Phospholipase Dδ (PLDδ) is involved in promoting basal, post-penetration resistance against powdery mildew through a potentially novel pathway(s) independent of EDS1/PAD4, SA, and JA, whereas PLDα1 negatively modulates this process.
Keywords: Arabidopsis thaliana, EDS1, Hyaloperonospora arabidopsidis, jasmonic acid, phospholipase D, plant defense signaling, post-penetration resistance, powdery mildew, salicylic acid
Abstract
Plants use a tightly regulated immune system to fight off various pathogens. Phospholipase D (PLD) and its product, phosphatidic acid, have been shown to influence plant immunity; however, the underlying mechanisms remain unclear. Here, we show that the Arabidopsis mutants pldα1 and pldδ, respectively, exhibited enhanced resistance and enhanced susceptibility to both well-adapted and poorly adapted powdery mildew pathogens, and a virulent oomycete pathogen, indicating that PLDα1 negatively while PLDδ positively modulates post-penetration resistance. The pldα1δ double mutant showed a similar infection phenotype to pldα1, genetically placing PLDα1 downstream of PLDδ. Detailed genetic analyses of pldδ with mutations in genes for salicylic acid (SA) synthesis (SID2) and/or signaling (EDS1 and PAD4), measurement of SA and jasmonic acid (JA) levels, and expression of their respective reporter genes indicate that PLDδ contributes to basal resistance independent of EDS1/PAD4, SA, and JA
signaling. Interestingly, while PLDα1–enhanced green fluorescent protein (eGFP) was mainly found in the tonoplast before and after haustorium invasion, PLDδ–eGFP’s focal accumulation to the plasma membrane around the fungal penetration site appeared to be suppressed by adapted powdery mildew. Together, our results demonstrate that PLDα1 and PLDδ oppositely modulate basal, post-penetration resistance against powdery mildew through a non-canonical mechanism that is independent of EDS1/PAD4, SA, and JA.
Introduction
Many fungal and oomycete pathogens penetrate the plant cell wall and extract nutrients from host cells by a similar feeding structure called the haustorium. Plant defense against haustorium-forming pathogens exhibits clear spatiotemporal characteristics that can be conveniently divided into two distinct layers: penetration resistance (cell wall-based; the first layer) and post-penetration resistance (haustorium-targeted; the second layer). Penetration resistance is usually sufficient to stop non-adapted pathogens from entering the host cell by forming a papilla, which is cell wall thickening with deposition of callose (1,3-β-glucan) and other defense chemicals at the penetration site. This process is contributed by at least two independent mechanisms in Arabidopsis. One involves focal exocytosis of antimicrobial materials mediated by PENETRATION1 (PEN1), a syntaxin, and its SNARE partners (Collins et al., 2003; Kwon et al., 2008); the other engages the production of glucosinolates by PEN2 myrosinase and subsequent transport of such antifungal chemicals by the PEN3 ATP-binding cassette transporter (Lipka et al., 2005; Stein et al., 2006; Bednarek et al., 2009). Both mechanisms are probably activated upon recognition of conserved pathogen-associated molecular patterns (PAMPs) by cell surface pattern recognition receptors (PRRs), and thus may be part of PAMP-triggered immunity (PTI) (Jones and Dangl, 2006; Hückelhoven and Panstruga, 2011).
Adapted fungi or oomycetes that can overcome penetration resistance face the second layer of plant defense. Despite successful penetration, early stage haustorial development and/or function can be inhibited by stage I post-penetration resistance which may continue to engage PTI and other defense mechanisms. However, once stage I post-penetration resistance is suppressed by effector proteins secreted from better-adapted pathogens, haustoria can establish function, and disease ensues. Plants have evolved stage II post-penetration resistance to defeat these better adapted pathogens through the action of plant resistance (R) proteins. Most characterized R proteins are intracellular immune receptors belonging to the nucleotide-binding site–leucine-rich repeat (NB-LRR) superfamily that detects the presence or activity of specific effector proteins termed avirulence factors (Avrs). Thus, stage II post-penetration resistance in many cases is equivalent to effector-triggered immunity (ETI), which often exhibits race specificity and features with rapid cell death at the infection site, namely the hypersensitive response (HR) (Jones and Dangl, 2006). Based on the N-terminal domains, NB-LRRs are divided into two major classes, Toll-interleukin 1 receptor (TIR)-NB-LRRs and coiled-coil (CC)-NB-LRRs. While characterized TIR-NB-LRRs require the nucleocytoplasmic lipase-like protein ENHANCED DISEASE SUSCEPTIBILITY 1 (EDS1) for signal transduction, most CC-NB-LRRs engage the plasma membrane (PM)-anchored integrin-like protein NON-RACE-SPECIFIC DISEASE RESISTANCE 1 (NDR1) for signaling (Cui et al., 2015).
Detection of pathogens triggers a conserved signaling network regulated by salicylic acid (SA), jasmonic acid (JA), and ethylene (ET), resulting in the activation of defense responses including pathogenesis-related (PR) gene expression, reactive oxygen species (ROS) production, and callose deposition (Bari and Jones, 2009; Pieterse et al., 2012). SA signaling plays a critical role in activation of local as well as systemic acquired resistance (SAR) to fight against biotrophic and hemi-biotrophic pathogens. Depending on the context of specific plant–pathogen interactions, the SA pathway could act antagonistically or synergistically with the JA/ET pathways, which are mainly effective against necrotrophic pathogens (Glazebrook, 2005; Robert-Seilaniantz et al., 2011). EDS1 and its interacting homologous partner PHYTOALEXIN-DEFICIENT 4 (PAD4) are both required for adequate SA synthesis and signaling, and play a role in the antagonism between SA- and JA/ET-dependent defense pathways (Zhou et al., 1998; Falk et al., 1999; Feys et al., 2001; Wiermer et al., 2005). Furthermore, EDS1 and PAD4 have also been shown to regulate SA-independent defense responses (Feys et al., 2005; Venugopal et al., 2009; Zhu et al., 2011; Wagner et al., 2013; Cui et al., 2017).
Two non-NB-LRR Arabidopsis R proteins, RPW8.1 and RPW8.2, confer broad-spectrum resistance to powdery mildew fungi (Xiao et al., 2001), which requires EDS1, PAD4, and SA signaling (Xiao et al., 2003; Xiao et al., 2005). RPW8.2 is specifically targeted to the host-derived extra-haustorial membrane (EHM) encasing the fungal haustorium to activate on-site defenses including the formation of callose-enriched haustorial encasement and interface-focused H2O2 production to constrain the haustorium (Wang et al., 2009; Berkey et al., 2017). Previous studies suggest that a specific protein trafficking pathway is engaged for targeting RPW8.2 to the EHM (Wang et al., 2013; Zhang et al., 2015). However, how RPW8.2 achieves haustorium-targeted defense remains to be determined. A tempting speculation is that RPW8.2 may interact with a signaling lipid(s) to realize its specific targeting. In an effort to test this speculation, we instead found that two phospholipase D (PLD) enzymes play opposing roles in plant defense against powdery mildew fungi, but neither of them seems to be involved in RPW8-mediated resistance
PLD and its product phosphatidic acid (PA) have been implicated in modulating plant immunity. Exogenous SA treatment could induce higher PA levels as a result of PLD activity (Kalachova et al., 2013; Rodas-Junco et al., 2015), suggesting a positive role for PLD-derived PA; however, a limited number of genetic studies on PLD genes suggest that the outcome varies depending on the PLD isoforms involved and/or pathosystems examined. This is not surprising since there are 12 identified PLD isoforms [PLDα (3), PLDβ (2), PLDγ (3), PLDδ (1), PLDε (1), and PLDζ (2)] in Arabidopsis (Zhao, 2015; Zhang and Xiao, 2015; Hong et al. 2016). For example, Zhao et al. showed that genetic depletion of PLDβ1 led to elevated levels of SA, ROS, SA-inducible gene expression, and enhanced resistance to the virulent bacterial strain Pseudomonas syringae tomato DC3000, indicating a negative role for PLDβ1 in the SA signaling pathway (Zhao et al., 2013). In contrast, Pinosa et al. reported that loss of PLDδ in Arabidopsis resulted in a higher penetration rate from two non-adapted powdery mildew fungi, barley mildew Blumeria graminis f.sp. hordei (Bgh) and pea mildew Erysiphe pisi, suggesting a positive role for PLDδ in penetration resistance (Pinosa et al., 2013). However, despite the fact that repression of PLD-produced PA by n-butanol in Arabidopsis strongly inhibited the HR during ETI, not a single PLD gene was found to be responsible for this (Johansson et al., 2014). Together, these studies suggest that PLDs play important roles in plant defenses with functional redundancy among family members. However, whether and how PLDs (or PLD-derived PA)-mediated signaling intersects with the well-defined SA and/or JA/ET signaling pathways is poorly understood (Zhao, 2015; Zhang and Xiao, 2015; Hong et al., 2016).
In this study, we screened a panel of Arabidopsis mutants with T-DNA insertions in PLD, pPLA (patatin-related phospholipase), PLC (phospholipase C), DGK (diacylglycerol kinase), and PIP5K (phosphatidylinositol 4-phosphate 5-kinase) genes for an altered infection phenotype to adapted powdery mildew fungi. We found that while PLDδ knockout plants showed enhanced susceptibility, PLDα1 knockout plants displayed enhanced resistance, suggesting that PLDδ and PLDα1 play opposing roles in post-penetration resistance against powdery mildew. We thus conducted a detailed analysis to determine the genetic relationships between these two PLD genes, their possible involvement in PRW8.2’s localization and function, and the defense pathways they might modulate.
Materials and methods
Plant lines and growth conditions
All mutants used in this study were in the Arabidopsis thaliana accession Col-0 background. Sequence data of the genes in this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases. The accession numbers of all genes used in this study are listed in Supplementary Table S1 at JXB online. Mutants sid2-2 (Wildermuth et al., 2001), eds1-2 (Bartsch et al., 2006), pad4-1 (Jirage et al., 1999), dde2-2 (von Malek et al., 2002), coi1-1 (Xie et al., 1998), pad4-1sid2-2 (Tsuda et al., 2009), and eds1-2pad4-1 (Kim et al., 2014) have been described previously. The phospholipase-related mutants used for infection tests with Golovinomyces cichoracearum (Gc) UCSC1 are listed in Supplementary Table S1. The homozygous double (sid2-2pldα1, eds1-2pldα1, pad4-1pldα1, sid2-2pldδ, eds1-2pldδ, and pad4-1pldδ), triple (pad4-1sid2-2pldα1, pad4-1sid2-2pldδ, eds1-2pad4-1pldδ, and eds1-2pad4-1sid2-2), and quadruple (eds1-2pad4-1sid2-2pldδ) mutants were generated by genetic crosses and identified by PCR genotyping. S5/pldα1 and S5/pldδ homozygous plants were made by crossing pldα1 and pldδ to S5 (Xiao et al., 2005) and subsequent PCR genotyping. All genotyping primers are listed in Supplementary Table S2.
