Abstract
Motivation
Virus phylogeographers rely on DNA sequences of viruses and the locations of the infected hosts found in public sequence databases like GenBank for modeling virus spread. However, the locations in GenBank records are often only at the country or state level, and may require phylogeographers to scan the journal articles associated with the records to identify more localized geographic areas. To automate this process, we present a named entity recognizer (NER) for detecting locations in biomedical literature. We built the NER using a deep feedforward neural network to determine whether a given token is a toponym or not. To overcome the limited human annotated data available for training, we use distant supervision techniques to generate additional samples to train our NER.
Results
Our NER achieves an F1-score of 0.910 and significantly outperforms the previous state-of-the-art system. Using the additional data generated through distant supervision further boosts the performance of the NER achieving an F1-score of 0.927. The NER presented in this research improves over previous systems significantly. Our experiments also demonstrate the NER’s capability to embed external features to further boost the system’s performance. We believe that the same methodology can be applied for recognizing similar biomedical entities in scientific literature.
1 Introduction
The steady increase in global travel over the past decades has led to a great concern for public health officials, and recent events like Zika and Ebola outbreaks make it even more important to track the origin and spread of infectious diseases, both geographically and over time. In order to model the spread of the virus, phylogeographers utilize DNA sequences of the virus as well as additional metadata describing the virus and the infected host. The National Center for Biotechnology Information (NCBI) maintains GenBank®, one of the largest open access and publicly available databases of biological information that includes viral nucleotide sequences (https://www.ncbi.nlm.nih.gov/genbank/ Accessed: 20 March 2018). The database is organized by records, and each record’s metadata contains information such as organism, strain, host, gene, date and location of collection and when available, a link to the PubMed Central® article describing the research that produced the virus sequence (https://www.ncbi.nlm.nih.gov/pubmed/ Accessed: 20 March 2018). While the record metadata usually contains country name, a more precise geolocation of the infected host is often unavailable, making it unsuitable for localized phylogeography studies. Previous analyses have shown that the percentage of GenBank records that have insufficient location information range from 64% to 80% (Scotch et al., 2011; Tahsin et al., 2014). In such cases, the articles associated with the records have to be parsed to extract a more precise location of the virus. Due to the exponential increase in GenBank data each year (Lathe et al., 2008), it is not feasible to manually curate the location metadata.
In this article, we present a named entity recognizer (NER) to detect toponym mentions, i.e. names of places, in scientific articles associated with GenBank records. While the ultimate goal of the end-to-end pipeline is to find the precise location associated with a given record, a goal which involves the disambiguation of the location (Paris in France versus Paris in Texas) and its association with the event of interest i.e. virus collection and virus isolation, the proposed work focuses solely on the toponym detection task. The toponym detection task is defined as the automatic identification of the boundaries of all toponyms mentions in selected articles. Like many Natural Language Processing (NLP) tasks, detection of toponyms is challenging due to the inherent ambiguity of the natural language. For instance, words like ‘May’ which appear in ‘was extracted in May, Russia’ needs to be tagged as toponym, but not in ‘found in May 2013’
Previous solutions for toponym detection have included dictionary lookups, rule based and machine learning (ML) based approaches but they suffer from well known limitations, such as coverage or scalability among others (Piskorski and Yangarber, 2013). Dictionary based approaches are unable to resolve correctly the ambiguities between phrases in documents and entries in the dictionary, resulting in many false positives. Rule based techniques encode the contexts where toponyms appear to solve these ambiguities. However, the rules, written manually, never describe all possible contexts, resulting in many false negatives (Tamames and de Lorenzo, 2010; Weissenbacher et al., 2015). ML systems, classifiers or sequence labelers, are able to learn the rules from annotated examples. With better performances, they have been dominant over rule-based approaches in recent times. ML systems rely on features describing the examples to learn the rules. Features, which commonly include orthographic, lexical, syntactic and semantic information about the phrase and its context, are typically manually selected and encoded. Features are valuable in decision making in NLP systems, but feature engineering can be challenging because it is never known in advance if a feature or a combination of features contribute to increased performance of the ML system (Tang et al., 2014). Moreover, many basic features are often computed from other NLP systems that are individually error-prone (e.g. part-of-speech taggers or dependency parsers) and, as a consequence, can be susceptible to adding noise when combined. Noisy features make the inferences of ML systems harder during their training and quickly degrade their deductions at runtime (Goldman and Sloan, 1995; Zhu and Wu, 2004).
Our NER relies on classification with deep neural networks and word embeddings. NERs based on deep learning (DL) have been shown to be effective at selecting and computing the features required for their tasks directly from vectors representing words. In this representation, also known as word embedding, each word of a predefined vocabulary is represented by, or embedded in, a vector of n floating point numbers. n is often called the dimensionality of the word embeddings and it is the length of the word vector. n is fixed for all words in the vocabulary. Each vector encodes the position of the word it embeds in a high dimensional space. Word embeddings are initialized randomly and trained on a large unlabeled corpus to adjust the values based on the idea that words which are used in similar contexts must have vectors with similar values. Hence, in a pre-trained word embedding, the vectors for words in the vocabulary are clustered such that words with similar meaning lie close to each other in the n dimensional space (Kusner et al., 2015; Li et al., 2015a).
Word embeddings have been shown to capture morphological, lexical, syntactical and shallow semantic properties of phrases in their raw representation of the vectors (Mikolov et al., 2013; Pennington et al., 2014). The use of word embedding removes the need to encode manually basic features into the architecture and limits the errors caused by noisy features during their inference. Leveraging this knowledge representation has shown to improve performance in a multitude of NLP tasks that rely on semantics (dos Santos and Guimarães, 2015). Many advanced neural network architectures like convolutional neural networks (Xu et al., 2016), recurrent neural networks (RNNs; Socher et al., 2013) and long short term memory (LSTM; Lample et al., 2016) systems have since been explored to accomplish state-of-the-art performances in NLP tasks. However, their optimal performances are limited by the availability of human annotated data for training. We propose a solution to this problem by using distant supervision to generate additional training instances for greater coverage.
