Abstract
Motivation
The effective representation of proteins is a crucial task that directly affects the performance of many bioinformatics problems. Related proteins usually bind to similar ligands. Chemical characteristics of ligands are known to capture the functional and mechanistic properties of proteins suggesting that a ligand-based approach can be utilized in protein representation. In this study, we propose SMILESVec, a Simplified molecular input line entry system (SMILES)-based method to represent ligands and a novel method to compute similarity of proteins by describing them based on their ligands. The proteins are defined utilizing the word-embeddings of the SMILES strings of their ligands. The performance of the proposed protein description method is evaluated in protein clustering task using TransClust and MCL algorithms. Two other protein representation methods that utilize protein sequence, Basic local alignment tool and ProtVec, and two compound fingerprint-based protein representation methods are compared.
Results
We showed that ligand-based protein representation, which uses only SMILES strings of the ligands that proteins bind to, performs as well as protein sequence-based representation methods in protein clustering. The results suggest that ligand-based protein description can be an alternative to the traditional sequence or structure-based representation of proteins and this novel approach can be applied to different bioinformatics problems such as prediction of new protein–ligand interactions and protein function annotation.
Availability and implementation
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
The aging population is putting drug design studies under pressure as we see an increase in the incidence of complex diseases. Multiple proteins from different protein families or protein networks are usually implicated in these complex diseases such as cancer, cardiovascular, immune and neurodegenerative diseases (Hu et al., 2016; Poornima et al., 2016; Santiago and Potashkin, 2014). Reliable representation of proteins plays a crucial role in the performance of many bioinformatics tasks such as protein family classification and clustering, prediction of protein functions and prediction of the interactions between protein–protein and protein–ligand pairs. Proteins are usually represented based on their sequences (Cai et al., 2003; Chou, 2001; Iqbal et al., 2013). A recent study adapted Word2Vec (Mikolov et al., 2013), which is a widely used word-embeddings model in natural language processing (NLP) tasks, into the genomic space to describe proteins as real-valued continuous vectors using their sequences, and utilized these vectors to classify proteins (Asgari and Mofrad, 2015). However, even though the structure of a protein is determined by its sequence, sequence alone is usually not adequate to completely understand its mechanism. Furthermore, the relationship between fold or architecture and function was shown to be weak, while a strong correlation was reported for architecture and bound ligand (Martin et al., 1998). Semantic features such as functional categories and annotations and gene ontology classes (Cao and Cheng, 2016; Frasca and Cesa-Bianchi, 2017; Nascimento et al., 2016; Shi et al., 2015) have been suggested to support the functional understanding of proteins, nevertheless these features are usually described in the form of binary vectors preventing the direct use of the provided information. Therefore, a novel approach that defines proteins by integrating functional characterizations can provide important information toward understanding and predicting protein structure, function and mechanism. Ligand-centric approaches are based on the chemical similarity of compounds that interact with similar proteins (Peón et al., 2016) and have been successfully adopted for tasks such as target fishing, off-target effect prediction and protein-clustering (Chiu et al., 2014; Schenone et al., 2013) following the pioneering works that proposed to measure protein similarity using their ligands (Hert et al., 2008; Keiser et al., 2007). The use of chemical similarity of the interacting ligands of proteins to group them resulted in both biologically and functionally related protein clusters (Keiser et al., 2007; Öztürk et al., 2015). Motivated by these results, we propose to describe proteins using their interacting ligands.
In order to define a protein with a ligand-centric approach, the description of the ligands is critical. Ligands can be represented in many different forms including knowledge-based fingerprints, graphs, or strings. Simplified molecular input line entry system (SMILES), which is a character-based representation of ligands, has been used for QSAR studies (Cao et al., 2012; Schwartz et al., 2013) and protein–ligand interaction prediction (Jastrzębski et al., 2016; Öztürk et al., 2016). Even though it is a string-based representation form, use of SMILES performed as well as powerful graph-based representation methods in protein–ligand interaction prediction and has been proven to be computationally less expensive (Öztürk et al., 2016). A recent study that employed recurrent neural networks-based model to describe compound properties also used SMILES to predict chemical properties. However, such deep-learning-based approaches require more computational power.
An advantage of SMILES is that it provides a promising environment for the adoption of NLP approaches because it is character based. Distributed word representation models have been widely used in recent studies of NLP tasks, especially with the introduction of Word2Vec (Mikolov et al., 2013). The model requires a large amount of text data to learn the representations of words to describe them in low-dimensional space as real-valued vectors. These vectors comprise the syntactic and semantic features of the words, e.g. the vectors of words with similar meanings are also similar. A recent study, Mol2Vec (Jaeger et al., 2018), adopts Word2vec to learn representations for compounds and uses these to predict their properties such as toxicity and mutagenicity. Rather than representing the compounds with their SMILES strings directly, Mol2Vec represents them with the identifiers of the corresponding atoms obtained by using the Morgan Algorithm (Rogers and Hahn, 2010).
In this study, we introduce SMILESVec, in which we adopted the word-embeddings approach to define ligands by utilizing their SMILES strings. Ligands are represented by learning features from a large SMILES corpus via Word2Vec (Mikolov et al., 2013), instead of using manually constructed ligand features. We then describe each protein using the average of its interacting ligand vectors that are built by SMILESVec. We followed a similar pipeline for evaluation that is presented in (Bernardes et al., 2015), in which the authors compared the performances of different clustering algorithms on the task of detecting remote homologous protein families. We measured how well SMILESVec-based protein representation describes proteins within a protein clustering task by using two state-of-the-art clustering algorithms; transitive clustering (TransClust) (Wittkop et al., 2010) and Markov clustering algorithm (MCL) (Enright et al., 2002).
The performance of clustering using SMILESVec-based protein representation was compared with that using the traditional basic local alignment tool (BLAST), MACCS-based (Willighagen et al., 2017) and extended-connectivity fingerprint (ECFP)-based protein representations as well as the recently proposed distributed protein vector representation, which is called ProtVec (Asgari and Mofrad, 2015). ASTRAL dataset (A-50) of structural classification of protein (SCOP) database was used as benchmark (Chandonia et al., 2017; Murzin et al., 1995).
The results showed that the representation of proteins with their ligands is a promising method with competitive F-scores in the protein clustering task, even though no sequence or structure information is used. SMILESVec can be an alternative approach to binary-vector-based fingerprint models for ligand-representation. The ligand-based protein representation might be useful in different bioinformatics tasks such as identifying new protein–ligand interactions and protein function annotations.
