Abstract
The immune response to pathogens varies substantially among people. While both genetic and non-genetic factors contribute to inter-person variation, their relative contributions and potential predictive power have remained largely unknown. By systematically correlating host factors in 534 healthy volunteers, including baseline immunological parameters and molecular profiles (genome, metabolome and gut microbiome), with cytokine-production capacity after stimulation with 20 pathogens, we identified distinct patterns of co-regulation. Among the 91 different cytokine–stimulus pairs, 11 categories of host factors together explained up to 67% of inter-individual variation in cytokine production induced by stimulation. A computational model based on genetic data predicted the genetic component of stimulus-induced cytokine-production (correlation 0.28-0.89), while non-genetic factors influenced cytokine production as well.
Variability in baseline immune response influences an individual’s susceptibility to immune-mediated diseases such as infection, autoimmune and inflammatory diseases, as well as their severity1–5. Both environmental and host factors are responsible for this variation in immune response6–9, which makes deciphering their interaction crucial for understanding their influence on susceptibility and instrumental for building quantitative predictors of disease. The Human Functional Genomics Project (HFGP) aims to identify the factors responsible for variability in immune response in the general population and upon perturbations, such as disease state. Within the HFGP, the 500 Human Functional Genomics (500FG) consortium has collected extensive molecular and phenotypic measurements from approximately 500 healthy volunteers of Western-European descent. Earlier 500FG studies assessed the separate impacts of host-related factors, genetic variation or microbiome on cytokine-production capacity7–9. However, an integrated understanding of the effect of these factors and of additional host-related factors, such as endocrine hormones, circulating metabolites, platelet-mediated effects or transcriptional profiles of immune cells on stimulus induced cytokine levels has been lacking.
Here, we used a comprehensive systems biology approach to integrate the large-scale genomic, metagenomic and metabolomic data available within the 500FG consortium with the immune cell composition, hormone levels and platelet activation profiles of each person analyzed. This allowed us to describe the baseline heterogeneity of immunological parameters, identify inter-correlated immune components, infer functional connections within the immune system and build predictive models of cytokine-production capacity upon stimulation. Using transcriptome data from a subset of samples, we showed that expression of genes after stimulation explained the variation in cytokine-production better than baseline expression. By integrating multi-omics layers, we showed that cytokine production was regulated by multiple genetic and non-genetic host factors, that production of cytokines after stimulation could be moderately predicted using multiple baseline profiles and that inter-individual variation in immune responses correlated with an individual’s genetic risk for (auto)immune disease.
Results
Baseline immune parameters are inter-correlated
To understand inter-individual variation in human immune response, we previously generated a database of immunological measurements, multi-omics data (cytokine response profiles, genetics, gene expression, immune cell frequencies, immune modulators, immunoglobulins, hormone levels, blood platelets, circulating metabolites, gut microbiome composition) and classical phenotypes (age, gender and BMI) from volunteers in the 500FG cohort (Supplementary Fig. 1 a,b and Supplementary Table 1). Cytokine production capacity of individuals was assessed using previously generated ELISA profiles on the production of 6 cytokines (IL-1β, IL-17, IL-22, IL-6, TNF-α and IFN-γ), by peripheral blood mono-nuclear cells (PBMC), whole blood and PBMC derived macrophages derived from blood after stimulation with 20 pathogens (Supplementary Table 2) 7–9. IL-1β, IL-6 and TNF-α levels were measured 24 hours after stimulation and IL-22, IL-17 and IFN-γ seven days after stimulation in PBMC and PBMC derived macrophages. In whole blood IL-1β, IL-6 and TNF-α levels were measured 48 hours after stimulation.
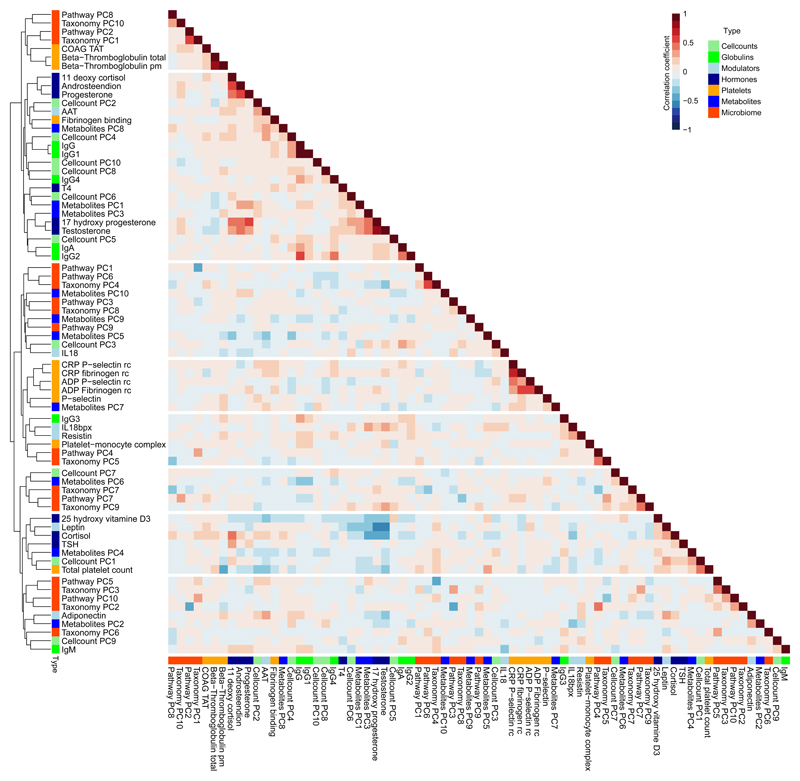
To map the relationships between these different molecular and immune parameters, we first performed clustering analysis of all immunological measurements besides cytokine production. To reduce the dimensionality of the dataset, the first ten principal components (PCs), covering >75% of variance in each dataset, were individually extracted from the cell count, metabolite and microbiome datasets. These PCs were then combined with the measurements of immune modulators (IL-18, IL-18BP, resistin, leptin, adiponectin, α-1 antitripsyn), immunoglobulins (IgG1-4, IgA, IgM), platelet activation profiles (p-selectin expression, fibrinogen binding, coagulation markers, β-Thromboglobulin) and hormone levels (androsteendion, cortisol, 11 deoxy cortisol, 17 hydroxy progesterone, progesterone, testosterone, 25 hydroxy vitamin D3, TSH, T4) (Supplementary Table 1). Subsequent unsupervised clustering analysis revealed several clusters (Fig. 1) that were consistent with previous observations, validating the current correlations. As such, we observed a negative correlation between the amount of the hormone leptin and the levels of progesterone and testosterone in peripheral blood (Fig. 1), consistent with an inhibitory effect of leptin on progesterone and on testosterone in humans10–13. We also observed a negative correlation of expression of p-selectin (whole blood flow cytometry) and fibrinogen activation profiles in peripheral blood (Fig. 1), consistent with evidence that they are under shared control14,15. Similarly, the hormone levels of 17 hydroxy-progesterone and testosterone were positively correlated with progesterone, androsteendion and 11 deoxy cortisol levels in peripheral blood (Fig. 1), consistent with these molecules having a common synthesis pathways. Finally, we observed the cluster of α1- antitrypsin with adiponectin and the association of 2 immune cell frequency PC’s with total platelet count, as well as a negative association between IL-18 and IgM abundance (Fig. 1). These results show that baseline immune parameters in healthy individuals are correlated and likely to be influenced by co-regulatory pathways.
Figure 1. Analysis of baseline immune parameters and molecular profiling shows baseline parameters are inter-correlated.
Spearman’s Rank correlations between both immune traits and baseline molecular profiles show that they are inter-correlated (n = 282). For the cell count and omics datasets, the first 10 principal components were extracted and used for calculating the correlation. Colors beside the cluster dendrogram indicate the type of measurements. Every sample represents an individual.
Baseline molecular profiles show substantial variation
Next, we examined the baseline (unstimulated) inter-individual variation in the immunological and molecular profiles described above and found a wide range of variation for the majority of immunological parameters analyzed (Supplementary Fig. 1c-e). Because some variation is known to result from differences in age, gender and season9,16–19, we corrected for these effects, when applicable. Among the immune-cell populations with high variability, effector T cell subpopulations showed the largest inter-individual variation compared to the other immune cell subpopulations (Supplementary Fig. 1c), in agreement with previous observations6. Baseline transcript abundance in whole blood also showed substantial inter-individual variation (Supplementary Fig. 1d). The top 75 most-variable transcripts were significantly enriched in 23 innate immune gene ontology (GO) terms (P <0.05 using an online tool20) (Supplementary Table 3), suggesting that the innate immune response was a major contributor to variations in transcript abundance. This analysis demonstrates that the baseline molecular profiles vary substantially between healthy individuals.
