Abstract
Lipids are critical for the function of membrane proteins. NADPH-cytochrome-P450-reductase, the sole electron transferase for microsomal oxygenases, possesses a conformational dynamics entwined with its topology. Here, we use peptide-nanodiscs to unveil cytochrome-P450-reductase’s lipid boundaries, demonstrating a protein-driven enrichment of ethanolamine lipids (by 25%) which ameliorates by 3-fold CPR’s electron-transfer ability.
Keywords: NADPH-Cytochrome P450 Reductase, Nanodiscs, Electron Transfer, Lipid ordering, Membrane protein
Graphical abstract
NADPH-Cytochrome P450 reductase (CPR) is the electron transferase of the microsomal monooxygenase system. CPR is the obligate electron donor to a variety of enzymatic systems, including cytochrome P450s and heme oxygenases, among others.1 Being the only reducing partner in the endoplasmic reticulum, it is a key component of human detox and biosynthetic machineries, and its physiology and pharmacology have emerged in the last decade.2, 3 CPR binds NADPH, which can reduce the enzyme by transferring electrons to both the FAD and FMN domains within the protein.1 The NADPH/FAD domain is connected to the FMN domain by a hinge region that closes when NADPH binds to CPR and transfers two electrons through a hydride ion transfer, followed by interflavin electron transfer.4-6 The open form of the enzyme is the responsible for delivering the electrons to the acceptor protein (i.e. cytochrome P450).1, 7, 8 CPR is found in the cytoplasmic side of the endoplasmic reticulum (ER) of eukaryotic cells, and possesses a hydrophobic N-terminal ER membrane-binding domain. The topology of CPR in ER membrane is dramatically altered by its functional conformational changes.8 Several studies have demonstrated that CPR catalytic activity can be reduced or enhanced by the interaction with specific phospholipids.9 CPR‘s interactions with anionic phospholipids have been controversial,10, 11 although an efficient tuning on CPR’s redox potential has been demonstrated.9 The importance of CPR’s stability on catalysis for the physiology of the endoplasmic reticulum has been recently recognized,3 although the role played by the membrane has been surprisingly neglected. It is imperative to fill this scientific gap and provide critical knowledge on the molecular factors regulating CPR’s stability and interprotein electron transfer. Recently, we introduced the use of peptide-based nanodiscs to overcome such limitations by studying lipid-protein interactions at nanometric scale and unveil protein-dependent lipid segregation in a microsomal P450.12 Here, for the first time, we report the CPR’s interaction with a biomimetic ER (Table S1) and its preference for lipid composition using peptide based nanodiscs and biophysical experiments.5, 12, 13 The findings of this study contribute to the ongoing efforts of exploring and characterizing the important roles of lipids in biological functions.14
Peptide-based nanodiscs allow detergent-free protein incorporation, and avoids post-reconstitution purification steps.15, 16 Size-exclusion chromatography (SEC, Figure S1a,b), dynamic light scattering (DLS, Figure S1c,d) and transmission electron microscopy (Figure S1d) indicate the formation of homogeneous nanodiscs. The size of 4F peptide-nanodiscs can be tuned by altering the lipid:peptide ratio;12 at 1:0.8 w/w lipid:peptide ratio (for all preparations), the diameter of the nanodisc was 8±1 nm, which is suitable for protein monomerization. We were