2 Materials and methods
2.1 Dataset
The ASTRAL datasets are part of SCOPs collection and classified under folds, families and super-families (Fox et al., 2014). A family denotes a group of proteins with typically distinct functionalities but also with high sequence similarities, whereas a super-family is a group of protein families with structural and functional similarities amongst families. The ASTRAL datasets are named based on the minimum sequence similarity of the proteins that they comprise. For instance, A-50 dataset includes proteins with at most 50% sequence similarity (http://scop.berkeley.edu/astral/subsets/ver=1.75&seqOption=1). In this study, we used A-50 dataset from SCOP 1.75 version to demonstrate the performance of the protein representation methods and considered clustering into families and super-families for evaluation. Families and super-families with single protein were removed while preparing the data (Bernardes et al., 2015). We used the same protein pairs that Bernardes et al. (2015) used for A-50 to compute similarity scores (http://www.lcqb.upmc.fr/julianab/software/cluster/).
2.2 Collection of protein–ligand interactions
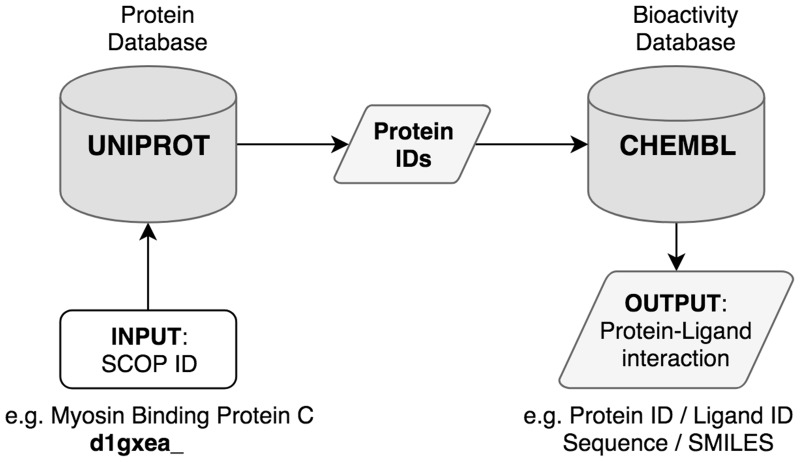
First, the corresponding UniProt identifiers were extracted for each protein in A-50 dataset using Bioservices Python package (Cokelaer et al., 2013). Then, the interacting ligands with their corresponding canonical SMILES were retrieved from ChEMBL (Gaulton et al., 2011) using ChEMBL web services (Davies et al., 2015) (Data collected on December 30, 2017). The workflow of protein-ligand interaction extraction is illustrated in Figure 1. The collected interactions were used to build the proposed SMILESVec-based protein representations.
Fig. 1.
Extraction of protein–ligand interactions. As an example protein, Cardiac Myosin Binding Protein C is provided as input with its corresponding SCOP ID: d1gxea_
2.3 Distributed representation of proteins and ligands
The Word2Vec model, which is based on feed-forward neural networks, has been previously adopted to represent proteins using their sequences (Asgari and Mofrad, 2015). The approach that we will refer to as ProtVec throughout the article, improved the performance for the protein classification problem. In this study, we used the Word2Vec model with the Skip-gram approach to consider the order of the surrounding words. In the biological context, we can use the string representations of proteins/ligands (e.g. FASTA sequence for proteins and SMILES for ligands) in textual format and define words as sub-sequences of these representations.
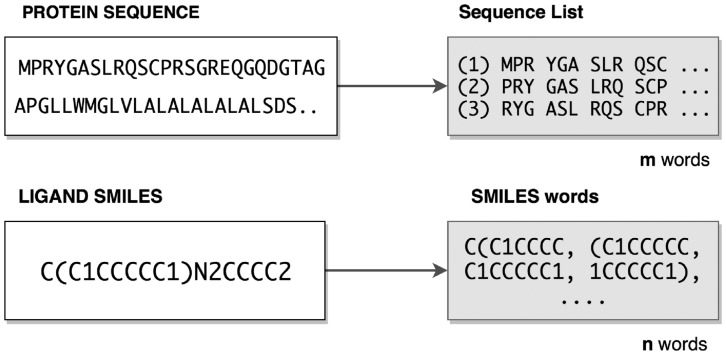
Figure 2 illustrates a sample protein sequence and its sequence list (biological words) as well as a sample ligand SMILES and its corresponding sub-sequences (chemical words). The biological words which are referred to as sequence-lists are created with a set of three characters of non-overlapping sub-sequences for each list that starts from the character indices 1, 2 and 3, respectively, therefore leading to three sequence lists (Asgari and Mofrad, 2015). The chemical words were created as eight-character long overlapping substrings of SMILES with sliding window approach. As shown in Figure 2, the SMILES string ‘C(C1CCCCC1)N2CCCC2’ is divided into the following chemical words: ‘C(C1CCCC’, ‘(C1CCCCC’, ‘C1CCCCC1’, ‘1CCCCC1)’, ‘CCCCC1)N’, ‘CCCC1)N2’, …, ’)N2CCCC2’. We performed several experiments in which word size varied in the range of 4–12 characters and eight-charactered chemical words obtained the best results.
Fig. 2.
Representation of biological and chemical words
With the use of the Word2Vec model we were able to describe complex structures using their simplified representations. For each subsequence (word) that was extracted from protein sequence/ligand SMILES, Word2Vec produced a real-valued vector that is learned from a large training set. The vector learning is based on the context of each subsequence (e.g. its surrounding subsequences) and can detect some important subsequences that usually occur in the same contexts. Therefore, with the help of the neural-network-based nature of Word2Vec, every subsequence of a protein sequence/ligand SMILES was described in a semantically meaningful way. The Word2Vec model defined a vector representation for each of the three-residue subsequences of the proteins. Protein vectors were constructed as the average of these subsequence vectors as described in Equation (1), where refers to the 100D real-valued vector for the kth subsequence and m is equal to the total number of sub-sequences that can be extracted from a protein sequence. For proteins, 550 K protein sequences from UniProt were used for training.