Genetics contributes the most to immune variation
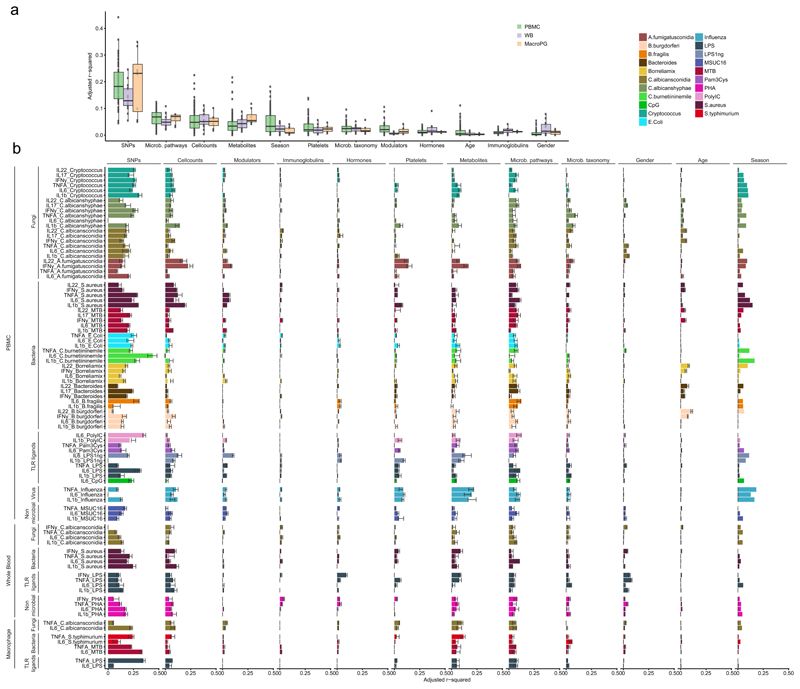
To address to what extent responses to a perturbation were affected by the pre-existing immune status, we first assessed the effect of host factors at baseline on cytokine production. Using a multivariate linear model (MVLM) to examine the percent of variance explained by these factors21, we found that genetic variation, as measured by single nucleotide polymorphisms (SNP), collectively explained most of the variation in stimulated cytokine production (avg. adj. R2 = 0.18) (Fig. 2a). In contrast, the gut microbiome, immune-cell counts, circulating metabolites and seasons displayed only moderate effects (avg. adj. R2 = 0.061, 0.057, 0.047 and 0.041, respectively) on most cytokine-stimulation pairs (Fig. 2a), while the concentration of circulating immunoglobulins, inflammatory mediators or hormones, and platelet activation (whole blood flow cytometry) generally had negligible effects (Fig. 2a,b). To evaluate the significance of the estimates of variation explained by genetics (VG), we performed 1000 permutations of sample labels in the cytokine data and applied the analysis pipeline on the permuted data to obtain the empirical distribution of the estimates of VG (null distribution). We subsequently compared the estimate of VG from the 500FG data with the estimate of VG from the permuted data. In total the estimates of VG in the 500FG were significant in 59 of 91 cases (P <0.05, Supplementary Table 4). For example, we found that the cytokine stimulation pairs explained the best by genetics (Poly I:C and C.Burnetti induced IL-6 levels in PBMC) showed significance.
Figure 2. Contribution of baseline immune parameters and multi-omics to cytokine variation.
(a) Percentage of variation in stimulated cytokine production explained by each category of measurements. The distribution indicates the adjusted R2 of a set multivariate linear models (MVLM) representing cytokine stimulation pairs from PBMC (n=67 models), whole blood (n=16 models) and PBMC derived macrophages (n=8 models). Each dot represents the adjusted R2 of a MVLM for a specific cytokine stimulation pair. (b) Contribution of each category to inter-individual cytokine variation. X-axis denotes the adjusted R2 values for the MVLMs. Bars indicate the adjusted R2 estimated on the full dataset. Error bars indicate the standard deviation in adjusted R2 of 10 MVLMs trained on a random subset of samples from the full data (90% of all samples). Y-axis denotes the cytokine-stimulation pairs. Colors indicate different stimulations applied in the experiments. Sample sizes differ between the different categories with the platelet, immune modulator, immunoglobulin and classical phenotypes having n = 489, the immune cell counts n = 472, the metabolites n = 377, microbial pathways n = 384, microbial taxonomy n = 411, hormones n = 486 and SNPs n = 392 samples. Every sample represents an individual.
Furthermore, we assessed several specific baseline categories that show cytokine- or pathogen-specificity in explaining the inter-individual variation (Fig. 2b). We observed that the abundance of circulating metabolites, including acetate and HDL cholesterol, showed a moderate negative effect on influenza-stimulated cytokine production by PBMC (avg. adjusted R2 = 0.19) (Fig. 2b), suggesting that these factors modulate susceptibility to viral infections. The production of the lymphocyte-derived cytokines IL-17, IL-22 and IFN-γ by PBMC in response to Aspergillus fumigatus (A. fumigatus) conidia was driven more by non-genetic host factors (cell counts, platelet amounts, circulating metabolites, gut microbiome composition and season) than by genetic factors (Fig. 2b), which was in contrast to the genetic-component-driven cytokine production in response to all other stimulations used (Fig. 2b). More specifically, individuals with high concentration of HDL cholesterol or α1- antitrypsin in the circulation showed lower cytokine production in response to A. fumigatus. To validate the link between HDL cholesterol and cytokine production, we cultured PBMCs collected from 6 healthy volunteers in medium containing lipoprotein-deficient plasma (LPDP) and LPDP+HDL cholesterol and measured cytokine production for TNF-α, IL-1β and IL-6 in response to A. fumigatus conidia after 24 hours. We observed lower production of all the cytokines assessed in PBMCs cultured with HDL compared to the LPDP control (Supplementary Fig. 2 a), indicating that HDL cholesterol modulates immune responses to A. fumigatus conidia.
Next, we compared the stimulus-dependent cytokine production data from the three different types of stimulation assays (PBMC, whole blood and PBMC derived macrophages) from the same individuals. We found that season, platelet-activation profiles, concentration of immune modulators, and age had a higher impact on stimulus-dependent cytokine production in PBMCs than in macrophages (Fig. 2a,b). In contrast, stimulus-dependent cytokine production correlated less with baseline metabolite levels in PBMC and whole blood then it did in macrophages (Fig. 2a,b).
This analysis shows that genetics contribute substantially to the observed inter-individual variation in cytokine level upon stimulation, and the non-genetic molecular profiles and immune parameters contribute as well.
