal P = 6.9 × 10−4, UKBEC, Supplementary Fig. 9).
As expected from the correlations reported in the ‘Effect of APOE4 status on quantitative traits’ section, for hippocampal volume and amyloid, once the number of APOE4 alleles was not included as covariate in the association studies, significant association was found for the APOE locus with both hippocampal volume (rs429358 P = 1.34 × 10−10) and cortical amyloid (rs429358 P = 2.54 × 10−50) (Supplementary Fig. 10), in excellent agreement with previously published results (Schuff et al., 2008; Drzezga et al., 2009). Similarly, a strong association between the APOE locus and DPS can be seen when both the corrections for APOE4 status and baseline cortical amyloid are dropped (rs429358 P = 4.43 × 10−33; Supplementary Fig. 10D). The inclusion of correlated traits as confounders in the DPS GWAS did not introduce any bias in the estimated effect size for rs6850306 (Wald test statistic W = 0.14, P = 1).
SNPs effect on risk of conversion
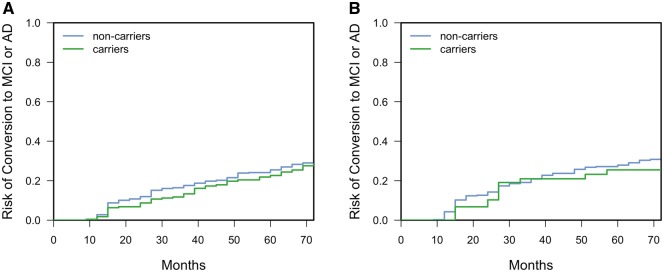
Competing risks regression analysis in the NACC dataset was conducted for the two top SNPs rs6850306 (GWAS β = −0.07; P = 1.03 × 10−8) and rs114368656 (GWAS β = −0.05; P = 1.70 × 10−6). Given the relatively low minor allele frequency of both rs6850306 and rs114368656, the analysis was conducted on minor allele carriers (recessive homozygotes and heterozygotes) versus non-carriers (major allele homozygotes). rs6850306 appears to be protective against conversion [hazard ratio (HR) = 0.593, 95% CI = 0.387–0.907, n = 911, PBonf = 0.032], conferring decreased risk to homozygous and heterozygous carriers of the minor allele A and confirming the negative direction of effect reported in the GWAS (Fig. 5A and Supplementary Table 3). However, results suggest that rs114368656 does not modulate conversion risk between carriers and non-carriers of the minor allele T (HR = 1.08, 95% CI = 0.737–1.57, n = 480, PBonf = 1; Fig. 5B).
Figure 5.
Cumulative distribution functions (complementary survival functions) for risk of conversion to MCI or Alzheimer’s disease against months from baseline for NACC study participants. (A) Results stratified by rs6850306 minor allele carriers versus non-carriers (A/A and A/G versus G/G); (B) stratified by rs114365686 minor allele carriers versus non-carriers (T/T and T/C versus C/C). AD = Alzheimer’s disease.
Discussion
We assessed the potential of a compound phenotype comprising cortical amyloid and hippocampal volume to discriminate disease stages in Alzheimer’s disease. The DPS exploits the different temporal properties of the individual biomarkers: brain amyloid accumulation begins during the prodromal phase where no cognitive decline is manifest; hippocampal atrophy tends to be detectable later during the disease course closer to clinical symptoms (Jack et al., 2010). The resulting DPS increased with increasing severity of the clinical diagnosis (Fig. 2B), was related to longitudinal disease progression (Supplementary Fig. 3) and was superior to the individual biomarkers in separating Alzheimer’s disease subjects from healthy controls or MCI subjects (Supplementary Fig. 4). In addition, the DPS showed a stronger association with genome-wide polygenic Alzheimer’s disease risk than the individual biomarkers (Fig. 4B).
Having shown the advantage of the DPS over individual biomarkers in tracking disease progression, the DPS was used as a quantitative phenotype in a GWAS. Genome-wide significant rs6850306 and its haplotype fall in the LCORL gene on chromosome 4p15.31. For this variant we identified a tissue-specific effect on LCORL expression levels in the hippocampus, with expression levels increasing with each copy of the minor allele; this cis-eQTL behaviour was consistently found in two independent gene expression databases.
