Abstract
Background
The aim of this study was to assess the expression profile of the KIF26B gene, its effects on ovarian cancer cell behavior in vitro, the clinical prognostic value of expression of the KIF26B gene and associated signaling pathways, from bioinformatics data analysis.
Material/Methods
The Cancer Genome Atlas-Ovarian Cancer (TCGA-OV) database and the online Kaplan-Meier plotter for ovarian cancer were analyzed. Human ovarian cancer cell lines A2780 and SKOV3 were used to study the in vitro effects of KIF26B gene expression on cell proliferation and cell invasion. Genes that interacted with, co-localized to, and were co-expressed with the KIF26B gene (Pearson’s r ≥0.6) were identified and underwent Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis.
Results
Increased expression of the KIF26B gene increased the proliferation and migration of A2780 and SKOV3 cells in vitro. KIF26B protein expression was upregulated in ovarian cancer tissue and was associated with lymphatic and venous invasion. TCGA-OV data analysis showed that increased expression of the KIF26B gene was significantly associated with reduced 3-year, 5-year, and 10-year overall survival (OS), progression-free survival (PFS), and post-progression survival (PPS). The genes that interacted with, co-localized to, and were co-expressed with KIF26B were enriched in several KEGG pathways; upregulation of the KIF26B gene was associated with TGF-β signaling pathway.
Conclusions
Upregulation of KIF26B enhanced proliferation and migration of ovarian cancer cells in vitro. Bioinformatics analysis supported the association between increased expression of the KIF26B gene and reduced clinical outcome in patients with ovarian carcinoma.
MeSH Keywords: Lymphatic Metastasis, Ovarian Neoplasms, Prognosis
Background
Epithelial ovarian cancer is a common gynecological malignancy and is one of the leading causes of cancer-related death among women [1]. Ovarian cancers can be derived from the ovarian surface epithelium, inclusion cysts, or the Fallopian tube and are a group of heterogeneous tumors [2–4]. The molecular processes that underlie the onset and progression of ovarian cancer remain poorly understood [2,5,6]. Therefore, to support future development of individualized treatment regimens, it is necessary to explore the genetic and epigenetic dysregulations involved in ovarian carcinoma [7].
The human kinesin family member 26B (KIF26B) gene, which maps to 1q44, encodes a member of the kinesin family protein [8]. In a mouse model of embryogenesis, the KIF26B gene is specifically involved in the development of limbs, face, and somites [8]. In embryonic kidney development, KIF26B plays a critical role by regulating the adhesion of mesenchymal cells in contact with ureteric buds [9]. Some recent studies also found that upregulated the KIF26B gene contribute to oncogenesis and confers malignant behaviors of some cancers. Increased expression of KIF26B was correlated with larger tumor size, higher risk of metastases and poor prognosis of breast cancer [10], gastric cancer [11], and colorectal cancer [12]. Overexpression of the KIF26B gene can also increase chemoresistance in osteosarcoma [13].
In ovarian cancer, the role of some kinesin family members has been gradually revealed. For example, high KIF14 expression in ovarian cancer might be an independent predictor of worse outcomes [14]. Increased expression of the KIFC1 gene has been proposed to be a potential prognostic biomarker for metastatic disease, of increased stage, in patients with ovarian cancer [15]. However, the expression profiles, prognostic value, and functional role of the KIF26B gene in ovarian cancer are still poorly characterized.
The aim of this study was to assess the expression profile of the kinesin family member 26B (KIF26B) gene, and its effects on ovarian cancer cell behavior in vitro, and the clinical prognostic value the KIF26B gene, and the possible signaling pathways involved in its effects, from bioinformatics data analysis.
Material and Methods
Data analysis from the Human Protein Atlas
The kinesin family member 26B (KIF26B) gene expression at the RNA and protein level in normal and malignant ovarian tissue was reviewed by data mining in the Human Protein Atlas (http://www.proteinatlas.org) [16,17]. Images of KIF26B protein immunohistochemistry (IHC) staining in normal ovarian tissues and ovarian cancer tissues were downloaded from this database.
Data analysis from The Cancer Genome Atlas-Ovarian Cancer (TCGA-OV)
Secondary analysis of the data from The Cancer Genome Atlas-Ovarian Cancer (TCGA-OV) was performed using the University of California Santa Cruz (UCSC) Xena Browser (http://xena.ucsc.edu). Expression of the KIF26B gene mRNA in normal ovarian tissues and in ovarian cancer tissues was compared. Online Kaplan-Meier plotting data of overall survival (OS) of patients with primary serous ovarian cancer were generated. The patients were classified into high and low KIF26B expression groups according to the median KIF26B gene expression levels.
Bioinformatics analysis using the online Kaplan-Meier plotter
The association between the expression levels of the KIF26B gene and overall survival (OS), first progression-free survival (FPS), or post-progression survival (PPS) in ovarian cancer patients were investigated by using the online Kaplan-Meier plotter, which is an online database for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data of 1,287 ovarian cancer patients (http://kmplot.com/analysis/) [18]. Median KIF26B mRNA expression was used as the cutoff. The hazard ratio (HR) with 95% confidence intervals (CI), log-rank p-value, as well as median survivals, were calculated.
