Abstract
Recently, we demonstrated the feasibility of a chemical synthetic lethality screen in cultured human cells. We now demonstrate the principles for a genetic synthetic lethality screen. The technology employs both an immortalized human cell line deficient in the gene of interest, which is complemented by an episomal survival plasmid expressing the wild-type cDNA for the gene of interest, and the use of a novel GFP-based double-label fluorescence system. Dominant negative genetic suppressor elements (GSEs) are selected from an episomal library expressing short truncated sense and antisense cDNAs for a gene likely to be synthetic lethal with the gene of interest. Expression of these GSEs prevents spontaneous loss of the GFP-marked episomal survival plasmid, thus allowing FACS enrichment for cells retaining the survival plasmid (and the GSEs). The dominant negative nature of the GSEs was validated by the decreased resident enzymatic activity present in cells harboring the GSEs. Also, cells mutated in the gene of interest exhibit reduced survival upon GSE expression. The identification of synthetic lethal genes described here can shed light on functional genetic interactions between genes involved in normal cell metabolism and in disease.
INTRODUCTION
The rapid accumulation of human cDNA sequences over the last few years and completion of the human genome sequence have created an urgent need to develop additional functional genomic techniques for the elucidation of gene function (1).
One of the classical approaches for studying the function of a protein is to identify other proteins that interact with it. Over the last two decades, several important biochemical and genetic technologies have evolved leading to direct identification and cloning of mammalian genes encoding interacting proteins (2,3). These techniques have led to the isolation of novel genes encoding proteins that associate with a known protein of interest or to the elucidation of previously unidentified interactions between known proteins. However, it is noteworthy that most of these powerful methods rely on direct physical interaction of the gene products, excluding indirect functional links between proteins.
Identification of a genetic interaction between a mutation in a gene coding for the protein of interest and a mutation in an unlinked gene is an established method for finding functional links between proteins. A genetic interaction can be manifested as a novel phenotype that is not displayed by either mutation alone. Some of the most powerful genetic interaction screens are based on either loss or gain of viability as a phenotype. In yeast, among those widely used are second site suppression, dosage suppression (4) and synthetic lethality screens (5,6).
The synthetic lethality screen in yeast identifies non-allelic and non-lethal mutations that are lethal in combination with a non-lethal mutation in a gene of interest (i.e. synthetic lethality). This genetic method is very powerful as it can reveal not only interactions between gene products with direct physical contacts, but also interactions along the same or parallel pathways. This is accomplished by random chemical mutagenesis of the entire yeast genome in a cell culture carrying a mutation in the gene of interest. The occurrence of synthetic lethality between the mutated gene of interest and a randomly inactivated gene enforces maintenance of an otherwise unstable episomal plasmid expressing a wild-type copy of the gene of interest (i.e. survival plasmid).
Recently, we reported the establishment of a chemical synthetic lethality screen in cultured human cells (7; reviewed in 8). The method employs as a recipient the pseudo-diploid HT1080 immortal human fibrosarcoma cell line (9) carrying mutations in both alleles of the gene for hypoxanthine-guanine phosphoribosyl transferase (HPRT1; EC 2.4.2.8) serving as the gene of interest. A variant green fluorescent protein (GFP) gene (sphGFP) is stably integrated into the chromosomal DNA of these cells. A chimeric EBV-based episomal survival plasmid expressing both the wild-type cDNA for HPRT1 and a second GFP variant (tpzGFP) is also introduced, allowing measurement of the double-label fluorescence ratio between the two GFPs. The survival plasmid carries the EBV origin of DNA replication (OriP) and a nuclear antigen gene (EBNA1) enabling its autonomous replication in the host cells. Yet, its maintenance as an episome is unstable and requires selective pressure. In a large scale blind test, addition of any one of several chemical reagents inhibitory to a gene product that is synthetic lethal with the mutated gene of interest prevented spontaneous loss of the episomal survival plasmid. This was detected by measuring the double-label fluorescence ratio between the two GFPs, while human cells were grown in microtiter plates. We propose that application of the method should permit high throughput screening of drugs that are synthetically lethal with any mutant human gene of interest, whose normal counterpart can be expressed in culture. As the method selects for a lethal phenotype, it is particularly suitable for the identification of drugs that kill malignant cells in a gene-specific manner, based on their predetermined cellular genotype.