able to reconstitute CPR in 4F-nanodiscs in homogeneous (POPC) and heterogeneous (POPC:POPS 8:2 and ER) lipid compositions. As recently demonstrated, peptide-based nanodiscs allow lipid “exchange” between nanodiscs.13, 15-17 The presence of a membrane protein in a fraction of nanodiscs perturbs the mass equilibrium in a way that reflects the ability of the protein itself to recruit or exclude specific membrane components in its proximate boundaries. CPR was reconstituted in excess 4F-nanodiscs (Figure 1a), the fraction containing the protein was subsequently purified by SEC (Figure 1b), and phospholipids and cholesterol were quantified (Figure 2a,b). The lipid composition uniquely reflects the perturbation induced by CPR. Since the beginning of P450 research, studies have reported that specific lipids preferably associate with CPR function. Balvers and coworkers10 found residual PI and PS bound to CPR purified from rat microsomes, whereas others have reported an enrichment of PE.11 Both studies used detergents to extract CPR from microsomal membrane.18 The lipid-exchange nanodiscs approach described here surmounts the limitations of detergents in characterizing protein-dependent membrane organization. The first observation is that, compared to the initial ER lipid composition (“ER” in the rest of the manuscript), the phospholipid profiles were altered by CPR (Figure 2b and Table S1). After lipid exchange, the nanodiscs (denoted as “ERex”) did not contain any detectable PS, but instead a higher amount of zwitterionic PE (+6%, p<0.05), and PI, although this is not statistically significant (+4%, p<0.28). On the contrary, the empty nanodiscs fraction was enriched with PS and depleted of PE (Figure S2). Cytb5 – whose cytosolic does not interact with membrane19 – was used as a control, and as expected, it did not perturb the nanodiscs’s lipid composition (Figure 2b). Preferential binding of PS to CPR has been justified by the 46Lys-Lys-Lys triplet in the transmembrane domain,9 which also contains an acidic 49Glu-Glu-x-x-Glu sequence20 that precedes the cytosolic side of the protein.21 In a recent report, super-resolution imaging of fluorescently labeled CPR in a 2:1 PC:PS mole ratio supported lipid bilayer showed its exclusion from PS-rich domains22 corroborating our findings. PE has a cone-shaped structure favoring a negative-curvature strain of the membrane,23 which might adapt CPR to the lipid bilayer,10, 24 thus the observed enrichment may be functional to CPR membrane topology.5, 21 These findings underlined that detergent-based approaches are inappropriate for studying membrane-protein topology, and most significant observations can be attained using nanodiscs and other biomimetics which better resemble physiological membranes.12, 16, 25
Figure 1. Peptide-based nanodiscs as a tool to unveil lipid-boundary regions of CPR.
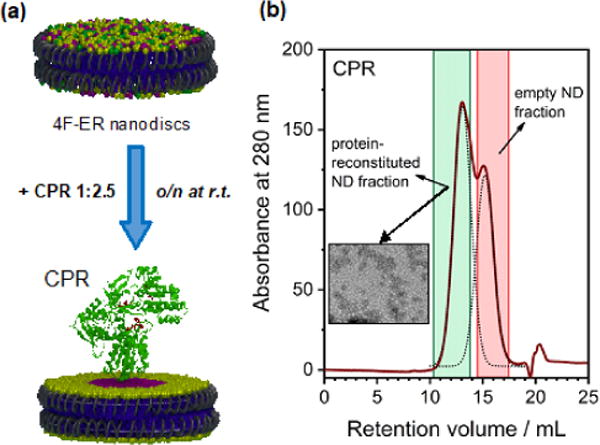
4F-ER nanodiscs allow detergent-free incorporation and monomerization of CPR. (a) Schematic of the lipid-exchange experiment. CPR was reconstituted with a 2.5-fold excess of empty 4F peptide based nanodiscs containing ER membrane lippids and incubated overnight to enable lipid exchange between nanodiscs. (b) SEC profiles showing two partially overlapped peaks, representing the reconstituted and empty fractions; the insert depicts a TEM image of the reconstituted fraction.
Figure 2. CPR lipid boundaries differ from membrane bulk composition and determine topological and structural fluctuations.