The protein encoded by LCORL is a direct interaction partner of CTBP1 (Szklarczyk et al., 2015), which mediates activity-dependent synapse-to-nucleus communication (Ivanova et al., 2015). Furthermore, the CTBP1/BARS complex co-localizes with amyloid precursor protein (APP) in macropinosomes at the cell surface (Tang et al., 2015). Previous GWAS have linked LCORL to human height (Wood et al., 2014). Interestingly, human height exhibits a negative genetic correlation with Alzheimer’s disease (Bulik-Sullivan et al., 2015).
Among the suggestive variants, a haplotype on chromosome 22 falls in the SYN3/TIMP3 gene. Of note, SYN3/TIMP3 was also identified as susceptibility locus for age-related macular degeneration (Chen et al., 2010; Fritsche et al., 2016), a late-onset neurodegenerative disease of the retina involving amyloid-β pathology (Ratnayaka et al., 2015). It has also been suggested that upregulation of TIMP3 may occur at an early disease stage, playing an important role in the development of neurofibrillary tangles (Dunckley et al., 2006). A number of interesting association signals to neurodegenerative diseases have been reported in this region on chromosome 22 (Lachman et al., 2006; Otaegui et al., 2009; Zaltieri et al., 2015). However, some caution is required when interpreting these association signals due to scarce coverage on some genotyping platforms and lack of information on LD structure in the 1000 Genomes reference panel and all resources based on it. In fact, the ADNI whole-genome sequencing data called no variants in the 5 kb region surrounding rs114368656. We believe this is due to the general architecture of the locus, that is characterized by several insertions/deletions and copy-number variations of different lengths.
We demonstrated the first successful application of a multimodal DPS as quantitative phenotype in a GWAS. There remain, however, some limitations. First, the DPS relies heavily on the choice of considered biomarkers. In this study, amyloid burden and hippocampal volume were used owing to their specificity to Alzheimer’s disease pathology. However, larger or different sets of biomarkers may provide more granular estimates of disease progression, which may translate into increased statistical power for reaching genome-wide significance. Second, the multimodal nature of the DPS complicates validation in independent cohorts. In fact, for this work we could not secure a sufficiently sized cohort with genotyping, longitudinal amyloid and structural imaging data to validate our findings. Although lacking a formal replication cohort, a supporting evidence for our genome-wide significant association in chromosome 4 comes from the conversion risk analysis performed on an independent dataset (NACC), confirming the protective role of rs6850306 revealed through GWAS and linking rs6850306 genotype, lower DPS values and decreased risk for conversion. Additionally, there is supporting evidence for the two top associated loci in the literature; linking one locus to age-related macular degeneration, which shares many genetic similarities with Alzheimer’s disease including APOE as a risk locus. Third, despite their inherent appeal for genetic studies, imaging-derived phenotypes may also be a source of undesired variation. For instance, our analysis of cross-sectional amyloid burden (Fig. 3B) did not show a significant association at rs509208, which was identified in an earlier study using a subset of these data (Ramanan et al., 2014). The discrepancy may originate from slightly different sample sizes and the use of a different pipeline to compute brain amyloid burden, highlighting the need for robust imaging biomarkers such as the DPS presented here. Lastly, despite the several lines of evidence here presented (statistical association, cis-eQTL, modulation of conversion risk), these are not sufficient to claim the identification of a protective gene for Alzheimer’s disease: this can only be achieved through fine-mapping of the genomic region of interest and follow-up with functional studies and animal models. Without confirmation from these studies, any other gene in the locus can potentially be the causal gene.
In summary, we performed the first successful genetic study of a progression score for Alzheimer’s disease derived from multiple imaging modalities. We believe that this integrative approach has great potential for identifying common genetic variation related to the multiple, interlinked pathogenic pathways involved in Alzheimer’s disease and other complex diseases.