Here, we have replaced the chemicals used in the chemical screen (7) by a library of truncated sense and antisense cDNAs prepared from a gene synthetic lethal with the chosen gene of interest. The genetic synthetic lethality screen in this system uncovered dominant negative genetic suppressor elements (GSEs) of truncated sense and antisense polarity from the candidate gene. These were identified among the library clones by their ability to impose a selective pressure for retention of the unstable survival plasmid, whose tpzGFP expression allowed isolation of their host cells by FACS. The GSEs were confirmed for their dominant negative mode of action by inhibiting activity of the corresponding endogenous gene. Thus, the methodology for a genetic synthetic lethality screen in cultured human cells has been established in this work. We expect that this approach will add an important new tool for analysis of gene function in humans and may lead to the identification of new drug targets in human diseases, in particular, cancer.
MATERIALS AND METHODS
Construction of plasmids
pIS was constructed by replacing the BamHI fragment encoding CD20 from pCMV-CD20 with a blunt-ended HindIII–BamHI fragment containing the coding sequence of sphGFP from the pGFPsph-b [R] vector (Packard Instruments). The dominant selectable marker encoding resistance to zeocin (zeoR) was initially excised (together with a synthetic bacterial promoter) from pVgRXR (Invitrogen) by the restriction enzymes PstI and SalI. The zeoR gene was introduced into a mammalian transcription unit by substitution of the neoR gene present in pREP9 (Invitrogen) in between the BglII and RsrII restriction sites. The generated plasmid product was cleaved by restriction endonucleases PflMI and BstEII and the excised zeoR transcription unit replaced a HindIII and XbaI flanked neoR gene present in the original pIS vector (7). The episomal HPRT1-tpzGFP survival plasmid was previously described (7).
Construction of the truncated sense and antisense cDNA libraries
The guanosine monophosphate synthetase (GMPS, EC 6.3.5.2) cDNA coding region was obtained by PCR amplification of cDNA from the SL.1NFLS human fetal spleen/liver cDNA library (Research Genetics). The primers for GMPS cDNA were 5′-ACATCCCATGGCTCTGTGCAACGG-3′ and 5′-GCATCCCGGGTTACTCCCACTCAGTAG-3′. The 2082 bp GMPS cDNA PCR product was subcloned into pBluescript SK+ (Stratagene). An aliquot of 5 µg of this insert was excised and treated with DNase I (Worthington) in 50 mM Tris–HCl pH 8, 10 mM MnCl2 and 0.005 U DNase I in a 50 µl reaction, until fragments were estimated to be of an average length of 300 bp. The reaction was stopped by adding an equal volume of phenol/chloroform, extraction and ethanol precipitation. The DNA fragments were then blunted using T4 DNA polymerase (New England Biolabs).
The adaptor encoding an initiating ATG in all three reading frames and a HindIII recognition sequence was prepared by annealing the oligonucleotides P1 (5′-AAACAAGCTTACCATGGATGGATGG-3′) and P2 (5′-CCATCCATCCATGGTAAGCTTG-3′). The adaptor with a translation termination codon in all three reading frames and a XhoI recognition sequence was prepared by annealing oligonucleotides P3 (5′-TAGTTAGTTAGCTCGAGTGC-3′) and P4 (5′-AAAGCACTCGAGCTAACTAACTA-3′). Ligation of the cDNA fragments to the adaptors was carried out overnight at 16°C with T4 DNA ligase (New England Biolabs). The ligated fragments were then PCR amplified using primers P1 and P4. PCR products were digested with XhoI and HindIII, electrophoresed, and products that were larger than 100 bp in length were excised/extracted from agarose gels and then subcloned into pREP9 (Invitrogen), the former site being proximal to the RSV promoter. Library DNA was prepared from several thousand bacterial colonies.
Transfection and expression of constructs in cells
HPRT1-deficient HT1080 fibrosarcoma cells (9) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum (FCS) and 4 mM l-glutamine. All transfections were carried out using Lipofectamine Plus (Life Technologies) according to the manufacturer’s instructions. For pIS, selection in zeocin (Cayla) was at 500 µg/ml. For the HPRT1-tpzGFP survival plasmid, selection in hygromycin B (Sigma) was carried out at 450 µg/ml, while maintenance was at 50 µg/ml. Selection and maintenance of the pREP9-containing cDNA fragments was at 500 and 400 µg/ml G418, respectively. During the selection for GSEs, or verification of their action, besides G418, guanine was added at 100 µM concentration and the hygromycin B selection for the survival plasmid removed.
Fluorescent scanning of microtiter plates and FACS sorting
For fluorescent scanning, cells were trypsinized and distributed at 30 000 cells/well into 96-well microplates (TPP). Growth medium was changed twice a week and plates were maintained for up to 75 days. Cells remained viable over the entire time period. Prior to scanning, the medium in the microplates was replaced with Hank’s balanced salt solution without phenol red. This procedure greatly minimized background fluorescence from the growth medium while maintaining maximal viability. Plates were scanned with a Fluoroskan Ascent CF microplate fluorescence reader using the Ascent software (Labsystems). Excitation for sphGFP was at 390 nm, while emission was measured at 510 nm. Excitation of tpzGFP was at 485 nm, while emission was measured at 527 nm. Integrated sphGFP was used as an internal control for the number of cells. This was achieved by dividing the relative fluorescence resulting from the episomal tpzGFP vector by the relative fluorescence from sphGFP for each well. Cells were returned to growth medium immediately following scanning.