| (1) |
Similarly, the Word2Vec model produced a real-valued vector for each SMILES word and the corresponding ligand vector is constructed as the average of the SMILES word vectors as described in Equation (2). represents the Word2Vec output for the eight-character long kth subsequence of the SMILES string and n indicates the total number of these SMILES subsequences (words). We will refer to ligand vectors as SMILESVec throughout the article. For learning, 1.7 M canonical SMILES from CHEMBL database (ftp.ebi.ac.uk/pub/databases/chembl/ChEMBLdb/releases/chembl_23) were retrieved. We used the Gensim implementation (Řehůřek and Sojka, 2010) of Word2Vec and the size of the vectors was set to the default value of 100. The Skip-Gram approach was employed.
| (2) |
We also used the Word2Vec model to learn embeddings for the characters in the SMILES alphabet. Therefore instead of word-level, we created char-level embeddings for the unique characters that appear in SMILES in ChEMBL23 dataset (58 chars). Equation (3) describes SMILESVecchar where n in this case represents the total number of the characters in a SMILES.
| (3) |
We further investigated an important aspect when working with SMILES representation, since there are several valid SMILES for a single molecule. Canonicalization algorithms were coined for the purpose of generating an unique SMILES for a molecule; however, couldn’t prevent the diversity that came with different canonicalization algorithms. Thus, it is not that surprising that canonical SMILES definition can differ from database to database. ChEMBL uses Accelrys’s Pipeline Pilot that uses an algorithm derived from Daylight’s (Papadatos and Overington, 2014), whereas Pubchem (Bolton et al., 2008) uses OpenEye software (https://www.eyesopen.com/) for canonical SMILES generation (Balakin, 2009). The most evident difference between the canonical SMILES of two databases is that ChEMBL includes isomeric information, whereas Pubchem does not. Therefore, even though we collected the SMILES of the interacting ligands from the ChEMBL database, we both experimented learning chemical words and characters from ChEMBL and Pubchem canonical SMILES corpora both separately and together (combined).
We can represent a protein/ligand vector as the output of the maximum or minimum functions, where m is the total number of the subsequences that are created from the protein/ligand sequence and d is the dimensionality of the vector (i.e. the number of features). MINi represents the minimum value of the ith feature among m subsequences (Equation 4). To obtain a protein vector of minimum, MINi is selected for each feature as defined in Equation (5). Similarly, MAXi represents the maximum value of the ith feature among m subsequences (Equation 6) and protein vector of maximum is created as in Equation (7) for d number of features. The concatenation of these minimum and maximum protein vectors results in a vector with twice the dimensionality of the original vectors (De Boom et al., 2016). The min/max representation is described in Equation (8).
| (4) |
| (5) |
| (6) |
| (7) |
| (8) |
2.4 Protein similarity computation
We used BLAST and ProtVec-based methods as baseline to compare to the ligand-centric protein representation that we proposed.
2.4.1 Basic local alignment tool
BLAST reports the similarity between protein sequences using local alignment (Altschul et al., 1990). For the ASTRAL datasets, we used both BLAST sequence identity values and BLAST e-values that were previously obtained (Bernardes et al., 2015) with all-versus-all BLAST with e-value threshold of 100.
2.4.2. Word frequency-based protein similarity
Word frequency-based protein similarity method uses three-charactered protein words that are created as explained in Section 2.3. However, instead of a learning process, we simply count the occurrences of the protein words in a protein sequence. In order to compute the similarity between two proteins, we used the formula depicted in Equation (9) (Vidal et al., 2005):
| (9) |
where m is the total number of unique words created from protein sequences P1 and P2, is the frequency of words of type i in protein P1 and is the frequency of words of type i in protein P2.
2.4.3. ProtVec-based protein similarity
In ProtVec-based clustering, protein vectors were constructed as defined in Section 2.3, either with the average or minmax method. The cosine similarity function was used to compute the similarity between two protein vectors P1 and P2 as in Equation (10), where d denotes the size (dimensionality) of the vectors.
| (10) |
2.4.4. SMILESVec-based protein similarity
First, the ligand vectors were constructed by the SMILESVec approach described in Section 2.3. Then, each protein was represented as the average of the vectors of the ligands they interact with. Equation (11) describes the construction of a protein vector from its binding ligands, where SMILESVec represents the ligand vector and nl represents the total number of ligands that the protein interacts with.
| (11) |
Similarly, protein similarity is computed using the cosine similarity function.
2.4.5. Fingerprint-based protein similarity
We used two fingerprint-based compound representation methods as an alternative to SMILESVec, namely MACCS and ECFP (Rogers and Hahn, 2010). MACCS is a structural fingerprint where each bit represents a specific substructure. ECFP, on the other hand, is a hash-based representation that describes features of substructures based on the atoms and their circular neighbors within a radius range (Sawada et al., 2014). MACCS and ECFP are represented with 166 and 1024 bit vectors, respectively. We used the default settings of chemical development kit (Willighagen et al., 2017) to obtain the MACCS and ECFP representations of the ligands, and for ECFP we chose the value of 6 as the maximum diameter (ECFP6). The proteins were represented as described in Equation (12) in which fingerprints were used to represent each interacting ligand.
| (12) |
Fingerprints were used in order to compare how competitive a text-based data-driven approach (SMILESVec) is against the widely adopted chemical descriptors.
2.4.6. SMILES word frequency-based protein similarity
For each interacting ligand of a protein, eight-character-long SMILES words were created as explained in Section 2.3. Then, the similarity between two proteins was computed as in Equation (9) using the collection of chemical words of their respective interacting ligands.
2.5 Clustering algorithms
We evaluated the effectiveness of the different protein representation approaches for the task of protein clustering. TransClust, which has been shown to produce the best F-measure score amongst several other algorithms in protein clustering (Bernardes et al., 2015) and the commonly used MCL were used as the protein clustering algorithms.
2.5.1. Transitivity clustering
TransClust is a clustering method that is based on the weighted transitive graph projection problem (Wittkop et al., 2010). The main idea behind TransClust is to construct transitive graphs by adding or removing edges from an intransitive graph using a weighted cost function. The weighted cost function is calculated as the distance between a user-defined threshold and a pairwise similarity function. TransClust connects two proteins on the network if their similarity is greater than the user-defined threshold. The graph is expanded by adding or removing edges until it becomes a disjoint union of cliques (Bernardes et al., 2015). TransClust requires a user-defined threshold to identify clusters. Therefore, in order to choose the best threshold value, we computed the F-measure values for similarity threshold range of [0, 1] with 0.001 step-size for the similarity computation methods that output similarity values in the range of [0, 1]. For BLAST, range of [0, 100] with step-size value of 0.05 was tested for similarity threshold. We chose the similarity thresholds that gave the best F-measure for super-family and family to decide the final clusters.
