Abstract
λ genes O and P are required for replication initiation from the bacteriophage λ origin site, oriλ, located within gene O. Questions have persisted for years about whether O-defects can indeed be complemented in trans. We show the effect of original null mutations in O and the influence of four origin mutations (three are in-frame deletions and one is a point mutation) on complementation. This is the first demonstration that O proteins with internal deletions can complement for O activity, and that expression of the N-terminal portion of gene P can completely prevent O complementation. We show that O-P co-expression can limit the lethal effect of P on cell growth. We explore the influence of the contiguous small RNA OOP on O complementation and P-lethality.
Keywords: bacteriophage lambda (λ), bi-directional replication initiation from oriλ, O and P initiator proteins, oriλ interaction site, O complementation by oriλ-defective alleles, influence of O:P interactions on cell growth and O activity
1. Introduction
Bacteriophage λ prophage is maintained within the chromosome of Escherichia coli cells by its CI repressor protein, which prevents the transcription of λ genes positioned leftward and rightward from promoters pL and pR that straddle cI (Figure 1A). CI binds to operator sites that overlap these promoters. Upon inactivation of CI, the derepressed prophage genes N-int are expressed from pL, and genes cro-cII-O-P-Q are expressed from pR. Transcription initiated from pR requires gpN activity to proceed effectively past the rho-dependent tR1 termination site positioned between cro and cII (reviewed in [1,2,3,4]).
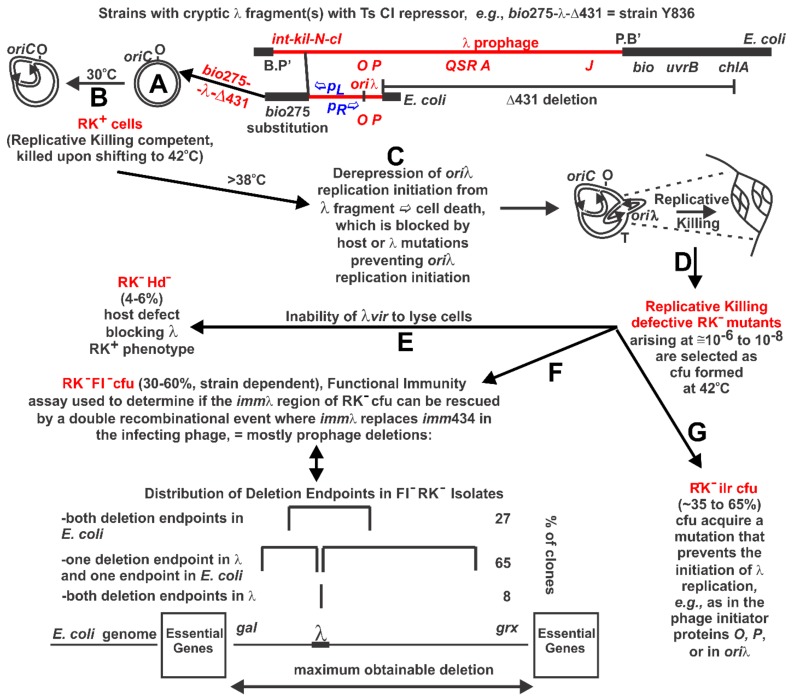
Figure 1.
Replicative Killing, RK+ phenotype and selection for RK− mutants. (A) Defective prophage strains were made where the int-kil or int-ral genes of λ were substituted with the bio275 or bio10 regions of specialized transducing phage to remove a phenotype termed “killing to the left”, dependent on kil [23]. The starting cells included the chlA deletion Δ434 that removed all of the late genes, i.e., cell lysis, head and tail for λ [11]. These constructs include (i) an active immλ region with gene cI[Ts]857 encoding a repressor that blocks transcription from promoters pL and pR along with the cro repressor just right of pR, and (ii) the repλ region that includes genes O and P and the oriλ target for replication initiation from the λ genome. The genome for strain Y836, shown, has the bio+ operon to the left of the λ fragment and Δ431deletion to the right; (B) As long as strain Y836 maintains CI repressor activity the cells can grow normally without gene expression from the repressed λ fragment; (C) When the cells are shifted to growth conditions where the CI[Ts] repressor loses its ability to block transcription from pL and pR the remaining λ genes become derepressed, the phage replication initiation genes O and P are expressed and rounds of replication initiation arise from oriλ. The λ replication forks extend bidirectionally into the adjacent regions of the E. coli genome, likely colliding with E. coli replication forks. The event is highly lethal to the cell because the λ fragment has no mechanism for excision from the genome and was termed Replicative Killing [18]; (D) When cells with a conditionally repressible defective λ prophage are shifted from growth at 30 °C to 42 °C the Replicative Killing, RK+, phenotype is triggered, resulting in cell death. Rare mutations that suppress the loss of λ replication control are selected as RK− clones capable of colony formation at 42 °C. These survivor CFU have lost the capacity for λ replication. This strategy is based on the PDS selection [15], where an intact prophage is made N-defective, so that expression of int-xis and late/cell lysis gene expression is limited without N-antitermination of pL and pR transcription upon prophage induction. There are many possibilities for RK− mutants; (E) Cells acquiring defects in host genes participating λ replication are termed RK− Hd−. For example, the GrpD55 mutation in dnaB is of this type, though not isolated as shown [24,25]; (F) A marker rescue recombination assay is used to determine if the immλ regions genes and target sites remain functional (i.e., FI+) when substituted for the imm434 region of a hybrid phage. The FI assay scores for the activity of the pR promoter, but in practice it is a good indication of whether the λ fragment in Y836 cells was partially or fully deleted. An example of the deletion endpoints of RK− FI− mutants from Y836 is shown [26,27,28]; (G) It was found that brief pretreatment RK+ of cells held at 30 °C with a mutagenic substance, prior to shifting them to 42 °C increases the frequency of RK− mutants. This assay, termed the RK Mutatest, proved very sensitive due to the rather large target potential for RK− mutants [29,30,31].
The mechanism for bi-directional initiation of λ DNA replication involves a complex interaction of phage proteins gpO and gpP (designated herein as O and P) with E. coli host DNA replication proteins. In brief, O acts to bind the replicator site, oriλ, or origin for replication initiation that is situated midway within the O sequence [5,6]. The P protein recruits DnaB, the major replicative helicase for unwinding double-stranded DNA, bringing it to oriλ-bound O to form a DnaB:P:O:oriλ preprimosomal complex. P can commandeer DnaB away from its cellular equivalent, DnaC [7]. Since the interaction of P with DnaB inactivates the helicase activity of DnaB, the dissociation of P bound to DnaB in the preprimosomal complex is required to restore DnaB activity, which involves E. coli heat shock proteins DnaK, DnaJ, and GrpE [8,9]. By inhibiting transcription from pR, CI blocks a cis requirement for replication initiation described as transcriptional activation, explaining why providing O and P from a superinfecting heteroimmune phage will not stimulate replication initiation from an integrated resident immλ prophage [10,11]. This requirement for pR transcription can be suppressed by riC (replicative-inhibition constitutive) mutations which lie outside of oriλ [12,13].
The excision of a λ prophage from the host chromosome between B.P’ and P.B’ sites (Figure 1A) is dependent upon λ genes int and xis (reviewed in [1,4,14]. Their expression requires that gpN antiterminate transcription at tL terminator signals positioned between pL-N and ahead of xis. The PDS selection (refer to Abbreviations, Unique λ Terminology) for cell survivors of λ N cI ([Ts], temperature sensitive) prophage, named for its inventor [15], takes advantage of the induced prophage being unable to excise from the chromosome or lyse its host cell. In addition, the N null mutation reduces late λ gene expression and cell lysis, which depend on N for transcriptional antitermination at several tR sites upstream of the late genes. Examinations for E. coli cell survival using the PDS selection led to the suggestion that mutations preventing the initiation of λ replication suppress cell killing [15,16,17,18,19,20,21].
The induction lethality phenotype for non-excisable prophage was termed Replicative Killing [18]. Accordingly, starting cells with de-repressible, but non-excisable prophage, as in Figure 1A and B, are termed RK+ (Replicative Killing competent) cells and the selected survivor cells that form CFU at 42 °C were named Replicative Killing defective (RK−) mutants (Figure 1C–G). The concept evolved that the starting cells possess the capacity for λ replication initiation upon prophage induction, whereas the survivor clones do not. The results of these early studies were reviewed [22]. Only a few mutations conferring the RK− phenotype for survivor CFU derived from the PDS selection or those from the N+ λ fragment strains (an example is shown in Figure 1) have been characterized by DNA sequence analysis. Most all the RK− mutants were obtained before the possibility for PCR amplification of a mutated region of the chromosome, which enables direct sequence determination of the RK− mutation. Hence, those RK− mutations that have been characterized depended mainly upon genetic analysis using phage mapping and complementation.