Sorting was carried out using a FACS Star Plus (Becton Dickinson) flow cytometer. Cells were maintained in Hank’s balanced salt solution to maintain viability. Sorted cells were immediately lysed in order to extract low molecular weight DNA (10). Plasmids from the extract were transformed into bacteria and selected for ampicillin resistance.
In vitro enzyme activity
Cell lysates were prepared by resuspension of cells in 200 mM Tris–HCl pH 7.5, 200 mM NaCl, 5 mM MgCl2, 1 mM EGTA, 2 mM DTT and 1 mM PMSF, followed by three cycles of freeze–thaw and centrifugation in a microfuge at 14 000 r.p.m. at 4°C. Protein concentration of supernatants was measured using the Bradford method where the average of three separate samples of each lysate was determined.
GMPS activity was measured by a modification of the method of Nakamura and Lau (11). This was carried out with 40 µg of protein in 30 µl reactions containing 75 mM Tris–HCl pH 7.8, 10 mM MgCl2, 2 mM ATP, 5 mM l-glutamine, 10 mM DTT, 0.266 mM XMP and 33 µM [14C]8-XMP (Moravek). Reactions were carried out at 40°C for 1 h and terminated by adding 6 µl of 0.25 mM EDTA. Aliquots of 4 µl of the reactions were diluted in 25 ml of water and 2 µg each of XMP and GMP were added. This mixture was spotted onto PEI–cellulose UV254 TLC plates (Machery-Nagel). Plates were chromatographed in 2 M formic acid, dried, and scanned with a Fuji phosphorimager. Enzyme activity was calculated as percent conversion of XMP to GMP.
RESULTS
Design of the genetic synthetic lethality screening system
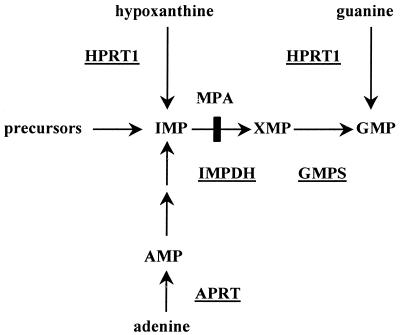
As a model system for the establishment of genetic synthetic lethality, the purine biosynthesis pathway was chosen (Fig. 1). This pathway was successfully utilized in the chemical synthetic lethality system (7). Biosynthesis of the essential metabolite GMP is achieved in fibroblasts by either the de novo pathway or, when needed, via the human salvage enzyme HPRT1. In the HPRT1-deficient variant of the HT1080 cell line (9), chemicals such as mycophenolic acid which inhibit the IMP dehydrogenase enzyme (IMPDH; EC 1.1.1.205) disrupt the de novo pathway, thus leading to synthetic lethality. We have shown that such selective pressure imposes the retention of an otherwise unstable EBV-based episomal plasmid expressing the HPRT1 cDNA, serving as the gene of interest (7). In order to test whether the screening for synthetic lethal chemicals could be extended to a genetic screen, we sought to disrupt the activity of GMPS (12), which should be synthetic lethal with HPRT1 deficiency. In yeast, mutant genes that are synthetic lethal to a mutant gene of interest are identified by a two-step process: first, mutagenizing the endogenous chromosomal genes with a chemical mutagen, which leads to retention of the survival plasmid when synthetic lethality occurs; second, ectopic expression in the latter recipients of the wild-type counterparts for the synthetic lethal genes (present in a normal yeast genomic library), leading to relief of survival plasmid retention (5). In human cells, we attempted an alternative method, in which abrogation of cellular gene activity is achieved by expression of dominant negative GSEs (13,14) contained within a library made of truncated sense or antisense cDNAs for human GMPS. The library itself was incorporated into an episomal expression vector that can be easily rescued. Selection should be achieved if a dominant negative GSE decreases the enzymatic activity of the resident GMPS, leading to retention of the HPRT1-encoding episomal survival plasmid marked with tpzGFP. Efficient isolation of cells displaying high tpzGFP-emitted flourescence is enabled by FACS sorting. As compared with the yeast method, this system couples the ‘mutagenesis’ step with sequence identification of the affected target in one step.
Figure 1.
De novo and salvage pathways of purine biosynthesis. Key enzymes are underlined. Arrows indicate the action of the enzymes. The site of inhibition by mycophenolic acid (MPA) is marked.