Cell culture and transfection
Human ovarian cancer cell lines A2780 and SKOV3 were obtained from American Type Culture Collection (ATCC) (Manassas, VA, USA). The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum, 100 U/ml penicillin and 100 U/ml streptomycin, in a humidified incubator with 5% CO2 at 37°C.
Full-length KIF26B cDNA was cloned into a pcDNA3.1 vector, and the recombinant plasmids were termed as pcDNA3.1-KIF26B. The A2780 and SKOV3 cells were transfected with the recombinant plasmids using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Lentiviral KIF26B shRNA was obtained from OriGene (Rockville, MD, USA). The A2780 and SKOV3 cells were infected with the lentiviral particles, or the empty controls, in the presence of polybrene, 24 hours after transfection or infection, and the cells then underwent cell counting kit-8 (CCK-8) or transwell assays.
Western blot analysis
Cell samples were lysed using RIPA buffer containing protease and phosphatase inhibitors. Protein concentrations were determined with a BCA assay (Cat. No. 23227) (Thermo Scientific, Waltham, MA, USA). Then, 30 μg of protein was separated in a 10% SDS-PAGE gel and was transferred to nitrocellulose membranes. The membranes were blocked, washed, and incubated overnight with primary antibodies, including anti-KIF26B (1: 200) (ab121992) (Abcam, Cambridge, UK), anti-p-SMAD2 (1: 1000) (ab53100) (Abcam, Cambridge, UK), anti-SMAD2 (1: 2000) (ab40855) (Abcam, Cambridge, UK), or anti-β-actin (1: 1000) (sc-47778) (OriGene, Rockwell, MD, USA). After 24 hours, the membranes were washed three times with TBST, incubated with appropriate secondary antibody conjugated to horseradish peroxidase (HRP) for 30 min at room temperature. The membranes were then washed three times with TBST. The signals were developed using SuperSignal West Femto Maximum Sensitivity Substrate, which is an ultra-sensitive enhanced chemiluminescent (ECL) substrate (Thermo Scientific).
CCK-8 assay of cell proliferation
The cell counting kit-8 (CCK-8) assay (Dojindo, Kumamoto, Japan) was used to assess cell proliferation, according to the manufacturer’s instruction. Briefly, transfected cells (1×104 cells/well) were seeded onto a 96-well plate and incubated for 24, 48, 72, or 96 hours, respectively. At the indicated time points, 10 μl of the CCK-8 solution was added to each well and incubated for another 4 hours. Absorbance at 450 nm was measured on a microplate reader.
Transwell assay of cell invasion
Transwell assay of cell invasion was conducted using standardized conditions with BD BioCoat Matrigel invasion chambers (BD Biosciences, San Jose, CA, USA) according to the manufacturer’s protocol. Briefly, 20% FBS in the lower chamber was used as the chemoattractant. Non-invading cells on the upper surface of the membrane were removed, while the invading cells were fixed and stained with 0.1% crystal violet, 24 hours after seeding. The cells that invaded through the filter were quantified by counting the entire area of each filter using a grid and a light microscope at ×20 magnification.
Bioinformatic analysis using Genemania, cBioPortal for Cancer Genomics, and ClueGo
Data mining was performed using Genemania (http://genemania.org/) to identify the known proteins with interactions or co-localization with KIF26B.The genes strongly co-expressed with the KIF26B gene (Pearson’s r ≥0.6) in TCGA-OV were identified by using cBioPortal for Cancer Genomics (www.cbioportal.org). Then, the genes were loaded into the ClueGo in Cytoscape plug-in [19], to analyze their enrichment in the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (p≤0.05).
Statistical analysis
Continuous variables were reported as the mean ± standard deviation (SD). Comparisons were performed by two-tailed Student’s t-test. The log-rank test was used to assess the significance of the difference between survival curves. A p-value <0.05 was considered as statistically significant.
Results
The KIF26B gene was not expressed in normal ovarian tissues but was upregulated in ovarian cancer
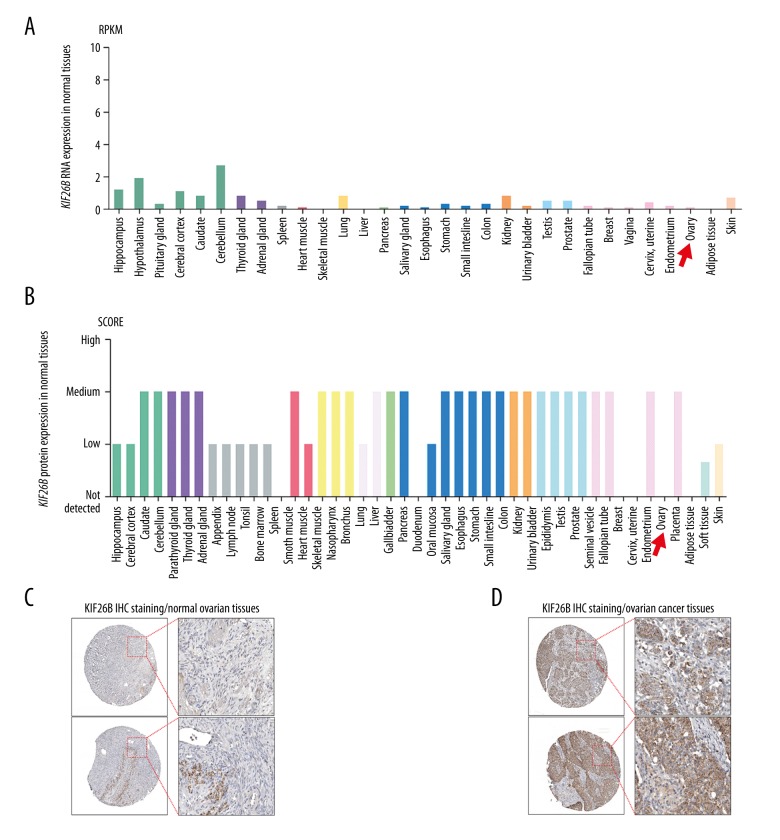
Using data in the Human Protein Atlas, the expression of the kinesin family member 26B (KIF26B) gene at both RNA and protein levels in normal human tissues were characterized. The expression of KIF26B RNA was low, and there was no or minimal expression of the KIF26B protein in normal ovarian tissues (Figure 1A–1C; red arrows).