2.5.2 Markov clustering algorithm
MCL is a network clustering algorithm that considers the weights of the edges (flows) in the network (Enright et al., 2002). MCL finds the clusters of a network by first, transforming similarity measures into probabilities and then, computing the probabilities of random-walks. The algorithm utilizes expansion and inflation operators to alternate between set of probabilities (https://micans.org/mcl/index.html?sec_description1). Inflation decides the granularity of the predicted clusters whereas expansion is responsible from reducing the occurrences of higher length paths. We used the default value (2.0) of inflation the in MCL package which is described as the only parameter that the user might need to change.
2.6 Evaluation
In order to evaluate the performance of the proposed methods, we utilized the F-measure, precision and recall metrics. These metrics are widely used in the evaluation of classification methods. To adapt these metrics for the assessment of the clustering task, we followed the formulation explained by Bernardes et al. (2015).
For a dataset of n proteins, let us assume nf represents the number of proteins that belong to the fth family or class, ng is the number of proteins that are placed in the gth cluster and nfg represents the number of proteins that belong to the fth family and are placed in the gth cluster. Precision of cluster g with respect to the fth family is computed as , whereas recall is defined as . Finally we can define F-measure as in Equation (13):
| (13) |
maxg indicates that for each family f, we compute precision and recall values for each cluster g, and choose the maximum resulting F-score.
The weighted mean precision and recall are described in Equations (14) and (15), respectively (Bernardes et al., 2015).
| (14) |
| (15) |
3 Results
We evaluated the performance of five different protein similarity computation approaches in clustering of the A-50 dataset. The similarity approaches were BLAST, ProtVec, SMILESVec, MACCS and ECFP, the first two of which are protein sequence-based similarity methods, whereas the latter three utilize the ligands to which proteins bind. We took word frequency-based protein similarity methods that use protein sequences and compound SMILES strings, respectively, as the baseline. Average (avg) and minimum/maximum (min/max) of the vectors were taken to build combined vectors for ProtVec and SMILESVec from their subsequence vectors.
We performed our experiments on the A-50 dataset using two different clustering algorithms, TransClust and MCL. The ligand-based (SMILESVec, MACCS and ECFP) protein representation approaches require a protein to bind to at least one ligand in order to define a ligand-based vector for that protein. Therefore, we removed the proteins with no ligand-binding information from the dataset. Table 1 provides a summary of the A-50 dataset before and after filtering.
Table 1.
Distribution of families and super-families in A-50 dataset before and after filtering
| Dataset | Number of Sequences | Super-families | Families |
|---|---|---|---|
| Before filtering | 10 816 | 1080 | 2109 |
| After filtering | 1639 | 425 | 652 |
When the set of proteins that remain in our dataset are examined, we see that some of the superfamilies/families that were initially in the top-10 most frequent family and super-family lists are replaced by others (Supplementary Table S1). Among the superfamilies that are no longer in the most frequent list are ‘Winged helix’ DNA-binding domain and thioredoxin-like superfamilies because the number of known ligands is lower. On the other hand, super-families and families that weren’t initially in the top-10 list such as Protein-kinase like (d.144.1) super-family and nuclear-receptor binding domain (a.123.1) and their respective descendant families make it to the frequent set of proteins when ligand interactions are taken into account. Table 2 summarizes the top-10 most frequent family and super-families with known ligand interactions.
Table 2.
Distribution of the top-10 most frequent super-families and families with known ligand interactions
| Super-family | No. of prots. | Family | No. of prots. | |
|---|---|---|---|---|
| 1 | Protein kinase-like (d.144.1) | 47 | Protein kinases, catalytic subunit (d.144.1.7) | 39 |
| 2 | P-loop containing nucleoside triphosphate hydrolases (c.37.1) | 43 | Fibronectin type III (b.1.2.1) | 28 |
| 3 | Immunoglobulin (b.1.1) | 41 | Eukaryotic proteases (b.47.1.2) | 25 |
| 4 | NAD(P)-binding Rossmann-fold domain (c.2.1) | 32 | EGF-type module (g.3.11.1) | 24 |
| 5 | Trypsin-like serine proteases (b.47.1) | 31 | Immunoglobulin I set (b.1.1.4) | 23 |
| 6 | Fibronectin type III (b.1.2) | 28 | SH2 domain (d.93.1.1) | 22 |
| 7 | EGF/Laminin (g.3.11) | 27 | Nuclear receptor ligand-binding domain (a.123.1.1) | 18 |
| 8 | SH2 domain (d.93.1) | 22 | Cyclin (a.74.1.1) | 15 |
| 9 | Cysteine proteinases (d.3.1) | 20 | Pleckstrin-homology domain (b.55.1.1) | 15 |
| 10 | Nuclear receptor ligand-binding domain (a.123.1) | 19 | Tyrosine-dependent oxidoreductases (c.2.1.2) | 15 |
In the filtered dataset in which all proteins have an interacting ligand, there are 1057 proteins with fewer than 200 ligands (64% of all proteins) and 101 proteins with single ligands (0.6% of all proteins). There are 67 proteins with more than 10 000 interacting ligands (0.4%), thus increasing the mean number of the interacting ligands to 1791. The protein with the highest number of interacting ligands is d2dpia2 (DNA polymerase iota), a protein involved in DNA repair (Jain et al., 2017) and implicated in esophageal squamous cell cancer (Zou et al., 2016) and breast cancer (Yang et al., 2004), with 115 018 ligands.
We assessed the performance of the clustering algorithms with F-measure values for two different clustering scenarios, family and super-family clustering. We also provided Precision and Recall values for each of the methods. In clustering, high recall indicates that the method assigns a high number of proteins from the same family/super-family to the same cluster. High precision, on the other hand, means the assigned clusters contain high percentage of proteins that belong to the same family/super-family. Higher precision values indicate that the clusters are more homogeneous, i.e., mostly contain proteins from the same families/supefamilies.
Tables 3 and 4 report the Precision, Recall and F-measure values for family and super-family clustering and the number of clusters that are detected with the TransClust and MCL algorithms, respectively. Between TransClust and MCL, TransClust produced better F-measure values in all representation methods on the A-50 dataset. The results obtained by both clustering algorithms were better in family clustering than in super-family clustering, which was an expected outcome, since detection of relationships between distantly related proteins is a much harder task.
Table 3.