Genetic mapping of RK− mutations within O or P requires that both O and P initiator gene products can complement and function in trans. When a cell is infected with two phages, one defective in O and the other in P, complementation is observed suggesting that the products of these genes are diffusible [32]. However, Rao and Rodgers [33] were unable to demonstrate that cells carrying a ColE1 plasmid expressing O+ could complement, i.e., support the efficient plating of an infecting λimm21 Oam29 phage, even though the plasmid copy number varied between 50 copies at 32 °C and 260 copies at 42 °C. However, they could demonstrate trans complementation for phages with amber mutations in N or P by plasmids that can express these genes. Kleckner [34] suggested that O might act in cis or be poorly complemented under N defective conditions, and other experiments suggesting that O functions in cis were reported in [22]. These findings throw into question whether it is possible to designate using a phage complementation assay whether an induced RK− mutant has an O+ or O− phenotype. Alternatively, these divergent observations suggest that the ability of O to complement is more complex than initially assumed. A complicating problem in addressing this historical issue is that the mutant λ phages used in these early complementation assays were never subjected to DNA sequence analysis, so that in many cases their designations depend only upon unreported phage mapping studies, without accompanying proof of mutational site determination.
In this report, we examine the sequences of some early O mutations provided by A. Campbell from his original collection [35], and phages we have acquired over the years from laboratories that have participated in studies on O. We have cloned out O alleles from the chromosomes of RK− mutants and inserted them into a plasmid where the expression of the allele is regulated by CI[Ts], and is repressed in cells growing at 30 °C, or can be slightly to fully induced at growth temperatures between 37 °C to 42 °C. Each of these alleles was examined for their ability to complement the growth of a λ O amber mutant(s) in trans. We have explored the influence of O:P interactions on cell growth and toxicity, since the expression of P by itself, using the same system, is highly toxic [25,36]. These studies reveal that some alleles of O with internal deletions can complement as well or better than O+ and that the co-expression of O-P, or of O with portions of the N-terminal end of P, prevents an ability of O to complement in trans.
2. Results
2.1. Taking Stock of O Mutations in Phage and Prophage Collections
DNA sequence characterization of O mutations in phage and prophage collections available to us is summarized, Table 1. Campbell (AC) described and mapped Oam mutations 8, 29 and 125 [35]. Furth genetically mapped Oam mutations and ordered them (N- to C-terminal) 905, 29, 1005, 8, 125, 205 by marker rescue using six prophage strains, each with a deletion designated as extending into O [37,40]. The original AC prophage in strain R573 representing Oam125 included two missense mutations in addition to an amber mutation at 39511 bpλ. These three mutations were carried on a phage (our lysate #1024, Table 1) from LT designated MMS254. In contrast, isolate #1023 for Oam29 designated LT-MMS99 included a silent mutation in addition to the amber mutation at 39511. Of relevance, four phage lysates (designated as carrying Oam8 or Oam29 mutations) that were obtained from researchers were found WT for λ genes cII-O-P through base 40712 in orf ninB, but they did include a nonsense mutation somewhere in λ since they grew well on a supE host but not on a supo host. These results can explain why the initial assignment of an O+ phenotype to some RK− ilr mutants proved incorrect (Figure 2).
Table 1.
Collection of sequenced phage mutants in O and P.
| Collection Isolate | Mutated Base in λ, Mutation(s), Comment, Strain Source a |
|---|---|
| Phage lysates (#) | |
| Mutations in O | |
| λcI857 Oam905 (#1022) | 38797 G to T GAG to TAG), LT; 37 AA at N-terminal of O |
| λ [Ts] Oam29 (#51) | 38914 G to T (GAG to TAG), λ induced from AC 1966 slant R473, sc1,2; 76 AA from N-terminal of O |
| λcI857 Oam317 (#52) | 39166 G to T (GAG to TAG), LT; 160 AA at N-terminal of O |
| λ Oam8 (#50) | 39301 A to T (AAG to TAG), λ induced from AC 1966 slant R377, sc1,2; 205 AA at N-terminal of O |
| λcI857 Oam8 (#1025) | 39301 A to T (AAG to TAG), LT |
| λcI857 Oam205 (#586, 630) | 39570, C to G, (TAC to TAG), WD; 294 AA from N-terminal of O |
| Mutations in P | |
| λimm434cI Pam3 (#664) | 39786 (CAG to TAG), WD |
| λcI+prm116 Pam902 (#719,722) | 39894 (CAG to TAG), GG |
| Host b (prophage), strain # | |
| C600(λ[Ts] Oam29) #1170 | 38914 G to T (GAG to TAG) AC 1966 slant R473 (both sc’s) |
| C600(λ Oam8) #Y1169 | 39301 A to T (AAG to TAG), AC 1966 slant R377 (both sc’s) |
| C600(λ Nam7 cI857[Ind] Oam8) #Y239 | 39301 G to T (GAG to TAG), WD |
| M72 su+ (λ Nam7 cI857 r95) #Y85 c | 12 bpΔ λ bases 39123–39134, WD |
| M72 su+ (λ Nam7 cI857 r96) #Y84 c | 15 bpΔ λ bases 39139–39153, WD |
| M72 su+ (λ Nam7 cI857 r98) #Y88 | 24 bpΔ λ bases 39096–39119, WD |
| 594(λ Nam7 cI857 ti12) #188 | 39122 C to A (ACA to AAA), WD |
| Aberrant designations or with additional mutations, as received (lysate or strain #) | |
| λcI857 Oam8 (#16) | has nonsense mutation, but WT for cII-O-P-40712 in ninB; WS |
| λimm434 Oam8 (#656) | has nonsense mutation, but WT for cII-O-P-40712 in ninB; WD |
| λcI857 Oam8 (#518) | no mutation in O; 39786 in P, (CAG to TAG), same as Pam3; WS |
| λcI857 Oam29 (#582) | has nonsense mutation, WT for cII-O-P-40712 in ninB; WS |
| λcI857 Oam29 (#1023) | 38914 G to T (GAG to TAG); 38713 T to C (TTC to TCC); LT-MMS99 |
| λcI857 Oam125 (#1024) | 39511 C to T (CAG to TAG); 39182 C to T (TCC to TTC); and 39510 A to T (CAA to CAT); LT-MMS254 |
| 594(λcI857 Oam8) #Y49, Y52 | 38914 G to T (GAG to TAG), really is Oam29; WS |
| C600(λ Oam125) #1171 | 39182 C to T (TCC to TTC); and 39510 A to T (CAA to CAT); and 39511 C to T (CAG to TAG), AC 1966 slant R573 |
a Known strain sources: AC, A. Campbell; LT, L. Thomason; GG, G. Gussin, WD, W. Dove; WS, W. Szybalski. Furth [37] reported the original sources of the Oam mutations as: 8, 29 and 125 from [35], 905 from P. Toothman and I. Herskowitz, 1005 from I. Herskowitz, and 205 [38]. “AA” = amino acid(s). b Host C600 is SupE; The Pm− hosts 594 and M72 are supo. c Transcription from pL and pR, the lack of replication arising from oriλ, and the absence of any increase in phage titer following prophage induction were reported for these strains in [39].
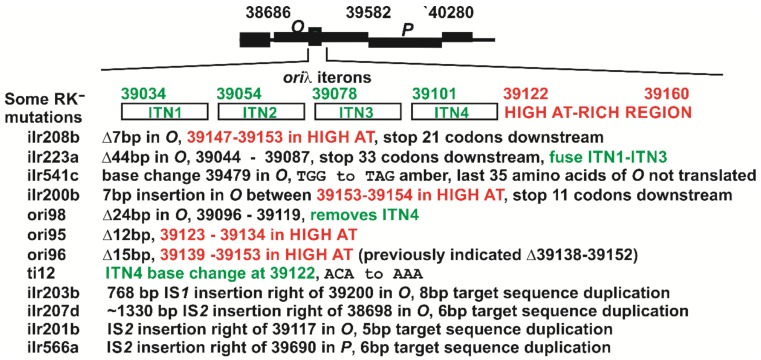
Figure 2.
RK− ilr mutations characterized within λ genes O-P. The minimal oriλ size was suggested to include a HIGH-AT-rich region to the right of four iteron sequences, ITN’s1-4 [43,44], which each contain an 18 bp inverted repeat of hyphenated symmetry, joined by adenine residues that can cause oriλ to assume a bent structure [45]. The mutants shown designated ori95, ori96 and ori98 were obtained from WD from prophage with r-mutants r-95, r-96, and r-98. Note that Denniston-Thompson, et al., [46] sequenced the r-mutants r-99, r-96 and r93 which represent Δ12 bp (39120–39131), Δ15 bp (39138–39152) and Δ24 bp (39092–39115) [42]. Our sequence localization for ori96 (r96) differs by one bp from that assigned in [42].