Figure 1.
The KIF26B gene is not expressed in normal ovarian tissues, but is upregulated in ovarian cancer. (A, B) The expression of the KIF26B gene at the mRNA (A) and protein (B) levels in all human tissues. RNA-seq data are reported as median reads per kilobase per million mapped (RPKM) reads, and were generated by the Genotype-Tissue Expression (GTEx) project. (C, D) Representative images of immunohistochemistry (IHC) staining of KIF26B protein in normal ovarian tissues (C), and ovarian cancer tissues (D). Data were obtained from the Human Protein Atlas: (www.proteinatlas.org/ENSG00000162849-KIF26B/tissue) and (www.proteinatlas.org/ENSG00000162849-KIF26B/pathology/tissue/ovarian+cancer).
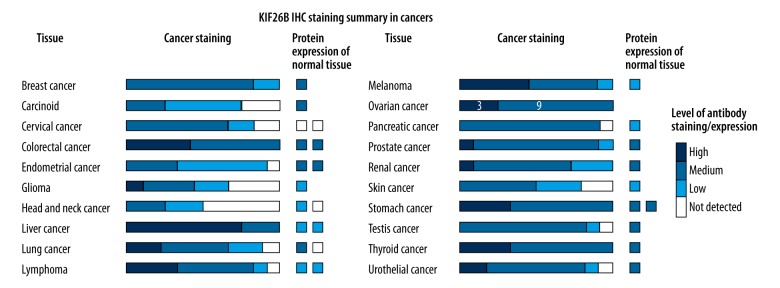
Of the cases of ovarian cancer from the Human Protein Atlas, that were 11 cases that showed mildly positive immunohistochemical staining for expression of the KIF26B protein product, nine cases of that showed medium immunohistochemical staining for expression of the KIF26B protein product, and three cases that showed strong immunohistochemical staining for expression of the KIF26B protein product (Figure 2). These findings suggest that the KIF26B gene was upregulated in ovarian cancer compared with normal ovarian tissue.
Figure 2.
Immunohistochemistry for detection of KIF26B protein product staining in ovarian cancer tissue. Of the cases of ovarian cancer from the Human Protein Atlas, that were 11 cases that showed mild immunohistochemical staining for expression of the KIF26B protein product, nine cases of that showed medium immunohistochemical staining for expression of the KIF26B protein product, and three cases that showed strong immunohistochemical staining for expression of the KIF26B protein product. Data were obtained from the Human Protein Atlas: (www.proteinatlas.org/ENSG00000162849-KIF26B/pathology).
Upregulation of the KIF26B gene was associated with lymphatic and venous invasion
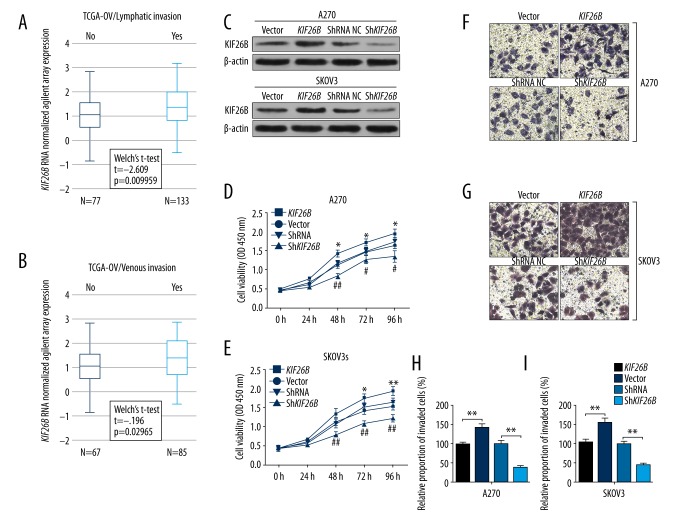
Data mining and analysis of the Cancer Genome Atlas-Ovarian Cancer (TCGA-OV) included the clinicopathologic features of patients with primary ovarian serous cystadenocarcinoma, which is the major type of ovarian cancer. KIF26B gene expression was significantly greater in cases with lymphatic invasion (N=133) compared with those without lymphatic invasion (N=77) (p=0.01) (Figure 3A). Also, cases with venous invasion were associated with significantly increased KIF26B gene expression compared with cases without venous invasion (p=0.03) (Figure 3B). These findings might provide support for the role of the KIF26B gene in vascular invasion of ovarian cancer cells, which is required for tumor metastasis to occur.