Funding
M.A.S. acknowledges financial support by the EPSRC-funded UCL Centre for Doctoral Training in Medical Imaging (EP/L016478/1). M.D.G. was supported by the NIH (P50 AG047366). J.M.S. acknowledges the support of the National Institute for Health Research Queen Square Dementia Biomedical Research Unit, the NIHR UCL/H Biomedical Research Centre, Wolfson Foundation, Engineering and Physical Sciences Research Council (EP/J020990/1), Medical Research Council (CSUB19166), Alzheimer's Research UK (ARUK-Network 2012-6-ICE; ARUK-PG2014-1946), Brain Research Trust (UCC14191) and European Union’s Horizon 2020 research and innovation programme (Grant 666992). S.O. receives funding from the EPSRC (EP/H046410/1, EP/J020990/1, EP/K005278), the Medical Research Council (MR/J01107X/1), the EU-FP7 project VPH-DARE@IT (FP7-ICT-2011-9-601055), the National Institute for Health Research Biomedical Research Unit (Dementia) at UCL and the National Institute for Health Research University College London Hospitals Biomedical Research Centre (NIHR BRC UCLH/UCL High Impact Initiative- BW.mn.BRC10269). A.A. holds an Medical Research Council eMedLab Medical Bioinformatics Career Development Fellowship. This work was supported by the Medical Research Council [grant number MR/L016311/1].
Supplementary material
Supplementary material is available at Brain online.
Appendix 1
Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
We thank the International Genomics of Alzheimer's Project (IGAP) for providing summary results data for these analyses. The investigators within IGAP contributed to the design and implementation of IGAP and/or provided data but did not participate in analysis or writing of this report. IGAP was made possible by the generous participation of the control subjects, the patients, and their families. The i–Select chips was funded by the French National Foundation on Alzheimer's disease and related disorders. EADI was supported by the LABEX (laboratory of excellence program investment for the future) DISTALZ grant, Inserm, Institut Pasteur de Lille, Université de Lille 2 and the Lille University Hospital. GERAD was supported by the Medical Research Council (Grant n° 503480), Alzheimer's Research UK (Grant n° 503176), the Wellcome Trust (Grant n° 082604/2/07/Z) and German Federal Ministry of Education and Research (BMBF): Competence Network Dementia (CND) grant n° 01GI0102, 01GI0711, 01GI0420. CHARGE was partly supported by the NIH/NIA grant R01 AG033193 and the NIA AG081220 and AGES contract N01–AG–12100, the NHLBI grant R01 HL105756, the Icelandic Heart Association, and the Erasmus Medical Center and Erasmus University. ADGC was supported by the NIH/NIA grants: U01 AG032984, U24 AG021886, U01 AG016976, and the Alzheimer's Association grant ADGC–10–196728.
The NACC database is funded by NIA/NIH Grant U01 AG016976. NACC data are contributed by the NIA-funded ADCs: P30 AG019610 (PI Eric Reiman, MD), P30 AG013846 (PI Neil Kowall, MD), P50 AG008702 (PI Scott Small, MD), P50 AG025688 (PI Allan Levey, MD, PhD), P50 AG047266 (PI Todd Golde, MD, PhD), P30 AG010133 (PI Andrew Saykin, PsyD), P50 AG005146 (PI Marilyn Albert, PhD), P50 AG005134 (PI Bradley Hyman, MD, PhD), P50 AG016574 (PI Ronald Petersen, MD, PhD), P50 AG005138 (PI Mary Sano, PhD), P30 AG008051 (PI Thomas Wisniewski, MD), P30 AG013854 (PI M. Marsel Mesulam, MD), P30 AG008017 (PI Jeffrey Kaye, MD), P30 AG010161 (PI David Bennett, MD), P50 AG047366 (PI Victor Henderson, MD, MS), P30 AG010129 (PI Charles DeCarli, MD), P50 AG016573 (PI Frank LaFerla, PhD), P50 AG005131 (PI James Brewer, MD, PhD), P50 AG023501 (PI Bruce Miller, MD), P30 AG035982 (PI Russell Swerdlow, MD), P30 AG028383 (PI Linda Van Eldik, PhD), P30 AG053760 (PI Henry Paulson, MD, PhD), P30 AG010124 (PI John Trojanowski, MD, PhD), P50 AG005133 (PI Oscar Lopez, MD), P50 AG005142 (PI Helena Chui, MD), P30 AG012300 (PI Roger Rosenberg, MD), P30 AG049638 (PI Suzanne Craft, PhD), P50 AG005136 (PI Thomas Grabowski, MD), P50 AG033514 (PI Sanjay Asthana, MD, FRCP), P50 AG005681 (PI John Morris, MD), P50 AG047270 (PI Stephen Strittmatter, MD, PhD).