Setting up of the experimental system
Cells for this screen were prepared by stable integration of pIS (Fig. 2A), which encodes the sapphire green fluorescent protein (sphGFP) and the bacterial zeocin resistance (zeoR) DNA, into the HPRT1–/– variant of the HT1080 fibrosarcoma cell line. Stable cell clones were examined by fluorescent light microscopy and using a fluorescence plate reader. sphGFP fluorescence is used as the internal marker, normalizing the fluorescence readings originating from the episomal survival vector to cell number. After selection of a stable cell clone, the HPRT1-tpzGFP survival plasmid (Fig. 2B) was transfected into the chosen clone. This episomal vector carries ColE1 ori, AmpR, cDNA encoding HPRT1, tpzGFP, the hygromycin B resistance gene (hygR) and Epstein–Barr viral protein EBNA-1 that, together with the viral oriP element, is necessary for the replication and episomal state of the plasmid. A stable cell clone, clone 13, was chosen as the recipient of choice for the GSE library. This clone retains tpzGFP fluorescence when grown under hygromycin B selection and this fluorescence decays to <1% when cells are grown without hygromycin B for a period of 1 month (data not shown).
Figure 2.
Structure of the double-labeled GFP plasmid system and of the library expression vector. (A) The pIS integrating sphGFP vector with a zeoR dominant selectable marker. (B) The HPRT1-tpzGFP survival plasmid. (C) The truncated sense and antisense cDNA library vector. P-CMV, P-RSV and P-TK indicate the promoters of CMV, RSV and herpes TK1, respectively. IVS represents the rabbit globin second intron, while PA stands for the polyadenylation signal. zeoR, hygR, neoR and ampR identify the resistance genes for zeocin, hygromycin B, G418 and ampicillin, respectively.
We constructed a library consisting of random DNase I- generated fragments of the coding region of the GMPS cDNA. These fragments were ligated to adaptors that encode for an initiating AUG in all three reading frames and an adaptor encoding a termination codon in all three reading frames. These adaptors also carried restriction sites for insertion of the library into an episomal vector pREP9 that also encodes for neomycin resistance (neoR). The library fragments were then amplified using PCR, size selected for length greater than ∼100 bp, cut with the appropriate restriction enzymes and inserted into the episomal vector (Fig. 2C). Library DNA represented several thousand separate bacterial colonies.
Selection for GSEs via a synthetic lethality screen
Clone 13 cells were transfected with DNA from the library and hygromycin B selection was removed, allowing spontaneous loss of the survival plasmid. Selection of stable library transfected cells was carried out with G418 together with 100 µM of guanine. The presence of guanine in the growth medium is necessary so that the salvage enzyme HPRT1, encoded by the survival plasmid, could efficiently produce GMP, in case a GSE blocked the activity of GMPS. After the initial selection period the cells were grown as a pool of transfectants in the presence of G418 and guanine. Pools of cells were maintained by trypsinization and reseeding over a period of 7 weeks. tpzGFP fluorescence was periodically monitored using a fluorescence microscope and by FACS.
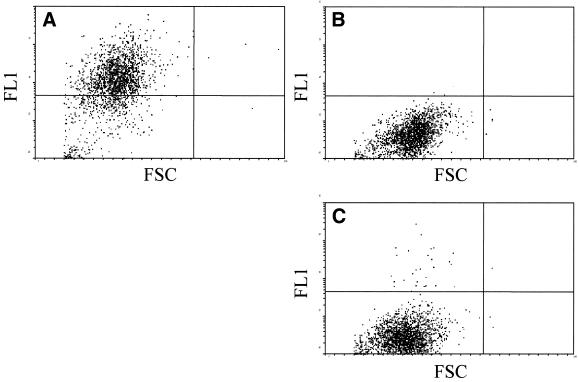
Highly fluorescent cells were then sorted by FACS. In contrast to clone 13 cells kept continuously under hygromycin B selection pressure (Fig. 3A), the population of library transfected cells retaining tpzGFP fluorescence was ∼2% of the total population (Fig. 3C). This small percentage of fluorescent cells was not detectable in clone 13 cells that were grown for the same period of time without hygromycin B (Fig. 3B), where virtually all cells had lost fluorescence. A total of 240 000 cells were separated by FACS sorting (Fig. 3C).
Figure 3.
FACS analysis of retention of the tpzGFP-expressing episomal survival plasmid. (A) Clone 13 cells maintained under selection with hygromycin B, the resistance gene encoded by the survival plasmid. (B) Clone 13 cells grown without hygromycin B selection for the entire time period. (C) Clone 13 cells transfected with the truncated sense and antisense cDNA library and grown in the presence of G418 in order to retain the library plasmid. TpzGFP fluorescence is shown on the y-axis while forward scatter (FSC) is shown on the x-axis.