Figure 3.
Upregulation of the KIF26B gene is associated with lymphatic and venous invasion. (A, B) Box plots of expression of the KIF26B gene in patients with or without lymphatic invasion (A) or venous invasion (B). (C) Western blot analysis of KIF26B expression in A2780 and SKOV3 cells, 24 hours after transfection of KIF26B gene expression vectors or KIF26B short hairpin RNA (shRNA). (D, E) Cell proliferation of A2780 (D) and SKOV3 (E) cells after transfection, was evaluated using the cell counting kit-8 (CCK-8) assay. * p<0.05 and ** p<0.01 versus the vector control group, # p<0.05 and ## p<0.01 versus the shRNA control group. (F–I) Representative images (F, G) and quantitation (H, I) of transwell assay of invasion capability of A2780 (F, H) and SKOV3 (G and I) cells after transfection. * p<0.05 and ** p<0.01.
Upregulation of the KIF26B gene enhanced cell proliferation and migration of ovarian cancer cells in vitro
To investigate the regulatory effect of the KIF26B gene on cancer cell behavior, the A2780 and SKOV3 ovarian cancer cells were transfected with the pcDNA3.1-KIF26B expression vector, or lentiviral short hairpin RNA (shRNA) particles, respectively (Figure 3.C). After confirmation of transfection, the cells underwent the cell counting kit-8 (CCK-8) assay.
Overexpression the KIF26B gene significantly increased proliferation of both A2780 and SKOV3 cells (Figure 3D, 3E). In comparison, knockdown of the KIF26B gene significantly reduced the proliferation of cells in vitro (Figure 3D, 3E). Also, the results of transwell assay showed that increased KIF26B gene expression significantly enhanced the invasion of both A2780 and SKOV3 cells, while KIF26B gene knockdown reduced the invasive potential of the ovarian cancer cells in vitro (Figure 3F–3I).
Increased expression levels of the KIF26B gene were associated with reduced overall survival (OS) in patients with ovarian cancer
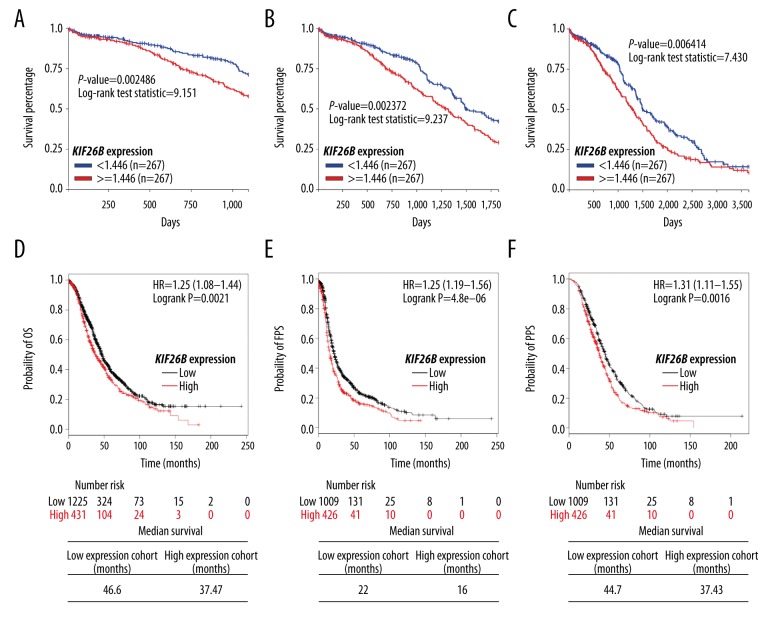
The expression levels of the KIF26B gene and survival outcomes in ovarian cancer patients were analyzed, including overall survival (OS), progression-free survival (PFS), and post-progression survival (PPS). Data mining was performed in both TCGA-OV and the online Kaplan-Meier plotter. In 534 patients with the KIF26B gene expression measured by Agilent RNA array in TCGA-OV, high KIF26B expression was associated with a significantly reduced 3-year, 5-year, and 10-year OS (p=0.002, p=0.002, and p=0.006, respectively) (Figure 4A–4C).
Figure 4.
High KIF26B gene expression is associated with poor overall survival (OS), progression-free survival (PFS), and post-progression survival (PPS) in patients with ovarian cancer. (A–C) Online Kaplan-Meier plotting of 3-year (A), 5-year (B), and 10-year (C) overall survival (OS) in ovarian cancer patients with high or low KIF26B gene expression. Data were obtained from The Cancer Genome Atlas-Ovarian Cancer (TCGA-OV) database. (D–F) Online Kaplan-Meier plotting (upper) of OS (D), FPS (E), and PPS (F) in patients with ovarian cancer with high/low KIF26B gene expression. Median survival was calculated (lower). Data were obtained from online Kaplan-Meier plotting.