2.2. Replicative Killing Selection and Mutants
The selection of RK− mutants with defects in λ replication initiation were categorized (Figure 1) as follows: RK− clones designated Hd− (host defects), representing about 4% to 6% of selected spontaneous RK− clones [22], are not lysed by λvir. These mutants are arbitrarily considered to have an altered host gene whose product participates in vegetative λ growth, or a λ-fragment mutation whose effect is to complement negatively for the growth of λvir. The RK− clones that are lysed by λvir, retain the immλ phenotype at 30 °C, and were FI+, were designated RK− ilr (initiation of λ replication defective). The FI, or functional immunity assay [11,41], represents a stab of an RK− CFU to a lawn of cells lysogenized with λimm434T to which is added free λimm434cI. If immλ double recombinant phage can be generated (indicated by a lysis area forming around the RK− clone stabbed to the overlay plate), this is taken to indicate that the immλ region encoding oL/pL -cI- oR/pR is functional both in the RK− mutant and in the immλ recombinant. The RK− FI− Imm− isolates mainly have had large deletions (>10 Kb) [26,27,28]. In an examination of the spontaneous RK− mutants/mutations arising from four RK+ N+ selector strains, 256/650 RK− isolates were the RK− ilr type [22].
Figure 2 shows eight sequences for RK− ilr mutants falling within O-P that were derived from induced N+ prophage, along with mutants derived from induced N− prophage, including ori-95, -96, -98 obtained by Rambach [20] and ti12 from the Dove laboratory [18]). The sequences for ori95 and ori98 were not previously reported [42]. The ori96 mutation was a 15 bp deletion of λ bases 39139–39153 (not 39138–39152 as reported [42]). Mutation ori98 removed the entire iteron-ITN4 region, and ori95 and ori96 each deleted part of the High-AT rich region within O. Except for ilr541c, which included a stop codon that eliminated translation of the last 35 codons of O, the remaining RK− ilr O mutations (Figure 2) represented small deletions within O or insertions that could exert a polar effect on downstream P expression. Each of the ilr mutants were initially scored as being O+, which clearly was not proved correct by sequence analysis. None of the ilr mutations had in-frame deletions within O as with those obtained in Rambach’s λ N− selection.
2.3. Complementation for O Activity in Trans
The wild type O protein is 299 amino acids (AA) [43]. Alleles of O were cloned into an expression plasmid, Figure 3. Each allele was from an RK− mutant for which complementation analysis had suggested was O+.
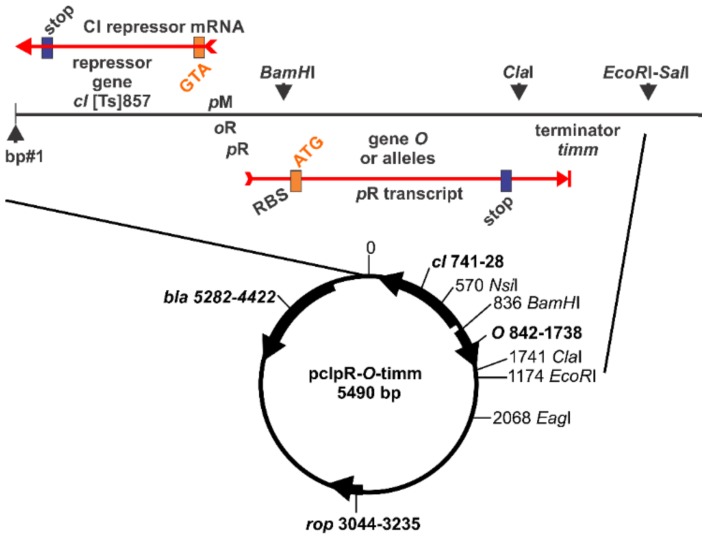
Figure 3.
Expression vector pcIpR-(O alleles)-timm. Expression of gene O or an allele occurs upon inactivation of the CI repressor by raising cells grown at 30 °C to 42 °C. Immediately following the 299 codons of O is an ochre stop codon, where the last base in TAA represents the first base of the ClaI restriction site ATCGAT.
Each plasmid was transformed into 594 cells, creating strains as 594[pcIpR-O-timm] that were used as hosts for λ Oam plating. The assays for O complementation were incubated at 30, 37, 39 and 42 °C. The results for plating assays at 42 °C, where the O allele is fully expressed, are shown in Table 2. Phages whose O allele produced 76 or 294 AA’s of O were weakly complemented by O+, whereas the Oam905 mutation expressing 37 AA of O was not complemented by O+. Complementation was improved 5- to 6-fold in two of the three λ Oam suppression assays by the addition of a SPA tag to the COOH end of the wild type O sequence, i.e., the addition of a seven AA linker (GGSGAPM) joined to the 69 AA SPA tag [47] sequence. The O-ori:98 mutation removing ITN4 within O was incapable of complementing for O. Remarkably, O alleles with the ori:ti12 point mutation in ITN4 and those with ori:95 and ori:96 in-frame deletions, respectively, of 12 and 15 bp’s within the High AT-rich region of oriλ, were each able to complement all three Oam mutants. Any condition where P, or a portion of the N-terminal end of P was expressed, completely prevented O-complementation. Induced prophage strains with insertions within P, but with sequenced intact O genes that would be fully derepressed when shifted to 42 °C were incapable of providing for O complementation. The RK− ilr mutants, previously designated phenotypically as O+ [11], proved to have insertions or deletions in O and should not complement, as was found for the cloned prophage O genes from mutants 208b, 223a and 541c (each of which could complement for P [36]).
Table 2.
Complementation of λ Oam mutants by alleleic forms of O expressed from plasmids.
| Host Strains and [Plasmid] # a | EOP of λ Phage with Oam Mutants b | ||
|---|---|---|---|
| λ cI857 Oam905 c (37 AA of O) |
λ cI857 Oam29 c (76 AA of O) |
λ cI857 Oam205 c (294 AA of O) |
|
| Pm+ SupE d | 1.0 | 1.0 | 1.0 |
| Pm− Supo | 0 | 0 | 0 |
| Complementation by O variations | |||
| O+ combinations | |||
| [O] e, p465 | 0 | 0.2 | 0.1 |
| [O-SPA] f, p472 | 0 | 1.0 | 0.6 |
| [oop-O] g, p677 | 0.05 | 0.1 | 0.1 |
| O null mutations h | |||
| [O-ilr208b], p488 | 0 | 0 | 0 |
| [O-ilr223a], p486 | 0 | 0 | 0 |
| [O-ilr541c], p485 | 0 | 0 | 0 |
| O-origin (oriλ) mutations i | |||
| [O-ori:98], p489 | 0 | 0 | 0 |
| [O-ori:95], p491 | 0.2 | 0.3 | 0.2 |
| [O-ori:96], p492 | 0.3 | 1.0 | 0.3 |
| [O-ori:ti12], p493 | 0.3 | 0.1 | 0.3 |
| O-P combinations j | |||
| [O-36P], p565 | 0 | 0.01 | 0 |
| [O-63P], p566 | 0 | 0 | 0 |
| [O-P], p569 | 0 | 0 | 0 |
| [oop-O-P] k p567, p568 | 0 | 0 | 0 |
| RK− O+ P− prophage derived from Y836 transduced into 594 | |||
| ilr 566a l | 0 | 0 | 0.002 |
| P::kan m, Bib11t | 0 | 0 | 0.001 |
a All of the complementation studies were undertaken in strains 594[pcIpR-Ovariant-timm] or with 594 cells transduced for the λ fragment mutants (ilr566a, BiB11t) from the original RK+ strain Y836. The host strain 594 is designated as being nonpermissive, Pm− (supo), without an amber suppressor). The precise sequences for each of the allelic forms of O were amplified by PCR, cloned into plasmid pcIpR-(‥)-timm, between BamHI and ClaI sites (designated by the internal brackets), and the inserted O-variant fragments were each verified by DNA sequence analysis. Each plasmid includes an allele of O positioned as is gene cro in WT λ, just downstream of promoter pR and the consensus ribosomal binding site (RBS) for cro. The initiation of transcription from pR is regulated by the Ts CI857 λ repressor encoded on the plasmid via binding to the operator site, oR that overlaps pR. For cells grown at 30 °C the CI Ts repressor remains active, binds to oR and blocks transcription initiation from pR. When the cells are shifted to 39–42 °C the CI Ts repressor transitions from partially to fully-inactive, allowing transcription initiation from pR and the expression of the downstream O allele. b The EOP value for “0” was set to <0.001. Thus, the difference between “0” and 0.1 is more than 100-fold. The efficiency of plating (EOP) was assessed at 30, 39 and 42 °C on cell lines containing plasmids. At 30 °C the results were all 0, i.e., the EOP was <0.001. In every situation, the plating results obtained at 42 °C showed an equivalent, or somewhat higher EOP than at 39 °C. All EOP data were calculated as: titer of Oam phage on indicated host with plasmid containing O allele/titer of the same phage on the Pm+ SupE host at same temperature. The phage titer on the Pm+ SupE strain was set as EOP = 1.0. All values are rounded up and are relative so that standard error is not shown but represents ±10–20% of the values indicated. Each of the plating phage were sequenced