The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The data used for the analyses described in this manuscript were obtained from: the GTEx Portal on 08/05/2017 and dbGaP accession number phs000424.v6.p1 on 08/05/2017.
Supplementary Material
Glossary
Abbreviations
- ADNI
Alzheimer’s Disease Neuroimaging Initiative
- DPS
disease progression score
- GRS
genetic risk score
- GWAS
genome-wide association study
- IGAP
International Genomics of Alzheimer’s Project
- MAF
minor allele frequency
- MCI
mild cognitive impairment
- NACC
National Alzheimer’s Coordinating Center
- SNP
single nucleotide polymorphism
- SUVR
standardized uptake value ratio
References
- Aguet F, Brown AA, Castel SE, Davis JR, He Y, Jo B, et al. Genetic effects on gene expression across human tissues. Nature 2017; 550: 204–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmann A, Modat M, Ourselin S. Genome-wide polygenic risk for Alzheimer’s disease is associated with rate of metabolic decline but not with rate of amyloid deposition. Alzheimers Dement 2016; 12: P5–6. [Google Scholar]
- Altshuler DM, Gibbs RA, Peltonen L, Altshuler DM, Gibbs RA, Peltonen L, et al. Integrating common and rare genetic variation in diverse human populations. Nature 2010; 467: 52–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aschard H, Vilhjálmsson BJ, Joshi AD, Price AL, Kraft P. Adjusting for heritable covariates can bias effect estimates in genome-wide association studies. Am J Hum Genet 2015; 96: 329–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet 2015; 47: 1236–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso MJ, Modat M, Wolz R, Melbourne A, Cash D, Rueckert D, et al. Geodesic information flows: spatially-variant graphs and their application to segmentation and fusion. IEEE Trans Med Imaging 2015; 34: 1976–88. [DOI] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 2015; 4: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Pollack S, Hunter DJ, Hirschhorn JN, Kraft P, Price AL. Improved ancestry inference using weights from external reference panels. Bioinformatics 2013; 29: 1399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Stambolian D, Edwards AO, Branham KE, Othman M, Jakobsdottir J, et al. Genetic variants near TIMP3 and high-density lipoprotein-associated loci influence susceptibility to age-related macular degeneration. Proc Natl Acad Sci USA 2010; 107: 7401–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruchaga C, Kauwe JSK, Harari O, Jin SC, Cai Y, Karch CM, et al. GWAS of cerebrospinal fluid tau levels identifies risk variants for Alzheimer’s disease. Neuron 2013; 78: 256–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawber TR, Meadors GF, Moore FE. Epidemiological approaches to heart disease: the Framingham Study*. Am J Public Health Nations Health 1951; 41: 279–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods 2011; 9: 179–81. [DOI] [PubMed] [Google Scholar]
- Deming Y, Xia J, Cai Y, Lord J, Holmans P, Bertelsen S, et al. A potential endophenotype for Alzheimer’s disease: cerebrospinal fluid clusterin. 2016; Neurobiol Aging 2016; 37: 208.e1–e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohue MC, Jacqmin-Gadda H, Le Goff M, Thomas RG, Raman R, Gamst AC, et al. Estimating long-term multivariate progression from short-term data. Alzheimers Dement 2014; 10: S400–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drzezga A, Grimmer T, Henriksen G, Mühlau M, Perneczky R, Miederer I, et al. Effect of APOE genotype on amyloid plaque load and gray matter volume in Alzheimer disease. Neurology 2009; 72: 1487–94. [DOI] [PubMed] [Google Scholar]
- Dunckley T, Beach TG, Ramsey KE, Grover A, Mastroeni D, Walker DG, et al. Gene expression correlates of neurofibrillary tangles in Alzheimer’s disease. Neurobiol Aging 2006; 27: 1359–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin R. Efficient haplotype matching and storage using the positional Burrows-Wheeler transform (PBWT). Bioinformatics 2014; 30: 1266–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin RM, Abecasis GR, Altshuler DL, Bentley DR, Chakravarti A, Clark AG, et al. A map of human genome variation from population-scale sequencing. Nature 2010; 467: 1061–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escott-Price V, Sims R, Bannister C, Harold D, Vronskaya M, Majounie E, et al. Common polygenic variation enhances risk prediction for Alzheimer’s disease. Brain 2015; 138: 3673–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MA, Purcell SM. A multivariate test of association. Bioinformatics 2009; 25: 132–3. [DOI] [PubMed] [Google Scholar]