Baseline molecules associate differentially to cytokine response
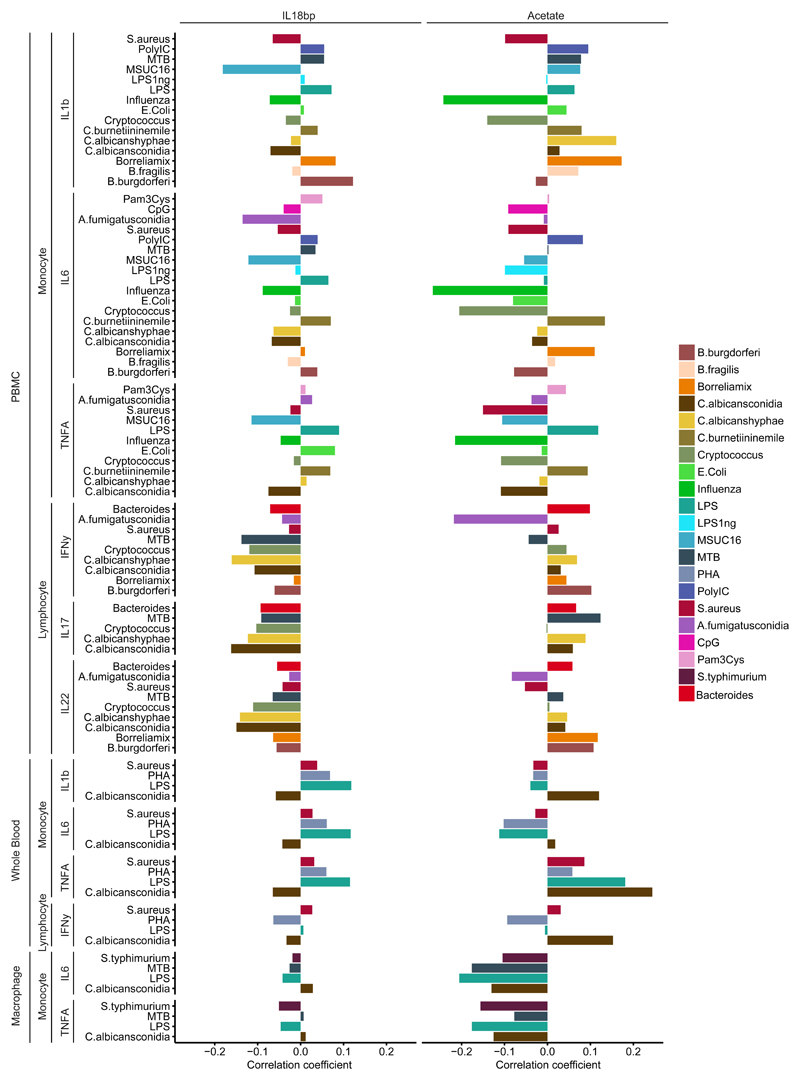
We next assessed which baseline immune and molecular components contribute most to variation in stimulus-induced cytokine production. We extracted the top five immune modulators (i.e. A-1 antitrepsin, IL18-BP, adiponecting, resistin and leptin) and metabolites (i.e. the total cholesterol level in HDL3, glutamine, free cholesterol and α-1 acid glycoprotein) in the analysis of explained variance. They are the molecules that show strong association with most of the cytokine measurements in the analysis of explained variance (Fig. 3, Supplementary Fig. 3). For example, circulating IL-18BP concentrations negatively correlate with lymphocyte-derived cytokine production (IL-17, IL-22, and IFN-γ) by PBMC after stimulation, but this pattern is not observed for the monocyte-derived cytokine production (IL-1β, IL-6, and TNF-α) by PBMC after stimulation (Fig. 3). IL-18BP is an inhibitor of IL-1822 and IL-18 induces cytokine production in natural killer (NK) cells and T helper cells23. The known function of IL-18BP in vitro and the observed correlations suggested IL-18BP could potentially be a biomarker for reduced T cell activity in vivo. To validate the divergent effect between IL-18BP concentrations and cytokine production by lymphocytes, we tested for this association in an independent cohort of 300 volunteers of Western-European descent with BMI >25 (300OB), for which we have obtained cytokine production profiles (ELISA) after stimulation of PBMC using the same pathogens and protocols as used in 500FG. In addition, circulating baseline (unstimulated) measurements for IL-18BP were determined. Because this cohort is comprised of mainly obese (BMI >25) and older (age >55) individuals, we limited the analysis to a subset of (n=51) 300-OB volunteers with BMI <28, to bring this distribution more in line with the 500FG cohort. We tested for association (Spearman correlation) between the cytokine production profiles after stimulation and circulating IL-18BP levels (Supplementary Fig. 2 b). We could replicate the negative effect of IL-18BP on lymphocyte cytokines.
Figure 3. Examples of baseline molecules which associate differentially to cytokine responses.
IL-18BP, a circulating inhibitor of IL-18, displays negative Spearman correlations with general cytokine production capacity of lymphocytes after correcting for age and gender effects (n=489). The metabolite acetate positively correlates with stimulated cytokine production in response to influenza and displays a mostly positive effect on lymphocyte-derived cytokines after correcting for age and gender effects (n=377). Each sample represents an individual.
The short chain fatty acid (SCFA) acetate showed the strongest correlation (negative correlation between -0.25 and -0.20) with influenza-induced monocyte–derived IL-1β, IL-6 and TNF-α cytokine production capacity (Fig. 3). Cytokine response to bacterial and fungal stimulations showed either positive or negative effects for monocyte-derived cytokine production capacity. In contrast, lymphocyte-derived IL-17, IL-22 and IFN-γ cytokine production showed consistently positive effects in response to most of the bacterial and fungal stimulations. This agrees with previous findings that SCFAs, including acetate, influence cytokine production capacity24–26. The negative correlation between acetate and stimulus-induced production of IL-1β, IL-6 and TNF-α was also observed when assessed in PBMC derived macrophages, but not in whole blood (Fig. 3). To further investigate the association between acetate and stimulus-induced cytokine production, we cultured PBMC derived macrophages obtained from whole blood of 6 healthy Dutch volunteers in vitro in the presence of acetate in the medium, stimulated them with MTB, C. albicans, S. aureus and E. coli, and assessed the cytokine production of TNF-α and IL-6 after 24 hours. We observed an association between acetate and cytokine production in macrophages where the production of TNF-α and IL-6 in PBMC derived macrophages upon two of the stimuli (E. coli and S. aureus) were lower in the presence of acetate, but this effect was not observed for C. albicans (Supplementary Fig. 2 c).
Glutamine is known to negatively regulate IL-6 production in human intestinal mucosa27 and decreases IL-6, TNF-α and IL-1β production in biopsies from Crohn’s disease patients28. We observed that glutamine, consistently correlated negatively with all monocyte- and lymphocyte-derived cytokines assessed after stimulation (Supplementary Fig. 3), suggesting it could be used as an anti-inflammatory biomarker. These results show that baseline molecules are differentially associated with cytokine production between stimuli, as well as between cell types.
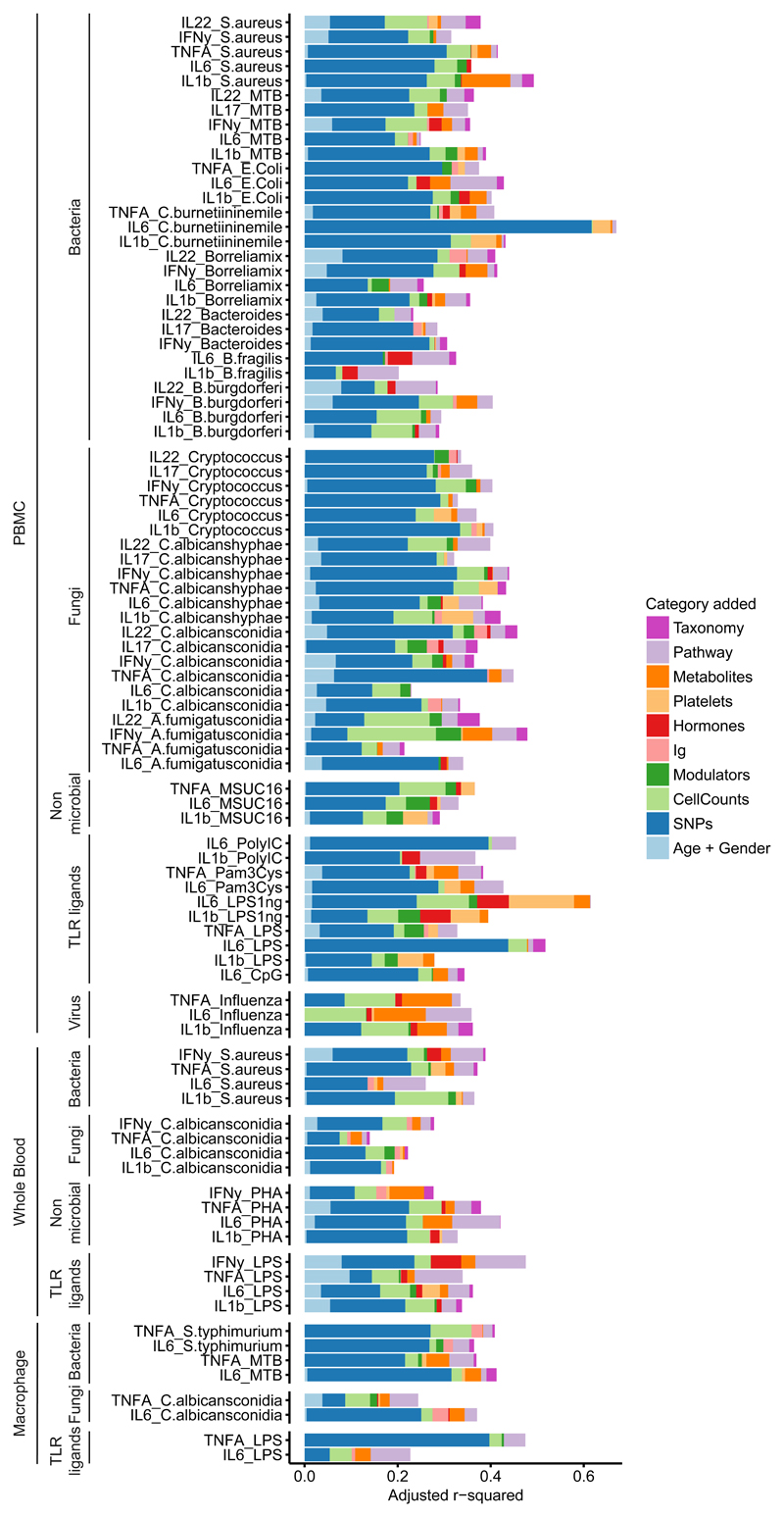
Host factors explain up to 67% variation in cytokine level
To determine the collective contribution of genetic variation and immune components at baseline to cytokine production in response to pathogens, a multivariate linear model was used. We constructed a MVLM for each cytokine stimulation pair where we added relevant features from each category of dataset sequentially and subsequently evaluated the increase in variance explained by each added dataset. This integrated approach indicated that a combination of genetic, baseline molecular profiles and immune parameters can explain up to 67% of the inter-individual variation in cytokine production capacity (Fig. 4). Because cytokine production is a highly complex phenotype, and many factors that influence it are associated to each other, we tested if changing the order in which specific datasets were added into the models generated different results. When we compared MVLM containing all datasets, to the partial MVLMs, in which each of the 10 datasets were omitted once, we found similar estimates of explained variation as in the sequential analysis (Supplementary Fig. 4). For example, regardless of the order the factors were added, genetics remained the largest individual contributor to explaining inter-individual variation (Supplementary Fig. 4). This indicated that the order in which various factors were added into the model did not influence the results to a large extent.