Episomal DNA was extracted from the sorted cells and subsequently transformed into DH10B bacteria. In order to distinguish between rescued survival plasmid and plasmids harboring putative GSEs, ampicillin-resistant bacterial colonies were hybridized to the GMPS cDNA.
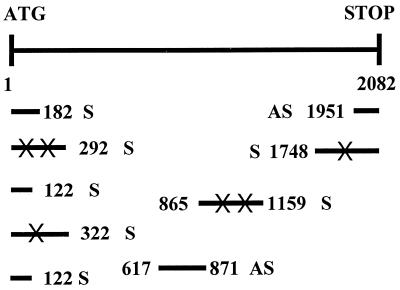
The inserts of nine plasmids were sequenced. Figure 4 shows the distribution of the putative GSEs derived from the GMPS library. The distribution of the putative GSEs revealed a certain bias to the N- and C-termini of the GMPS open reading frame (ORF). However, two fragments also originated from the center of the ORF. A number of single base pair mutations were present in the putative GSE sequences. These mutations were most likely introduced by Taq polymerase during the PCR amplification step of library production. However, we have no knowledge as to whether these mutations contributed to the dominant negative activity (see below) of the particular GSEs.
Figure 4.
Schematic representation showing localization of the rescued putative GSEs within the GMPS coding region. ATG and STOP symbols indicate the translation initiation and termination codons for the human GMPS cDNA, respectively. The numbers show the base pair position of each fragment within the GMPS cDNA. Sense (S) or antisense (AS) orientation is also shown. X indicates a single base pair substitution from the wild-type sequence.
Validation of dominant negative activity of the putative GSEs was determined by reintroducing separately each of the rescued GSE-containing plasmids into clone 13 cells. These secondary transfections were carried out under similar conditions to the primary library transfection, and pools of stable transfectants were grown as separate cell lines. Verification that these putative GSEs could force retention of the HPRT1-encoding survival plasmid was accomplished by seeding cells into 96-well microtiter plates in medium with or without G418. In the presence of G418, the episome-encoded GSE is retained. If the GSE encodes for a dominant negative GMPS activity, one would expect retention of the survival plasmid and a concomitant high tpzGFP:sphGFP fluorescence ratio. Without G418, loss of the putative GSE should allow loss of the survival plasmid and decay of the tpzGFP fluorescence. All clones except one retained high tpzGFP fluorescence over time, while tpzGFP fluorescence was lost without G418 (data not shown). One putative GSE, truncated sense 5 (182 S in Fig. 4), did not cause retention of the survival plasmid and therefore tpzGFP fluorescence decayed both with and without G418. The kinetics of tpzGFP loss in GSE 5 were similar to clone 13 when removed from hygromycin B selection.
In order to further verify the dominant negative nature of the GSEs, separate transfections of the plasmids were carried out again, so that fresh, stably transfected pools of cells could be measured for in vitro GMPS enzyme activity. Cells maintained with G418 and guanine were harvested 3 weeks after transfection and protein lysates were prepared. Table 1 shows the average of three GMPS activity assays performed with cell lines of pooled stable transformants expressing the GMPS GSEs. Values are expressed as percent activity when compared to GSE 5, which was used as a negative baseline control. GMPS activity was significantly low in all cell lines, with activity values in the range 34–73% of that displayed by GSE 5. In the GMPS activity assays, enzyme activity of the HT1080 parental cells and clone 13 recipient cells was also examined (data not shown), and was comparable to that found in GSE 5.
Table 1. GMPS activity in lysates of clone 13 cells stably transfected with GMPS GSEs.
| GSE |
Typea |
Position n GMPS |
Average activity (%)b |
SD (%) |
| 9 | S | 1–122 | 52.57 | 7.16 |
| 11 | S | 1–122 | 41.76 | 9.57 |
| 8 | S | 1–292 | 56.03 | 13.69 |
| 3 | S | 1–322 | 59.23 | 12.57 |
| 1 | AS | 617–871 | 33.89 | 5.22 |
| 10 | S | 865–1159 | 39.71 | 16.93 |
| 4 | S | 1748–2082 | 73.33 | 10.07 |
| 2 | As | 1951–2082 | 41.45 | 6.85 |
aType refers to sense (S) or antisense (AS) orientation of the GSE.
bAverage activity is the average of three independent experiments and is expressed as a percentage of GSE 5 (bp 1–182 sense) activity.