- Fine JP, Gray RJ. A proportional hazards model for the subdistribution of a competing risk. J Am Stat Assoc 1999; 94: 496–509. [Google Scholar]
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, et al. Whole brain segmentation: neurotechnique automated labeling of neuroanatomicalstructures in the human brain. Neuron 2002; 33: 341–55. [DOI] [PubMed] [Google Scholar]
- Fonteijn HM, Modat M, Clarkson MJ, Barnes J, Lehmann M, Hobbs NZ, et al. An event-based model for disease progression and its application in familial Alzheimer’s disease and Huntington’s disease. Neuroimage 2012; 60: 1880–9. [DOI] [PubMed] [Google Scholar]
- Fritsche LG, Igl W, Bailey JN, Grassmann F, Sengupta S, Bragg-Gresham JL, et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat Genet 2016; 48: 134–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Chang D, Biddanda A, Ma L, Guo Y, Zhou Z, et al. XWAS: a software toolset for genetic data analysis and association studies of the X chromosome. J Hered 2015; 106: 666–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Starmer J, Martin ER. A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet Epidemiol 2008; 32: 361–9. [DOI] [PubMed] [Google Scholar]
- Ivanova D, Dirks A, Montenegro-Venegas C, Schone C, Altrock WD, Marini C, et al. Synaptic activity controls localization and function of CtBP1 via binding to Bassoon and Piccolo. EMBO J 2015; 34: 1056–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, et al. The Alzheimer’s Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging 2008; 27: 685–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Knopman DS, Jagust WJ, Petersen RC, Weiner MW, Aisen PS, et al. Tracking pathophysiological processes in Alzheimer’s disease: an updated hypothetical model of dynamic biomarkers. Lancet Neurol 2013; 12: 207–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Knopman DS, Jagust WJ, Shaw LM, Aisen PS, Weiner MW, et al. Hypothetical model of dynamic biomarkers of the Alzheimer’s pathological cascade. Lancet Neurol 2010; 9: 119–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagust WJ, Bandy D, Chen K, Foster NL, Landau SM, Mathis CA, et al. The Alzheimer’s Disease Neuroimaging Initiative positron emission tomography core. Alzheimers Dement 2010; 6: 221–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachman HM, Stopkova P, Papolos DF, Pedrosa E, Margolis B, Aghalar MR, et al. Analysis of synapsin III –196 promoter mutation in schizophrenia and bipolar disorder. Neuropsychobiology 2006; 53: 57–62. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet 2013; 45: 1452–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau SM, Fero A, Baker SL, Koeppe R, Mintun M, Chen K, et al. Measurement of longitudinal-amyloid change with 18F-florbetapir PET and standardized uptake value ratios. J Nucl Med 2015; 56: 567–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet 2010; 11: 499–511. [DOI] [PubMed] [Google Scholar]
- Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet 2007; 39: 906–13. [DOI] [PubMed] [Google Scholar]
- McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet 2016; 48: 1279–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GRS, Thormann A, et al. The ensembl variant effect predictor. Genome Biol 2016; 17: 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly PF, Hoggart CJ, Pomyen Y, Calboli FCF, Elliott P, Jarvelin MR, et al. MultiPhen: joint model of multiple phenotypes can increase discovery in GWAS. PLoS One 2012; 7: e34861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otaegui D, Zuriarrain O, Castillo-Triviño T, Aransay A, Ruíz-Martinez J, Olaskoaga J, et al. Association between synapsin III gene promoter SNPs and multiple sclerosis in Basque patients. Mult Scler J 2009; 15: 124–8. [DOI] [PubMed] [Google Scholar]
- Ourselin S, Roche A, Subsol G, Pennec X, Ayache N. Reconstructing a 3D structure from serial histological sections. Image Vis Comput 2001; 19: 25–31. [Google Scholar]