Figure 4. Cumulative contribution of multiple baseline traits to the variation in stimulated cytokine production.
Adjusted R2 values (x-axis) obtained from multivariate linear models (MVLM) increase when measurements from 10 categories are added sequentially. Each colored bar represents how much additional variation (on top of the preceding colors) the MVLM for that category explains. The order in which features from a dataset were added is from left to right. The combined dataset consisted of 266 samples. Each sample represents an individual. Gene expression was not included in this analysis because of the relatively small sample size of the RNA-seq experiment after overlapping with the other datasets (n = 69). X-axis denotes adjusted R2 values. Y-axis denotes different cytokine-stimulation pairs.
Gene expression correlates with cytokine response
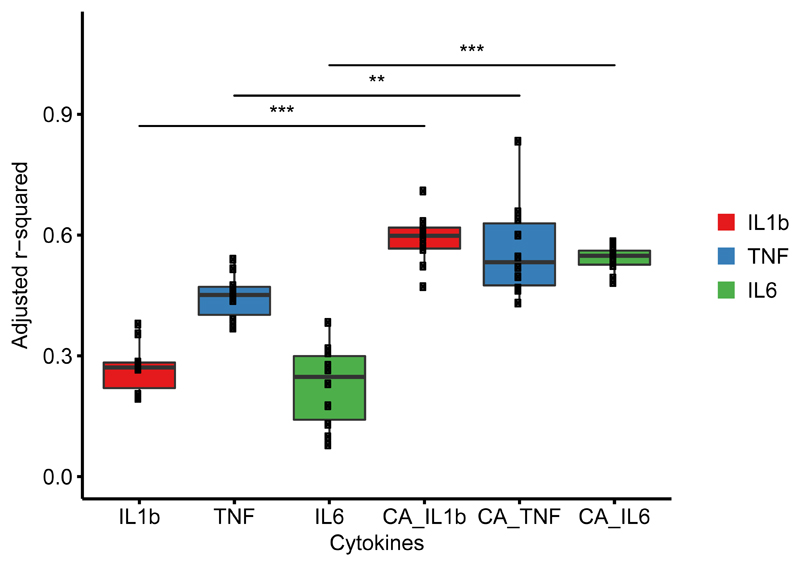
Next we integrated baseline transcript abundance with stimulus-induced cytokine expression. We made use of whole genome gene expression profiles obtained using RNA-Seq both before and after stimulation of peripheral blood with C.albicans conidia from a subset of volunteers (n = 64) from an independent Dutch cohort (Genome of The Netherlands cohort29). We used measurements of the production of TNF-α, IL-6 and IL-1β by PBMC upon stimulation with C.albicans conidia after 24 hours in the same individuals. We then applied the same MVLM based analysis approach used earlier to obtain estimates of how much inter-individual variation in cytokine production capacity could be explained by gene expression. We observed that baseline gene expression could explain a substantial portion of the inter-individual variation in production of TNF-α, IL-6 and IL-1β (Fig. 5). Production of TNF-α, IL-6 and IL-1β by PBMC stimulated with C. albicans conidia showed significantly higher correlations with gene expression induced by stimulation (adj. R2 reaching up to 0.75) than with baseline gene expression (Wilcox test, P=1.08e-05, P=8.93e-03, P=1.08e-05, for TNF-α, IL-6 and IL-1β respectively). Using GO enrichment (online tool20), we found that the genes selected during modelling (Supplementary Table 5) showed enrichment for several GO terms related to immune responses. For example the genes associated to C.albicans induced TNF-α levels were nominally enriched for negative regulation of mast cell cytokine production (P=1.28e-3), negative regulation of isotype switching to IgE isotypes (P=1.71e-3) and negative regulation of T-helper 2 cell differentiation (P=2.15e-3). These results imply a strong correlation between gene expression and functional responses upon stimulation by pathogens, and thus they present gene expression as a target for future studies into the prediction of immune responses.
Figure 5. Integrating gene expression profiles and cytokine production in response to C. albicans.
Percentage of inter-individual variation (y-axis, adjusted R2) in stimulated cytokine level of TNF-α, IL-6 and IL-1β explained by gene expression measured at baseline and upon C. albicans stimulation (denoted by CA) is significantly (Wilcox rank sum test, * P < 0.05, ** P < 0.01, *** P < 0.001) higher in the multivariate linear models (MVLM) fitted on stimulated gene expression data. Exact P values of the Wilcox rank sum test are as follows: IL-1β (P = 1.08e-05), TNF-α (P = 8.93e-04) and IL6 (P = 1.08e-05). The distribution shows adjusted R2 (y-axis) of 10 MVLMs fitted after re-sampling using a random subset of samples (90% of all samples each time). Each dot represents the adjusted R2 of a MVLM. The dataset consisted of 64 samples from the GoNL cohort. Each sample represents an individual.
Immune disease risk is associated with stimulated cytokine level
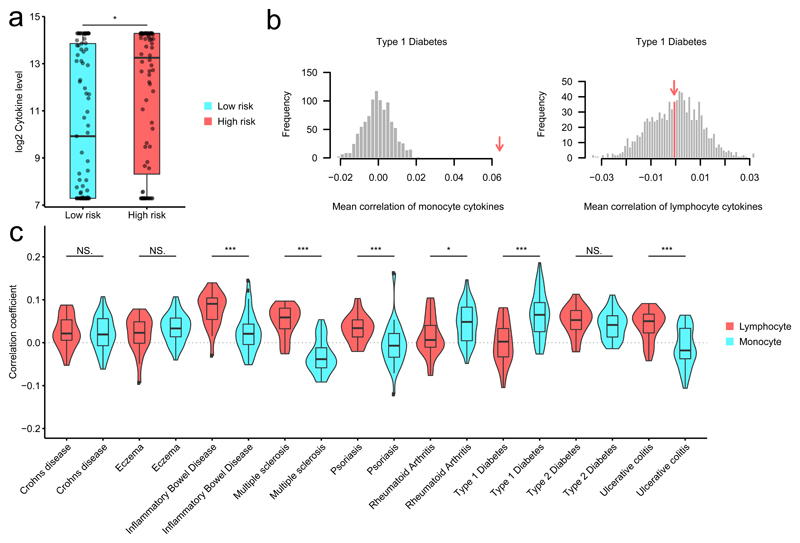
Many complex diseases appear to result from multiple genetic variants exerting small effects on disease risk30, which implies that complex diseases conform closely to a classical polygenic model. Using publicly available summary statistics from GWAS we calculated polygenic risk scores (PRS) for 15 immune mediated diseases (Supplementary Table 6) for all the volunteers in the 500FG cohort as a measure of relative disease risk between individuals We then tested whether volunteers with a higher risk for an immune mediated disease displayed higher or lower stimulus-induced cytokine production compared to the lower risk individuals. For this analysis, we focused those immune mediated diseases that showed both a significant change (two tailed, two sample t-test, Bonferroni P <0.05, Supplementary Table 7) compared to a permutation-based null distribution, and a consistent pattern at different thresholds used for PRS calculation (Fig. 6a-c, Supplementary Fig. 5a,b). We found that volunteers with higher risk for inflammatory bowel disease, multiple sclerosis, psoriasis and ulcerative colitis had significantly higher (P < 0.05) stimulus-induced production of lymphocyte-derived IL-17, IL-22 and IFN-γ compared to monocyte-derived TNF-α, IL-6 and IL-1β cytokines (Fig. 6). In contrast, higher risk for type 1 diabetes (T1D) and rheumatoid arthritis was associated with increased stimulus-induced production of monocyte-derived TNF-α, IL-6 and IL-1β compared to lymphocyte-derived cytokines (Fig. 6c). Higher risk for Crohn’s disease, eczema and type 2 diabetes was associated with a significant increase (compared to their respective null distributions, P < 0.05) in both monocyte- and lymphocyte-derived cytokines compared to the permutation-based null distribution, with no significant differences between the monocyte and lymphocyte derived groups (Fig. 6b). These observations suggest that the genetic basis for immune-mediated diseases could influence the functionality of the immune system even in otherwise healthy individuals.