Data from the online Kaplan-Meier plotter also showed that increased KIF26B gene expression was associated with a reduced OS (HR: 1.25; 95% CI, 1.08–1.44; p=0.0021), FPS (HR: 1.36; 95% CI, 1.19–1.56; p<0.001), and PPS (HR: 1.31; 95% CI, 1.11–1.55; p=0.0016) in patients with ovarian cancer (Figure 4D–4F). The median OS, FPS, and PPS in the low KIF26B expression group were 46.6 months, 22 months, and 44.7 months, respectively (Figure 4D–4F), compared with median OS, FPS, and PPS in the high KIF26B expression group of 37.47 months, 16 months, and 37.43 months, respectively (Figure 4D–4F).
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of genes that interacted with, co-localized to, or were co-expressed with the KIF26B gene
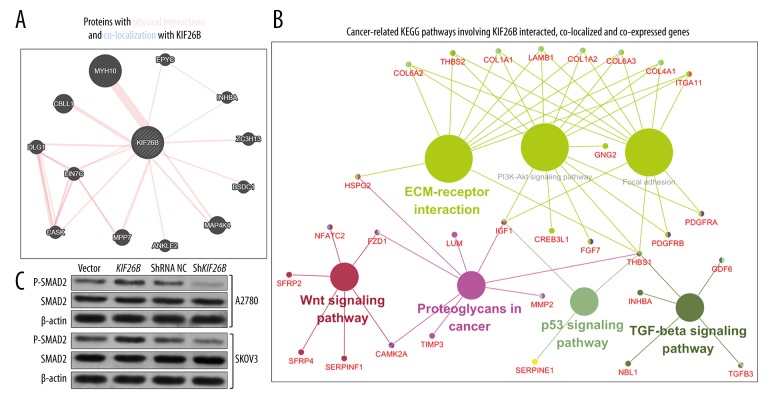
Using Genemania, the proteins that interacted with and co-localized to the KIF26B gene were identified, and included: MYH10, EPYC, ZC3H13, BSDC1, MAP4K4, ANKLE2, MPP7, CASK, LIN7C, DLG1, and CBLL1 (Figure 5A).
Figure 5.
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of KIF26B and its co-expressed genes in The Cancer Genome Atlas-Ovarian Cancer (TCGA-OV) database. (A) The proteins that interacted and co-localize with KIF26B. (B) Cancer-related Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways involving genes that interacted with, co-localized to, and were co-expressed with the KIF26B gene. (C) Western blot analysis of p-SMAD2 and SMAD2 expression in A2780 and SKOV3 cells, 46 hours following transfection of the KIF26B expression vectors or KIF26B shRNA.
Using cBioPortal for Cancer Genomics, there were 199 identified genes that were co-expressed with the KIF26B gene in TCGA-OV (Pearson’s r ≥0.6). To provide further insights to provide direction for future exploration of the regulative network of the KIF26B gene in ovarian cancer, the genes that interacted with, co-localized to, and co-expressed with KIF26B were subjected to the KEGG pathway analysis (Table 1).
Table 1.
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of the KIF26B gene and the interactive, co-localized, and co-expressed genes in breast cancer.
| GO ID | GO term | Term P value | % associated genes | Nr. genes | Associated genes found |
|---|---|---|---|---|---|
| GO: 0004512 | ECM-receptor interaction | 1.5E-9 | 13.41 | 11.00 | [COL1A1, COL1A2, COL4A1, COL6A2, COL6A3, HSPG2, ITGA11, ITGA5, LAMB1, THBS1, THBS2] |
| GO: 0004974 | Protein digestion and absorption | 57.0E-9 | 11.11 | 10.00 | [COL11A1, COL1A1, COL1A2, COL3A1, COL4A1, COL5A1, COL5A2, COL6A2, COL6A3, ELN] |
| GO: 0004510 | Focal adhesion | 310.0E-9 | 6.53 | 13.00 | [COL1A1, COL1A2, COL4A1, COL6A2, COL6A3, IGF1, ITGA11, ITGA5, LAMB1, PDGFRA, PDGFRB, THBS1, THBS2] |
| GO: 0004151 | PI3K-Akt signaling pathway | 1.1E-6 | 4.68 | 16.00 | [COL1A1, COL1A2, COL4A1, COL6A2, COL6A3, CREB3L1, FGF7, GNG2, IGF1, ITGA11, ITGA5, LAMB1, PDGFRA, PDGFRB, THBS1, THBS2] |
| GO: 0004933 | AGE-RAGE signaling pathway in diabetic complications | 120.0E-6 | 7.07 | 7.00 | [COL1A1, COL1A2, COL3A1, COL4A1, MMP2, SERPINE1, TGFB3] |
| GO: 0004390 | Hippo signaling pathway | 340.0E-6 | 5.19 | 8.00 | [DLG1, FRMD6, FZD1, GDF6, LATS2, SERPINE1, SNAI2, TGFB3] |
| GO: 0004270 | Vascular smooth muscle contraction | 420.0E-6 | 5.79 | 7.00 | [ACTA2, ACTG2, CALD1, EDNRA, MRVI1, PRKG1, PTGIR] |
| GO: 0005205 | Proteoglycans in cancer | 460.0E-6 | 4.43 | 9.00 | [CAMK2A, FZD1, HSPG2, IGF1, ITGA5, LUM, MMP2, THBS1, TIMP3] |
| GO: 0005146 | Amoebiasis | 740.0E-6 | 6.25 | 6.00 | [COL1A1, COL1A2, COL3A1, COL4A1, LAMB1, TGFB3] |
| GO: 0005144 | Malaria | 2.2E-3 | 8.16 | 4.00 | [LRP1, TGFB3, THBS1, THBS2] |
| GO: 0004350 | TGF-beta signaling pathway | 2.5E-3 | 5.95 | 5.00 | [GDF6, INHBA, NBL1, TGFB3, THBS1] |
| GO: 0004360 | Axon guidance | 3.6E-3 | 4.00 | 7.00 | [CAMK2A, CXCL12, NFATC2, SEMA3D, SEMA6B, SEMA7A, SLIT2] |