Distant supervision is a form of weak supervision where the idea is to leverage weakly structured data to obtain labeled data (Liu et al., 2003; Mintz et al., 2009). As most ML systems have the potential to improve their performance with more training data, distant supervision techniques have been used for multiple relation extraction tasks where labeled data for training ML systems are limited or not available (Krause et al., 2012; Nguyen and Moschitti, 2011; Takamatsu et al., 2012). In NER tasks, labeled data are also difficult or expensive to obtain (Purver and Battersby, 2012; Roth et al., 2013). To overcome limited labeled data available for training our NER, we employ distant supervision to generate additional positive and negative examples from publicly available articles on PubMed Central that are linked to GenBank articles. We rely on distant supervision data within the domain as opposed to annotated geographic mentions in other domains (Richman and Patrick, 2008) for multiple reasons. Firstly, the differences in effective vocabulary between the domains can be quite large (as shown later) and such differences can affect the performance of the NER task. Secondly, our method to generate the examples uses the geographic location of the infected host i.e. the virus location in GenBank metadata. Hence, we hypothesize that this method may prioritize the identification of geographic locations that helps the eventual task for resolving the geographic location of the infected host.
Sequence labelers such as Conditional Random Fields (CRF) and most recently recurrent neural models such as RNNs (Li et al., 2015b), LSTMs (Lample et al., 2016; Limsopatham and Collier, 2016) and Gated Recurrent Units (Yang et al., 2016), are popularly used for NER due to their fundamental design to factor in previous decisions into the current decision, a design well adapted to fit the sequential nature of the natural language. However, in this work we use a feedforward neural network (also known as multi-layer perceptron) to make use of a very large volume of training data obtained from distant supervision. A choice uncommon but not unprecedented, deep neural networks have been previously used for NER tasks (Godin et al., 2015) including works in the biomedical domain (Wu et al., 2015). The distant supervision method used in this paper reveals only some of the toponyms contained in sentences whereas the others remain unlabeled. This prevents the use of sequence labelers which require all toponyms to be labeled during the training phase.
Previous work on the dataset evaluated in this paper such as Weissenbacher et al. (2015) and Weissenbacher et al. (2017) have used rule based and CRF based NER systems, respectively. The first paper introduces the dataset and provides baseline performance scores using a rule based classifier. The second improves over the previous classifier using a CRF labeler that uses handcrafted lexical, morphological and semantic features to improve the performance. The second paper suggests the use of distant supervision data for improving the performance of the labeler through additional training and lists the steps involved in creating a distant supervision dataset. It uses a naive bayes classifier to evaluate the quality of the distant supervision examples and reports a poor performance when tested on the gold-standard annotations. The paper stops short of evaluating the contribution of distant supervision examples in conjunction with gold-standard annotations on the overall NER task using the CRF labeler. In this work, we propose a new NER model with significantly better performance, make improvements in generating the distant supervision examples and perform a comprehensive evaluation of multiple NER systems.
The contributions of this paper are as follows: (i) We present a NER system based on deep neural networks which performs significantly better than previously developed ML systems that use hand-crafted features. (ii) For a robust comparison, we run experiments with other variants of classifiers and off-the-shelf NER systems and compare the methods and results of previous NERs trained on this dataset. (iii) We demonstrate the capability of our system to add handcrafted features into the neural network. (iv) We show how improved distant supervision techniques can be used to generate more training examples and further boost the performances of our NER. The rest of the document is structured as follows. Section 2 describes the deep neural network architecture of our NER, the training procedure for the NER and the distant supervision technique to generate additional examples. The Results and Discussion Sections details the NER task results as well as the error analysis and the efficacy of distant supervision.
2 Materials and methods
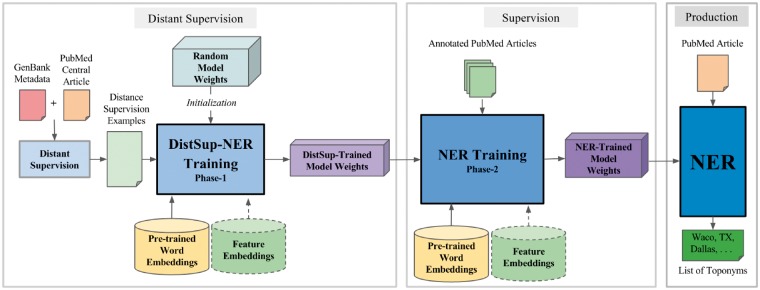
In Figure 1, we show the architecture of our NER system. As illustrated in the figure, there are three different phases of operation for the NER: distant supervision, supervision and testing. At the core of each phase is a deep neural network that forms the NER. The first two phases involve training the NER to detect toponyms and the last phase, the testing phase, uses a trained system to detect toponyms. We begin by describing the components and steps involved in training our NER.
Fig. 1.
The NER architecture with distant supervision. The NER model is first trained on distant supervision data followed by human annotated data to obtain the final model
2.1 Input
The annotated data consists of scientific articles in which toponyms have been tagged by either human annotators or using distant supervision. Training instances created from the annotated data are used as input during the NER’s training phase. Each training instance consists of an input word, the word’s context and a label indicating if the word is in a phrase which is a toponym. The context of the word is formed by the words in its neighborhood, i.e. a window of words where the given word is at the center. The size of the window is fixed. For instance, the sentence ‘AIV H9N2 was detected in domestic ducks in Hong Kong until 1985’ contains 13 tokens including the period, thereby forming 13 training instances. For the word Hong, the words ‘ducks in Hong Kong until’ form its context when the window size is 5. We use the context of a word because it helps in determining if the word is or is not in a toponym phrase. Words in the beginning and end of the document that lack neighbors are padded with the required number of start words or end words.