Seeds were sown in Metro Mix 360 (Maryland Plant and Suppliers) and cold treated (4 °C for 2 d), and seedlings were grown under 22 °C, 65% relative humidity, short day (8 h light at 125 µmol m−2 s−1, 16 h dark).
DNA constructs, plant transformation, and microscopy
For genetic complementation, the genomic sequences of PLDα1 and PLDδ were amplified by PLDα1-F/PLDα1-R2 and PLDδ-F/PLDδ-R primers (Supplementary Table S2), respectively, using Q5 DNA polymerase (New England Biolabs, M0491L), cloned into pCX-SN (Chen et al., 2009) containing the 35S promoter, and introduced into pldα1 and pldδ, respectively, via Agrobacterium-mediated transformation using the A. tumefaciens strain GV3101 (Clough and Bent, 1998).
For determining subcellular localizations of PLDα1 and PLDδ, the p35S-pPLDα1:PLDα1-eGFP (a 2 kb PLDα1 untranslated promoter region and genomic sequence is amplified by the PLDα1-pF/PLDα1-R1 primer pairs), p35S:PLDδ- enhanced green fluorescent protein (eGFP), and pPLDδ:PLDδ-eGFP fusion constructs were made according to a previous report (Pinosa et al., 2013). p35S-pPLDα1:PLDα1-eGFP was introduced into pldα1 and Col-0, while p35S:PLDδ-eGFP and pPLDδ:PLDδ-eGFP were introduced into both pldδ and Col-0 via Agrobacterium-mediated transformation (Clough and Bent, 1998).
The expression and localization of the PLDα1–eGFP and PLDδ–eGFP fusion proteins were examined by confocal microscopy using a Zeiss LSM710 microscope (Wang et al., 2013). Confocal images were processed using the ZEN software (2009 edition) from Carl Zeiss (http://www.well.ox.ac.uk/_asset/file/zeiss-elyra-quick-start-guide-pdf-2.pdf; last accessed 24 April 2018) and Adobe Photoshop CC.
Pathogen infection, disease phenotyping, and quantification
Isolate Gc UCSC1 was maintained on Col-nahG plants, Gc UMSG1 on sow thistle plants (Wen et al., 2011), and Gc UMSG3, a new isolate purified in the Xiao lab, on tobacco plants for fresh inocula. Inoculation, visual scoring of disease reaction phenotypes, and conidiophore quantification were done as previously described (Xiao et al., 2005). Briefly, for conidiophore quantification, ~6 leaves per genotype were collected from sparsely and evenly inoculated 6-week-old plants at 4 days post-inoculation (dpi), cleared in a clearing solution (ethanol:phenol:acetic acid:glycerol=8:1:1:1, v/v/v/v), and stained by trypan blue solution (250 μg ml−1 in lactic acid:glycerol:water=1:1:1, v/v/v) for visualizing the fungal structure under the microscope. For each experiment, the total number of conidiophores per fungal colony was counted for at least 20 colonies per genotype. Data combined from three independent experiments were presented in a boxplot. For spore quantification, 4–6 leaf samples (~150 mg leaves per sample) per genotype from 6- to 7-week-old plants at 10–13 dpi were collected. A spore suspension of each sample was made by vortexing the leaves for 1 min in 40 ml of H2O (0.02% Silwet L-77) and used (diluted if necessary for susceptible genotypes) for spore counting using a hemocytometer under a dissecting microscope. Spore counts were normalized to the fresh weight of the corresponding leaf samples. All data analyses were done in R (R Core Team, 2014), and graphics were generated using ‘ggplot2’ (Wickham, 2009).
Assays with oomycete strains Hyaloperonospora arabidopsidis Noco2 and Emwa1, and bacterial strains Pseudomonas syringae pv. maculicola (Pma) ES4326, Pma avrRpm1, Pma avrRps4, and Pma ∆hrcC were done according to previous reports (Bonardi et al., 2011; Tornero and Dangl, 2001).
In situ detection of H2O2 accumulation and callose deposition
In situ H2O2 production and accumulation in the haustorium-invaded epidermal cells were stained and assessed using DAB (3,3'-diaminobenzidine) solution (Thordal-Christensen et al., 1997). Callose deposition at the fungal penetration sites and around the haustorium was detected and evaluated by aniline blue staining. Light microscopy images were viewed using Zeiss Imager A1.
Determination of endogenous SA, JA, and ABA concentrations
Three leaf samples of 6- to 7-week-old plants (~150 mg per sample) per genotype were harvested before and at 5 dpi with Gc UCSC1 for determining endogenous SA, JA, and abscisic acid (ABA) concentrations simultaneously. Phytohormone analyses were done as described previously for auxins (Novák et al., 2012; Blakeslee and Murphy, 2016), with the following modifications for the analysis of SA, JA, and ABA: ~40 mg of the tissue/sample ground in liquid nitrogen was extracted with 1.00 ml of 40 mM sodium phosphate buffer (pH 7.0). A 10 ng aliquot of d4-SA (C/D/N Isotopes Inc., Quebec, Canada, part #D-1156), 50 ng of d5-JA (C/D/N Isotopes Inc., part #D-6936), and 50 ng of d6-ABA (OlChemIm, Ltd., Olomouc, Czech Rebuplic, part #0342722) were added into each sample as internal standards. Samples were buffer-extracted at 4 °C on a lab rotator for 20 min, centrifuged at 12000 g for 15 min, and supernatants were collected and transferred to fresh 1.7 ml centrifuge tubes. The pH of supernatants was then adjusted using HCl, and samples were further purified via solid-phase extraction. Eluted samples were dried under nitrogen gas, re-dissolved in 100 µl of methanol, and filtered through 0.2 µm PTFE filters (Fisher Scientific, Pittsburgh, PA, USA part #03-391-4E).
For LC-MS/MS analysis, 1 µl of each re-dissolved sample was injected into an Agilent 1260 infinity LC system. Compounds were separated using an Agilent Poroshell 120EC-C18 (3.5 × 50 mm, 2.7 µm) column and an acidified water:methanol buffer system (Buffer A: 0.1% acetate, 5% methanol in water; Buffer B: 0.1% acetate in methanol). Gradient conditions were as follows: hold at 2% B for 1.5 min, 2 min at 2–60% B, 4.5 min at 60–98% B, hold at 98% B for 3.5 min, and then back to 2% B in 1 min. Eluted samples were further separated and quantified through the coupled Agilent 6460 triple quadrupole dual mass spectrometer equipped with an electrospray ionization (ESI) source. Compounds were quantified in negative ion mode. ESI source parameters were set as follows: gas temperature at 250 °C, gas flow rate at 10 L min–1, nebulizer at 60 psi, sheath gas temperature at 400 °C, sheath gas flow at 12 L min–1, capillary at 4500 V, nozzle voltage at 500 V. Retention and mass transitions for SA, JA, and ABA were verified using authentic standards. Specific mass transitions (precursor ion→product ion pairs, m/z) monitored for each phytohormone were: ABA, 263→153, 263→203; JA, 209→59; and SA, 137→93, 137→65.
qRT-PCR analysis
Three leaf samples of 6- to 7-week-old plants (~100 mg) per genotype were harvested before and at 5 dpi with Gc UCSC1 infection. Total RNA was isolated for each sample using TRIzol® Reagent and reverse transcribed using SuperScript™ III Reverse Transcriptase (Invitrogen, Thermo Fisher Scientific Inc.). For each experiment, qRT-PCR was performed with three biological replicates per treatment and three technical replicates per sample using an Applied Biosystems 7300 Real-Time PCR System with SYBR™ Green PCR Master Mix (Thermo Fisher Scientific Inc.). The transcript levels of the target genes were normalized to that of UBC9 (Ubiquitin conjugating enzyme 9, AT4G27960). Data were analyzed using the Applied Biosystems 7300 Real-Time PCR System Software and comparative ∆∆Ct method (Livak and Schmittgen, 2001). Primers are listed in Supplementary Table S2.
JA sensitivity assay
The assay for Arabidopsis root response to MeJA was adapted from a previous report (Xiao et al., 2004). Images of the seedlings were taken at day 10, and root length was measured using ImageJ (Schneider et al., 2012).
Results
PLDα1 and PLDδ play opposing roles in post-penetration resistance
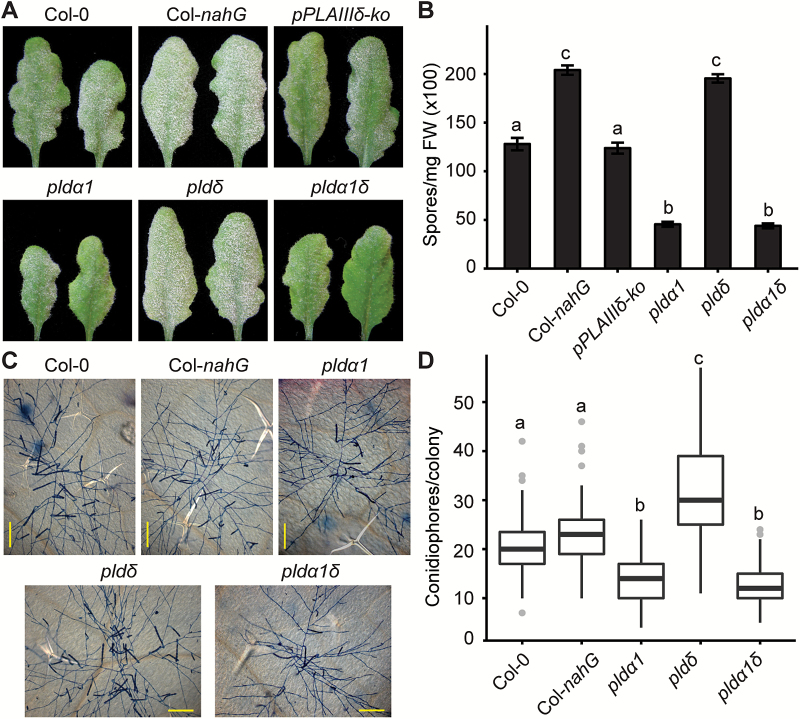
We tested a panel of T-DNA insertion lines (Supplementary Table S1) including six PLD knockout mutants (pldα1, pldδ, pldβ1, pldα1δ, pldα1δα3, and pldα1δε) with Gc UCSC1, a well-adapted powdery mildew isolate. Interestingly, we found that the pldδ mutant with compromised penetration resistance (Pinosa et al., 2013) showed clear enhanced disease susceptibility (‘eds’) while pldα1 defective in ABA signaling (Zhang et al., 2004) and pldα1-containing mutants (pldα1δ, pldα1δα3, and pldα1δε) exhibited enhanced disease resistance (‘edr’) to Gc UCSC1 (Fig. 1A, B; Supplementary Fig. S1). The ‘edr’ phenotype of pldα1δ led us to speculate that PLDα1 may act genetically downstream of PLDδ to modulate plant immunity negatively. Visual scoring of fungal mass on the leaf surface at 12 dpi and quantification of fungal spore production showed that the level of the ‘eds’ of pldδ was almost comparable with that of Col-nahG, a Col-0 transgenic line defective in SA signaling due to conversion of SA to catechol by the bacterial SA hydrolase encoded by nahG as a transgene (Fig. 1A, B). All other mutants tested exhibited levels of disease susceptibility similar to those of the Col-0 wild type (Fig. 1A, B; Supplementary Fig. S1). Consistent with the results at 12 dpi, pldδ supported significantly more conidiophores per colony while pldα1 and pldα1δ had fewer conidiophores per colony than Col-0 during early infection stage at 4 dpi when the fungus begins asexual reproduction (Fig. 1C, D). Interestingly, Col-nahG supported a similar amount of conidiophores to Col-0 at 4 dpi (Fig. 1D), suggesting that PLDδ-mediated defense against Gc UCSC1 probably occurs earlier than SA-mediated defense. This raises an intriguing question as to whether PLDδ (and PLDα1) functions in a signaling pathway distinct from the SA-dependent pathway.