| GO: 0004392 | Hippo signaling pathway -multiple species | 4.2E-3 | 10.34 | 3.00 | [DCHS1, FRMD6, LATS2] |
| GO: 0004310 | Wnt signaling pathway | 5.6E-3 | 4.20 | 6.00 | [CAMK2A, FZD1, NFATC2, SERPINF1, SFRP2, SFRP4] |
| GO: 0005214 | Glioma | 5.9E-3 | 6.25 | 4.00 | [CAMK2A, IGF1, PDGFRA, PDGFRB] |
| GO: 0005218 | Melanoma | 7.7E-3 | 5.80 | 4.00 | [FGF7, IGF1, PDGFRA, PDGFRB] |
| GO: 0005414 | Dilated cardiomyopathy | 19.0E-3 | 4.44 | 4.00 | [IGF1, ITGA11, ITGA5, TGFB3] |
| GO: 0004115 | p53 signaling pathway | 43.0E-3 | 4.35 | 3.00 | [IGF1, SERPINE1, THBS1] |
Some of the identified pathways were recognized to be important for cancer initiation and progression and included extracellular matrix (ECM)-receptor interaction, focal adhesion, PI3K-Akt signaling pathway, proteoglycans in cancer, the TGF-beta signaling pathway, the Wnt signaling pathway and p53 signaling pathway (Figure 5B). COL1A1, COL1A2, COL4A1, COL6A2, COL6A3, HSPG2, ITGA11 and ITGA5, LAMB1, THBS1, and THBS2 were enriched in the ECM-receptor interaction pathway. COL1A1, COL1A2, COL4A1, COL6A2, COL6A3, IGF1, ITGA11, ITGA5, LAMB1, PDGFRA, PDGFRB, THBS1, and THBS2 were enriched in the focal adhesion pathway. COL1A1, COL1A2, COL4A1, COL6A2, COL6A3, CREB3L1, FGF7, GNG2, IGF1, ITGA11, ITGA5, LAMB1, PDGFRA, PDGFRB, THBS1 and THBS2 were enriched in the PI3K-Akt signaling pathway. GDF6, INHBA, NBL1, TGFB3 and THBS1 were enriched in the TGF-beta signaling pathway. CAMK2A, FZD1, NFATC2, SERPINF1, SFRP2 and SFRP4 are enriched in the Wnt signaling pathway. IGF1, SERPINE1, and THBS1 were enriched in the p53 signaling pathway (Table 1).
The INHBA gene was the only gene that was co-localized and co-expressed with the KIF26B gene in ovarian cancer. Because the INHBA gene activates the TGF-β signaling pathway in ovarian cancer [20], an assessment was undertaken as to whether the KIF26B gene had a similar function to KIF26B. Western blot data showed that overexpression of the KIF26B gene induced upregulation of p-SMAD2 levels, while knockdown of the KIF26B gene decreased p-SMAD2 levels, in both A2780 and SKOV3 cells in vitro (Figure 5C).
Discussion
The kinesin family member 26B (KIF26B) gene has been recognized to be an oncogene in several human cancers, including breast cancer [10], gastric cancer [11], and colorectal cancer [12]. In these human cancers, upregulation of the KIF26B gene is consistently associated with malignant clinicopathologic features, including increased tumor size, increased tumor grade, lymph node metastasis, and distant metastasis, all of which are associated with reduced tumor prognosis [10–12].
In this study, with the use of bioinformatics data mining in two large databases, the expression of the KIF26B gene and KIF26B protein was not present in normal ovarian tissues, but was upregulated in ovarian cancer. Upregulation of the KIF26B gene was significantly associated with lymphatic and venous invasion, based on data from Cancer Genome Atlas-Ovarian Cancer (TCGA-OV). In in-vitro cell models using the ovarian carcinoma cell lines, A2780 and SKOV3, cells with overexpression of the KIF26B gene had an increased rate of proliferation and increased cell migration and invasion properties. In comparison, knockdown of the KIF26B gene impaired cell proliferation and invasion in vitro. These findings suggest that the KIF26B gene might be a key gene in the regulation of the metastatic potential of ovarian cancer. The prognostic value of the KIF26B gene in ovarian cancer was further investigated in this study using the online Kaplan-Meier plotter survival analysis. This analysis method showed that high levels of expression of the KIF26B gene were associated with reduced overall survival (OS), progression-free survival (PFS), and post-progression survival (PPS) in patients with ovarian cancer.