2.1.1. Word embeddings
Each word is represented by its word embedding obtained from unsupervised pre-training. A word embedding consists of a vector formed by a set of real numbers that represents its position in a multi-dimensional space. A word’s context is represented by the concatenation of individual word embeddings of the words in the window to form a long input vector. We use a randomly initialized vector to represent all words not present in the vocabulary of the pre-trained word embeddings used during our experiments.
2.1.2. Feature embeddings
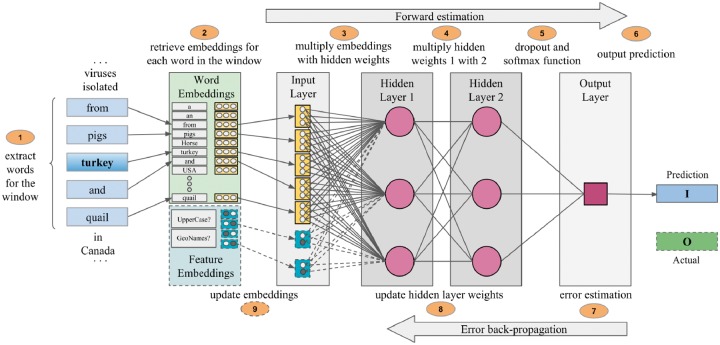
In addition to the word’s context, features describing properties of the word, its context or properties of the document that may help in decision making can also be concatenated into the input vector. For instance, features could include information about the section of the article the word was taken from (i.e. abstract, introduction, body, table), or information if the word was found in a database of city names. A feature is represented by a one-hot vector, (e.g. for binary features, the corresponding index of either ‘Yes’ or ’No’ is set to 1 and the other is set to 0). To demonstrate the capability of embedding features, we implement two simple word based binary features: the word’s presence in a publicly available toponym dictionary, for our experiments we used GeoNames (http://www.geonames.org/Accessed: 20 March 2018) and the presence of full uppercase letters in the word. For example, for the phrase ‘isolated from pigs, turkey and quail in Canada’ in Figure 2, the feature to detect if ‘turkey’ is an abbreviation will check if all letters of the word are uppercase and since it is not, the index for ‘No’ is set to 1 and added to the input vector. In the architecture proposed, embedding features are optional but we introduce them to demonstrate the NER’s capability of using them.
Fig. 2.
The training procedure of the NER’s neural network with two hidden layers
2.2 Training
The NER model consists of weight matrices, where the weights are real numbers initialized randomly and optimized during the training procedure. The training phase of the NER involves a series of matrix multiplication operations between the input matrix and the NER model’s weight matrices in the hidden and output layers of the model. The output of each training phase includes the collection of matrices that form the NER model’s weights that have been optimized during training and ready to be used in the NER system. The model outputs from the training phase are used to initialize the final NER system that processes articles and extracts toponyms from the text.
The first two phases of the architecture in Figure 1 involve training the NER that consists of two parts: forward estimation to determine the probability of a word being in a toponym, and error back-propagation to adjust the model weights and embeddings to reduce error in future predictions. The testing phase involves only the forward estimation part. A representation of the training phase in a neural network with two hidden layers based on a window size of 5 is shown in Figure 2.
2.2.1. Forward-estimation
The text from the input PubMed articles are tokenized into words and punctuations which form an input stream of training data processed as windows of words. The input vector is constructed as described earlier. From the training data, the tokens occurring in a phrase labeled as toponym, i.e. inToponym(I), are encoded to the value of 1 while others tokens, outToponym(O), are encoded to 0. Hence, for the previous example the encodings will be ‘AIV = 0 H9N2 = 0 was = 0 detected = 0 in = 0 domestic = 0 ducks = 0 in = 0 Hong = 1 Kong = 1 until = 0 1985 = 0.=0’.
The overall transformations for the two layer feedforward neural network are shown following equations:
| (1) |
| (2) |
| (3) |
Here, , and represents the first, second and output layer weights, respectively, where d is the number of dimensions of the hidden layer. represents the input layer vector, where w is the number of words in the window and n is the number of dimensions in the word embeddings. and represents the bias terms of the first, second and final layer. After evaluating available activation functions such as tanh and sigmoid, rectified linear units (ReLU) were found to be most efficient. We use a dropout function at layer 2 with a probability of 0.5 to prevent the model to overfit the data, leading to poor generalization. More hidden layers (depth) can be added to the architecture by repeating Equation (2). At the output layer, a softmax function is used to decide the label of the word.
2.2.2. Error back-propagation
During the training phase, we computed the error for each prediction using the cross entropy function. This loss function computes a score reflecting the scale of the difference between the expected output value y and the probability estimated by our system for the encoded label values 0 (for O) or 1 (for I). To minimize the loss, the system uses stochastic gradient descent (Bottou, 1991) to determine the values for U, b3, W2, b2, W1, b1 that maximizes the likelihood of the predictions. We do not update or fine-tune the pre-trained word embeddings during training as they did not show a significant increase in performance. For purposes of brevity, the objective function and derivations of the equations are left out of the paper, but they can be inferred from previous works (Collobert et al., 2011; LeCun et al., 1998, 2012).
In addition to the word embeddings, handcrafted feature embeddings can be concatenated to the input layer along with the word embeddings and be trained. Post-training, the matrices of the hidden layers (i.e. U, b3, W2, b2, W1 and b1) form the model of the NER system. The NER system can now be used to identify toponyms in unseen articles by following the first six steps shown in Figure 2.
2.3 Corpus
To evaluate the performance of the system, we trained the system on annotated data obtained from two different sources, manual annotation and automatic generation with distant supervision, Ddist.