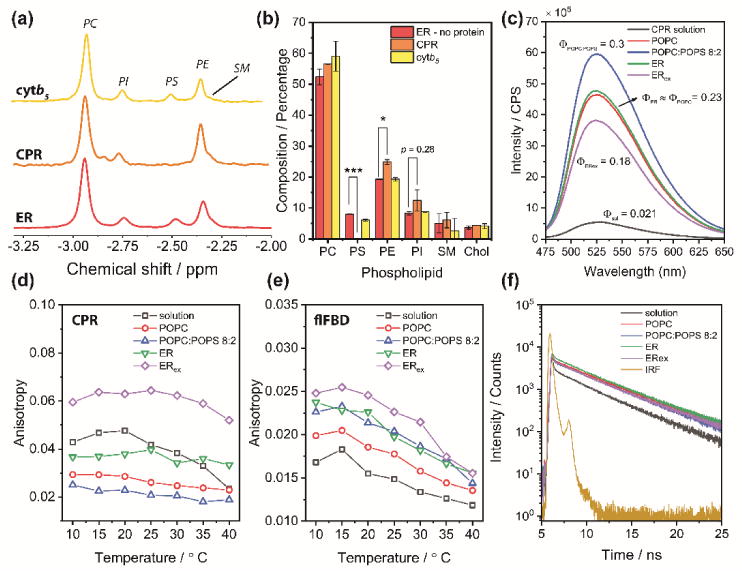
(a) 31P-NMR on detergent-treated nanodiscs was used to assess the phospholipid composition on the protein-containing fractions after overnight lipid exchange. (b) Percentage of lipids and cholesterol contents in empty, CPR, and cytb5 4F-ER nanodiscs after lipid exchange, as measured by 31P-NMR (phospholipids) and GC-MS (cholesterol). Data are expressed as average ± standard deviation (n = 3). * p<0.05, ** p<0.01, *** p<0.001. (c) Steady-state fluorescence emission of CPR in the 4F-nanodiscs (λex=450 nm). Φi is the relative quantum yield (to flavin) measured as described in the Experimental Section. (d–e) Steady-state fluorescence anisotropy of CPR and flFBD in solution and 4F-nanodiscs (λex=450 nm, λem = 525 nm). (f) Fluorescence lifetime decays measurements of CPR in solution or in 4F-nanodiscs nanodiscs (λex=450 nm, λem = 525 nm). IRF, instrument response function.
Next, we investigated CPR’s rotational mobility as well as structural fluctuations that can be attributed to the differences in membrane immersion due to preferential lipid boundaries. For this, we used the native flavin to study both steady-state and time-dependent fluorescence emissions. CPR possesses both an FMN and a FAD moiety, located in different domains. Flavin in the FAD domain is mostly silent,26 so the observed fluorescence can be attributed almost entirely to the isoalloxazine ring belonging to the FMN binding domain (FBD). When excited at the absorbance maximum (450 nm), CPR showed a typical fluorescence emission with a peak at 525 nm. Differences were observed between membrane-free and nanodiscs samples. CPR’s relative quantum yield (Φ) was higher in membrane (Figure 2c), with a ~10-fold increase in POPC:POPS; however, lipid exchange (ERex) led to a comparatively lower emission. This is a direct consequence of protein/membrane interactions: if CPR is more embedded into the lipid bilayer, the FBD microenvironment would be consequently more rigid and less solvent exposed, allowing flavin’s radiationless deactivation pathways to be reduced and the relative higher. Φ Temperature-dependent steady-state anisotropy was also measured (Figure 2d). The anisotropy experiments unveil that: (i) CPR possesses a significant higher anisotropy in ERex compared to POPC or POPC-POPS membranes, and (ii) CPR’s rotational mobility in the membrane is not affected by the thermally increased membrane fluidity.5, 8 Both results indicate that the preferential membrane allows the FBD domain to better interact with the lipid bilayer, decreasing its rotational mobility. Similar experiments performed with the isolated full-length FBD (flFBD, Figure 2e),27 which contains the entire transmembrane domain and the FMN-binding domain.28 For the flFBD, the observed anisotropy values were significantly lower, due to reduced mass (ca. 17-kDa) compared to full-length CPR (ca. 77-kDa). Going from POPC to the preferential ERex, a progressive reduction of rotational mobility was observed, indicating that membrane can indeed exert a significant effect on the immersion of FBD domain. Contrary to CPR, flFBD’s anisotropy behavior was temperature-dependent, suggesting that CPR’s membrane topology likely differs from that of flFBD. When flFBD was studied in lipid bicelles, it was reported A combination of solution and solid-state NMR experiments has been used to reveal distinct time scales of motion for the transmembrane and cytosolic domains and also shown to be dependent on the lipid composition.28 Unfortunately, no structural studies have been performed on the full-length CPR, although it is believed that extended fragments of CPR’s cytosolic domain also interact with the lipid bilayer,21 explaining the different anisotropic behavior.