Figure 6. Stimulated cytokine production correlates with genetic risk score for autoimmune diseases.
(a) Example individuals with high genetic risk for (auto)immune disease tend to be high producers of cytokines in response to pathogens. * indicates the significance of the Wilcox rank sum test between low- and high-risk groups for T1D (P=0.011). Low- and high-risk groups (x-axis) were selected by taking the top and bottom quantile of the PRS for T1D. Y-axis indicates the IL-6 level after stimulation of PBMCs with influenza. (b) Distribution mean correlations between T1D risk in monocyte-derived cytokines (left panel) and lymphocyte cytokines (right panel) for 1000 permutations. The measured estimate is indicated by the red arrow. T1D shows significance for monocyte derived cytokines (left) but not for the lymphocyte derived cytokines (right). (c) Distribution of Spearman correlation coefficients between stimulated cytokine production and genetic risk score for immune disease in 430 individuals, shown for PBMC. Genetic risk scores calculated based on genome-wide association studies for different diseases. Significant differences in mean correlation between the lymphocyte- and monocyte-derived cytokines are shown by Wilcox rank sum test (* P < 0.05, ** P < 0.01, *** P < 0.001). Exact p-values are as follows Crohns disease P=7.28E-01, Eczema P=2.55E-01, Inflammatory Bowel Disease P=9.34E-06, Multiple sclerosis P=4.85E-11, Psoriasis P=1.40E-04, Rheumatoid Arthritis P=1.41E-02, Type P=1 Diabetes P=1.00E-05, Type P=2 Diabetes P=1.65E-01, Ulcerative colitis P=1.34E-05.
Stimulated cytokine level predicted by genetics
Finally, we integrated both genetics and other molecular features to construct MVLMs to predict each cytokine stimulation pair in PBMC, whole blood and macrophages. To achieve the best prediction of ex vivo stimulus-induced cytokine production, we tested several linear prediction methods (Elastic Net, RR-BLUP and PLS) and compared them using both genetic and non-genetic factors to train the MVLMs for each cytokine stimulation pair. Predictive performance was quantified by Spearman’s correlation between the measured and the predicted stimulus-induced cytokine production in multiple randomly selected subsets of the volunteers from 500FG. While the prediction performances of the different methods are similar (Supplementary Fig. 6a-c), Elastic Net marginally outperformed the others, so we used it for subsequent analyses.
We first tested if SNP data could predict cytokine production. Among the 91 stimulation-cytokine pairs, the correlations between predicted and measured stimulus-induced cytokine production were, on average, 0.69 (range 0.28-0.89) (Fig. 7a). Inclusion of the baseline immune parameters and multi-omics data significantly increased the predictive power and stability of the model (two tailed student t-test, P=1.36e-09, t-statistic=6.09, degrees of freedom = 1792) and most predictions for cytokine production increased to, on average, 0.72 (range 0.35-0.90) (Fig. 7b). Additional inclusion of the gene expression data from the RNA-seq analysis decreased the predictive power (avg. 0.60, range 0-1) (Supplementary Fig. 6d), most likely due to the reduced number of samples for which both RNA-seq and the other factors were available (n = 69).
Figure 7. Cytokine production in response to pathogens can be predicted using genetics and baseline immune profiles.
Spearman correlation between predicted and measured cytokine levels (y-axis) are shown for each of the 10 multivariate linear models from cross validation for all available cytokine stimulation pairs. Cytokine production in response to pathogens can be predicted using SNPs (n = 392 individuals). Prediction accuracy increases when baseline immune parameters and molecular profiles (immune cell frequencies, immune modulators, immunoglobulins, hormone levels, blood platelets, circulating metabolites, gut microbiome composition) are added to the model (n = 353 individuals).
We then tested the predictive capabilities of the Elastic net trained MVLMs using only SNPs as input and applying it to independent subset of 500FG individuals were new cytokine stimulation experiments were performed (50FG). We found prediction accuracies up to 0.56 for some cytokine stimulation pairs (Fig.8), although the MVLMs performed poorly for most stimulations. Among the best-performing stimulus-cytokine pairs, C.burnetti stimulated IL-1β and Poly I:C-stimulated IL-6 gave prediction accuracies of on average 0.56 and 0.46 respectively (Fig. 8). Because both pathways are known to have a large genetic component31 this indicated that the MVLMs could predict cytokine production for stimulus-induced cytokines whose mechanism of induction are primarily driven by genetics.
Figure 8. Prediction using the genetic model in an independent dataset. shows some cytokine stimulation pairs can be predicted successfully.
Spearman correlations between predicted cytokine level by the multivariate linear models (MVLM) built using genetics (n = 336) and the measured values in an independent set of stimulation experiments (n = 56). The boxplots show the variation in Spearman correlations from each of the 10 MVLMs predictions from the cross validation strategy.
By applying MVLMs to genetics data, we were able to predict the cytokine production upon stimulation, with varying degrees of accuracy.
Discussion
In this study we assessed the combined contribution of genetic and non-genetic factors to the inter-individual variation in cytokine production in response to pathogens by examining the cytokine production of immune cells following stimulation with 20 different pathogens or TLR ligands ex vivo in PBMC, whole blood and PBMC derived macrophages. This analysis identified new modulators of cytokine production, including circulating inflammatory mediators and metabolites. We found that volunteers with increased genetic risk for immune mediated diseases were more likely to be high responders in terms of stimulus-induced cytokine production. Finally, we trained MVLMs that could predict human stimulus-induced cytokine production for Poly I:C induced IL-6 and C.burnetti IL-1β levels in PBMC using only the genetic profiles or a combination of genetic and other molecular profiles.
A recent study on the heritability of immune phenotypes in 210 twins suggested that variations in circulating cytokine concentrations are mostly driven by non-heritable influences32. Although we observed here that genetics was the largest single contributor to inter-individual variation (avg. adj. R2 = 0.18), this still leaves room for the majority of the variation to be explained by non-genetic influences. Any differences we observed in estimates of heritability are likely due to differences in the experimental design between the two studies. As such, we assessed cytokine profiles upon stimulation ex vivo, whereas the above study32 measured baseline circulating concentrations in vivo. This strongly suggests that it is the response to pathogens during infection that is under stronger genetic pressure rather than the background level of mediators in the circulation. Our study thus agrees with the idea that infections have a strong selective impact on the genetic control of immune responses33–40.
The present study has potentially important implications for our understanding of the human immune response. We found out that acetate, a circulating metabolites, was associated with changes in stimulus-induced cytokine production and especially in the modulation of TH1 and TH17 responses. SCFA such as acetate, propionate and succinate are released by the gut microbiome and current literature suggests that SCFA have important immunomodulatory properties24–27. We show here that acetate has similar effects in humans in vivo. It appears important to further investigate the broader impact of SCFA and identify which microbiome profiles modify their concentration in the circulation. We found a strong inhibitory effect of acetate on influenza-stimulated cytokine production, a phenomenon that deserves further scrutiny. Another important metabolic pathway that strongly influenced cytokine responses was the cholesterol and lipoprotein synthesis pathway. Cholesterol pathways have been described to have important immune-modulating effects, with the levels of cholesterol sulfate, a derivative of membrane cholesterol, shown to influence immune processes such as TCR signalling and thymic selection41. Here we showed that HDL cholesterol negatively impacted influenza and Aspergillus-stimulated cytokine production, possibly with important effects on the pathophysiology of these infections.