2.3.1. Distant supervision
The performance of deep neural networks have shown to improve with increase in training size even when the training data may contain a small amount of noise (Amodei et al., 2016; Chilimbi et al., 2014). Distant supervision uses heuristic rules to generate both positive and negative training examples. Positive examples for NER tasks refers to word windows where the center word is in a toponym (e.g. ‘several regions of Spain, and infection’) and negative examples are ones where the center word is not in a toponym (e.g. ‘samples collected in December 2009 and January’). Distant supervision was used to generate 8 million training examples that could be used to train the NER in addition to the 260 000 instances from manually annotated data. We estimated the quality of the distant supervision examples generated by manually analyzing a random sample of 200 positive and 200 negative examples to find 19 false positives and 6 false negatives. The false positives were dominated by tokens that were part of an organization, institution or strain. Due to the sparsity of toponym mentions in large documents, we restricted the ratio of positive/negative examples to its ratio observed in the training set.
2.3.2 Generating positive examples
The following steps were used to generate positive examples: (i) Find GenBank records for which a location in the location field of the metadata and a link to the full text article are both available. (ii) Annotate as toponyms in the article all phrases which match the locations in the metadata of the records. (iii) Include the annotated locations’ word windows as positive examples for training. A manual inspection of positive examples generated revealed that the positive examples included many false positives which we needed to eliminate. (iv) Analyze the false positives and manually create a list of words called blacklistPOS that contains frequent words that are collocated with the false positives. For instance, blacklistPOS will contain words that indicate organization entities such as University, Department, or Center and words that refer to organism entities such as virus, isolate and strain. (v) Check for presence of blacklistPOS words in positive examples from step 3 and move them to negative examples because they are crucial in eliminating similar false positives during NER training.
2.3.3 Generating negative examples
Negative examples were generated using similar steps as documented previously in Weissenbacher et al. (2017). We summarize them: (i) Manually compile a list of words called whitelistNEG that contain words collocated with toponyms in the word windows by analyzing word windows from human annotated training data. The whitelistNEG will contain words such as ‘isolated’, ‘locations’, ‘near’ or ‘from’. (ii) Process articles and select sentences that contain phrases matching with toponyms in a dictionary based on case-sensitive lookups. Sentences such as ‘Gene UL111A encodes viral interleukin-10 (Lockridge et al., 2000)’ are selected where Lockridge is a phrase matching a toponym in our dictionary, GeoNames. (iii) Create negative examples by generating word windows from the sentences where no words from whitelistNEG appear in the examples.
2.3.4 Human annotated data
The second type of annotated data that the NER was trained on was a publicly available annotated corpus of articles from PubMed Central (Weissenbacher et al., 2015). The dataset contains 60 articles manually annotated with 1881 toponym mentions and an inter-annotator agreement of 97%. While the dataset also list the toponym’s GeoNameID, latitude and longitude information, we do not use this information for the NER task proposed in this work. Among the 1881 toponym mentions, 343 toponyms are composed of more than 1 token and the average token length per toponym was found to be 1.21. For purposes of comparison, the proposed system uses the same 48 articles (containing 1596 toponym mentions) for training, data Dtrain and 12 articles (containing 285 toponym mentions) for testing data, Dtest, as used in those tasks (Weissenbacher et al., 2015, 2017). Of the 48 articles available for training, 5 articles (containing 159 toponym mentions) were initially separated as held-out data for validation and tuning the hyperparameters of the model. Although the BIO schemes of annotation is popular in multiple word named entities (e.g. […]in(O) Papua(B) New(I) Guinea(I) and(O)[…]), we use the IO scheme because it reduces the NER task from choosing between three labels to a binary classification problem. In the annotated corpus containing 1881 toponym instances, there was only one occurrence (0.0005%) where a toponym immediately followed a multi-word toponym i.e. a B-I-B sequence.
2.4 Pre-trained word embeddings and model hyperparameters
In our experiments, we used publicly available pre-trained word embeddings from two different data sources: glove (Pennington et al., 2014) uses text gathered by CommonCrawl (http://commoncrawl.org/Accessed: 20 March 2018), and wiki-pm-pmc uses a collection of abstracts and articles from PubMed and Wikipedia (Pyysalo et al., 2013). We observed that dimensions of the word embeddings and the effective vocabulary (i.e. the set of different words found in the word embedding vocabulary) for the annotated dataset vary greatly, 300 and 152 786 for glove, and 200 and 201 380 for wiki-pm-pmc. We also compose a baseline word embedding with random numbers using the largest vocabulary and the largest dimension among the embeddings that are updated during the training.
The performance of the proposed NER model depends on the tuning of hyperparameters of the deep neural network during the training phase. We limit the architecture to use two hidden layers because additional hidden layers did not improve the performance significantly. We set the number of dimensions of both hidden layers to 150 and learning rate was set to 0.001. For initializing the weight matrices in the hidden layers, U, W1 and W2, random numbers from a uniform distribution in the range (–r,) were used, where and m and n are the dimensions of the said matrix. The bias terms, b1, b2 and b3 are all initialized to zeros.
The deep neural network based NER was built using the TensorFlow (https://www.tensorflow.org/Accessed: 20 March 2018) framework and trained on a Dell Precision T3610 workstation equipped with an Intel Xeon Processor E5-1620 v2 with 8 cores and NVIDIA Titan Xp GPU for faster training time.
2.5 Comparison with other classifiers
For the purpose of comparison, we train additional models using the random forest and support vector machine (SVM) (Vapnik, 2013) classifiers which use the same concatenated input of word embeddings and custom features. For these models, we train on the entire training dataset under 10-fold cross-validation (by training instances) to pick the best model and evaluate them on Dtest. The random forests classifier (Breiman, 2001) works by constructing multiple decision trees on sub-samples of the training data that optimize the decisions for the labels given the inputs (i.e. the concatenated word embeddings and features). In the final model, the labels are chosen by averaging predictions from the individual decision trees. In our experiment with the random forest classifier we construct 10 individual trees where the minimum number of samples i.e. leaves required for a split is 1. The SVM classifier on the other hand is fundamentally very similar to the single layered feedforward neural network, in that both classifiers try to find a linear separation between the classes (I and O) in high dimensional vector space. However, the key difference lies in the usage of kernel functions in the SVM classifier to assist linear separations for non-linear classification problems. Feedforward neural networks typically do not employ kernel functions although they could be added into the network. In our experiment with the SVM classifier, we use the radial basis function as the kernel function.