Although the findings of this study identified the expression profile and potential prognostic value of expression of the KIF26B gene in ovarian cancer, the signaling pathways involved remain unknown. In gastric cancer, expression of KIF26B has been shown to enhance the vascular endothelial growth factor (VEGF) signaling pathway, via inducing the expression of VEGFA, PXN, FAK, PIK3CA, BCL2, and CREBI [11]. Another study found that expression of the KIF26B gene was positively correlated with increased tissue expression of the proliferation marker, Ki67, in colorectal cancer cells [12].
This study used bioinformatics analysis using Genemania and cBioPortal for Cancer Genomics, which identified the genes that interacted with, co-localized to, and were co-expressed with the KIF26B gene, and analyzed their enrichment in Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. Among the identified pathways, the ECM-receptor interaction, the focal adhesion, the PI3K-Akt signaling pathway, the proteoglycans in cancer, the TGF-beta signaling pathway, the Wnt signaling pathway, and the p53 signaling pathway were closely related to the pathological development of ovarian cancer. This study showed that the key genes involved in these pathways included: COL1A1, COL1A2, COL4A1, COL6A2, COL6A3, HSPG2, ITGA11, ITGA5, LAMB1, THBS1, THBS2, IGF1, PDGFRA, PDGFRB, CREB3L1, FGF7, GNG2, GDF6, INHBA, NBL1, TGFB3, CAMK2A, FZD1, NFATC2, SERPINF1, SFRP2, SFRP4 and SERPINE1. Previous studies have shown that the genes COL1A1, COL1A2, LAMBI, and FZD1 were significantly upregulated in multi-drug resistant ovarian cancer cells when compared with untreated cells [21,22]. Also, COL6A2 and THBS2 are in a ten-gene signature that is associated with poor OS in patients with high-grade serous ovarian cancer [23].
DNA duplication of the FGF7 gene has been reported to occur in ovarian cancer and to have a significant role in the development of ovarian cancer [24]. The SFRP4 gene is a Wnt gatekeeper modulating epithelial-mesenchymal transition (EMT), cell migration, and downstream Wnt signaling in serous ovarian cancer cells [25]. The INHBA gene was the only gene that was found to be co-localized and co-expressed with the KIF26B gene in ovarian cancer in this study. The upregulation of the INHBA gene has previously been shown to contribute to EMT in both normal and ovarian cancer cells, and also promotes the invasive behavior of ovarian cancer cells [26]. The INHBA gene has also been previously reported to be part of the metastasis-associated gene expression signature of ovarian cancer, together with the THBS2 gene [20].
The TGF-β signaling pathway plays a critical role in the pathogenesis of ovarian cancer, including in DNA methylation during the EMT associated with ovarian cancer cells [27]. The findings of this study showed that upregulation of the KIF26B gene enhanced the TGF-β signaling pathway in ovarian cancer cells, which might be one of the mechanisms contributing to the oncogenic properties of the KIF26B gene. However, current understanding of the association between the expression of KIF26B and other genes in ovarian cancer remains unclear. Further future studies are needed to explore the effects of KIF26B gene expression in the regulatory and signaling networks in human breast cancer.
Conclusions
The findings of this study showed that, in ovarian carcinoma, upregulation of the KIF26B gene enhanced cell proliferation, cell migration, and cell invasion of ovarian cancer cells in vitro, and was associated with poor survival outcomes when bioinformatics data were analyzed.
Footnotes
Conflict of interest
None.