The ability to calculate prediction scores for specific immune mediated diseases and to link them to cytokine production shows that certain stimulus-induced cytokine profiles may contribute to particular diseases, e.g. the capacity to release high amounts of monocyte-derived cytokines in T1D. Although we acknowledge that our power to detect these smaller associations is relatively limited, our approach can be used to link any given phenotype to disease scores when individual-level data is available. This offers the opportunity to identify immune pathways important in disease, which may represent new therapeutic targets.
A second limitation of the 500FG cohort is that it contains a higher proportion of young people than the general population9, which could introduce age bias into the MVLM’s predictions. While we acknowledge that the performance of the MVLMs prediction may vary in a population with a different range in age, BMI or ancestry, our study represents a proof-of-concept that stimulus-induced cytokine production can be moderately predicted. Future studies in larger general population cohorts with greater ranges of age and ethnicity will contribute to the generation of models with improved predictive potential for a general population. Future studies should also aim to extend the current analysis, which was limited to common SNP polymorphisms (MAF >0.1), to include rare variants and mutations, a broadening of scope likely to further increase the observed impact of genetics on cytokine production upon stimulation.
In conclusion, we present the most comprehensive assessment to date of the host factors that influence cytokine production. We show that genetics was a major contributor to the inter-individual variation in cytokine production upon pathogen stimulation. However, other non-genetic factors also influenced cytokine production in response to most stimuli, including gut microbiome composition, immune cell numbers in circulation and circulating metabolite concentrations. Individuals with increased genetic risk for a given immune disease tended to have increased cytokine production, and stimulus-induced cytokine production could be predicted for Poly I:C induced IL-6 and C. burnetti IL-1β levels. This study provides the fundamentals for predicting components of cytokine production based on genetics and baseline host factor profiles, paving the way towards personalized immune-based therapies.
Methods
Study cohort
The main analyses were performed in the 500FG cohort, which is part of the Human Functional Genomics Project. This cohort consists of 534 healthy individuals (237 males and 296 females) of Caucasian origin. Volunteers range from 18 to 75 years of age, with the majority (421 individuals) being 30 years or younger (Supplementary Fig. 1A). BMI is within normal limits (15 to 35) with the majority (380 individuals) having a BMI between 20 and 25 (Supplementary Fig. 1B). Of these 534 original volunteers, 45 were excluded based on genetic background and questionnaire results (medication usage, chronic disease) leaving 489 individuals.
Replication cohort
Validation experiments were performed in the 300-OB cohort. This cohort consists of ~300 Dutch individuals. All individuals had a BMI >25, with an average BMI of 31, and range in age from 55 to 80 years, with an average age of 67 years. Validations were performed in a subset of the 300-OB cohort with an BMI <28 (N=55). Circulating metabolites and mediators as well as stimulated cytokine levels were measured in the same way as in 500FG.
Experimental procedures
The experimental procedures used to measure levels of cytokines, modulators, immunoglobulins and hormones have been described previously9. Genotyping, metagenomic sequencing of the gut microbiome, FACS sorting of PBMCs and determination of platelet activation profiles have also been described previously7,8,42. We selected a representative subset of 89 samples from the 500FG cohort for RNASeq (balanced for age and sex to match the original distribution in the cohort). These samples were processed for sequencing using the Illumina TruSeq version 2 library preparation kit. Paired-end sequencing of 2×50-bp reads was performed using the Illumina HiSeq 2000 platform. The quality of the raw reads was checked using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Read alignment was performed using STAR 2.3.043, and aligned reads sorted using SAMTools. Gene level quantification of reads was done using HTSeq44. Circulating metabolites were measured and analysed using the BrainShake Biomarker Analysis Platform that is based on nuclear magnetic resonance (NMR) spectroscopy (BrainShake, Finland)
Statistical methods
Data pre-filtering
After pre-processing, the gene expression, SNP, metabolite and microbiome datasets were filtered to remove any non-significantly-associated features. This was done to increase the efficiency of downstream analysis. The gene expression metabolite and microbiome datasets were correlated to all of the cytokine measurements, and all features showing a Spearman correlation with a Benjamini-Hochberg adjusted P <0.05 to at least one cytokine were kept. This resulted in a dataset of 4,499 genes, 205 metabolites, 509 microbial pathways and 162 microbial taxonomies. The genetic variants were filtered using previously generated cytokine QTL profiles7 by setting the P-value cut-off at various thresholds depending on the application. To calculate the variance explained by genetics, a P-value threshold of P <5×10-6 was chosen. For prediction using the Elastic Net model, various thresholds were evaluated after which all SNPs with a P <5×10-5 were included in the analysis.
Estimation of explained variance
The estimation of variance explained by each of the data levels on the different stimulated cytokine production profiles was performed by applying a correlation-based feature selection approach. In this approach, we built a model for each stimulated cytokine measurement in which only features associated to this measurement are included in the model. We select these features by first regressing out the effects of age and gender, then associating the features in a data level to the current cytokine stimulation pair. If a feature showed a significant association (Spearman P-value <0.05), the feature was included in the set of potential predictors. Once all the associations had been computed, the set of potential predictors was correlated to itself to identify collinearity among this predictor set. If features within this predictor set showed an association (Spearman correlation >0.4), the feature which showed the least association (based on the correlation P-values) to the cytokine stimulation pair is removed. This yielded a unique set of predictors for every cytokine stimulation pair, which was then used to fit a multivariate linear model to estimate the variance explained by these features for that cytokine stimulation pair. To account for the inflation that adding predictors has on the explained variation, the adjusted R2 was taken as the measure of explained variance.
Permutation of cytokine GWAS
The baseline cytokine GWAS was performed as described previously 7. We randomly permuted the cytokine and covariate datasets 1000 times then ran the GWAS using these datasets to obtain 1000 random profiles for each cytokine stimulation pair. For each run we obtained the QTL profile and estimated the explained variance using the permuted cytokine and covariate dataset and the pipeline described above. This yielded a distribution of 1000 estimates of explained variance for each cytokine stimulation pair. A measured estimate was considered significant if it was in the top 5% of the permuted distribution of estimates for that cytokine stimulation pair.
Estimation of age and gender effects
Age and gender effects on cytokine production were assessed by fitting univariate linear models for each cytokine stimulation pair with age and gender as the independent variables, respectively. The R2 was taken as the measure of explained variation of these models.
Estimation of seasonal effect
The effect of season on stimulated cytokine production was assessed using a linear combination of sine and cosine terms with the same period (equation 1) as described by ter Horst et al9:
| (1) |
Where y represents the response (cytokine level), β the estimated intercept, α the estimated predictor effect, x the day of the year the sample was taken in, and ϵ the residual effect.
Estimation of cumulative explained variance
To assess the proportion of variance that can be explained by all levels cumulatively, individual levels were added to a multivariate linear model one by one, and the total model adjusted R2 calculated for each step. If adding a level showed an increase in the total adjusted R2 of the model, this value was extracted. To assess the contribution of each level conditional upon the others, the full model was fit first. Subsequently several reduced models were fit where one data level was missing. The adjusted R2 for this full model was then compared against the model with the missing level. The difference between the reduced model and the full model was taken as a measure of the variance explained by that level when accounting for the effects of the other levels.
Cytokine level prediction
Our objectives were to investigate whether genetic variants can reveal predictive insights into the cytokine production upon stimulation and whether baseline immune parameters, which are treated as quantitative phenotypes that are continuously distributed over a population, can improve predictive power for cytokine production upon stimulation. Using our population-based study, we searched for those subsets of genetic variants and immune components that are most predictive of the various stimulated cytokine production profiles, rather than using exclusively those variants meeting a stringent level of statistical significance.
We assessed the validity of this approach by applying multiple methods, each of which is discussed in detail below. In total three datasets were evaluated: one for predicting stimulated cytokine production using only SNPs, one containing all levels except gene expression, and one with all levels including gene expression. Firstly, features with little association with cytokine production levels (Spearman P >0.05) were removed for building the prediction models. For the SNP dataset, all SNPs with an association to a cytokine stimulation pair with P <5x10-5 were used as input for feature selection. No filtering for collinearity was applied because Elastic Net accounts for potential collinearity among predictors45.
Elastic Net
Prediction of the cytokine levels was facilitated by training an Elastic Net model. A 2×10-fold cross-validation approach was used, where the data was first split up into 10 random training and test sets to validate the prediction, and the training set was then split up once more for feature selection. Prediction accuracy was evaluated by calculating Spearman correlations between the measured cytokine levels and the predictions of the Elastic Net model on the test sets.