3 Results
We evaluate our NER on Dtest containing 12 manually annotated articles. For our experiments, the NER model was trained for 50 epochs with each of the three word embeddings described above and the one with the highest accuracy on the validation set was selected. The results for the models running under the three different configurations in addition to the random forest and SVM classifiers are shown in Table 1.
Table 1.
Precision, Recall and F1 scores using strict tokenwise evaluation for toponym detection where the NER was trained on Dtrain and tested on Dtest
| Configuration | Word embedding | P | R | F1 |
|---|---|---|---|---|
| FFNN 1-layer | No pre-training | 0.97 | 0.65 | 0.779 |
| Glove | 0.89 | 0.87 | 0.883 | |
| Wiki-pm-pmc | 0.92 | 0.82 | 0.878 | |
| FFNN 2-layers | Glove | 0.92 | 0.86 | 0.891 |
| Wiki-pm-pmc | 0.93 | 0.88 | 0.906 | |
| FFNN 2-layers + features | Glove | 0.94 | 0.87 | 0.903 |
| Wiki-pm-pmc | 0.96 | 0.86 | 0.910 | |
| Random forest + features | Wiki-pm-pmc | 0.82 | 0.91 | 0.862 |
| SVM + features | Wiki-pm-pmc | 0.83 | 0.92 | 0.875 |
Bold indicates highest scores in the performance measure.
For comparison with previous systems on this dataset, the strict tokenwise scheme of evaluation (Tsai et al., 2006) was used, i.e. the predictions of the system were evaluated only on words in toponyms and words predicted as toponyms, words outside of toponyms and correctly predicted with the value 0 (for O) were ignored. In standard NER tasks where an entity can span across tokens, tokenwise evaluation may not be a suitable evaluation scheme because partially extracted entities such as ‘Hong’ in ‘Hong Kong’ may not be sufficient in disambiguating geographic locations. Hence, the phrasal evaluation scores are used for measuring performance. In this evaluation, a multi-token entity is counted as a true positive only when all tokens in the entity exactly match the gold standard entity. We report the phrasal evaluation scores on the best model in the following sub-section for future comparisons.
We observe a significant improvement in performance when using pre-trained word embeddings over randomly initialized word embeddings. We also observe that there is an increase in the performance of the deep (two layer) neural network over a simple (one layer) feedforward network that demonstrates the need for non-linear classification models for the task. The wiki-pm-pmc word embeddings performs consistently better with its high coverage on vocabulary despite having low dimensionality. The glove word embeddings perform equally well under all models despite being from a generic domain and having less coverage on the vocabulary compared to wiki-pm-pmc. We believe that its high dimensionality i.e. 300 as compared to wiki-pm-pmc’s 200 is the reason behind such good performance. This motivates the creation of pre-trained word embeddings of higher dimensionality from the same domain for better performance. The basic handcrafted features implemented in this model provided a combined boost of 0.46% on the best model. The GeoNames lookup feature and capitalization feature individually provided 0.32% and 0.25% increase in F1-score, respectively, to the 2-layer feedforward model.
Both Random Forest and SVM classifiers trained on similar features on the wiki-pm-pmc word embeddings achieve F1-scores marginally lower than the single layer feedforward neural network. We find that repeated experiments with various combinations of kernel functions may be necessary to draw strong conclusions when comparing the performance of the SVM classifier and the single layer feedforward model. While we only use binary features in this implementation for the sake of demonstration, advanced orthographic, semantic features and domain-specific pragmatic features can be encoded in vector format both at the word and context level as described by Limsopatham and Collier (2016).
4 Discussion
4.1 Error analysis
To understand the nature of the errors, we analyze errors found in the predictions in Dtest from the model built on the wiki-pm-pmc word embeddings with features. In Table 2, we show examples of some of these errors. In total, 255 out of 285 toponyms in the test data were fully matched and there were 32 false positives and 30 false negatives. A majority of the errors were associated with multi-token entities where the entity was matched only partially. Such partial matches lead to both false positives and false negatives in a strict phrasal evaluation. Among the false positives and false negatives, 16 such errors were associated with partial matches as shown in examples 1–4 in the table. Among the remaining 16 false positives, 10 instances were names of places that were used as part of names of organizations, group of countries, gene pools, or strains as shown by examples 5–7. Among the false positives, three were toponyms that seemed to be wrongly or partially annotated. For instance, example 8 in the table shows that ‘BJ’ and ‘Bei’ could have been annotated as geographic locations as both refer to the location ‘Beijing’. The remaining three errors were associated with capitalized tokens confused as abbreviated toponyms. The 14 false negatives seemed to belong in two categories. The first class is toponyms not recognized due to their presence in tables which do not follow natural language syntaxes and semantics. Example 9 in the table shows three out of eight such errors. The remaining six toponyms belonged to the second class where they seemed to stay unrecognized and untagged because their contexts were not present in annotated training data. Examples 10 and 11 show such examples.
Table 2.