Performance of the TransClust algorithm in super-family and family clustering for all protein similarity computation methods with Precision, Recall and F-measure values
| Super-family |
Family |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| No. Clusters | Precision | Recall | F-measure | No. Clusters | Precision | Recall | F-measure | ||
| Protein sequence based | |||||||||
| Blast (e-value) | A-50 | 1596 | 0.997 | 0.261 | 0.350 | 1636 | 1.0 | 0.399 | 0.500 |
| Blast (identity) | A-50 | 606 | 0.861 | 0.550 | 0.595 | 660 | 0.781 | 0.668 | 0.631 |
| Protein Word frequency | A-50 | 708 | 0.952 | 0.621 | 0.686 | 688 | 0.844 | 0.777 | 0.744 |
| ProtVec Avg (word) | A-50 | 655 | 0.927 | 0.620 | 0.681 | 704 | 0.845 | 0.757 | 0.739 |
| ProtVec Avg (char) | A-50 | 707 | 0.940 | 0.603 | 0.674 | 707 | 0.842 | 0.746 | 0.729 |
| ProtVec MinMax (word) | A-50 | 586 | 0.891 | 0.623 | 0.667 | 704 | 0.829 | 0.741 | 0.718 |
| Ligand based | |||||||||
| SMILES Word frequency | A-50 | 801 | 0.951 | 0.548 | 0.624 | 957 | 0.934 | 0.658 | 0.704 |
| SMILESVec (word, chembl) | A-50 | 621 | 0.921 | 0.621 | 0.677 | 730 | 0.855 | 0.744 | 0.735 |
| SMILESVec (word, pubchem) | A-50 | 573 | 0.888 | 0.627 | 0.668 | 692 | 0.839 | 0.751 | 0.730 |
| SMILESVec (word, combined) | A-50 | 617 | 0.923 | 0.627 | 0.675 | 764 | 0.873 | 0.732 | 0.735 |
| SMILESVec (char, chembl) | A-50 | 636 | 0.920 | 0.621 | 0.678 | 710 | 0.844 | 0.743 | 0.729 |
| SMILESVec (char, pubchem) | A-50 | 714 | 0.941 | 0.600 | 0.671 | 715 | 0.845 | 0.744 | 0.729 |
| SMILESVec (char, combined) | A-50 | 712 | 0.949 | 0.602 | 0.675 | 712 | 0.850 | 0.749 | 0.739 |
| MACCS | A-50 | 589 | 0.909 | 0.629 | 0.679 | 683 | 0.839 | 0.757 | 0.736 |
| ECFP6 | A-50 | 611 | 0.917 | 0.627 | 0.679 | 725 | 0.860 | 0.746 | 0.733 |
Note: The best F-measure values for the Protein sequence- and Ligand-based methods are shown in bold.
Table 4.
Performance of the MCL algorithm in super-family and family clustering for all protein similarity computation methods with Precision, Recall and F-measure values
| Super-family |
Family |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| No. Clusters | Precision | Recall | F-measure | No. Clusters | Precision | Recall | F-measure | ||
| Protein sequence based | |||||||||
| Blast (e-value) | A-50 | 728 | 0.792 | 0.271 | 0.290 | 728 | 0.687 | 0.406 | 0.379 |
| Blast (identity) | A-50 | 783 | 0.882 | 0.496 | 0.540 | 783 | 0.803 | 0.622 | 0.592 |
| Protein Word frequency | A-50 | 411 | 0.769 | 0.625 | 0.590 | 411 | 0.643 | 0.767 | 0.606 |
| ProtVec avg (word) | A-50 | 1001 | 0.964 | 0.514 | 0.596 | 1001 | 0.909 | 0.639 | 0.665 |
| ProtVec avg (char) | A-50 | 1017 | 0.964 | 0.508 | 0.590 | 1017 | 0.910 | 0.633 | 0.662 |
| ProtVec MinMax (word) | A-50 | 1014 | 0.964 | 0.508 | 0.590 | 1014 | 0.909 | 0.634 | 0.662 |
| Ligand based | |||||||||
| SMILES Word frequency | A-50 | 312 | 0630 | 0.550 | 0.470 | 312 | 0.497 | 0.686 | 0.475 |
| SMILESVec (word, chembl) | A-50 | 867 | 0.937 | 0.544 | 0.608 | 867 | 0.870 | 0.672 | 0.667 |
| SMILESVec (word, pubchem) | A-50 | 857 | 0.931 | 0.544 | 0.604 | 857 | 0.861 | 0.673 | 0.664 |
| SMILESVec (word, combined) | A-50 | 894 | 0.940 | 0.540 | 0.607 | 894 | 0.877 | 0.666 | 0.668 |
| SMILESVec (char, chembl) | A-50 | 999 | 0.962 | 0.514 | 0.596 | 999 | 0.908 | 0.641 | 0.668 |
| SMILESVec (char, pubchem) | A-50 | 977 | 0.958 | 0.514 | 0.595 | 977 | 0.900 | 0.643 | 0.667 |
| SMILESVec (char, combined) | A-50 | 1006 | 0.963 | 0.514 | 0.595 | 1006 | 0.909 | 0.641 | 0.669 |
| MACCS | A-50 | 874 | 0.936 | 0.540 | 0.606 | 874 | 0.866 | 0.668 | 0.667 |
| ECFP6 | A-50 | 618 | 0.863 | 0.582 | 0.599 | 618 | 0.762 | 0.710 | 0.631 |
Note: The best F-measure values for the Protein sequence- and ligand-based methods are shown in bold.
Both clustering algorithms relied on similarity scores in order to group proteins. Among the protein sequence-based similarity methods, the poorest clustering F-measure performance in super-family/family (0.350/0.500) belonged to BLAST with e-value, the baseline. Protein word frequency obtained the best performance on the A-50 dataset in super-family and family clustering (0.686/0.744). The performance of the ProtVec Avg (0.681/0.739) and the ligand-based protein representation methods followed the best result closely. Bringing in a semantic aspect with learning through the Word2Vec model, ProtVec-based similarity (avg and minmax), was outperformed by the straightforward word frequency-based approach.
The results also showed that the average-based combination method (ProtVec avg) was better than the min/max-based combination method (ProtVec minmax) to build a single protein vector from subsequence vectors in the protein clustering task. Since min/max-based combination method did not perform well in sequence-based protein similarity, we did not test the technique for SMILES-based protein similarity approaches.