RR BLUP
To show that the prediction results are not influenced to a large extent by the methodology, a mixed linear model (equation 2), as implemented in the package rrBLUP 46, was applied:
| (2) |
Where y represents the response (cytokine level), 1 a vector of 1s, μ the overall mean of the training set, Z the matrix of predictors (traits), u the random effect of the predictors, and ϵ a vector of residual effects. Predictions were made using 10-fold cross-validation. Spearman correlation was then calculated between predicted and measured values. We applied this model as was described previously47.
Partial least squares regression
In addition to the Elastic Net and rrBLUP a partial least squares model was applied. Models were validated using 10-fold cross-validation. Prediction of cytokine levels on the test set was done using a linear model (equation 3):
| (3) |
Where y represents the response (cytokine level), β the intercept, α a vector containing the coefficients from the model, X the matrix of predictors (immune traits), and ϵ the residual error.
Polygenic risk scores
We carried out polygenic scoring of disease risk using publically available GWAS results. Quantitative scores were computed for each trait in this study based on the set of SNPs with P-values lower than predefined P-value thresholds (pT) in the GWAS. Multiple pT were evaluated (pT <5e--8, 1e-5, 1e-4, 1e-3, and pT < 1e-2). Throughout this work, we refer to the scores defined at pT <1e-5 as Polygenic Risk Scores (PRS). Full association summary statistics were downloaded from several publicly available resources indicated in Supplementary Table 6 48–60,. Studies done exclusively in non-European cohorts were omitted. Filters applied to the separate data sources are indicated below. All the dbSNP rs numbers were standardized to match GIANT 1kG p1v3 and the directions of the effects were standardized to correspond to the GIANT 1kG p1v3 minor allele. SNPs with different opposite-strand alleles compared to GIANT alleles were flipped. SNPs with A/T and C/G SNPs and SNPs with different alleles GIANT 1kG p1v3 (tri-allelic SNPs, indels, unknown alleles) were removed from the analysis. Genomic control was applied to all P-values for the datasets not genotyped by Immunochip or Metabochip. We calculated PRS by first clumping variants based on the threshold pT, linkage-disequilibrium (R2 < 0.2) and a 250kb window using the PLINK 1.9 option “clump” and exclusively European samples from 1000 genomes data as a reference for linkage disequilibrium calculation. PRS were subsequently obtained for each threshold pT by calculating them using the linkage-disequilibrium-clumped subset of SNPs using the PLINK 1.9 option “score”.
Association between polygenic risk scores and cytokine production
The association between the PRS and cytokine production capacity upon stimulation was determined by calculating the Spearman correlation between each of the PRS profiles and each of the stimulated cytokine profiles. To evaluate the statistical significance of association, a permutation method was used. The cytokine data was permuted 1000 times and the correlation was calculated for each of these permuted datasets. Both the measured and permuted distributions were separated into the lymphocyte and monocyte groups, and a student t-test was applied between the measured distribution and the permuted distribution. When either the monocyte or lymphocyte group showed a significant deviation from the permuted distribution (Bonferroni adjusted two sample t-test P <0.05) the disease was selected for interpretation.
Supplementary Material
Acknowledgements
We thank all of the volunteers in the 500FG and GoNL cohorts for their participation. We thank T.A. Wassenaar and L. Steenhuis of the Hanze University of Applied Science for providing input on the project and helping with the analyses. We thank K. Mc Intyre for editing the text. We thank the EAGLE eczema consortium for making their summary statistics publicly available. The HFGP is supported by a European Research Council (ERC) Consolidator grant (ERC 310372). This study was further supported by an IN-CONTROL CVON grant (CVON2012-03) and a Netherlands Organization for Scientific Research (NWO) Spinoza prize (NWO SPI 94-212) to M.G.N.; an ERC advanced grant (FP/2007-2013/ERC grant 2012-322698) and an NWO Spinoza prize (NWO SPI 92-266) to C.W.; a European Union Seventh Framework Programme grant (EU FP7) TANDEM project (HEALTH-F3-2012-305279) to C.W. and V.K.; and an NWO VENI grant (NWO 863.13.011) and an ZonMw-OffRoad-91215206 grant to Y.L. M.O. was supported by an NWO VENI grant (016.176.006). RJX was supported by Nation Institutes of Health (NIH) grants - DK43351, AT009708, AI137325.
Footnotes
Data availability
The data that support the findings of this study are available at https://hfgp.bbmri.nl/ were it has been meticulously catalogued and archived at BBMRI-NL aiming for maximum reuse following the FAIR principles, i.e., Findability, Accessibility, Interoperability, and Reusability. Individual level genetic data as well as other privacy sensitive datasets are available upon request at http://www.humanfunctionalgenomics.org/site/?page_id=16. These datasets are not publicly available because they contain information that could compromise the research participants privacy. The central data stewardship and access has been implemented using MOLGENIS open source platform for scientific data that enables flexible data upload, management and querying, including sufficiently rich metadata and interfaces for machine processing and custom (R statistics) visualization for human processing (see http://molgenis.org). Also summaries of the study have been submitted to BBMRI central catalogues https://catalogue.bbmri.nl (Netherlands) and http://www.bbmri-eric.eu/news-events/bbmri-eric-directory-2-0/ (EU).
Author contributions
Y.L., C.W. and M.G.N. designed this study. M.O., S.P.S., M.J., R.T.N.-M., H.J.P.M.K., I.J., R.J.X., and L.A.B.J. performed the experiments and processed the data. U.V. collected and pre-processed public summary statistics. O.B.B. performed statistical analysis assisted by R.A.-G., S.S., U.V. and L.F.. O.B.B., M.Z., Y.L., S.W., V.K., M.G.N, and C.W interpreted the data. Y.L., C.W., M.G.N. and O.B.B. wrote the manuscript with input from all authors.
Competing Financial Interests
The authors declare no competing interests.
Ethics statement
The HFGP study was approved by the ethical committee of Radboud University Nijmegen (no. 42561.091.12). Experiments were conducted according to the principles expressed in the Declaration of Helsinki. Samples of venous blood were drawn after informed consent was obtained.