Examples of errors made by the NER trained on supervised annotated data
| Error type | No | Category | Examples |
|---|---|---|---|
| Partial match | 1 | Tagged prefix | Probable person to person transmission of novel avian influenza A (H7N9) virus in EasternChina, 2013. |
| 2 | Tagged suffix | Surveillance was conducted in live poultry markets in Fujian, Guangdong, Guangxi, Guiyang, Hunan and YunnanProvinces. | |
| 3 | Tagged suffix | University of Ibadan, OyaState, Ibadan and Nigeria. | |
| 4 | Unrecognized token | The overwhelming majority (94.2%) of H9N2 influenza viruses were isolated in Asia, with > 65% coming from mainland and Hong Kong of China | |
| False positive | 5 | Other entities | Phylogenetic analyses show that it is a recombinant virus containing genome segments derived from the Eurasia and North America gene pools. |
| 6 | Other entities | Thus, current G1-like viruses in southern China might have originally been introduced from Middle Eastern countries, or it is also likely that the virus spread the other way around, similar to the transmission of FIG. | |
| 7 | Other entities | This work was supported by a Natural Sciences and Engineering Research Council of Canada discovery grant. | |
| 8 | Partial annotation | Abbreviations: BJ and Bei, Beijing; Ck, chicken; Dk, duck. | |
| False negative | 9 | Table entries | Virus Group State of isolation Date of isolation A/chicken/Nigeria/1071-1/2007 EMA1/EMA2-2: 6-R07 Plateau Jan 2 A/chicken/Nigeria/1071-3/2007 EMA2 Sokoto Jan 5. |
| 10 | Unrecognized toponym | The characterization of the swH3N2 / pH1N1 reassortant vi- ruses from swine in the province of Quebec indicates that reassortment of gene segments had occurred between the North American swine H3N2. | |
| 11 | Unrecognized toponym | Centers for Disease Control and Prevention, Atlanta, Ga. |
Note: Underlined tokens indicate entities recognized by the NER. Italicized tokens are human annotated gold standard entities.
4.2 Improving supervised NER with distant supervision
The error analysis reveals the need to increase the number of training examples to present additional contexts to the NER for reducing false negatives. For this reason, we trained our NER on examples generated with distant supervision. The resulting model shown in Figure 1 used both Ddist and Dtrain for training, and achieved the best F1 score of 0.927, as indicated in Table 3. The 1.7 p.p. (percentage points) improvement over the NER trained only on manually annotated examples Dtrain showed the results to be significantly different at 95% confidence level () when a McNemar’s test (McNemar, 1947) was performed on the individual words. The positive and negative examples from the distant supervision were used to train the NER and subsequently reinforced with the human-annotated training examples to achieve the best performance. The training examples from Ddist provide extended coverage for the contexts appearing around the toponyms, thus shaping the weights for the NER task. In comparison to the error analysis in the earlier section, from the 285 toponyms in the test set, the number of true positives increased from 255 to 265, false positives decreased from 32 to 29 and false negatives decreased significantly from 30 to 20. Overall, it was observed that errors associated with multi-token entities were greatly reduced.
Table 3.
Tokenwise scores for performance comparison of NERs
| Implementation | P | R | F1 |
|---|---|---|---|
| Knowledge-based | 0.58 | 0.88 | 0.70 |
| CRF-All | 0.85 | 0.76 | 0.80 |
| Stanford-NER | 0.89 | 0.85 | 0.872 |
| Train and Test | 0.96 | 0.86 | 0.910 |
| Train and Test | 0.97 | 0.89 | 0.927 |
Bold indicates highest scores in the performance measure.
The training on distant supervision data improved the recall by 3%. Table 3 shows the performance comparison of the proposed NER system with previous NERs developed on the same dataset: (i) a rule based approach (Weissenbacher et al., 2015), and (ii) a CRF based NER system (Weissenbacher et al., 2017) that used handcrafted features. For a robust comparison, we also train (iii) the Stanford NER on the entire training set. While both classifiers 2 (CRF-All) and 3 (Stanford-NER) are based on the CRF classifier that looks for the best sequence of tokens given the input features for each word sequence, there are significant differences between the number and type of features used in the models. The ‘CRF-All’ model applied previously on this dataset combines features such as N-grams (up to 4), capitalization, POS-tags, dictionary lookups and k-means clustered word vectors that total approximately 80 000 features per token. However, the ‘Stanford-NER’ combines features such as N-grams (upto 6), word shape features and a multitude of sequence features that total approximately 230 000 features per token. The sequence features implemented in ‘Stanford-NER’ alone contributed to a 5 p.p. improvement out of the 7.2 p.p. total performance increase over ‘CRF-All’. In comparison, the features used in the feedforward models used in this work are merely around 1000 per token (i.e. 5 concatenated 200 dimensional word vectors along with binary shape and knowledge features). The ‘CRF-All’ classifier uses similar word embeddings used in this work, hence we speculate that the factors affecting the performance could be attributed to k-means clustered word vectors, the noisy or redundant features, or a combination of both.
All NERs proposed in this paper (F1 = 0.88 to 0.927) outperform the previous best system ‘CRF-All’ (F1 = 0.80) and the ‘Stanford-NER’ (F1 = 0.87). We confirm the findings of previously proposed DL based NER architectures (Lample et al., 2016) that it is possible to obtain state-of-the-art results without the use of handcrafted features. For future comparison as per standard NER tasks, we find the phrasal classification scores for the best model with distant supervision to be P/R/F1 scores of 0.901, 0.929 and 0.915.
4.3 Generalizability
Although our research specifically looks at geographic location extraction, we find that the approach can be used for named entities across other domains where the availability of human annotated data is very limited. In contrast to human annotated data, the cost and manual effort involved in generating weakly supervised data is significantly lower and the volume of data obtained is much higher. Although, this data comes at the cost of quality, we find that it is possible to boost the performance of a NER using using such weakly supervised data.
Entities like geographic locations that have millions of entries in a database like Geonames.org, can contain numerous words such as The and of that are part of a smaller but a widely used English vocabulary. These along with ambiguous proper nouns like Turkey and May make it challenging for generating valid distant supervision examples. In this work, we demonstrate that it is possible to effectively improve the NER’s performance by adopting distant supervision, even for such challenging named entities. Other named entities such as organisms, genes, drugs and diseases that contain comparatively fewer terms in common with the general domain English vocabulary do not demand extensive disambiguation measures using blacklistPOS and whitelistNEG. Hence, we believe that distant supervision can contribute to significant improvements in NER tasks for recognizing such entities with minimal effort.