throughout the oop-O-P-ren genes and shown to contain only the designated Oam mutation. c Sequence designations for the O mutations are shown in Table 1 with mutations introducing amber stop codons: Oam905 at 38797, Oam29 at 38914, and Oam205 at 39579. To have all phages include the cI857 mutation the lysate #1023 was used for Oam29 which includes a silent mutation at 38713 (Ser to Ser). The phage nonsense mutations in O truncate gene expression, producing polypeptides of the length shown in the heading of Table 2, each with the N-terminal end of the WT protein. d Permissive, Pm+, strain was TC600 SupE, where the efficiency for amber suppression (not complementation) was used as the baseline for full complementation. e Sequence for O (λ WT bases 38686–39582), representing 299 codons, plus an ochre stop codon was inserted to make plasmid pcIpR-O-timm. f Fusion construct represents WT O-(7 amino acid linker GGSGAPM)-69 amino acid SPA tag sequence-ochre stop codon. The SPA tag sequence at COOH end of O includes a calmodulin binding site, a TEV (tobacco etch virus) protease cleavage site and 3X FLAG sequence [47]. g Results approximate data for two plasmid constructs oop#1-O, representing λ WT bases 38559–39582, and oop#2-O, representing λ bases 38546–39582, each inserted between the BamHI and ClaI sites in pcIpR-(‥)-timm. h Replicative-Killing defective (RK−) mutants in gene O (see Figure 2) isolated as survivors from induced cryptic λ prophage strain Y836 as described in [11,48]. i Removed O fragments from replicator mutants of M72(λ Nam7am53 cI857 r95), M72(λ Nam7am53 cI857 r96), and M72(λ Nam7am53 cI857 r98) lysogens from Rambach [20] via WD. j Plasmids were described in [25]. Each plasmid includes the intact sequence of O (λ bases 38686–39582 plus the N-terminal portions of gene P followed by ochre stop codon: 38686–39687 = O-P36, 38686–39768 = O-P63, 38686–40280 = O-P. Note that O and P are in different reading frames. k Plasmids p567 oop#1-O-P and p568 oop#2-O-P include λ bases 38559–40280 and 38546–40280, respectively, cloned between the BamHI and ClaI sites in the pcIpR-(‥)-timm plasmid [25]. The oop-O-P line represents equivalent data for plasmids oop#1-O-P and oop#2-O-P. l The RK− ilr mutant 566a derived from strain Y836 was transduced using P1 with the marker nad57::Tn10 that was inserted contiguous to the chromosomal λ fragment. Then the TetR immλ region was transduced into strain 594. m The kan marker was introduced into gene P in strain Y836 by recombineering (=strain Bib11t) and the defective λ fragment was transduced into 594 cells as indicated for moving mutation 566a (see [25] for additional details).
2.4. Influence of O-P Co-Expression on Cell Growth, P-Lethality, and Plasmid Loss
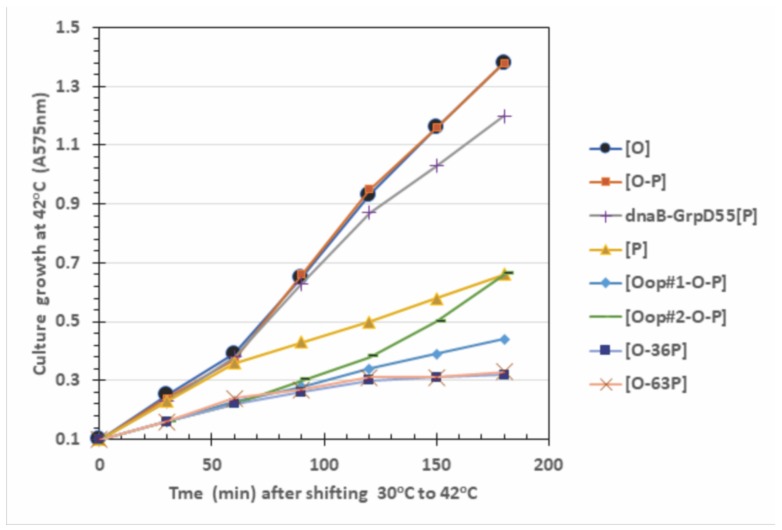
The expression of P, or N-terminal fragments of P, block O complementation (Table 2). If O and P expression can influence O complementation, does their co-expression negate P-lethality? Figure 4 shows that O expression alone, or the co-expression of WT genes O and P over a span of four doublings in culture absorbance (with 45 min per doubling) did not perturb cell growth for cultures shifted from growth at 30 °C to 42 °C. In contrast, expressing P alone, or constructs expressing oop-O-P, or constructs that were WT for O but could express a portion of the N-terminal region of P were each highly inhibitory to cell growth. Table 3 shows the effect of O-P constructs on cell viability and plasmid loss. We re-examined several of the observations reported in [25], where the expression of P, even in trace levels at 37 °C, kills about 99% of the transformed cells and all plasmids were lost in survivor CFU’s. The lethality of P is completely suppressed by two missense mutations in dnaB that comprise the allele dnaB-GrpD55. Expressed by itself, O is not toxic and does not cause plasmid loss. Co-expression of O-P reduces the cellular toxicity of P expression alone by 12 to 15-fold at 37 and 39 °C. Co-expression of oop-O-P significantly prevents plasmid loss at 37 and 39 °C but exerts a minimal effect on cell viability. The inclusion of portions of the N-terminal end of P plus O significantly reduces cell viability and plasmid retention at 39 and 42 °C, compared to the expression of only O.
Figure 4.
Influence of induced O, P gene expression on cell growth at 42 °C. All the strains were made by transforming hosts 594 or 594 dnaB-GrpD55 with pcIpR-(‥)-timm plasmids that included the cloned O, P DNA fragment, and selecting the transformants on LBAmp50 agar (medium composition is described in footnote to Table 3). The plasmid inserts in each CFU employed were verified by DNA sequence analysis. Cells were inoculated from overnight cultures grown up overnight in LBAmp50 broth and then 0.4 mL of culture was added to triplicate 20 mL fresh LB cultures that were incubated at 30 °C for ~30 min to reach an A575 = 0.1. Upon reaching an absorbance of 0.1 the cultures were transferred to a shaking 42 °C water bath. Aliquots were sampled every 30 min for 3 h. The average absorbance is shown, with a standard error for each culture time of less than 5% the averaged absorbance value.
Table 3.
Does O-P co-expression temper P-lethality and plasmid loss?
| Plasmid in 594 Host Cells | Cell Viability and (Plasmid Retention per CFU Assayed; %) at CFU Growth Temperature a | |||
|---|---|---|---|---|
| 30 °C | 37 °C | 39 °C | 42 °C | |
| Only P expression | ||||
| [P] | 1.0 (35/35; 100%) | 0.01 (0/35; 0%) | 0.008 (0/35; 0%) | 0.07 (0/35; 0%) |
| dnaB-GrpD55 [P] | 1.0 (35/35; 100%) | 1.0 (35/35; 100%) | 1.0 (35/35; 100%) | 0.98 (35/35; 100%) |
| Only O expression | ||||
| [O] | 1.0 (30/30; 100%) | 1.0 (30/30; 100%) | 1.0 (30/30; 100%) | 0.61 (29/30; 97%) |
| O-P expression combinations | ||||
| [O-P] | 1.0 (62/70; 89%) | 0.12 (0/70; 0%) | 0.12 (0/70; 0%) | 0.022 (0/36; 0%) |
| [oop#1-O-P] | 1.0 (120/120; 100%) | 0.20 (98/101; 97%) | 0.005 (115/120; 96%) | 0.002 (0/36; 0%) |
| [oop#2-O-P] | 1.0 (117/120; 98%) | 0.055 (76/154; 49%) | 0.048 (62/120; 52%) | 0.008 (0/36; 0%) |
| [O-36P] | 1.0 (30/30; 100%) | 0.79 (30/30; 100%) | 0.012 (1/30; 3%) | 0.0005 (0/36; 0%) |
| [O-63P] | 1.0 (30/30; 100%) | 0.90 (30/30; 100%) | 0.055 (14/40; 35%) | 0.0023 (0/36; 0%) |
a The 594 cultures with indicated plasmids were grown to stationary phase in LB (10 g Bacto-Tryptone, 10 g Bacto-Yeast Extract, 5 g NaCl per liter) plus 50 µg/mL ampicillin (=LBAmp50) for 48 h at 30 °C, diluted, spread on LB agar (includes the addition of 11 g/liter Bacto-Agar) plates (no ampicillin) that were incubated at 30, 37, 39, or 42 °C for 30 h and the average survivor titer for CFU per mL was determined for each plating temperature. Isolated survivor CFU were stabbed to LB and to LBAmp50 agar plates to estimate the proportion of CFU retaining the AmpR plasmid. We tried to assay all the CFU per plate to avoid colony size discrimination, and minimally 30 CFU. The cell viability results shown in each column entry were determined by dividing the cell titer obtained for each incubation by the cell titer at 30 °C, and each value represents the average of duplicate plasmid isolates for each single experiment, with plating variations of about 10%. The results in parentheses represent the sum of results for all the CFU’s assayed from isolates. These results represent independent determinations by KR to those reported in [25], where it was shown that 594[P] cells lost 100% of their plasmids when grown at 36 °C and higher, indicating that trace levels of P expression (where the CI[Ts] repressor still retains some ability to block pR-promoted transcription below about 38–39 °C) will cure cells of the plasmid.