Fig. 1.
Arabidopsis PLDα1 negatively modulates while PLDδ positively modulates post-penetration resistance against well-adapted powdery mildew Gc UCSC1. (A) Representative images of Arabidopsis leaves of the indicated genotypes infected with Gc UCSC1 at 12 dpi. Note, pldα1 and pldα1δ were less susceptible while pldδ was more susceptible than Col-0. (B) Quantification of spore production in the indicated genotypes at 10 dpi normalized to leaf FW. Data represent the mean ±SEM of three samples (n=3, four leaves each) from one experiment, which was repeated three times with similar results. (C) Representative microscopic images of single colonies of Gc UCSC1 on leaves of the indicated genotypes at 4 dpi. Fungal structures were stained by trypan blue. Scale bars=200 μm. (D) Total number of conidiophores per colony on leaves of the indicated genotypes at 4 dpi. The boxplot shows combined data from three independent experiments (at least 20 colonies were counted for each genotype per experiment). The bold line within the box represents the median. The bottom and top edge of the box represent the first and third quartile, respectively. Ends of whiskers represent the minimum and maximum of data points. Gray dots represent outliers. Different lower case letters indicate statistically different groups (P<0.01) as determined by multiple comparisons using one-way ANOVA, followed by Tukey’s HSD test.
To test whether the ‘edr’ phenotype of pldα1 and the ‘eds’ phenotype of pldδ are indeed due to the loss of PLDα1 and PLDδ, respectively, multiple pldα1 and pldδ lines expressing the respective wild-type genes were generated and tested with Gc UCSC1. These lines displayed similar disease phenotypes to Col-0 (Supplementary Fig. S2), indicating genetic complementation of these two genetic mutations by their respective wild-type genes. Thus, our genetic data established a positive role for PLDδ and a negative role for PLDα1 in basal, stage II post-penetration resistance against well-adapted powdery mildew in Arabidopsis.
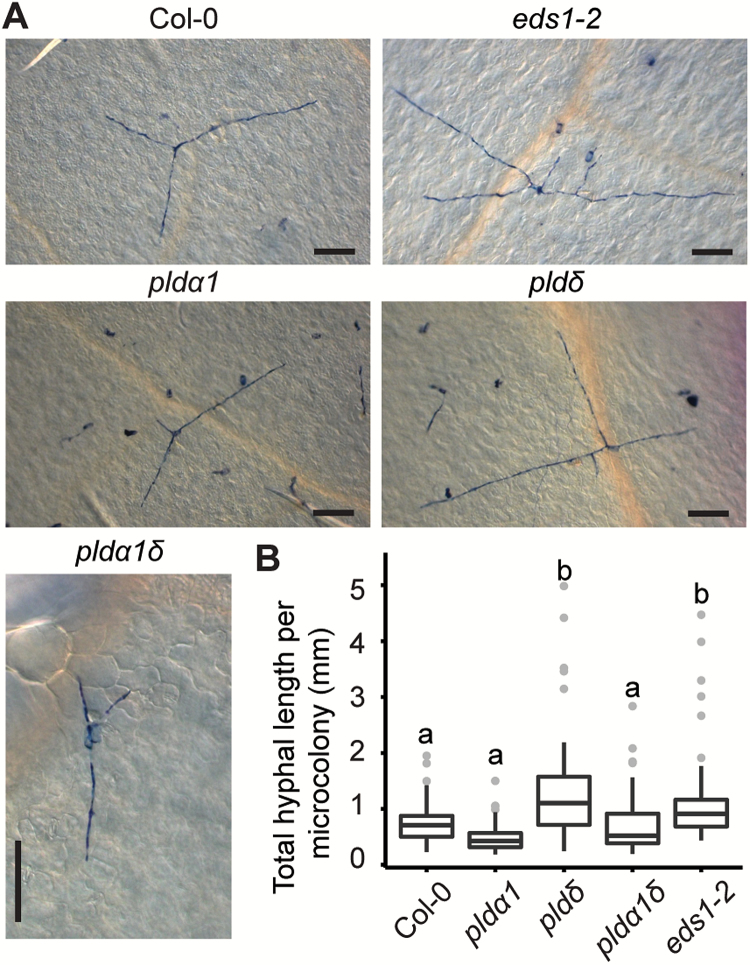
To test if the PLD genes are also involved in stage I post-penetration resistance, we inoculated the pld mutants with Gc UMSG1. Gc UMSG1 is a powdery mildew fungus infectious on sow thistle. It has largely overcome penetration resistance of 25 Arabidopsis accessions examined and is capable of forming initial haustoria but arrested before sporulation by stage I post-penetration resistance in Arabidopsis (Wen et al., 2011). We assessed the growth of Gc UMSG1 on the pld mutants by measuring the total hyphal length of each microcolony at 5 dpi. Not surprisingly, pldδ supported significantly more hyphal growth than Col-0 (Fig. 2B), which is similar to eds1-2 (in Col-0; Bartsch et al., 2006), known to support better growth of Gc UMSG1 (Wen et al., 2011). However, while limited sporulation of Gc UMSG1 can occasionally be seen on eds1-2, indicating breakdown of non-host resistance, it has never been observed on pldδ, suggesting that PLDδ acts differently from EDS1 and is not as critical as EDS1 in stage I post-penetration resistance defined by this pathosystem. However, hyphal growth in pldα1 and pldα1δ showed no significant difference from that in Col-0 (Fig. 2).
Fig. 2.
PLDδ in Arabidopsis contributes to post-penetration resistance against a non-adapted powdery mildew Gc UMSG1. (A) Representative microscopic images of typical Gc UMSG1 fungal microcolonies grown on leaves of the indicated genotypes at 5 dpi. Scale bars=100 μm. (B) Total hyphal length per microcolony of the indicated genotypes at 5 dpi. The boxplot shows combined data from three independent experiments (n >60). Different lower case letters indicate statistically different groups as determined by multiple comparisons using one-way ANOVA, followed by Tukey’s HSD test (P<0.01).
The subcellular defense responses such as powdery mildew-induced H2O2 production and callose deposition were investigated in the pld mutants. Because Gc UCSC1 can largely suppress the production of H2O2 in Col-0 (Xiao et al., 2005), the non-adapted isolate Gc UMSG1 was used to challenge the plants, and in situ H2O2 production was visualized by DAB staining (Thordal-Christensen et al., 1997). We divided the haustorium–epidermal cell interaction in terms of H2O2 production into three types: (i) H2O2 is undetectable; (ii) H2O2 accumulates in the haustorial complex; and (iii) H2O2 is found in both the haustorial complex and the whole cell (Supplementary Fig. S3A). Of >750 interaction sites evaluated in Col-0, 39.5, 25.7, and 34.7% were (i), (ii), and (iii), respectively, and the pld mutants showed a similar frequency distribution for the three interaction types (Supplementary Fig. S3B). This suggests that H2O2 production induced by haustorium invasion is not affected due to loss of PLDα1 or PLDδ, or both. Next, we examined callose deposition at the fungal penetration sites (i.e. papillae) or around the haustorium (i.e. haustorial encasement) by aniline blue staining after Gc UCSC1 inoculation. Again, callose deposition was grossly unaffected in the pld mutants compared with that in Col-0 based on visual scoring (Supplementary Fig. S3C). These suggest that the ‘eds’ phenotype of pldδ and the ‘edr’ phenotype of pldα1 are not apparently associated with these two typical subcellular defense responses.
Loss of PLDα1 or PLDδ affects basal resistance against an oomycete but not ETI
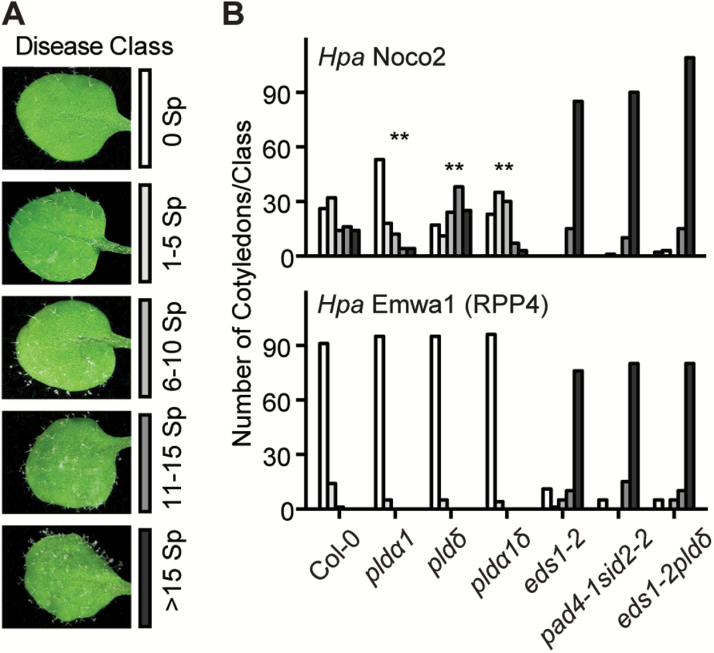
Hyaloperonospora arabidopsidis (Hpa) is a fungus-like oomycete pathogen of Arabidopsis. To test if post-penetration resistance to Hpa is also altered in the pld mutants, we inoculated 10-day-old seedlings of Col-0, pldα1, pldδ, pldα1δ, and two known ‘eds’ mutant lines, eds1-2 and pad4-1sid2-2, with Hpa isolate Noco2 (virulent on Col-0). While pldα1 and pldα1δ were significantly less susceptible, pldδ was significantly more susceptible (albeit not as susceptible as eds1-2 and pad4-1sid2-2) to this pathogen than Col-0 (P<0.01) (Fig. 3B, upper panel). These further support the distinct roles of PLDα1 and PLDδ in post-penetration resistance against haustorium-forming pathogens.