Two additional controls have been performed. In one, we have shown that stable introduction of the characterized GMP synthetase GSEs into another human cell line (U2-OS) resulted in decreased enzymatic activity of the resident enzyme. Secondly, expression of the same GSEs in HPRT1-deficient HT1080 cells (in the absence of the survival plasmid) led to cell death/growth inhibition (data not shown). The latter strongly supports our contention for GMPS GSE-mediated synthetic lethality with HPRT1 deficiency.
DISCUSSION
This report describes the development of a genetic synthetic lethality screen in cultured human cells. The methodology is an extension of the chemical synthetic lethality screen, which we reported recently (7). The chemicals used in the chemical screen were replaced here by dominant negative GSEs. Cells maintaining the survival plasmid due to expression of a GMPS GSE exhibit a high tpzGFP expression, allowing their enrichment from the total cell population by FACS sorting. Putative GSE-containing episomes were recovered from the low molecular weight DNA fraction. Individual DNA clones were retransfected into the recipient cell system and pools of secondary transfectants displaying a high tpzGFP:sphGFP ratio over time were spotted by periodic readings with a microplate fluorescence reader. Retention of the HPRT1 survival plasmid is dependent on continued selection with G418 for the GSE-expressing episomal vector. As expected for GSEs, irrespective of whether their polarity was sense or antisense, they had a dominant negative effect on the activity of the GMPS endogenous gene. Notably, GSE-containing clones exhibiting as much as 52–73% of the normal GMPS activity were still picked up by our screen (Table 1). Similarily, the dose–response characteristics of IMPDH inhibitors such as mycophenolic acid, mizoribine and ribavirin (7, and figures 4A and 5 within) indicate that partial inhibition of IMPDH enzyme activity is accompanied by sub-saturated tpzGFP fluorescence levels, reflecting a lower requirement for the survival plasmid-encoded HPRT1 enzyme.
We have therefore demonstrated that using the method described here, one could identify a gene (GMPS) synthetic lethal to the gene of interest (HPRT1). In comparison to the yeast screen (5), this method combines ‘mutagenesis’ together with rescue of the synthetic lethal GSEs into a single step. This system allowed isolation of dominant negative mutants, which should be useful on their own. To the best of our knowledge these mutants are the first dominant negative suppressors to be described for the GMPS gene.
Initially, the GSE methodology was used to isolate dominant negative mutants of single given genes, based on the ability of such constructs to disrupt a drug-induced apoptotic response (13,14) or to abrogate cell transformation (14). A further extension to this basic theme has been the use of random libraries of either antisense RNA alone (15–18) or GSEs (19) for isolation of apoptotic genes via actual selection for survival. Because of the high complexity of random GSE libraries, where each cDNA is usually represented by tens or hundreds of antisense and truncated sense short DNA fragments, enrichment steps for cDNAs of interest have been employed prior to the construction of such libraries (20). As might have been anticipated, some of the apoptotic genes selected by this approach turned out to be tumor suppressors (21–23). The present report demonstrates that incorporation of the GSE method together with a FACS sorting step into a genetic synthetic lethality screen leads to isolation of dominant negative mutants for the gene of interest. Clearly, this approach should enable the isolation of new survival/anti-apoptotic genes, some of which may turn out to be dominant oncogenes, such as Bcl2 (24–26), Survivin (27) and others (reviewed in 28).
To accomplish a genetic synthetic lethality screen at the multi-gene level, several modifications to the current method seem to be required. The bacterial dominant selectable markers tagging the episomal survival plasmid and the GSE library episomal vector should be different, so that recovery of the latter via bacterial transformation would eliminate the former. Transfection of the GSE library into the recipient human cells should be done at a DNA:cell ratio leading to one plasmid species per cell. The recovery of GSE 5 (which eventually served as a negative control) resulted from either the presence of more than one plasmid species in a fluorescent colony, or a rare integration event of the survival plasmid into chromosomal DNA, making the resident plasmid a likely false positive. The large complexity of a GSE library, coupled with the low efficiency of gene ablation by a GSE, suggest the use of either an enriched cDNA pool and/or the employment of several rounds of sorting by FACS, in which DNA from GSE-containing plasmids is reintroduced, after amplification in bacteria, into recipient cells and subject to decay and selection for fluorescent cells by FACS.
Enrichment for human cDNAs, serving as the source for production of the GSE library, could be performed by any one of a number of methods, such as subtractive hybridization, differential display or assaying for gene activity by DNA microarrays. In comparison to a GSE expressed by a retroviral vector, present usually as one copy per cell, our method has the advantage of the employment of a multi-copy episomal vector, whose GSE activity is not subject to modulation by neighboring chromosomal sequences.
The genetic synthetic lethality screen need not be confined to the use of GSEs. Libraries of ribozymes (29), RNA aptamers (30) or peptide aptamers (31,32) could be used as well.