Among the ligand-based representation methods, we examined the performance of the word-based embeddings and character-based embeddings as well as the effect of the source of the training dataset on the embeddings. We collected canonical SMILES from both ChEMBL (∼1.7 M) and Pubchem (∼2.3 M) databases. The SMILES strings of the interacting ligands were only collected from ChEMBL as explained in Section 2.2. The main difference between these two databases is that ChEMBL allows the isomeric information of the molecule to be encoded within SMILES. The results indicated that the choice of the SMILES corpus in which the word-embeddings are trained on should be considered carefully, since even slight changes in the notation of SMILES, affects the formation of the chemical words directly. In our case, since the SMILES of the interacting ligands of the A-50 dataset were collected from the ChEMBL database, the performance of SMILESVec in which embeddings were learned from training with ChEMBL SMILES rather than Pubchem SMILES was notably better.
We also investigated whether using the combination of the SMILES corpus of ChEMBL and Pubchem can improve the performance of SMILESVec embeddings. We indeed reported an improvement on character-based embedding in family clustering (0.739 F-measure) whereas word-based embedding produced F-measure values higher than the Pubchem-based learning and lower than the ChEMBL-based learning. We can suggest that the increase in the performance of the character-based learning with the combination of two different SMILES corpora might be positively correlated with the increase in SMILES samples, while the number of unique letters that appear in the SMILES did not significantly change between databases (e.g. absence/presence of the few characters that represent isometry information). However, with the word-based learning, we observed that there was significant increase in the variety of the chemical words, thus the combined SMILES corpus model did not work as well as it did in character-based learning. This result suggests that the size of the learning corpus may affect the representation of the embeddings, and a larger SMILES corpus could lead to better character-based embeddings for SMILESVec.
Considering only ChEMBL trained SMILESVec, word-based approach was slightly better than character-based SMILESVec in terms of F-measure in family clustering. In super-family clustering however, character-based approach performs as well as word-based SMILESVec. Similarly, ProtVec is also better represented in word-level rather than character-level.
The ligand-based protein representation methods, SMILESVec and MACCS-based approach performed almost as well as ProtVec in family and super-family clustering with TransClust algorithm, even though no protein sequence information was used. A lower clustering performance was obtained with MCL than with TransClust, and both SMILESVec and MACCS-based method produced slightly better F-measure than ProtVec Avg in both super-family and family clustering. Since ligand-based protein representation methods capture indirect function information through ligand binding, they were recognizably better at detecting super-families than families compared with sequence-based ProtVec on a relatively distant dataset. Furthermore, SMILESVec, a text-based unsupervised learning model, produced comparable F-measure values to MACCS and ECFPs, which are binary vectors based on human-engineered and hash-based feature descriptions, respectively.
Table 5 reports the Pearson correlations (Pearson, 1895) among the protein similarity computation methods. Comparison with BLAST e-value resulted in a negative correlation, as expected, since e-values closer to zero indicate high match (similarity). Ligand-based protein representation methods had higher correlation values with BLAST e-value than protein sequence-based methods. We also observed strong correlation among the ligand-based protein representation methods, suggesting that, regardless of the ligand representation approach, the use of interacting ligands to represent proteins provides similar information.
Table 5.
Pearson correlation between protein similarity methods
| Method | Method | Pearson correlation |
|---|---|---|
| BLAST (e-value) | BLAST (identity) | −0.109 |
| BLAST (e-value) | Protein word frequency | −0.250 |
| BLAST (e-value) | ProtVec (avg) | −0.291 |
| BLAST (e-value) | SMILESVec (word, chembl) | −0.335 |
| BLAST (e-value) | SMILESVec (char, chembl) | −0.207 |
| BLAST (e-value) | MACCS | −0.336 |
| SMILESVec (word, chembl) | MACCS | 0.895 |
| SMILESVec (char, pubchem) | MACCS | 0.590 |
| SMILESVec (word, chembl) | SMILESVec (char, pubchem) | 0.682 |
| SMILESVec (word, chembl) | ECFP6 | 0.933 |
| ECFP6 | MACCS | 0.898 |
We further investigated a case in which similar super-family clusters were produced with SMILESVec-based protein similarity and ProtVec protein similarity using the TransClust algorithm. We chose one of the medium-sized clusters for manual inspection. We observed that Fibronectin Type III proteins (seven proteins) were clustered together when SMILESVec was used, whereas using ProtVec placed them into four different clusters; one cluster contained four of those proteins, another cluster contained a single protein and the other two proteins were part of other clusters. The protein that was clustered by itself (SCOP ID: d1n26a3, Human Interleukin-6 Receptor alpha chain) had two interacting ligands (CHEMBL81; Raloxifene and CHEMBL46740; Bazedoxifene) that were also shared by a protein (SCOP ID: d1bqua2, Cytokine-binding region of GP130) clustered separately with ProtVec. Thus, we can suggest that using information on common interacting ligands, SMILESVec achieved to combine these seven proteins into a single cluster, while ProtVec failed to do so with a sequence-based approach.
We would like to mention that ASTRAL datasets contain domains rather than full length proteins, while CHEMBL collects protein–ligand interaction information based on the whole protein sequence from UniProt. A multidomain protein may have multiple and diverse chemotypes of ligands binding to each domain and retrieving ligand information based on the full length protein may lump this disparate information together, leading to loss of information on domain specific ligand interactions. The performance of domain sequence-based methods is therefore at an advantage because family/superfamily assignment in SCOP is also based on domain sequence, while the ligand-based approach we use in SMILESVec uses more noisy data. Despite this disadvantage, ligand-based approach performs as well as the sequence-based approaches.
Due to the domain-based nature of the ASTRAL datasets, clustering based on the full protein sequence can lead to a reduction in performance because of the presence of multidomain proteins. Similarly, we hypothesized that the ligand-based methods might not show their true performance, since the interactions collected from ChEMBL are based on protein–ligand interactions and not domain–ligand interactions. For instance, the domains d2nxyb1 and d2nxyb2 belong to different families, b.1.1.1 and b.1.1.3, respectively. If the ligands that bind to each of these domains were known, the performance of the ligand-based models might have improved. However, in our current setting, for each of these domains, we collected the same interacting ligands from ChEMBL, since their target identifiers are the same. Therefore, as expected we observed that these two domains were clustered together with ligand-based protein representation methods leading to a decrease in F-measure.