Fig. 3.
Loss of PLDα1 and/or PLDδ affects basal resistance against oomycetes, but not ETI mediated by RPP4. (A) Representative cotyledons showing disease phenotypes of the indicated disease classes at 7 dpi. Ten-day-old seedlings were inoculated with virulent Hyaloperonospora arabidopsidis (Hpa) isolate Noco2 or avirulent isolate Emwa1. Sporangiophores (Sp) per cotyledon were assessed at 7 dpi, and categorized into five classes as indicated by the corresponding figure keys. (B) Quantification of the number of cotyledons (n >100 for each of the indicated genotypes) per class of the indicated genotypes infected with Hpa isolate Noco2 (upper panel) or avirulent isolate Emwa1 (lower panel) based on categorization of leaf infection defined in (A). χ2 test was used to test statistical significance for disease degree between Col-0 and the indicated mutant lines at 7 dpi (**P<0.01).
To test if loss of PLDα1 or PLDδ impacts ETI, we tested the mutants with an avirulent oomycete strain Hpa Emwa1 (recognized by RPP4, a TIR-NB-LRR; van Der Biezen et al., 2002), and Pseudomonas syringae pv. maculicola (Pma) ES4326 strains expressing either AvrRpm1 (recognized by RPM1, a CC-NB-LRR; Grant et al., 1995) or AvrRps4 (recognized by RPS4/RRS1, a pair of TIR-NB-LRR immune receptors; Narusaka et al., 2009), since no NB-LRR-mediated resistance against powdery mildew has been defined in Arabidopsis. While eds1-2 and pad4-1sid2-2 were compromised in resistance against Hpa Emwa1, the pld mutants displayed similar levels of resistance to that seen in Col-0 (Fig. 3), indicating that loss of PLDα1 and/or PLDδ does not seem to affect RPP4-dependent ETI. Similarly, no significant difference was detected between pldα1, pldδ, pldα1δ, and Col-0 (Supplementary Fig. S4C, D) in defense against Pma, further supporting that PLDα1 or PLDδ individually or together do not play a significant role in ETI. In addition, the pld mutants remained resistant like Col-0 to Pma ∆hrcC, which is unable to inject type III effectors to suppress PTI, implying that the PTI against bacterial pathogens is not affected by the loss of PLDα1 and/or PLDδ (Supplementary Fig. S4B). This could be due to functional redundancy among the PLD enzymes in defense against bacterial pathogens as suggested in an earlier study since there are 12 PLD isoforms in Arabidopsis (Johansson et al., 2014).
PLDδ is dispensable for RPW8-mediated resistance
RPW8.1 and RPW8.2 (referred to as RPW8 in later text unless otherwise indicated) confer post-penetration, haustorium-targeted resistance to powdery mildew (Xiao et al., 2001; Wang et al., 2009). To examine whether PLDα1 and/or PLDδ contribute to RPW8-mediated resistance, we first stably expressed the RPW8.2-RFP (red fluorescent protein) transgene from the native RPW8.2 promoter in pldα1 and pldδ. Confocal microscopy showed that the localization of RPW8.2–RFP to the EHM was unchanged in pldα1 or pldδ (as represented by RPW8.2–RFP’s localization in pldδ; Supplementary Fig. S5A), indicating that neither PLDα1 nor PLDδ is required for precise EHM targeting of RPW8.2 (Wang et al., 2009). Next, we individually introduced these two mutations into S5 (a Col-gl line expressing RPW8; Xiao et al., 2005). Both S5/pldα1 and S5/pldδ displayed the same levels of resistance to Gc UCSC1 (Supplementary Fig. S5C, D) and H2O2 production as S5 in haustorium-invaded cells (as represented by H2O2 production in S5/pldδ,; Supplementary Fig. S5B). Given that RPW8’s defense function requires SA signaling (Xiao et al., 2005), these results support that the PLDα1/PLDδ pair most probably function via an SA-independent signaling pathway.
PLDα1 and PLDδ have distinct subcellular localizations
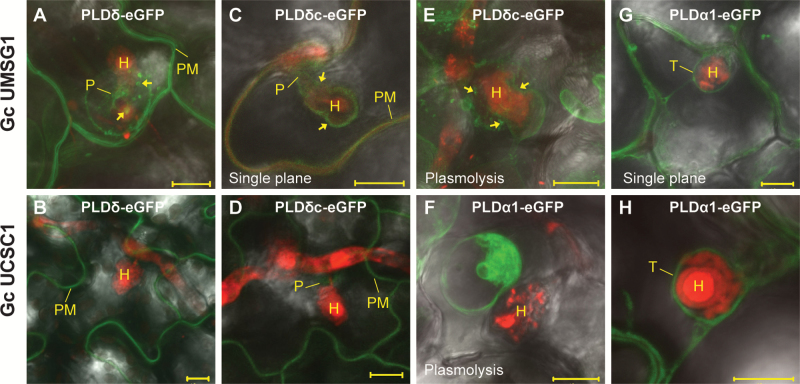
Since there is active membrane trafficking and biogenesis (of the EHM) in haustorium-invaded cells (Berkey et al., 2017), we wondered whether the contrasting defense responses of pldα1 and pldδ to adapted powdery mildew are due to possible differential subcellular enzymatic activities of PLDα1 and PLDδ in haustorium-invaded cells. To test this, we fused eGFP to the C-termini of the genomic DNA of the two PLD genes and expressed the fusion constructs from 35S plus the native promoter (for PLDα1-eGFP) or 35S (for PLDδ-eGFP) in pldα1 or pldδ, respectively, since the GFP signal from the native promoter-driven PLDδ cDNA (PLDδc) in fusion with eGFP was reported to be too weak for imaging (Pinosa et al., 2013). PLDδ-eGFP could fully, while PLDα1-eGFP could partially, rescue the respective mutant phenotypes (Supplementary Fig. S6), indicating that these fusion proteins are (partially) functional. We then used leaves of the respective transgenic lines infected with Gc UMSG1 or Gc UCSC1 at 2 dpi for subcellular localization analysis using confocal microscopy. When examining leaves infected with Gc UMSG1, we detected PLDδ–eGFP in the PM of all epidermal cells and in two or more concentric rings around the penetration site forming the ‘bull’s eye’ domain (Assaad et al., 2004; Koh et al., 2005) often with small dots or bulbs within or nearby (Fig. 4A). However, it was rarely seen in the Gc UCSC1 penetration site (Fig. 4B), implying that the adapted pathogen suppresses the recruitment of PLDδ–eGFP to the probably perturbed PM around the papilla. PLDδc–eGFP was reported to exhibit focal accumulation around the Bgh penetration site in Arabidopsis epidermal cells (Pinosa et al., 2013). We thus examined the subcellular localization of the PLDδc–eGFP expressed from 35S in our pathosystems. In the case of Gc UMSG1, PLDδc–eGFP was often more preferentially detected in the ‘bull’s eye’ domain (Fig. 4A) or in an EHM-like membrane surrounding the constrained haustorium than PLDδ–eGFP (Fig. 4C). After plasmolysis (0.5 M NaCl for 20 min), GFP signal was retained around the haustorium in small dots or bulbs (Fig. 4E), similar to those in the papilla at the penetration site (Fig. 4A), indicating that PLDδc–eGFP is not at the EHM because the EHM largely remains intact within 30 min of such plasmolysis treatment (Berkey et al., 2017). In the case of Gc UCSC1, the PLDδc–eGFP signal was much weaker at the penetration site (Fig. 4D), suggesting that recruitment of PLDδc–eGFP to the penetration site is also similarly suppressed by the adapted powdery mildew pathogen. The slight discrepancy in localization between PLDδ–eGFP and PLDδc–eGFP may be attributable to alternative splicing of PLDδ (Wang and Wang, 2001) which is pertinent to the PLDδ-eGFP construct but irrelevant to the PLDδc-eGFP construct for which a full-length PLDδ cDNA was used (Pinosa et al., 2013).
Fig. 4.
Differential subcellular localization of PLDα1 and PLDδ in powdery mildew-infected epidermal cells. Stable transgenic lines were inoculated with Gc UMSG1 or Gc UCSC1. At 2 dpi, sections of infected leaves were immersed in propidium iodide (PI, 0.5% aqueous solution) for 40–60 min for staining haustoria (H, red) and mycelia (red) before confocal imaging. All representative images shown are merged (GFP, PI, and bright field) Z-stack projections of 15–20 optical sections unless otherwise indicated. (A, B) Localization of PLDδ–eGFP (from the PLDδ genomic sequence translationally fused with eGFP) in a Gc UMSG1-invaded cell (A) or a Gc UCSC1-invaded cell (B). Arrows: concentric ring and dots. (C–E) Localization of PLDδc-eGFP (from the PLDδ full-length coding sequence translationally fused with eGFP; Pinosa et al., 2013) in a Gc UMSG1-invaded cell before (C; arrows, peri-haustorial membrane) or after plasmolysis (E; 0.5 M NaCl for 20 min; arrows indicate dots and membrane retained around the haustorium), or a Gc UCSC1-invaded cell (D). (F–H) Localization of PLDα1–eGFP in a Gc UMSG1-invaded cell (G), or a Gc UCSC1-invaded cell before (H) or after plasmolysis (F). Scale bars=10 μm. PM, plasma membrane; P, penetration site; T, tonoplast.
A strong fluorescence signal of PLDα1–eGFP was found in a peri-haustorium membrane similar to the EHM (Fig. 4G, H), which could be completely separated from the haustorium after plasmolysis (Fig. 4F). This indicates that PLDα1–eGFP is not localized to the EHM but rather it may be in the tonoplast that tightly wraps around the haustorium.