Because the method is based on identification of a lethal phenotype, it is particularly relevant to the search for human genes acting in the same essential pathway or along two parallel ones. Therefore, such genetic synthetic lethality screens should have a major impact on human functional genomics. Moreover, the genetic synthetic lethality screen, when applied to human tumor cell lines having known primary genetic alterations, could lead to identification of new, perhaps even unexpected, secondary targets for cancer therapy. As targets identified by this approach would result in cellular synthetic lethality only when the primary tumor mutation is present, a high selectivity towards the tumor is ensured. In this regard, Hartwell et al. (33) suggested the use of genetic screens in yeasts, fruit flies or nematodes for identification of secondary targets which are synthetic lethal with mutations modeling primary alterations found in human tumors. The next challenge in the human system described here would be to demonstrate a synthetic lethality screen at the multi-gene level, so that significant anticancer drug targets may be discovered.
Acknowledgments
ACKNOWLEDGEMENTS
We thank C. Kahana for reagents. This work was supported by a biotechnological infrastructure grant in the field of functional genomics to D.C. and Y.O. from the Israeli Ministry of Science and Technology.
References
- 1.Brent R. (2000) Genomic biology. Cell, 100, 169–183. [DOI] [PubMed] [Google Scholar]
- 2.Fields S. (1997) The future is function. Nature Genet., 15, 325–327. [DOI] [PubMed] [Google Scholar]
- 3.Mendelsohn A.R. and Brent,R. (1999) Protein interaction methods—toward an endgame. Science, 284, 1948–1950. [DOI] [PubMed] [Google Scholar]
- 4.Rine J. (1991) Gene overexpression in studies of Saccharomyces cerevisiae. Methods Enzymol., 194, 239–250. [DOI] [PubMed] [Google Scholar]
- 5.Bender A. and Pringle,J.R. (1991) Use of a screen for synthetic lethal and multicopy suppressee mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol., 11, 1295–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guarente L. (1993) Synthetic enhancement in gene interaction: a genetic tool come of age. Trends Genet., 9, 362–366. [DOI] [PubMed] [Google Scholar]
- 7.Simons A., Dafni,N., Dotan,I., Oron,Y. and Canaani,D. (2001) Establishment of a chemical synthetic lethality screen in cultured human cells. Genome Res., 11, 266–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tlsty T. (2001) Searching for targets: the power of somatic cell genetics. Genome Res., 11, 187–188. [DOI] [PubMed] [Google Scholar]
- 9.Benedict W.F., Weissman,B.E., Mark,C. and Stanbridge,E.J. (1984) Tumorigenicity of human HT1080 fibrosarcoma × normal fibroblast hybrids: chromosome dosage dependency. Cancer Res., 44, 3471–3479. [PubMed] [Google Scholar]
- 10.Hirt B. (1967) Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol., 26, 365–369. [DOI] [PubMed] [Google Scholar]
- 11.Nakamura J. and Lau,L. (1995) Biochemical characterization of human GMP synthetase. J. Biol. Chem., 270, 7347–7353. [DOI] [PubMed] [Google Scholar]
- 12.Page T., Bakay,B. and Nyhan,W.L. (1984) Human GMP synthetase. Int. J. Biochem., 16, 117–120. [DOI] [PubMed] [Google Scholar]
- 13.Gudkov A.V., Zelnick,C., Kazarov,A.R., Thimmapaya,R., Suttle,D.P., Beck,W.T. and Roninson,I.B. (1993) Isolation of genetic suppressor elements, inducing resistance to topoisomerase II-interactive cytotoxic drugs, from human topoisomerase II cDNA. Proc. Natl Acad. Sci. USA, 90, 3231–3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ossovskaya V.S., Mazo,I.A., Chernov,M.V., Chernova,O.B., Strezoska,Z., Kondratov,R., Stark,G., Chumakov,P.M. and Gudkov,A.V. (1996) Use of a genetic suppressor elements to dissect distinct biological effects of separate p53 domains. Proc. Natl Acad. Sci. USA, 93, 10309–10314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deiss L.P. and Kimchi,A. (1991) A genetic tool used to identify thioredoxin as a mediator of a growth inhibitory signal. Science, 252, 117–120. [DOI] [PubMed] [Google Scholar]