References
- 1.Fairfax BP, et al. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science. 2014;343:1246949. doi: 10.1126/science.1246949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar V, Wijmenga C, Xavier RJ. Genetics of immune-mediated disorders: from genome-wide association to molecular mechanism. Current Opinion in Immunology. 2014;31:51–57. doi: 10.1016/j.coi.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee MN, et al. Common genetic variants modulate pathogen-sensing responses in human dendritic cells. Science. 2014;343:1246980. doi: 10.1126/science.1246980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Netea MG, Wijmenga C, O’Neill LAJ. Genetic variation in Toll-like receptors and disease susceptibility. Nat Immunol. 2012;13:535–542. doi: 10.1038/ni.2284. [DOI] [PubMed] [Google Scholar]
- 5.Ye CJ, et al. Intersection of population variation and autoimmunity genetics in human T cell activation. Science. 2014;345:1254665. doi: 10.1126/science.1254665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brodin P, Davis MM. Human immune system variation. Nat Rev Immunol. 2017;17:21–29. doi: 10.1038/nri.2016.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y, et al. A Functional Genomics Approach to Understand Variation in Cytokine Production in Humans. Cell. 2016;167:1099–1110.e14. doi: 10.1016/j.cell.2016.10.017. [DOI] [PubMed] [Google Scholar]
- 8.Schirmer M, et al. Linking the Human Gut Microbiome to Inflammatory Cytokine Production Capacity. Cell. 2016;167:1125–1136.e8. doi: 10.1016/j.cell.2016.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.ter Horst R, et al. Host and Environmental Factors Influencing Individual Human Cytokine Responses. Cell. 2016;167:1111–1124.e13. doi: 10.1016/j.cell.2016.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brannian JD, Zhao Y, McElroy M. Leptin inhibits gonadotrophin-stimulated granulosa cell progesterone production by antagonizing insulin action. Hum Reprod. 1999;14:1445–1448. doi: 10.1093/humrep/14.6.1445. [DOI] [PubMed] [Google Scholar]
- 11.Härle P, et al. Possible role of leptin in hypoandrogenicity in patients with systemic lupus erythematosus and rheumatoid arthritis. Annals of the Rheumatic Diseases. 2004;63:809–816. doi: 10.1136/ard.2003.011619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blum WF, et al. Plasma Leptin Levels in Healthy Children and Adolescents: Dependence on Body Mass Index, Body Fat Mass, Gender, Pubertal Stage, and Testosterone. J Clin Endocrinol Metab. 1997;82:2904–2910. doi: 10.1210/jcem.82.9.4251. [DOI] [PubMed] [Google Scholar]
- 13.Behre HM, Simoni M, Nieschlag E. Strong association between serum levels of leptin and testosterone in men. Clinical Endocrinology. 1997;47:237–240. doi: 10.1046/j.1365-2265.1997.2681067.x. [DOI] [PubMed] [Google Scholar]
- 14.Xu T, et al. P-Selectin Cross-Links PSGL-1 and Enhances Neutrophil Adhesion to Fibrinogen and ICAM-1 in a Src Kinase-Dependent, but GPCR-Independent Mechanism. Cell Adh Migr. 2007;1:115–123. doi: 10.4161/cam.1.3.4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gawaz M, Langer H, May AE. Platelets in inflammation and atherogenesis. J Clin Invest. 2005;115:3378–3384. doi: 10.1172/JCI27196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furman D, et al. Apoptosis and other immune biomarkers predict influenza vaccine responsiveness. Mol Syst Biol. 2013;9:659. doi: 10.1038/msb.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furman D, et al. Cytomegalovirus infection improves immune responses to influenza. Sci Transl Med. 2015;7:281ra43. doi: 10.1126/scitranslmed.aaa2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furman D, et al. Systems analysis of sex differences reveals an immunosuppressive role for testosterone in the response to influenza vaccination. PNAS. 2014;111:869–874. doi: 10.1073/pnas.1321060111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis MM, Tato CM, Furman D. Systems immunology: just getting started. Nat Immunol. 2017;18:725–732. doi: 10.1038/ni.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ashburner M, et al. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shah S, et al. Improving Phenotypic Prediction by Combining Genetic and Epigenetic Associations. Am J Hum Genet. 2015;97:75–85. doi: 10.1016/j.ajhg.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Novick D, et al. Interleukin-18 Binding Protein. Immunity. 1999;10:127–136. doi: 10.1016/s1074-7613(00)80013-8. [DOI] [PubMed] [Google Scholar]
- 23.Okamura H, et al. Cloning of a new cytokine that induces IFN-gamma production by T cells. Nature. 1995;378:88–91. doi: 10.1038/378088a0. [DOI] [PubMed] [Google Scholar]
- 24.Vinolo MAR, Rodrigues HG, Nachbar RT, Curi R. Regulation of Inflammation by Short Chain Fatty Acids. Nutrients. 2011;3:858–876. doi: 10.3390/nu3100858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tedelind S, Westberg F, Kjerrulf M, Vidal A. Anti-inflammatory properties of the short-chain fatty acids acetate and propionate: a study with relevance to inflammatory bowel disease. World Journal of Gastroenterology. 2007;13:2826. doi: 10.3748/wjg.v13.i20.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cavaglieri CR, et al. Differential effects of short-chain fatty acids on proliferation and production of pro- and anti-inflammatory cytokines by cultured lymphocytes. Life Sciences. 2003;73:1683–1690. doi: 10.1016/s0024-3205(03)00490-9. [DOI] [PubMed] [Google Scholar]
- 27.Coëffier M, Marion R, Ducrotté P, Déchelotte P. Modulating effect of glutamine on IL-1β-induced cytokine production by human gut. Clinical Nutrition. 2003;22:407–413. doi: 10.1016/s0261-5614(03)00040-2. [DOI] [PubMed] [Google Scholar]
- 28.Lecleire S, et al. Combined Glutamine and Arginine Decrease Proinflammatory Cytokine Production by Biopsies from Crohn’s Patients in Association with Changes in Nuclear Factor-κB and p38 Mitogen-Activated Protein Kinase Pathways. J Nutr. 2008;138:2481–2486. doi: 10.3945/jn.108.099127. [DOI] [PubMed] [Google Scholar]
- 29.The Genome of the Netherlands Consortium. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat Genet. 2014;46:818–825. doi: 10.1038/ng.3021. [DOI] [PubMed] [Google Scholar]
- 30.Lvovs D, Favorova OO, Favorov AV. A Polygenic Approach to the Study of Polygenic Diseases. Acta Naturae. 2012;4:59–71. [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y, et al. Inter-individual variability and genetic influences on cytokine responses to bacteria and fungi. Nature Medicine. 2016;22:952–960. doi: 10.1038/nm.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brodin P, et al. Variation in the human immune system is largely driven by non-heritable influences. Cell. 2015;160:37–47. doi: 10.1016/j.cell.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barreiro LB, Quintana-Murci L. From evolutionary genetics to human immunology: how selection shapes host defence genes. Nat Rev Genet. 2010;11:17–30. doi: 10.1038/nrg2698. [DOI] [PubMed] [Google Scholar]
- 34.Casals F, et al. Genetic adaptation of the antibacterial human innate immunity network. BMC Evolutionary Biology. 2011;11:202. doi: 10.1186/1471-2148-11-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andrés AM, et al. Targets of Balancing Selection in the Human Genome. Mol Biol Evol. 2009;26:2755–2764. doi: 10.1093/molbev/msp190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pickrell JK, et al. Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 2009;19:826–837. doi: 10.1101/gr.087577.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang K, Thornton KR, Stoneking M. A New Approach for Using Genome Scans to Detect Recent Positive Selection in the Human Genome. PLOS Biology. 2007;5:e171. doi: 10.1371/journal.pbio.0050171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A Map of Recent Positive Selection in the Human Genome. PLOS Biology. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang ET, Kodama G, Baldi P, Moyzis RK. Global landscape of recent inferred Darwinian selection for Homo sapiens. PNAS. 2006;103:135–140. doi: 10.1073/pnas.0509691102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williamson SH, et al. Localizing Recent Adaptive Evolution in the Human Genome. PLOS Genetics. 2007;3:e90. doi: 10.1371/journal.pgen.0030090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang F, Beck-García K, Zorzin C, Schamel WWA, Davis MM. Inhibition of T cell receptor signaling by cholesterol sulfate, a naturally occurring derivative of membrane cholesterol. Nat Immunol. 2016;17:844–850. doi: 10.1038/ni.3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aguirre-Gamboa R, et al. Differential Effects of Environmental and Genetic Factors on T and B Cell Immune Traits. Cell Reports. 2016;17:2474–2487. doi: 10.1016/j.celrep.2016.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zou H, Hastie T. Regularization and variable selection via the elastic net. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2005;67:301–320. [Google Scholar]
- 46.Endelman JB. Ridge Regression and Other Kernels for Genomic Selection with R Package rrBLUP. The Plant Genome Journal. 2011;4:250. [Google Scholar]
- 47.Riedelsheimer C, et al. Genomic and metabolic prediction of complex heterotic traits in hybrid maize. Nat Genet. 2012;44:217–220. doi: 10.1038/ng.1033. [DOI] [PubMed] [Google Scholar]
- 48.Morris AP, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paternoster L, et al. Multi-ancestry genome-wide association study of 21,000 cases and 95,000 controls identifies new risk loci for atopic dermatitis. Nat Genet. 2015;47:1449–1456. doi: 10.1038/ng.3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Putov NV, Bulatov PK, Gorovenko GG, Fedoseev GB, Brusilovskiĭ BM. [Classification of unspecific diseases of the bronchopulmonary system] Vrach Delo. 1977:52–56. [PubMed] [Google Scholar]
- 51.Köttgen A, et al. Genome-wide association analyses identify 18 new loci associated with serum urate concentrations. Nat Genet. 2013;45:145–154. doi: 10.1038/ng.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu JZ, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trynka G, et al. Dense genotyping identifies and localizes multiple common and rare variant association signals in celiac disease. Nat Genet. 2011;43:1193–1201. doi: 10.1038/ng.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hinks A, et al. Dense genotyping of immune-related disease regions identifies 14 new susceptibility loci for juvenile idiopathic arthritis. Nat Genet. 2013;45:664–669. doi: 10.1038/ng.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.International Multiple Sclerosis Genetics Consortium et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cordell HJ, et al. International genome-wide meta-analysis identifies new primary biliary cirrhosis risk loci and targetable pathogenic pathways. Nat Commun. 2015;6:8019. doi: 10.1038/ncomms9019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsoi LC, et al. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat Genet. 2012;44:1341–1348. doi: 10.1038/ng.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Okada Y, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bentham J, et al. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat Genet. 2015;47:1457–1464. doi: 10.1038/ng.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Onengut-Gumuscu S, et al. Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat Genet. 2015;47:381–386. doi: 10.1038/ng.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.