These results in general agree with the subcellular localizations of PLDα1 and PLDδ inferred by protein localization and fractionation analyses in earlier studies (Wang, 2000; Wang and Wang, 2001; Pinosa et al., 2013). The distinct localization patterns of these two PLDs may in part contribute to their opposing roles in post-penetration resistance against powdery mildew pathogens.
PLDδ contributes to resistance independent of EDS1/PAD4, SA, and JA signaling pathways
Our earlier results (Fig. 1C, D; Supplemenary Figs S3, S5) suggest that PLDδ and perhaps PLDα1 may function through an SA-independent pathway. To define this pathway further, we made double and triple mutants by crossing pldα1 or pldδ to well-characterized SA-dependent (sid2-2) (Wildermuth et al. 2001, Dewdney et al. 2000) or both SA-dependent and -independent signaling (eds1-2 and pad4-1) mutants (Bartsch et al., 2006; Venugopal et al., 2009).
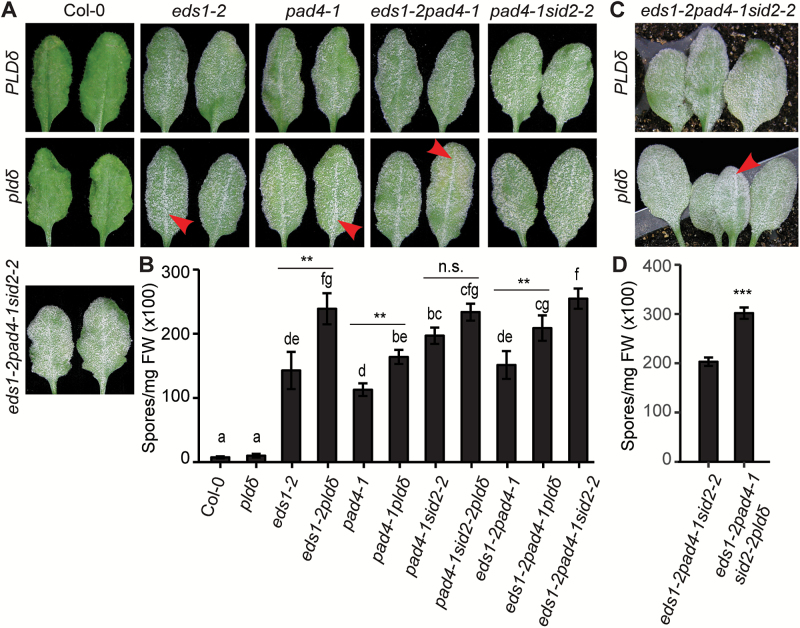
We first examined if pldδ-mediated ‘eds’ is additive to the ‘eds’ phenotypes of eds1-2 or pad4-1 in response to the well-adapted Gc UCSC1 isolate and found that eds1-2pldδ and pad4-1pldδ were not statistically more susceptible than the single mutants (Supplementary Fig. S7A, B). We then made pad4-1sid2-2pldδ, eds1-2pad4-1pldδ, and eds1-2pad4-1sid2-2 triple mutants, and compared the disease phenotypes between these and the two double mutants. No significant differences were detected between the mutants except pad4-1sid2-2pldδ versus pad4-1sid2-2 (Supplementary Fig. S7A, B), suggesting that either PLDδ somehow acts in the SA pathway or the pldδ-mediated ‘eds’ phenotype may be masked in the various double or triple mutants because Gc UCSC1 is too aggressive on these mutants to allow reliable detection of any phenotypic differences.
To test the latter possibility, we used Gc UMSG3, a powdery mildew isolate from tobacco which can only weakly sporulate on Col-0, to resolve subtle infection phenotypic differences between different genotypes. Sporulation of Gc UMSG3 was found to be very weak on both Col-0 and pldδ; however, a whitish fungal mass was more easily discernible on pldδ at 11 dpi (Fig. 5A, B). Interestingly, eds1-2, pad4-1, eds1-2pad4-1, and pad4-1sid2-2 all supported profuse sporulation (Fig. 5A), suggesting that EDS1 and/or PAD4 make a major contribution to stage II post-penetration resistance to Gc UMSG3 probably via both SA-dependent and SA-independent mechanisms.
Fig. 5.
PLDδ in Arabidopsis contributes to post-penetration resistance via an SA- and EDS1/PAD4-independent pathway.(A, C) Representative leaves of the indicated genotypes (defined by name IDs from both x- and y-axes) infected with Gc UMSG3 at 11 dpi. Note that fungal mass is more noticeable on leaves, especially the mid-vein area (arrowheads), from eds1-2pldδ, pad4-1pldδ, eds1-2pad4-1pldδ, and eds1-2pad4-1sid2-2pldδ than the corresponding leaves from eds1-2, pad4-1, eds1-2pad4-1, and eds1-2pad4-1sid2-2 (upper panel). (B, D) Quantification of spore production in the indicated genotypes in (A, C), respectively, at 11 dpi normalized to leaf FW. Data represent the mean ±SEM of four samples (n=4, 4–5 leaves each) from one experiment, which was repeated three times with similar results. Different lower case letters indicate statistically different groups as determined by multiple comparisons using one-way ANOVA, followed by Tukey’s HSD test (B, **P<0.01), or by Student’s t-test (D, ***P<0.001). n.s., not significant.
Notably, eds1-2pldδ and pad4-1pldδ supported significantly more fungal growth (white powder around the mid-vein in particular) than eds1-2 and pad4-1 visually (Fig. 5A) and quantitatively (Fig. 5B), indicating that PLDδ contributes to resistance against Gc UMSG3 through a mechanism(s) that is at least partially EDS1 or PAD4 independent. Interestingly, pad4-1sid2-2 was as susceptible as pad4-1pldδ (Fig. 5B), which seemingly implies that PLDδ and SID2 may act in the same signaling pathway. Yet, pad4-1sid2-2pldδ was significantly more susceptible than pad4-1pldδ to Gc UMSG1 (Fig. 5A, B) and pad4-1sid2-2 to Gc UCSC1 (Supplementary Fig. S7A, B). Similarly, eds1-2pad4-1pldδ exhibited an even higher level of susceptibility than eds1-2pad4-1 and pad4-1pldδ (Fig. 5A, B). Finally, eds1-2pad4-1sid2-2pldδ exhibited significantly higher susceptibility to Gc UMSG3 than eds1-2pad4-1sid2-2 (Fig. 5C, D). These observations together support that PLDδ acts through a yet to be characterized pathway to limit fungal infection at the post-penetration stage. It is worth pointing out that eds1-2pldδ showed a similar level of susceptibility to eds1-2pad4-1pldδ (Fig. 5A, B), implying that EDS1 and PAD4 are both required for resistance against Gc UMSG3. Supporting this inference, eds1-2pad4-1 was not statistically more susceptible than eds1-2 or pad4-1 (Fig. 5A, B).
To assess if PLDδ functions through the JA pathway, the Gc UCSC1 infection phenotype of pldδ was compared with that of dde2-2, which is impaired in JA biosynthesis (von Malek et al., 2002). The susceptibility of dde2-2 was similar to that of Col-0 (Supplementary Fig. S7C, D), consistent with our earlier finding that the JA signaling receptor mutant coi1 did not show ‘eds’ to Gc UCSC1 (Xiao et al., 2005), suggesting that the JA pathway has little or very limited contribution to defense against Gc UCSC1. Taken together, PLDδ is unlikely to act through the JA pathway.
Next, we investigated if the ‘edr’ phenotype of the pldα1 mutant is affected by the sid2-2, eds1-2, or pad4-1 mutations by first crossing pldα1 to the three single and pad4-1sid2-2 double mutants and then testing their infection phenotypes. Intriguingly, eds1-2pldα1, pad4-1pldα1, sid2-2pldα1, and pad4-1sid2-2pldα1 all displayed similar ‘eds’ to Gc UCSC1 to the respective single or double mutants with wild-type PLDα1 (Supplementary Fig. S8). This suggests that pldα1-mediated ‘edr’ is completely neutralized/suppressed when the SA- and/or EDS1/PAD4-mediated signaling is defective, genetically placing PLDα1 upstream of EDS1, PAD4, and SID2, which is in sharp contrast to the epistatic effect of pldα1-mediated ‘edr’ over pldδ-caused ‘eds’. A mechanistic model is proposed to explain the distinct yet related roles of PLDδ and PLDα1 (see the Discussion; Supplementary Fig. S9).
Loss of PLDα1 and/or PLDδ has no significant impact on SA, JA, and ABA levels and signaling
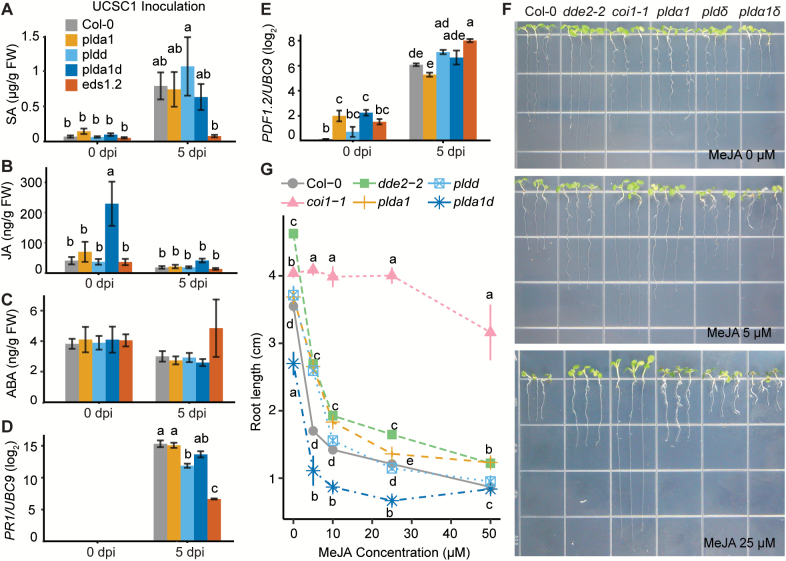
To investigate if PLDα1- and/or PLDδ-mediated defense mechanisms are connected with defense-related phytohormones, we first measured levels of endogenous SA, ABA, and JA in pldα1, pldδ, and pldα1δ along with Col-0 and eds1-2 prior to and at 5 dpi with Gc UCSC1 using LC-MS/MS. Compared with naïve plants, SA levels increased by 5- to 16-fold in mildew-infected Col-0 and pld mutants, but remained low in eds1-2 (Fig. 6A), indicating that pathogen-induced SA biosynthesis is intact in the pld mutants. To see if SA signaling is affected in the pld mutants, the expression of the marker gene PR1 (Wiermer et al., 2005) was measured and found to be induced to a level similar to that in Col-0, suggesting that SA signaling was not affected by any of the pld mutations (Fig. 6D). These results support the inference from our genetic data that PLDα1 and PLDδ oppositely modulate post-penetration resistance via an SA-independent pathway. No significant changes in ABA levels were observed in Col-0 and the pld mutants before and after powdery mildew infection (Fig. 6C).