4.4 Limitations
In spite of the considerable performance improvement, there are a few limitations to the NER and the distant supervision system proposed. Although the number of errors are reduced in the system after the adoption of a deep neural network for NER and additional training on distant supervision data, many errors remain when the NER is tested on Dtest. Most of the errors were due to unrecognized tokens, many of which were present in a table structure in the source literature. Text extraction from such scientific articles flattens out the table entries into individual tokens that lack the typical syntactic structure found in natural language. Since the majority of training instances (including distant supervision instances) contain some syntactic structure in the context windows, recognizing entities in tables often result in errors. Such errors are consistent with similar statistical models where syntactic features are used for NER or text classification tasks.
While the NER itself can be treated like a black-box for use in similar applications, we find that there can be some challenges in adoption of distant supervision for improving the NER’s performance. Firstly, distant supervision requires some amount of domain expertise to recognize the named entities and contexts of interest. In our experiments, we found that it is necessary to populate the blacklistPOS and whitelistNEG based on training instances in the gold standard annotations and the accompanying annotation guideline. Secondly, the quality of the distant supervision examples and its contribution to performance improvements may demand some manual modifications the blacklistPOS and whitelistNEG depending on the type of named entities. One good approach would be to iteratively train on Ddist and test on Dtrain to recognize false positives and false negatives. And finally, training the NER on weakly supervised data increases the training time, especially if the model hyperparameters have to be tuned during the process. However, once the NER is trained and tuned for performance, it’s execution time remains constant.
5 Conclusion and future work
The location metadata in a GenBank record is crucial in virus phylogeography as it enables for estimates of migration. We attempt to improve the geographic scope of this metadata by developing a NLP pipeline that automatically scans the journal article associated with the record in order to identify more localized toponyms. This study presents a deep neural network based NER for toponym detection in biological publications which outperforms the previous state-of-the-art systems without the use of any handcrafted features. All proposed models are evaluated across two publicly available pre-trained word embeddings. The paper shows how distant supervision can be used to generate more training data to boost the NER’s performance. All models presented in this research achieved a high performance, with the best tokenwise F1-score being 0.927 and a phrasal F1-score of 0.915. The proposed deep neural network based NER is general enough to be used reliably for detecting similar named entities in biomedical texts such as host, organism, date of collection or genes. The dataset containing 60 annotated PubMed articles is available at (https://healthlanguageprocessing.org/software-and-downloads/Accessed: 20 March 2018). The source code of the generic feedforward NER is made available on github (https://github.com/amagge/ner-topo-ff Accessed: 20 March 2018).
As future work, we intend to employ appropriate heuristics (Limaye et al., 2010; Shen et al., 2012) to deal with toponyms in the table data, an important cause of errors. Similar to the idea proposed in this work, we intend to run additional experiments where we use our current best NER to label additional full-text articles and use such annotations in combination with gold-standard annotations on RNN models such as LSTMs to test if they contribute to increased performance. Having demonstrated the capability to embed handcrafted features, a bigger goal of this research is to be able to leverage character-level embeddings to capture orthographic features and embed domain specific pragmatic knowledge features in addition to features from external knowledge databases. Using such techniques will be necessary to further boost the performance of the NER close to human performance (97%).
Acknowledgements
AM designed and trained the neural network, ran the experiments, performed the error analysis and wrote most of the manuscript. DW proposed the idea of using of distant supervision for improving the CRF NER’s performance in the previous manuscript, created the distant supervision dataset, supervised the experiments and wrote revisions of the manuscript. AS reviewed, restructured and contributed many sections and revisions of the manuscript. MS and GG provided overall guidance on the work and edited the final manuscript. The authors would also like to acknowledge Karen O’Connor, Megan Rorison and Briana Trevino for their efforts in the annotation processes. The authors are grateful to the anonymous reviewers for their valuable feedback and comments to improve the quality of the paper. The authors gratefully acknowledge the support of NVIDIA Corporation with the donation of the Titan Xp GPU used for this research.
Funding
Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health (NIH) under grant number R01AI117011. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Conflict of Interest: none declared.
References
- Amodei D. et al. (2016) Deep speech 2: end-to-end speech recognition in english and mandarin. In: International Conference on Machine Learning, pp. 173–182. Journal of Machine Learning Research, New York, NY, USA.
- Bottou L. (1991) Stochastic gradient learning in neural networks. Proc. Neuro-Nimes, 91, 687–696. [Google Scholar]
- Breiman L. (2001) Random forests. Mach. Learn., 45, 5–32. [Google Scholar]
- Chilimbi T.M. et al. (2014) Project adam: building an efficient and scalable deep learning training system. In: OSDI, Vol. 14, pp. 571–582. USENIX Association, Broomfield, CO, USA.
- Collobert R. et al. (2011) Natural language processing (almost) from scratch. J. Mach. Learn. Res., 12, 2493–2537. [Google Scholar]
- dos Santos C.N., Guimarães V. (2015) Boosting named entity recognition with neural character embeddings. CoRR, abs/1505.05008.
- Godin F. et al. (2015) Multimedia lab@ acl w-nut ner shared task: named entity recognition for twitter microposts using distributed word representations. ACL-IJCNLP, 2015, 146–153. [Google Scholar]
- Goldman S.A., Sloan R.H. (1995) Can pac learning algorithms tolerate random attribute noise?. Algorithmica, 14, 70–84. [Google Scholar]
- Krause S. et al. (2012) Large-scale learning of relation-extraction rules with distant supervision from the web. In: The Semantic Web–ISWC 2012, pp. 263–278. Springer, Boston, MA, USA.
- Kusner M.J. et al. (2015) From word embeddings to document distances. In: ICML, Vol. 15, pp. 957–966. Journal of Machine Learning Research, Lille, France.
- Lample G. et al. (2016) Neural architectures for named entity recognition. In: HLT-NAACL. Association for Computational Linguistics, San Diego, California, USA.