The oop DNA sequence encodes a 77-base noncoding small RNA that is transcribed in an antisense orientation to O. Part of the oop sequence overlaps the sequence for gene cII preceding O, with the pO promoter for oop transcription overlapping the N-terminal end of the O sequence. The #1 and #2 oop-O or oop-O-P constructs are deleted for the N-terminal end of cII. Since O or O-P are directly transcribed from pR on the plasmid, we asked if the antisense oop transcript would influence O or P activity expressed from the plasmid. Except for its ability to support some low-level suppression of λOam905 plating, OOP RNA expression does not significantly influence O complementation (Table 2); however, it exhibits a profound effect on the ability of P expression to evoke plasmid loss (Table 3), over and above the quenching influence of O-P co-expression on cellular P-lethality. A hypothesis is that OOP RNA hybridization to the pR-oop-O-P mRNA expressed from the induced plasmid reduces downstream P translation/accumulation within the cell.
3. Discussion
3.1. O-Complementation
Our sequencing of Oam29 reveals that this O allele encodes 294 of 299 amino acids. Thus, the last five amino acids of O are essential for O activity, and yet 76 amino acids, i.e., a linker and the SPA tag sequence, can be added to its COOH-terminal end, with the effect of improving the ability of O to complement.
Until this report, no one appears to have determined if in-frame deletions within O can influence its ability to complement, or will simply nullify its activity. We show that the 24 bp deletion in ori98 (r98) removing ITN4 nullifies the ability of O to complement; however, the 12 and 15 bp deletions in ori95 and ori96 (r95, r96), each falling within the High-AT rich region of oriλ improved the ability of O to complement. In addition, the oriλ ti12 mutation, representing a mismatch changing threonine to lysine within the ITN4 interval seemed to improve, rather than reduce, O complementation.
We were unable to demonstrate O complementation or saw extremely poor complementation for two sequenced O+ prophages in N+ RK− cells, each of which had acquired insertions within P, i.e., strains ilr566a and Bib11t. This result leads us to question whether it is possible to demonstrate complementation where the prophage for the RK− cells carries a N− mutation and would poorly express O, e.g., Rambach’s conclusion that the r96 mutant isolated from an N− prophage complemented for O. Indeed, full O+ expression from the pcIpR-O-timm plasmid in cells plated at 42 °C did not complement (i.e., support plaque formation of) a phage with an Oam905 mutation.
3.2. O:P Interaction Effects
The functional cooperation of O and P in λ replication initiation was suggested by genetic studies [49]. The N-terminal region of O was suggested to contain a DNA binding domain and the COOH-terminal region to contain a P-binding domain [40,50,51], with the domains separated by a flexible linker region [52]. Tsurimoto and Matsubara [5] showed that O protein binds to each ITN as a dimer, thus oriλ should bind four dimers, with higher order binding suggested [53] to form an O-some that produces torsional stress on the adjacent AT rich region causing the double-stranded DNA to become slightly destabilized and partially unwound [54]. The N-terminal portion of P was assumed to contain an O-binding domain [55], while its COOH-terminal domain was suggested to interact with the host DnaB replicative helicase [55,56]. It has been suggested that a complex between O and P is formed that can be independent of DnaB [51,57].
In essence, the idea was advanced that O bound to oriλ is a display platform that is recognized by P:DnaB. However, we show that the co-expression of O-P results in several phenotypic effects which suggest that this idea is too simplistic. The co-expression of O-P nullifies the inhibitory effect of P expression on cell growth, for over four hours, and it reduces cell killing caused by prolonged expression of P (i.e., when expressed at 39 °C). In contrast, the co-expression of O-P nullifies the ability of O to complement. These opposed activities suggest that O and P physically interact without having O bound to oriλ. Combining O expression with the possibility for expression of the N-terminal portion of P eliminates O complementation, suggesting that O binding to the N-terminal portion of P prevents its useful binding to oriλ, which is presumably a requirement for O complementation activity. However, the co-expression O-P does not temper the ability of P to cause plasmid loss. Thus, while the expression of P, or N-terminal portions of P, can obviate O complementation, coordinate O expression does not fully nullify all the P-lethality phenotypes.
3.3. RK− Mutant Selection Considerations
Dove and Blattner’s laboratories collaborated in mapping [40] and sequencing [46] some of the nine r mutants selected by Rambach [20], who based his selection on the assumption that the “replicator” gene was different from initiator genes O or P. They concluded that regions of the initiator gene, i.e., O, overlap the replicator site, now termed oriλ. We show herein that regions of oriλ are not essential for activity of the O initiator protein. We previously demonstrated [39] that λ Nam7am53 cI857 r95, or r96 prophages in su+ hosts (hence the prophages were phenotypically N+) were defective in oriλ replication initiation, were 7- to 17-fold reduced in pR-Q transcription, and did not yield any increase in phage titer after prophage induction. Five of the nine r mutants have now been sequenced and each has a small, in-frame deletion within O. In contrast, among hundreds of RK− ilr mutants isolated from a defective N+ prophage (Figure 1), none were identified with in-frame deletions in O. Nor, have other instances involving use of the PDS, or similar selections resulted in small in-frame deletions within O being reported [12,16,17,58,59]. The major theme of those reports was that perturbing the expression of pR-O-P can influence replication initiation. The recent documentation on the lethal effect of P expression (see [25,36] and included references) may help to explain the selection differences, i.e., constitutive P expression is lethal to a cell, even if there is no replication initiation from oriλ. Rambach’s study required several other unstated assumptions: (i) replication initiation will occur from an induced N mutant prophage with reduced transcription of O-P (we note above that this was not observed for the r95 and r96 mutants); (ii) in induced N-defective prophage, sufficient rightward transcription occurs across (or near to) the replicator (oriλ) site to provide the requirement for transcriptional activation (as noted above, even when the r-mutant prophages were made su+ rightward transcription across pR-Q was significantly reduced); and (iii) the constitutive expression of the replication initiation proteins O and P or other de-repressed λ gene products will not be lethal to the host cell. Assumptions (i) and (ii) may still require additional study. A previous characterization of RK− ilr survivor mutations from N+ prophage, revealed that all were defective in P or had insertions in O that could limit downstream P expression [48], suggesting that assumption (iii) is unlikely.
4. Materials and Methods
4.1. Complementation Assays and Initial Strategy for Characterizing RK− Mutants
Past studies have generated hundreds of RK− mutants capable of colony formation at 42 °C. Since almost all these mutants were selected prior to an ability to combine PCR with rapid DNA sequence analysis of the generated PCR fragment, the characterization of the genetic defect that permitted cell survival and growth at 42 °C required genetic analysis. This, in principle, involved complementation analysis for expression of genes N, cI, cro, cII, O, and P. Of those mutants that retained an active immλ region encoding a Ts CI repressor, the cells grown at 30 °C expressed an immune response to plating by immλ phage, but not to the heteroimmune phages as λimm434. Shifting the cells to growth at 42 °C inactivated the Ts CI repressor and permitted the expression of N, cro, cII, O, and P. RK− clones were inoculated into 1.5 mL tryptone broth (TB: 10 g of Bacto Tryptone, 5 g of NaCl per liter) and grown to stationary phase at 30 °C. One-tenth mL of each culture was mixed with dilutions high titer lysates immλ or imm434 phages carrying an amber mutation in genes N, O, or P (none of which—at the time—were characterized by DNA sequence analysis) plus 2.5 mL of TB top agar (0.65%) agar. The mixture was poured on TB agar (1.1%) plates that were incubated at 42 °C. In the present study lysates of λ phage with Oam mutants were freshly prepared. A single colony of E. coli strain 594 or these cells transformed with different versions of O plasmids were grown in LBAmp50 broth (see footnote “a” Table 3) at 30 °C overnight. Then a mixture of cells and soft agar (3 mL of warm top agar, 0.25 mL of cells and 0.25 mL of 0.01 M MgCl2) was poured on the top of LB plates. After agar solidification, diluted Oam λ phage lysates were spotted on the agar, allowed to dry, the plates were incubated inverted at 30, 37, 39 and 42 °C overnight and plaque forming units were counted. The appearance of plaques at elevated plating efficiency indicated complementation for the defective gene carried on the infecting phage by the thermally induced prophage in the RK− mutant cells or expressed from the plasmid. 100% plating efficiency was equated to the titer of the amber phage mutant on E. coli cells with a suppressor tRNA, e.g., on strain TC600 supE. Very low plating efficiency (<10−4) suggested phage-prophage marker rescue. The ability of RK− cells to complement for the wild type functions expressed from N or P has always been very simple to assess. However, the interpretation of whether O expressed from the induced prophage was able to complement an Oam infecting phage proved problematic.