Fig. 6.
Impact of the pldα1 and pldδ single and double mutations on the levels and signaling of SA and JA before and after powdery mildew infection. (A–C) Levels of the plant hormones SA (A), JA (B), and ABA (C) were measured by LC-MS/MS in leaves of 6-week-old plants of the indicated genotypes prior to (0 dpi) and post- (5 dpi) Gc UCSC1 inoculation. Notably, before inoculation, the JA level of pldα1δ was higher than that of the two single mutants and was reduced by ~4-fold at 5 dpi. Bars represent the mean ±SEM of three independent experiments combined (n=3 for each experiment). (D, E) Log2-fold changes of PR1 (D) or PDF1.2 (E) relative to UBC9 encoding ubiquitin conjugating enzyme 9. Bars represent the mean ±SEM of three biological replicates. (F) Representative pictures showing 10-day-old seedlings of the indicated genotypes grown on MS-agar medium without or with 5 μM and 25 μM MeJA. (G) Dose–response curve of root growth of the indicated genotypes upon MeJA treatment. Root lengths of 10-day-old seedlings growing on MS-agar medium supplemented with exogenous MeJA at 0, 5, 10, 25, or 50 μM were measured and are presented as the mean ±SEM at each MeJA dosage. The line graph shows combined data from two independent experiments (n >15 for each experiment). Different lower case letters indicate statistically different groups (P<0.05) as determined by multiple comparisons using one-way ANOVA, followed by Tukey’s HSD test.
Surprisingly, the JA level in uninfected pldα1δ was higher (3- to 6-fold) than that in all other genotypes (Fig. 6B), and the expression of its marker gene PDF1.2 was significantly higher in unchallenged pldα1 and pldα1δ compared with that in Col-0 (Fig. 6E), suggesting that PLDα1 and PLDδ may act together to repress JA production/signaling in the absence of pathogens. At 5 dpi with Gc UCSC1, JA in pldα1δ was reduced to a level that is only slightly higher (~2-fold) than that in other plants (Fig. 6B), which is probably caused by an antagonistic effect from enhanced SA biosynthesis and signaling in the mildew-infected plants. However, despite a slight decrease in JA levels in all the genotypes at 5 dpi, expression levels of PDF1.2 showed a similar increase (2.5- to 12-fold ) in all the plants, with no significant difference between the pld mutants and Col-0 (Fig. 6E). Together these results indicate that (i) although well-adapted powdery mildew infection does not induce JA biosynthesis, it can still induce JA signaling; and (ii) the altered defense phenotypes in pldα1 and pldα1δ do not correlate with the changes in JA levels and/or JA signaling.
It is known that high JA levels inhibit root growth (Staswick et al., 1992). To test the results concerning the endogenous JA levels further, we examined root growth of pldα1δ along with Col-0, pldα1, pldδ, and two JA mutants, dde2-2 (defective in JA synthesis; von Malek et al., 2002) and coi1-1 (insensitive to JA; Xie et al., 1998) in Murashige and Skoog (MS)-agar medium without or with supplement of exogenous methyl jasmonate (MeJA). Consistent with the results from the JA level measurements, only roots of pldα1δ grown in MeJA-free MS-agar medium were significantly shorter (~76.9% of Col-0) (Fig. 6F, G). Roots of all genotypes, except those of coi1-1, showed similar rates of growth inhibition in MS-agar medium supplemented with different concentrations of MeJA (5, 10, 25, and 50 µM) (Fig. 6G). This indicates that JA signaling in the pld mutants is not affected. Taken together, our results further demonstrate that PLDα1 and PLDδ oppositely modulate defense in an SA-independent manner but may act together to curb JA accumulation in naïve plants.
Discussion
In this study, we collected genetic evidence to demonstrate that Arabidopsis PLDα1 and PLDδ oppositely modulate basal, post-penetration resistance against powdery mildew, and oomycete pathogens via an EDS1/PAD4-, SA-, and JA-independent pathway.
PLDδ and PLDα1 modulate post-penetration resistance against powdery mildew
Pinosa et al. previously reported that the loss-of-function pldδ mutant is compromised in penetration resistance against the non-adapted barley mildew Bgh (Pinosa et al., 2013). Here, we show that the same pldδ mutant exhibited ‘eds’ to a well-adapted powdery mildew isolate Gc UCSC1 (Fig. 1) and supported more hyphal growth of the non-adapted powdery mildew isolate Gc UMSG1 that has overcome penetration resistance (Wen et al., 2011) (Fig. 2). This implies that the PLDδ-based defense mechanism operates throughout the entire infection cycle of powdery mildew and apparently has not been (fully) suppressed by even aggressive powdery mildew pathogens such as Gc UCSC1. To determine if PLDδ-mediated defense is effective against other pathogens, we tested pldδ with the fungus-like oomycete Hpa Noco2 that also employs a haustorium-based nutrient acquisition strategy. Notably, pldδ was significantly more susceptible than Col-0 but not as susceptible as eds1-2 or pad4-1sid2-2 to Hpa (Fig. 3B). Given that powdery mildew fungi only invade host epidermal cells while oomycete pathogens invade both epidermal and mesophyll cells (Takemoto et al., 2003), it is possible that PLDδ-mediated defense is more effective in epidermal cells compared with mesophyll cells. It is also possible that oomycete pathogens may be able to suppress PLDδ-mediated defense more effectively than powdery mildew. In addition, PLDδ-mediated defense may be attenuated under higher humidity (>90%) conditions necessary for infection of Hpa Noco2. High humidity-caused suppression of resistance has been reported for several different defense mechanisms (Xiao et al., 2003; Zhou et al., 2004; Wang et al., 2007). Similar to what was reported earlier (Johansson et al., 2014), we did not observe any difference in growth of virulent bacteria between Col-0 and pldδ, suggesting that PLDδ is specifically involved in defense against cell wall-breaching pathogens. Notably, among all reported genes involved in penetration and post-penetration resistance, PLDδ is unique in that it contributes to both penetration and post-penetration resistance against powdery mildew fungi. In contrast to pldδ, both the pldα1 single and the pldα1δ double mutant exhibited ‘edr’ to virulent powdery mildew and oomycete pathogens (Figs 1, 3). This suggests that genetically PLDα1 and PLDδ function oppositely in the same pathway with PLDα1 acting downstream of PLDδ. We reported earlier that loss of PLDβ1 resulted in ‘edr’ to virulent bacterial pathogens and ‘eds’ to a necrotrophic fungal pathogen Botrytis cinerea (Zhao et al., 2013), suggesting a positive role for PLDβ1 in the JA pathway and a negative role in the SA pathway. We found in this study that pldβ1 showed slight ‘edr’ to Gc UCSC1 based on our visual scoring of the infection phenotypes (Supplementary Fig. S1A), supporting a role for PLDβ1 in modulating SA–JA signaling. Whether PLDα1 and PLDβ1 share similar regulatory mechanisms and/or have overlapping function remains to be determined.
PLDα1 and PLDδ may modulate defense via a potentially novel pathway
Three lines of genetic evidence collectively support our conclusion that PLDδ functions through an SA-independent pathway. First, RPW8-mediated resistance, which is known to engage SA signaling, is intact in pldδ (Fig. S5C); secondly, adding the pldδ mutation to the SA signaling mutants eds1-2 and pad4-1, or the SA biosynthesis mutant sid2-2, resulted in increased ‘eds’ to the poorly adapted isolate Gc UMSG3 (Fig. 5); lastly, pldδ showed similar elevation of SA levels and induction of PR1 expressions to Col-0 upon powdery mildew infection (Fig. 6A, D).
Because EDS1 and PAD4 are believed to function upstream of SA and modulate defense via both SA-dependent and SA-independent pathways (Bartsch et al., 2006; Venugopal et al., 2009), the increased ‘eds’ of eds1-2pldδ, pad4-1pldδ, eds1-2pad4-1pldδ, and eds1-2pad4-1sid2-2pldδ to Gc UMSG3 (Fig. 5) also provide clear genetic evidence to support a role for PLDδ in defense through an EDS1- and/or PAD4-independent pathway. However, based on our genetic analyses alone, we could not exclude the possibility that PLDδ also contributes to EDS1/PAD4-dependent resistance. It is possible that the defense pathways mediated by EDS1, PAD4, and PLDδ may be interconnected or partially overlapping, since the phenotypic differences among the single and double mutants concerning these three genes were largely diminished when they were tested with the aggressive isolate Gc UCSC1 (Supplementary Fig. S7).
We also evaluated whether PLDα1 and PLDδ function via the JA pathway. Our results from genetic analysis (Supplementary Fig. S7C, D; Xiao et al., 2005), measurements of JA levels (Fig. 6B), and PDF1.2 expression (Fig. 6E) showed that the altered defense phenotypes of the pld mutants could be uncoupled from the changes in the JA levels and signaling, thus excluding the possibility that PLDα1 and PLDδ modulate defense through the JA pathway.
Taken together, our results indicate that PLDα1 and PLDδ play opposing roles in modulating resistance against powdery mildew via a pathway that is independent of the EDS1/PAD4, SA, and JA pathways. Notably, mlo-based durable and broad-spectrum resistance against powdery mildew has recently been shown to be independent of all the known defense pathways (Kuhn et al., 2017). Therefore, it will be interesting for future studies to determine if PLDα1 and PLDδ have a mechanistic connection with MLO or other known defense pathways such as the ET and mitogen-activated protein (MAP) kinase signaling pathways (Tsuda et al., 2013; Kim et al., 2014; Hillmer et al., 2017; Kuhn et al., 2017).