Differences in Φ cannot be accounted only to the extent of protein/membrane interface, so we investigated if structural alterations can alter flavin time-dependent fluorescence decays. CPR’s fluorescence lifetime in its soluble truncated form has been characterized29 and has been shown to be multiexponential, as for other flavoproteins, including the cytosolic nitric oxide synthase.30 The observed complex fluorescence behavior is attributable to the structural heterogeneity of reductase with respect to the flavin environment.29 If the fluctuations of the protein architecture occurs on a time scale comparable with the fluorescence lifetime, multiple decays will be observed. When in solution, CPR’s fluorescent decay was biphasic, with a slower component comparatively matching free-FMN (Table S2). This can represent an “open” form population, with FAD distant from the FBD.30, 31 When incorporated into nanodiscs, the decay curves became more complex, and a good fitting was achieved only by considering a tri-exponential function (Figure 2e). When in PE-containing membrane, CPR had lower average lifetime 〈τ〉, and the ~6 ns decay was split into two components (Table S2). A short lifetime population (~2 ns) indicates that membrane facilitates either FAD/FMN domains interactions or π-stacking of the isoalloxazine ring with aromatic residues (Figure S3). A third slower decay component (~8-10 ns), representing ~2% of the total decay, was not observed in previous reports,29 and is likely attributable to the lipid environment which reshapes flavin photophysics and delays non-radiative decay (Fig. 2c). As for anisotropy, fluorescence decays were also measured for flFBD (Table S2 and Figure S4). Solution flFBD decays as free FMN, with a shorter component τ1 of ~4 ns (Table S2). Interestingly, membrane reconstitution significantly decreased both τ1 and 〈τ〉, indicating that indeed the lipid environment alters flFBD’s topology and flavin’s microenvironment.
Fluorescence experiments clearly insinuate lipid-dependent structural fluctuations. The conformational plasticity of CPR is key for its catalytic abilities, thus we expected differences in electron transfer rates. For this, we measured the CPR‘s ability to reduce cyt c (Figure 3). Cyt c is a soluble hemeprotein, which has been extensively used as P450’s surrogate for its high homology in the redox interface.32 The catalytic behavior strongly supports the spectroscopic observations. A progressive increase in kcat values was observed from solution (52 s−1) to ER (110 s−1); however, after lipid exchange and PE-enrichment the electron transfer ability was further improved (175 s−1, p<0.001). NADPH-consumption rates mirrored cyt c reduction (Figure S5). A recent study in MSP-nanodiscs found that CPR was more active towards cyt c in a ER lipid mixture;33 however, the highest catalysis was obtained using a native lipid extract. Our findings support that CPR’s preference for PE and exclusion of PS is indeed linked to its function of an improved electron transfer ability. Membrane-induced structural fluctuations revealed by a decrease of 〈τ〉 and the appearance of a shorter decay component (Table S2) supports a more “compact” CPR in ERex.31 If more compact, intermolecular electron transfer between the FAD and FBD domains will be facilitated, favoring a faster reduction rate (Figure 3).
Figure 3. The preferential lipid boundaries increase CPR’s ability to transfer electrons to cyt c.
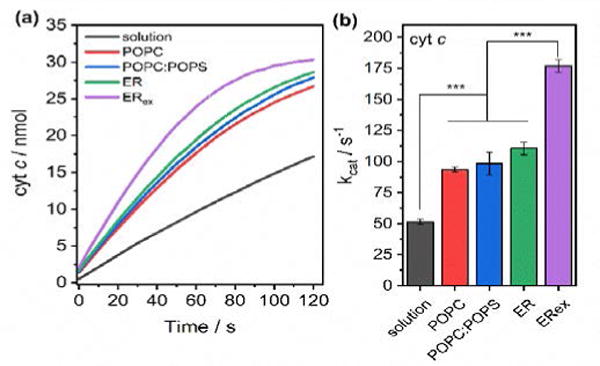
a) Kinetic traces of cyt c reduction as measured by UV-vis absorption spectroscopy. b) Calculated kcat for cyt c reduction by CPR in solution or in 4F-nanodiscs. Data are expressed as average ± standard deviation (n = 3). * p<0.05, ** p<0.01, *** p<0.001.