4.2. DNA Sequence Analysis of λ Phage, Prophage and Plasmid Constructs
The DNA sequencing results reported herein, for each plasmid construct and phage isolate were obtained by us using methods for colony PCR, plaque PCR, and PCR amplification of cloned regions from isolated plasmid constructs, as previously described [60]. The actual sequencing results were obtained from sequencing services at the NRC National Biotechnology Institute, Saskatoon, or were submitted to Eurofins Genomics. The oligonucleotide primers employed, Table 4, were obtained from Integrated DNA Technologies, Inc. Coralville, IA, USA. In every case, at minimum, four individual representative colony, plaque or plasmids were sequenced per construct or isolate.
Table 4.
Oligonucleotide primers employed for DNA sequence analysis and plasmid constructions.
| Name | λ Map Position | Sequence (5′ to 3′) a |
|---|---|---|
| L-37904+18 | 37904–37922 | GCTGCTCTTGTGTTAATGG |
| L-MH29 | 37905–37922 | CGTCCTCAAGCTGCTCTTGTGTTAATGG |
| L20 | 39465–39484 | ACTCCGCGATAAGTGGACCC |
| L-22 | 38517–38534 | TGCTGCTTGCTGTTCTTG |
| L-PG30 | 38530–38547 | TTGGAACTGAGAAGACAG |
| L-PG1 | 38784–38801 | AAATATGCTGCTTGAGGC |
| L-38985p20 | 38985–39005 | GCAGCAAGGCGGCATGTTTGG |
| L-MH32 | 39531–39550 | CACAGATCTATAGCAAACCAAAACTCGACCTGA |
| L-18 | 39980–39996 | TTGCCGGAAGCGAGGCC |
| L-21 | 40360–40377 | CGCAACAGTAACCAGCAT |
| R-PG2 | 40747–40764 | GGTTGCGTTCCTGAATGG |
| L-Bam-O | 38686–38718 | ATATGGATCCATGACAAATACAGCAAAAATACTCAACTTCGGC |
| L-Bam-P | 39582–39606 | ATATGGATCCATGAAAAACATCGCCGCACAGATGG |
| L-Bam-OOP#1 | 38559–38580 | ATATGGATCCTGGCTCGATTGGCGCGACAAGT |
| L-Bam-OOP#2 | 38546–38577 | ATATGGATCCGTTGACGACGACATGGCTCGAT |
| L-Bam-O | 38686–38718 | ATATGGATCCATGACAAATACAGCAAAAATACTCAACTTCGGC |
| L-Bam-P | 39582–39606 | ATATGGATCCATGAAAAACATCGCCGCACAGATGG |
| R-40769m22 | 40747–40769 | GCTGCGGTTGCGTTCCTGAATGG |
| R-MH33 | 40315–40295 | GCGACGTCCCCAGGTAATGAATAATTGC |
| R-17 | 40018–40002 | TAAGACTCCGCATCCGG |
| R-MH25 | 39626–39609 | CTGCTCACGGTCAAAGTT |
| R-39280m21 | 39259–39280 | CTGCGGCGGTCAGGTCTTCTGC |
| R9+1 | 39191–39175 | TGGTCAGAGGATTCGCC |
| R-PG6 | 38569–38552 | CAATCGAGCCATGTCGTC |
| R-1536-19 | pcIpR-()-timm | GAAGACAGTCATAAGTGCGG |
| R-ClaI-P | 40280–40259 | ATATATCGATTATACACTTGCTCCTTTCAGTCCG |
| R-ClaI-O | 39582–39559 | ATATATCGATTATAGATCCACCCCGTAAATCCAGTC |
| R-ClaI-36P | 39687–39662 | ATATATCGATTACCTGCTGTACCTGCGGCTTTTCGTCG |
| R-ClaI-63P | 39768–39746 | ATATATCGATTACTTCGTTCTGGTCACGGTTAGCC |
| R-AscI-O | 39582–39559 | ATATGGCGCGCCGCTGCCGCCTAGATCCACCCCGTAAATCCAGTC |
a The portion of primer sequences shown in smaller font size contain restrictions sites used for cloning into plasmids and are not included within λ map sequence shown.
4.3. Plasmid Constructs
The O gene alleles were amplified from E. coli strains with a prophage (e.g., each of the RK− mutants shown in Figure 2) or λ phage DNA, using PCR primers L-Bam-O and R-ClaI-O. The PCR fragments were cloned just downstream of promoter pR between the BamHI and ClaI restriction sites in the pcIpR-(…)-timm plasmid isolated from a dam host strain, as drawn in Figure 3. The R-ClaI-O primer introduces an ochre stop codon at the COOH-terminal end of O. Primers L-Bam-P and R-ClaI-P were used for to clone gene P. Primers L-Bam-O and R-ClaI-P were used to clone genes O-P, which include the natural TGA stop codon for O and an ochre codon terminating P. The construction of plasmids pcIpR-P-timm, pcIpR-O-timm, pcIpR-O-P-timm, pcIpR-O-36P-timm, pcIpR-O-63-P-timm, pcIpR-oop#1-timm, and pcIpR-oop#2-timm was as reported in [25]. The plasmids oop-O and oop-O-P, Table 2, were constructed using primers L-Bam-oop#1 or L-Bam-oop#2 and R-ClaI-O to make oop-O and R-ClaI-P to make oop-O-P and the PCR fragments were cloned between the BamHI and ClaI restriction sites in the unmethylated pcIpR-(…)-timm plasmid. Plasmid O-SPA was constructed by removing the BamHI-P-AscI fragment from pcIpR-P-SPA-timm [25] and inserting the fragment BamHI-O-AscI prepared using primers L-Bam-O and R-O-AscI. This construct is described in footnote “f” of Table 2. The DNA template used for amplifying wild type alleles of oop-O-P was from λcI857 [25].
4.4. Bacterial and Phage Strains
The genotype, source and laboratory reference number for bacterial strains 594 (Pm−), TC600 (Pm+), 594 dnaB-grpD55 is described in Table 10 in reference [25] as are the reference phages (see also [41]). Y836, Y836 P:kan (Bib11t), Y836 RK− ilr566a are described in Table 8 in reference [36]. Examples showing the characterization of RK− mutants can be found in [11,48].
Acknowledgments
This work was supported by NSERC Canada Discovery grant 138296 to Sidney Hayes. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations
Unique λ Terminology:
Replicative Killing (RK+) competent phenotype: A property of E. coli cells that possess a “defective” λ prophage that is blocked by one of several means for chromosomal excision. The initiation of bi-directional replication from the prophage, which is normally prevented by an action of the prophage CI repressor—until repression is relieved, results in replication forks that move outward from the defective prophage into the E. coli chromosome. Massive cellular killing occurs, likely due to collisions between E. coli-initiated and λ-initiated opposing replication forks. Replicative Killing (RK−) defective phenotype: Mutants originating in RK+ cells that have lost the RK+ phenotype. There are numerous possibilities for RK− mutants, but they all relate to being defective in a phage or host function required for the initiation of λ bi-directional replication. RK− Hd− mutants are defective in a host function required for λ replication initiation. RK+ ilr mutants are defective in a λ function required for the initiation of λ bi-directional replication, where ilr designates initiation of λ replication defective RK− FI− mutants represent RK− mutants where the immλ region of the defective prophage cannot be rescued using a phage-prophage marker rescue recombination assay. The hundreds of mutants characterized have resulted from large chromosomal deletions where the deletion endpoints straddle or partially straddle the defective prophage; but they could also represent defects in pR, preventing pR-cro-cII-O-P transcription. immλ region: part of the λ genetic map between genes N and cII that includes the genetic elements oL/pL-rexB-rexA-cI[Ts857]-oR/pR-cro. imm434 region: the region of a λimm434 hybrid phage where the immλ region of λ is replaced by DNA from phage 434, but all the remaining portions of the phage genetic map are the same as for λ. FI (functional immunity) assay: a marker rescue assay where the immλ region of a prophage (in RK+ cells) is rescued by an infecting λimm434 phage, where the recombinant λimmλ phage released can form plaques on cells lysogenized by a λimm434 prophage. oriλ region: A region within λ gene O that contains four iteron (or ITN) sequences each containing an 18 bp inverted repeat of hyphenated symmetry (each bound by two O proteins) and an adjacent region of 39 bp termed the High-AT-RICH region that is sensitive to DNA unwinding. PDS selection: a selection for RK− survivor mutants starting with lysogenic cells with an intact λ prophage that encodes a temperature sensitive cI[Ts857] repressor and is defective for N, such that prophage excision is prevented because genes int-xis are not expressed from the induced prophage, i.e., when the cells are shifted from growth at 30 °C (where the TS CI repressor is active, binds operator sites oL and oR and prevents transcription initiation from promoters pL and pR) to 42 °C where the TS CI repressor is thermally inactivated and transcription is de-repressed from promoters pL and pR. RK− selection: same as the PDS selection except that the starting cells include an N+ cryptic λ prophage deleted for genes int-xis-exo-bet-gam-kil left of immλ (encoding a cI Ts857 repressor), and all λ late genes for cell lysis and phage morphogenesis. riC mutation: putative new promoters arising left or right of oriλ that enable transcription near to oriλ, thus suppressing “replicative inhibition” caused by the loss of a cis-requirement for transcription from pR (or in the vicinity of oriλ), which, in addition to the activities of λ replication initiator genes O and P is a requirement for bi-directional λ replication initiation.