PLDα1 may repress PLDδ-mediated defense signaling
We previously reported that PLDα1 promotes H2O2 production whereas PLDδ facilitates downstream H2O2 signaling in guard cells to regulate stomatal closure positively during drought stress (Zhang et al., 2009; Guo et al., 2012). However, our genetic data from this study position PLDα1 as a negative regulator downstream of PLDδ-mediated defense. Consistent with this, powdery mildew haustorium-induced H2O2 production was not affected in any of the pld mutants (Supplementary Fig. S3A, B). Given that drought response relies on the movement of guard cells, whereas plant defense against powdery mildew pathogens mostly occurs in the leaf pavement cells, it is possible that the proteins interacting with these two PLDs and/or their substrates during drought stress and pathogen infection are different. Hence, it is conceivable that PLDα1 and PLDδ probably participate in distinct signaling networks in these two different types of cells in response to abiotic and biotic stresses.
It is unclear to us how PLDδ positively modulates while PLDα1 negatively modulates post-penetration resistance against powdery mildew pathogens. One possible mechanism is that PLDα1 and PLDδ exert their opposing roles in defense by producing distinct pools of PA to modulate distinct cellular processes by targeting spatiotemporally restricted proteins at different subcellular localizations (Supplementary Fig. S9). Our confocal microscopy show that while PLDδ–eGFP is localized at the PM, around the penetration site and peri-haustorium, PLDα1–eGFP is most likely to be associated with the tonoplast and other intracellular membranes (Fig. 4), which are compatible with results previously reported (Wang, 2000; Wang and Wang, 2001; Pinosa et al., 2013). Notably, the eGFP signal of PLDδ–eGFP was the strongest around the penetration site of non-host barley mildew (Pinosa et al., 2013), weaker around the penetration site and/or the haustorial complex of the non-adapted Gc UMSG1, and almost undetectable in such subcellular compartments induced by the well-adapted Gc UCSC1 (Fig. 4A–D). This suggests that PLDδ is recruited to the PM around the penetration site to produce PA to (in)activate target proteins locally, and adapted powdery mildew pathogens may suppress this recruitment to interfere with PLDδ’s role in defense activation. As for PLDα1, its tonoplast localization may be related to vacuole-based removal of defense molecules to prevent inappropriate activation of defense in the absence of pathogens. However, its suppression is relieved by PLDδ-triggered signaling once pathogens invade. Future work will be directed to identifying relevant immunity proteins that are modulated by the two functionally distinct PLDs.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Disease reaction phenotypes of pPLA, PLD, PLC, DGK, and PIP5K T-DNA insertion mutants infected with Gc UCSC1.
Fig. S2. Genetic complementation of the pldα1 and pldδ mutant genes by their respective wild-type genes.
Fig. S3. Loss of PLDα1, PLDδ, or both does not impact H2O2 production and callose deposition in the haustorium-invaded epidermal cells.
Fig. S4. Loss of PLDα1 and/or PLDδ does not affect ETI against bacterial pathogens.
Fig. S5. PLDα1 and PLDδ are not required for RPW8-mediated resistance to Gc UCSC1.
Fig. S6. The PLDδ–eGFP and PLDα1–eGFP fusion proteins are functional.
Fig. S7. Gc UCSC1 infection phenotypes of pldδ-containing double and triple mutants and relevant controls.
Fig. S8. The ‘edr’ phenotype of pldα1 to Gc UCSC1 is suppressed by the eds1-2, sid2-2, and/or pad4-1 mutations.
Fig. S9. A working model for the roles of PLDα1 and PLDδ in plant immunity.
Table S1. Arabidopsis T-DNA insertion mutants screened in this study.
Table S2. Primers used in this study.
Acknowledgements
We thank Shauna Somerville for the G. cichoracearum UCSC1 isolate; Hua Lu for the bacterial strains; Jane Parker for the eds1-2 mutant in the Col-0 background; Fumiaki Katagiri for the pad4-1sid2-2 and eds1-2pad4-1 double mutants; and Franker Coker for maintaining the plant growth facilities. This project was supported by a National Science Foundation grant (IOS1457033) to SX, and a scholarship from the China Scholarship Council to QZ.
Glossary
Abbreviations:
- Avr
avirulence factor
- Bgh
Blumeria graminis f.sp. hordei
- CC-NB-LRRs
coiled-coil–nucleotide-binding site–leucine-rich repeat
- DAB
3,3'-diaminobenzidine
- DGK
diacylglycerol kinase
- EDS1
ENHANCED DISEASE SUSCEPTIBILITY 1
- EHM
extra-haustorial membrane
- ET
ethylene
- ETI
effector-triggered immunity
- Gc
Golovinomyces cichoracearum
- Hpa
Hyaloperonospora arabidopsidis
- HR
hypersensitive response
- JA
jasmonic acid
- NB-LRR
nucleotide-binding site–leucine-rich-repeat
- NDR1
NON-RACE-SPECIFIC DISEASE RESISTANCE 1
- PA
phosphatidic acid
- PAD4
PHYTOALEXIN-DEFICIENT 4
- PAMP
pathogen-associated molecular pattern
- PEN1
PENETRATION1
- PIP5K
phosphatidylinositol 4-phosphate 5-kinase
- PLC
phospholipase C
- PLD
phospholipase D
- PM
plasma membrane
- Pma
Pseudomonas syringae pv. maculicola
- pPLA
patatin-related phospholipase
- PR
pathogenesis-related
- PRR
pattern recognition receptor
- PTI
PAMP-triggered immunity
- SA
salicylic acid
- TIR-NB-LRRs
Toll-interleukin 1 receptor–NB–LRRs
- UBC9
ubiquitin conjugating enzyme 9.
References
- Assaad FF, Qiu JL, Youngs H, et al. . 2004. The PEN1 syntaxin defines a novel cellular compartment upon fungal attack and is required for the timely assembly of papillae. Molecular Biology of the Cell 15, 5118–5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R, Jones JD. 2009. Role of plant hormones in plant defence responses. Plant Molecular Biology 69, 473–488. [DOI] [PubMed] [Google Scholar]
- Bartsch M, Gobbato E, Bednarek P, Debey S, Schultze JL, Bautor J, Parker JE. 2006. Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. The Plant Cell 18, 1038–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarek P, Pislewska-Bednarek M, Svatos A, et al. . 2009. A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323, 101–106. [DOI] [PubMed] [Google Scholar]
- Berkey R, Zhang Y, Ma X, King H, Zhang Q, Wang W, Xiao S. 2017. Homologues of the RPW8 resistance protein are localized to the extrahaustorial membrane that is likely synthesized de novo. Plant Physiology 173, 600–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakeslee JJ, Murphy AS. 2016. Microscopic and biochemical visualization of auxins in plant tissues. Methods in Molecular Biology 1398, 37–53. [DOI] [PubMed] [Google Scholar]
- Bonardi V, Tang S, Stallmann A, Roberts M, Cherkis K, Dangl JL. 2011. Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proceedings of the National Academy of Sciences, USA 108, 16463–16468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Songkumarn P, Liu J, Wang GL. 2009. A versatile zero background T-vector system for gene cloning and functional genomics. Plant Physiology 150, 1111–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Collins NC, Thordal-Christensen H, Lipka V, et al. . 2003. SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425, 973–977. [DOI] [PubMed] [Google Scholar]
- Cui H, Gobbato E, Kracher B, Qiu J, Bautor J, Parker JE. 2017. A core function of EDS1 with PAD4 is to protect the salicylic acid defense sector in Arabidopsis immunity. New Phytologist 213, 1802–1817. [DOI] [PubMed] [Google Scholar]
- Cui H, Tsuda K, Parker JE. 2015. Effector-triggered immunity: from pathogen perception to robust defense. Annual Review of Plant Biology 66, 487–511. [DOI] [PubMed] [Google Scholar]
- Dewdney J, Reuber TL, Wildermuth MC, Devoto A, Cui J, Stutius LM, Drummond EP, Ausubel FM. 2000. Three unique mutants of Arabidopsis identify eds loci required for limiting growth of a biotrophic fungal pathogen. The Plant Journal 24, 205–218. [DOI] [PubMed] [Google Scholar]
- Falk A, Feys BJ, Frost LN, Jones JDG, Daniels MJ, Parker JE. 1999. EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proceedings of the National Academy of Sciences, USA 96, 3292–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys BJ, Moisan LJ, Newman MA, Parker JE. 2001. Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. EMBO Journal 20, 5400–5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys BJ, Wiermer M, Bhat RA, Moisan LJ, Medina-Escobar N, Neu C, Cabral A, Parker JE. 2005. Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. The Plant Cell 17, 2601–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook J. 2005. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annual Review of Phytopathology 43, 205–227. [DOI] [PubMed] [Google Scholar]
- Grant MR, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes RW, Dangl JL. 1995. Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science 269, 843–846. [DOI] [PubMed] [Google Scholar]
- Guo L, Devaiah SP, Narasimhan R, Pan X, Zhang Y, Zhang W, Wang X. 2012. Cytosolic glyceraldehyde-3-phosphate dehydrogenases interact with phospholipase Dδ to transduce hydrogen peroxide signals in the Arabidopsis response to stress. The Plant Cell 24, 2200–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillmer RA, Tsuda K, Rallapalli G, Asai S, Truman W, Papke MD, Sakakibara H, Jones JDG, Myers CL, Katagiri F. 2017. The highly buffered Arabidopsis immune signaling network conceals the functions of its components. PLoS Genetics 13, e1006639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y, Zhao J, Guo L, Kim SC, Deng X, Wang G, Zhang G, Li M, Wang X. 2016. Plant phospholipases D and C and their diverse functions in stress responses. Progress in Lipid Research 62, 55–74. [DOI] [PubMed] [Google Scholar]
- Hückelhoven R, Panstruga R. 2011. Cell biology of the plant–powdery mildew interaction. Current Opinion in Plant Biology 14, 738–746. [DOI] [PubMed] [Google Scholar]
- Jirage D, Tootle TL, Reuber TL, Frost LN, Feys BJ, Parker JE, Ausubel FM, Glazebrook J. 1999. Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proceedings of the National Academy of Sciences, USA 96, 13583–13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson ON, Fahlberg P, Karimi E, Nilsson AK, Ellerström M, Andersson MX. 2014. Redundancy among phospholipase D isoforms in resistance triggered by recognition of the Pseudomonas syringae effector AvrRpm1 in Arabidopsis thaliana. Frontiers in Plant Science 5, 639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. 2006. The plant immune system. Nature 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kalachova T, Iakovenko O, Kretinin S, Kravets V. 2013. Involvement of phospholipase D and NADPH-oxidase in salicylic acid signaling cascade. Plant Physiology and Biochemistry 66, 127–133. [DOI] [PubMed] [Google Scholar]