In conclusion, this study reveals for the first time that CPR topology in an ER biomimetic is defined by its ability to exclude the negatively charged phospholipid PS and increase the cone-shaped PE. The protein-induced lipid segregation enhances CPR’s catalytic activity, when compared to homogeneous lipid environment,5, 33, 34 probably due to conformational fluctuations that favour proximity of the intra-protein ET pathway. Previous findings on residual PS binding to CPR may probably be influenced due to detergent-mediated solubilization. This study also demonstrated that CPR‘s lipid surrounding are somehow complementary to that of P450, suggesting monomerization into the ER occurs via protein-induced lipid-ordering.12 Co-existence of P450 and CPR in rafts domain has also been proposed;34 however, this view collides with CPR promiscuous activity towards numerous oxidative partners, which can be achieved only assuming a more disordered and distinct lipid environment for CPR. We expect that the observed differences can be potentially exacerbated in the highly heterogeneous endoplasmic reticulum. These findings add novel information regarding the factors that control the stability and dynamics of CPR, and the reported experimental strategies can be used for characterizing the topology of CPR’s polymorphic variants2, 3 and their relation with function deficiencies. We believe that the approach reported in this study will be valuable to study the structure, dynamics and function of membrane proteins. Particularly, the ability to undergo lipid exchange via collisions by the self-assembled peptide-based lipid-nanodiscs would be useful to probe protein-protein, protein-membrane and protein-drug interactions.
Supplementary Material
Acknowledgments
This study was supported by NIH (GM084018 to A. R.). We thank Matthew Schweiss for assistance with GC-MS, the SMART center (NSF DBI-0959823 to Dr. Nils Walter) and Damon Hoff for assistance with Alba Confocal Microscope.
Footnotes
Conflicts of interest: There are no conflicts to declare.
Notes and references
- 1.Murataliev MB, Feyereisen R, Walker FA. Biochimica et biophysica acta. 2004;1698:1–26. doi: 10.1016/j.bbapap.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 2.Riddick DS, Ding X, Wolf CR, Porter TD, Pandey AV, Zhang QY, Gu J, Finn RD, Ronseaux S, McLaughlin LA, Henderson CJ, Zou L, Fluck CE. Drug metabolism and disposition: the biological fate of chemicals. 2013;41:12–23. doi: 10.1124/dmd.112.048991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCammon KM, Panda SP, Xia C, Kim JJ, Kranendonk M, Auchus RJ, Lafer EM, Ghosh D, Martasek P, Kar R, Masters BS, Roman LJ. J Biol Chem. 2016;291:20487–20502. doi: 10.1074/jbc.M116.716019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang WC, Ellis J, Moody PC, Raven EL, Roberts GC. Structure. 2013;21:1581–1589. doi: 10.1016/j.str.2013.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnaba C, Martinez MJ, Taylor E, Barden AO, Brozik JA. J Am Chem Soc. 2017;139:5420–5430. doi: 10.1021/jacs.7b00663. [DOI] [PubMed] [Google Scholar]
- 6.Galiakhmetov AR, Kovrigina EA, Xia C, Kim JJP, Kovrigin EL. J Biomol NMR. 2018;70:21–31. doi: 10.1007/s10858-017-0152-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gentry KA, Zhang M, Im SC, Waskell L, Ramamoorthy A. Chem Comm. 2018 doi: 10.1039/c8cc02525h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barnaba C, Taylor E, Brozik JA. J Am Chem Soc. 2017;139:17923–17934. doi: 10.1021/jacs.7b08750. [DOI] [PubMed] [Google Scholar]
- 9.Das A, Sligar SG. Biochemistry. 2009;48:12104–12112. doi: 10.1021/bi9011435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balvers WG, Boersma MG, Vervoort J, Ouwehand A, Rietjens IMCM. Eur J Biochem. 1993;218:1021–1029. doi: 10.1111/j.1432-1033.1993.tb18461.x. [DOI] [PubMed] [Google Scholar]
- 11.Narayanasami R, Otvos JD, Kasper CB, Shen A, Rajagopalan J, McCabe TJ, Okita JR, Hanahan DJ, Masters BSS. Biochemistry. 1992;31:4210–4218. doi: 10.1021/bi00132a009. [DOI] [PubMed] [Google Scholar]