Author Contributions
Performed the experiments and helped analyze the data: Connie Hayes and Karthic Rajamanickam. Conceived, designed the experiments, and wrote the paper: Sidney Hayes.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Casjens S.R., Hendrix R.W. Bacteriophage lambda: Early pioneer and still relevant. Virology. 2015;479–480:310–330. doi: 10.1016/j.virol.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedman D.I., Gottesman M. Lytic mode of lambda development. In: Hendrix R.W., Roberts J.W., Stahl F.W., Weisberg R.A., editors. Lambda II. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY, USA: 1983. pp. 21–51. [Google Scholar]
- 3.Hendrix R.W., Casjens S. Bacteriophage lambda and its genetic neighborhood. In: Calendar R., editor. The Bacteriophages. 2nd ed. Oxford University Press; Oxford, UK: 2006. pp. 409–447. [Google Scholar]
- 4.Court D.L., Oppenheim A.B., Adhya S.L. A new look at bacteriophage lambda genetic networks. J. Bacteriol. 2007;189:298–304. doi: 10.1128/JB.01215-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsurimoto T., Matsubara K. Purified bacteriophage lambda O protein binds to four repeating sequences at the lambda replication origin. Nucleic Acids Res. 1981;9:1789–1799. doi: 10.1093/nar/9.8.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsurimoto T., Matsubara K. Purification of bacteriophage lambda O protein that specifically binds to the origin of replication. Mol. Gen. Genet. 1981;181:325–331. doi: 10.1007/BF00425606. [DOI] [PubMed] [Google Scholar]
- 7.Biswas S.B., Biswas E.E. Regulation of dnaB function in DNA replication in Escherichia coli by dnaC and lambda P gene products. J. Biol. Chem. 1987;262:7831–7838. [PubMed] [Google Scholar]
- 8.Alfano C., McMacken R. Ordered assembly of nucleoprotein structures at the bacteriophage lambda replication origin during the initiation of DNA replication. J. Biol. Chem. 1989;264:10699–10708. [PubMed] [Google Scholar]
- 9.Zylicz M., Ang D., Liberek K., Georgopoulos C. Initiation of lambda DNA replication with purified host- and bacteriophage-encoded proteins: The role of the dnaK, dnaJ and grpE heat shock proteins. EMBO J. 1989;8:1601–1608. doi: 10.1002/j.1460-2075.1989.tb03544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas R., Bertani L.E. On the Control of the Replication of Temperate Bacteriophages Superinfecting Immune Hosts. Virology. 1964;24:241–253. doi: 10.1016/0042-6822(64)90163-1. [DOI] [PubMed] [Google Scholar]
- 11.Hayes S., Hayes C. Spontaneous lambda OR mutations suppress inhibition of bacteriophage growth by nonimmune exclusion phenotype of defective lambda prophage. J. Virol. 1986;58:835–842. doi: 10.1128/jvi.58.3.835-842.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furth M.E., Dove W.F., Meyer B.J. Specificity determinants for bacteriophage lambda DNA replication. III. Activation of replication in lambda ric mutants by transcription outside of ori. J. Mol. Biol. 1982;154:65–83. doi: 10.1016/0022-2836(82)90417-X. [DOI] [PubMed] [Google Scholar]
- 13.Moore D.D., Blattner F.R. Sequence of lambda ric5b. J. Mol. Biol. 1982;154:81–83. doi: 10.1016/0022-2836(82)90418-1. [DOI] [PubMed] [Google Scholar]
- 14.Echols H., Guarneros G. Control of Integration and Excision. In: Hendrix R.W., Roberts J.W., Stahl F.W., Weisberg R.A., editors. Lambda II. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY, USA: 1983. pp. 75–92. [Google Scholar]
- 15.Pereira da Silva L., Eisen H., Jacob F. Sur la replication du bacteriophage. C. R. Acad. Sci. Paris. 1968;266:926–928. [PubMed] [Google Scholar]
- 16.Brachet P., Eisen H., Rambach A. Mutations of coliphage lambda affecting the expression of replicative functions O and P. Mol. Gen. Genet. 1970;108:266–276. doi: 10.1007/BF00283357. [DOI] [PubMed] [Google Scholar]
- 17.Castellazzi M., Brachet P., Eisen H. Isolation and characterization of deletions in bacteriophage lambda residing as prophage in E. coli K 12. Mol. Gen. Genet. 1972;117:211–218. doi: 10.1007/BF00271648. [DOI] [PubMed] [Google Scholar]
- 18.Dove W.F., Inokuchi H., Stevens W.F. Replication control in phage lambda. In: Hershey A.D., editor. The Bacteriophage Lambda. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY, USA: 1971. pp. 747–771. [Google Scholar]
- 19.Lieb M. Studies of heat-inducible lambda-phage. 3. Mutations in cistron N affecting heat induction. Genetics. 1966;54:835–844. doi: 10.1093/genetics/54.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rambach A. Replicator mutants of bacteriophage lambda: Characterization of two subclasses. Virology. 1973;54:270–277. doi: 10.1016/0042-6822(73)90136-0. [DOI] [PubMed] [Google Scholar]
- 21.Sly W.S., Eisen H.A., Siminovitch L. Host survival following infection with or induction of bacteriophage lambda mutants. Virology. 1968;34:112–127. doi: 10.1016/0042-6822(68)90014-7. [DOI] [PubMed] [Google Scholar]
- 22.Hayes S. Mutations suppressing loss of replication control: Genetic analysis of bacteriophage lambda-dependent replicative killing, replication initiation, and mechanisms of mutagenesis. In: Moses R.E., Summers W.C., editors. DNA Replication and Mutagenesis. American Society for Microbiology; Washington, DC, USA: 1988. pp. 367–377. [Google Scholar]
- 23.Greer H. The kil gene of bacteriophage lambda. Virology. 1975;66:589–604. doi: 10.1016/0042-6822(75)90231-7. [DOI] [PubMed] [Google Scholar]
- 24.Bull H.J., Hayes S. The grpD55 locus of Escherichia coli appears to be an allele of dnaB. Mol. Gen. Genet. 1996;252:755–760. doi: 10.1007/BF02173984. [DOI] [PubMed] [Google Scholar]
- 25.Hayes S., Erker C., Horbay M.A., Marciniuk K., Wang W., Hayes C. Phage Lambda P Protein: Trans-Activation, Inhibition Phenotypes and their Suppression. Viruses. 2013;5:619–653. doi: 10.3390/v5020619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayes S., Duincan D., Hayes C. Alcohol treatment of defective lambda lysogens is deletionogenic. Mol. Gen. Genet. 1990;222:17–24. doi: 10.1007/BF00283017. [DOI] [PubMed] [Google Scholar]
- 27.Hayes S. Mapping ethanol-induced deletions. Mol. Gen. Genet. 1991;231:139–149. doi: 10.1007/BF00293831. [DOI] [PubMed] [Google Scholar]
- 28.Hayes S. Ethanol-induced genotoxicity. Mutat. Res. 1985;143:23–27. doi: 10.1016/0165-7992(85)90099-5. [DOI] [PubMed] [Google Scholar]
- 29.Hayes S., Hayes C., Taitt E., Talbert M. A simple, forward selection scheme for independently determining the toxicity and mutagenic effect of environmental chemicals: Measuring replicative killing of Escherichia coli by an integrated fragment of bacteriophage lambda DNA. In: Kolber A.R., Wong T.K., Grant L.D., DeWoskin R.S., Hughes T.J., editors. In Vitro Toxiciry Testing of Environmental Agents, Part A. Plenum Publishing Corp.; New York, NY, USA: 1883. [Google Scholar]
- 30.Hayes S., Gordon A., Sadowski I., Hayes C. RK bacterial test for independently measuring chemical toxicity and mutagenicity: Short-term forward selection assay. Mutat. Res. 1984;130:97–106. doi: 10.1016/0165-1161(84)90109-2. [DOI] [PubMed] [Google Scholar]
- 31.Hayes S., Gordon A. Validating RK test: Correlation with Salmonella mutatest and SOS chromotest assay results for reference compounds and influence of pH and dose response on measured toxic and mutagenic effects. Mutat. Res. 1984;130:107–111. doi: 10.1016/0165-1161(84)90110-9. [DOI] [PubMed] [Google Scholar]
- 32.Kaiser A.D. In: Lambda DNA Replication. Hershey A.D., editor. Cold Spring Harbor Press; Cold Spring Harbor, NY, USA: 1971. [Google Scholar]