- Kim Y, Tsuda K, Igarashi D, Hillmer RA, Sakakibara H, Myers CL, Katagiri F. 2014. Mechanisms underlying robustness and tunability in a plant immune signaling network. Cell Host and& Microbe 15, 84–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh S, André A, Edwards H, Ehrhardt D, Somerville S. 2005. Arabidopsis thaliana subcellular responses to compatible Erysiphe cichoracearum infections. The Plant Journal 44, 516–529. [DOI] [PubMed] [Google Scholar]
- Kuhn H, Lorek J, Kwaaitaal M, et al. . 2017. Key components of different plant defense pathways are dispensable for powdery mildew resistance of the Arabidopsis mlo2 mlo6 mlo12 triple mutant. Frontiers in Plant Science 8, 1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C, Neu C, Pajonk S, et al. . 2008. Co-option of a default secretory pathway for plant immune responses. Nature 451, 835–840. [DOI] [PubMed] [Google Scholar]
- Lipka V, Dittgen J, Bednarek P, et al. . 2005. Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 310, 1180–1183. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Narusaka M, Shirasu K, Noutoshi Y, Kubo Y, Shiraishi T, Iwabuchi M, Narusaka Y. 2009. RRS1 and RPS4 provide a dual resistance-gene system against fungal and bacterial pathogens. The Plant Journal 60, 218–226. [DOI] [PubMed] [Google Scholar]
- Novák O, Hényková E, Sairanen I, Kowalczyk M, Pospíšil T, Ljung K. 2012. Tissue-specific profiling of the Arabidopsis thaliana auxin metabolome. The Plant Journal 72, 523–536. [DOI] [PubMed] [Google Scholar]
- Pieterse CM, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SC. 2012. Hormonal modulation of plant immunity. Annual Review of Cell and Developmental Biology 28, 489–521. [DOI] [PubMed] [Google Scholar]
- Pinosa F, Buhot N, Kwaaitaal M, Fahlberg P, Thordal-Christensen H, Ellerström M, Andersson MX. 2013. Arabidopsis phospholipase dδ is involved in basal defense and nonhost resistance to powdery mildew fungi. Plant Physiology 163, 896–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2014. R: a language and environment for statistical computing.Vienna, Austria, R Foundation for Statistical Computing. [Google Scholar]
- Robert-Seilaniantz A, Grant M, Jones JD. 2011. Hormone crosstalk in plant disease and defense: more than just jasmonate–salicylate antagonism. Annual Review of Phytopathology 49, 317–343. [DOI] [PubMed] [Google Scholar]
- Rodas-Junco BA, Muñoz-Sánchez JA, Vázquez-Flota F, Hernández-Sotomayor SM. 2015. Salicylic-acid elicited phospholipase D responses in Capsicum chinense cell cultures. Plant Physiology and Biochemistry 90, 32–37. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH image to ImageJ: 25 years of image analysis. Nature Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Su W, Howell SH. 1992. Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proceedings of the National Academy of Sciences, USA 89, 6837–6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein M, Dittgen J, Sánchez-Rodríguez C, Hou BH, Molina A, Schulze-Lefert P, Lipka V, Somerville S. 2006. Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. The Plant Cell 18, 731–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto D, Jones DA, Hardham AR. 2003. GFP-tagging of cell components reveals the dynamics of subcellular re-organization in response to infection of Arabidopsis by oomycete pathogens. The Plant Journal 33, 775–792. [DOI] [PubMed] [Google Scholar]
- Thordal-Christensen H, Zhang Z, Wei Y, Collinge DB. 1997. Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley–powdery mildew interaction. The Plant Journal 11, 1187–1194. [Google Scholar]
- Tornero P, Dangl JL. 2001. A high-throughput method for quantifying growth of phytopathogenic bacteria in Arabidopsis thaliana. The Plant Journal 28, 475–481. [DOI] [PubMed] [Google Scholar]
- Tsuda K, Mine A, Bethke G, Igarashi D, Botanga CJ, Tsuda Y, Glazebrook J, Sato M, Katagiri F. 2013. Dual regulation of gene expression mediated by extended MAPK activation and salicylic acid contributes to robust innate immunity in Arabidopsis thaliana. PLoS Genetics 9, e1004015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Sato M, Stoddard T, Glazebrook J, Katagiri F. 2009. Network properties of robust immunity in plants. PLoS Genetics 5, e1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Biezen EA, Freddie CT, Kahn K, Parker JE, Jones JD. 2002. Arabidopsis RPP4 is a member of the RPP5 multigene family of TIR-NB-LRR genes and confers downy mildew resistance through multiple signalling components. The Plant Journal 29, 439–451. [DOI] [PubMed] [Google Scholar]
- Venugopal SC, Jeong RD, Mandal MK, et al. . 2009. Enhanced disease susceptibility 1 and salicylic acid act redundantly to regulate resistance gene-mediated signaling. PLoS Genetics 5, e1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Malek B, van der Graaff E, Schneitz K, Keller B. 2002. The Arabidopsis male-sterile mutant dde2-2 is defective in the ALLENE OXIDE SYNTHASE gene encoding one of the key enzymes of the jasmonic acid biosynthesis pathway. Planta 216, 187–192. [DOI] [PubMed] [Google Scholar]
- Wagner S, Stuttmann J, Rietz S, Guerois R, Brunstein E, Bautor J, Niefind K, Parker JE. 2013. Structural basis for signaling by exclusive EDS1 heteromeric complexes with SAG101 or PAD4 in plant innate immunity. Cell Host and Microbe 14, 619–630. [DOI] [PubMed] [Google Scholar]
- Wang C, Wang X. 2001. A novel phospholipase D of Arabidopsis that is activated by oleic acid and associated with the plasma membrane. Plant Physiology 127, 1102–1112. [PMC free article] [PubMed] [Google Scholar]
- Wang W, Devoto A, Turner JG, Xiao S. 2007. Expression of the membrane-associated resistance protein RPW8 enhances basal defense against biotrophic pathogens. Molecular Plant-Microbe Interactions 20, 966–976. [DOI] [PubMed] [Google Scholar]
- Wang W, Wen Y, Berkey R, Xiao S. 2009. Specific targeting of the Arabidopsis resistance protein RPW8.2 to the interfacial membrane encasing the fungal haustorium renders broad-spectrum resistance to powdery mildew. The Plant Cell 21, 2898–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Zhang Y, Wen Y, Berkey R, Ma X, Pan Z, Bendigeri D, King H, Zhang Q, Xiao S. 2013. A comprehensive mutational analysis of the Arabidopsis resistance protein RPW8.2 reveals key amino acids for defense activation and protein targeting. The Plant Cell 25, 4242–4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. 2000. Multiple forms of phospholipase D in plants: the gene family, catalytic and regulatory properties, and cellular functions. Progress in Lipid Research 39, 109–149. [DOI] [PubMed] [Google Scholar]
- Wen Y, Wang W, Feng J, Luo MC, Tsuda K, Katagiri F, Bauchan G, Xiao S. 2011. Identification and utilization of a sow thistle powdery mildew as a poorly adapted pathogen to dissect post-invasion non-host resistance mechanisms in Arabidopsis. Journal of Experimental Botany 62, 2117–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. 2009. ggplot2: elegant graphics for data analysis. New York: Springer-Verlag. [Google Scholar]
- Wiermer M, Feys BJ, Parker JE. 2005. Plant immunity: the EDS1 regulatory node. Current Opinion in Plant Biology 8, 383–389. [DOI] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM. 2001. Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414, 562–565. [DOI] [PubMed] [Google Scholar]
- Xiao S, Brown S, Patrick E, Brearley C, Turner JG. 2003. Enhanced transcription of the Arabidopsis disease resistance genes RPW8.1 and RPW8.2 via a salicylic acid-dependent amplification circuit is required for hypersensitive cell death. The Plant Cell 15, 33–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Calis O, Patrick E, Zhang G, Charoenwattana P, Muskett P, Parker JE, Turner JG. 2005. The atypical resistance gene, RPW8, recruits components of basal defence for powdery mildew resistance in Arabidopsis. The Plant Journal 42, 95–110. [DOI] [PubMed] [Google Scholar]
- Xiao S, Dai L, Liu F, Wang Z, Peng W, Xie D. 2004. COS1: an Arabidopsis coronatine insensitive1 suppressor essential for regulation of jasmonate-mediated plant defense and senescence. The Plant Cell 16, 1132–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Ellwood S, Calis O, Patrick E, Li T, Coleman M, Turner JG. 2001. Broad-spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science 291, 118–120. [DOI] [PubMed] [Google Scholar]
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG. 1998. COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Berkey R, Pan Z, Wang W, Zhang Y, Ma X, King H, Xiao S. 2015. Dominant negative RPW8.2 fusion proteins reveal the importance of haustorium-oriented protein trafficking for resistance against powdery mildew in Arabidopsis. Plant Signaling and Behavior 10, e989766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Xiao S. 2015. Lipids in salicylic acid-mediated defense in plants: focusing on the roles of phosphatidic acid and phosphatidylinositol 4-phosphate. Frontiers in Plant Science 6, 387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Qin C, Zhao J, Wang X. 2004. Phospholipase Dα1-derived phosphatidic acid interacts with ABI1 phosphatase 2C and regulates abscisic acid signaling. Proceedings of the National Academy of Sciences, USA 101, 9508–9513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhu H, Zhang Q, Li M, Yan M, Wang R, Wang L, Welti R, Zhang W, Wang X. 2009. Phospholipase Dα1 and phosphatidic acid regulate NADPH oxidase activity and production of reactive oxygen species in ABA-mediated stomatal closure in Arabidopsis. The Plant Cell 21, 2357–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J. 2015. Phospholipase D and phosphatidic acid in plant defence response: from protein–protein and lipid–protein interactions to hormone signalling. Journal of Experimental Botany 66, 1721–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Devaiah SP, Wang C, Li M, Welti R, Wang X. 2013. Arabidopsis phospholipase Dβ1 modulates defense responses to bacterial and fungal pathogens. New Phytologist 199, 228–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F, Menke FL, Yoshioka K, Moder W, Shirano Y, Klessig DF. 2004. High humidity suppresses ssi4-mediated cell death and disease resistance upstream of MAP kinase activation, H2O2 production and defense gene expression. The Plant Journal 39, 920–932. [DOI] [PubMed] [Google Scholar]
- Zhou N, Tootle TL, Tsui F, Klessig DF, Glazebrook J. 1998. PAD4 functions upstream from salicylic acid to control defense responses in Arabidopsis. The Plant Cell 10, 1021–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Jeong RD, Venugopal SC, Lapchyk L, Navarre D, Kachroo A, Kachroo P. 2011. SAG101 forms a ternary complex with EDS1 and PAD4 and is required for resistance signaling against turnip crinkle virus. PLoS Pathogens 7, e1002318. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.