- 12.Barnaba C, Sahoo BR, Ravula T, Medina-Meza IG, Im SC, Anantharamaiah GM, Waskell L, Ramamoorthy A. Angewandte Chemie. 2018;57:1–6. doi: 10.1002/anie.201713167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ravula T, Ishikuro D, Kodera N, Ando T, Anantharamaiah GM, Ramamoorthy A. Chem Mater. 2018 doi: 10.1021/acs.chemmater.8b00946. [DOI] [Google Scholar]
- 14.Harayama T, Riezman H. Nat Rev Mol Cell Biol. 2018 doi: 10.1038/nrm.2017.138. [DOI] [PubMed] [Google Scholar]
- 15.Midtgaard SR, Pedersen MC, Kirkensgaard JJ, Sorensen KK, Mortensen K, Jensen KJ, Arleth L. Soft Matter. 2014;10:738–752. doi: 10.1039/c3sm51727f. [DOI] [PubMed] [Google Scholar]
- 16.Ravula T, Barnaba C, Mahajan M, Anantharamaiah GM, Im SC, Waskell L, Ramamoorthy A. Chem Comm. 2017;53:12798–12801. doi: 10.1039/c7cc07520k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arenas RC, Danielczak B, Martel A, Porcar L, Breyton C, Ebel C, Keller S. Sci Rep. 2017;7:45875. doi: 10.1038/srep45875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lichtenberg D, Goñi FM, Heerklotz H. Trends in Biochem Sci. 2005;30:430–436. doi: 10.1016/j.tibs.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 19.Barnaba C, Gentry K, Sumangala N, Ramamoorthy A. F1000 Research. 2017;6:662. doi: 10.12688/f1000research.11015.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang M, Roberts DL, Paschke R, Shea TM, Masters BSS, Kim JJP. Proc Natl Acad Sci USA. 1997;94:8411–8416. doi: 10.1073/pnas.94.16.8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mast N, Liao WL, Pikuleva IA, Turko IV. Arch Biochem Biophys. 2009;483:81–89. doi: 10.1016/j.abb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barden AO, Goler AS, Humphreys SC, Tabatabaei S, Lochner M, Ruepp MD, Jack T, Simonin J, Thompson AJ, Jones JP, Brozik JA. Neuropharmacol. 2015;98:22–30. doi: 10.1016/j.neuropharm.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 23.Epand RM. Biochimica et biophysica acta. 1998;1376:353–368. doi: 10.1016/s0304-4157(98)00015-x. [DOI] [PubMed] [Google Scholar]
- 24.Zimmerberg J, Kozlov MM. Nat Rev Mol Cell Biol. 2006;7:9. doi: 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]
- 25.Denisov IG, Sligar SG. Chem Rev. 2017;117:4669–4713. doi: 10.1021/acs.chemrev.6b00690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brunner K, Tortschanoff A, Hemmens B, Andrew PJ, Mayer B, Kungl AJ. Biochemistry. 1998;37:17545–17553. doi: 10.1021/bi981138l. [DOI] [PubMed] [Google Scholar]
- 27.Prade E, Mahajan M, Im SC, Zhang M, Anantharamaiah GM, Waskell L, Ramamoorthy A. Angewandte Chemie. 2018 doi: 10.1002/anie.201802210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang R, Yamamoto K, Zhang M, Popovych N, Hung I, Im SC, Gan Z, Waskell L, Ramamoorthy A. Biophys J. 2014;106:2126–2133. doi: 10.1016/j.bpj.2014.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bastiaens PI, Bonants PJ, Mueller F, Visser AJ. Biochemistry. 1989;28:8416–8425. doi: 10.1021/bi00447a022. [DOI] [PubMed] [Google Scholar]
- 30.Ghosh DK, Ray K, Rogers AJ, Nahm NJ, Salerno JC. FEBS J. 2012;279:1306–1317. doi: 10.1111/j.1742-4658.2012.08525.x. [DOI] [PubMed] [Google Scholar]
- 31.Bavishi K, Li DR, Eiersholt S, Hooley EN, Petersen TC, Moller BL, Hatzakis NS, Laursen T. Sci Rep. 2018;8:6817. doi: 10.1038/s41598-018-24922-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gentry KA, Prade E, Barnaba C, Zhang M, Mahajan M, Im SC, Anantharamaiah GM, Nagao S, Waskell L, Ramamoorthy A. Sci Rep. 2017;7:7793. doi: 10.1038/s41598-017-08130-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu KC, Hughes JM, Hay S, Scrutton NS. FEBS J. 2017;284:2302–2319. doi: 10.1111/febs.14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brignac-Huber L, Reed JR, Backes WL. Mol Pharm. 2011;79:549–557. doi: 10.1124/mol.110.068817. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