To test our methodology on single domain proteins of the A-50 dataset, we created a subset that contains only single domains and another that contains the rest of the sequences. SCOP stable domain identifier (sid) uses seven-charactered system in which the last character defines the domains uniquely (e.g. d2sqca1, d2sqca2 for several domain or d1n4qb_ when there is no need for domain specification). The single domain subset comprised sequences with sid ending with the ‘_’ character. Using the predicted clusters, we measured how accurately proteins of the single-domain were assigned by computing the percentage of True positives (TPs) (NTP/N) where N is the number of the samples in the subset and NTP is equal to the number of the correctly clustered samples of the subset. As expected, when only single domains were considered, we observed that both Protvec and SMILESVec had higher percentage of TPs. The performance of SMILESVec was increased from 0.743 for all proteins to 0.82 for single domain proteins. ProtVec had a slightly less pronounced increase from 0.757 to 0.829. On the other hand, when multidomain proteins were taken into account, the TP percentage reduced to 0.671 (SMILESVec) and 0.689 (ProtVec). These results suggest that taking domain information into account can enhance the performance of these representation methods.
4 Conclusion
In this study, we first propose a ligand-representation method, SMILESVec, which uses a word-embeddings model. Then, we represent proteins using their interacting ligands. In this approach, the interacting ligands of each protein in the dataset are collected. Then, the SMILES string of each ligand is divided into fixed-length overlapping substrings. These created substrings are then used to build real-valued vectors with the Word2vec model and then the vectors are combined into a single vector to represent the whole SMILES string. Finally, protein vectors are constructed by taking the average of the vectors of their ligands. The effectiveness of the proposed method in describing the proteins was measured by performing clustering on the A-50 dataset from the SCOP database using two different clustering algorithms, TransClust and MCL. Both of these clustering algorithms use protein similarity scores to identify cliques. SMILESVec-based protein representation was compared with other protein representation methods, namely BLAST and ProtVec, both of which depend on protein sequence to measure protein similarity, and the MACCS and ECFP binary fingerprint-based ligand-centric protein representation approaches. The performance of the clustering algorithms, as reported by F-measure, showed that protein word frequency-based similarity model was a better alternative to BLAST e-value or sequence identity to measure protein similarity. Furthermore, ligand-based protein representation methods also produced comparable F-measure scores to ProtVec.
Using SMILESVec, we were able to define proteins based on their interacting ligands even in the absence of sequence or structure information. SMILESVec-based protein representation had better clustering performance than BLAST and comparable clustering performance to protein word frequency-based method, both of which use protein sequences. We should emphasize that SCOP datasets were constructed based on protein similarity, thus high performance with the protein sequence-based models in family/super-family clustering is no surprise. However, the fact that ligand-based protein representation methods, either learning from SMILES or represented with binary compound features, perform as well as protein sequence-based models is quite intriguing and promising.
SMILESVec, MACCS and ECFP representations performed similarly in the task of protein clustering, suggesting that the word-embeddings approach that learns representations from a large SMILES corpus in an unsupervised manner is as accurate as widely adopted Fingerprint models. We propose that the ligand-based representation of proteins might reveal important clues especially in protein–ligand interaction related tasks like drug specificity or identification of proteins for drug targeting. The similarity between a candidate ligand and the SMILESVec for a protein can be used as an indicator for a possible interaction.
The study we conducted here also showed that SMILES description is sensitive to the database definition conventions; therefore, the use of SMILES strings requires careful consideration. Since we collected the protein–ligand interaction and ligand SMILES information from ChEMBL database to represent proteins, building SMILESVec vectors from the chemical words trained in ChEMBL SMILES corpus yielded better F-measure than the model in which the Pubchem SMILES corpus was used for training of the chemical words.
We showed that ligand-centric protein representation performed at least as well as protein sequence-based representations in the clustering task even in the absence of sequence information. Ligand-centric protein representation is only available for proteins with at least one known ligand interaction, while a sequence-based approach can miss key functional/mechanistic properties of the protein. The orthogonal information that can be obtained from the two approaches has been previously recognized (O’meara et al., 2016). As future work, we will investigate combining both sequence and ligand information in protein representation. We believe that this approach will provide a deeper understanding of protein function and mechanism toward the use of these representations in clustering and other bioinformatics tasks such as function annotation and prediction of novel protein–drug interactions.
Supplementary Material
Acknowledgements
TUBITAK-BIDEB 2211-E Scholarship Program (to H.O.) and BAGEP Award of the Science Academy (to A.O.) are gratefully acknowledged. We thank Prof. Kutlu O. Ulgen and Mehmet Aziz Yirik for helpful discussions.
Funding
This work was supported by Bogazici University Research Fund (BAP) [Grant Number 12304].
Conflict of Interest: none declared.