Source of support: Departmental sources
References
- 1.Qiu HL, Deng SZ, Li C, et al. High expression of KIF14 is associated with poor prognosis in patients with epithelial ovarian cancer. Eur Rev Med Pharmacol Sci. 2017;21:239–45. [PubMed] [Google Scholar]
- 2.Mccluggage WG. Morphological subtypes of ovarian carcinoma: A review with emphasis on new developments and pathogenesis. Pathology. 2011;43:420–32. doi: 10.1097/PAT.0b013e328348a6e7. [DOI] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong L, Che H, Li M, Li X. Sam68 is overexpressed in epithelial ovarian cancer and promotes tumor cell proliferation. Med Sci Monit. 2016;22:3248–56. doi: 10.12659/MSM.899980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong L, Hui L. HOTAIR promotes proliferation, migration, and invasion of ovarian cancer SKOV3 cells through regulating PIK3R3. Med Sci Monit. 2016;22:325–31. doi: 10.12659/MSM.894913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yiwei T, Hua H, Hui G, et al. HOTAIR Interacting with MAPK1 regulates ovarian cancer skov3 cell proliferation, migration, and invasion. Med Sci Monit. 2015;21:1856–63. doi: 10.12659/MSM.893528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yao Y, Yu L, Su X, et al. Synthesis, characterization and targeting chemotherapy for ovarian cancer of trastuzumab-SN-38 conjugates. J Control Release. 2015;220:5–17. doi: 10.1016/j.jconrel.2015.09.058. [DOI] [PubMed] [Google Scholar]
- 8.Marikawa Y, Fujita TC, Alarcon VB. An enhancer-trap LacZ transgene reveals a distinct expression pattern of Kinesin family 26B in mouse embryos. Dev Genes Evol. 2004;214:64–71. doi: 10.1007/s00427-003-0377-x. [DOI] [PubMed] [Google Scholar]
- 9.Uchiyama Y, Sakaguchi M, Terabayashi T, et al. KIF268, a kinesin family gene, regulates adhesion of the embryonic kidney mesenchyme. Proc Natl Acad Sci USA. 2010;107:9240–45. doi: 10.1073/pnas.0913748107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Q, Zhao ZB, Wang G, et al. High expression of KIF26B in breast cancer associates with poor prognosis. PLoS One. 2013;8:e61640. doi: 10.1371/journal.pone.0061640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang H, Ma RR, Wang XJ, et al. KIF26B, a novel oncogene, promotes proliferation and metastasis by activating the VEGF pathway in gastric cancer. Oncogene. 2017;36(40):5609–19. doi: 10.1038/onc.2017.163. [DOI] [PubMed] [Google Scholar]
- 12.Wang J, Cui F, Wang X, et al. Elevated kinesin family member 26B is a prognostic biomarker and a potential therapeutic target for colorectal cancer. J Exp Clin Cancer Res. 2015;34:13. doi: 10.1186/s13046-015-0129-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pu Y, Yi Q, Zhao F, et al. MiR-20a-5p represses multi-drug resistance in osteosarcoma by targeting the KIF26B gene. Cancer Cell Int. 2016;16:64. doi: 10.1186/s12935-016-0340-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Theriault BL, Pajovic S, Bernardini MQ, et al. Kinesin family member 14: An independent prognostic marker and potential therapeutic target for ovarian cancer. Int J Cancer. 2012;130:1844–54. doi: 10.1002/ijc.26189. [DOI] [PubMed] [Google Scholar]
- 15.Pawar S, Donthamsetty S, Pannu V, et al. KIFCI, a novel putative prognostic biomarker for ovarian adenocarcinomas: Delineating protein interaction networks and signaling circuitries. J Ovarian Res. 2014;7:53. doi: 10.1186/1757-2215-7-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uhlen M, Oksvold P, Fagerberg L, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28:1248–50. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 17.Thul PJ, Akesson L, Wiking M, et al. A subcellular map of the human proteome. Science. 2017;356(6340) doi: 10.1126/science.aal3321. pii: eaal3321. [DOI] [PubMed] [Google Scholar]
- 18.Gyorffy B, Lanczky A, Szallasi Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr Relat Cancer. 2012;19:197–208. doi: 10.1530/ERC-11-0329. [DOI] [PubMed] [Google Scholar]
- 19.Bindea G, Galon J, Mlecnik B. CluePedia Cytoscape plugin: Pathway insights using integrated experimental and in silico data. Bioinformatics. 2013;29:661–63. doi: 10.1093/bioinformatics/btt019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim H, Watkinson J, Varadan V, Anastassiou D. Multi-cancer computational analysis reveals invasion-associated variant of desmoplastic reaction involving INHBA, THBS2 and COL11A1. BMC Med Genomics. 2010;3:51. doi: 10.1186/1755-8794-3-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Januchowski R, Swierczewska M, Sterzynska K, et al. Increased expression of several collagen genes is associated with drug resistance in ovarian cancer cell lines. J Cancer. 2016;7:1295–310. doi: 10.7150/jca.15371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Januchowski R, Zawierucha P, Rucinski M, et al. Extracellular matrix proteins expression profiling in chemoresistant variants of the A2780 ovarian cancer cell line. Biomed Res Int. 2014;2014:365867. doi: 10.1155/2014/365867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheon DJ, Tong Y, Sim MS, et al. A collagen-remodeling gene signature regulated by TGF-beta signaling is associated with metastasis and poor survival in serous ovarian cancer. Clin Cancer Res. 2014;20:711–23. doi: 10.1158/1078-0432.CCR-13-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yasuhara T, Okamoto A, Kitagawa T, et al. FGF7-like gene is associated with pericentric inversion of chromosome 9, and FGF7 is involved in the development of ovarian cancer. Int J Oncol. 2005;26:1209–16. [PubMed] [Google Scholar]
- 25.Ford CE, Jary E, Ma SS, et al. The Wnt gatekeeper SFRP4 modulates EMT, cell migration and downstream Wnt signalling in serous ovarian cancer cells. PLoS One. 2013;8:e54362. doi: 10.1371/journal.pone.0054362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Basu M, Bhattacharya R, Ray U, et al. Invasion of ovarian cancer cells is induced byPITX2-mediated activation of TGF-beta and Activin-A. Mol Cancer. 2015;14:162. doi: 10.1186/s12943-015-0433-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cardenas H, Vieth E, Lee J, et al. TGF-beta induces global changes in DNA methylation during the epithelial-to-mesenchymal transition in ovarian cancer cells. Epigenetics. 2014;9:1461–72. doi: 10.4161/15592294.2014.971608. [DOI] [PMC free article] [PubMed] [Google Scholar]