- Piccio L, Deming Y, Del-Águila JL, Ghezzi L, Holtzman DM, Fagan AM, et al. Cerebrospinal fluid soluble TREM2 is higher in Alzheimer disease and associated with mutation status. Acta Neuropathol 2016; 131: 925–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potkin SG, Guffanti G, Lakatos A, Turner JA, Kruggel F, Fallon JH, et al. Hippocampal atrophy as a quantitative trait in a genome-wide association study identifying novel susceptibility genes for Alzheimer’s disease. PLoS One 2009; 4: e6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince M, Wimo A, Guerchet M, Ali GC, Wu YT, Prina M. World Alzheimer report 2015: the global impact of dementia: an analysis of prevalence, incidence, cost and trends. London: Alzheimer’s Disease International, 2015. [Google Scholar]
- Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 2011; 26: 2336–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: a language and environment for statistical computing. The R Foundation for Statistical Computing, 2005. [Google Scholar]
- Ramanan VK, Risacher SL, Nho K, Kim S, Shen L, McDonald BC, et al. GWAS of longitudinal amyloid accumulation on 18F-florbetapir PET in Alzheimer’s disease implicates microglial activation gene IL1RAP. Brain 2015; 138: 3076–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanan VK, Risacher SL, Nho K, Kim S, Swaminathan S, Shen L, et al. APOE and BCHE as modulators of cerebral amyloid deposition: a florbetapir PET genome-wide association study. Mol Psychiatry 2014; 19: 351–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy A, Trabzuni D, Guelfi S, Varghese V, Smith C, Walker R, et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci 2014; 17: 1418–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnayaka JA, Serpell LC, Lotery AJ. Dementia of the eye: the role of amyloid beta in retinal degeneration. Eye 2015; 29: 1013–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, Patterson N, Campbell D, Tandon A, Mazieres S, Ray N, et al. Reconstructing native American population history. Nature 2012; 488: 370–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roshyara NR, Kirsten H, Horn K, Ahnert P, Scholz M, Huang J, et al. Impact of pre-imputation SNP-filtering on genotype imputation results. BMC Genet 2014; 15: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunder M, Strittmatter WJ, Schmechel D, George-Hyslop PH, Pericak-Vance MA, Joo SH, et al. Association of apolipoprotein E allele epsilon 4 with late-onset familial and sporadic Alzheimer’s disease. Neurology 1993; 43: 1467–72. [DOI] [PubMed] [Google Scholar]
- Saykin AJ, Shen L, Foroud TM, Potkin SG, Swaminathan S, Kim S, et al. Alzheimer’s Disease Neuroimaging Initiative biomarkers as quantitative phenotypes: genetics core aims, progress, and plans. Alzheimers Dement 2010; 6: 265–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuff N, Woerner N, Boreta L, Kornfield T, Shaw LM, Trojanowski JQ, et al. MRI of hippocampal volume loss in early Alzheimer’s disease in relation to ApoE genotype and biomarkers. Brain 2008; 132: 1067–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrucca L, Santucci A, Aversa F. Regression modeling of competing risk using R: an in-depth guide for clinicians. Bone Marrow Transplant 2010; 45: 496–509. [DOI] [PubMed] [Google Scholar]
- Servin B, Stephens M. Imputation-based analysis of association studies: candidate regions and quantitative traits. PLoS Genet 2007; 3: 1296–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Kim S, Risacher SL, Nho K, Swaminathan S, West JD, et al. Whole genome association study of brain-wide imaging phenotypes for identifying quantitative trait loci in MCI and AD: a study of the ADNI cohort. Neuroimage 2010; 53: 1051–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein JL, Hua X, Lee S, Ho AJ, Leow AD, Toga AW, et al. Voxelwise genome-wide association study (vGWAS). Neuroimage 2010; 53: 1160–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015; 43: D447–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Tam JHK, Seah C, Chiu J, Tyrer A, Cregan SP, et al. Arf6 controls beta-amyloid production by regulating macropinocytosis of the amyloid precursor protein to lysosomes. Mol Brain 2015; 8: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AR, Esko T, Yang J, Vedantam S, Pers TH, Gustafsson S, et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet 2014; 46: 1173–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaltieri M, Grigoletto J, Longhena F, Navarria L, Favero G, Castrezzati S, et al. α-synuclein and synapsin III cooperatively regulate synaptic function in dopamine neurons. J Cell Sci 2015; 128: 2231–43. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.