- 33.Rao R.N., Rogers S.G. A thermoinducible lambda phage-ColE1 plasmid chimera for the overproduction of gene products from cloned DNA segments. Gene. 1978;3:247–263. doi: 10.1016/0378-1119(78)90035-5. [DOI] [PubMed] [Google Scholar]
- 34.Kleckner N. Amber mutants in the O gene of bacteriophage lambda are not efficiently complemented in the absence of phage N function. Virology. 1977;79:174–182. doi: 10.1016/0042-6822(77)90343-9. [DOI] [PubMed] [Google Scholar]
- 35.Campbell A. Sensitive mutants of bacteriophage lambda. Virology. 1961;14:22–32. doi: 10.1016/0042-6822(61)90128-3. [DOI] [PubMed] [Google Scholar]
- 36.Hayes S., Wang W., Rajamanickam K., Chu A., Banerjee A., Hayes C. Lambda gpP-DnaB Helicase Sequestration and gpP-RpoB Associated Effects: On Screens for Auxotrophs, Selection for Rif(R), Toxicity, Mutagenicity, Plasmid Curing. Viruses. 2016;8:172. doi: 10.3390/v8060172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Furth M.E. Specificity Determinants for Bacteriophage Lambda DNA Replication, and Structure of the Origin of Replication. University of Wisconsin; Madison, WI, USA: 1978. [Google Scholar]
- 38.Thomas R., Leurs C., Dambly C., Parmentier D., Lambert L., Brachet P., Lefebvre N., Mousset S., Porcheret J., Szpirer J., et al. Isolation and characterization of new sus (amber) mutants of bacteriophage lambda. Mutat. Res. 1967;4:735–741. doi: 10.1016/0027-5107(67)90082-6. [DOI] [PubMed] [Google Scholar]
- 39.Hayes S. Initiation of coliphage lambda replication, lit, oop RNA synthesis, and effect of gene dosage on transcription from promoters PL, PR, and PR. Virology. 1979;97:415–438. doi: 10.1016/0042-6822(79)90352-0. [DOI] [PubMed] [Google Scholar]
- 40.Furth M.E., Blattner F.R., McLeester C., Dove W.F. Genetic structure of the replication origin of bacteriophage lambda. Science. 1977;198:1046–1051. doi: 10.1126/science.929186. [DOI] [PubMed] [Google Scholar]
- 41.Hayes S., Asai K., Chu A.M., Hayes C. NinR- and red-mediated phage-prophage marker rescue recombination in Escherichia coli: Recovery of a nonhomologous immlambda DNA segment by infecting lambdaimm434 phages. Genetics. 2005;170:1485–1499. doi: 10.1534/genetics.105.042341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Daniels D.L., Schroeder J.L., Szybalski W., Sanger F., Blattner F.R. Appendix I. A molecular map of coliphage lambda. In: Hendrix R.W., Roberts J.W., Stahl F.W., Weisberg R.A., editors. Lambda II. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY, USA: 1983. pp. 469–517. [Google Scholar]
- 43.Scherer G. Nucleotide sequence of the O gene and of the origin of replication in bacteriophage lambda DNA. Nucleic Acids Res. 1978;5:3141–3156. doi: 10.1093/nar/5.9.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moore D.D., Denniston K., Kruger K.E., Furth M.E., Williams B.G., Daniels D.L., Blattner F.R. Proceedings of the Cold Spring Harbor Symposium Quantitative Biology. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY, USA: 1979. Dissection and comparative anatomy of the origins of replication in lambdoid coliphages; pp. 155–163. [DOI] [PubMed] [Google Scholar]
- 45.Zahn K., Blattner F.R. Sequence-induced DNA curvature at the bacteriophage lambda origin of replication. Nature. 1985;317:451–453. doi: 10.1038/317451a0. [DOI] [PubMed] [Google Scholar]
- 46.Denniston-Thompson K., Moore D.D., Kruger K.E., Furth M.E., Blattner F.R. Physical structure of the replication origin of bacteriophage lambda. Science. 1977;198:1051–1056. doi: 10.1126/science.929187. [DOI] [PubMed] [Google Scholar]
- 47.Zeghouf M., Li J., Butland G., Borkowska A., Canadien V., Richards D., Beattie B., Emili A., Greenblatt J.F. Sequential Peptide Affinity (SPA) system for the identification of mammalian and bacterial protein complexes. J. Proteome Res. 2004;3:463–468. doi: 10.1021/pr034084x. [DOI] [PubMed] [Google Scholar]
- 48.Hayes S., Hayes C., Bull H.J., Pelcher L.A., Slavcev R.A. Acquired mutations in phage lambda genes O or P that enable constitutive expression of a cryptic lambdaN+cI[Ts]cro- prophage in E. coli cells shifted from 30 degreesC to 42 degreesC, accompanied by loss of immlambda and Rex+ phenotypes and emergence of a non-immune exclusion-state. Gene. 1998;223:115–128. doi: 10.1016/s0378-1119(98)00363-1. [DOI] [PubMed] [Google Scholar]
- 49.Tomizawa J. Functional cooperation of genes O and P. In: Hershey A.D., editor. The Bacteriophage Lambda. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY, USA: 1971. pp. 549–552. [Google Scholar]
- 50.Furth M.E., Yates J.L. Specificity determinants for bacteriophage lambda DNA replication. II. Structure of O proteins of lambda-phi80 and lambda-82 hybrid phages and of a lambda mutant defective in the origin of replication. J. Mol. Biol. 1978;126:227–240. doi: 10.1016/0022-2836(78)90360-1. [DOI] [PubMed] [Google Scholar]
- 51.Wickner S.H., Zahn K. Characterization of the DNA binding domain of bacteriophage lambda O protein. J. Biol. Chem. 1986;261:7537–7543. [PubMed] [Google Scholar]
- 52.Gonciarz-Swiatek M., Wawrzynow A., Um S.J., Learn B.A., McMacken R., Kelley W.L., Georgopoulos C., Sliekers O., Zylicz M. Recognition, targeting, and hydrolysis of the lambda O replication protein by the ClpP/ClpX protease. J. Biol. Chem. 1999;274:13999–14005. doi: 10.1074/jbc.274.20.13999. [DOI] [PubMed] [Google Scholar]
- 53.Dodson M., Echols H., Wickner S., Alfano C., Mensa-Wilmot K., Gomes B., LeBowitz J., Roberts J.D., McMacken R. Specialized nucleoprotein structures at the origin of replication of bacteriophage lambda: Localized unwinding of duplex DNA by a six-protein reaction. Proc. Natl. Acad. Sci. USA. 1986;83:7638–7642. doi: 10.1073/pnas.83.20.7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alfano C., McMacken R. The role of template superhelicity in the initiation of bacteriophage lambda DNA replication. Nucleic Acids Res. 1988;16:9611–9630. doi: 10.1093/nar/16.20.9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reiser W., Leibrecht I., Klein A. Structure and function of mutants in the P gene of bacteriophage lambda leading to the pi phenotype. Mol. Gen. Genet. 1983;192:430–435. doi: 10.1007/BF00392186. [DOI] [PubMed] [Google Scholar]
- 56.Wickner S.H. DNA replication proteins of Escherichia coli and phage lambda. Cold Spring Harb. Symp. Quant. Biol. 1979;43:303–310. doi: 10.1101/SQB.1979.043.01.037. [DOI] [PubMed] [Google Scholar]
- 57.Zylicz M., Gorska I., Taylor K., Georgopoulos C. Bacteriophage lambda replication proteins: Formation of a mixed oligomer and binding to the origin of lambda DNA. Mol. Gen. Genet. 1984;196:401–406. doi: 10.1007/BF00436186. [DOI] [PubMed] [Google Scholar]
- 58.Eisen H., Barrand P., Spiegelman W., Reichardt L.F., Heinemann S., Georgopoulos C. Mutants in the y region of bacteriophage lambda constitutive for repressor synthesis: Their isolation and the characterization of the Hyp phenotype. Gene. 1982;20:71–81. doi: 10.1016/0378-1119(82)90088-9. [DOI] [PubMed] [Google Scholar]
- 59.Fiandt M., Szybalski W., Malamy M.H. Polar mutations in lac, gal and phage lambda consist of a few IS-DNA sequences inserted with either orientation. Mol. Gen. Genet. 1972;119:223–231. doi: 10.1007/BF00333860. [DOI] [PubMed] [Google Scholar]
- 60.Hayes S., Horbay M.A., Hayes C. A CI-Independent Form of Replicative Inhibition: Turn Off of Early Replication of Bacteriophage Lambda. PLoS ONE. 2012;7:e36498. doi: 10.1371/journal.pone.0036498. [DOI] [PMC free article] [PubMed] [Google Scholar]