- 16.Deiss L.P., Feinstein,E., Berissi,H., Cohen,O. and Kimchi,A. (1995) Identification of a novel serine/threonine kinase and a novel 15-kD protein as potential mediators of the γ-interferon-induced cell death. Genes Dev., 9, 15–30. [DOI] [PubMed] [Google Scholar]
- 17.Levy-Strumpf N. and Kimchi,A. (1998) Death associated proteins (DAPs): from gene identification to the analysis of their apoptotic and tumor suppressive functions. Oncogene, 17, 3331–3340. [DOI] [PubMed] [Google Scholar]
- 18.Vito P., Lacana,E. and D’Adamio,L. (1996) Interfering with apoptosis: Ca(2+)-binding protein ALG-2 and Alzheimer’s disease gene ALG-3. Science, 26, 521–525. [DOI] [PubMed] [Google Scholar]
- 19.Gudkov A.V., Kazarov,A.R., Thimmapaya,R., Axenovich,S.A., Mazo,I.A. and Roninson,I.B. (1994) Cloning mammalian genes by expression selection of genetic suppressor elements: association of kinesin with drug resistance and cell immortalization. Proc. Natl Acad. Sci. USA, 91, 3744–3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garkavtsev I., Kazarov,A., Gudkov,A.V. and Riabowol,K. (1996) Suppression of the novel growth inhibitor p33ING1 promotes neoplasic transformation. Nature Genet., 14, 415–420. [DOI] [PubMed] [Google Scholar]
- 21.Kissil J.L., Feinstein,E., Cohen,O., Jones,P.A., Tsai,Y.C., Knowles,M.A., Eydmann,M.E. and Kimchi,A. (1997) DAP-kinase loss of expression in various carcinoma and B-cell lymphoma cell lines: possible implications for role as tumor suppressor gene. Oncogene, 15, 403–407. [DOI] [PubMed] [Google Scholar]
- 22.Inbal B., Cohen,O., Polak-Charcon,S., Kopolovic,J., Vadai,E., Eisenbach,L. and Kimchi,A. (1997) DAP kinase links the control of apoptosis to metastasis. Nature, 390, 180–184. [DOI] [PubMed] [Google Scholar]
- 23.Garkavtsev I., Grigorian,I.A., Ossovskaya,V.S., Chernov,M.V., Chumakov,P.M. and Gudkov,A.V. (1998) The candidate tumor suppressor p33ING1 cooperates with p53 in cell growth control. Nature, 391, 295–298. [DOI] [PubMed] [Google Scholar]
- 24.Tsujimoto Y., Finger,L.R., Yunis,J., Nowell,P.C. and Croce,C.M. (1984) Cloning of the chromosome breakpoint of neoplastic B cells with the t(14;18) chromosome translocation. Science, 226, 1097–1099. [DOI] [PubMed] [Google Scholar]
- 25.Vaux D.L., Cory,S. and Adams,J.M. (1988) Bcl-2 gene promotes haematopoietic cell survival and cooperates with c-myc to immortalize pre-B cells. Nature, 335, 440–442. [DOI] [PubMed] [Google Scholar]
- 26.McDonnell T.J., Deane,N., Platt,F.M., Nunez,G., Jaeger,U., McKearn,J.P. and Korsmeyer,S.J. (1989) Bcl-2-immunoglobulin transgenic mice demonstrate extended B cell survival and follicular lymphoproliferation. Cell, 57, 79–88. [DOI] [PubMed] [Google Scholar]
- 27.Ambrosini G., Adida,C. and Altieri,D. (1997) A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nature Med., 3, 917–921. [DOI] [PubMed] [Google Scholar]
- 28.Lowe S.W. and Lin,A.W. (2000) Apoptosis in cancer. Carcinogenesis, 21, 485–495. [DOI] [PubMed] [Google Scholar]
- 29.Breaker R.R. and Joyce,G.F. (1994) Inventing and improving ribozyme function: rational design versus iterative selection methods. Trends Biotechnol., 12, 268–275. [DOI] [PubMed] [Google Scholar]
- 30.Thomas M., Chedin,S., Carles,C., Riva,M., Famulok,M. and Sentenac,A. (1997) Selective targeting and inhibition of yeast RNA polymerase II by RNA aptamers. J. Biol. Chem., 272, 27980–27986. [DOI] [PubMed] [Google Scholar]
- 31.Colas P., Cohen,B., Jessen,T., Grishina,I., McCoy,J. and Brent,R. (1996) Genetic selection of peptide aptamers that recognize and inhibit cyclin-dependent kinase 2. Nature, 380, 548–550. [DOI] [PubMed] [Google Scholar]
- 32.Geyer C.R., Colman-Lerner,A. and Brent,R. (1999) “Mutagenesis” by peptide aptamers identifies genetic network members and pathway connections. Proc. Natl Acad. Sci. USA, 96, 8567–8572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hartwell L.H., Szankasi,P., Roberts,C.J., Murray,A.W. and Friend,S.H. (1997) Integrating genetic approaches into the discovery of anticancer drugs. Science, 278, 1064–1068. [DOI] [PubMed] [Google Scholar]