- Lathe W. et al. (2008) Genomic data resources: challenges and promises. Nat. Educ., 1, 2. [Google Scholar]
- LeCun Y. et al. (1998) Gradient-based learning applied to document recognition. Proc. IEEE, 86, 2278–2324. [Google Scholar]
- LeCun Y.A. et al. (2012) Efficient backprop In: Neural Networks: Tricks of the Trade. Springer, London, UK, pp. 9–48. [Google Scholar]
- Li J. et al. (2015a) Visualizing and understanding neural models in nlp. arXiv preprint arXiv: 150601066.
- Li L. et al. (2015b) Biomedical named entity recognition based on extended recurrent neural networks. In: 2015 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), pp. 649–652.
- Limaye G. et al. (2010) Annotating and searching web tables using entities, types and relationships. Proc. VLDB Endowment, 3, 1338–1347. [Google Scholar]
- Limsopatham N., Collier N. (2016) Learning orthographic features in bi-directional lstm for biomedical named entity recognition. BioTxtM 2016, p. 10.
- Liu B. et al. (2003) Building text classifiers using positive and unlabeled examples. In: Third IEEE International Conference on Data Mining, 2003, ICDM 2003, pp. 179–186. IEEE Computer Society, Washington, DC, USA.
- McNemar Q. (1947) Note on the sampling error of the difference between correlated proportions or percentages. Psychometrika, 12, 153–157. [DOI] [PubMed] [Google Scholar]
- Mikolov T. et al. (2013) Distributed representations of words and phrases and their compositionality. In: Advances in neural information processing systems, pp. 3111–3119. Curran Associates Inc., Lake Tahoe, Nevada, USA.
- Mintz M. et al. (2009) Distant supervision for relation extraction without labeled data. In: Proceedings of the Joint Conference of the 47th Annual Meeting of the ACL and the 4th International Joint Conference on Natural Language Processing of the AFNLP, Vol. 2, pp. 1003–1011. Association for Computational Linguistics, Suntec, Singapore.
- Nguyen T.-V.T., Moschitti A. (2011) End-to-end relation extraction using distant supervision from external semantic repositories. In: Proceedings of the 49th Annual Meeting of the Association for Computational Linguistics: Human Language Technologies: short papers, Vol. 2, pp. 277–282. Association for Computational Linguistics, Portland, Oregon, USA.
- Pennington J. et al. (2014) Glove: global vectors for word representation. In EMNLP, Vol. 14, pp. 1532–1543. ACL, Doha, Qatar.
- Piskorski J., Yangarber R. (2013) Information extraction: past, present and future In: Multi-Source, Multilingual Information Extraction and Summarization. Springer, pp. 23–49. [Google Scholar]
- Purver M., Battersby S. (2012) Experimenting with distant supervision for emotion classification. In: Proceedings of the 13th Conference of the European Chapter of the Association for Computational Linguistics, pp. 482–491. Association for Computational Linguistics, Avignon, France.
- Pyysalo S. et al. (2013) Distributional semantics resources for biomedical text processing.
- Richman A., Patrick S. (2008) Mining wiki resources for multilingual named entity recognition. In: 46th Annual Meeting of the Association for Computational Linguistics: Human Language Technologies, pp. 1–9.
- Roth B. et al. (2013) A survey of noise reduction methods for distant supervision. In: Proceedings of the 2013 workshop on Automated knowledge base construction, pp. 73–78. ACM, San Francisco, California, USA.
- Scotch M. et al. (2011) Enhancing phylogeography by improving geographical information from genbank. J. Biomed. Informatics, 44, S44–S47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W. et al. (2012) Liege: link entities in web lists with knowledge base. In: Proceedings of the 18th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, pp. 1424–1432. ACM, Beijing, China.
- Socher R. et al. (2013) Recursive deep models for semantic compositionality over a sentiment treebank. In: Proceedings of the Conference on Empirical Methods in Natural Language Processing (EMNLP), Vol. 1631, pp. 1642. Citeseer, Seattle, Washington, USA.
- Tahsin T. et al. (2014) Natural language processing methods for enhancing geographic metadata for phylogeography of zoonotic viruses. In: AMIA Summits on Translational Science Proceedings, 2014, p. 102. [PMC free article] [PubMed]
- Takamatsu S. et al. (2012) Reducing wrong labels in distant supervision for relation extraction. In: Proceedings of the 50th Annual Meeting of the Association for Computational Linguistics: Long Papers, Vol. 1, pp. 721–729. Association for Computational Linguistics, Jeju Island, Korea.
- Tamames J., de Lorenzo V. (2010) Envmine: a text-mining system for the automatic extraction of contextual information. BMC Bioinformatics, 11, 294.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J. et al. (2014) Feature selection for classification: a review. Data Classification: Algorithms and Applications, p. 37.
- Tsai R.T.-H. et al. (2006) Various criteria in the evaluation of biomedical named entity recognition. BMC Bioinformatics, 7, 92.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vapnik V. (2013) The Nature of Statistical Learning Theory. Springer Science and Business Media, New York, NY, USA. [Google Scholar]
- Weissenbacher D. et al. (2015) Knowledge-driven geospatial location resolution for phylogeographic models of virus migration. Bioinformatics, 31, i348–i356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenbacher D. et al. (2017) Extracting geographic locations from the literature for virus phylogeography using supervised and distant supervision methods. American Medical Informatics Association, Translational Bioinformatics. [PMC free article] [PubMed]
- Wu Y. et al. (2015) Named entity recognition in chinese clinical text using deep neural network. Studies in Health Technology and Informatics, 216, 624.. [PMC free article] [PubMed] [Google Scholar]
- Xu H. et al. (2016) Text classification with topic-based word embedding and convolutional neural networks. In: Proceedings of the 7th ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics, pp. 88–97. ACM, Seattle, WA, USA.
- Yang Z. et al. (2016) Multi-task cross-lingual sequence tagging from scratch. arXiv preprint arXiv: 1603.06270.
- Zhu X., Wu X. (2004) Class noise vs. attribute noise: a quantitative study. Artif. Intel. Rev., 22, 177–210. [Google Scholar]