References
- Altschul S.F. et al. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Asgari E., Mofrad M.R. (2015) Continuous distributed representation of biological sequences for deep proteomics and genomics. PloS One, 10, e0141287.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakin K.V. (2009) Pharmaceutical Data Mining: Approaches and Applications for Drug Discovery, Vol. 6 John Wiley & Sons, New Jersey, USA. [Google Scholar]
- Bernardes J.S. et al. (2015) Evaluation and improvements of clustering algorithms for detecting remote homologous protein families. BMC Bioinformatics, 16, 34.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton E.E. et al. (2008) PubChem: integrated platform of small molecules and biological activities. Annu. Rep. Comput. Chem., 4, 217–241. [Google Scholar]
- Cai C. et al. (2003) Svm-prot: web-based support vector machine software for functional classification of a protein from its primary sequence. Nucleic Acids Res., 31, 3692–3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D.-S. et al. (2012) In silico toxicity prediction by support vector machine and smiles representation-based string kernel. SAR QSAR Environ. Res., 23, 141–153. [DOI] [PubMed] [Google Scholar]
- Cao R., Cheng J. (2016) Integrated protein function prediction by mining function associations, sequences, and protein–protein and gene–gene interaction networks. Methods, 93, 84–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandonia J.-M. et al. (2017) Scope: manual curation and artifact removal in the structural classification of proteins–extended database. J. Mol. Biol., 429, 348–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu Y.-Y. et al. (2014) Homopharma: a new concept for exploring the molecular binding mechanisms and drug repurposing. BMC Genomics, 15, S8.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou K.-C. (2001) Prediction of protein cellular attributes using pseudo-amino acid composition. Proteins, 43, 246–255. [DOI] [PubMed] [Google Scholar]
- Cokelaer T. et al. (2013) Bioservices: a common python package to access biological web services programmatically. Bioinformatics, 29, 3241–3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies M. et al. (2015) Chembl web services: streamlining access to drug discovery data and utilities. Nucleic Acids Res., 43, W612–W620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boom C. et al. (2016) Representation learning for very short texts using weighted word embedding aggregation. Pattern Recogn. Lett., 80, 150–156. [Google Scholar]
- Enright A.J. et al. (2002) An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res., 30, 1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox N.K. et al. (2014) Scope: structural classification of proteins–extended, integrating scop and astral data and classification of new structures. Nucleic Acids Res., 42, D304–D309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasca M., Cesa-Bianchi N. (2017) Multitask protein function prediction through task dissimilarity. IEEE/ACM Trans. Comput. Biol. Bioinformatics, doi: 10.1109/TCBB.2017.2684127. [DOI] [PubMed]
- Gaulton A. et al. (2011) ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res, 40, D1100–D1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hert J. et al. (2008) Quantifying the relationships among drug classes. J. Chem. Inform. Model., 48, 755–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J.X. et al. (2016) Network biology concepts in complex disease comorbidities. Nat. Rev. Genet., 17, 615–629. [DOI] [PubMed] [Google Scholar]
- Iqbal M.J. et al. (2013) A distance-based feature-encoding technique for protein sequence classification in bioinformatics In: Computational Intelligence and Cybernetics (CYBERNETICSCOM), 2013 IEEE International Conference on, pp.1–5. IEEE, Yogyakarta, Indonesia. [Google Scholar]
- Jaeger S. et al. (2018) Mol2vec: unsupervised machine learning approach with chemical intuition. J. Chem. Inform. Model., 58, 27–35. [DOI] [PubMed] [Google Scholar]
- Jain R. et al. (2017) Mechanism of error-free dna synthesis across n1-methyl-deoxyadenosine by human dna polymerase-ι. Sci. Rep., 7, 43904.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jastrzębski S. et al. (2016) Learning to SMILE (S). In: International Conference on Learning Representations, ICLR 2016 - Workshop Track. May 2, 2016 - May 4, 2016. San Juan, Puerto Rico.
- Keiser M.J. et al. (2007) Relating protein pharmacology by ligand chemistry. Nat. Biotechnol., 25, 197.. [DOI] [PubMed] [Google Scholar]
- Martin A.C. et al. (1998) Protein folds and functions. Structure, 6, 875–884. [DOI] [PubMed] [Google Scholar]
- Mikolov T. et al. (2013) Distributed representations of words and phrases and their compositionality In: Advances in Neural Information Processing Systems. pp. 3111–3119. Lake Tahoe, Nevada, USA. [Google Scholar]
- Murzin A.G. et al. (1995) SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol., 247, 536–540. [DOI] [PubMed] [Google Scholar]
- Nascimento A.C. et al. (2016) A multiple kernel learning algorithm for drug-target interaction prediction. BMC Bioinformatics, 17, 46.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’meara M.J. et al. (2016) Ligand similarity complements sequence, physical interaction, and co-expression for gene function prediction. PloS One, 11, e0160098.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Öztürk H. et al. (2015) Classification of beta-lactamases and penicillin binding proteins using ligand-centric network models. PloS One, 10, e0117874.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Öztürk H. et al. (2016) A comparative study of smiles-based compound similarity functions for drug-target interaction prediction. BMC Bioinformatics, 17, 128.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadatos G., Overington J.P. (2014) The chembl database: a taster for medicinal chemists. Future, 6, 361–364. [DOI] [PubMed] [Google Scholar]
- Pearson K. (1895) Note on regression and inheritance in the case of two parents. Proc. Roy. Soc. Lond., 58, 240–242. [Google Scholar]
- Peón A. et al. (2016) How reliable are ligand-centric methods for target fishing? Front. Chem., 4, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poornima P. et al. (2016) Network pharmacology of cancer: from understanding of complex interactomes to the design of multi-target specific therapeutics from nature. Pharmacol. Res., 111, 290–302. [DOI] [PubMed] [Google Scholar]
- Řehůřek R., Sojka P. (2010) Software Framework for Topic Modelling with Large Corpora. In: Proceedings of the LREC 2010 Workshop on New Challenges for NLP Frameworks, pp. 45–50. ELRA, Valletta, Malta.
- Rogers D., Hahn M. (2010) Extended-connectivity fingerprints. J. Chem. Inform. Model., 50, 742–754. [DOI] [PubMed] [Google Scholar]
- Santiago J.A., Potashkin J.A. (2014) A network approach to clinical intervention in neurodegenerative diseases. Trends Mol. Med., 20, 694–703. [DOI] [PubMed] [Google Scholar]
- Sawada R. et al. (2014) Benchmarking a wide range of chemical descriptors for drug-target interaction prediction using a chemogenomic approach. Mol. Inform., 33, 719–731. [DOI] [PubMed] [Google Scholar]
- Schenone M. et al. (2013) Target identification and mechanism of action in chemical biology and drug discovery. Nat. Chem. Biol., 9, 232–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz J. et al. (2013) Smifp (smiles fingerprint) chemical space for virtual screening and visualization of large databases of organic molecules. J. Chem. Inform. Model., 53, 1979–1989. [DOI] [PubMed] [Google Scholar]
- Shi J.-Y. et al. (2015) Predicting drug–target interaction for new drugs using enhanced similarity measures and super-target clustering. Methods, 83, 98–104. [DOI] [PubMed] [Google Scholar]
- Vidal D. et al. (2005) Lingo, an efficient holographic text based method to calculate biophysical properties and intermolecular similarities. J. Chem. Inform. Model., 45, 386–393. [DOI] [PubMed] [Google Scholar]
- Willighagen E.L. et al. (2017) The chemistry development kit (cdk) v2. 0: atom typing, depiction, molecular formulas, and substructure searching. J. Cheminform., 9, 33.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittkop T. et al. (2010) Partitioning biological data with transitivity clustering. Nat. Methods, 7, 419–420. [DOI] [PubMed] [Google Scholar]
- Yang J. et al. (2004) Altered dna polymerase ι expression in breast cancer cells leads to a reduction in dna replication fidelity and a higher rate of mutagenesis. Cancer Res., 64, 5597–5607. [DOI] [PubMed] [Google Scholar]
- Zou S. et al. (2016) Dna polymerase iota (pol ι) promotes invasion and metastasis of esophageal squamous cell carcinoma. Oncotarget, 7, 32274.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.